Abstract
Anti-vascular endothelial growth factor (VEGF) agents are widely regarded as the first line of therapy for diabetic macular edema (DME) but are not universally effective. An automatic method that can predict whether a patient is likely to respond to anti-VEGF therapy can avoid unnecessary trial and error treatment strategies and promote the selection of more effective first-line therapies. The objective of this study is to automatically predict the efficacy of anti-VEGF treatment of DME in individual patients based on optical coherence tomography (OCT) images. We performed a retrospective study of 127 subjects treated for DME with three consecutive injections of anti-VEGF agents. Patients’ retinas were imaged using spectral-domain OCT (SD-OCT) before and after anti-VEGF therapy, and the total retinal thicknesses before and after treatment were extracted from OCT B-scans. A novel deep convolutional neural network was designed and evaluated using pre-treatment OCT scans as input and differential retinal thickness as output, with 5-fold cross-validation. The group of patients responsive to anti-VEGF treatment was defined as those with at least a 10% reduction in retinal thickness following treatment. The predictive performance of the system was evaluated by calculating the precision, sensitivity, specificity, and area under the receiver operating characteristic curve (AUC). The algorithm achieved an average AUC of 0.866 in discriminating responsive from non-responsive patients, with an average precision, sensitivity, and specificity of 85.5%, 80.1%, and 85.0%, respectively. Classification precision was significantly higher when differentiating between very responsive and very unresponsive patients. The proposed automatic algorithm accurately predicts the response to anti-VEGF treatment in DME patients based on OCT images. This pilot study is a critical step toward using non-invasive imaging and automated analysis to select the most effective therapy for a patient’s specific disease condition.
1. Introduction
Diabetic macular edema (DME) is a major cause of central vision loss in patients with diabetic retinopathy if untreated [1]. Current options for DME treatment include intravitreal injections of anti-vascular endothelial growth factor (anti-VEGF) agents such as bevacizumab, ranibizumab, and aflibercept, injections of corticosteroid drugs, or use of a macular thermal laser [2]. Intravitreal anti-VEGF agents are the most common first line of therapy for DME, but not every patient responds to them [3] and other forms of treatment are often required [4,5].
In addition, previous randomized clinical trials have demonstrated that a subset of patients responds well to any given treatment modality [6]. However, the challenge of selecting the optimal treatment modality a priori remains a clinically unmet need, and many clinicians utilize a trial and error approach in which anti-VEGF therapy is first-line for all patients with alternatives utilized following treatment failure.
Frequent injections of anti-VEGF agents are costly and burdensome for both patients and physicians. It is of great interest to predict a priori whether anti-VEGF treatment will be effective for a particular patient. In a previous study, we developed a semi-automatic method to predict treatment outcomes in DME patients based on invasive fluorescein angiography imaging.[3] Here we use a fully automatic and non-invasive method to predict treatment outcomes in DME patients after three consecutive monthly anti-VEGF injections based solely on analysis of pretreatment optical coherence tomography (OCT) images. The single-shot term in our study emphasizes that the proposed method predicts the response to therapy by analyzing a single timepoint pre-treatment OCT volume, without the need for longitudinal treatment information such as time-series OCT images, patient records, or other metadata.
Algorithms developed previously for automatic retinal OCT image analysis include retinal layer segmentation [7,8], classification [9–17], and biomarker detection [18,19]. A few studies have assessed differential effects of anti-VEGF treatment on retinal diseases using OCT images [20–24]. Most of these studies have made predictions based on longitudinal analysis of a series of retinal OCT images from patients who received multiple anti-VEGF injections.
Bogunovic et al. predicted responses to anti-VEGF treatment of age-related macular degeneration (AMD) [25] by extracting features from a longitudinal series of OCT images followed by a support vector machine classifier. The method showed a success rate of 87% (n = 30) in predicting whether a subject would respond to treatment at the next visit. In a subsequent study, they predicted responses to anti-VEGF therapy for 317 AMD patients based on automatic analysis of macular OCT microstructures as well as best-corrected visual acuity (BCVA) and demographic characteristics, with an area under the receiver operating characteristic curve (AUC) of 0.735 [26].
Vogl et al. predicted the recurrence of central retinal vein occlusion (CRVO) or branch retinal vein occlusion (BRVO) within one year based on retinal thickness features extracted from longitudinal SD-OCT scans [27]. They based predictions on three initial SD-OCT images and evaluated the predictive performance using a dataset of monthly images of 155 CRVO patients and 92 BRVO patients over one year. Two algorithms were used for predictions: extra trees and sparse logistic regression. The extra trees algorithm achieved an AUC of 0.83 for predicting the recurrence of BRVO and an AUC of 0.76 for CRVO; the logistic regression method achieved an AUC of 0.78 for BRVO and 0.79 for CRVO.
Prahs et al. developed a deep learning algorithm to distinguish retinal OCT B-scans from patients with or without an anti-VEGF injection, achieving 95.5% accuracy [28].
In this work, we address whether a DME patient’s response to anti-VEGF therapy can be predicted prior to treatment based on pretreatment OCT images. Our study is novel with respect to algorithm design and application. Specifically, our main contributions are the following: (1) Single-shot prediction of response to intravitreal anti-VEGF treatment based on automatic retinal OCT image analysis. (2) An attention-based [29,30] convolutional neural network (CNN) model which preserves and highlights global structures in OCT images while enhancing local features from fluid/exudate-affected regions to efficiently use retinal thickness information. (3) An additional feature selection step to efficiently mine CNN-encoded features that have high correlations with the anti-VEGF response for optimal decision making. We demonstrate the predictive ability of the algorithm and its superior performance versus other competitive deep learning methods.
The remainder of this paper is structured as follows. Section (2) describes material and methods in detail, including the clinical dataset, pre-processing, and CNN-based classification methods. Experimental results on OCT dataset are reported in Section (3). Sections (4) and (5) discuss and conclude this study and suggest our prospective research lines and future works.
2. Material and methods
2.1. Dataset
This study was approved by the Duke University Medical Center Institutional Review Board and was conducted in compliance with the Health Insurance Portability and Accountability Act (HIPAA) and the Declaration of Helsinki. Subjects who underwent three consecutive anti-VEGF injections were included in the study.
