Abstract
Endometrial cancer (EC) is the second leading type of cancer among women, and its progression is dependent on several factors. The aim of the present study was to examine the effect of solute carrier organic anion transporter family member 4C1 (SLCO4C1) on human EC and determine the underlying molecular mechanism. A total of 57 differentially expressed genes associated with advanced stage and survival were identified in The Cancer Genome Atlas database. In addition, gene ontology analysis indicated that SLCO4C1 was highly expressed in cell differentiation and integral component of plasma membrane. High SLCO4C1 expression in EC tissues was verified by immunohistochemistry. The results demonstrated that the downregulation of SLCO4C1 could significantly suppress the viability, sphere formation, migration and invasion abilities of cells, but enhance apoptosis in EC cell lines. Furthermore, the present results demonstrated that SLCO4C1 had effects on the epithelial-mesenchymal transition (EMT) phenotype in EC cells and regulated the expression of EMT-related proteins. Mechanistically, the present study revealed that SLCO4C1 regulated the biological functions of EC cells by inactivating the PI3K/Akt signaling pathway. Collectively, it was demonstrated that the SLCO4C1/PI3K/Akt pathway may play an important role in EC progression and metastasis and serve as a potential biomarker and target for EC diagnosis and treatment.
Keywords: SLCO4C1, PI3K/Akt, apoptosis, migration, invasion, endometrial cancer
Introduction
Endometrial cancer (EC), the most common gynecological malignant cancer, can be clinically classified into 2 types (1,2). Type I, the most common clinically, is most likely caused by the long-term stimulation of estrogen, obesity, hypertension and diabetes. Generally, the Type I EC population is younger with a higher degree of tumor differentiation and better prognosis (3). By contrast, type II EC is usually observed in women aged >65 years, who have experienced pregnancy (2,4). Although most EC cases are diagnosed at an early stage, which is correlated with a better outcome, women at an intermediate and late stage of EC are at a highest risk of recurrence and poor prognosis. In addition, the lowly differentiated ones can hardly be cured, resulting in death rates of ~20%. In particular, EC cases with regional or distal metastasis tend to recur, and the effect of systemic therapies is limited (5–8). Therefore, it is important to further explore the molecular mechanisms responsible for EC tumorigenesis and progression, and to develop novel diagnostic and therapeutic strategies.
According to The Cancer Genome Atlas (TCGA) datasets, solute carrier organic anion transporter family member 4C1 (SLCO4C1) upregulation was revealed to be strongly associated with the survival time and clinical stage of patients with EC. SLCO4C1 is a human kidney-specific organic anion transporting polypeptide (9,10). In addition, SLCO4C1 was revealed to excrete uremic toxins, which led to reduced blood pressure and decreased kidney inflammation (11,12). In addition, SLCO4C1 also served as a tumor suppressor gene in head and neck cancer (HNC) (13). These results revealed that SLCO4C1 is involved in tumorigenesis, however, there is no study on the relationship between SLCO4C1 and EC.
In the present study, it was revealed that SLCO4C1 interference regulated biological behaviors of EC cells, such as proliferation, apoptosis, sphere formation, migration, invasion and EMT phenotype by inactivating the PI3K/Akt signaling pathway. These results shed lights on the mechanisms underlying EC occurrence and development, and provided evidence that the SLCO4C1/PI3K/Akt signaling pathway may be a potential therapeutic target for the treatment of EC.
Materials and methods
SLCO4C1 expression analysis in the TCGA dataset
TCGA_UCEC_exp_RNA-seq, a TCGA dataset containing 177 Uterine Corpus Endometrial Carcinoma and 24 para-tumor tissue specimens was downloaded from https://tcga-data.nci.nih.gov/tcga/. The clinical features of patients, including survival time and clinical stage, were collected retrospectively, based on the records of patients. The significance threshold of the differentially expressed mRNA was set at P<0.05 following t-tests. The correlation of clinical characteristics with SLCO4C1 expression was analyzed using χ2 test. The prognostic value of SLCO4C1 in EC was analyzed using ggsurv R package (http://www.sthda.com/english/rpkgs/survminer/), the clinical stage of SLCO4C1 in EC was assessed using Spearman's rank correlation coefficient method.
Gene ontology (GO) enrichment analysis
GO analysis has been recognized as a helpful approach to annotating genes and gene sets that possess relevant biological features for high-throughput genomic or transcriptomic data. The Database for Annotation, Visualization and Integrated Discovery (DAVID) has provided simple and efficient annotation tools for researchers, aiding them in identifying the biological meanings of different genes. In the present study, GO enrichment analysis of the differentially expressed genes (DEGs) was performed using the aforementioned DAVID database, and FDR<0.05 was deemed as the threshold.
Patients and tissues
A total of 20 EC and 10 normal endometrial tissues were obtained from female Chinese patients who had undergone surgical treatment from January 2017 to December 2019 at the Shanghai First Maternity and Infant Hospital (Shanghai, China). No patient had undergone endocrine therapy, radiotherapy or chemotherapy prior to surgery. The present research was approved by the Human Investigation Ethics Committee of the Shanghai First Maternity and Infant Hospital. Samples were collected from patients after receiving their written informed consent.
Immunohistochemistry (IHC)
Endometrial tissues were sliced into 5-µm thick sections and embedded in paraffin. Next, the sections were mounted on poly-L-lysine-coated slides and then deparaffinized in xylene and dehydrated with gradient ethanol. Antigen retrieval was carried out in 10 mM citrate buffer (pH 6.0) for 10 min. Endogenous peroxidase activity was blocked with 3% hydrogen peroxide for 10 min at room temperature. Following blocking with 1% horse serum albumin for 20 min at room temperature, the sections were incubated with rabbit polyclonal antibody against SLCO4C1 (cat. no. 203261-T10; 1:100 dilution; Sino Biological, Inc.) overnight at 4°C and then incubated with rabbit polyclonal secondary antibody (cat. no. VC003; 1:200 dilution; R&D Systems, Inc.) at 37°C for 30 min. The sections were stained with DAB for 5 min and re-stained with hematoxylin for 2 min.
Cell culture
Several human EC cell lines, including HEC-1A, HEC-1B, Ishikawa 3H12, RL95-2 and KLE, were provided by the Shanghai Institute of Cell Biology, Chinese Academy of Sciences. HEC-1A cells were cultured in McCoy'5A (Thermo Fisher Scientific, Inc.) containing 10% fetal bovine serum (FBS; GE Healthcare) and 1% penicillin/streptomycin (P/S; Thermo Fisher Scientific, Inc.). HEC-1B, Ishikawa 3H12 and RL95-2 cells were cultured in Dulbecco's Modified Eagle's Medium (DMEM)/Nutrient Mixture F-12 (Biological Industries) containing 10% FBS and 1% P/S. KLE cells were cultured in high-glucose DMEM (Thermo Fisher Scientific, Inc.) containing 10% FBS as well as 1% P/S. Cells were placed in an incubator supplemented with 5% CO2 at 37°C.
