Abstract
Humans and African great apes (AGAs) are naturally infected with several species of closely related malaria parasites. The need to understand the origins of human malaria as well as the risk of zoonotic transmissions and emergence of new malaria strains in human populations has markedly encouraged research on great ape Plasmodium parasites. Progress in the use of non-invasive methods has rendered investigations into wild ape populations possible. Present knowledge is mainly focused on parasite diversity and phylogeny, with still large gaps to fill on malaria parasite ecology. Understanding what malaria infection means in terms of great ape health is also an important, but challenging avenue of research and has been subject to relatively few research efforts so far. This paper reviews current knowledge on African great ape malaria and identifies gaps and future research perspectives.
Introduction
African great apes (AGAs) have been known to naturally host malaria parasites since the early 1920s, when researchers first described various Plasmodium species infecting chimpanzees (Pan troglodytes) and gorillas (Gorilla spp.), which are closely related to those infecting humans (Reichenow, 1920). To this day, it remains difficult to date the actual origins of the malaria parasites which infect humans (Sharp et al., 2013) as well as their emergence in human populations. However, the expansion of P. falciparum for instance, the most virulent Plasmodium parasite (White, 2003), is believed to have started in Africa at least thousands or even tens of thousands of years ago (Carter and Mendis, 2002; Rich et al., 2009; Tanabe et al., 2010; Volkman et al., 2001) and to have played a significant role in recent human evolution (Carter and Mendis, 2002). Malaria is currently geographically widespread and constitutes a heavy burden for human public health (WHO, 2015). Malaria in great apes, our closest evolutionary relatives, has thus increasingly been subject to research as this leads to a better understanding of human malaria and the significance and implications of malaria parasite infection for great ape population health.
Research on AGA malaria parasites was initially limited to the study of captive and laboratory individuals (Bray, 1960; Contacos et al., 1970; Garnham et al., 1956; Rodhain and Dellaert, 1943; Rodhain, 1936, 1940; Schwetz, 1934). However, the development of molecular diagnostic tools and, later on, the ability to use non-invasive samples, i.e. the discovery that malaria parasite genetic material can be found in faecal samples of primates (Prugnolle et al., 2010), opened up opportunities for further research in wild ape populations. This allowed for more accurate identifications, the discovery of additional Plasmodium species in chimpanzees and gorillas, and the study of their phylogenetic relationships (Kaiser et al., 2010; Liu et al., 2010, 2014). Most of the current knowledge concerns malaria parasite diversity, host specificity and evolutionary history. There is still relatively little understanding of the epidemiology, ecology and biology of great ape malaria, but with increasing efforts being made in recent years to investigate these aspects. Here, we review up-to-date knowledge about the natural history of malaria parasite infection in wild AGAs, highlight the gaps in the knowledge and conclude with some perspectives for further investigation.
Parasite diversity, distribution and prevalence in African great apes
Malaria parasites naturally infecting AGAs were initially described in the 1920s by Reichenow (Reichenow, 1920) and then studied further and experimented on by various researchers such as Blacklock and Adler (Adler, 1923; Blacklock and Adler, 1922), Schwetz (Schwetz, 1934), Rodhain (Rodhain and Dellaert, 1943; Rodhain, 1936, 1940), Brumpt (Brumpt, 1939), Garnham (Garnham et al., 1956), Bray (Bray, 1958a, 1960) and Contacos (Contacos et al., 1970). Based on morphological features, three distinct Plasmodium species, considered similar to the human parasites, were identified in West and Central Africa: P. falciparum-like and P. ovale- or vivax-like parasites in chimpanzees and gorillas and P. malariae-like parasites in chimpanzees (Reichenow, 1920). These three species were subsequently named P. reichenowi (P. falciparum-like parasites) (Blacklock and Adler, 1922), P. schwetzi (P. ovale or vivax-like parasites) (Brumpt, 1939) and P. rodhaini (P. malariae-like parasites) (Brumpt, 1939). P. falciparum and P. reichenowi, considered relatively distinct from other known Plasmodium species, were grouped separately under the Laverania subgenus (Bray, 1958b).