One hundred twenty-seven subjects with DME and who met inclusion criteria were identified from the retina practices of MJA, PSM, and SWC via retrospective chart review. Inclusion criteria were center involving DME defined as central subfield thickness greater than 320 µm for men or 305 µm for women. Subjects were required to have had OCT before and after three consecutive anti-VEGF injections spaced 4 to 6 weeks apart. Exclusion criteria were macular edema due to causes other than DME, treatment with anti-VEGF within three months, treatment with intravitreal triamcinolone or dexamethasone intravitreal implant within one year, any history of treatment with fluocinolone intravitreal implant or macular photocoagulation or pan-retinal photocoagulation within one year. Potential subjects with incomplete macular volume scan at either time point were excluded by the retina specialist. The software development and analysis team did not participate in data selection. No image, regardless of quality, was excluded from analysis after inclusion by the retinal specialist.
All participants underwent three intravitreous anti-VEGF injections (ranibizumab 0.3 mg, aflibercept 2.0 mg, or bevacizumab 1.25 mg) where 54.3% (69/127), 34.7% (44/127), and 11.0% (14/127) of subjects received bevacizumab, aflibercept, and ranibizumab, respectively. We chose to examine treatment response following three anti-VEGF injections based on the fact that prior analyses of large randomized controlled trials have evaluated response to treatment after the first three injections [31–33] and because this reflects our clinical practice patterns.
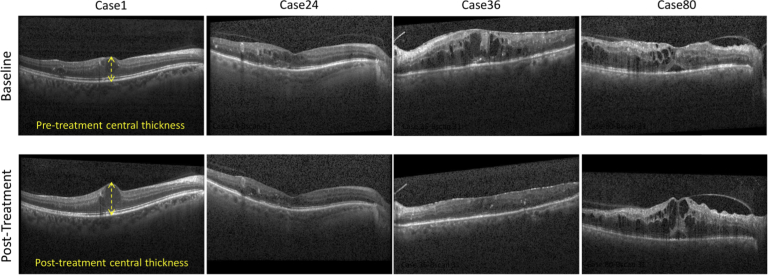
For each subject, we used the image acquired on the same day as the first anti-VEGF injection as the baseline, and one post-treatment image acquired at the clinic visit following three anti-VEGF injections. All images were acquired at Duke University between May 2013 and February 2019 using a Spectralis SD-OCT imaging system (Heidelberg Engineering Inc., Heidelberg, Germany). OCT images included 61 B-scans of 768×496 pixels with average axial, lateral, and azimuthal scanning pixel pitches of 3.8 µm, 11.4 µm, and 121.9 µm, respectively. Figure 1 displays foveal B-scans depicting different responses to anti-VEGF treatment after three months of therapy.
Fig. 1.
Example foveal SD-OCT images from pre-treatment (row 1) and post-treatment (row 2) acquisition sets. Only the patients in the second and third columns showed signs of response to treatment.
2.2. Data preparation and ground truth generation
We selected the central 49 B-scans of each SD-OCT volume to obtain a uniform region of analysis that was unaffected by artifacts that can appear in peripheral scans. Region of interest (ROI) images were obtained by cropping the central 496 A-scans in these B-scans and were resized to 256×256 pixels to reduce computational complexity.
We delineated the inner borders of the retinal pigment epithelium (RPE) layer and internal limiting membrane (ILM) in each B-scan semi-automatically, resulting in binary masks separating the retina from non-retina regions. The retinal thickness at the center of each B-scan was estimated using pixel pitch information in the SD-OCT metadata (See Fig. 1).
Differential retinal thicknesses (DRT) between baseline and post-treatment ROI images served as the ground truth values. DRT values were assigned to each pair of ROI images for registered B-scans from a given subject. In total, 6223 (127×49) baseline ROIs and corresponding DRTs were included to build a deep learning-based predictive system.
2.3. Data distribution
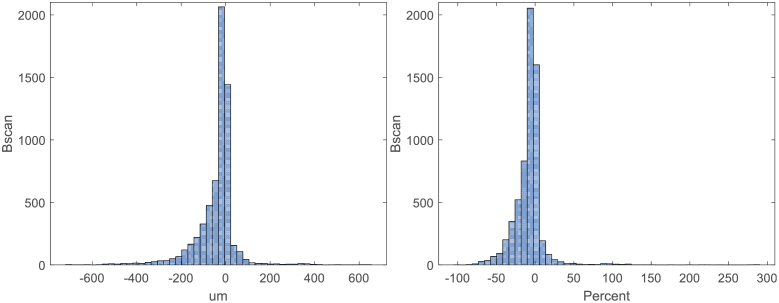
Patients were divided into responsive and non-responsive groups based on average DRT for all B-scans, with a 10% reduction in retinal thickness as the threshold for defining the groups. Figure 2 shows histograms of absolute DRT values and percent change DRT values. The mean, median, and standard deviation of absolute DRT over all B-scans were -41.7 µm, -19.4 µm, and 93.9 µm, respectively; the mean, median, and standard deviation of the percent change in retinal thickness were -8.1%, -5.2%, and 20.1%, respectively. These results indicate that our dataset—which was selected without bias from clinical data—was significantly more populated by subjects that had some retinal thickness reduction after treatment. This is expected given the known efficacy of anti-VEGF agents in treating DME and demonstrates that our subject cohort (including 80 responsive and 47 non-responsive cases) was reflective of a typical patient population.
Fig. 2.
Histogram of retinal thickness changes in pre-treatment OCT B-scans. The horizontal axis indicates the central thickness difference between post-treatment and baseline screenings. (Left) Differential thickness (µm). (Right) Percentage change in differential thickness.