Small interfering RNA (siRNA) transfection
The siRNAs targeting SLCO4C1 (siSLCO4C1-1 and siSLCO4C1-2) and the negative control (siNC) were purchased from Hanbio Biotechnology Co., Ltd. The control (Ctrl) was a blank control group. HEC-1A and RL95-2 cells were transfected with the siRNAs, siNC or Ctrl in Opti-MEM (Thermo Fisher Scientific, Inc.) using Lipofectamine 3000 (Thermo Fisher Scientific, Inc.), according to the manufacturer's instructions. The concentration of siRNA transfected in the cells reached 1 nM in a 12-well plate. The sequences of the RNA oligonucleotides were as follows: siSLCO4C1-1 forward, 5′-CAGGCUCAUCAGAGUAAUAdTd-3′ and reverse, 3′-UAUUACUCUGAUGAGCCUGdTd-5′; siSLCO4C1-2 forward, 5′-CAUAUGCUAGUAGCCAUAAdTd-3′ and reverse, 3′-UUAUGGCUACUAGCAUAUGdTd-5′; siNC forward, 5′-UUCUCCGAACGUGUCACGUdTd-3′ and reverse, 3′-ACGUGACACGUUCGGAGAAdTd-5.
Cell viability assay
Cell viability was assessed by CCK-8 assay (Dojindo Molecular Technologies, Inc.). HEC-1A and RL95-2 cells were inoculated into 96-well plates (4×103 cells/well) for 24 h, followed by transfection with siNC, siSLCO4C1-1 or siSLCO4C1-2. In the rescue experiment, an equal number of cells from the siSLCO4C1-1 or siSLCO4C1-2 group was treated with 740 Y-P (PI3K signaling activator, 100 µg/ml and incubation for 24 h; MedChemExpress), comprising groups siSLCO4C1-1+740 Y-P or siSLCO4C1-2+740 Y-P, respectively. After 24, 48 and 72 h, 10 µl CCK-8 solution was added to each well, followed by further incubation of the plates at 37°C for 4 h. Subsequently, a microplate reader was used to measure the ultraviolet absorbance of all samples at 450 nm.
Apoptosis assay
Apoptosis was detected using the Annexin V-FITC apoptosis detection kit I (BD Biosciences). HEC-1A and RL95-2 cells were washed with cold phosphate-buffered saline (PBS; Biological Industries) twice and resuspended in Annexin V binding buffer at a density of 0.5×107 cells/ml. Subsequently, 100 µl cell suspension was transferred into the 1 ml test tube, followed by the addition of Annexin V- FITC (5 µl) and propidium iodide solution (10 µl). Then, cells were subjected to gentle vortex and 15 min of incubation in the dark under ambient temperature (25°C). Subsequently, Annexin V binding buffer (400 µl) was added to each tube, followed by flow cytometry (Accuri C6; BD Biosciences) in 1 h. Early apoptotic cells were stained Annexin V-positive and PI-negative, whereas late apoptotic cells were stained both Annexin V- and PI-positive. The sum ratio of early and late apoptosis represented in Q3 and Q2 quadrants, respectively, was calculated for apoptosis rate analysis.
Sphere-formation assay
A total of 1,000 single cells were inoculated into each well of the 24-well ultra-low attachment plate (Corning Inc.), which was filled with serum-free high-glucose DMEM. The N2 plus media supplement (Thermo Fisher Scientific, Inc.), epidermal growth factor (20 ng/ml; Thermo Fisher Scientific, Inc.), basic fibroblast growth factor (20 ng/ml; Thermo Fisher Scientific, Inc.), and heparin (4 mg/ml; Merck KGaA) were also added to the medium. The number of formed spheres >50 µm was calculated 10 days after culture using the Leica DMi8 inverted microscope (magnification, ×200; Leica Microsystems GmbH).
Wound healing assay
HEC-1A and RL95-2 cells (from the Ctrl, siNC, siSLCO4C1-1, siSLCO4C1-2, siSLCO4C1-1+740 Y-P and siSLCO4C1-2+740 Y-P groups) were inoculated into the 6-well plate until they reached full confluence. A 200-µl pipette tip was used to create scratch wounds on the cells. Cells were further cultured in the serum-free high-glucose DMEM, and images of the scratches were captured using the Leica DMi8 inverted microscope (magnification, ×50) at 0 and 48 h respectively.
Cell invasion assay
HEC-1A and RL95-2 cells were grown until they had reached 50–70% convergence, followed by transfection as aforementioned. The transfected cells were inoculated into the upper Boyden chamber covered with Matrigel 24 h later, as previously described (14). Next, 24 h after incubation, the cells that had not invaded the Matrigel were gently eliminated using a cotton swab. The invasive cells on the lower chamber surface were stained using Giemsa (prepared by mixing 10 drops of Giemsa stock solution in 10 ml of PBS; Leagene Biotechnology) at room temperature for 15 min and counted under the Leica DMi8 inverted microscope (magnification, ×200). The relative cell invasion activity was indicated as the fold change relative to that of the respective controls.
RNA extraction and RT-qPCR
TRIzol reagent (Thermo Fisher Scientific, Inc.) was used to extract the total RNA from cells. Next, the SYBR FAST one-Step RT-qPCR kit (Merck KGaA) was used for RT-qPCR assays with the CFX96 Real Time-PCR Detection System (Bio-Rad Laboratories, Inc.). RT-qPCR was performed with following cycling conditions: 30 sec at 95°C; 40 cycles of 5 sec at 95°C, 20 sec at 60°C and 10 sec at 72°C; 1 min at 95°C, 30 sec at 60°C; and 30 sec at 95°C. The primer sequences used were as follows: SLCO4C1 (128 bp) forward, 5′-CAGACATGAAGAGCGCCAAA-3′ and reverse, 5′-AAGCTCCTGTGGCTGAGAAT-3′; GAPDH (124 bp) forward, 5′-ACCCAGAAGACTGTGGATGG-3′ and reverse, 5′-TCAGCTCAGGGATGACCTTG-3′. The SLCO4C1 expression was normalized to that of GAPDH, and the relative miRNA expression level was measured using the 2−ΔΔCq method (15).
