Dear Editor
We read with interest the article by Kamboj et al., who demonstrated that low Clostridium difficile real-time polymerase chain reaction (PCR) cycle thresholds of detection (CT) were predictive of toxin enzyme immunoassay positivity and disease severity in oncology patients who showed a positive C. difficile nucleic acid amplification test (NAAT) result.1 These findings are consistent with those reported in other studies that found C. difficile PCR CT (i.e., ≤ 26.0–28.0) may be similar to the results obtained from the cell cytotoxicity neutralization assay (CCNA) and superior to those obtained from toxin enzyme immunoassay in differentiating clinical C. difficile infection (CDI).2–5
We previously reported the use of a computerized clinical decision support (CCDS) tool that led to significantly reduced NAAT testing and National Healthcare Safety Network (NHSN) surveillance CDI events in our institution.6 On the basis of the report by Kamobj et al., we sought to determine whether CT data contributed to the identification of patients with lower probability of the disease.
Positive GeneXpert (Cepheid, Sunnyvale, CA) NAAT results were analyzed retrospectively between January 2014 and June 2018. CT values obtained from tests that were ordered appropriately, according to the CCDS tool, were compared with those obtained from tests categorized as inappropriate. Inappropriate orders were defined as patients identified by the provider through the CCDS tool (post-CCDS) as lacking diarrhea or signs/symptoms of CDI or automatically flagged as a duplicate test (pre- or post-CCDS). A very high CT value was defined as > 30.85, which is shown to have a 98.7% negative predictive value of a negative CCNA and toxin EIA, and thus, it likely reflects colonization with low organism burden.4
We found that CT values were significantly higher in the inappropriate test group than in the appropriate test group (median: 26.7 versus 24.8 cycles, Table 1). The strongest predictor of an increased CT value was a duplicate of a negative test. Fig. 1 demonstrates that CT values were increased in the inappropriate test group, with a clustering of very high CT results.
Table 1.
CT values by order appropriateness
| Total | Appropriate | Inappropriate | P | |||
|---|---|---|---|---|---|---|
| n (%) | Median CT (IQR) | n (%) | Median CT (IQR) | |||
| CCDS question | ||||||
| Presence of diarrhea? (Appropriate Response = “Yes”) | 460 | 453 (98.5%) | 24.9 (22.1–30.4) | 7 (1.5%) | 25.0 (22.2–29.9) | .847 |
| Signs/Symptoms of CDI? (Appropriate Response = “Yes”) | 460 | 375 (81.5%) | 24.9 (22.1–30.3) | 85 (18.5%) | 25.6 (22.3–31.2) | .393 |
| Duplicate test* | ||||||
| Duplicate of positive | 1839 | 1799 (97.8%) | 25.4 (22.4–30.7) | 40 (2.2%) | 27.3 (23.5–32.4) | .087 |
| Duplicate of negative | 1839 | 1825 (99.2%) | 25.4 (22.4–30.7) | 14 (0.8%) | 31.6 (27.7–34.4) | .003 |
| Inappropriate CCDS response or duplicate Test** | 505 | 361 (71.5%) | 24.8 (22.1–30.3) | 144 (28.5%) | 26.7 (22.8–32.2) | .023 |
P values were obtained by the Mann–Whitney U test. Duplicate of negative is defined as a negative result within 3 days of a previous negative result. Duplicate of positive is defined as a positive result within 14 days of a previous positive result.
Three of the duplicate of negative and six of the duplicate of positive tests were performed post-CCDS implementation.
Compared to all appropriate positive tests post-CCDS implementation.
Fig. 1.