2.4. Deep learning system and treatment response prediction
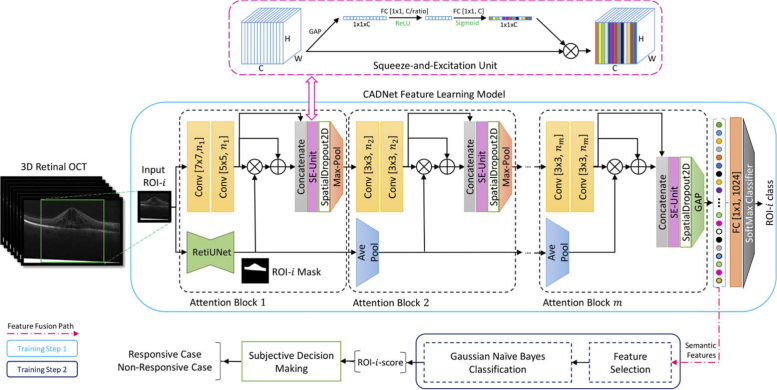
Our treatment response prediction method, the Convolutional Attention-to-DME Network (CADNet) (Fig. 3), is a novel modification of the VGG network [34]. CADNet benefits from multiple convolutional, pooling, and concatenation layers, along with two attention mechanisms: (1) A thickness-aware attention mechanism, which performs multiple pooling processes on the input mask image and softly weights the output feature maps of the CNN blocks. This mechanism allows highlighting of retinal thicknesses and weighting of extracted local feature maps. (2) A self-attention mechanism using a Squeeze-and-Excitation-Unit (SE-Unit) [35], which improves attention to informative feature maps generated by the convolutional layers and the thickness-aware attention mechanism. We used six attention blocks for the CADNet as the feature learner model at the ROI level. The number of kernels for the convolutional (Conv) layers was set to 16, 32, 64, 128, 256, and 512 for the subsequent attention blocks. The CADNet structure was trained and applied for data analysis and case-level decision making in the stages below.
Fig. 3.
Overview of the CADNet predictive framework with m = 6 attention blocks. The SE-Unit is demonstrated in detail. Values inside the bracket indicate the kernel size and the number of feature maps according to the block number, respectively. RetiUnet is a developed and pre-trained UNet model used as a non-trainable layer of CADNet for total retina segmentation. The sub-sampling factor and squeeze ratio of the pooling layers and SE-Units were 2 and 8, respectively. The symbols ⊗ and ⊕ indicate element-wise multiplication and summation operations, respectively. (GAP: global average pooling layer; FC: fully connected; ReLU: rectified linear units)
Fig. 3 demonstrates the architecture of the CADNet model with m = 6 attention blocks and the SE-Unit in detail. Values inside the bracket indicate the kernel size and the number of feature maps according to the block number, respectively. RetiUnet is a developed and pre-trained UNet model [36] used as a non-trainable layer of CADNet for total retina segmentation. The sub-sampling factor and squeeze ratio of the pooling layers and SE-Units were 2 and 8, respectively.
Stage I. ROI labeling
We used DRT data to partition patients into responsive and non-responsive classes. We defined responsive as and non-responsive as Using this stringent threshold, the rather than zero, only patients with showing significantly (more than 10%) reduced retinal thickness were counted as responsive, while patients showing minimally improved or increased retinal thickness were counted as non-responsive.
Stage II. ROI feature learning and selection schemes
ROI feature learning and extraction: The final block in CADNet is a fully connected (FC) layer with two active neurons and a SoftMax activation function. The CADNet model is optimized in a training process to learn discriminative image features and to map input ROIs to the corresponding class labels. The model’s parameters are then kept fixed and used for the representative feature fusion step. Two additional feature selection and classification steps were used to assess the redundancy level of the features learned by the model. For this aim, we considered 1024 output codes (the semantic feature in Fig. 3) from the last attention block in the model, which are processed by the global averaging pooling (GAP) layer.
ROI feature selection: We focused on the recursive feature elimination (RFE) method and the Elastic-Net (EN) estimator [37]. For the EN, a linear regression model with combined and priors was considered. The following objective function was used and minimized for the EN model:
| (1) |
Here y, X, and w are desired outputs, input samples, and parameter vector of the model, respectively. For controlling and weighting the and penalty terms, this loss function is equivalent to ; where , , and n is the number of input samples. In this study, we used the coefficients (parameter vector ) of the EN estimator in the RFE method. While the EN estimator assigns weights to features (the coefficients), the RFE method selects features by recursively considering smaller and pruned sets of coefficients.
We also evaluated two alternatives for the feature selection/reduction step for the comparison purpose: univariate feature selection (UFS) and principal component analysis (PCA) [38]. All the feature selection methods were considered in conjunction with a Gaussian Naïve Bayes (GNB) classifier [39]. The parameters for the feature selectors and the GNB classifier were optimized using the grid search method over the training subsets.
Stage III. Subjective decision making
Stages I and II were performed at the B-scan ROI level. To obtain the final diagnosis decision for test subjects, the majority voting rule was used for previously categorized ROIs in Stage II. That is, if more than 50 percent of ROIs in a patient were predicted to be responsive, the subject was assigned to the responsive group.
3. Experimental design and results
3.1. Cross-validation-based data partitioning
We used unbiased 5-fold cross-validation to evaluate and generalize the performance of the deep learning framework. SD-OCT volumes in the dataset were shuffled and partitioned into five subsets. Subsequently, in each of five iterations, four subsets were used for training and the remaining subset was held out for testing. Folding was applied at the subject level to ensure that ROIs from the same subjects were not used in both training and testing sets in each iteration. The final performance of the model was obtained by averaging the evaluation metrics on testing sets across the iterations.
3.2. Performance measures
Classification performance was quantified using AUC [40] and the following criteria: precision (positive predictive value), sensitivity (recall), and specificity, which were defined as:
| (2) |
| (3) |
| (4) |
where TP, TN, FP, and FN indicate the number of true positives, true negatives, false positives, and false negatives, respectively. P and N refer to the total number of non-responsive and responsive samples in the dataset, respectively.
3.3. Comparing CADNet with other baselines and deep learning methods
To attain a benchmark, following Vogel et al. [27], we used the pre-injection central retinal thickness (CRT) to predict the treatment response. The proposed classifiers in [27], i.e., Sparse Logistic Regression and Extra Trees, were trained and evaluated on our dataset based on CRT values. Moreover, we compared the performance of CADNet with the performance of other competitive CNN methods, including VGG16 [34], GoogLeNet InceptionV3 [41], ResNet50 [42], and Xception [43]. To do so, a transfer-learning (TL) method [44] was used where all convolutional layer weights were fixed, and only the last FC layer was replaced by a multi-layer perceptron (MLP) network and retrained to classify input images into the target categories. Furthermore, ablation studies were conducted to investigate the impact and contributions of the RetiUNet and SE layers on the classification performance of the CADNet model. Specifically, we designed a series of experiments where the RetiUNet and/or SE layers were added in the model. To get a benchmark for the RetiUNet segmentation performance, we conducted an experiment to evaluate the pre-trained RetiUNet on our OCT dataset, where we randomly selected 500 B-scans and their corresponding manual segmentations.