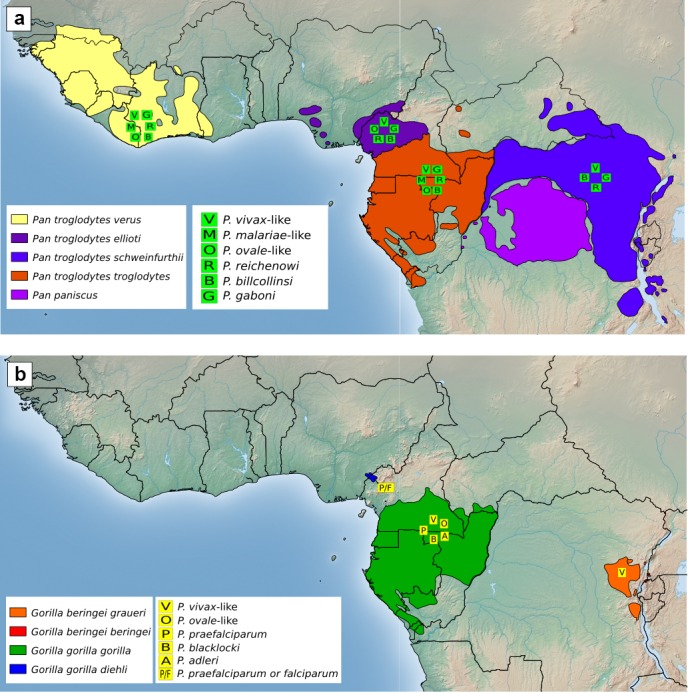
Distribution of Plasmodium species detected in wild chimpanzees and gorillas. Plasmodium species symbols placed on the geographical range of chimpanzee subspecies (a) and gorilla species and subspecies (b) indicate that the assigned Plasmodium species was, to this date, detected in at least one sample of the relevant great ape species and subspecies. Positions of the Plasmodium symbols do not represent specific field site locations.
From 2009 onwards, with the increased use of molecular diagnostic tools (deoxyribonucleic acid, DNA, amplification and genome sequencing) and, later on, the development of non-invasive methods to detect malaria infection through the amplification of parasite DNA from faecal samples, large-scale studies on malaria parasites infecting wild great apes flourished. These studies led to the description of a higher diversity of parasites than previously thought and gave clearer insight into prevalence rates and the possible origins and evolutionary history of malaria infection in humans. To date, at least six Plasmodium species have been shown to be endemic in wild-living chimpanzees: three Laverania species – P. reichenowi (Boundenga et al., 2015; Kaiser et al., 2010; Liu et al., 2010, 2016; Prugnolle et al., 2010; Rich et al., 2009) and two newly discovered Laverania species, P. gaboni (Boundenga et al., 2015; Kaiser et al., 2010; Liu et al., 2010, 2016; Prugnolle et al., 2010; Rich et al., 2009) and P. billcollinsi (Boundenga et al., 2015; Krief et al., 2010; Liu et al., 2010, 2016); and three non-Laverania species – P. ovale- (Kaiser et al., 2010; Liu et al., 2010), vivax- (Kaiser et al., 2010; Liu et al., 2010, 2014) and malariae-like (Kaiser et al., 2010; Liu et al., 2010) parasites (putative nomenclature was proposed by Rayner et al., 2011, and used in the present review). All four chimpanzee subspecies are naturally infected with all known chimpanzee Laverania parasites and with at least one of the non-Laverania parasites (Fig. 1a). Western lowland gorillas (Gorilla gorilla gorilla) also naturally harbour three parasite species belonging to the Laverania subgenus, i.e. P. praefalciparum, P. adleri and P. blacklocki (Liu et al., 2010, 2016; Prugnolle et al., 2010), and two non-Laverania species, i.e. P. vivax- (Liu et al., 2014) and ovale-like parasites (Mapua et al., 2015) (Fig. 1b). A P. praefalciparum, or falciparum-like, sequence (an undistinguishable sequence) was also detected in Cross River gorillas (Gorilla gorilla diehli) (Prugnolle et al., 2010), and P. vivax-like parasites were detected in eastern lowland gorillas (G. b. graueri) (Liu et al., 2014). Despite significant search efforts (861 samples tested by Liu et al., 2010, 2014), natural infections of wild-living bonobos (Pan paniscus) have not been reported so far (Fig. 1).