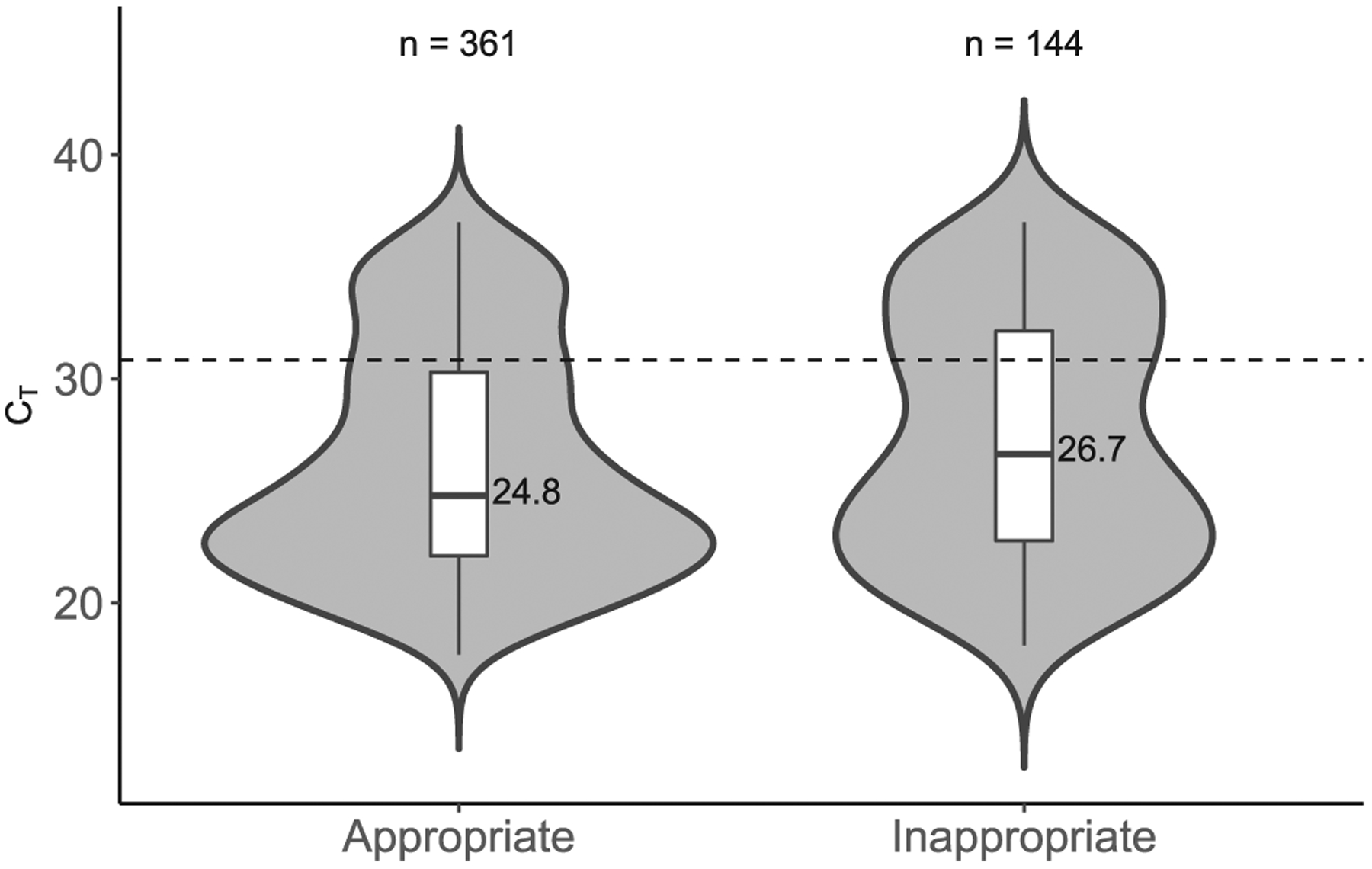
Violin and box plots comparing CT values between appropriate and inappropriate positive C. difficile NAATs. The dotted line depicts very high threshold = 30.85.
These results support the use of our current CCDS-based strategy. It is difficult to ascertain whether the result of 22.2% of very high CT values (> 30.85) obtained from the appropriate test (compared to the result of 34.0% of very high CT values obtained from the inappropriate test) is acceptable or not. We hypothesize that refinement of the CCDS may further reduce the proportion of tests with very high CT values. In addition, it should be noted that 35% of CT values categorized as inappropriate (excluding duplicates of positives) were < 26.0, thus suggesting that the patients were mis-classified as being at a low pretest probability for the disease. We feel that this supports the use of CCDS tools for diagnostic guidance during test ordering, while allowing clinicians to bypass the tool and order tests on the basis of their clinical judgment.
Although the absolute difference in the median CT value between groups is relatively small, this likely reflects the prevalence of C. difficile colonization described among hospitalized patients (~4–29%) and the fact that colonized patients outnumber infected patients as 5 to 1.7 Although we have not validated CT values with CCNA at our institution, we found a similar association between CT and toxin EIA described by Kamboj et al. and others, using a small (70 positive NAAT samples) set of historical internal validation samples (data not shown).
