Abstract
Lung cancer is the leading cause of death in cancer patients, and microwave ablation (MWA) has been extensively used in clinical treatment. In this study, we characterized the spectra of MWA-treated and untreated lung squamous cell carcinoma (LSCC) tissues, as well as healthy lung tissue, and conducted a preliminary analysis of spectral variations associated with MWA treatment. The results of characteristic spectral analysis of different types of tissues indicated that MWA treatment induces an increase in the content of nucleic acids, proteins, and lipid components in lung cancer tissues. The discriminant model based on the principal component analysis - linear discriminant analysis (PCA-LDA) algorithm together with leave-one-out cross validation (LOOCV) method yield the sensitivities of 90%, 80%, and 96%, and specificities of 86.2%, 93.8%, and 100% among untreated and MWA-treated cancerous tissue, and healthy lung tissue, respectively. These results indicate that Raman spectroscopy combined with multivariate analysis techniques can be used to explore the biochemical response mechanism of cancerous tissue to MWA therapy.
1. Introduction
Lung cancer is the most common type of cancer and the leading cause of cancer death [1]. In 2015, the number of new lung cancer cases and deaths reached 733,300 and 610,200 in China, respectively [2]. Current treatment modalities for lung cancer, such as radiotherapy, chemotherapy, adjuvant and surgical resection [3], are highly traumatic procedures that carry a high therapeutic risk for some patients with poor cardiorespiratory function. Therefore, a safer, more effective and minimally invasive treatment method for improving the treatment of lung cancer patients is required. Thermal ablation with microwave or radiofrequency energy has been used as an effective treatment modality for unresectable lung tumors, as well as primary and secondary thoracic malignancies [4–6]. Microwave ablation (MWA) is a local thermal ablation technique that transmits high-frequency electromagnetic waves (915 and 2450 MHz) to the tumor tissue through the antenna, causing oscillation of the electric dipole of water molecules [7–9]. Kinetic energy from the water molecules is then transferred neighboring tumor molecules, resulting in a rapid increase in the tissue temperature to above 60°C, thus inducing irreversible coagulation necrosis of the tissue in the central ablation zone [7,10]. This ablation technique has significant advantages of flexible application in different treatment strategies, good tolerance, and the potential to create necrotic areas reproducibly [9]. Moreover, MWA may be less affected by the heat-sink effect than other thermal ablation methods [11].
The biochemical responses of cancerous tissue to MWA treatment can be further elucidated by studying the molecular changes induced by the thermal effects by comparison of tissues pre- and post-therapy. Raman spectroscopy is an ideal technique for this type of study in that it can provide a “fingerprint” of the structure and conformation of molecule [12–16]. This information can be used to improve the pathological and biochemical interpretation of tissue samples, as well as the ability to identify subtle biochemical variations with high sensitivity and specificity [17–24]. To clarify the biochemical mechanisms of cancer treatment, Fox et al. used Raman spectroscopy combined with multivariate analysis to classify normal and cancerous skin tissue, and evaluated the differences between tissues treated with high energy CO2 laser and untreated tissue [25]. In studies of the spectral variations of breast cancer tissues pre- and post-photodynamic therapy in animal models, Fontana et al. revealed the underlying mechanisms for the variations of biochemical components in cancerous tissues after PDT treatment [26]. Raman spectroscopy has also been used for prognostic evaluations, such as in vitro radiation responses in human tumor cells, radiotherapy responses in patients with cervical cancer, and radiation damage in mouse brain [27–29].
Followed by our previous studies [30], we used Raman spectroscopy combined with multivariate analysis techniques to elucidate the biochemical response of lung squamous cell carcinoma (LSCC) tissue to MWA therapy. Compositional changes were analyzed by identifying significant spectral variations between the cancerous tissues in the pre-therapy group (Pre-MWA group) and post-therapy group (Post-MWA group); healthy lung tissues in Healthy Group were used for further comparisons. The spectral dataset obtained was further analyzed using principal component analysis-linear discriminant analysis (PCA-LDA) to explore the biochemical response of the cancerous tissue to MWA treatment. The results revealed a novel and objective method for the evaluation of the multiple biochemical variabilities in the responses of lung cancer tissue during MWA treatment. This method may represent the basis of new Raman-based analytical techniques for improved clinical efficacy.
2. Experimental
2.1. Tissue sample preparation
Tissue sections in Healthy Group (n = 6, 20 µm thick) were bought from Alenabio (Xi’an, Shaanxi, China), which was collected from clinical autopsy and kept in −80°C frozen environment. The commercial usage is approved under Institution Review Board (IRB) and Health Insurance Portability and Accountability Act (HIPAA) approved protocols. LSCC tissues (n = 12) were obtained from Jiangsu Cancer Hospital (Nanjing, China), from six male patients undergoing MWA treatment in the clinic. All experiments were performed in accordance with the guidance from Institutional Review Board/Independent Ethical Committee (IRB/IEC) in both Northwest University and Jiangsu Cancer Hospital, and approved by the ethics committee at Jiangsu Cancer Hospital. Informed consents were obtained from all of human participants in this study. Tissues in the Pre-MWA group were acquired by needle biopsy. After histopathological confirmation, patients received clinical MWA treatment (2145 MHz at 30 W for 5 min) delivered using the ablation system from Nanjing Weijing Jiuzhou Medical Equipment Research Institute. Approximately 24 h later, tissues in Post-MWA group were acquired by needle biopsy from the location of MWA treatment and prepared for histopathological analysis and Raman spectroscopy investigations.
Immediately after tissue collection, the LSCC tissue samples obtained from the untreated and MWA-treated groups were embedded in an optimal cutting temperature (OCT) medium (Surgipath FSC 22, Leica Biosystems, USA) and frozen with liquid nitrogen for better preservation of its native morphology. Transverse sections (20 µm thick) were prepared on gold-coated glass substrates (BioGold 63479-AS, Electron Microscopy Sciences, USA), which is used to eliminate the background fluorescence from the microscope slides and optics for spectroscopic measurement. Consecutive sections (5 µm thick) were hematoxylin and eosin (H&E) stained to facilitate comparison of the spectral measurement results with known histopathology. The frozen sections were immersed in an acetone cooling bath for dehydration and stored at −20°C until they were transported to Northwest University, Xi’an, Shaanxi, China for spectroscopy studies. The tissue sections were thawed 30 min prior to spectroscopic analysis or further histological processing.
