Abstract
Radioresistance hinders the therapeutic outcomes of radiotherapy in non-small cell lung cancer (NSCLC). Although long non-coding RNAs (lncRNAs) have been demonstrated to participate in the regulation of multiple cell behaviors, whether they can modulate the radiosensitivity of NSCLC and the underlying molecular mechanisms have not been well investigated. In the present study, it was revealed that NSCLC NCI-H460 cells were more sensitive to ionizing radiation (IR) than A549 cells. Using the RNA-Seq method, four highly differentially expressed lncRNAs were identified, including the growth arrest-specific transcript 5 (GAS5), syntaxin binding protein 5 antisense RNA 1 (STXBP5-AS1), metastasis associated lung adenocarcinoma transcript 1 (MALAT1) and X-inactive specific transcript (XIST), which were predicted to play roles in the acquisition of radiosensitivity. Using real-time quantitative PCR (qPCR), it was demonstrated that lncRNA GAS5 was significantly upregulated in NCI-H460 cells but not in A549 cells during IR. Mechanistically, it was demonstrated that overexpression of lncRNA GAS5 decreased the level of microRNA-21 (miR-21). Overexpression of lncRNA GAS5 or suppression of miR-21 markedly increased the IR-induced cell apoptosis of A549 cells. It was also demonstrated that overexpression of lncRNA GAS5 increased PTEN expression and suppressed Akt phosphorylation through the modulation of miR-21. Notably, it was revealed that IR enhanced the interaction between lncRNA GAS5 and the miR-21/PTEN/Akt axis. In summary, the present findings revealed that lncRNA GAS5 has a radiosensitization effect on NSCLC, indicating the potential application of lncRNA GAS5 in NSCLC radiotherapy.
Keywords: non-coding RNA GAS5, non-small cell lung cancer, miR-21, PTEN, Akt
Introduction
Lung cancer remains the leading cause of cancer incidence and mortality worldwide. The latest statistics revealed that there were 2.1 million newly diagnosed lung cancer cases and 1.8 million deaths predicted in 2018 (1). NSCLC is the most common lung cancer form, representing 80–85% of all lung cancer cases (2). Radiotherapy plays a key role in the treatment of both early stage and locally advanced NSCLC (3). However, due to the intrinsic or acquired radioresistance, the therapeutic outcomes of radiotherapy in NSCLC patients are not entirely satisfactory. Therefore, to improve the efficiency of radiotherapy, enhancing radiosensitivity and overcoming radioresistance have great practical significance in the clinical treatment of NSCLC.
Noncoding RNAs (ncRNAs) are a class of RNA molecules that are not translated into proteins (4). It has been demonstrated that >98% of the human genome is transcribed into ncRNA, including microRNA (miRNA), circular RNA(circRNA) and lncRNA (5–7). lncRNAs are largely polyadenylated transcripts and of >200 nts in length (8). Extensive studies have demonstrated that lncRNAs regulate gene expression at both the transcriptional and post-transcriptional levels and are involved in multiple cellular activities including cell development, differentiation, proliferation, apoptosis, invasion and migration (9,10). It is also suggested that lncRNAs participate in the regulation of the IR-induced apoptotic process. For example, Lai et al reported that downregulation of lncRNA CCAT1 improved the radiosensitivity of breast cancer cells (11) and Wu et al reported that knockdown of lncRNA PVT1 enhanced the radiosensitivity of NSCLC (12).
lncRNA growth arrest-specific transcript 5 (GAS5), located at chromosome 1q25.1, was originally isolated from a screen for potential tumor suppressor genes during cancer cell growth arrest and apoptosis (13). Recently, lncRNA GAS5 was revealed to be aberrantly expressed in various cancerous tissues (14,15) and modulate chemo- and radio-responses (16,17). However, little is known concerning the functional role of lncRNA GAS5 and its underlying molecular mechanism in promoting radiosensitivity of NSCLC.
In the present study, it was determined that lncRNA GAS5 was differentially expressed between the radiosensitive NSCLC cell line NCI-H460 and the radioresistant cell line A549. The effects of lncRNA GAS5 and its binding target microRNA-21 (miR-21) on IR-induced cell apoptosis were investigated and the interactions between lncRNA GAS5, miR-21 and the PTEN/Akt pathway were explored.
Material and methods
Cell line selection and cell culture
Two human NSCLC cell lines A549 and NCI-H460 were selected for this study. These two lines were selected because they share common genetic features, e.g. both the two cell lines are wild-type in TP53 and therefore, compared to the mutated lines, they are less likely to exhibit genomic instability over the course of lncRNA screening (18). Another reason is that they exhibit markedly different responses to IR (19).
The NCI-H460 cell line was obtained from the American Type Culture Collection (ATCC). A549 and 239T cells were purchased from the Type Culture Collection of the Chinese Academy of Sciences (#SCSP-503 and #GNHu17; Shanghai) and maintained in our laboratory. Cells were grown in DMEM medium with 10% fetal bovine serum and penicillin/streptomycin (all from Hyclone; GE Healthcare Life Sciences) at 37°C in 95% air/5% CO2.
Ionizing radiation
A549 and NCI-H460 cells were cultured in 75 cm2 cell culture flasks (Corning, Inc.). For IR, the cells were received up to a total dose of 8 Gy X-ray at a dose rate of 1 Gy/min in X-RAD 320 (Precision X-RAD; Precision X-Ray). After IR, the tissue culture medium was refreshed and the cells were continually cultured in the same condition until the subsequent experiments were performed.
Cell viability assay
Cell viability was evaluated by WST-1 assay (Roche Diagnostics). A549 and NCI-H460 cells (5×103) were seeded in 96-well plates. After 24 h, the cells were divided into five groups and irradiated with 0, 2, 4, 6 or 8 Gy X-ray. According to the manufacturer's instructions, at 12 h post-IR, WST-1 was added to cell supernatants and incubated at 37°C for 3 h in the dark. The absorbance of 450–630 nm was measured with a microplate reader (Thermo Labsystem MK3; Thermo Fisher Scientific, Inc.).
Cell apoptosis assay
Cell apoptosis was analyzed using an Annexin V-FITC Apoptosis Detection Kit (cat. no. 556547; BD Biosciences). Briefly, 1×105 A549 and NCI-H460 cells were digested and washed with 1X binding buffer and centrifuged for 5 min at 200 × g. The cell pellet was suspended and stained with 50 µl staining solution containing 5 µl PI and 5 µl Annexin V-FITC. Data were acquired on a FACScan flow cytometer (BD Biosciences) and analyzed with FlowJo software (version 10.0; FlowJo LLC).
RNA extraction and reverse transcriptional reaction
Total RNA was isolated using EasyPure RNA Purification Kit (cat. no. ER701; TransGen Biotech Co., Ltd.). For microRNA experiments, EasyPure miRNA Kit (cat. no. ER601; TransGen Biotech Co., Ltd.) was used specially for <200 nt RNA purification. For reverse transcriptional reaction, 800–1,000 ng isolated RNA was reverse-transcribed with PrimeScript RT reagent Kit (cat. no. RR037, Takara Biotechnology, Co, Ltd.). Reverse transcriptional primers were as follows: miR-21, 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTCAACA-3′ (16); miR-23a, 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGGAAAT-3′; and miR222, 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACACCCAG-3′; for the cDNA library construction and PCR reaction oligo(dT)15 plus random primers were used.