Western blotting
RIPA buffer (Beyotime Institute of Biotechnology) was used to extract the total protein from HEC-1A and RL95-2 cells in the Ctrl, siNC, siSLCO4C1-1, siSLCO4C1-2, siSLCO4C1-1+740 Y-P and siSLCO4C1-2+740 Y-P groups. The concentration of the extracted protein was determined using BCA Protein Assay Kit (Thermo Fisher Scientific, Inc.). The protein samples were mixed with loading buffer and then boiled for 10 min. Total 20 µg of protein were loaded onto each lane of an 10% SDS-PAGE gel for protein separation, and transferred onto polyvinylidene fluoride membranes (EMD Millipore). The membranes were blocked with 5% bovine serum albumin for 2 h and incubated with antibodies against SLCO4C1 (cat. no. 24584-1-AP; 1:600 dilution; ProteinTech Group, Inc.), GAPDH (product code ab9485; 1:2,500 dilution; Abcam), E-cadherin (product no. 3195; 1:1,000 dilution), vimentin (product no. 5741; 1:1,000 dilution) and N-cadherin (product no. 13116; 1:1,000 dilution; all from Cell Signaling Technology, Inc.), Ki67 (product code ab243878; 1:1,000 dilution) and cleaved caspase-3 (product code ab2302; 1:1,000 dilution) and cleaved caspase-9 (product code ab32539; 1:1,000 dilution; all from Abcam), p-PI3K (product no. 4228; 1:1,000 dilution), PI3K (product no. 4257; 1:1,000 dilution), p-Akt (product no. 4060; 1:1,000 dilution) and Akt (product no. 9272; 1:1,000 dilution; all from Cell Signaling Technology, Inc.) at 4°C overnight. The membranes were incubated with peroxidase-linked secondary anti-rabbit antibodies (product no. 7074; 1:2,000 dilution; Cell Signaling Technology, Inc.) for 1 h at room temperature to detect the bound primary antibodies, and the blotted proteins were visualized using an enhanced chemiluminescence kit (Thermo Fisher Scientific, Inc.). The intensity of the protein bands was quantified using ImageJ software (v1.8.0; National Institutes of Health). The relative expression of target proteins was described as a ratio relative to the expression of GAPDH, and statistical data from at least three experiments were graphed.
Statistical analysis
In the present study, SPSS 19.0 (IBM Corp.) was used for all statistical analysis, and images were edited using GraphPad Prism 6.0 version (GraphPad Software, Inc.). A Student's t-test was used for pairwise comparisons of differences, whereas one-way analysis of variance (ANOVA) and Tukey's post hoc test were used to analyze the difference among groups. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of DEGs in EC based on TCGA datasets
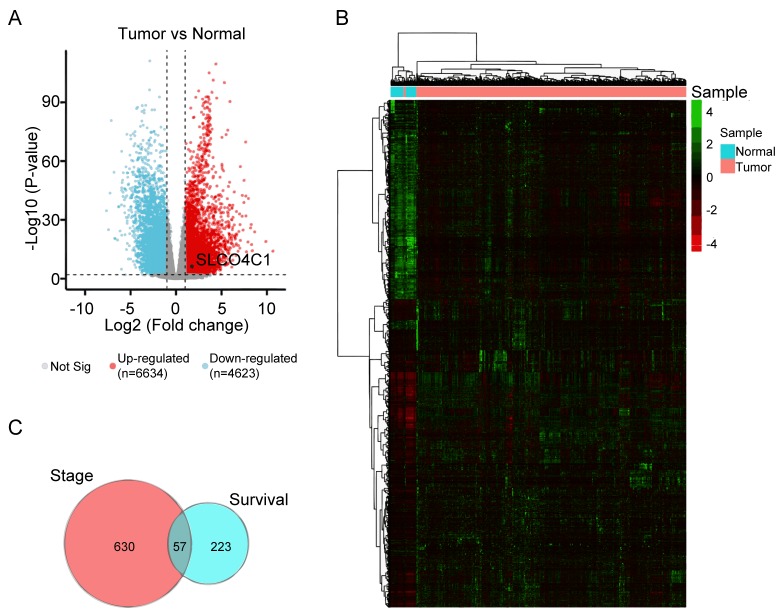
The TCGA RNA-seq dataset of EC patients was selected, and DEGs were analyzed using Deseq2. First, the RNA-seq was analyzed to perform the volcano plot. Analysis of RNA-seq expression profiling data from the TCGA patient cohort was plotted into upregulated (red dots) or downregulated genes (blue dots) to compare EC to normal cohorts, as revealed in Fig. 1A. Subsequently, 6,634 upregulated and 4,623 downregulated genes were examined, and their expression levels were determined and presented in a heatmap; it was revealed that these genes were well clustered into the non-endometrial and EC tissues. The significantly altered genes (fold change ≥2 or ≤-2 and P<0.01) were defined as DEGs (Fig. 1B). The prognostic value and clinical stage of these DEGs in EC were analyzed using ggsurv R package and Spearman's rank correlation coefficient, respectively, in order to identify the molecular signatures affecting the survival time and status of EC. In addition, Venn analysis was used to obtain the intersection of the DEG profiles, as revealed in Fig. 1C. A total of 57 DEGs were revealed to be strongly associated with survival time and clinical stage in EC.
Figure 1.
DEG expression in EC based on TCGA datasets. (A) Volcano plot showing the gene expression differences between normal and EC patient cohorts. Blue dots, downregulated genes. Red dots, upregulated genes. Black dot, SLCO4C1. (B) Heatmap of the expression of 6,634 upregulated and 4,623 downregulated genes, when comparing normal to EC tissues from TCGA database. (C) Venn analysis of 57 overlapping genes associated with clinical stage and survival time. DEG, differentially expressed genes; EC, endometrial cancer; TCGA, The Cancer Genome Atlas; SLCO4C1, solute carrier organic anion transporter family member 4C1.
Identifying the specific gene signatures and the key SLCO4C1 gene
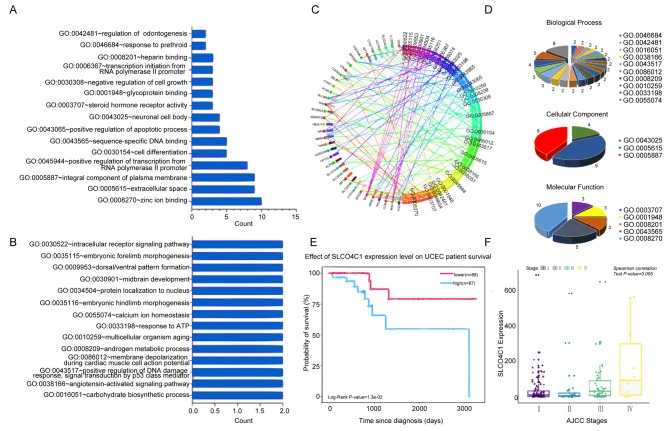
To predict the functional category of these 57 DEGs, GO analysis was performed and it was revealed that 57 DEGs were associated with various biological processes, molecular functions and cellular components. Among them, 37 DEGs were found to mainly participate in 18 significant GOs (P<0.05; Fig. 2A-D). Specific genes were revealed to participate in zinc ion binding, integral component of plasma membrane, extracellular space, positive transcriptional regulation promoted by RNA polymerase II, sequence-specific DNA binding and cell differentiation. Notably, SLCO4C1 was significantly highly expressed in 2 terms, including integral component of plasma membrane and cell differentiation, which was correlated with poor prognosis and advanced clinical stage in EC (Fig. 2E and F). Thus, the function of SLCO4C1 and its molecular mechanism were next explored.
Figure 2.