In addition to the above experiments, the statistical significance of the precision performance difference of selected predictors was determined using nonparametric statistical analysis.
3.4. Optimization method and implementation settings
All CNN models were optimized using Adam optimizer [45] and the cross-entropy objective function. For all CNNs, the mini-batch size, number of epochs, initial learning rate, and weight regularization factor were set to 256, 50, 2e-4, and 1e-3, respectively. We used an FC layer with 1024 neurons before the SoftMax classifier for all CNNs, and the exponential linear unit (ELU) [46] activation function was selected for all hidden layers. To improve the robustness of the CNNs, data augmentation strategies were performed on ROIs by vertical and horizontal transitions, horizontal flipping, and a range of random rotations [-10°, +10°].
CNN models were implemented and optimized using TensorFlow v1.13 [47] and Keras v2.2.4 [48] frameworks with NVIDIA CUDA v10.0, and a cuDNN v7.3 accelerated library and were coded in Python 3.6. All experiments were performed under a Win10 × 64 operating system on a machine with CPU Intel Xeon E5-2643@2 × 3.40 GHz, 4-GPU NVIDIA TITAN V, and 128 GB of RAM. Statistical analyses were performed using open-source libraries, including Scikit-learn v0.20.3, SciPy v1.2.1 in Python and also the open-source JAVA package available at http://sci2s.ugr.es/sicidm.
3.5. Results
Table 1 summarizes the performance of CADNet versus other baseline models for OCT B-scan classification on the ROI level, using a threshold of -10% to define responsive and non-responsive ROIs, with a dataset consisting of 2159 responsive and 4064 non-responsive ROIs. CADNet’s learned features were also analyzed by evaluating three feature selection/extraction methods (UFS, RFE, and PCA) and the GNB classifier. For the UFS method, Chi-square (χ2) and mutual information (MI) statistics were used to examine each feature individually to determine the dependency between features and labels. For the UFS.kBest, UFS.MI, and PCA methods, a range of [1%, 2%, 5%, 10%, 20%, 50%] of total features was explored for the selected subset of features. An EN estimator () was implemented for the RFE method to grid search on the optimal number of features according to the above range.
Table 1. Evaluation of different classification algorithms using the 5-fold cross-validation method at T=-10% (mean ± std).
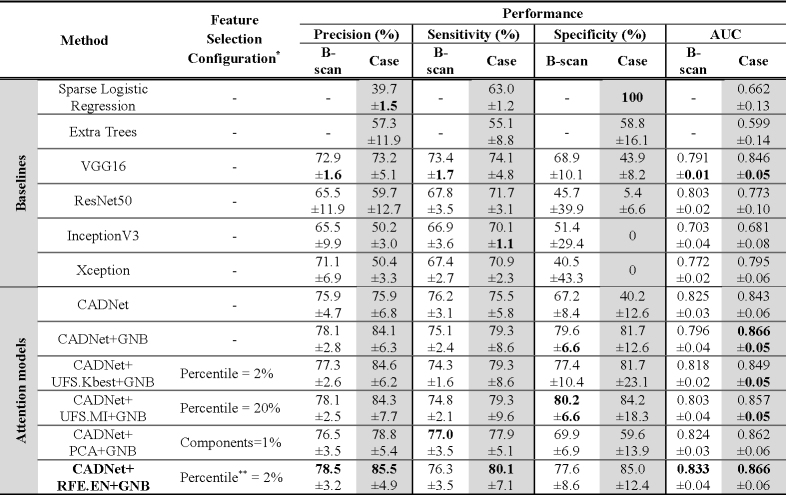 |
Feature selection configurations and hyperparameters were determined and optimized based on a subset of subjects in the training set of one fold and then used for all experiments.
The method uses two percent of the learned features by the CADNet model for the prediction purpose. The final feature set was selected based on the RFE.EN method.
The ablation study results are also shown in Table 2. In this table, we have reported the contributions of the RetiUNet and SE layers to the performance of the CADNet model.
Table 2. Performance contributions of the RetiUNet and SE layers in the CADNet model using the 5-fold cross-validation method.
| CADNet Model .Configuration |
B-scan Level Performance |
||||
|---|---|---|---|---|---|
| Precision (%) | Recall (%) | Specificity (%) | AUC | ||
| RetiUNet Layer | SE Layer | ||||
| ✗ | ✗ | 69.8 ± 3.3 | 72.3 ± 3.7 | 60.1 ± 8.8 | 0.777 ± 0.04 |
| ✗ | ✓ | 74.6 ± 5.5 | 74.9 ± 4.1 | 64.8 ± 9.1 | 0.818 ± 0.09 |
| ✓ | ✗ | 73.9 ± 3.6 | 77.1 ± 5.0 | 65.3 ± 8.9 | 0.824 ± 0.08 |
| ✓ | ✓ | 75.9 ± 4.7 | 76.2 ± 3.1 | 67.2 ± 8.4 | 0.825 ± 0.03 |
The segmentation performance analysis showed that the pre-trained RetiUNet’s Dice-coefficient [49], weighted accuracy, sensitivity, and Jaccard index [50] measures were 97.6%, 99.0%, 99.3%, and 97.2%, respectively.
In the RFE.EN method, first, the EN estimator was trained on the initial set of CNN features, and the importance of each feature was obtained through the coefficient attribute of the EN model. Then, the least important features were pruned from the nominated set of features. This procedure was recursively repeated on the pruned set (5 percent of features were removed at each iteration) until the desired percentiles of the learned CADNet features were eventually reached. Finally, based on a nested grid search on (a range between 0 and 1 in increments of 0.1) and the number of selected features, the and two percent of the learned CADNet features were selected.