2.2. Raman spectroscopy
The equipment used for Raman spectroscopy has been described in detail in previous studies [31–33]. Briefly, the WITec Alpha500 confocal micro-Raman spectroscopy system (WITec GmbH, Germany) was used for single spectrum acquisition with a 633 nm He-Ne laser (35 mW, Research Electro-Optics, Inc., USA) as the excitation source. A 100× microscope objective (NA = 1.25, EC Epiplan-Neofluar, Zeiss, Germany) was used for spectral excitation and measurements. A total of twenty-five spectra were used for averaging and normalization in healthy lung tissue sections, while forty spectra were used in the Pre-MWA and Post-MWA groups, respectively. Such spectral information is recorded by a spectrometer (UHTS300, WITec GmbH, Germany) with a resolution of approximately 3 cm−1 incorporating a 600 mm−1 grating blazed at 500 nm with a back-illuminated deep-depletion charge coupled device camera (Du401A-BR-DD-352, Andor Technology, UK) working at −60°C with a 1.5 s acquisition time for a single spectrum.
2.3. Data analysis
The acquired spectral data were pre-processed using WITec Project 4 (WITec GmbH, Germany) following our previously generated protocols [31–33], including a nine-order polynomial fit to subtract the spectral background and a five-order Savitzky-Golay smoothing. Each pre-processed spectrum was then normalized by the integrated area under the curve in the range from 600 to 1800 cm−1, and from 2800 to 3000 cm−1 to eliminate interference from unrelated factors and provide a superior comparison of the spectral features of different tissues. Subsequently, the data matrices containing all spectra were mean-centered and used for PCA and LDA in multivariate analysis. To improve the effectiveness of the analysis and reduce the complexity of the interpretation of our results, one-way ANOVA was used to identify the most diagnostically significant PCs (P < 0.01) from the first 10 significant PCs to better classify the three types of tissue samples [34]. In addition, Fisher’s least significant difference (LSD) post-hoc multiple tests were used to analyze the pairwise differences between different types of tissue [35]. LDA is a supervised method of analysis adopted to identify an optimal projection direction that maximizes differences between samples in different groups and minimizes differences between samples within the same group [36–38]. Significant PC scores were used as the input variable for LDA to generate a diagnostic algorithm [39]. To verify the performance of the discriminant model based on the PCA-LDA algorithm for different types of tissues classification, we used leave-one-out cross validation (LOOCV) method to further classify all spectra, while reducing the risk of over-fitting. The discriminant results based on Raman spectroscopy and the PCA-LDA algorithm was compared with the standard histopathological examination results to determine the sensitivity, specificity and overall accuracy of the discriminant model. The receiver operating characteristic (ROC) curves were used to further evaluate the overall discriminatory performance of Raman spectroscopy together with multivariate analysis method for different types of tissues. All statistical analyses were performed using MATLAB R2015b software (Mathworks. Inc., Natick, MA, USA).
3. Results and discussion
3.1. Raman spectral analysis
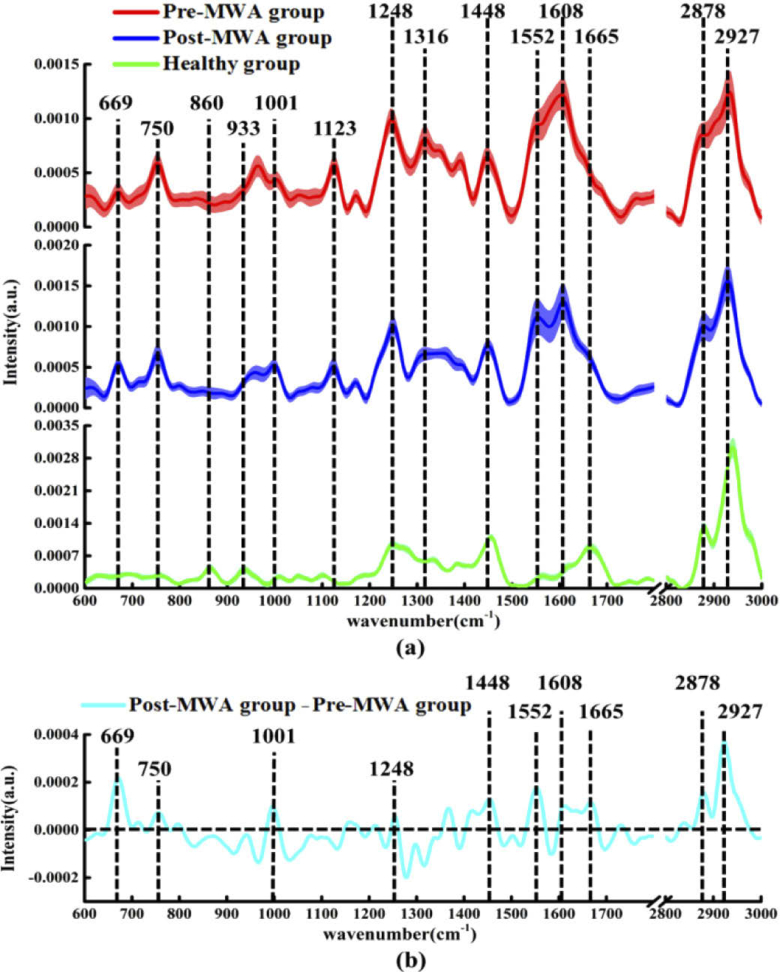
The mean spectra ± standard deviation (SD) acquired from the cancerous tissues in the Pre-MWA group and Post-MWA groups, as well as those obtained from healthy lung tissue are shown in Fig. 1(a) . The main characteristic peaks observed in both the Pre-MWA group and Post-MWA groups at the following locations (with their respective tentative biochemical assignments shown in parentheses) [38,40–47]: 669 cm−1 (T, G of nucleic acids); 750 cm−1 (symmetric breathing of tryptophan); 1002 cm−1 (C-C symmetric ring breathing of phenylalanine); 1123 cm−1 (C-N stretching of proteins); 1248 cm−1 (β-sheet conformation of collagen and proline); 1316 cm−1 (CH2CH3 bending of collagen); 1448 cm−1 (CH2CH3 bending of collagen, CH2 bending of phospholipids); 1552 cm−1 (C = C stretching of tryptophan); 1608 cm−1 (C = C stretching of phenylalanine); 2878 cm−1 (CH2 asymmetric stretch of lipids and proteins); and 2927 cm−1 (CH2 anti-symmetric stretching of lipids). However, the characteristic peaks in healthy lung tissues were observed at the following locations: 860 cm−1 (C = O stretching of phosphatidic acid) [32]; 933 cm−1 (C-C stretching skeletal of collagen backbone, hydroxyproline, proline) [42]; 1002 cm−1, 1248 cm−1, and 1454 cm−1 (overlapping asymmetric CH3 bending and CH2 scissoring of collagen and phospholipids) [48]; 1552 cm−1, and 1665 cm−1 (amide I band of collagen) [42]; and 2878 cm−1 and 2940 cm−1 (C-H vibrations in lipids and proteins) [49]. Furthermore, the normalized intensity of the nucleic acid peak at 669 cm−1, tryptophan at 750 and 1552 cm−1, phenylalanine at 1002 and 1608 cm−1, collagen at 1248 cm−1, and the lipids peaks at 1448, 2878 and 2927 cm−1 in the Post-MWA group were significantly higher than those in the Pre-MWA group.
