Abstract
Models of Titan predict that there is a subsurface ocean of water and ammonia under a layer of ice. Such an ocean would be important in the search for extraterrestrial life since it provides a potentially habitable environment. To evaluate how Earth-based proteins would behave in Titan's subsurface ocean environment, we used molecular dynamics simulations to calculate the properties of proteins with the most common secondary structure types (alpha helix and beta sheet) in both Earth and Titan-like conditions. The Titan environment was simulated by using a temperature of 300 K, a pressure of 1000 bar, and a eutectic mixture of water and ammonia. We analyzed protein compactness, flexibility, and backbone dihedral distributions to identify differences between the two environments. Secondary structures in the Titan environment were found to be less long-lasting, less flexible, and had small differences in backbone dihedral preferences (e.g., in one instance a pi helix formed). These environment-driven differences could lead to changes in how these proteins interact with other biomolecules and therefore changes in how evolution would potentially shape proteins to function in subsurface ocean environments.
Key Words: Biophysics, Titan, Molecular dynamics
1. Introduction
Despite the lack of sunlight, extreme pressures, and high temperatures, chemotrophic bacteria take advantage of the products of water-rock interactions to survive at Earth's deep-sea vents (Nisbet and Fowler, 1999). Earth may not be the only body to host such habitable niches. Subsurface oceans are considered confirmed (Hendrix et al., 2018) on the jovian moons Ganymede, Callisto, and Europa (Kivelson et al., 1996; Carr et al., 1998; Khurana et al., 1998; Iess et al., 2012, 2014; Čadek et al., 2016) and the saturnian moons Enceladus and Titan (Iess et al., 2012, 2014). Hidden under ice crusts tens to hundreds of kilometers thick, the liquid water environments of these ocean worlds interact with rocky cores, albeit to different extents (Sohl et al., 2010), making them excellent targets in the search for life elsewhere in the Solar System due to the potential confluence of liquid water, chemical building blocks, and energy sources (Lunine, 2017; Hendrix et al., 2018).
Titan's subterranean oceans may be seeded with the hydrogen and carbon necessary for Earth-like biochemistry. Evidence for this possibility comes from measurements of Titan's atmosphere. Nitrogen and methane dominate the bulk composition at 98% and 1.8% respectively, with a multitude of trace gases probably including some oxygen species (Coustenis, 2005; Hörst, 2017). Ultraviolet photons from the Sun, energetic particles from Saturn's magnetosphere, and galactic cosmic rays drive photolysis of methane and nitrogen throughout Titan's atmosphere, leading to various chemical reactions that create a variety of complex organic compounds (Waite et al., 2007; Crary et al., 2009; Vuitton et al., 2009; Niemann et al., 2010). Eventually, the products of this chemistry form haze particles about a micron in size (Tomasko et al., 2008) that fall to the surface, blanketing Titan's terrain in an organic-rich layer estimated at over 2 × 105 km3 (Lorenz et al., 2008). However, the current rate of photolysis could not have been sustained over the lifetime of the Solar System without exhausting the current atmospheric inventory of methane. Forward modeling suggests that all the methane in Titan's atmosphere today would be consumed within 30 Myr (Yung et al., 1984; Wilson and Atreya, 2004). One way to balance this consumption with the ∼1 Gyr age suggested by the 12C/13C ratio observed by both Cassini (Mandt et al., 2012; Nixon et al., 2012) and Huygens (Niemann et al., 2010) is through continuous or episodic replenishing of methane from Titan's interior (Tobie et al., 2006; Castillo-Rogez and Lunine, 2010). In such a scenario, methane from the rocky interior dissolves in the subsurface ocean before outgassing (Lunine and Stevenson, 1987) and could be delivered to the surface via cyrovolcanoes (Tobie et al., 2008). The possibility of organics going down into the ocean is compelling—especially given the evidence that amino acids can quickly form when liquid water interacts with lab analogs of Titan's haze (Neish et al., 2008, 2010)—but beyond the scope of this paper and will be left to future research. Additionally, Titan's subsurface ocean is thought to contain ammonia to ensure the ocean remains liquid over the age of Titan (Choukroun et al., 2010; Grasset et al., 2010; Fortes, 2012). Thus, Titan's subsurface ocean may contain hydrogen, carbon, nitrogen. (The presence of ammonia in Titan's subsurface ocean should not prevent Earth-like biochemistry, as terrestrial extremophiles can thrive in high pH [9.0–11.6] solvents [Preiss et al., 2015].)
At 5150 km in diameter and with a bulk density of 1881 kg/m3, a high-pressure ice (ice VI) likely separates Titan's ocean from the rocky core today (Lunine and Stevenson, 1987; Sotin and Tobie, 2004; Grasset and Pargamin, 2005); however, this layer may not inhibit water-rock interactions. Journaux et al., for example, demonstrated how brines at certain high temperatures and pressures can be transported through an ice VI layer to the ocean (Journaux et al., 2013; Vance et al., 2018). It is also possible that earlier in Titan's history the ocean may have both been in direct contact with the rocky core (facilitating hydrothermal reactions) as well as received exogenic cometary and chondritic material from meteorite falls and impacts (Shock and McKinnon, 1993). Fortes (2000) outlines several similarities between Titan's evolving ocean environment and extreme environments on Earth.