Identification of the specific gene signatures and the key SLCO4C1 gene by GO annotation analysis. (A and B) The bar chart displays the 29 GO terms by GO analysis. (C and D) A pie graph was used to reveal the distribution of the biological processes, cell components and molecular functions. (E) SLCO4C1 was significantly associated with the survival rate in EC. (F) SLCO4C1 was significantly associated with cancer stage in EC. SLCO4C1, solute carrier organic anion transporter family member 4C1; GO, Gene Ontology; EC, endometrial cancer.
SLCO4C1 is highly expressed in EC tissues
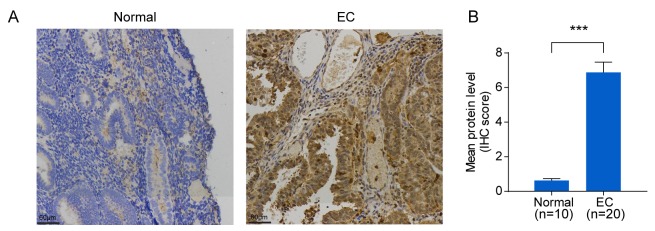
To verify SLCO4C1 expression in EC tissues, immunochemistry was carried out in EC (n=20) and normal endometrium (n=10) tissues. Immunochemistry revealed that the SLCO4C1 protein was predominantly localized in the membrane and cytoplasm of endometrial epithelial cells. Only weak or no staining was observed in normal endometria, whereas strong SLCO4C1 immunostaining was observed in EC tissues (P<0.001; Fig. 3). These data indicated that SLCO4C1 expression was significantly higher in EC tissues.
Figure 3.
SLCO4C1 expression in normal endometrium and EC tissues. (A) Immunohistochemical analysis of SLCO4C1 expression in normal endometrium and EC tissues. No or weak expression of SLCO4C1 was observed in normal endometrium tissues. Strong cell membrane and cytoplasm expression of SLCO4C1 was observed in the majority of EC tissues. (B) Immunostaining scoring results in normal endometrium (n=10) and EC (n=20) tissues. The results were determined from triplicate experiments and the error bars represent the mean ± SD. ***P<0.001. SLCO4C1, solute carrier organic anion transporter family member 4C1; EC, endometrial cancer.
SLCO4C1 downregulation inhibits proliferation and sphere formation, and induces apoptosis in HEC-1A and RL95-2 cells
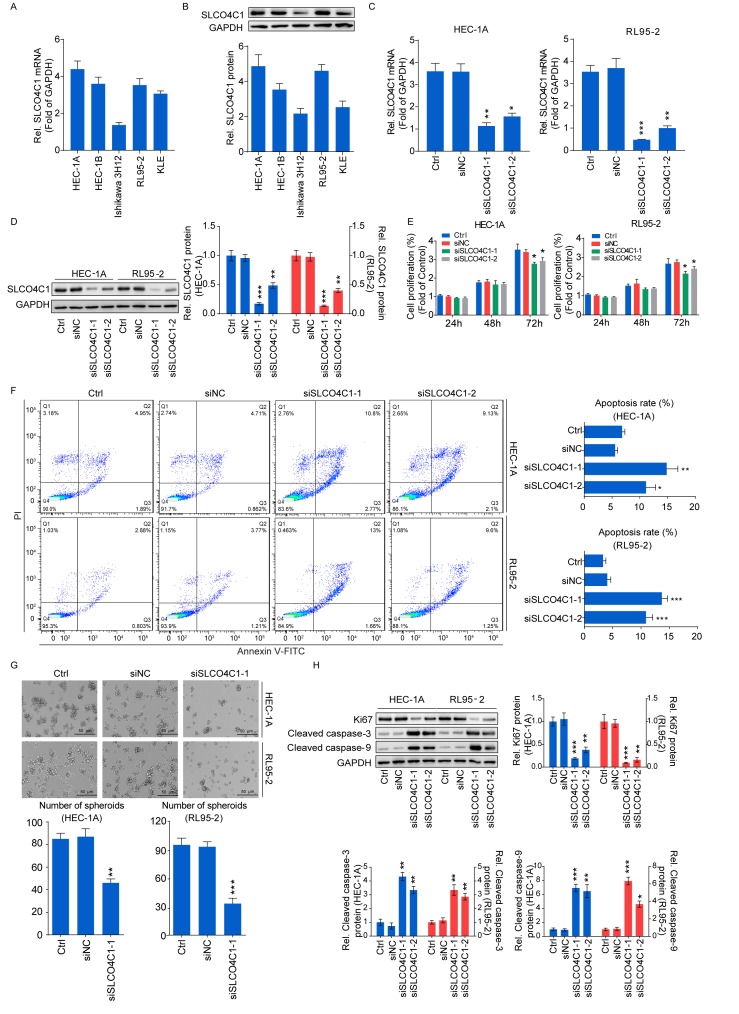
A high SLCO4C1 expression was detected in the EC cell lines, as verified by RT-qPCR and western blotting (Fig. 4A and B). HEC-1A and RL95-2 cells with a relatively higher SLCO4C1 expression were used for subsequent experiments. siSLCO4C1-1 and siSLCO4C1-2 were successfully transfected into HEC-1A and RL95-2 cells, respectively (Fig. 4C and D). To examine whether SLCO4C1 would affect cell proliferation, apoptosis and sphere formation in EC, a CCK-8 assay, flow cytometry and a sphere formation assay were performed. As revealed in Fig. 4E, the proliferation of HEC-1A and RL95-2 cells was significantly decreased by SLCO4C1 interference at 72 h. Concurrently, HEC-1A and RL95-2 cells transfected with siSLCO4C1-1 and siSLCO4C1-2 were associated with increased apoptosis, compared to Ctrl and siNC cells (Fig. 4F). The downregulation of SLCO4C1 could inhibit the sphere forming ability of HEC-1A and RL95-2 cells while suspending their growth (Fig. 4G). In addition, the knockdown of SLCO4C1 significantly downregulated the expression of proliferation-related protein Ki67 and enhanced the expression of apoptotic-related protein cleaved caspase-3/9, as revealed by western blotting (Fig. 4H).
Figure 4.
Downregulation of SLCO4C1 affects the proliferation, apoptosis and sphere forming abilities of HEC-1A and RL95-2 cells. (A) The expression of SLCO4C1 in EC cell lines was detected by RT-qPCR. (B) The expression of SLCO4C1 in EC cell lines was detected by western blotting. (C) The expression of SLCO4C1 in HEC-1A and RL95-2 cells transfected with siNC, siSLCO4C1-1 or siSLCO4C1-2 was detected by RT-qPCR analysis. (D) The expression of SLCO4C1 in HEC-1A and RL95-2 cells transfected with siNC, siSLCO4C1-1 or siSLCO4C1-2 was detected by western blotting. (E) Cell proliferation was detected by CCK-8 assay in HEC-1A and RL95-2 cells transfected with siNC, siSLCO4C1-1 or siSLCO4C1-2. (F) Apoptosis analysis of HEC-1A and RL95-2 cells transfected with siNC, siSLCO4C1-1 or siSLCO4C1-2 by flow cytometry. (G) Sphere forming ability of HEC-1A and RL95-2 cells transfected with siNC or siSLCO4C1-1 by sphere-formation assay. (H) Expression of proliferation-related protein Ki67 and apoptotic-related proteins cleaved caspase-3/9 in HEC-1A and RL95-2 cells transfected with siNC, siSLCO4C1-1 or siSLCO4C1-2 by western blotting. GAPDH was used as a loading control. The results were determined from triplicate experiments and the error bars represent the mean ± SD. *P<0.05, **P<0.01, ***P<0.001 vs. the Ctrl group and siNC group. SLCO4C1, solute carrier organic anion transporter family member 4C1; EC, endometrial cancer.