Fig. 1.
(a) The mean ± standard deviations (SD) of normalized spectra in cancerous tissue of the Pre-MWA group, Post-MWA group and healthy group, shading area represents standard deviations. (b) The differential spectra calculated from the mean Raman spectra between the cancerous tissue of Pre-MWA group and Post-MWA groups.
To more intuitively compare the biochemical variations of cancerous tissues in the Pre-MWA group and Post-MWA groups, the normalized spectrum of the Pre-MWA group was subtracted from the normalized spectrum of the Post-MWA group as shown in Fig. 1(b). The positive peaks appearing in the difference spectrum were found to be basically consistent with the peak positions in Fig. 1(a), including those at 669 cm−1 (nucleic acid), 750, 1002, 1248, 1552, 1608, and 1665 cm−1 (proteins) and 1448, 2878, and 2927 cm−1 (lipids). These positive peaks in the differential spectrum indicated that the contents of nucleic acid, tryptophan, phenylalanine, collagen, and lipid components in the Post-MWA group were higher than those in the Pre-MWA group.
The features of both the characteristic spectra and the differential spectra indicated that MWA treatment induced an increase in the contents of nucleic acid, proteins and lipid components in cancerous tissues. Significant increases in nucleic acid and protein levels in the cancerous tissues in the Post-MWA group may be associated with the destruction of cancer architecture and malignance in the ablation zone [10,26,30]. MWA treatment causes mechanical cell damage, affects the tumor microenvironment, and releases a variety of immunogenic intracellular substrates (such as RNA, DNA, heat shock proteins and high mobility group protein B1) that activate innate immune system [10]. Recent studies have confirmed that circulating histones act as highly tissue-damaging mediators released into the extracellular space following cell death caused by thermal ablation [50]. This suggests that the increased protein levels in the Post-MWA group may also be associated with the biological events such as the release of circulating histones induced by thermal ablation. The presence of large amounts of protein in the Post-MWA group can also be explained by the tumor cell destruction pathways activated by MWA therapy. In particular, the significant increase in the collagen content (1248 cm−1 and 1665 cm−1) in the cancerous tissue in the Post-MWA group as the positive response of MWA therapy. The degradation and redeposition of collagen and structural changes will promote tumor invasion, angiogenesis and migration, and collagen changes will trigger a series of biological events by releasing biochemical signals [51]. Thus, change (increase) in collagen content may a positive indicative of MWA efficacy.
3.2. PCA-LDA analysis
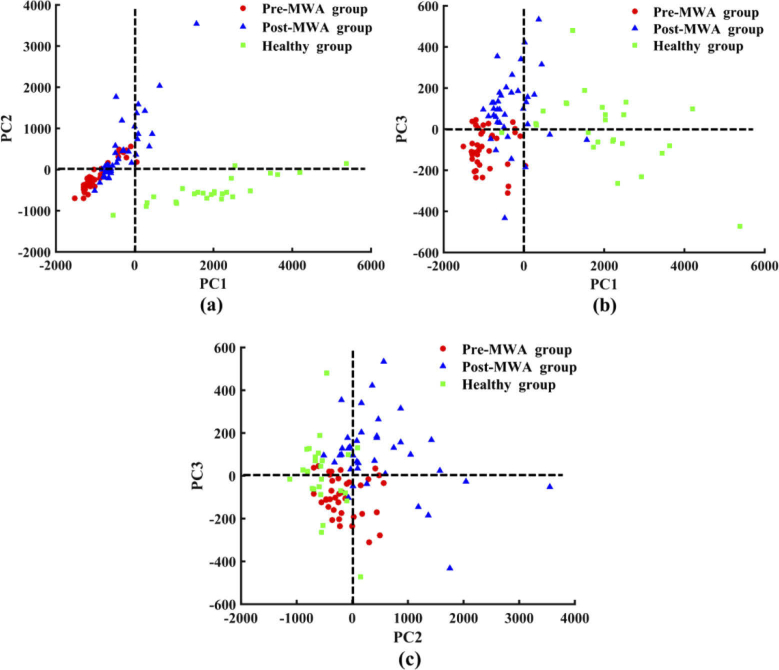
To capture all significant Raman spectral features for MWA treatment evaluation, we conducted a multivariate analysis including all recorded biochemical variations to distinguish tissue types under investigation. The PCA-LDA algorithm was used to determine the most diagnostically significant features in the spectral dataset and further investigation of biochemical variations in different tissues. The Raman spectra of the fingerprint region (600–1800 cm−1) and the high-wavenumber region (2800–3000 cm−1) acquired from the three types of tissue samples were calculated by PCA to obtain the corresponding PC scores and loadings. One-way ANOVA and Fisher’s LSD post-hoc multiple tests showed that PC1, PC2, and PC3 were the most diagnostically significant PCs at the 99% confidence level. These scores were then plotted to visualize the spectral distribution differences between the tissue types. The scatter plot of PC1 and PC2 shown in Fig. 2(a) revealed a clear separation between cancerous tissue and healthy lung tissue. Basically, the cancerous tissues in the Pre-MWA group and Post-MWA groups were present on the negative axis of PC1, whereas the healthy lung tissue was located predominantly on the the positive axis. In the same way, the scatter plots of PC1 and PC3, PC2 and PC3 shown in Fig. 2(b) and (c) were also used to separate different tissue spectra. The vast majority of the spectra on the positive axis of PC2 belonged to cancerous tissue, whereas the spectra of healthy lung tissue and cancerous tissue in the Pre-MWA group were located mainly on the negative axis. It is worth noting that the zero line of PC3 is mainly used to distinguish the cancerous tissue spectrum of the Pre-MWA group and Post-MWA groups. Most of the spectra in the Post-MWA group were located on the positive axis of PC3, while most of the spectra in the Pre-MWA group were distributed on the negative axis.
Fig. 2.
Scatter plots of the diagnostically significantly principal components scores for the three types of tissue, (a) PC1 versus PC2; (b) PC1 versus PC3; and (c) PC2 versus PC3.