Previous work has been done to look at the effects of pressure and temperature on protein flexibility (Sawle and Ghosh, 2011; Karshikoff et al., 2015; Ichiye, 2016) and adaptation (Campanaro et al., 2008; Siddiqui and Thomas, 2008; Michoud and Jebbar, 2016). Piezophiles, organisms that live in high pressures, are not well understood, but the high pressure limit of known life is around 1.1 kbar (Simonato et al., 2006; Meinhold et al., 2007; Ichiye, 2018). Alkaliphiles, organisms that typically live in pH values between 9 and 12, can thrive in a variety of environments as long as the basicity is optimum (Kevbrin et al., 1998; Krulwich and Ito, 2013; Kulkarni et al., 2019). Combining the traits of these two extremophiles would result in an organism able to live and thrive in an environment similar to that of the subsurface ocean of Titan.
In this work, we build on the hypotheses of Fortes (2000). Instead of answering whether molecules can form in Titan's ocean (today or in some earlier epoch), we explore how biologically relevant molecules might behave in Titan's ocean. Some studies have been done that show the folding of proteins in different organic solvents can lower folding free energy (Yu et al., 2016) or can function similarly to water in an aqueous-organic solvent (Soares et al., 2003). This study does not analyze the folding or binding of proteins but rather the integrity of a protein over the course of a simulation. In Section 2, we describe the molecular dynamics simulations that we used to study Earth-based proteins in both Earth and Titan-like conditions. In Section 3, we analyze the results, measuring protein compactness, flexibility, and backbone dihedral angle distributions. Section 4 discusses the implications of the results for Titan and other Ocean Worlds, and we conclude in Section 5.
2. Materials and Methods
2.1. Protein folding
Inside biological cells on Earth, ribosomes manufacture proteins by sequentially adding individual amino acids in a chain according to instructions in messenger RNA as copied from the cell's nuclear DNA. Knowledge of the amino acid sequence alone does not allow us to directly infer the resulting protein's chemical behavior, however. After their manufacture, proteins fold in on themselves in a manner governed by their own self-affinity and electrical interactions with the surrounding solvent. Therefore, in Titan's high-pressure water-ammonia subsurface ocean proteins might behave differently than they do in biological systems on Earth.
Those different shapes and the variability thereof would necessarily alter the functionality of the protein. We see similar situations in extreme environments on Earth, where, for instance, high-temperature single-celled organisms living in deep-sea vents have evolved distinctly different versions of common proteins that specifically tailor their amino acid sequence to match their environment (at high temperature, for instance, deletions of large swaths of protein helps those proteins behave similarly to non-deleted ones at room temperature [Barnes, 1992]). In that high-temperature situation, proteins typically work better with fewer large ancillary residues so as to not have excessive conformal variation with high internal kinetic energy.
As a first step toward understanding how the conditions in Titan's subsurface ocean affect protein folding and movement, we simulate the behavior of representative proteins computationally. The goal of these initial calculations is not to design an alien biochemistry compatible with Titan's ocean but rather to broadly determine the effects of aspects of that ocean such as high pressure and ammonia content.
2.2. Protein selections
Three types of proteins were chosen to simulate the two most common secondary structures on Earth: alpha helices and beta sheets (Berman et al., 2000). These proteins were chosen from the CATH protein database (Sillitoe et al., 2015; Lam et al., 2016) by selecting the most common motif of that secondary structure. For example, one of our chosen motifs containing primarily alpha helices was selected by using the following steps: (1) Opened the website http://www.cathdb.info/browse/tree. (2) Chose “1 Mainly Alpha.” (3) Chose the largest number of folds, “1.10 Orthogonal Bundle.” (4) Chose the largest number of superfamilies, “1.10.287 Helix Hairpins.” (5) Chose the largest number of Domains, “1.10.287.610 Helix hairpin bin.” (6) Chose example protein, “3uq8.” (7) Looked at sequence to ensure that chain A amino acids 3–61 is the appropriate fragment. (8) Downloaded protein structure from the RCSB protein data bank and edited file to include only the amino acids that are within the appropriate fragment. The proteins chosen were 3uq8 and 1xmk (mainly alpha), 3ulj and 4unu (mainly beta), and 4g1q and 2gxq (mixed alpha/beta).
2.3. System preparation
The idea is to create a system that replicates an Earth environment at sea level and a Titan environment at the bottom of its subsurface ocean. We chose a practical temperature of 300 K and a pressure of 1 bar. For Titan we also chose 300 K. This decision was made based on an assumption that there would be hydrothermal vents, noting that a similar assumption has been made about Titan's neighbor, Enceladus (Hsu et al., 2015). The pressure chosen for the Titan environment was 1000 bar. This would correspond to a depth of about 10 km below Earth's ocean's surface. On Titan, using an aqueous ammonia solution density of 0.89 g/cm3 (O'Neil, 2013) and a gravitational constant of 1.35 m/s2 (Jacobson et al., 2006), we would get a pressure of 1 kbar at a depth of about 80 km below Titan's surface. This does not consider several variables such as ice thickness, ice density, or compressibility. However, this shows that a plausible pressure and temperature were used for the Titan environment.
Therefore, setting up the simulations, the Earth environment was set to a temperature of 300 K, pressure of 1 bar, and pure water. The deep Titan ocean environment used the same temperature as Earth but with 3 orders of magnitude larger pressure (1000 bar) and a eutectic water-ammonia mixture (Fortes, 2000). The three protein types were separately placed in each environment for a total of six simulations.