Next-generation sequencing
cDNAs from the irradiated and control cells were sent for sequencing. RNA-Seq was performed on a HiSeq4000 (Illumina, Inc.) and yielded ~35 million reads with a length of 150 bp per sample (Beijing Honor Tech Co., Ltd.). Gene counts were normalized to the values of fragments per kilobase of transcript per million mapped reads (FPKM).
Plasmid construction
To overexpress lncRNA GAS5 in cells, the full-length of GAS5 gene was synthesized by Shanghai Sangon Biotech Co., Ltd. and cloned into a GFP-deleted pGreenPuro lentiviral vector (SBI) using XbaI and EcoRV to construct the pGreenPuro-GAS5 plasmid.
Lentivirus packaging and transduction
293 T cells were transfected with a total amount of 2 µg of lentiviral expression construct including 800 ng psPAX2, 400 ng pMD2.G and 800 ng pGreenPuro-GAS5. Transfections were performed in 6-well plates using Lipofectamine 2000 according to the manufacturer's instructions (Invitrogen; Thermo Fisher Scientific, Inc.). Viral supernatants were collected twice at 24 and 48 h post-transfection. For the transduction, A549 cells were incubated in lentiviral supernatants for 48 h until the transduction efficiency was ~100%.
siRNA and miRNA transfection
siRNA targeting GAS5 (si-GAS5) and siRNA negative control (si-NC) were synthesized by Shanghai GenePharma Co., Ltd. The si-GAS5 sense sequence was: 5′-UCUUCAAUCAUGAAUUCUGAG-3′ and the antisense sequence was: 5′-CAGAAUUCAUGAUUGAAGAAA-3′. The si-NC sense sequence was: 5′-UUCUCCGAACGUGUCACGUTT-3′ and the antisense sequence was: 5′-ACGUGACACGUUCGGAGAATT-3′ (16). miR-21 inhibitor and inhibitor negative control molecules (inhibitor-NC) were purchased from Guangzhou Ribobio Co., Ltd. The sequence of miR-21 inhibitor was 5′-UCAACAUCAGUCUGAUAAGCUA-3′ and the sequence of inhibitor-NC was 5′-CAGUACUUUUGUAGUACAA-3′. Briefly, 2×105 A549 cells in one well of a 6-well plate were transfected with 50 nM siRNA/si-NC or miR-21 inhibitor/inhibitor-NC mixed in 2 ml Opti-MEM plus 10% FBS using Lipofectamine 2000. Subsequently, 24 h after transfection, the cells were prepared for the subsequent experiments.
qPCR reaction
qPCR was conducted based on the instruction of RealStar Green Fast Mixture (GenStar). Primers for qPCR are listed as follows: GAS5 forward, 5′-CCTGTGAGGTATGGTGCTGG-3′ and reverse, 5′-CTGTGTGCCAATGGCTTGAG-3′ (20); syntaxin binding protein 5-antisense RNA 1 (STXBP5-AS1) forward, 5′-AGGGACTTGCCTTGTCGCTGAT-3′ and reverse, 5′-GAGATTTAGGTGGGGACGCTGC-3′ (21); metastasis associated lung adenocarcinoma transcript 1 (MALAT1) forward, 5′-CATAACCCTGAGATTCTTACTAC-3′ and reverse, 5′-TTGTGGTTATAGCTTGACAAGCA-3′; X-inactive specific transcript (XIST) forward, 5′-CTAGCTAGCTTTTGTAGTGAGCTTGCTCCT-3′ and reverse, 5′-TGCTCTAGAATGTCTCCATCTCCATTTTGC-3′ (22); miR-21 forward, 5′-GCACtagcttatcagactga-3′; miR-23a forward, 5′-TCACatcacattgccagG-3′; miR-222 forward, 5′-GCACagctacatctggct-3′. The universal reverse primer for miRNA amplification was 5′-GTGCAGGGTCCGAGGT-3′ (16). Glyceraldehyde phosphate dehydrogenase (GAPDH) and U6 small nuclear RNA were used as the internal control. The primers for GAPDH were forward, 5′-TGTGGGCATCAATGGATTTGG-3′ and reverse, 5′-ACACCATGTATTCCGGGTCAAT-3′. The primers for U6 were forward, 5′-AAAGCAAATCATCGGACGACC-3′ and reverse, 5′-GTACAACACATTGTTTCCTCGGA-3′ (20). The PCR program consisted of 10 sec at 95°C followed by 40 cycles at 95°C for 5 sec and 60°C for 35 sec, which was performed on an ABI 7500 qPCR instrument (Applied Biosystems; Thermo Fisher Scientific, Inc.). The relative expression of the aforementioned target gene was calculated based on the 2−ΔΔCq method (23).
Western blotting
RIPA buffer (KeyGen Biotech. Co., Ltd.) was used to collect total protein. Protein concentration was determined by BCA reagent (KeyGen Biotech). Spacer gel (5%) and 10% separation gel were prepared using an SDS-PAGE gel kit (Beijing, Solarbio Science & Technology Co., Ltd.). An equal amount of each protein (30 µg) was loaded to the gel and electrophoresis was performed on a Bio-Rad Mini-PROTEAN system (Bio-Rad Laboratories, Inc.). Proteins were transferred to 0.45 µm PVDF membranes (EMD Millipore) using a semi-dry transfer system (Bio-Rad Laboratories, Inc.). Membranes were blocked with 5% BSA at room temperature for 1 h and incubated with the following rabbit monoclonal primary antibodies (Cell Signaling Technology, Inc.) at 4°C overnight: PTEN (1:1,000; product no. 9188), Akt (1:1,000; product no. 4691), p-Akt (1:1,000; Ser473; product no. 4060) and β-actin (1:3,000; product no. 4970). After membranes were incubated with an HRP-conjugated secondary antibody (1:3,000; cat. no. sc-2357; Santa Cruz Biotechnology, Inc.) for 45 min at room temperature, blotting signals were visualized with ECL substrates (EMD Millipore). Densitometric analysis was conducted using Quantity One software (version 4.6; Bio-Rad Laboratories) and protein expression was normalized to endogenous β-actin.
Statistical analysis
All data were analyzed with the SPSS software 17.0 (SPSS, Inc.). Measurement data were expressed as the arithmetic mean ± standard deviation. Statistical significance between two or more groups were compared using Student's t-test or one-way analysis of variance (ANOVA) followed by an LSD or Dunnett's post hoc test. P<0.05 and P<0.01 were considered as statistically significant. All data were obtained from at least 3 independent experiments.
Results
Differential radiosensitivity between A549 and NCI-H460 cells
We used 2/4/6/8 Gy X-ray to irradiate A549 and NCI-460 cells and compared the response of these two cell lines. As revealed in Fig. 1, compared with the unirradiated group, the relative cell viability of A549 vs. NCI-H460 in 2/4/6/8 Gy IR groups were 87 vs. 75%, 77 vs. 42%, 68 vs. 33%, 29 vs. 18% (P<0.05), respectively. Notably, NCI-H460 cells were more radiosensitive than A549 cells, especially in the 4/6 Gy IR groups.