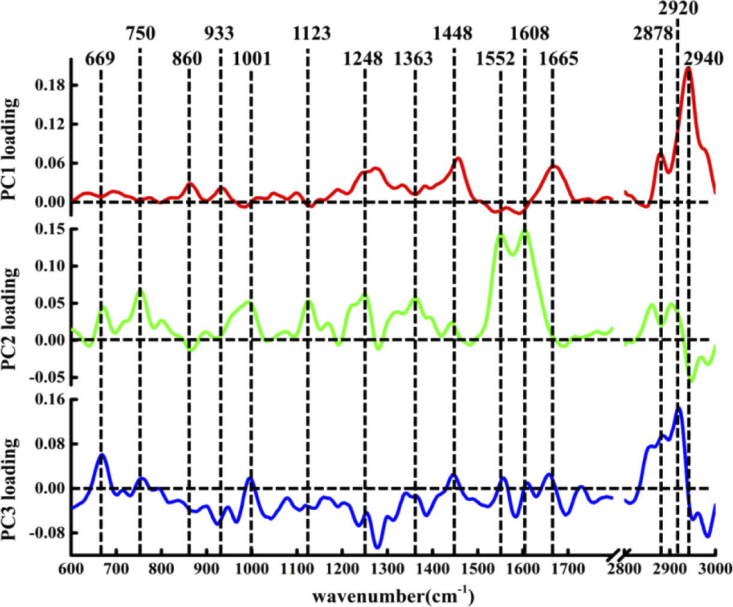
In addition to distinguishing the spectra of different types of tissues, PCA can also be used to extract molecular feature information related to classification based on the loading corresponding to each PC [52]. The loadings of PCA contain both positive and negative features, which are meaningful only for the assignment of the corresponding PCA scores [53]. The loadings of PC1, PC2, and PC3 are shown in Fig. 3 . By comparison with the existing Raman peaks in the single spectrum of the tissue, the negative and positive features in the loading of each PC can be considered to be derived from variations in the biochemical molecules in the tissue. The first most significant PC accounts for the largest variance (76.9%) in the Raman spectral dataset. The loading of PC1 was basically above the zero line, with obvious peak positions in the loadings at 860, 933, 1248, 1454, 1665, 2878, and 2940 cm−1, which can be attributed to the characteristic bands of phosphatidic acid, collagen, and lipid components. The features of the PC1 loadings are extremely similar to the spectral characteristics of healthy lung tissue, suggesting that PC1 can be used primarily to differentiate the healthy group to other two groups. This is consistent with the distribution the spectra of healthy lung tissue shown in the scatter plot in Fig. 2(a) and (b).
Fig. 3.
The PCA loading of PC1, PC2 and PC3.
PC2 explained the total variance of 18.7% of the Raman spectral dataset. The positive peaks of the corresponding loading can be attributed to biochemical components such as nucleic acids at 669 cm−1, tryptophan at 750 and 1552 cm−1, phenylalanine at 1002 and 1608 cm−1, protein at 1123 cm−1, collagen at 1248 cm−1, Guanine (N7, B, Z-marker) at 1363 cm−1 [54], while the negative peaks can be attributed to phosphatidic acid at 860 cm−1 and lipids at 2940 cm−1. Comparison of the loading of PC2 with the characteristic spectrum of cancerous tissue revealed marked similarities between the two groups of spectra in the low-wavenumber region of 600–1800 cm−1. In addition, the two negative characteristic peaks at 860 and 2940 cm−1 in the loading were derived from healthy lung tissue. These observations indicate that the positive features of PC2 extraction are derived mainly from cancerous tissues, whereas the negative features are due mainly to the spectral contribution of healthy lung tissue.
PC3 represented 1.2% of the total variance in the spectral dataset. Although the loadings of PC3 are distributed on both sides of the zero line, the most diagnostically significant features are located in the positive loading, which are 669, 750, 1002, 1448, 1552, 2878, and 2920 cm−1 (C-H stretching bands mostly found in lipids) [55]. These peaks indicate that the positive characteristics of PC3 loading are mainly contributed by nucleic acid, protein, and lipid components. Combining the positive and negative score distribution of PC3 shown in Fig. 2 revealed that the cancerous tissue of Post-MWA group contains more nucleic acid, protein and lipid biochemical components. Moreover, this also shows that PCA loading can represent underlying biochemical differences between different tissues.
To gain a more comprehensive understanding of the information contained in the PCA loading, a simple comparison was applied. The loading of PC3 was compared to the differential spectrum obtained by comparison of the spectra of the cancerous tissue in the Pre-MWA group and Post-MWA groups shown in Fig. 1(b). There was a high degree of similarity between the two groups of spectra, which indicated that the content of nucleic acid, protein and lipid components in cancerous tissues increased after MWA treatment. This further confirms the reliability of PCA in describing the subtle differences in biochemical molecules between the cancerous tissues in the Pre-MWA group and Post-MWA groups.
Therefore, our findings indicate a correlation between PCA loading and the spectral distribution in the score scatter plot. PCA loading contains both positive and negative features, which can be assigned to different biochemical groups through the reference spectrum. The positive characteristics in the loading represent the relatively large contribution of the relevant biochemical components (e.g., nucleic acid at 669 cm−1, proteins at 750, 1001, 1123, 1248, 1552, 1608 cm−1 and lipids at 1448, 1665, 2878, 2920, 2940 cm−1) in the loading of the corresponding PCs, which also could be found in the negative peak of loading. Thus, PCA can be used effectively to analyze the molecular composition and underlying biochemical differences in spectral datasets, and can provide analysis results similar to the difference spectrum.