2.4. Molecular dynamics simulations
The software package GROMACS 5.1.2 was used for all molecular dynamics simulations with the Amber99sb*-ildnp forcefield (Hess et al., 2008). The Earth system was placed in a dodecahedral box of TIP3P water (Jorgensen et al., 1983). A TIP3P water model was chosen, as it is a good balance of accuracy and computational efficiency. The Titan system was placed in a dodecahedral box of a eutectic mixture of TIP3P water and 32% weight ammonia at a density of 0.89 g/cm3 (Fortes, 2000). Each system's energy was minimized by using steepest descent for 1000 steps. To allow for some equilibration of the water around the proteins, each system was then simulated for 1 ns with the positions of all heavy atoms in the complex restrained via a harmonic potential, and then simulated for another 1 ns with no restraints. During the restrained simulations, the temperature of the system was increased linearly from 100 to 300 K, and the pressure was maintained at 1 atm using the Berendsen algorithm. Production simulations for each system were then carried out for 100 ns with pressure maintained using Parrinello-Rahman coupling. For all simulations, the LINCS algorithm (Hess et al., 1997) was used to constrain all bonds to their ideal lengths allowing for a timestep of 2 fs. The temperature was controlled using the v-rescale option. Reaction-Field-zero was used for electrostatics with a real-space cutoff of 1.2 nm. The Van der Waals interaction cutoff was set to 1.2 nm with the Potential-shift-Verlet method for smoothing interactions.
2.5. Analyzing data
Radius of gyration of a protein is a measure of its compactness; smaller values correspond to more compact protein shape. The radius of gyration measurements were obtained using the GROMACS command gmx gyrate, and histograms were then generated using python. The root-mean-square fluctuation (RMSF) is a measure of how much an atom fluctuates about its average position, that is, the secondary structure flexibility. RMSF plots were obtained by running the GROMACS command gmx rmsf. RMSF values were converted to log form and used to color the protein using PyMOL 1.7 (blue for low RMSF values and red for high). Secondary structures plots as a function of simulation time were generated using the GROMACS command gmx do_dssp (Mclendon et al., 2015; Touw et al., 2015). Ramachandran plots show the distribution of backbone dihedrals that largely determine a protein's conformation and secondary structure preference during simulation. These results were generated using the GROMACS command gmx rama, and histograms were generated by subtracting the Earth measurements from the Titan measurements.
3. Results
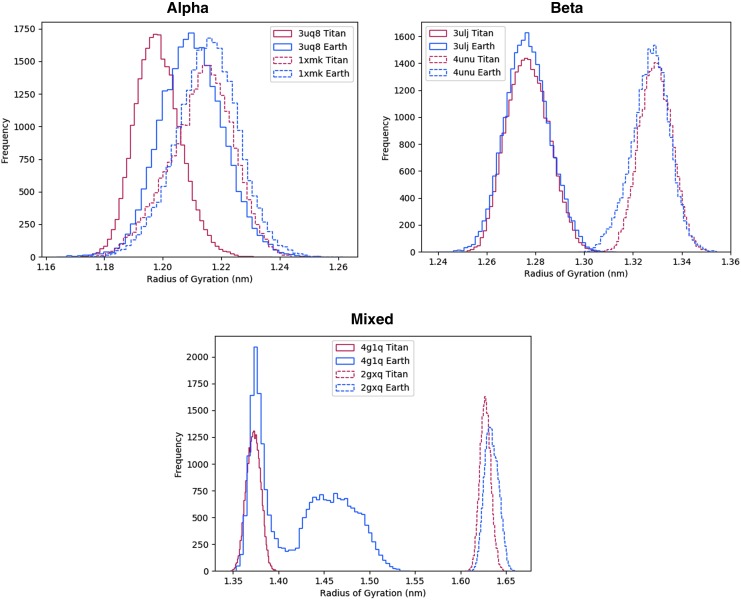
Radius of gyration histograms are shown in Fig. 1 and illustrate the compactness of the proteins. The peaks of each plot represent the most likely radius of gyration value for each protein in each environment. A difference in peak location represents either a more or a less compact protein (smaller radius of gyration is more compact). Our results show that proteins in the Titan environment experience a shift in the peaks toward a lower radius of gyration value in both the alpha and mixed alpha/beta as compared to the Earth environment. The beta shows no or negligible shifting of its peaks. The shifted plateau in the mixed alpha/beta 4g1q Earth environment is caused by the C-terminus of the protein moving to a new conformation.
FIG. 1.
Radius of gyration for all proteins over the simulation. A shift to the left is a more compact protein, while a shift to right is less compact. The Titan environment experiences a shift in its peaks toward a lower radius of gyration value in both the alpha and mixed alpha/beta compared to the Earth environment. The beta shows no or negligible shifting of its peaks. The shifted plateau in the mixed alpha/beta 4g1q Earth environment is caused by the C-terminus of the protein moving to a new conformation.