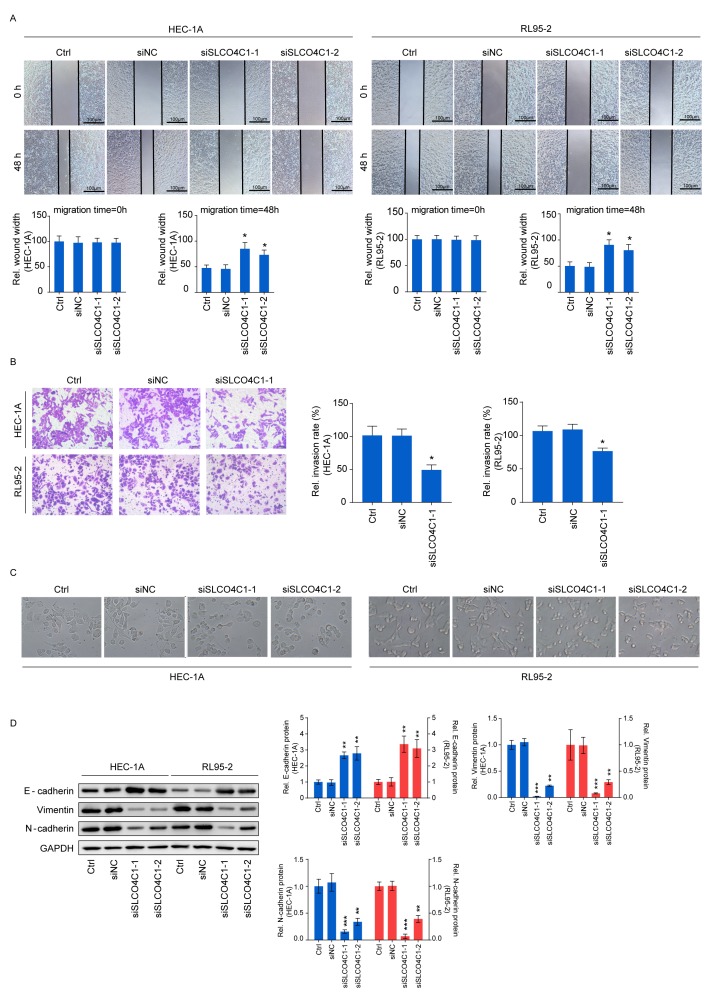
Downregulation of SLCO4C1 suppresses the migration and invasion capacities of HEC-1A and RL95-2 cells and affects the epithelial-mesenchymal transition (EMT) phenotype in EC cells
Wound healing and Transwell assays were carried out in order to determine cell migration and invasion, thus further exploring the influence of SLCO4C1 on the behavior of EC cells. As revealed in Fig. 5A and B, the migration ability of HEC-1A and RL95-2 cells was significantly reduced by SLCO4C1 interference at 48 h, while these cells transfected with siSLCO4C1-1 displayed a markedly decreased invasion ability, compared to Ctrl and siNC cells. The functions of SLCO4C1 in migration and invasion prompted us to examine whether SLCO4C1 knockdown could induce morphological changes. siSLCO4C1-1 and siSLCO4C1-2 cells were morphologically transformed toward epithelial, compared to Ctrl and siNC cells (Fig. 5C). This change was characterized by a transition from the spindle-like morphology of the parental cells to a more differentiated keratinocyte-like morphology, indicating a phenotypic transition from mesenchymal to epithelial. Processes involved in EMT are closely correlated with cancer metastasis. To further determine if this transformation represented an EMT, the levels of several characteristic epithelial and mesenchymal proteins were analyzed by western blotting in Ctrl, siNC, siSLCO4C1-1 and siSLCO4C1-2 cells. As revealed in Fig. 5D, the expression of E-cadherin was significantly upregulated while that of vimentin and N-cadherin was significantly downregulated in HEC-1A and RL95-2 cells transfected with siSLCO4C1-1 and siSLCO4C1-2, compared to Ctrl and siNC cells. Collectively, these findings indicated that SLCO4C1 knockdown was accompanied by the loss of mesenchymal and gain of epithelial markers, and that SLCO4C1 is likely to play a crucial role in the regulation of EMT and metastasis in EC cells.
Figure 5.
SLCO4C1 downregulation suppresses the migration and invasion abilities of HEC-1A and RL95-2 cells and affects the EMT phenotype in EC cells. (A) Migration of HEC-1A and RL95-2 cells transfected with siNC, siSLCO4C1-1 or siSLCO4C1-2 by wound healing assay at 0 and 48 h. (B) Invasion of HEC-1A and RL95-2 cells transfected with siNC or siSLCO4C1-1 by Transwell assay. (C) Epithelial morphology induced by the suppression of SLCO4C1 in HEC-1A and RL95-2 cells (magnification, ×400). (D) Expression of EMT markers in HEC-1A and RL95-2 cells transfected with siNC, siSLCO4C1-1 or siSLCO4C1-2 by western blotting. GAPDH was used as a loading control. The results were determined from triplicate experiments and the error bars represent the mean ± SD. *P<0.05, **P<0.01, ***P<0.001 vs. the Ctrl group and siNC group. SLCO4C1, solute carrier organic anion transporter family member 4C1; EMT, epithelial-mesenchymal transition; EC, endometrial cancer.