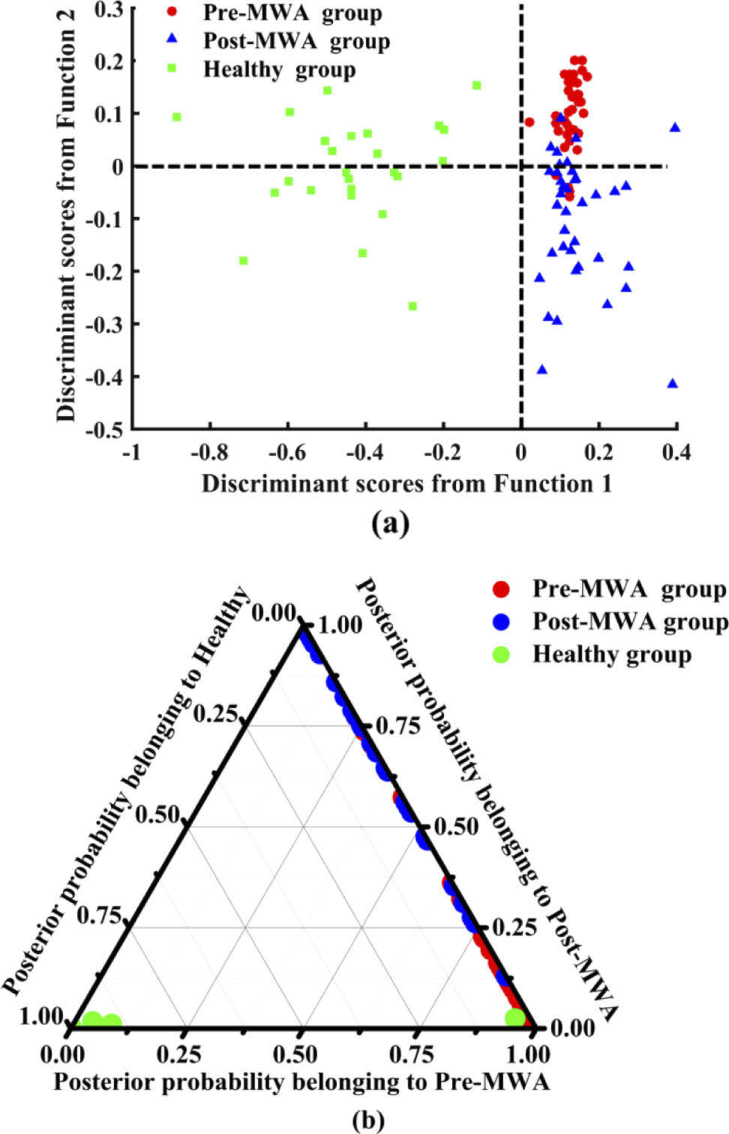
To incorporate the most significant spectral features into the evaluation of lung cancer MWA therapy, several significant PCs identified by one-way ANOVA were used as the input variables for LDA to generate an effective tissue discriminant model. Figure 4 shows the linear discriminant scores and posterior probabilities of different types of tissue obtained by the tissue classification model based on the PCA-LDA algorithm. Figure 4(a) shows that the spectra of healthy lung tissue are distributed in the negative side of the first discriminant function, and all the spectral distributions of cancerous tissue are in the positive side. The zero line of the second discriminant function clearly separates the Post-MWA group from the Pre-MWA group. The spectra of the Post-MWA group tissue are mainly in the negative axis, and the spectral distribution in the positive axis is dominated by the spectra from the Pre-MWA group. The posterior probability values were calculated and are shown as a 2-D ternary scatter plot in Fig. 4(b).
Fig. 4.
(a) represents the scatter plot of linear discriminant scores for three types of tissue; (b) displays two-dimensional ternary plot of the posterior probabilities belonging to the Pre-MWA group, Post-MWA group and healthy lung tissue calculated from the Raman spectral data set, using the PCA-LDA discriminant model combined with LOOCV method.
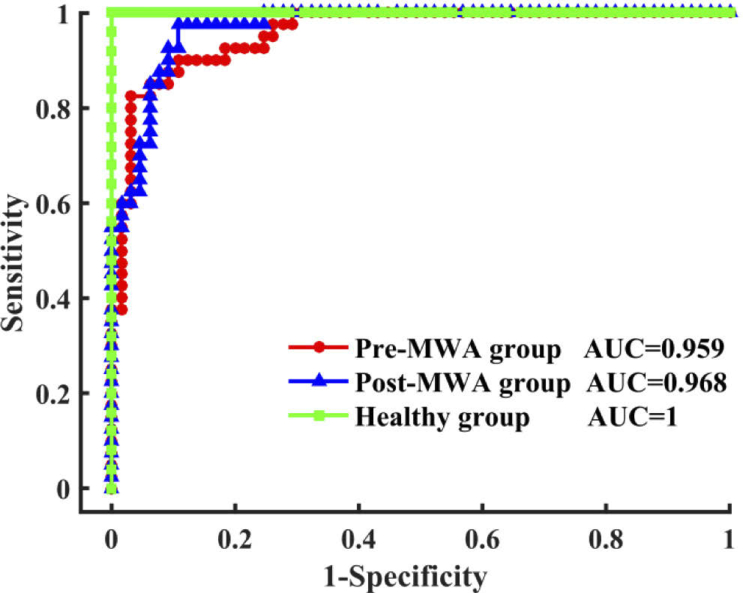
In the LOOCV confusion matrix (Table 1 ), there was a partial misclassified spectrum between the cancerous tissue in the Pre-MWA group and Post-MWA groups that indicated a spectral similarity between the two groups of only 10%–20%. The misclassification of the spectra between the Pre-MWA group and Post-MWA groups may be caused by the reason that the randomly acquired spectra is from the tissue of interest measured areas of the post MWA treated sample that were less affected or treated at all. The differential sensitivity of the discriminant model for the Pre- and Post-MWA groups and healthy lung tissue was 90%, 80%, and 96%, respectively, and the specificity was 86.2%, 93.8%, and 100%, respectively. The overall classification accuracy was 87.6%, indicating that the discriminant model has good performance in separating different types of tissues. Finally, to further evaluate the classification performance of the discriminant model using spectral data of different types of tissues, the ROC curves of the three types of tissues generated by varying the threshold of the posterior probability (Fig. 5 ). The integrated areas under the ROC curves are 0.959, 0.968, and 1 for multiclass discrimination among the cancerous tissue of Pre-MWA group and Post-MWA groups, and healthy lung tissue, respectively. This result further demonstrates that Raman spectroscopy combined with multivariate analysis methods can be used to accurately distinguish different types of tissues.
Table 1. Discrimination results obtained by Raman spectroscopy of different types of tissues using the classification model based on PCA-LDA algorithms with leave-one-out cross validation method. Actual: Pathological diagnosis results. Predict: Raman spectra combined with the classification model obtained the diagnostic results.
| Actual\Predict | Pre-MWA group | Post-MWA group | Healthy group |
|---|---|---|---|
| Pre-MWA group | 36 | 4 | 0 |
| Post-MWA group | 8 | 32 | 0 |
| Healthy group | 1 | 0 | 24 |
Fig. 5.
The ROC curves of discrimination results for the Raman spectral classification of Pre-MWA group, Post-MWA group and healthy lung tissue based on PCA-LDA algorithms together with the LOOCV method. AUC: the areas under the ROC curves.