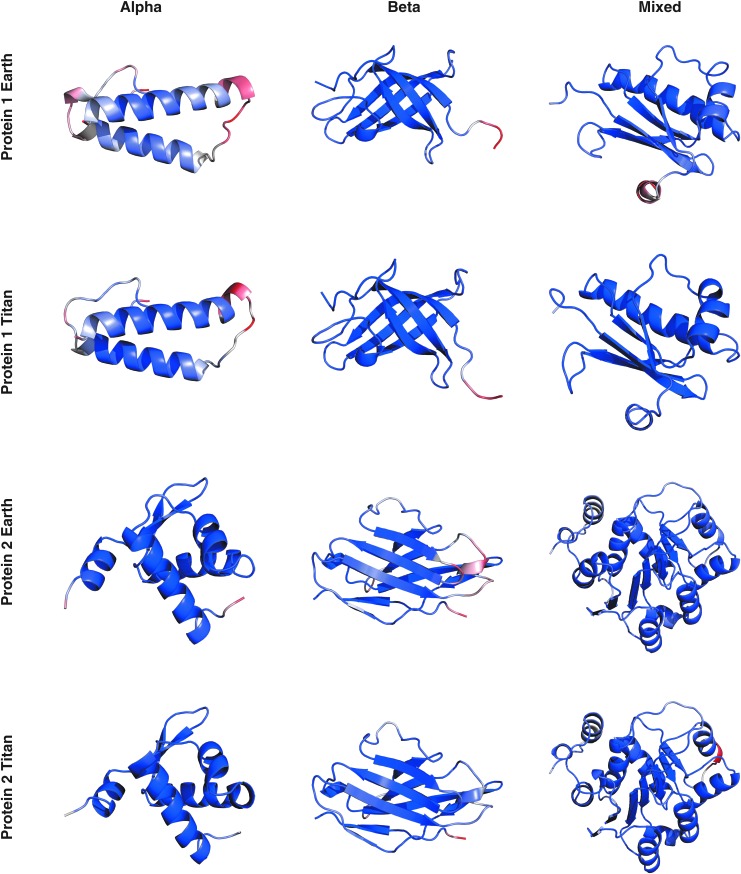
Root-mean-square fluctuation results are in Fig. 2 and show how much atoms fluctuate about their average position. The RMSF value was converted to a log form and overlaid onto a frame from the largest cluster of the simulation. The gradient ranges from blue to white to red, with blue being most stable and red being least stable. Proteins in the Titan environment experience larger maximum RMSF values as compared to the Earth environment but lower RMSF values on average.
FIG. 2.
Root-mean-square fluctuations for each amino acid in the protein systems. Larger fluctuations are shown in red and smaller in blue. Proteins in the Titan environment experience larger maximum RMSF values as compared to the Earth environment but lower RMSF values on average.
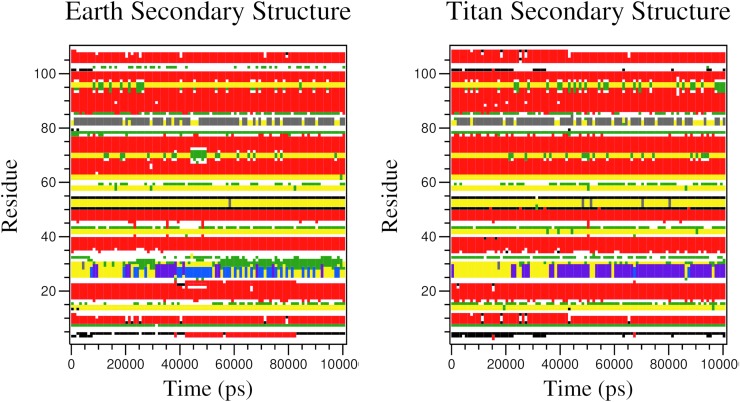
Secondary structure results are shown in Fig. 3 and demonstrate how the local structure of each amino acid in the protein changes over the course of the simulation. The vertical axis is the amino acids in the protein, the horizontal axis is the simulation time, and Table 1 shows the color definitions for each secondary structure type. The emphasized region highlights that the protein does not become a specific secondary structure type in the Earth environment, in contrast to the Titan environment where the same region becomes a pi helix.
FIG. 3.
Secondary structures for each amino acid of one of the protein systems over the simulation. Table 1 shows the color definitions used for each secondary structure type. The emphasized region highlights that the protein does not stabilize into a specific secondary structure type in the Earth environment in contrast to the Titan environment where the same region stabilizes into a pi helix.
Table 1.
Secondary Structures Legend
| Color | Structure |
|---|---|
| White | Coil |
| Red | Beta sheet |
| Black | Beta bridge |
| Green | Bend |
| Yellow | Turn |
| Blue | Alpha helix |
| Purple | Pi helix |
| Gray | 3-helix |
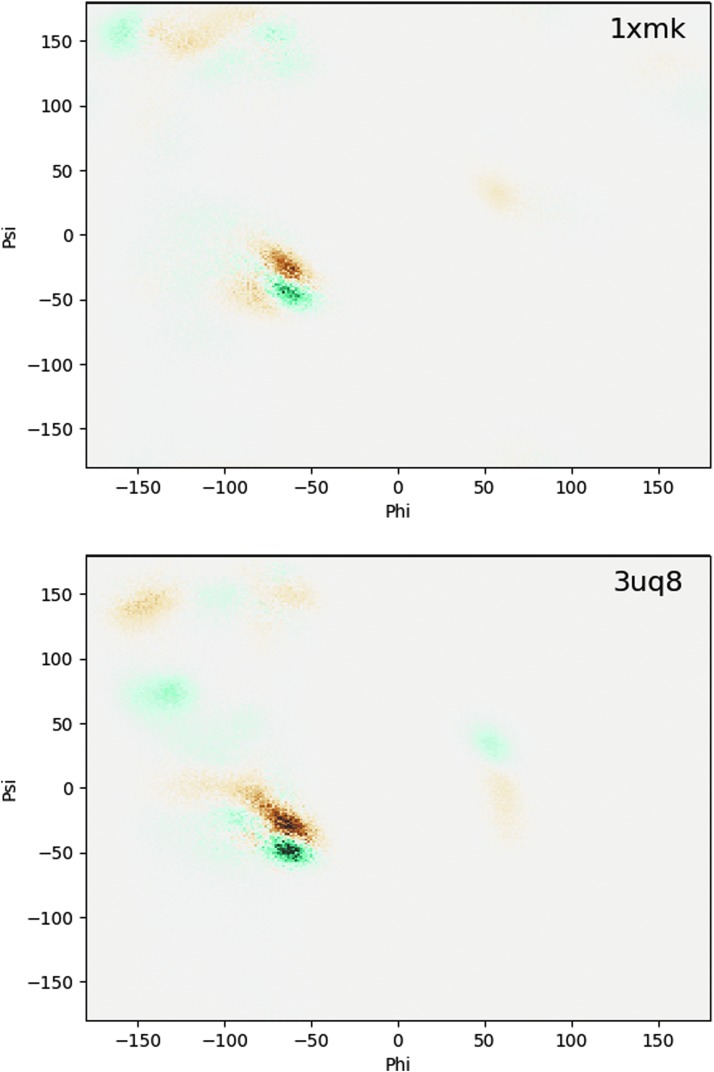
Ramachandran plots are shown in Fig. 4 and represent the distribution of backbone dihedral angles phi and psi that largely determine the protein's conformation for each protein as a two-dimensional histogram. These plots are made from subtracting the Earth dihedral distribution values from the Titan values. The gradient ranges from brown to blue-green; brown shows dihedrals that are favored in the Titan environment, and blue-green shows dihedrals that are favored in the Earth environment. In the Titan environment, the phi angle propensity is similar to the Earth environment, but the psi angle shifts from about -50 degrees to -25 degrees.