Before large-scale screenings on wild great ape populations were carried out, the natural prevalence of malaria parasites in some chimpanzee and gorilla populations was suspected to be high given the frequent detection in sporadic blood or tissue samples (Kaiser et al., 2010; Rich et al., 2009) as well as in faecal samples (Krief et al., 2010; Prugnolle et al., 2010) from wild-living chimpanzees (all four subspecies) and western gorillas. This was confirmed when prevalence levels of Plasmodium spp. infection in wild apes were estimated using detection rates from larger sets of faecal samples. Parasites were clearly found to be endemic in the four chimpanzee subspecies and in lowland gorillas (G. g. gorilla and G. b. graueri), with, according to different studies, estimated prevalence levels ranging from 32 % to 48 % for P. t. ellioti, P. t. troglodytes, P. t. schweinfurthii and G. g. gorilla, in which faecal detection rates were corrected for the sensitivity of the diagnostic test, sample degradation and redundant sampling (Liu et al., 2010); 15.42 % for P. t. troglodytes and 21.28 % for G. g. gorilla faecal detection rates (Boundenga et al., 2015); and 33 % for P. t. verus, in which the point prevalence rate was based on the proportion of positive individuals tested by faecal detection (De Nys et al., 2013, 2014). The vast majority of parasites harboured by AGAs belong to the Laverania clade with non-Laverania strains rarely found (Boundenga et al., 2015; Kaiser et al., 2010; Liu et al., 2010; De Nys et al., 2013, 2014). The specific prevalence rates of P. vivax-like parasites in chimpanzees (P. t. troglodytes and P. t. schweinfurthii) and lowland gorillas (G. g. gorilla and G. b. graueri) seem to range from 4 to 8 %, close to the infection rates in endemic human populations (Liu et al., 2014). Co-infection with two or more Plasmodium parasite species is prevalent (Krief et al., 2010; Liu et al., 2010), e.g. present in 55 % of positive chimpanzee samples tested by Liu et al., (2010). Bear in mind that these prevalence rates are likely to represent underestimates due to the lower sensitivity of faecal testing compared to blood testing (Liu et al., 2010, 2014). In some chimpanzee and gorilla communities, no malaria parasites were detected so far (Boundenga et al., 2015; Liu et al., 2010, 2014), which could be the result of the relatively low sensitivity of faecal detection combined with the small sample size (Liu et al., 2014). Additionally, low prevalence or absence of malaria parasites in certain communities could also be explained by environmental factors – for example, factors that lead to a scarcity of efficient vectors or demographic factors such as variations in host density.
Until the discovery of other Laverania parasites, P. reichenowi was known as the closest relative of P. falciparum (Escalante and Ayala, 1994; Escalante et al., 1995). Consequently, it was the first great ape Plasmodium species to be molecularly characterised. A number of phylogenetic analyses have led to various hypotheses on the origin of the human P. falciparum: (1) one is the co-speciation hypothesis, in which the two Plasmodium species diverged from a common ancestor at the same time as the chimpanzee and human lineages diverged, about 6 to 10 million years ago (Escalante and Ayala, 1994; Escalante et al., 1995; Hayakawa et al., 2008; Hughes and Verra, 2010); and (2) another is the hypothesis of a more recent chimpanzee origin through a single host transfer of P. reichenowi from chimpanzee to human. The latter theory was supported by the low genetic polymorphism of P. falciparum which was fully included into the broader genetic diversity of malaria parasite lineages, which the authors collectively considered as forming the species P. reichenowi (Prugnolle et al., 2010; Rich et al., 2009). However, recent broad-scale investigations based on the detection of malaria parasite DNA (mitochondrial, nuclear and apicoplast sequences) in faecal samples of wild AGA populations resulted in the genetic characterisation of a wide array of other Plasmodium species. This showed that the entire diversity of human-derived P. falciparum sequences is encompassed by the gorilla P. praefalciparum clade (Liu et al., 2010; Sundararaman et al., 2013) and led to the substantial indication that P. falciparum originated from a relatively recent zoonotic transmission of P. praefalciparum from gorillas to humans (Liu et al., 2010).