Figure 1.
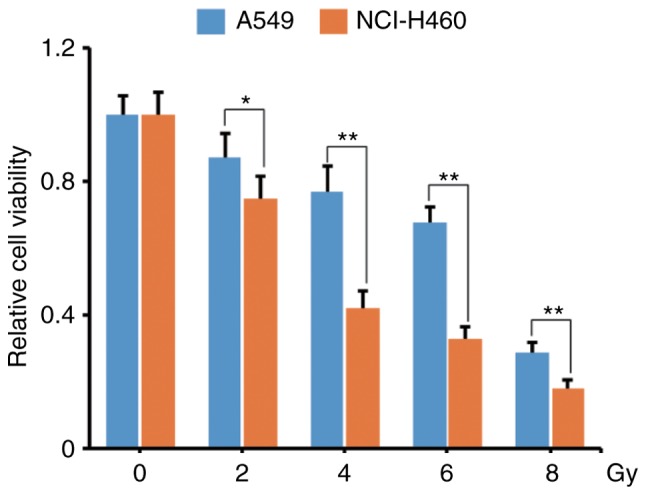
Differential radiosensitivity between A549 and NCI-H460 NSCLC cells. A549 and NCI-460 cells were irradiated with 2/4/6/8 Gy X-ray and the cell viability was analyzed by WST-1 at 12 h post-radiation. A549 cells had higher viability than NCI-H460 cells. *P<0.05 and **P<0.01. NSCLC, non-small cell lung cancer.
Screening of differentially expressed lncRNAs between A549 and NCI-460 cells after IR
Differentially expressed lncRNAs of the irradiated A549 and NCI-H460 cells were screened by RNA-Seq and analyzed. The following standards were set for the selection of lncRNA candidates: i) a FPKM fold-change (IR group/control group) of NCI-H460 cells >2; ii) a ratio of FPKM fold-change (FPKM fold-change of NCI-H460 cells/FPKM fold-change of A549 cells) >1.5; iii) having definite biological functions. Finally, four lncRNAs were identified: GAS5, STXBP5-AS1, MALAT1 and XIST. Their FPKM values are listed in Table I. The fold-change of each lncRNA and the ratios of fold-change between the two cell lines are presented and compared in Fig. 2.
Table I.
FPKM values of lncRNA GAS5, STXBP5-AS1, MALAT1 and XIST.
| FPKM (mean, n=3) | FPKM (mean, n=3) | |||||
|---|---|---|---|---|---|---|
| Gene | NCI-H460 (CT) | NCI-H460 (IR) | Fold-change | A549 (CT) | A549 (IR) | Fold-change |
| GAS5 | 19.1 | 256.3 | 13.4 | 34.7 | 55.2 | 1.6 |
| STXBP5-AS1 | 22.5 | 153.8 | 6.8 | 15.3 | 34.8 | 2.3 |
| MALAT1 | 253.4 | 1023.3 | 4.0 | 410.1 | 706.5 | 1.7 |
| XIST | 54.4 | 114.5 | 2.1 | 41.1 | 52.1 | 1.3 |
A549 and NCI-H460 cells were irradiated with 4 Gy X-ray. Total RNAs were extracted and RNA-Seq was performed. CT, 0 Gy group; IR, 4 Gy group. IR, ionizing radiation.
Figure 2.
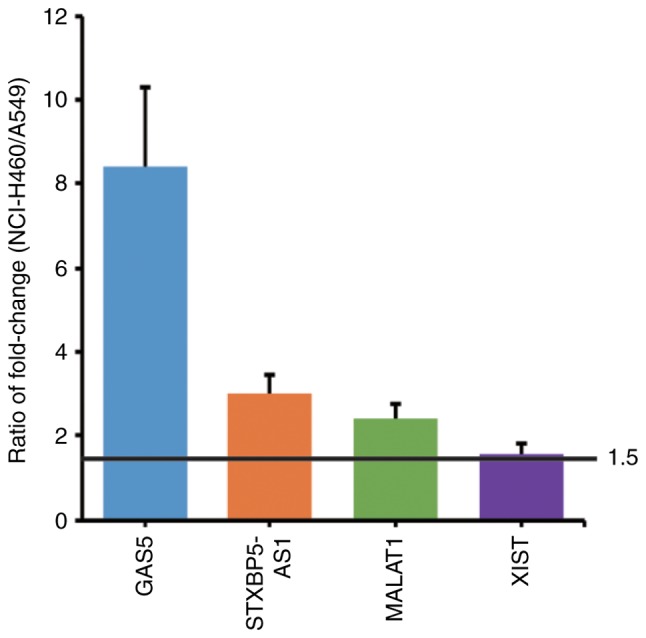
Screening of differentially expressed lncRNAs by RNA-Seq. A549 and NCI-H460 cells were irradiated with 4 Gy X-ray, and the differentially expressed lncRNAs were screened by RNA-Seq. The lncRNAs with an FPKM fold-change in NCI-H460 cells >2 and a ratio of FPKM fold-change >1.5 were selected. Fold-change=FPKM of the IR group/FPKM of the control group. The ratio of the fold-change=FPKM fold-change of NCI-H460 cells/FPKM fold-change of A549 cells. FPKM, fragments per kilobase of transcript per million mapped reads.
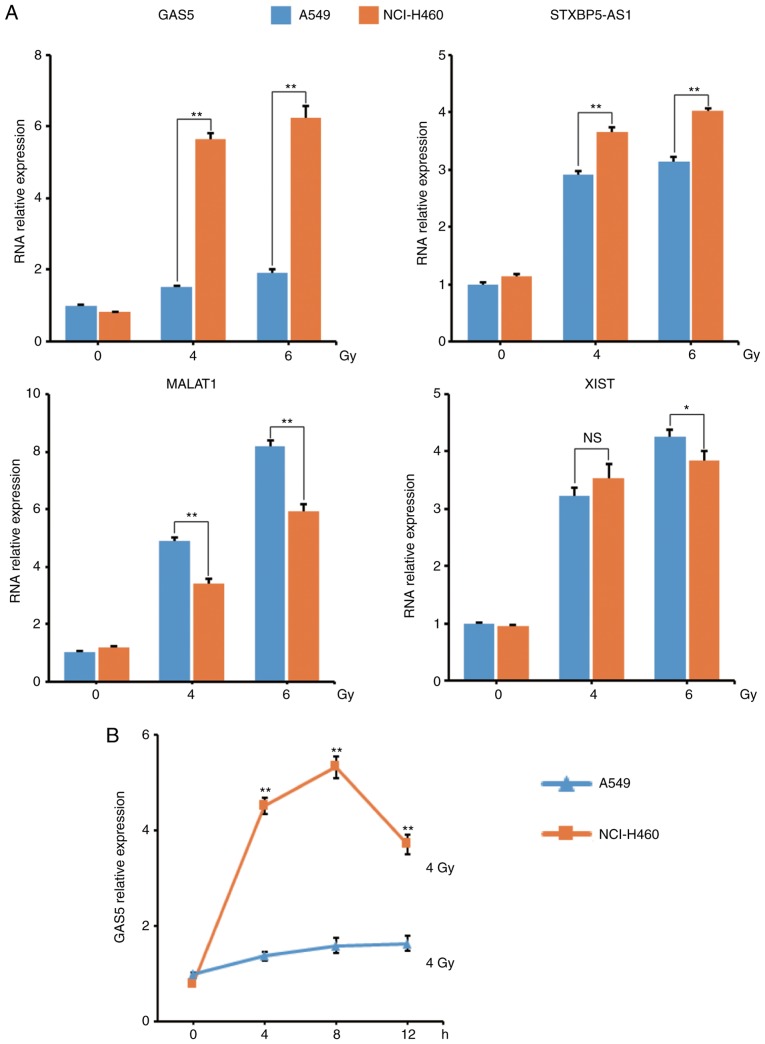
To verify the RNA-Seq result, the lncRNA levels in these two cell lines were assessed using qPCR at 8 h post-IR. The results revealed that lncRNA GAS5 had the most marked change and the qPCR result was in line with the RNA-Seq result (Fig. 3A). The level of lncRNA GAS5 was then monitored and it was revealed that the level of lncRNA GAS5 in NCI-H460 cells was significantly higher than in A549 cells at 4/8/12 h post-4 Gy-IR (Fig. 3B).