FIG. 4.
Ramachandran plots for all protein systems in the current study. Generally, the top left quadrant is beta sheets, and the bottom left quadrant is alpha helices. The gradient ranges from brown to blue-green; brown shows angles that are favored in the Titan environment, and blue-green shows angles that are favored in the Earth environment. In the Titan environment, the phi angle remains similar to the Earth environment, but the psi angle shifts from -50 degrees to -25 degrees.
4. Discussion
The radius of gyration results in Fig. 1 show that the beta sheet proteins simulated in this study are unaffected by the difference in environment between Titan and Earth. By contrast, our results show the Titan environment leads to slightly more compact alpha helix proteins. We believe that the beta sheet proteins are less affected due to the higher number of stabilizing hydrogen bonds present in beta secondary structures. The beta sheets in these proteins have on average five hydrogen bonds compared to alpha helices that have three hydrogen bonds.
The results in Fig. 2 show that proteins in the Titan environment have lower average RMSF values than the Earth environment likely due to a combination of the high pressure and water-ammonia of the Titan environment suppressing atom fluctuations. Even though alpha helices are generally less long-lasting in the Titan environment (as shown in Fig. 3), one protein had an alpha helix stabilize into a long-lasting pi helix (see Fig. 2, beta, protein 2). Further studies will be needed to understand the mechanisms behind these differences.
The secondary structure results in Fig. 3 show that the alpha helix region of one of the three proteins is more long-lasting in the Titan environment. In the Earth environment the helix region changes between different types of secondary structures: bends, turns, alpha helices, and pi helices. Turns and bends are similar in structure, but turns have hydrogen bonds whereas bends do not. In the Titan environment the helix region changes to a turn and then becomes a pi helix. The pi helix could be important because it shows Titan favoring a conformation that is not common for Earth conditions (Fodje and Al-Karadaghi, 2002). A pi helix is a helix with five hydrogen bonds compared to an alpha helix that has three. Due to the increase in hydrogen bonds, the pi helix is much more energetically favorable than an alpha helix. This suggests that proteins in the Titan environment may interact with other biomolecules with different biochemistry compared to Earth.
The Ramachandran plots in Fig. 4 show that the Titan environment favors slightly different angles for the alpha helices. These different angles prefer a psi angle of about -25 degrees, whereas the phi angle remains the same. This psi angle difference is likely the result of differences in the preferred secondary structure types in the Titan environment, for example, a pi helix instead of an alpha helix. From these results, it appears that life on Titan would have similar beta sheets as Earth, allowing comparable proteins to form on Titan as on Earth.
5. Conclusion
In summary, protein secondary structure elements have different properties in a Titan environment compared to Earth. In the Titan environment alpha helices tended to be less flexible (Fig. 2) and preferred slightly smaller psi dihedral backbone angles compared to an Earth environment (Fig. 4). Protein structures were more compact in the Titan environment compared to an Earth environment (Fig. 1). Most secondary structure elements were less long-lasting on Titan, but on rare occasions alpha helices were more long-lasting (see Fig. 3) compared to the Earth environment. This study should be considered a starting point for a larger study to understand how proteins, protein-ligand complexes, and protein-protein complexes could fold, interact, and function in subsurface oceans such as those on Titan.