As far as non-Laverania parasites are concerned, many questions remain on their evolutionary origins and the phylogenetic relationships between the parasites hosted by non-human primates (NHPs) and those hosted by humans. Phylogenetic analyses initially suggested that P. vivax originated in Asia from an ancestral macaque Plasmodium species (Escalante et al., 2005; Mu et al., 2005). P. simium, a New World monkey parasite, has also been designated as a potential ancestor of P. vivax given their high genetic similarity, which is suggestive of a recent host transfer between New World monkeys and humans, although the direction of this cross-species transmission remains uncertain (Escalante et al., 2005; Tazi and Ayala, 2011). Finally, the near fixation of the Duffy-negative phenotype in Sub-Saharan human populations, which confers resistance against P. vivax, supports the hypothesis of an African origin of this parasite with positive selection of the Duffy negativity through long-term exposure (Carter, 2003). The P. vivax strains found in captive AGAs are genetically distinct from those circulating in humans (Prugnolle et al., 2013). They include the monophyletic lineage formed by the human P. vivax parasites in their diversity, but they lack host specificity amongst the AGAs they infect, i.e. P. t. troglodytes, P. t. ellioti, P. t. schweinfurthii, G. g. gorilla and G. b. graueri (Liu et al., 2014). This supports the idea that the human P. vivax developed from within a Plasmodium species which also infected African apes and that all present human P. vivax parasites originated from an ancestor which spread out of Africa (Liu et al., 2014). For P. malariae- and ovale-like parasites, larger-scale investigations of infections in wild NHPs together with phylogenetic analyses are still needed to clarify the evolutionary relationship of these parasites with human strains.
Transmission of AGA malaria parasites
Vectors
It is well established that mosquitoes are responsible for the transmission of malaria between humans. These malaria vectors belong to the Anopheles genus (An.), of which approximately 40 species (out of 430) are thought to be competent for malaria parasite transmission (CDC, 2012). They have been studied extensively and mapped in detail (Sinka et al., 2010). The ecology of mosquito species, including habitat and host preferences, varies and determines their importance as a vector in given habitats (Sinka et al., 2010). For instance, An. gambiae is considered as one of the most efficient vectors in open landscapes in Sub-Saharan Africa (Coetzee, 2004; Sinka et al., 2010). Additionally, An. moucheti, also highly anthropophilic, is entirely restricted to forested habitats and considered a significant vector of human malaria in this type of environment (Sinka et al., 2010).
Until very recently, not a single vector responsible for AGA malaria parasite transmission had been identified. Early experiments had shown the susceptibility of some known human malaria vector species to P. reichenowi (Blacklock and Adler, 1922; Collins et al., 1986), with a notable exception being An. gambiae. Bray (1958a) had managed to infect An. gambiae with the formerly called P. schwetzi; however, the low levels of sporozoites in the salivary glands had suggested that these were not the natural vectors. Another experiment succeeded in infecting humans with P. schwetzi via Anopheles balabacensis bites (Contacos et al., 1970).
It is mainly since the discovery of the gorilla origin of P. falciparum that the question of AGA malaria vectors has been increasingly raised due to the concern for future zoonotic transfers of malaria parasites from NHPs to humans and thus the importance of identifying species that can act as bridges (Verhulst et al., 2012). At the present time, several studies have thus specifically searched for mosquito species responsible for the transmission of malaria parasites between chimpanzees or gorillas in their natural habitat. The first study tested female Anopheles () captured at chimpanzee (P. t. schweinfurthii) natural nesting and feeding sites in Uganda, but it did not detect any Plasmodium parasites (Krief et al., 2012). The failure of detection might, however, have been linked to the small number of specimens tested. A second study, which screened a larger number of female Anopheles () captured in areas hosting AGA populations in Gabon, found DNA from P. praefalciparum in one An. moucheti and from P. vivax-like parasites in one An. moucheti and one An. vinckei (Paupy et al., 2013). Finally, the analysis of 2415 female Anopheles captured in the same areas in Gabon led to the discovery of both chimpanzee and gorilla Laverania and non-Laverania parasites in An. vinckei, An. moucheti and An. marshallii (Makanga et al., 2016), with the highest prevalence rate in An. vinckei (). The detection of sporozoites in their salivary glands confirmed their role in natural transmission. These mosquito species appeared to be non-host-specific (they were shown to feed on humans and to carry malaria parasites from rodents, bats and antelopes) and were thus unlikely to be responsible for the host specificity of Laverania parasites.