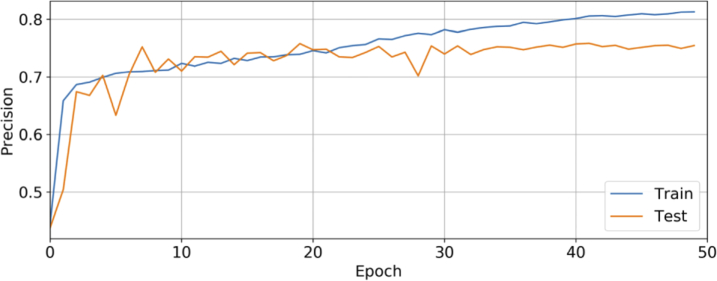
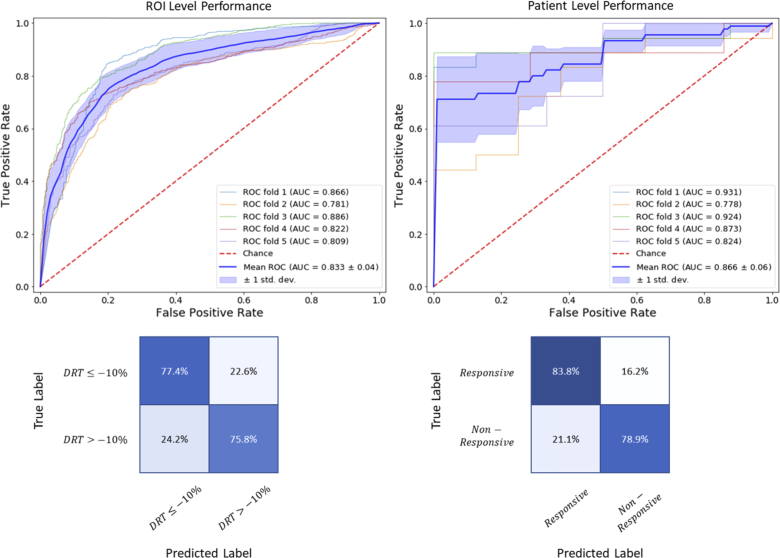
At the patient level, the diagnostic performance of different CADNet configurations was obtained according to the majority voting rule, summarized in Table 1. Figure 4 demonstrates ROI-level precision plot on training/testing sets versus epochs for the CADNet model. ROC curves and confusion matrices are also shown in Fig. 5 for the best CADNet framework configuration at the ROI and patient levels. The average training time for the CADNet model, with 7695602 trainable parameters, was approximately 137 millisecond per ROI.
Fig. 4.
Plot showing cross-validated precision performance against the epoch for the CADNet model. To avoid overfitting, we terminated the training process at the 50th epoch, at which point the validation precision shows lower performance. Due to our limited database and the wide range of DME manifestations on OCT in this prediction problem, our model is prone to overfitting.
Fig. 5.
Performance of the CADNet + RFE.EN + GNB framework. (Left column) Results at the ROI level. (Right column) Results at the patient level. (Top row) ROC curves. (Bottom row) Confusion matrices.
In addition to the reported measures in Table 1 and Fig. 5, the statistical analysis of the performance of the CADNet + RFE.EN + GNB method and the baselines (i.e., Extra Trees, VGG16, and CADNet classifiers) was performed considering non-parametric multiple significance tests [51,52]. For this purpose, 5 repetitions of the 5-fold CV method were executed at different seed points for data partitioning, and precision results at the patient level were analyzed using the open-source JAVA program developed in [53]. The software was used to calculate multiple comparison tests, including the Friedman ranking test and the Nemenyi post-hoc procedure test. The null hypothesis in this study states that all the evaluated machine learning algorithms perform equivalently on our dataset, and therefore their ranks should be equal. Here, the significance level of α=0.05 was used, so if the adjusted p-value for an individual hypothesis is less than 0.05, then the hypothesis is rejected. Moreover, the non-parametric Wilcoxon signed ranks test was computed for paired comparisons using the SciPy package.
The Friedman test, which uses the χ2 statistic, was performed to obtain average ranks. The calculated p-value of this statistic was 5.87e-11, thus rejecting the null hypothesis. Table 3 reports the average rankings of the algorithms by the Friedman test.
Table 3. Average rankings of the algorithms determined by the Friedman statistical test.
| Method | CADNet + RFE.EN + GNB | CADNet | VGG16 | Extra Trees |
|---|---|---|---|---|
| Ranking | 1-st (1.08) | 2-nd (2.20) | 3-rd (2.84) | 4-th (3.88) |
In Table 4, p-values for the Wilcoxon test and adjusted p-value for the Nemenyìs test for N×N comparisons are reported for all possible 6 pairs of algorithms.
Table 4. Adjusted p-values for N×N comparisons of diagnostic algorithms over 5 repetitions of 5-fold cross-validation method. The p-values below 0.05 demonstrate that the algorithms differ significantly (marked in italic font) in terms of precision values.
| Hypothesis | Wilcoxon p-value | Nemenyi p-value |
|---|---|---|
| VGG16 vs. Extra Trees | 4.07e-5 | 0.02638 |
| CADNet vs. Extra Trees | 1.39e-5 | 2.52e-5 |
| CADNet vs. VGG16 | 0.00024 | 0.47791 |
| CADNet + RFE.EN + GNB vs. Extra Trees | 1.23e-5 | 1.05e-13 |
| CADNet + RFE.EN + GNB vs. VGG16 | 1.39e-5 | 8.62e-6 |
| CADNet + RFE.EN + GNB vs. CADNet | 3.62e-5 | 0.01296 |
These tests show that the CADNet + RFE.EN + GNB has a significantly better performance than other algorithms.
For the sake of completeness, we also compared the effect of changing the threshold value (T). When comparing the CADNet + RFE.EN + GNB classification pipeline at the patient level for T = [-20%, -15%, -10%, -5%], the AUC values were 0.862 ± 0.15, 0.887 ± 0.04, 0.866 ± 0.06, and 0.761 ± 0.06, respectively.
A complementary analysis showed that the classification performance was significantly higher when differentiating between very responsive and very unresponsive groups (i.e., the top 20 cases with average DRT≤-40% or DRT≥+10%), with an AUC of 0.899.
4. Discussion
We investigated whether the response of DME patients to anti-VEGF treatment could be predicted from pretreatment OCT scans using modern machine learning algorithms. Automatic prediction of the response to anti-VEGF treatment is a step toward precision medicine, in which such predictions help clinicians better select first-line therapies for patients based on specific disease conditions. In contrast to most previous studies that used longitudinal series of OCTs for response trend prediction and classification [3,20,21,23,25,27], our algorithm required only pretreatment OCT scans to predict treatment outcome.