Supplementary Material
Acknowledgments
This research was supported by the Center for Modeling Complex Interactions sponsored by the National Institutes of Health (P20 GM104420), the National Science Foundation EPSCoR program (OIA-1736253), and the National Aeronautics and Space Administration Earth and Space Science Fellowship Program (NNX14AO30H). Computer resources were provided in part by the Institute for Bioinformatics and Evolutionary Studies Computational Resources Core sponsored by the National Institutes of Health (P30 GM103324). The funding agencies had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Abbreviation Used
- RMSF
root-mean-square fluctuation
Supplementary Material
Supplementary Material is available online at www.liebertonline.com/ast
References
- Barnes W.M. (1992) The fidelity of Taq polymerase catalyzing PCR is improved by an N-terminal deletion. Gene 112:29–35 [DOI] [PubMed] [Google Scholar]
- Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., and Bourne P.E. (2000) The protein data bank. Nucleic Acids Res 28:235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Čadek O., Tobie G., Van Hoolst T., Massé M., Choblet G., Lefèvre A., Mitri G., Baland R.-M., Běhounková M., Bourgeois O., and Trinh A. (2016) Enceladus's internal ocean and ice shell constrained from Cassini gravity, shape, and libration data. Geophys Res Lett 43, doi: 10.1002/2016GL068634 [DOI] [Google Scholar]
- Campanaro S., Treu L., and Valle G. (2008) Protein evolution in deep sea bacteria: an analysis of amino acids substitution rates. BMC Evol Biol 8, doi: 10.1186/1471-2148-8-313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr M.H., Belton M.J.S., Chapman C.R., Davies M.E., Geissler P., Greenberg R., McEwen A.S., Tufts B.R., Greeley R., Sullivan R., Head J.W., Pappalardo R.T., Klaasen K.P., Johnson T.V., Kaufman J., Senske D., Moore J., Neukum G., Schubert G., Burns J.A., Thomas P., and Veverka J. (1998) Evidence for a subsurface ocean on Europa. Nature 391:363–365 [DOI] [PubMed] [Google Scholar]
- Castillo-Rogez J.C. and Lunine J.I. (2010) Evolution of Titan's rocky core constrained by Cassini observations. Geophys Res Lett 37, doi: 10.1029/2010GL044398 [DOI] [Google Scholar]
- Choukroun M., Grasset O., Tobie G., and Sotin C. (2010) Stability of methane clathrate hydrates under pressure: influence on outgassing processes of methane on Titan. Icarus 205:581–593 [Google Scholar]
- Coustenis A. (2005) Formation and evolution of Titan's atmosphere. In The Outer Planets and Their Moons, Springer, Berlin, pp 171–184 [Google Scholar]
- Crary F.J., Magee B.A., Mandt K., Waite J.H., Westlake J., and Young D.T. (2009) Heavy ions, temperatures and winds in Titan's ionosphere: combined Cassini CAPS and INMS observations. Planet Space Sci 57:1847–1856 [Google Scholar]
- Fodje M.N. and Al-Karadaghi S. (2002) Occurrence, conformational features and amino acid propensities for the π-helix. Protein Eng 15:353–358 [DOI] [PubMed] [Google Scholar]
- Fortes A.D. (2000) Exobiological implications of a possible ammonia–water ocean inside Titan. Icarus 146:444–452 [Google Scholar]
- Fortes A.D. (2012) Titan's internal structure and the evolutionary consequences. Planet Space Sci 60:10–17 [Google Scholar]
- Grasset O. and Pargamin J. (2005) The ammonia-water system at high pressures: implications for the methane of Titan. Planet Space Sci 53:371–384 [Google Scholar]
- Grasset O., Coustenis A., Durham W.B., Hussmann H., Pappalardo R.T., Sasaki S., and Turrini D. (2010) Satellites of the outer Solar System: exchange processes involving the interiors. Space Sci Rev 153:5–9 [Google Scholar]
- Hendrix A.R., Hurford T.A., Barge L.M., Bland M.T., Bowman J.S., Brinckerhoff W., Buratti B.J., Cable M.L., Castillo-Rogez J., Collins G.C., Diniega S., German C.R., Hayes A.G., Hoehler T., Hosseini S., Howett C.J.A., McEwen A.S., Neish C.D., Neveu M., Nordheim T.A., Patterson G.W., Patthoff D.A., Phillips C., Rhoden A., Schmidt B.E., Singer K.N., Soderblom J.M., and Vance S.D. (2018) The NASA Roadmap to Ocean Worlds. Astrobiology 19:1–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess B., Bekker H., Berendsen H.J.C., and Fraaije J.G.E.M. (1997) LINCS: a Linear Constraint Solver for molecular simulations. J Comput Chem 18:1463–1472 [Google Scholar]
- Hess B., Kutzner C., Van Der Spoel D., and Lindahl E. (2008) GRGMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput 4:435–447 [DOI] [PubMed] [Google Scholar]
- Hörst S.M. (2017) Titan's atmosphere and climate. J Geophys Res Planets 122:432–482 [Google Scholar]
- Hsu H.-W.W., Postberg F., Sekine Y., Shibuya T., Kempf S., Horányi M., Juhasz A., Altobelli N., Suzuki K., Masaki Y., Kuwatani T., Tachibana S., Sirono S.I., Moragas-Klostermeyer G., and Srama R. (2015) Ongoing hydrothermal activities within Enceladus. Nature 519:207–210 [DOI] [PubMed] [Google Scholar]