It is important to note that the prevalence rates of different mosquito species might differ between geographical sites, suggesting a variability of vectors depending on the investigated sites. For example, mosquito trappings performed in and around the forest of the Taï National Park (TNP) in Côte d'Ivoire to study the distribution of mosquito genera in habitats varying from human settlements to a primary forest only identified one single Anopheles in the primary forest (at a camp site), whereas they were abundant in the neighbouring villages (Junglen et al., 2009). Mosquitoes of the Uranotaenia genus, however, were dominant in the primary forest but rarely found in other areas. This suggests that the malaria vectors responsible for the transmission of malaria parasites in the resident chimpanzee population differ from those transmitting parasites in humans and in other AGA populations. Obviously, other mosquito genera and other dipterans should not be excluded as possible vectors. Biting midges (genus Culicoides), for example, transmit Hepatocystis spp., which are bat and monkey malaria parasites (Blanquart and Gascuel, 2011), and are known to be abundant in the TNP (personal observation).
Interspecies transmission
Since the discovery of AGA malaria parasites, the marked similarities they share with those infecting humans have raised the question of whether the exchange of these parasites between AGAs and humans could occur, experimentally and/or naturally.
Early experiments involving captive chimpanzees and humans (volunteers and/or neurological patients for former therapeutic purposes) demonstrated that, for some strains, the infection of humans with chimpanzee parasites, or vice versa, is possible. This was the case for the chimpanzee-borne P. rodhaini (probably a P. malariae-like strain) (Bray, 1963; Rodhain and Dellaert, 1943; Rodhain, 1940), the human-borne P. malariae (Bray, 1960, 1963; Garnham et al., 1956), the chimpanzee-borne P. schwetzi (probably P. ovale- or vivax-like strains) (Bray, 1963; Coatney, 1968; Contacos et al., 1970; Rodhain and Dellaert, 1955) and the human-borne P. vivax (Bray, 1963; Garnham et al., 1956; Rodhain, 1939). In contrast, attempts to transfer the Laverania species P. reichenowi from chimpanzees to humans failed (Blacklock and Adler, 1922; Rodhain, 1939). The marked host specificity of P. reichenowi might be due to the human loss of the common primate Sia (sialic acid) N-glycolylneuraminic acid, which is the preferred target of P. reichenowi for erythrocyte binding, as opposed to P. falciparum, which prefers the precursor of the N-glycolylneuraminic acid, the Sia N-acetylneuraminic acid (Martin et al., 2005). In addition, the transfer of P. falciparum to chimpanzees only seemed possible in splenectomised individuals, or limited to the liver or to a very low parasitaemia in intact individuals (Bray, 1963; Taylor et al., 1985).
For certain human malaria parasites, natural (non-experimental) transmission to AGAs seems to occur. De facto, early experimental attempts at infecting chimpanzees with P. falciparum gave poor results, but captive bonobos (Krief et al., 2010) and chimpanzees (Duval et al., 2010; Ngoubangoye et al., 2016) living in areas of high human P. falciparum endemicity have been found to be infected with these parasites. These infections were probably acquired from humans (Rayner et al., 2011). There is, however, no information on the viability of such infections and thus the potential for vector-based transmission between apes. Similarly, P. malariae, which has never been detected in wild bonobos so far, was found in captive individuals (Krief et al., 2010), and a P. ovale strain, identical to the human P. ovale variant type, was found in a captive chimpanzee (Duval et al., 2009).
As for natural zoonotic transmission of AGA parasites to humans, it appears from the phylogenetic relationships between known malaria parasites that, historically, ape Plasmodium (P. praefalciparum) have managed to naturally cross the species barrier at least once (Liu et al., 2010). This, together with some of the old experimental results reported above, instinctively leads to the assumption that natural zoonotic transfers of AGA malaria parasites might still occur, leading to new host switches or just to regular transmission events in the same manner that a P. knowlesi infection in humans originates from regular zoonotic transfer events from Southeast Asian macaques (Lee et al., 2011; Sharp et al., 2013). Interestingly, the AGA malaria parasite vectors found by Makanga et al. (2016) are mosquito species that also seem to feed on other mammalian species, including humans. They could thus, in theory, act as vectors for interspecies transmissions. Studies led in Cameroon (Sundararaman et al., 2013) and Gabon (Délicat-Loembet et al., 2015) searched for evidence of zoonotic transfers of ape Laverania species by testing large numbers of human blood samples ( and ) from people living in the vicinity of chimpanzee and gorilla populations with high Laverania prevalence rates. No ape parasites (Laverania or non-Laverania) were detected in these samples, implying that zoonotic transfers might be rare events.