Figure 3.
Verification of lncRNA expression using qPCR. After A549 and NCI-H460 cells were irradiated with 4/6 Gy X-ray, the levels of the four identified lncRNAs were assessed by qPCR and compared. (A) qPCR results. (B) Expression of lncRNA GAS5 at 4/8/12 h post-IR. *P<0.05, **P<0.01; NS, no significance. IR, ionizing radiation.
Overexpression of lncRNA GAS5 increases the radiosensitivity of A549 cells
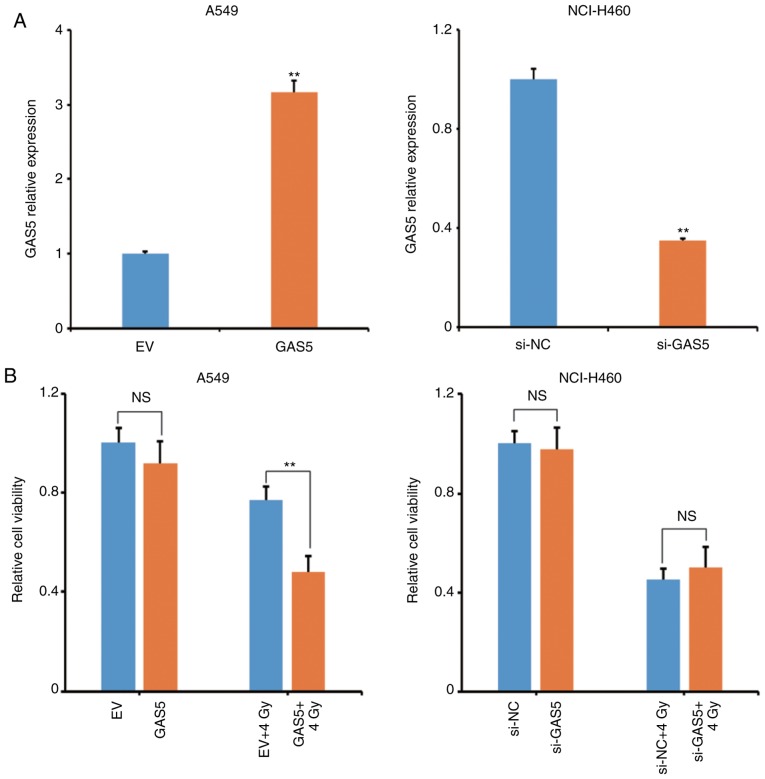
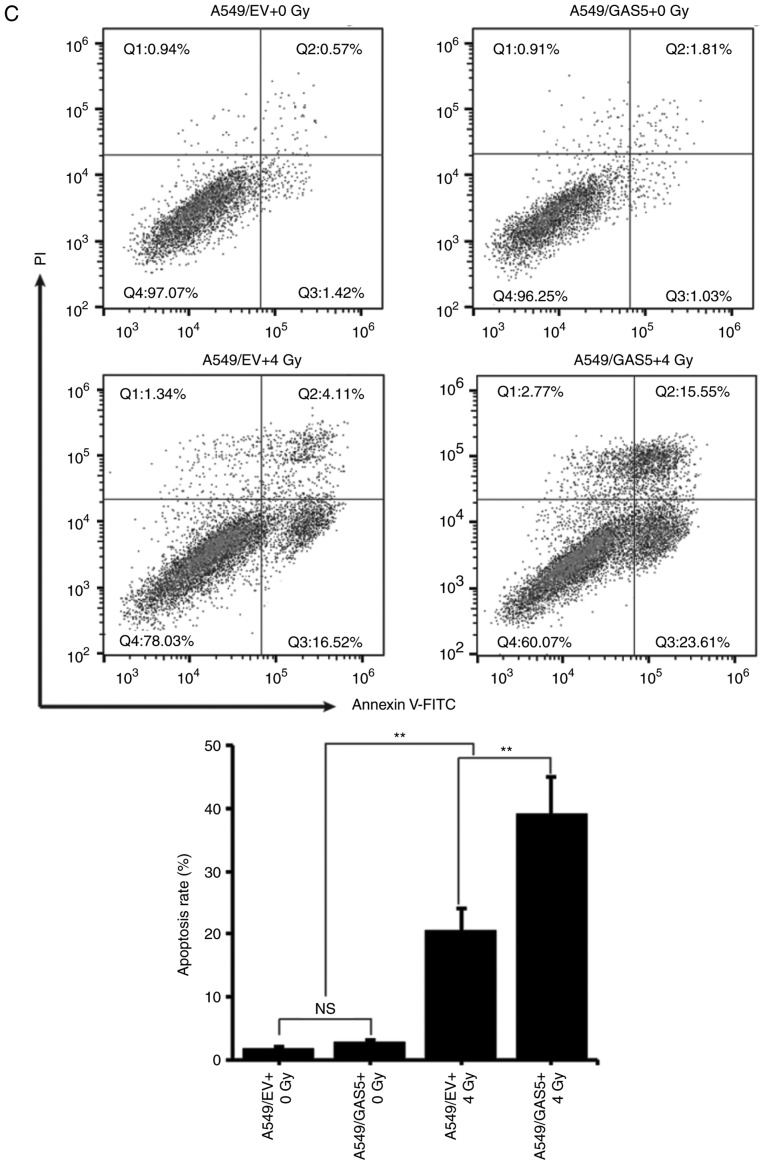
To investigate the effect of lncRNA GAS5 on IR-induced apoptosis, a GAS5-expressing plasmid was constructed and transfected to A549 cells to observe whether it would enhance the radiosensitivity. Concurrently, the lncRNA GAS5 siRNA was transfected to NCI-H460 cells to observe whether it would weaken the radiosensitivity. Fig. 4A revealed that the transfection efficiency was satisfactory. The level of lncRNA GAS5 in A549 cells was significantly increased and in NCI-H460 cells it was significantly decreased. The viability of transfectants at 12 h post-4 Gy-IR was then analyzed and it was revealed that overexpression of lncRNA GAS5 significantly decreased the viability of A549 cells (Fig. 4B, left panel). However, interfering of lncRNA GAS5 did not affect the viability of NCI-H460 cells after IR (Fig. 4B, right panel). The apoptosis of A549 transfectants was next analyzed and it was revealed that overexpression of lncRNA GAS5 enhanced the IR-induced cell apoptosis (Fig. 4C).