SLCO4C1 regulates the behavior of EC through the PI3K/Akt signaling pathway in HEC-1A and RL95-2 cells
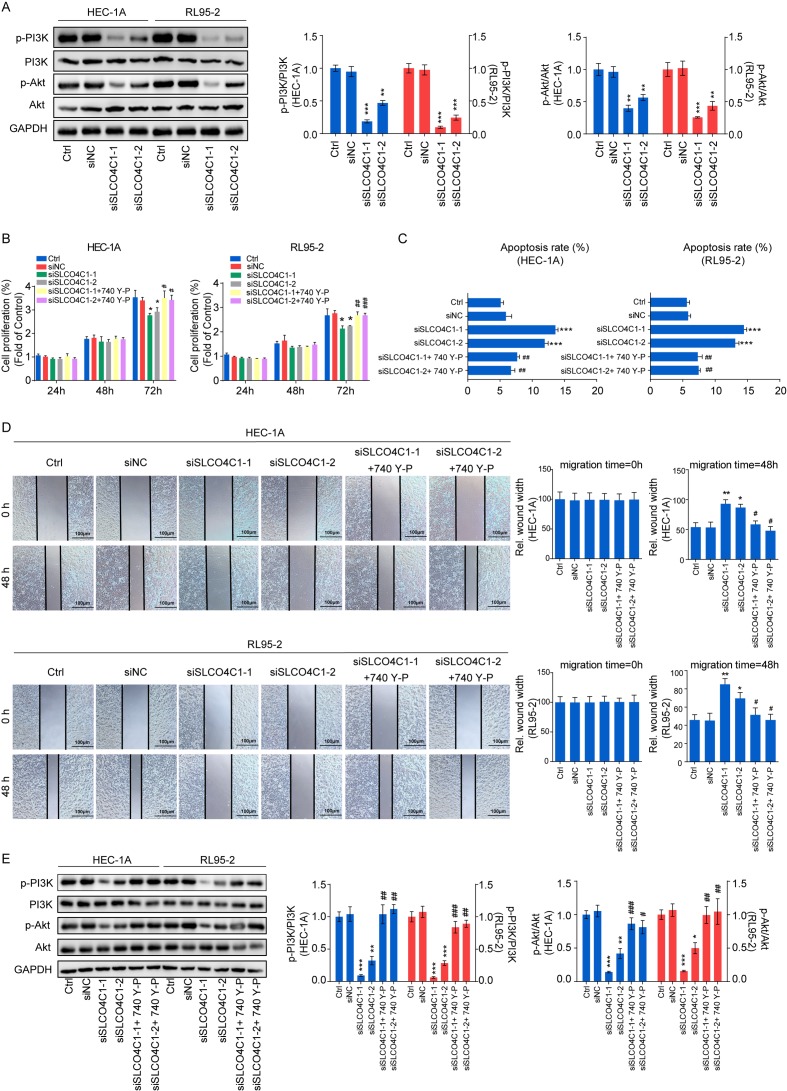
To explore the molecular mechanism underlying SLCO4C1, western blotting was performed in Ctrl, siNC, siSLCO4C1-1 and siSLCO4C1-2 cells, in order to detect the markers of the PI3K/Akt signaling pathway. It was revealed that SLCO4C1 interference could significantly inactivate the PI3K/Akt signaling pathway by decreasing the phosphorylated levels of PI3K and Akt proteins (Fig. 6A). In order to further verify the association between SLCO4C1 and the PI3K/Akt signaling pathway, an equal number of siSLCO4C1-1 or siSLCO4C1-2 cells were treated with 740 Y-P (PI3K signaling activator) for a rescue assay. The inhibition of proliferation caused by the knockdown of SLCO4C1 was reversed following 740 Y-P stimulation (Fig. 6B). The 740 Y-P treatment also prevented the enhancement of apoptosis mediated by siSLCO4C1 in HEC-1A and RL95-2 cells (Fig. 6C). In addition, the reduction of migration mediated by siSLCO4C1 was rescued with treatment of 740 Y-P (Fig. 6D). Furthermore, the PI3K/Akt signaling pathway was rescued with treatment of 740 Y-P in the presence of SLCO4C1 knockdown (Fig. 6E). These results indicated that SLCO4C1 played a pivotal role in EC through the PI3K/Akt signaling pathway.
Figure 6.
Downregulation of SLCO4C1 regulates the behavior of EC via the PI3K/Akt signaling pathway in HEC-1A and RL95-2 cells. (A) Expression of p-PI3K, PI3K, p-Akt and Akt in HEC-1A and RL95-2 cells from the Ctrl, siNC, siSLCO4C1-1 or siSLCO4C1-2 groups, as determined by western blotting. (B) Cell proliferation was detected by CCK-8 assay in HEC-1A and RL95-2 cells from the Ctrl, siNC, siSLCO4C1-1, siSLCO4C1-2, siSLCO4C1-1+740 Y-P (PI3K signaling activator) or siSLCO4C1-2+740 Y-P groups. (C) Cell apoptosis was detected by flow cytometry in HEC-1A and RL95-2 cells from the Ctrl, siNC, siSLCO4C1-1, siSLCO4C1-2, siSLCO4C1-1+740 Y-P or siSLCO4C1-2+740 Y-P groups. (D) Cell migration was detected by wound healing assay at 0 and 48 h in HEC-1A and RL95-2 cells from the Ctrl, siNC, siSLCO4C1-1, siSLCO4C1-2, siSLCO4C1-1+740 Y-P or siSLCO4C1-2+740 Y-P groups. (E) Expression of p-PI3K, PI3K, p-Akt and Akt in HEC-1A and RL95-2 cells from the Ctrl, siNC, siSLCO4C1-1, siSLCO4C1-2, siSLCO4C1-1+740 Y-P or siSLCO4C1-2+740 Y-P groups, as determined by western blotting. GAPDH was used as a loading control. The results were determined from triplicate experiments and the error bars represent the mean ± SD. Asterisks indicated a difference between siSLCO4C1 and siNC, whereas hashtags indicated a difference between siSLCO4C1+740 Y-P and siSLCO4C1. */#P<0.05, **/##P<0.01, ***/###P<0.001 vs. the Ctrl group and siNC group. SLCO4C1, solute carrier organic anion transporter family member 4C1; EC, endometrial cancer; p-PI3K, phosphorylated-PI3K; p-Akt, phosphorylated-Akt.
Discussion
Tumorigenesis and cancer development can be regulated by multiple genes, environmental factors and pathways in multiple ways. Certain genes have formed a complex regulatory network to induce pathogenesis (16). Each cancer type possesses a spectrum of tumor-specific molecular characteristics, including somatic mutations, copy number variations, changes in gene expression profiles and epigenetic changes. The life science field has entered the era of big data, which is characterized by mass multivariate omics data (17,18). In the present study, the newly sorted EC mRNA expression data, along with the corresponding clinical information, was downloaded from TCGA database, and 6,634 upregulated and 4,623 downregulated genes were detected in endometrial samples. According to the TCGA database and GO analysis, 57 DEGs were revealed to be associated with the survival time and clinical stage of tumor patients, and SLCO4C1 was revealed to be involved in cell differentiation and the composition of the cell membrane. Moreover, it has been reported in literature to be closely correlated with kidney disease, head and neck cancer and reproductive development (9–13). Therefore, the association of SLCO4C1 with EC was specifically investigated in the present study. Bioinformatics analysis results indicated that SLCO4C1 was highly expressed and the survival rate was reduced, accompanied by cancer procession. A high SLCO4C1 expression was also detected in EC tissues and cell lines.
Consequently, the SLCO4C1 expression in HEC-1A and RL95-2 cells was knocked down through siRNA transfection. In addition, proliferation, apoptosis, migration and invasion were detected by CCK-8, flow cytometry, and sphere-formation, wound healing and Transwell assays. The present results revealed that SLCO4C1 knockdown could suppress cell viability and promote apoptosis. It could also reduce the sphere formation, migration and invasion abilities of cells, indicating that SLCO4C1 could actually affect the biological behaviors of EC.