4. Conclusion
In conclusion, this study indicates that significant Raman biochemical differences could be observed among tissue in healthy, Pre-MWA and Post-MWA groups. Changes in the biochemical composition of tissues could be attributed to cellular mechanical necrosis caused by microwave thermal ablation. Combined with multivariate analysis techniques, the spectral character in the Pre-MWA group and Post-MWA groups were further discriminated by PCA loading and score plots, which highlight some major spectral variations after MWA treatment. We also confirmed that the tissue classification model based on the PCA-LDA algorithm together with LOOCV method could accurately discriminate between healthy lung tissue, and those of the cancerous tissues in the Pre-MWA group and Post-MWA groups for exploring the therapeutic response of tumor tissues accurately, and study related biochemical mechanisms based on specific compositional changes. Thus, the results of our study highlight the feasibility of Raman spectroscopy-based analytical approach for therapeutic monitoring assessments and pathological diagnostic studies.
Funding
International Cooperation and Exchange Programme10.13039/501100010896 (61911530695); Natural Science Foundation of Shaanxi Province10.13039/501100007128 (2018JM6033).
Disclosures
The authors declare that there are no conflicts of interest related to this article.
References
- 1.Chen W., Zheng R., Baade P. D., Zhang S., Zeng H., Bray F., Jemal A., Yu X. Q., He J., “Cancer statistics in China, 2015,” Ca-Cancer J. Clin. 66(2), 115–132 (2016). 10.3322/caac.21338 [DOI] [PubMed] [Google Scholar]
- 2.Zhang L., “SC17.02 Lung Cancer in China: Challenges and Perspectives,” J. Thorac. Oncol. 12(1), S113–S114 (2017). 10.1016/j.jtho.2016.11.100 [DOI] [Google Scholar]
- 3.Zappa C., Mousa S. A., “Non-small cell lung cancer: current treatment and future advances,” Transl. Lung Cancer Res. 5(3), 288–300 (2016). 10.21037/tlcr.2016.06.07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kodama H., Ueshima E., Gao S., Monette S., Paluch L.-R., Howk K., Erinjeri J. P., Solomon S. B., Srimathveeravalli G., “High Power Microwave Ablation of Normal Swine Lung: Impact of Duration of Energy Delivery on Adverse Event and Heat Sink Effects,” Int. J. Hyperthermia 34(8), 1186–1193 (2018). 10.1080/02656736.2018.1447149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ierardi A. M., Coppola A., Lucchina N., Carrafiello G., “Treatment of lung tumours with high-energy microwave ablation: a single-centre experience,” Med. Oncol. (N. Y., NY, U. S.) 34(1), 5 (2017). 10.1007/s12032-016-0861-6 [DOI] [PubMed] [Google Scholar]
- 6.Egashira Y., Singh S., Bandula S., Illing R., “Percutaneous High-Energy Microwave Ablation for the Treatment of Pulmonary Tumors: A Retrospective Single-Center Experience,” J. Vasc. Interv. Radiol. 27(4), 474–479 (2016). 10.1016/j.jvir.2016.01.001 [DOI] [PubMed] [Google Scholar]
- 7.Little M. W., Chung D. C., Boardman P., Gleeson F. V., Anderson E. M., “Microwave Ablation of Pulmonary Malignancies Using a Novel High-energy Antenna System,” CardioVascular and Interventional Radiology 36(2), 460–465 (2013). 10.1007/s00270-012-0465-2 [DOI] [PubMed] [Google Scholar]
- 8.Simon C. J., Dupuy D. E., MayoSmith W. W., “Microwave ablation: principles and applications,” 25 Suppl 1, S69 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Poggi G., Tosoratti N., Montagna B., Picchi C., “Microwave ablation of hepatocellular carcinoma,” World Journal of Hepatology 7(25), 2578–2589 (2015). 10.4254/wjh.v7.i25.2578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chu K. F., Dupuy D. E., “Thermal ablation of tumours: biological mechanisms and advances in therapy,” Nat. Rev. Cancer 14(3), 199–208 (2014). 10.1038/nrc3672 [DOI] [PubMed] [Google Scholar]
- 11.Wright A. S., Sampson L. A., Warner T. F., Mahvi D. M., Lee F. T., “Radiofrequency versus Microwave Ablation in a Hepatic Porcine Model,” Radiology 236(1), 132–139 (2005). 10.1148/radiol.2361031249 [DOI] [PubMed] [Google Scholar]
- 12.Xu J., Li C., Si H., Zhao X., Wang L., Jiang S., Wei D., Yu J., Xiu X., Zhang C., “3D SERS substrate based on Au-Ag bi-metal nanoparticles/MoS2 hybrid with pyramid structure,” Opt. Express 26(17), 21546–21557 (2018). 10.1364/OE.26.021546 [DOI] [PubMed] [Google Scholar]
- 13.Guo Y., Yu J., Li C., Li Z., Pan J., Liu A., Man B., Wu T., Xiu X., Zhang C., “SERS substrate based on the flexible hybrid of polydimethylsiloxane and silver colloid decorated with silver nanoparticles,” Opt. Express 26(17), 21784–21796 (2018). 10.1364/OE.26.021784 [DOI] [PubMed] [Google Scholar]
- 14.Yu J., Guo Y., Wang H., Su S., Zhang C., Man B., Lei F., “Quasi Optical Cavity of Hierarchical ZnO Nanosheets@Ag Nanoravines with Synergy of Near- and Far-Field Effects for in Situ Raman Detection,” J. Phys. Chem. Lett. 10(13), 3676–3680 (2019). 10.1021/acs.jpclett.9b01390 [DOI] [PubMed] [Google Scholar]
- 15.Zhang C., Jiang S., Huo Y., Liu A. H., Xu S., Liu X., Sun Z., Xu Y., Li Z., Man B., “SERS detection of R6G based on a novel graphene oxide/silver nanoparticles/silicon pyramid arrays structure,” Opt. Express 23(19), 24811–24821 (2015). 10.1364/OE.23.024811 [DOI] [PubMed] [Google Scholar]