We achieved this by using a new feature-learning and classification framework. At the heart of this framework is a novel CNN model called CADNet, which showed superior predictive ability versus other modern deep learning-based image classification architectures. The addition of feature selection and GNB classification steps to CADNet further improved classification performance. Of the CADNet configurations tested in this study, the CADNet + RFE.EN + GBN pipeline achieved the best results. By combining the results in Tables 1 and 4, we conclude that the proposed framework had a significantly higher performance than the baseline methods (Extra Trees and VGG16) and the basis model of CADNet, since the corresponding p-values were all less than 0.05 for 95% confidence level.
Overall, the experimental results supported the hypothesis that machine learning algorithms can use pretreatment retinal OCT images to accurately predict DME patient response to anti-VEGF therapy. The higher accuracy in discriminating patients who respond very positively or very negatively to anti-VEGF therapy also supports this hypothesis. Furthermore, the ability to accurately select highly responsive and very poorly responsive patients prior to treatment would be beneficial for practicing physicians and potentially for subject selection in clinical trials of novel therapies for DME.
Our study has the following limitations: First, it is limited by its retrospective nature and small sample size. While our network design was efficient in that it was capable of using only 127 patients for training and testing, it is reasonable to assume that the dataset did not cover all patterns of the disease. It is expected that a larger OCT dataset for training and testing would result in an even better prediction outcome. Second, treatment response may vary for different anti-VEGF agents such as for aflibercept, ranibizumab, or bevacizumab. Response differentiation for these agents was not feasible due to our limited dataset. Third, while our study considered only OCT images as input, a comprehensive algorithm that utilizes complementary features such as age, gender, genetic factors, and duration of diabetic disease in addition to OCT would likely result in better predictive performance. Fourth, our study did not identify anatomic and pathologic features that are significantly impactful in the prediction outcome. A detailed analysis of features that contribute most to the network outcome would help stratify DME patients into subgroups that reflect specific pathophysiological mechanisms. In turn, we expect that subgrouping per these mechanisms would facilitate a choice of therapy that is personalized for an individual’s specific disease condition. Part of our ongoing work is a careful occlusion analysis to address this question. Fifth, it is conceivable that better results could be attained by optimizing the hyperparameters over the whole dataset and then reassessing the performance of our algorithm, without further parameter tuning, based on an independently (and preferably prospectively) collected dataset, which is part of our ongoing work.
A larger prospective observational trial with a standardized imaging protocol is needed to allow us to confirm and extend these findings.
5. Conclusion
We present a deep learning method that accurately predicts retinal response to anti-VEGF treatment in patients with DME. The automatic image analysis framework uses pretreatment OCT scans to classify patients into responsive and non-responsive groups. This pilot study is a step toward automatic evaluation of electronic health record data to predict the effectiveness of various therapies (anti-VEGF, intravitreal corticosteroids, or thermal laser) to select the most effective first-line therapy for each patient.
Funding
Google10.13039/100006785 Faculty Research Award; 2018 Unrestricted Grant from Research to Prevent Blindness10.13039/100001818.
Disclosures
The authors declare that there is no conflict of interest.
References
- 1.Yau J., Rogers S. L., Kawasaki R., Lamoureux E. L., Kowalski J. W., Bek T., Chen S., Dekker J., Fletcher A., Grauslund J., “Meta-Analysis for Eye Disease (METAEYE) Study Group. Global prevalence and major risk factors of diabetic retinopathy,” Diabetes Care 35(3), 556–564 (2012). 10.2337/dc11-1909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Giuliari G. P., “Diabetic retinopathy: current and new treatment options,” Curr. Diabetes Rev. 8(1), 32–41 (2012). 10.2174/157339912798829188 [DOI] [PubMed] [Google Scholar]
- 3.Allingham M. J., Mukherjee D., Lally E. B., Rabbani H., Mettu P. S., Cousins S. W., Farsiu S., “A quantitative approach to predict differential effects of anti-VEGF treatment on diffuse and focal leakage in patients with diabetic macular edema: a pilot study,” Trans. Vis. Sci. Tech. 6(2), 7 (2017). 10.1167/tvst.6.2.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lazic R., Lukic M., Boras I., Draca N., Vlasic M., Gabric N., Tomic Z., “Treatment of Anti-Vascular Endothelial Growth Factor–Resistant Diabetic Macular Edema With Dexamethasone Intravitreal Implant,” Retina 34(4), 719–724 (2014). 10.1097/IAE.0b013e3182a48958 [DOI] [PubMed] [Google Scholar]