- Ichiye T. (2016) What makes proteins work: exploring life in P-T-X. Phys Biol 13, doi: 10.1088/1478-3975/13/6/063001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichiye T. (2018) Enzymes from piezophiles. Semin Cell Dev Biol 84:138–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iess L., Jacobson R.A., Ducci M., Stevenson D.J., Lunine J.I., Armstrong J.W., Asmar S.W., Racioppa P., Rappaport N.J., and Tortora P. (2012) The tides of Titan. Science 337:457–459 [DOI] [PubMed] [Google Scholar]
- Iess L., Stevenson D.J., Parisi M., Hemingway D., Jacobson R.A., Lunine J.I., Nimmo F., Armstrong J.W., Asmar S.W., Ducci M., and Tortora P. (2014) The gravity field and interior structure of Enceladus. Science 344:78–80 [DOI] [PubMed] [Google Scholar]
- Jacobson R.A., Antreasian P.G., Bordi J.J., Criddle K.E., Ionasescu R., Jones J.B., Mackenzie R.A., Meek M.C., Parcher D., Pelletier F.J., Owen W.M. Jr., Roth D.C., Roundhill I.M., and Stauch J.R. (2006) The gravity field of the saturnian system from satellite observations and spacecraft tracking data. Astron J 132:2520–2526 [Google Scholar]
- Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., and Klein M.L. (1983) Comparison of simple potential functions for simulating liquid water. J Chem Phys 79:926–935 [Google Scholar]
- Journaux B., Daniel I., Caracas R., Montagnac G., and Cardon H. (2013) Influence of NaCl on ice VI and ice VII melting curves up to GPa, implications for large icy moons. Icarus 226:355–363 [Google Scholar]
- Karshikoff A., Nilsson L., and Ladenstein R. (2015) Rigidity versus flexibility: the dilemma of understanding protein thermal stability. FEBS J 282:3899–3917 [DOI] [PubMed] [Google Scholar]
- Kevbrin V.V., Zhilina T.N., Rainey F.A., and Zavarzin G.A. (1998) Tindallia magadii gen. nov., sp. nov.: an alkaliphilic anaerobic ammonifier from Soda Lake deposits. Curr Microbiol 37:94–100 [DOI] [PubMed] [Google Scholar]
- Khurana K.K., Kivelson M.G., Stevenson D.J., Schubert G., Russell C.T., Walker R.J., and Polanskey C. (1998) Induced magnetic fields as evidence for subsurface oceans in Europa and Callisto. Nature 395:777–780 [DOI] [PubMed] [Google Scholar]
- Kivelson M.G., Khurana K.K., Russell C.T., Walker R.J., Warnecke J., Coroniti F.V., Polanskey C., Southwood D.J., and Schubert G. (1996) Discovery of Ganymede's magnetic field by the Galileo spacecraft. Nature 384:537–541 [Google Scholar]
- Krulwich T.A. and Ito M. (2013) Alkaliphilic Prokaryotes, Springer, Berlin [Google Scholar]
- Kulkarni S., Dhakar K., and Joshi A. (2019) Alkaliphiles: diversity and bioprospection. In Microbial Diversity in the Genomic Era, Academic Press, New York, pp 239–263 [Google Scholar]
- Lam S.D., Dawson N.L., Das S., Sillitoe I., Ashford P., Lee D., Lehtinen S., Orengo C.A., and Lees J.G. (2016) Gene3D: expanding the utility of domain assignments. Nucleic Acids Res 44:D404–D409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz R.D., Mitchell K.L., Kirk R.L., Hayes A.G., Aharonson O., Zebker H.A., Paillou P., Radebaugh J., Lunine J.I., Janssen M.A., Wall S.D., Lopes R.M., Stiles B., Ostro S., Mitri G., and Stofan E.R. (2008) Titan's inventory of organic surface materials. Geophys Res Lett 35, doi: 10.1029/2007GL032118 [DOI] [Google Scholar]
- Lunine J.I. (2017) Ocean worlds exploration. Acta Astronaut 131:123–130 [Google Scholar]
- Lunine J.I. and Stevenson D.J. (1987) Clathrate and ammonia hydrates at high pressure: application to the origin of methane on Titan. Icarus 70:61–77 [Google Scholar]
- Mandt K.E., Waite J.H., Teolis B., Magee B.A., Bell J., Westlake J.H., Nixon C.A., Mousis O., and Lunine J.I. (2012) The 12C/13C ratio on Titan from Cassini INMS measurements and implications for the evolution of methane. Astrophys J 749, doi: 10.1088/0004-637X/749/2/160 [DOI] [Google Scholar]
- Mclendon C., Opalko F.J., Illangkoon H.I., and Benner S.A. (2015) Solubility of polyethers in hydrocarbons at low temperatures. a model for potential genetic backbones on warm Titans. Astrobiology 15:200–206 [DOI] [PubMed] [Google Scholar]
- Meinhold L., Smith J.C., Kitao A., and Zewail A.H. (2007) Picosecond fluctuating protein energy landscape mapped by pressure temperature molecular dynamics simulation. Proc Natl Acad Sci USA 104:17261–17265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michoud G. and Jebbar M. (2016) High hydrostatic pressure adaptive strategies in an obligate piezophile Pyrococcus yayanosii. Sci Rep 6, doi: 10.1038/srep27289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neish C.D., Somogyi, Á., Imanaka H., Lunine J.I., and Smith M.A. (2008) Rate measurements of the hydrolysis of complex organic macromolecules in cold aqueous solutions: implications for prebiotic chemistry on the early Earth and Titan. Astrobiology 8:273–287 [DOI] [PubMed] [Google Scholar]
- Neish C.D., Somogyi, Á., and Smith M.A. (2010) Titan's primordial soup: formation of amino acids via low-temperature hydrolysis of tholins. Astrobiology 10:337–347 [DOI] [PubMed] [Google Scholar]
- Niemann H.B., Atreya S.K., Demick J.E., Gautier D., Haberman J.A., Harpold D.N., Kasprzak W.T., Lunine J.I., Own T.C., and Raulin F. (2010) Composition of Titan's lower atmosphere and simple surface volatiles as measured by the Cassini-Huygens probe gas chromatograph mass spectrometer experiment. J Geophys Res Planets 115, doi: 10.1029/2010JE003659 [DOI] [Google Scholar]