Cell proliferation and apoptosis are dynamically balanced processes, and an imbalance between them leads to the malignant transformation of cells and tumorigenesis (19). Ki67 is a proliferation-related protein; cleaved caspase-9 is the apoptotic initiator and cleaved caspase-3 is the executor of apoptosis (20,21). The present findings indicated that, following the knockdown of SLCO4C1, proliferation was inhibited and apoptosis was induced. Ki67 expression in cells was significantly downregulated, while cleaved caspase-3/9 expression was significantly upregulated, indicating that SLCO4C1 participated in the dynamic balance between cell proliferation and apoptosis in EC.
Cancer cells that disseminate from the primary tumor and invade distant organs are the leading causes of mortality in EC. EMT has been revealed to be of vital importance in the early events of cancer cell metastatic dissemination by endowing cells with a more motile, invasive potential (22). E-cadherin, vimentin and N-cadherin are the recognized molecular markers participating in EMT (23). During migration and invasion, the expression of E-cadherin is reduced while that of vimentin and N-cadherin is increased (24). The present study demonstrated that a reduction in SLCO4C1 expression could suppress migration and invasion, increase E-cadherin expression and reduce vimentin and N-cadherin expression, indicating that SLCO4C1 was responsible for the EMT process.
The associated molecular mechanism was further explored. Biological behaviors of tumor cells such as proliferation, apoptosis, migration and invasion are a result of the joint effect of multiple genes, and certain key proteins have been revealed to play crucial regulatory roles during these processes (25). The PI3K/Akt signaling pathway has been extensively identified among cancer cells and is an important signaling pathway involved in cancer cell proliferation, apoptosis, migration and invasion (25,26). Previous studies have identified that the activation of the PI3K/Akt signaling pathway is involved in EMT and processes of tumor progression and metastasis (26,27). PI3K-activated products can bind to the Akt PH domain, which can not only mediate the membrane translocation of Akt from the cytoplasm, but also promote its conformational change. Typically, such a process can expose the Ser473 and Thr308 phosphorylation sites of Akt, thus activating Akt (28,29). The activated Akt can further regulate cancer cell proliferation, apoptosis, migration and invasion (30). The present study revealed that SLCO4C1 knockdown could markedly suppress the phosphorylation levels of PI3K and Akt, indicating that decreasing SLCO4C1 expression could inactivate the signaling pathway. The rescue experiments with PI3K signaling activator 740 Y-P verified that the PI3K/Akt signaling pathway was essential in tumor progression and EMT mediated by SLCO4C1. Therefore, the present findings were the first, to the best of our knowledge, to demonstrate the role of SLCO4C1 interference in affecting apoptosis, proliferation, migration, invasion and the EMT phenotype in EC cells by alleviating the activation of the PI3K/Akt signaling pathway.
In conclusion, the present study revealed that SLCO4C1 is highly expressed in EC tissues, as compared to normal tissues. In addition, SLCO4C1 interference could promote apoptosis and suppress cell proliferation and metastasis in EC cells in vitro by inactivating the PI3K/Akt signaling pathway. Therefore, clinical diagnosis may benefit from SLCO4C1 assessment and the SLCO4C1/PI3K/Akt signaling pathway could serve as a potential therapeutic target for the treatment of EC.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Natural Science Foundation of China (grant nos. 81472427, 81672574 and 81702547), the Shanghai Sailing Program (grant no. 17YF1415300), the Shanghai New Frontier Technology Project (grant no. SHDC12015110) and the Shanghai Municipal Medical and Health Discipline Construction Projects (grant no. 2017ZZ02015).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
XH and YB wrote the original manuscript and analyzed the data. XH and TH undertook most of the experiments, manuscript rewriting or revising as well as figure and data processing. XH, TH, YB, HT and XWe performed the experiments and data analysis. YL and XWa designed the study and revised the manuscript critically for important intellectual content. All authors read and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
This research was approved by the Human Investigation Ethics Committee of the Shanghai First Maternity and Infant Hospital. Samples were collected from patients after receiving their written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Mazzocca A, Schönauer LM, De Nola R, Lippolis A, Marrano T, Loverro M, Sabbà C, Di Naro E. Autotaxin is a novel molecular identifier of type I endometrial cancer. Med Oncol. 2018;35:157. doi: 10.1007/s12032-018-1222-4. [DOI] [PubMed] [Google Scholar]
- 2.Setiawan VW, Yang HP, Pike MC, McCann SE, Yu H, Xiang YB, Wolk A, Wentzensen N, Weiss NS, Webb PM, et al. Type I and II endometrial cancers: Have they different risk factors? J Clin Oncol. 2013;31:2607–2618. doi: 10.1200/JCO.2012.48.2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou M, Liu C, Cao G, Gao H, Zhang Z. Expression of polymeric immunoglobulin receptor and its biological function in endometrial adenocarcinoma. J Cancer Res Ther. 2019;15:420–425. doi: 10.4103/jcrt.JCRT_536_18. [DOI] [PubMed] [Google Scholar]
- 4.Schlumbrecht M, Baeker Bispo JA, Balise RR, Huang M, Slomovitz B, Kobetz E. Variation in type II endometrial cancer risk by Hispanic subpopulation: An exploratory analysis. Gynecol Oncol. 2017;147:329–333. doi: 10.1016/j.ygyno.2017.09.002. [DOI] [PubMed] [Google Scholar]
- 5.Kiyohara T, Nakamaru S, Miyamoto M, Shijimaya T, Nagano N, Makimura K, Tanimura H. Metaplastic rectal adenocarcinoma into metastatic mucinous carcinoma on the buttock: The efficacy of immunohistochemistry. J Dermatol. 2019;46:e337–e339. doi: 10.1111/1346-8138.14873. [DOI] [PubMed] [Google Scholar]
- 6.Luo C, Cen S, Ding G, Wu W. Mucinous colorectal adenocarcinoma: Clinical pathology and treatment options. Cancer Commun (Lond) 2019;39:13. doi: 10.1186/s40880-019-0361-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maeda H, Yokomizo H, Okayama S, Yamada Y, Ida A, Satake M, Yano Y, Asaka S, Usui T, Shiozawa S, et al. A case of low-grade appendiceal mucinous neoplasm with cecum cancer. Gan To Kagaku Ryoho. 2019;46:518–520. (In Japanese) [PubMed] [Google Scholar]