Figure 4.
Overexpression of lncRNA GAS5 increases the radiosensitivity of A549 cells. To clarify the correlation between lncRNA GAS5 and radiosensitivity, lncRNA GAS5-overexpressing A549 cells and lncRNA GAS5-knockdown NCI-H460 cells were generated. Cell survival and apoptosis were assessed after 4 Gy IR. (A) Overexpression and knockdown efficacy in cell transfectants were confirmed by qPCR. (B) Cell viability of the transfectants was determined by WST-1 assay. **P<0.01; NS, no significance. IR, ionizing radiation. Overexpression of lncRNA GAS5 increases the radiosensitivity of A549 cells. To clarify the correlation between lncRNA GAS5 and radiosensitivity, lncRNA GAS5-overexpressing A549 cells and lncRNA GAS5-knockdown NCI-H460 cells were generated. Cell survival and apoptosis were assessed after 4 Gy IR. (C) Cell apoptosis of the transfectants was determined by flow cytometry. **P<0.01; NS, no significance. IR, ionizing radiation.
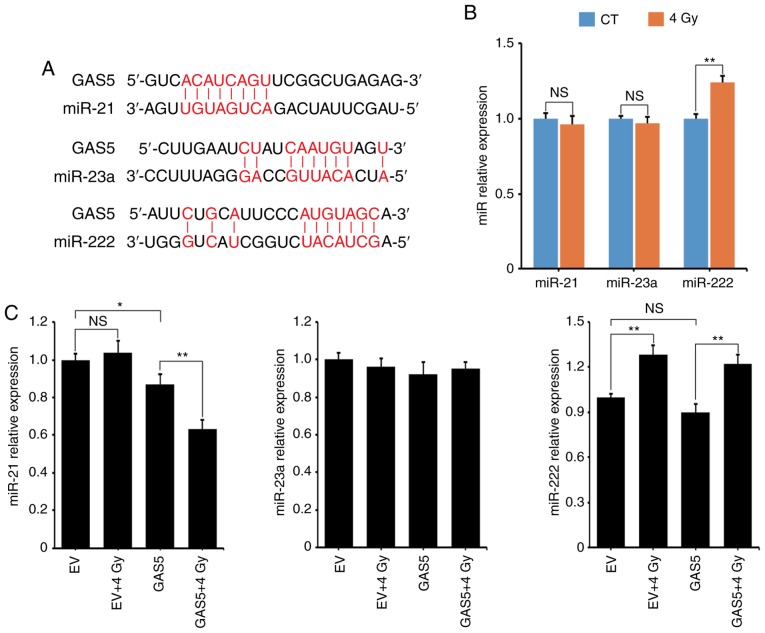
lncRNA GAS5 suppresses miR-21 expression during IR
It has been reported that lncRNA may serve as a ‘sponge’ to absorb miRNAs and regulate miRNA expression (24). In the present experiment, three miRNAs that may interact with lncRNA GAS5 were selected: miR-21, miR-23a and miR-222 (16,25,26). According to the references, the binding sites of these three miRNAs on lncRNA GAS5 are revealed in Fig. 5A. The expression levels of these miRNAs at 8 h post-4 Gy-IR was assessed using qPCR and fit was revealed that only miR-222 was upregulated (Fig. 5B). The expression levels of these miRNAs in A549 transfectants was next assessed and it was revealed that lncRNA GAS5 significantly suppressed the level of miR-21 during IR (Fig. 5C, left panel). Although the level of miR-222 increased after IR, it was not affected by the overexpression of lncRNA GAS5 (Fig. 5C, right panel). Additionally, neither IR nor lncRNA GAS5 affected the level of miR-23a (Fig. 5C, middle panel).
Figure 5.
Screening of miRNAs interacting with lncRNA GAS5. (A) Three predicted miRNAs and their binding sites on lncRNA GAS5. (B) After 4 Gy IR, the expression of miRNAs in A549 cells was assessed by qPCR. (C) The expression level of miRNAs in A549 transfectants. *P<0.05 and **P<0.01; NS, no significance. IR, ionizing radiation.
Inhibition of miR-21 increases the radiosensitivity of A549 cells
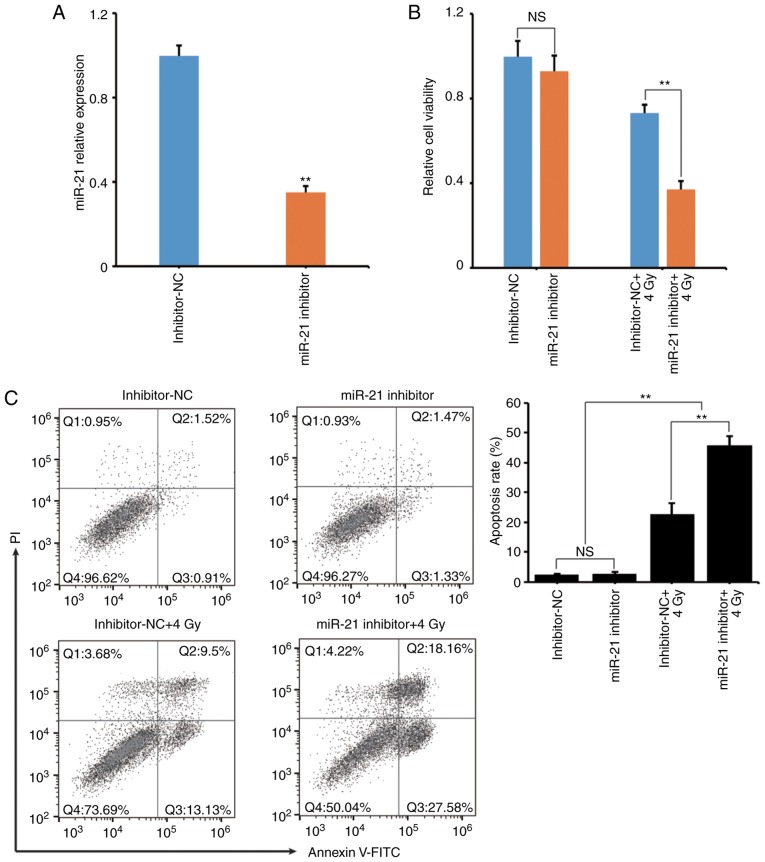
To clarify the role of miR-21 in lncRNA GAS5-induced cell apoptosis during IR, miR-21 expression was suppressed using a miR-21 inhibitor. Fig. 6A reveals that the transfection efficiency of the miR-21 inhibitor was confirmed to be successful and the endogenous miR-21 expression was significantly decreased. It was also revealed that suppression of miR-21 significantly decreased the viability and increased the IR-induced cell apoptosis rate (Fig. 6B and C) of A549 cells.
Figure 6.