Considering the gold standard among C. difficile diagnostics, the CCNA assay is technically complex and labor intensive and has a slow turnaround time, thus making it impractical for routine clinical use. Unfortunately, C. difficile EIAs lack sensitivity. For these reasons, > 70% of hospitals currently use NAAT for the diagnosis of CDI.8 The GeneXpert C. difficile PCR assay is highly sensitive at the manufacturer-set maximum CT (≤ 37.0), with an estimated detection limit of 1657 colony-forming units; however, a positive NAAT result alone may overdiagnose CDI up to half of the time.2
Analysis of CT may offer a means to tailor C. difficile NAAT sensitivity and specificity according to various patient populations and levels of risk by modulating the CT along a receiver operator characteristic curve. CT also allows valuable feedback for diagnostic stewards, as we have shown. Validation of CT for diagnostic and diagnostic stewardship purposes requires further research in various clinical settings before its clinical use can be widely applied.
Acknowledgments
We thank Frankie J. Brewster, BSMT(ASCP), for her help in data collection.
Funding
This research was supported by the National Institutes of Health Infectious Diseases Training Grant (no. 5T-32AI007046-41, Funder Id: 10.13039/10 000 0060).
Footnotes
Conflict of interests
None to declare.
Contributor Information
Gregory R. Madden, Division of Infectious Diseases & International Health, Department of Medicine, University of Virginia Health System, Charlottesville, VA, USA.
Melinda D. Poulter, Clinical Microbiology Laboratory, Department of Pathology, University of Virginia Health System, Charlottesville, VA, USA
Costi D. Sifri, Division of Infectious Diseases & International Health, Department of Medicine, University of Virginia Health System, Charlottesville, VA, USA Office of Hospital Epidemiology/Infection Prevention & Control, University of Virginia Health System, Charlottesville, VA, USA.
References
- 1.Kamboj M, Brite J, McMillen T, Robilotti E, Herrera A, Septowitz K, et al. Potential of real-time PCR threshold cycle (CT) to predict presence of free toxin and clinically relevant C. difficile infection (CDI) in patients with cancer. J Infect 2018;76(4):369–75. doi: 10.1016/jjinf.2017.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Polage CR, Gyorke CE, Kennedy MA, Leslie JL, Chin DL, Wang S, et al. Overdiagnosis of Clostridium difficile Infection in the Molecular Test Era. JAMA Intern Med 2015; 175(11):1792–801. doi: 10.1001/jamainternmed.2015.4114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garvey MI, Bradley CW, Wilkinson MAC, Holden E. Can a toxin gene NAAT be used to predict toxin EIA and the severity of Clostridium difficile infection? An-timicrob Resist Infect Control 2017;6(1):127. doi: 10.1186/s13756-017-0283-Z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Senchyna F, Gaur RL, Gombar S, Truong CY, Schroeder LF, Banaei N. Clostridium difficile PCR cycle threshold predicts free toxin. J Clin Microbiol 2017;55(9):2651–60. doi: 10.1128/JCM.00563-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hitchcock M, Holubar MInfectious LTOF PCR cycle-threshold-derived toxin identifies patients at low-risk for complications of C. difficile infection who do not require treatment. Open Forum Infect Dis 2017;4(Suppl):S395. doi: 10.1093/ofid/ofx163.985. [DOI] [Google Scholar]
- 6.Madden GR, German Mesner I, Cox HL, Mathers AJ, Lyman JA, Sifri CD, et al. Reduced Clostridium difficile tests and laboratory-identified events with a computerized clinical decision support tool and financial incentive. Infect Control Hosp Epidemiol 2018;39(6):737–40. doi: 10.1017/ice.2018.53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Furuya-Kanamori L, Marquess J, Yakob L, Riley TV, Paterson DL, Foster NF, et al. Asymptomatic Clostridium difficile colonization: epidemiology and clinical implications. BMC Infect Dis 2015;15(1):516. doi: 10.1186/s12879-015-1258-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taylor R Fecal transplantation, molecular testing among new recommendations. Clostridium difficile guidelines/Infectious Diseases Society of America; 2018. https://www.idsociety.org/New_Recommendations_Clostridium_Difficile_2018.aspx. Published February 15 Accessed August 21, 2018.