- 16.Zhang C., Li C., Yu J., Jiang S., Xu S., Yang C., Liu Y. J., Gao X., Liu A., Man B., “SERS activated platform with three-dimensional hot spots and tunable nanometer gap,” Sens. Actuators, B 258, 163–171 (2018). 10.1016/j.snb.2017.11.080 [DOI] [Google Scholar]
- 17.Huang E. Y., Chu S., Chen H. G., Chang W. Y., Kuo Y., Pan C., Chiu A. W., Lin A. T. L., Chiang H. K., “Raman spectral analysis of renal tissue: a novel application,” J. Raman Spectrosc. 45(9), 788–793 (2014). 10.1002/jrs.4546 [DOI] [Google Scholar]
- 18.Feng S., Chen R., Lin J., Pan J., Chen G., Li Y., Cheng M., Huang Z., Chen J., Zeng H., “Nasopharyngeal cancer detection based on blood plasma surface-enhanced Raman spectroscopy and multivariate analysis,” Biosens. Bioelectron. 25(11), 2414–2419 (2010). 10.1016/j.bios.2010.03.033 [DOI] [PubMed] [Google Scholar]
- 19.Yamazaki H., Kaminaka S., Kohda E., Mukai M., Hamaguchi H. O., “The diagnosis of lung cancer using 1064-nm excited near-infrared multichannel Raman spectroscopy,” Radiat. Med. 21, 1–6 (2003). [PubMed] [Google Scholar]
- 20.Oshima Y., Shinzawa H., Takenaka T., Furihata C., Sato H., “Discrimination analysis of human lung cancer cells associated with histological type and malignancy using Raman spectroscopy,” J. Biomed. Opt. 15(1), 017009 (2010). 10.1117/1.3316296 [DOI] [PubMed] [Google Scholar]
- 21.Mcgregor H. C., Short M. A., Mcwilliams A., Shaipanich T., Ionescu D. N., Zhao J., Wang W., Chen G., Lam S., Zeng H., “Real-time endoscopic Raman spectroscopy for in vivo early lung cancer detection,” J. Biophotonics 10(1), 98–110 (2017). 10.1002/jbio.201500204 [DOI] [PubMed] [Google Scholar]
- 22.Magee N. D., Villaumie J. S., Marple E. T., Ennis M., Elborn J. S., Mcgarvey J. J., “Ex vivo diagnosis of lung cancer using a Raman miniprobe,” J. Phys. Chem. B 113(23), 8137–8141 (2009). 10.1021/jp900379w [DOI] [PubMed] [Google Scholar]
- 23.Short M. A., Lam S., McWilliams A. M., Ionescu D. N., Zeng H., “Using Laser Raman Spectroscopy to Reduce False Positives of Autofluorescence Bronchoscopies: A Pilot Study,” J. Thorac. Oncol. 6(7), 1206–1214 (2011). 10.1097/JTO.0b013e3182178ef7 [DOI] [PubMed] [Google Scholar]
- 24.Short M. A., Lam S., Mcwilliams A., Zhao J., Lui H., Zeng H., “Development and preliminary results of an endoscopic Raman probe for potential in vivo diagnosis of lung cancers,” Opt. Lett. 33(7), 711–713 (2008). 10.1364/OL.33.000711 [DOI] [PubMed] [Google Scholar]
- 25.Fox S. A., Shanblatt A. A., Beckman H., Strasswimmer J., Terentis A. C., “Raman spectroscopy differentiates squamous cell carcinoma (SCC) from normal skin following treatment with a high-powered CO2 laser,” Lasers Surg. Med. 46(10), 757–772 (2014). 10.1002/lsm.22288 [DOI] [PubMed] [Google Scholar]
- 26.Bhattacharjee T., Fontana L. C., Raniero L., Ferreirastrixino J., “In vivo Raman spectroscopy of breast tumors prephotodynamic and postphotodynamic therapy,” J. Raman Spectrosc. 49(5), 786–791 (2018). 10.1002/jrs.5360 [DOI] [Google Scholar]
- 27.Matthews Q., Jirasek A., Lum J., Brolo A. G., “Biochemical signatures of in vitro radiation response in human lung, breast and prostate tumour cells observed with Raman spectroscopy,” Phys. Med. Biol. 56(21), 6839–6855 (2011). 10.1088/0031-9155/56/21/006 [DOI] [PubMed] [Google Scholar]
- 28.Vidyasagar M. S., Maheedhar K., Vadhiraja B. M., Fernendes D. J., Kartha V. B., Krishna C. M., “Prediction of radiotherapy response in cervix cancer by Raman spectroscopy: A pilot study,” Biopolymers 89(6), 530–537 (2008). 10.1002/bip.20923 [DOI] [PubMed] [Google Scholar]
- 29.Lakshmi R. J., Kartha V. B., Krishna C. M., Solomon J. G. R., Ullas G., Devi P. U., “Tissue Raman Spectroscopy for the Study of Radiation Damage: Brain Irradiation of Mice,” Radiat. Res. 157(2), 175–182 (2002). 10.1667/0033-7587(2002)157[0175:TRSFTS]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- 30.Song D., Chen T., Wang S., Chen S., Li H., Yu F., Zhang J., Zhang Z., “Study on the biochemical mechanisms of the micro-wave ablation treatment of lung cancer by ex vivo confocal Raman microspectral imaging,” Analyst (2020). [DOI] [PubMed] [Google Scholar]
- 31.Li J., Liang Z., Wang S., Wang Z., Zhang X., Hu X., Wang K., He Q., Bai J., “Study on the pathological and biomedical characteristics of spinal cord injury by confocal Raman microspectral imaging,” Spectrochim. Acta, Part A 210, 148–158 (2019). 10.1016/j.saa.2018.11.022 [DOI] [PubMed] [Google Scholar]
- 32.Gong Y., Wang S., Liang Z., Wang Z., Zhang X., Li J., Song J., Hu X., Wang K., He Q., “Label-Free Spectral Imaging Unveils Biochemical Mechanisms of Low-Level Laser Therapy on Spinal Cord Injury,” Cell. Physiol. Biochem. 49(3), 1127–1142 (2018). 10.1159/000493295 [DOI] [PubMed] [Google Scholar]
- 33.Wang S., Liang Z., Gong Y., Yin Y., Wang K., He Q., Wang Z., Bai J., “Confocal raman microspectral imaging of ex vivo human spinal cord tissue,” J. Photochem. Photobiol., B 163, 177–184 (2016). 10.1016/j.jphotobiol.2016.08.032 [DOI] [PubMed] [Google Scholar]
- 34.Lloyd G. R., Orr L. E., Christie-Brown J., McCarthy K., Rose S., Thomas M., Stone N., “Discrimination between benign, primary and secondary malignancies in lymph nodes from the head and neck utilising Raman spectroscopy and multivariate analysis,” Analyst 138(14), 3900–3908 (2013). 10.1039/c2an36579k [DOI] [PubMed] [Google Scholar]
- 35.Teh S. K., Zheng W., Ho K. Y., Teh M., Yeoh K. G., Huang Z., “Near-infrared Raman spectroscopy for early diagnosis and typing of adenocarcinoma in the stomach,” Br. J. Surg. 97(4), 550–557 (2010). 10.1002/bjs.6913 [DOI] [PubMed] [Google Scholar]