Suppression of miR-21 increases the radiosensitivity of A549 cells. To clarify the role of miR-21 in lncRNA GAS5-induced cell apoptosis, an inhibitor was used to suppress the level of miR-21, and cell survival and apoptosis were assessed after 4 Gy radiation. (A) The level of miR-21 in A549 transfectants was assessed by qPCR. (B) After 4 Gy radiation, the cell survival of the transfectants was determined by WST-1 assay. (C) After 4 Gy radiation, cell apoptosis of the transfectants was determined by flow cytometry. **P<0.01; NS, no significance.
lncRNA GAS5 regulates the radiosensitivity of A549 cells through the miR-21/PTEN/Akt axis
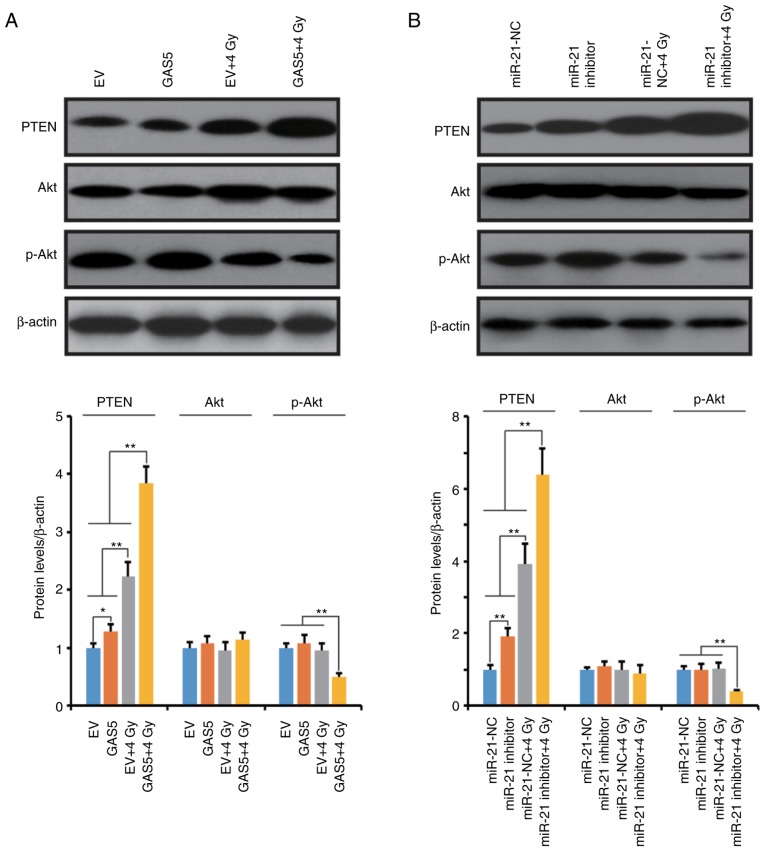
PTEN is one of the most important target proteins of miR-21, and the miR-21/PTEN/Akt axis has been revealed to play a crucial role in various cellular activities (27–30). In this experiment, the activation of the PTEN/Akt pathway was also assessed in our experimental model. In Fig. 7, western blot results revealed that overexpression of lncRNA GAS5 and suppression of miR-21 upregulated the expression level of PTEN but did not affect the phosphorylation of Akt in A549 cells. However, 4 Gy-IR significantly stimulated the expression of PTEN and suppressed the phosphorylation of Akt. Notably, overexpression of lncRNA GAS5 and inhibition of miR-21 exacerbated the IR-induced upregulation of PTEN and the phosphorylation of Akt in A549 cells. These data indicated that lncRNA GAS5 may function as a radiosensitizer through the regulation of the miR-21/PTEN/Akt axis.
Figure 7.
Overexpression of lncRNA GAS5 and suppression of miR-21 upregulate PTEN and activate the Akt pathway during IR. (A) Western blot results revealed that IR stimulated PTEN expression and suppressed Akt phosphorylation in A549 cells. Notably, overexpression of lncRNA GAS5 increased this effect. (B) Suppression of miR-21 generated a similar effect as the overexpression of lncRNA GAS5. Upper panel: Western blot bands; lower panels: Histograms, comparison of the relative band density. *P<0.05 and **P<0.01. IR, ionizing radiation.
Discussion
LncRNA GAS5 was originally identified from a serum starvation-induced cell growth arrest model (31). It was revealed to accumulate in cells and sensitize cells to apoptosis by suppressing the glucocorticoid-mediated induction of several responsive genes (32). In recent studies, lncRNA GAS5 has been revealed to be implicated in tumorigenesis and tumor development in a variety of cancers (33–36). However, the role of lncRNA GAS5 in NSCLC radiotherapy is still not fully investigated.
In the present study, it was revealed that NSCLC NCI-H460 cells were more prone to apoptosis than A549 cells during IR. The correlation between radiosensitivity and the unique lncRNAs was investigated. Using RNA-Seq, it was demonstrated that in NCI-H460 cells four well-characterized lncRNAs, GAS5, STXBP5-AS1, MALAT1 and XIST, were significantly upregulated after IR, among which lncRNA GAS5 was the most differentially expressed. It was also demonstrated that overexpression of lncRNA GAS5 could enhance apoptosis of A549 cells. However, knockdown of lncRNA GAS5 did not affect the radiosensitivity of NCI-H460 cells. These data indicated that lncRNA GAS5 was sufficient but not necessary for the radiosensitivity of NSCLC cells.
One important way that lncRNAs exert their regulatory function is to serve as competing endogenous RNAs to negatively regulate the biological activity of miRNAs. A number of studies have reported the interaction between lncRNAs and miRNAs in various physiological and pathological processes. For example, Lai et al demonstrated that downregulation of lncRNA CCAT1 improved the radiosensitivity of breast cancer cells by negatively regulating miR-148b (11). Wu et al demonstrated that knockdown of lncRNA PVT1 improved the radiosensitivity of NSCLC cells by directly interacting with miR-195 (12). In the present study, the expression of three miRNAs that may be targets of lncRNA GAS5 were assessed and it was demonstrated that only miR-21 was affected by lncRNA GAS5 inirradiated A549 cells. The direct interaction between lncRNA GAS5 and miR-21 has been revealed by several research groups using a luciferase assay (37,38). The present results indicated that although overexpression of lncRNA GAS5 could downregulate the level of miR-21 under normal conditions, this suppressive effect was markedly increased during IR. These data indicated that the interaction between lncRNA GAS5 and miR-21 may be an IR-specific response.
PTEN, a tumor suppressor, plays a critical role in ceRNA networks (39). PTEN mRNA is the competing target of many miRNAs, e.g. miR-624-3p, miR-130a, and miR-21 (40–42). There are several potential binding sites of miR-21 on the PTEN 3′UTR, such as GAUAAG and UCUG*UG*GCUA (43,44). PTEN is an inhibitor of the PI3K/Akt signaling pathway (45), and the PTEN/Akt axis is a pivotal regulatory pathway in IR-induced cell apoptosis. He et al demonstrated that overexpression of circVRK1 could reverse the radioresistance of esophageal squamous carcinoma cells by regulating the miR-624-3p/PTEN/PI3K/Akt signaling pathway (40). Wang et al reported that miR-29a regulated the radiosensitivity of human intestinal cells by activating the PTEN/PI3K/Akt signaling pathway (46). The present study indicated that overexpression of lncRNA GAS5 or suppression of miR-21 could increase the level of PTEN and decrease the phosphorylation of Akt, especially during the IR process. This indicated that lncRNA GAS5 enhanced the radiosensitivity of A549 cells by regulating the miR-21/PTEN/Akt pathway.