- 5.Wells J. A., Glassman A. R., Ayala A. R., Jampol L. M., Bressler N. M., Bressler S. B., Brucker A. J., Ferris F. L., Hampton G. R., Jhaveri C., “Aflibercept, bevacizumab, or ranibizumab for diabetic macular edema: two-year results from a comparative effectiveness randomized clinical trial,” Ophthalmology 123(6), 1351–1359 (2016). 10.1016/j.ophtha.2016.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elman M. J., Aiello L. P., Beck R. W., Bressler N. M., Bressler S. B., Edwards A. R., Ferris F. L., III, Friedman S. M., Glassman A. R., Miller K. M., “Randomized trial evaluating ranibizumab plus prompt or deferred laser or triamcinolone plus prompt laser for diabetic macular edema,” Ophthalmology 117(6), 1064–1077.e35 (2010). 10.1016/j.ophtha.2010.02.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fang L., Cunefare D., Wang C., Guymer R. H., Li S., Farsiu S., “Automatic segmentation of nine retinal layer boundaries in OCT images of non-exudative AMD patients using deep learning and graph search,” Biomed. Opt. Express 8(5), 2732–2744 (2017). 10.1364/BOE.8.002732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roy A. G., Conjeti S., Karri S. P. K., Sheet D., Katouzian A., Wachinger C., Navab N., “ReLayNet: retinal layer and fluid segmentation of macular optical coherence tomography using fully convolutional networks,” Biomed. Opt. Express 8(8), 3627–3642 (2017). 10.1364/BOE.8.003627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farsiu S., Chiu S. J., O’Connell R. V., Folgar F. A., Yuan E., Izatt J. A., Toth C. A., for the Age-Related Eye Disease Study 2 Ancillary Spectral Domain Optical Coherence Tomography Study Group , “Quantitative classification of eyes with and without intermediate age-related macular degeneration using optical coherence tomography,” Ophthalmology 121(1), 162–172 (2014). 10.1016/j.ophtha.2013.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lemaître G., Rastgoo M., Massich J., Cheung C. Y., Wong T. Y., Lamoureux E., Milea D., Mériaudeau F., Sidibé D., “Classification of SD-OCT volumes using local binary patterns: experimental validation for DME detection,” J. Ophthalmol. 2016, 1–14 (2016). 10.1155/2016/3298606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rasti R., Mehridehnavi A., Rabbani H., Hajizadeh F., “Wavelet-based Convolutional Mixture of Experts model: An application to automatic diagnosis of abnormal macula in retinal optical coherence tomography images,” in 10th Iranian Conference on Machine Vision and Image Processing (MVIP), (IEEE, 2017), 192–196. [Google Scholar]
- 12.De Fauw J., Ledsam J. R., Romera-Paredes B., Nikolov S., Tomasev N., Blackwell S., Askham H., Glorot X., O’Donoghue B., Visentin D., “Clinically applicable deep learning for diagnosis and referral in retinal disease,” Nat. Med. 24(9), 1342–1350 (2018). 10.1038/s41591-018-0107-6 [DOI] [PubMed] [Google Scholar]
- 13.Rasti R., Mehridehnavi A., Rabbani H., Hajizadeh F., “Automatic diagnosis of abnormal macula in retinal optical coherence tomography images using wavelet-based convolutional neural network features and random forests classifier,” J. Biomed. Opt. 23(03), 1 (2018). 10.1117/1.JBO.23.3.035005 [DOI] [PubMed] [Google Scholar]
- 14.Rasti R., Rabbani H., Mehridehnavi A., Hajizadeh F., “Macular OCT classification using a multi-scale convolutional neural network ensemble,” IEEE Trans. Med. Imaging 37(4), 1024–1034 (2018). 10.1109/TMI.2017.2780115 [DOI] [PubMed] [Google Scholar]
- 15.Fang L., Wang C., Li S., Rabbani H., Chen X., Liu Z., “Attention to Lesion: Lesion-Aware Convolutional Neural Network for Retinal Optical Coherence Tomography Image Classification,” IEEE Trans. Med. Imaging 38(8), 1959–1970 (2019). 10.1109/TMI.2019.2898414 [DOI] [PubMed] [Google Scholar]
- 16.Srinivasan P. P., Kim L. A., Mettu P. S., Cousins S. W., Comer G. M., Izatt J. A., Farsiu S., “Fully automated detection of diabetic macular edema and dry age-related macular degeneration from optical coherence tomography images,” Biomed. Opt. Express 5(10), 3568–3577 (2014). 10.1364/BOE.5.003568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rasti R., Mehridehnavi A., Rabbani H., Hajizadeh F., “Convolutional mixture of experts model: A comparative study on automatic macular diagnosis in retinal optical coherence tomography imaging,” J Med Signals Sens 9(1), 1–14 (2019). 10.4103/jmss.JMSS_27_17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Al-Sheikh M., Iafe N. A., Phasukkijwatana N., Sadda S. R., Sarraf D., “Biomarkers of neovascular activity in age-related macular degeneration using optical coherence tomography angiography,” Retina 38(2), 220–230 (2018). 10.1097/IAE.0000000000001628 [DOI] [PubMed] [Google Scholar]
- 19.Lai T.-T., Hsieh Y.-T., Yang C.-M., Ho T.-C., Yang C.-H., “Biomarkers of optical coherence tomography in evaluating the treatment outcomes of neovascular age-related macular degeneration: a real-world study,” Sci. Rep. 9(1), 529–539 (2019). 10.1038/s41598-018-36704-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Santos A. R., Gomes S. C., Figueira J., Nunes S., Lobo C. L., Cunha-Vaz J. G., “Degree of decrease in central retinal thickness predicts visual acuity response to intravitreal ranibizumab in diabetic macular edema,” Ophthalmologica 231(1), 16–22 (2013). 10.1159/000355487 [DOI] [PubMed] [Google Scholar]
- 21.Costa M., Santos A. R., Nunes S., Alves D., Schwartz C., Figueira J., Simao S. N., Cunha-Vaz J. G., “OCT retinal thickness response after first intravitreal injection is a predictor of visual acuity response to anti-VEGF treatment of DME,” Invest. Ophthalmol. Visual Sci. 57, 2085 (2016). [Google Scholar]
- 22.Shah A. R., Yonekawa Y., Todorich B., Laere L. V., Hussain R., Woodward M. A., Abbey A. M., Wolfe J. D., “Prediction of anti-VEGF response in diabetic macular edema after 1 injection,” J. Vitreoretin. Dis. 1(3), 169–174 (2017). 10.1177/2474126416682569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shiraya T., Kato S., Araki F., Ueta T., Miyaji T., Yamaguchi T., “Aqueous cytokine levels are associated with reduced macular thickness after intravitreal ranibizumab for diabetic macular edema,” PLoS One 12(3), e0174340 (2017). 10.1371/journal.pone.0174340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cho Y. J., Lee D. H., Kim M., “Optical coherence tomography findings predictive of response to treatment in diabetic macular edema,” J. Int. Med. Res. 46(11), 4455–4464 (2018). 10.1177/0300060518798503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bogunovic H., Abramoff M. D., Zhang L., Sonka M., “Prediction of treatment response from retinal OCT in patients with exudative age-related macular degeneration,” in Proceedings of the Ophthalmic Medical Image Analysis First International Workshop, 2014), 129–136. [Google Scholar]