- 36.Pichardomolina J. L., Fraustoreyes C., Barbosagarcia O., Huertafranco R., Gonzaleztrujillo J. L., Ramirezalvarado C. A., Gutierrezjuarez G., Medinagutierrez C., “Raman spectroscopy and multivariate analysis of serum samples from breast cancer patients,” Lasers Med. Sci. 22(4), 229–236 (2007). 10.1007/s10103-006-0432-8 [DOI] [PubMed] [Google Scholar]
- 37.Molckovsky A., Song L. M. W. K., Shim M. G., Marcon N. E., Wilson B. C., “Diagnostic potential of near-infrared Raman spectroscopy in the colon: Differentiating adenomatous from hyperplastic polyps,” Gastrointestinal Endoscopy 57(3), 396–402 (2003). 10.1067/mge.2003.105 [DOI] [PubMed] [Google Scholar]
- 38.Huang W., Wu S., Chen M., Sun L., Li Y., Huang M., Huang S., Xu Z., Chen R., Zeng H., “Study of both fingerprint and high wavenumber Raman spectroscopy of pathological nasopharyngeal tissues,” J. Raman Spectrosc. 46(6), 537–544 (2015). 10.1002/jrs.4684 [DOI] [Google Scholar]
- 39.Li Y., Pan J., Chen G., Li C., Lin S., Shao Y., Feng S., Huang Z., Xie S., Zeng H., “Micro-Raman spectroscopy study of cancerous and normal nasopharyngeal tissues,” J. Biomed. Opt. 18(2), 027003 (2013). 10.1117/1.JBO.18.2.027003 [DOI] [PubMed] [Google Scholar]
- 40.Notingher I., Verrier S., Haque S., Polak J. M., Hench L. L., “Spectroscopic study of human lung epithelial cells (A549) in culture: Living cells versus dead cells,” Biopolymers 72(4), 230–240 (2003). 10.1002/bip.10378 [DOI] [PubMed] [Google Scholar]
- 41.Huang Z., Mcwilliams A. H., Mclean D. I., Lam S., Zeng H., “Near-infrared Raman spectroscopy for optical diagnosis of lung cancer,” Int. J. Cancer 41, S50 (2003). 10.1016/S0169-5002(03)91823-X [DOI] [PubMed] [Google Scholar]
- 42.Cheng W., Liu M., Liu H., Lin S., “Micro-Raman spectroscopy used to identify and grade human skin pilomatrixoma,” Microsc. Res. Tech. 68(2), 75–79 (2005). 10.1002/jemt.20229 [DOI] [PubMed] [Google Scholar]
- 43.Kamemoto L. E., Misra A. K., Sharma S. K., Goodman M. T., Luk H., Dykes A. C., Acosta T., “Near-Infrared Micro-Raman Spectroscopy for in Vitro Detection of Cervical Cancer,” Appl. Spectrosc. 64(3), 255–261 (2010). 10.1366/000370210790918364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stone N., Kendall C., Smith J., Crow P., Barr H., “Raman spectroscopy for identification of epithelial cancers,” Faraday Discuss. 126, 141–157 (2004). 10.1039/b304992b [DOI] [PubMed] [Google Scholar]
- 45.García-Flores A. F., Raniero L., Canevari R. A., Jalkanen K. J., Bitar R. A., Martinho H. S., Martin A. A., “High-wavenumber FT-Raman spectroscopy for in vivo and ex vivo measurements of breast cancer,” Theor. Chem. Acc. 130(4-6), 1231–1238 (2011). 10.1007/s00214-011-0925-9 [DOI] [Google Scholar]
- 46.Abramczyk H., Brozekpluska B., Surmacki J., Jablonska J., Kordek R., “The label-free Raman imaging of human breast cancer,” J. Mol. Liq. 164(1-2), 123–131 (2011). 10.1016/j.molliq.2011.04.021 [DOI] [Google Scholar]
- 47.Dudala J., Bialas M., Surowka A. D., Berezabuziak M., Hubalewskadydejczyk A., Budzynski A., Pedziwiatr M., Kolodziej M., Wehbe K., Lankosz M., “Biomolecular characterization of adrenal gland tumors by means of SR-FTIR,” Analyst 140(7), 2101–2106 (2015). 10.1039/C4AN01891E [DOI] [PubMed] [Google Scholar]
- 48.Lau D. P., Huang Z., Lui H., Man C. S., Berean K., Morrison M., Zeng H., “Raman spectroscopy for optical diagnosis in normal and cancerous tissue of the nasopharynx-preliminary findings,” Lasers Surg. Med. 32(3), 210–214 (2003). 10.1002/lsm.10084 [DOI] [PubMed] [Google Scholar]
- 49.Sigurdsson S., Philipsen P. A., Hansen L. K., Larsen J., Gniadecka M., Wulf H. C., “Detection of skin cancer by classification of Raman spectra,” IEEE Trans. Biomed. Eng. 51(10), 1784–1793 (2004). 10.1109/TBME.2004.831538 [DOI] [PubMed] [Google Scholar]
- 50.Gu T., Wen T., Zhang Y., Zhang D., Hua H., Liu L., Zhang Y., Fu Z., Yuan Z., “Impact of Elevated Circulating Histones on Systemic Inflammation after Radiofrequency Ablation in Lung Cancer Patients,” BioMed Res. Int. 2017, 1–6 (2017). 10.1155/2017/6894832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fang M., Yuan J., Peng C., Li Y., “Collagen as a double-edged sword in tumor progression,” Tumor Biol. 35(4), 2871–2882 (2014). 10.1007/s13277-013-1511-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bonnier F., Byrne H. J., “Understanding the molecular information contained in principal component analysis of vibrational spectra of biological systems,” Analyst 137(2), 322–332 (2012). 10.1039/C1AN15821J [DOI] [PubMed] [Google Scholar]
- 53.Matthews Q., Jirasek A., Lum J., Duan X., Brolo A. G., “Variability in Raman Spectra of Single Human Tumor Cells Cultured in Vitro : Correlation with Cell Cycle and Culture Confluency,” Appl. Spectrosc. 64(8), 871–887 (2010). 10.1366/000370210792080966 [DOI] [PubMed] [Google Scholar]
- 54.Movasaghi Z., Rehman S., Rehman I. U., “Raman Spectroscopy of Biological Tissues,” Appl. Spectrosc. Rev. 42(5), 493–541 (2007). 10.1080/05704920701551530 [DOI] [Google Scholar]
- 55.Mignolet A., Wood B. R., Goormaghtigh E., “Intracellular investigation on the differential effects of 4 polyphenols on MCF-7 breast cancer cells by Raman imaging,” Analyst 143(1), 258–269 (2018). 10.1039/C7AN01460K [DOI] [PubMed] [Google Scholar]