In conclusion, the present study revealed that lncRNA GAS5 was associated with the distinct radiosensitivity of NSCLC. Overexpression of lncRNA GAS5 could increase the IR-induced cell apoptosis of A549 cells. Therefore, lncRNA GAS5 has a great potential of being used as a radiosensitizer or a radiosensitivity indicator in clinic radiotherapy. However, to draw a more solid conclusion, additional cell lines, in vivo animal experiments and clinical investigation should be carried out in future work.
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the Health Special Fund from Jilin Province Department of Finance no. 2018SCZWSZX-048 to XS), the Jilin International Collaboration Grant no. 20180414065GH to DY) and the Norman Bethune Program of Jilin University (Bethune plan to DY).
Availability of data and materials
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
XS and BG contributed to the conception and design of the study. XS and DY drafted the study. LC, PR and YZ contributed to the acquisition, analysis and interpretation of data. DY also revised this study critically for important intellectual content. All the authors have read and approve the final article to be submitted and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Theys J, Yahyanejad S, Habets R, Span P, Dubois L, Paesmans K, Kattenbeld B, Cleutjens J, Groot AJ, Schuurbiers OCJ, et al. High NOTCH activity induces radiation resistance in non small cell lung cancer. Radiother Oncol. 2013;108:440–445. doi: 10.1016/j.radonc.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker S, Dahele M, Lagerwaard FJ, Senan S. A critical review of recent developments in radiotherapy for non-small cell lung cancer. Radiat Oncol. 2016;11:115. doi: 10.1186/s13014-016-0693-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hsiao KY, Sun HS, Tsai SJ. Circular RNA-new member of noncoding RNA with novel functions. Exp Biol Med (Maywood) 2017;242:1136–1141. doi: 10.1177/1535370217708978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barbarotto E, Schmittgen TD, Calin GA. MicroRNAs and cancer: Profile, profile, profile. Int J Cancer. 2008;122:969–977. doi: 10.1002/ijc.23343. [DOI] [PubMed] [Google Scholar]
- 7.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: Insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 8.Wright MW, Bruford EA. Naming ‘junk’: Human non-protein coding RNA (ncRNA) gene nomenclature. Hum Genomics. 2011;5:90–98. doi: 10.1186/1479-7364-5-2-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 10.Wei S, Du M, Jiang Z, Hausman GJ, Zhang L, Dodson MV. Long noncoding RNAs in regulating adipogenesis: New RNAs shed lights on obesity. Cell Mol Life Sci. 2016;73:2079–2087. doi: 10.1007/s00018-016-2169-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lai Y, Chen Y, Lin Y, Ye L. Down-regulation of LncRNA CCAT1 enhances radiosensitivity via regulating miR-148b in breast cancer. Cell Biol Int. 2018;42:227–236. doi: 10.1002/cbin.10890. [DOI] [PubMed] [Google Scholar]
- 12.Wu D, Li Y, Zhang H, Hu X. Knockdown of Lncrna PVT1 enhances radiosensitivity in non-small cell lung cancer by sponging mir-195. Cell Physiol Biochem. 2017;42:2453–2466. doi: 10.1159/000480209. [DOI] [PubMed] [Google Scholar]
- 13.Ricciuti B, Mencaroni C, Paglialunga L, Paciullo F, Crinò L, Chiari R, Metro G. Long noncoding RNAs: New insights into non-small cell lung cancer biology, diagnosis and therapy. Med Oncol. 2016;33:18. doi: 10.1007/s12032-016-0731-2. [DOI] [PubMed] [Google Scholar]
- 14.Sun M, Jin FY, Xia R, Kong R, Li JH, Xu TP, Liu YW, Zhang EB, Liu XH, De W. Decreased expression of long noncoding RNA GAS5 indicates a poor prognosis and promotes cell proliferation in gastric cancer. BMC Cancer. 2014;14:319. doi: 10.1186/1471-2407-14-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li W, Huang K, Wen F, Cui G, Guo H, Zhao S. Genetic variation of lncRNA GAS5 contributes to the development of lung cancer. Oncotarget. 2017;8:91025–91029. doi: 10.18632/oncotarget.19955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cao L, Chen J, Ou B, Liu C, Zou Y, Chen Q. GAS5 knockdown reduces the chemo-sensitivity of non-small cell lung cancer (NSCLC) cell to cisplatin (DDP) through regulating miR-21/PTEN axis. Biomed Pharmacother. 2017;93:570–579. doi: 10.1016/j.biopha.2017.06.089. [DOI] [PubMed] [Google Scholar]
- 17.Xue Y, Ni T, Jiang Y, Li Y. Long noncoding RNA GAS5 inhibits tumorigenesis and enhances radiosensitivity by suppressing miR-135b expression in non-small cell lung cancer. Oncol Res. 2017;25:1305–1316. doi: 10.3727/096504017X14850182723737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cron KR, Zhu K, Kushwaha DS, Hsieh G, Merzon D, Rameseder J, Chen CC, D'Andrea AD, Kozono D. Proteasome inhibitors block DNA repair and radiosensitize non-small cell lung cancer. PLoS One. 2013;8:e73710. doi: 10.1371/journal.pone.0073710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim W, Youn H, Seong KM, Yang HJ, Yun YJ, Kwon T, Kim YH, Lee JY, Jin YW, Youn B. PIM1-activated PRAS40 regulates radioresistance in non-small cell lung cancer cells through interplay with FOXO3a, 14-3-3 and protein phosphatases. Radiat Res. 2011;176:539–552. doi: 10.1667/RR2609.1. [DOI] [PubMed] [Google Scholar]
- 20.Liu L, Wang HJ, Meng T, Lei C, Yang XH, Wang QS, Jin B, Zhu JF. lncRNA GAS5 inhibits cell migration and invasion and promotes autophagy by targeting miR-222-3p via the GAS5/PTEN-signaling pathway in CRC. Mol Ther Nucleic Acids. 2019;17:644–656. doi: 10.1016/j.omtn.2019.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Huang J, Xie N, Huang H, Yao J, Hu W. Long noncoding RNA STXBP5-AS1 inhibits cell proliferation, migration, and invasion via preventing the PI3K/AKT against STXBP5 expression in non-small-cell lung carcinoma. J Cell Biochem. 2018;18:28023. doi: 10.1002/jcb.28023. [DOI] [PubMed] [Google Scholar]