- 8.Pan J, Fan Z. Mucinous adenocarcinoma of the abdomen. Int Wound J. 2019;16:1045–1046. doi: 10.1111/iwj.13117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jang H, Choi Y, Yoo I, Han J, Kim M, Ka H. Expression and regulation of prostaglandin transporters, ATP-binding cassette, subfamily C, member 1 and 9, and solute carrier organic anion transporter family, member 2A1 and 5A1 in the uterine endometrium during the estrous cycle and pregnancy in pigs. Asian-Australas J Anim Sci. 2017;30:643–652. doi: 10.5713/ajas.16.0637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taghikhani E, Maas R, Fromm MF, Konig J. The renal transport protein OATP4C1 mediates uptake of the uremic toxin asymmetric dimethylarginine (ADMA) and efflux of cardioprotective L-homoarginine. PLoS One. 2019;14:e0213747. doi: 10.1371/journal.pone.0213747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suzuki T, Toyohara T, Akiyama Y, Takeuchi Y, Mishima E, Suzuki C, Ito S, Soga T, Abe T. Transcriptional regulation of organic anion transporting polypeptide SLCO4C1 as a new therapeutic modality to prevent chronic kidney disease. J Pharm Sci. 2011;100:3696–3707. doi: 10.1002/jps.22641. [DOI] [PubMed] [Google Scholar]
- 12.Toyohara T, Suzuki T, Morimoto R, Akiyama Y, Souma T, Shiwaku HO, Takeuchi Y, Mishima E, Abe M, Tanemoto M, et al. SLCO4C1 transporter eliminates uremic toxins and attenuates hypertension and renal inflammation. J Am Soc Nephrol. 2009;20:2546–2555. doi: 10.1681/ASN.2009070696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guerrero-Preston R, Michailidi C, Marchionni L, Pickering CR, Frederick MJ, Myers JN, Yegnasubramanian S, Hadar T, Noordhuis MG, Zizkova V, et al. Key tumor suppressor genes inactivated by ‘greater promoter’ methylation and somatic mutations in head and neck cancer. Epigenetics. 2014;9:1031–1046. doi: 10.4161/epi.29025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li LY, Yin KM, Bai YH, Zhang ZG, Di W, Zhang S. CTHRC1 promotes M2-like macrophage recruitment and myometrial invasion in endometrial carcinoma by integrin-Akt signaling pathway. Clin Exp Metastasis. 2019;36:351–363. doi: 10.1007/s10585-019-09971-4. [DOI] [PubMed] [Google Scholar]
- 15.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 16.Boyango I, Barash U, Fux L, Naroditsky I, Ilan N, Vlodavsky I. Targeting heparanase to the mammary epithelium enhances mammary gland development and promotes tumor growth and metastasis. Matrix Biol. 2018;65:91–103. doi: 10.1016/j.matbio.2017.08.005. [DOI] [PubMed] [Google Scholar]
- 17.Li X, Li MW, Zhang YN, Xu HM. Common cancer genetic analysis methods and application study based on TCGA database. Yi Chuan. 2019;41:234–242. doi: 10.16288/j.yczz.18-279. [DOI] [PubMed] [Google Scholar]
- 18.Liu Y, Nan F, Lu K, Wang Y, Liu Y, Wei S, Wu R, Wang Y. Identification of key genes in endometrioid endometrial adenocarcinoma via TCGA database. Cancer Biomark. 2017;21:11–21. doi: 10.3233/CBM-170164. [DOI] [PubMed] [Google Scholar]
- 19.Çıkla-Süzgün P, Küçükgüzel ŞG. Recent advances in apoptosis: The role of hydrazones. Mini Rev Med Chem. 2019;19:1427–1442. doi: 10.2174/1389557519666190410125910. [DOI] [PubMed] [Google Scholar]
- 20.Wan J, Zou S, Hu M, Zhu R, Xu J, Jiao Y, Fan S. Thoc1 inhibits cell growth via induction of cell cycle arrest and apoptosis in lung cancer cells. Mol Med Rep. 2014;9:2321–2327. doi: 10.3892/mmr.2014.2088. [DOI] [PubMed] [Google Scholar]
- 21.Rivera M, Ramos Y, Rodríguez-Valentín M, López-Acevedo S, Cubano LA, Zou J, Zhang Q, Wang G, Boukli NM. Targeting multiple pro-apoptotic signaling pathways with curcumin in prostate cancer cells. PLoS One. 2017;12:e0179587. doi: 10.1371/journal.pone.0179587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baek SH, Ko JH, Lee JH, Kim C, Lee H, Nam D, Lee J, Lee SG, Yang WM, Um JY, et al. Ginkgolic acid inhibits invasion and migration and TGF-β-induced EMT of lung cancer cells through PI3K/Akt/mTOR inactivation. J Cell Physiol. 2017;232:346–354. doi: 10.1002/jcp.25426. [DOI] [PubMed] [Google Scholar]
- 23.Shen M, Xu Z, Xu W, Jiang K, Zhang F, Ding Q, Xu Z, Chen Y. Inhibition of ATM reverses EMT and decreases metastatic potential of cisplatin-resistant lung cancer cells through JAK/STAT3/PD-L1 pathway. J Exp Clin Cancer Res. 2019;38:149. doi: 10.1186/s13046-019-1161-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee GA, Hwang KA, Choi KC. Roles of dietary phytoestrogens on the regulation of epithelial-mesenchymal transition in diverse cancer metastasis. Toxins (Basel) 2016;8:E162. doi: 10.3390/toxins8060162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y, Xiao X, Bossé Y, Gorlova O, Gorlov I, Han Y, Byun J, Leighl N, Johansen JS, Barnett M, et al. Genetic interaction analysis among oncogenesis-related genes revealed novel genes and networks in lung cancer development. Oncotarget. 2019;10:1760–1774. doi: 10.18632/oncotarget.26678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lv Q, Wang G, Zhang Y, Han X, Li H, Le W, Zhang M, Ma C, Wang P, Ding Q. FABP5 regulates the proliferation of clear cell renal cell carcinoma cells via the PI3K/AKT signaling pathway. Int J Oncol. 2019;54:1221–1232. doi: 10.3892/ijo.2019.4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sharma N, Nanta R, Sharma J, Gunewardena S, Singh KP, Shankar S, Srivastava RK. PI3K/AKT/mTOR and sonic hedgehog pathways cooperate together to inhibit human pancreatic cancer stem cell characteristics and tumor growth. Oncotarget. 2015;6:32039–32060. doi: 10.18632/oncotarget.5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haodang L, Lianmei Q, Ranhui L, Liesong C, Jun H, Yihua Z, Cuiming Z, Yimou W, Xiaoxing Y. HO-1 mediates the anti-inflammatory actions of Sulforaphane in monocytes stimulated with a mycoplasmal lipopeptide. Chem Biol Interact. 2019;306:10–18. doi: 10.1016/j.cbi.2019.04.007. [DOI] [PubMed] [Google Scholar]
- 29.Zong HY, Wang EL, Han YM, Wang QJ, Wang JL, Wang Z. Effect of miR-29b on rats with gestational diabetes mellitus by targeting PI3K/Akt signal. Eur Rev Med Pharmacol Sci. 2019;23:2325–2331. doi: 10.26355/eurrev_201903_17372. [DOI] [PubMed] [Google Scholar]
- 30.Zhang W, Liang X, Gong Y, Xiao C, Guo B, Yang T. The signal transducer and activator of transcription 5B (STAT5B) gene promotes proliferation and drug resistance of human mantle cell lymphoma cells by activating the Akt signaling pathway. Med Sci Monit. 2019;25:2599–2608. doi: 10.12659/MSM.914934. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.