- 26.Bogunović H., Waldstein S. M., Schlegl T., Langs G., Sadeghipour A., Liu X., Gerendas B. S., Osborne A., Schmidt-Erfurth U., “Prediction of anti-VEGF treatment requirements in neovascular AMD using a machine learning approach,” Invest. Ophthalmol. Visual Sci. 58(7), 3240–3248 (2017). 10.1167/iovs.16-21053 [DOI] [PubMed] [Google Scholar]
- 27.Vogl W.-D., Waldstein S. M., Gerendas B. S., Schmidt-Erfurth U., Langs G., “Predicting macular edema recurrence from spatio-temporal signatures in optical coherence tomography images,” IEEE Trans. Med. Imaging 36(9), 1773–1783 (2017). 10.1109/TMI.2017.2700213 [DOI] [PubMed] [Google Scholar]
- 28.Prahs P., Radeck V., Mayer C., Cvetkov Y., Cvetkova N., Helbig H., Märker D., “OCT-based deep learning algorithm for the evaluation of treatment indication with anti-vascular endothelial growth factor medications,” Graefe’s Arch. Clin. Exp. Ophthalmol. 256(1), 91–98 (2018). 10.1007/s00417-017-3839-y [DOI] [PubMed] [Google Scholar]
- 29.Treisman A. M., Gelade G., “A feature-integration theory of attention,” Cogn. Psychol. 12(1), 97–136 (1980). 10.1016/0010-0285(80)90005-5 [DOI] [PubMed] [Google Scholar]
- 30.Cortes C., Vapnik V., “Support-vector networks,” Mach. Learn. 20, 273–297 (1995). 10.1023/A:1022627411411 [DOI] [Google Scholar]
- 31.Maturi R. K., Glassman A. R., Liu D., Beck R. W., Bhavsar A. R., Bressler N. M., Jampol L. M., Melia M., Punjabi O. S., Salehi-Had H., “Effect of adding dexamethasone to continued ranibizumab treatment in patients with persistent diabetic macular edema: a DRCR network phase 2 randomized clinical trial,” JAMA Ophthalmol. 136(1), 29–38 (2018). 10.1001/jamaophthalmol.2017.4914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gonzalez V. H., Campbell J., Holekamp N. M., Kiss S., Loewenstein A., Augustin A. J., Ma J., Ho A. C., Patel V., Whitcup S. M., “Early and long-term responses to anti–vascular endothelial growth factor therapy in diabetic macular edema: analysis of protocol I data,” Am. J. Ophthalmol. 172, 72–79 (2016). 10.1016/j.ajo.2016.09.012 [DOI] [PubMed] [Google Scholar]
- 33.Bressler N. M., Beaulieu W. T., Maguire M. G., Glassman A. R., Blinder K. J., Bressler S. B., Gonzalez V. H., Jampol L. M., Melia M., Sun J. K., “Early Response to Anti–Vascular Endothelial Growth Factor and Two-Year Outcomes Among Eyes With Diabetic Macular Edema in Protocol T,” Am. J. Ophthalmol. 195, 93–100 (2018). 10.1016/j.ajo.2018.07.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simonyan K., Zisserman A., “Very deep convolutional networks for large-scale image recognition,” ArXiv preprint arXiv:1409.1556 (2014).
- 35.Hu J., Shen L., Sun G., “Squeeze-and-excitation networks,” in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 7132–7141 (2018). [Google Scholar]
- 36.Ronneberger O., Fischer P., Brox T., “U-net: Convolutional networks for biomedical image segmentation,” in International Conference on Medical Image Computing and Computer-assisted Intervention, (Springer, 2015), 234–241. [Google Scholar]
- 37.Zou H., Hastie T., “Regularization and variable selection via the elastic net,” J Royal Statistical Soc B 67(2), 301–320 (2005). 10.1111/j.1467-9868.2005.00503.x [DOI] [Google Scholar]
- 38.Jolliffe I., Principal component analysis, Springer Series in Statistics (Springer, 2002). [Google Scholar]
- 39.Zhang H., “The optimality of naive Bayes,” in Proceedings of the Seventeenth International Florida Artificial Intelligence Research Society Conference (FLAIRS), (AAAI Press, 2004), 3. [Google Scholar]
- 40.Fawcett T., “An introduction to ROC analysis,” Pattern Recognit. Lett. 27(8), 861–874 (2006). 10.1016/j.patrec.2005.10.010 [DOI] [Google Scholar]
- 41.Szegedy C., Vanhoucke V., Ioffe S., Shlens J., Wojna Z., “Rethinking the inception architecture for computer vision,” in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2818–2826 (2016). [Google Scholar]
- 42.He K., Zhang X., Ren S., Sun J., “Deep residual learning for image recognition,” in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 770–778 (2016). [Google Scholar]
- 43.Chollet F., “Xception: Deep learning with depthwise separable convolutions,” in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 1251–1258 (2017). [Google Scholar]
- 44.Shin H.-C., Roth H. R., Gao M., Lu L., Xu Z., Nogues I., Yao J., Mollura D., Summers R. M., “Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning,” IEEE Trans. Med. Imaging 35(5), 1285–1298 (2016). 10.1109/TMI.2016.2528162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kingma D. P., Ba J., “Adam: A method for stochastic optimization,” ArXiv preprint arXiv:1412.6980 (2014).
- 46.Clevert D.-A., Unterthiner T., Hochreiter S., “Fast and accurate deep network learning by exponential linear units (elus),” ArXiv preprint arXiv:1511.07289 (2015).
- 47.Abadi M., Barham P., Chen J., Chen Z., Davis A., Dean J., Devin M., Ghemawat S., Irving G., Isard M., “Tensorflow: A system for large-scale machine learning,” in 12th Symposium on Operating Systems Design and Implementation, 2016), 265–283. [Google Scholar]
- 48.Chollet F., “Keras” (2015), retrieved https://keras.io.
- 49.Dice L. R., “Measures of the amount of ecologic association between species,” Ecology 26(3), 297–302 (1945). 10.2307/1932409 [DOI] [Google Scholar]
- 50.Jaccard P., “The distribution of the flora in the alpine zone,” New Phytol. 11(2), 37–50 (1912). 10.1111/j.1469-8137.1912.tb05611.x [DOI] [Google Scholar]
- 51.Hollander M., Wolfe D. A., Chicken E., Nonparametric Statistical Methods, 3rd ed. (John Wiley & Sons, 2013), Vol. 751. [Google Scholar]
- 52.Trawiński B., Smętek M., Telec Z., Lasota T., “Nonparametric statistical analysis for multiple comparison of machine learning regression algorithms,” Int. J. Appl. Math. Comput. Sci. 22(4), 867–881 (2012). 10.2478/v10006-012-0064-z [DOI] [Google Scholar]
- 53.Garcia S., Herrera F., “An extension on ‘statistical comparisons of classifiers over multiple data sets’ for all pairwise comparisons,” Journal of Machine Learning Research 9 2677–2694 (2008). [Google Scholar]