- 22.Zhang YL, Li XB, Hou YX, Fang NZ, You JC, Zhou QH. The lncRNA XIST exhibits oncogenic properties via regulation of miR-449a and Bcl-2 in human non-small cell lung cancer. Acta Pharmacol Sin. 2017;38:371–381. doi: 10.1038/aps.2017.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Wei Y, Yan Z, Wu C, Zhang Q, Zhu Y, Li K, Xu Y. Integrated analysis of dosage effect lncRNAs in lung adenocarcinoma based on comprehensive network. Oncotarget. 2017;8:71430–71446. doi: 10.18632/oncotarget.19864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dong Z, Li S, Wang X, Si L, Ma R, Bao L, Bo A. lncRNA GAS5 restrains CCl4-induced hepatic fibrosis by targeting miR-23a through the PTEN/PI3K/Akt signaling pathway. Am J Physiol Gastrointest Liver Physiol. 2019;316:G539–G550. doi: 10.1152/ajpgi.00249.2018. [DOI] [PubMed] [Google Scholar]
- 26.Li Y, Gu J, Lu H. The GAS5/miR-222 axis regulates proliferation of gastric cancer cells through the PTEN/Akt/mTOR pathway. Dig Dis Sci. 2017;62:3426–3437. doi: 10.1007/s10620-017-4831-4. [DOI] [PubMed] [Google Scholar]
- 27.Wickramasinghe NS, Manavalan TT, Dougherty SM, Riggs KA, Li Y, Klinge CM. Estradiol downregulates miR-21 expression and increases miR-21 target gene expression in MCF-7 breast cancer cells. Nucleic Acids Res. 2009;37:2584–2595. doi: 10.1093/nar/gkp117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu HY, Zhang YY, Zhu BL, Feng FZ, Yan H, Zhang HY, Zhou B. MiR-21 regulates the proliferation and apoptosis of ovarian cancer cells through PTEN/PI3K/AKT. Eur Rev Med Pharm Sci. 2019;23:4149–4155. doi: 10.26355/eurrev_201905_17917. [DOI] [PubMed] [Google Scholar]
- 29.Liu H, Wang J, Tao Y, Li X, Qin J, Bai Z, Chi B, Yan W, Chen X. Curcumol inhibits colorectal cancer proliferation by targeting miR-21 and modulated PTEN/PI3K/Akt pathways. Life Sci. 2019;221:354–361. doi: 10.1016/j.lfs.2019.02.049. [DOI] [PubMed] [Google Scholar]
- 30.Zhu Y, Tang H, Zhang L, Gong L, Wu G, Ni J, Tang X. Suppression of miR-21-3p enhances TRAIL-mediated apoptosis in liver cancer stem cells by suppressing the PI3K/Akt/Bad cascade via regulating PTEN. Cancer Manag Res. 2019;11:955–968. doi: 10.2147/CMAR.S183328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schneider C, King RM, Philipson L. Genes specifically expressed at growth arrest of mammalian cells. Cell. 1988;54:787–793. doi: 10.1016/S0092-8674(88)91065-3. [DOI] [PubMed] [Google Scholar]
- 32.Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal. 2010;3:ra8. doi: 10.1126/scisignal.2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mei Y, Si J, Wang Y, Huang Z, Zhu H, Feng S, Wu X, Wu L. Long noncoding RNA GAS5 suppresses tumorigenesis by inhibiting miR-23a expression in non-small cell lung cancer. Oncol Res. 2017;25:1027–1037. doi: 10.3727/096504016X14822800040451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bian D, Shi W, Shao Y, Li P, Song G. Long non-coding RNA GAS5 inhibits tumorigenesis via miR-137 in melanoma. Am J Transl Res. 2017;9:1509–1520. [PMC free article] [PubMed] [Google Scholar]
- 35.Guo LJ, Zhang S, Gao B, Jiang Y, Zhang XH, Tian WG, Hao S, Zhao JJ, Zhang G, Hu CY, et al. Low expression of long non-coding RNA GAS5 is associated with poor prognosis of patients with thyroid cancer. Exp Mol Pathol. 2017;102:500–504. doi: 10.1016/j.yexmp.2017.05.008. [DOI] [PubMed] [Google Scholar]
- 36.Yan H, Zhang DY, Li X, Yuan XQ, Yang YL, Zhu KW, Zeng H, Li XL, Cao S, Zhou HH, et al. Long non-coding RNA GAS5 polymorphism predicts a poor prognosis of acute myeloid leukemia in Chinese patients via affecting hematopoietic reconstitution. Leuk Lymphoma. 2017;58:1948–1957. doi: 10.1080/10428194.2016.1266626. [DOI] [PubMed] [Google Scholar]
- 37.Wen Q, Liu Y, Lyu H, Xu X, Wu Q, Liu N, Yin Q, Li J, Sheng X. Long noncoding RNA GAS5, which acts as a tumor suppressor via microRNA 21, regulates cisplatin resistance expression in cervical cancer. Int J Gynecol Cancer. 2017;27:1096–1108. doi: 10.1097/IGC.0000000000001028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma N, Li S, Zhang Q, Wang H, Qin H, Wang S. Long non-coding RNA GAS5 inhibits ovarian cancer cell proliferation via the control of microRNA-21 and SPRY2 expression. Exp Ther Med. 2018;16:73–82. doi: 10.3892/etm.2018.6188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Poliseno L, Pandolfi PP. PTEN ceRNA networks in human cancer. Methods. 2015;77-78:41–50. doi: 10.1016/j.ymeth.2015.01.013. [DOI] [PubMed] [Google Scholar]
- 40.He Y, Mingyan E, Wang C, Liu G, Shi M, Liu S. CircVRK1 regulates tumor progression and radioresistance in esophageal squamous cell carcinoma by regulating miR-624-3p/PTEN/PI3K/AKT signaling pathway. Int J Biol Macromol. 2019;125:116–123. doi: 10.1016/j.ijbiomac.2018.11.273. [DOI] [PubMed] [Google Scholar]
- 41.Wei H, Cui R, Bahr J, Zanesi N, Luo Z, Meng W, Liang G, Croce CM. Mir-130a deregulates PTEN and stimulates tumor growth. Cancer Res. 2017;77:6168–6178. doi: 10.1158/0008-5472.CAN-17-0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu Y, Song Y, Xiong Y, Wang X, Xu K, Han B, Bai Y, Li L, Zhang Y, Zhou L. MicroRNA-21 (Mir-21) promotes cell growth and invasion by repressing tumor suppressor PTEN in colorectal cancer. Cell Physiol Biochem. 2017;43:945–958. doi: 10.1159/000481648. [DOI] [PubMed] [Google Scholar]
- 43.He C, Dong X, Zhai B, Jiang X, Dong D, Li B, Jiang H, Xu S, Sun X. MiR-21 mediates sorafenib resistance of hepatocellular carcinoma cells by inhibiting autophagy via the PTEN/Akt pathway. Oncotarget. 2015;6:28867–28881. doi: 10.18632/oncotarget.4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu H, Li H, Jin L, Li G, Hu S, Ning C, Guo J, Shuai S, Li X, Li M. Long noncoding RNA GAS5 suppresses 3T3-L1 cells adipogenesis through miR-21a-5p/PTEN signal pathway. DNA Cell Biol. 2018;37:767–777. doi: 10.1089/dna.2018.4264. [DOI] [PubMed] [Google Scholar]
- 45.Haddadi N, Lin Y, Travis G, Simpson AM, Nassif NT, McGowan EM. PTEN/PTENP1: ‘Regulating the regulator of RTK-dependent PI3K/Akt signalling’, new targets for cancer therapy. Mol Cancer. 2018;17:37. doi: 10.1186/s12943-018-0803-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang J, Xu J, Fu J, Yuan D, Guo F, Zhou C, Shao C. MiR-29a regulates radiosensitivity in human intestinal cells by targeting PTEN gene. Radiat Res. 2016;186:292–301. doi: 10.1667/RR14428.1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.