- Nisbet E.G. and Fowler C.M.R. (1999) Archaean metabolic evolution of microbial mats. Proc R Soc Lond B Biol Sci 266:2375–2382 [Google Scholar]
- Nixon C.A., Temelso B., Vinatier S., Teanby N.A., Bézard B., Achterberg R.K., Mandt K.E., Sherrill C.D., Irwin P.G.J., Jennings D.E., Romani P.N., Coustenis A., and Flasar F.M. (2012) Isotopic ratios in Titan's methane: measurements and modeling. Astrophys J 749, doi: 10.1088/0004-637X/749/2/159 [DOI] [Google Scholar]
- O'Neil M.J. (2013) The Merck Index: An Encyclopedia of Chemicals, Drugs, and Biologicals, 15th ed., Royal Society of Chemistry, Cambridge, UK
- Preiss L., Hicks D.B., Suzuki S., Meier T., and Krulwich T.A. (2015) Alkaliphilic bacteria with impact on industrial applications, concepts of early life forms, and bioenergetics of ATP synthesis. Front Bioeng Biotechnol 3, doi: 10.3389/fbioe.2015.00075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawle L. and Ghosh K. (2011) How do thermophilic proteins and proteomes withstand high temperature? Biophys J 101:217–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shock E.L. and McKinnon W.B. (1993) Hydrothermal processing of cometary volatiles—applications to Triton. Icarus 106:464–477 [DOI] [PubMed] [Google Scholar]
- Siddiqui K. and Thomas T. (2008) Protein Adaptation in Extremophiles, Nova Science, New York [Google Scholar]
- Sillitoe I., Lewis T.E., Cuff A., Das S., Ashford P., Dawson N.L., Furnham N., Laskowski R.A., Lee D., Lees J.G., Lehtinen S., Studer R.A., Thornton J., and Orengo C.A. (2015) CATH: comprehensive structural and functional annotations for genome sequences. Nucleic Acids Res 43, doi: 10.1093/nar/gku947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonato F., Campanaro S., Lauro F.M., Vezzi A., D'Angelo M., Vitulo N., Valle G., and Bartlett D.H. (2006) Piezophilic adaptation: a genomic point of view. J Biotechnol 126:11–25 [DOI] [PubMed] [Google Scholar]
- Soares C.M., Teixeira V.H., and Baptista A.M. (2003) Protein structure and dynamics in nonaqueous solvents: insights from molecular dynamics simulation studies. Biophys J 84:1628–1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohl F., Choukroun M., Kargel J., Kimura J., Pappalardo R., Vance S., and Zolotov M. (2010) Subsurface water oceans on icy satellites: chemical composition and exchange processes. Space Sci Rev 153:485–510 [Google Scholar]
- Sotin C. and Tobie G. (2004) Ice: from dislocations to icy satellites/La glace: des dislocations aux satellites de glace Internal structure and dynamics of the large icy satellites. C R Physique 5:769–780 [Google Scholar]
- Tobie G., Lunine J.I., and Sotin C. (2006) Episodic outgassing as the origin of atmospheric methane on Titan. Nature 440:61–64 [DOI] [PubMed] [Google Scholar]
- Tobie G., Choukroun M., Grasset O., Le Mouélic S., Lunine J., Sotin C., Bourgeois O., Gautier D., Hirtzig M., Lebonnois S., and Le Corre L. (2008) Evolution of Titan and implications for its hydrocarbon cycle. Philos Trans A Math Phys Eng Sci 367:617–631 [DOI] [PubMed] [Google Scholar]
- Tomasko M.G., Doose L., Engel S., Dafoe L.E., West R., Lemmon M., Karkoschka E., and See C. (2008) A model of Titan's aerosols based on measurements made inside the atmosphere. Planet Space Sci 56:669–707 [Google Scholar]
- Touw W.G., Baakman C., Black J., Te Beek T.A.H., Krieger E., Joosten R.P., and Vriend G. (2015) A series of PDB-related databanks for everyday needs. Nucleic Acids Res 43:D364–D368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance S.D., Panning M.P., Stähler S., Cammarano F., Bills B.G., Tobie G., Kamata S., Kedar S., Sotin C., Pike W.E., Lorenz R., Huang H.-H., Jacson J.M., and Benerdt B. (2018) Geophysical investigations of habitability in ice-covered ocean worlds. J Geophys Res Planets 123:180–205 [Google Scholar]
- Vuitton V., Lavvas P., Yelle R.V., Galand M., Wellbrock A., Lewis G.R., Coates A.J., and Wahlund J.-E. (2009) Negative ion chemistry in Titan's upper atmosphere. Planet Space Sci 57:1558–1572 [Google Scholar]
- Waite J.H., Young D.T., Cravens T.E., Coates A.J., Crary F.J., Magee B., and Westlake J. (2007) The process of tholin formation in Titan's upper atmosphere. Science 316:870–875 [DOI] [PubMed] [Google Scholar]
- Wilson E.H. and Atreya S.K. (2004) Current state of modeling the photochemistry of Titan's mutually dependent atmosphere and ionosphere. J Geophys Res 109, doi: 10.1029/2003JE002181 [DOI] [Google Scholar]
- Yu Y., Wang J., Shao Q., Shi J., and Zhu W. (2016) The effects of organic solvents on the folding pathway and associated thermodynamics of proteins: a microscopic view. Sci Rep 6, doi: 10.1038/srep19500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yung Y.L., Allen M., and Pinto J.P. (1984) Photochemistry of the atmosphere of Titan—comparison between model and observations. Astrophys J Suppl Ser 55:465–506 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.