There is, however, current evidence that cross-species transmission from wild apes to humans of the non-Laverania species P. vivax is ongoing. Since wild chimpanzees in West and Central Africa were confirmed hosts of P. vivax-like strains, it was hypothesised that wild apes could act as a natural reservoir (Rayner et al., 2011), thereby explaining the occasional infection of travellers despite the low prevalence of P. vivax in the human populations of these regions (Broderick et al., 2015; Culleton et al., 2008; Gautret et al., 2001; Mühlberger et al., 2004; Skarbinski et al., 2006). Moreover, a P. vivax-like parasite isolated from a traveller coming back from a Central African forest was recently shown to cluster with the AGA isolates outside of the human lineage, confirming the occurrence of cross-species transmissions (Liu et al., 2014; Prugnolle et al., 2013). This is supported by the fact that P. vivax-like parasite DNA was found in an anthropophilic mosquito species, Anopheles moucheti (Paupy et al., 2013). Given the close resemblance to their human counterparts and past experimental findings, it is likely that, similarly to P. vivax, zoonotic transfers of P. ovale- and malariae-like parasites from chimpanzees also take place. It would be useful to characterise more of these parasites in order to search for evidence of such transmissions (Sundararaman et al., 2013).
Transmission of host-specific parasites between different AGA genera has only, so far, been observed in captivity, where great ape species cohabit closely, e.g. transmission of Laverania parasites from chimpanzees to gorillas and vice versa (Ngoubangoye et al., 2016). Cross-species transmission between AGAs and other NHPs sharing their habitat is probably exceptional. So far, the only published finding showing such potential is that of P. praefalciparum detected in a single captive greater spot-nosed monkey, Cercopithecus nictitans (Prugnolle et al., 2011). Yet, during a subsequent study which tested blood from a significant number of wild spot-nosed monkeys (), only Hepatocystis spp. and one Plasmodium sp. previously described in birds and lizards were found (Ayouba et al., 2012). P. vivax has also specifically been searched for in blood samples () from 16 Old World monkey species; however, with the exception of Hepatocystis spp., no malaria parasites were found (Liu et al., 2014). Similar results were obtained after screening specimens from six monkey species from Uganda () (Thurber et al., 2013) and from three monkey species in the TNP, Côte d'Ivoire () (S. Calvignac-Spencer, personal communication, 2015). Hepatocystis represents one of the numerous genera of malaria parasites, just like the genus Plasmodium, which seems to be paraphyletic to Hepatocystis (Martinsen et al., 2008). Hepatocystis parasites comply with a broad pattern of host-specificity, with distinct lineages that specialise on monkeys and bats (Ayouba et al., 2012; Schaer et al., 2013). So far, there is no published evidence of transmission of NHP malaria parasites such as Hepatocystis spp. to AGAs. Moreover, amongst 30 chimpanzee carcasses from the TNP (Côte d'Ivoire) that were tested for malaria parasites, no Hepatocystis spp. were found (S. Calvignac-Spencer, personal communication, 2015). Interestingly, however, a liver sample from one of these chimpanzees tested positive for a saurian Plasmodium parasite.
Extrinsic and intrinsic determinants of Plasmodium infection
Extrinsic factors, such as environmental factors (habitat type and climate), group size and host density as well as possibly behavioural factors, such as sleeping site choice, most likely influence the exposure to malaria parasites and thus the frequency of infections in specific AGA communities. However, only very few studies so far have investigated their effects, which remain largely unknown.
The influence of monthly rainfall on the probability of Plasmodium detection in chimpanzee and gorilla faecal samples was tested in a single study so far, but no significant effect was found (Boundenga et al., 2015). Yet, it was shown that rainfall can affect Plasmodium parasite prevalence in the vectors of AGA malaria parasites, with increased prevalence and thus exposure during the rainy season (Makanga et al., 2016). Prevalence in mosquitoes was also shown to vary with canopy height, with higher rates at mid-height compared to ground and canopy level.
There is very limited information on the effects of host group size and sleeping site, mainly drawn from studies on New World primates. In contrast to the encounter-dilution effect, which predicts a decreased risk of exposure to an infected vector with increasing group size (Krebs et al., 2014; Mooring and Hart, 1992), malaria prevalence in New World primates was found to increase with average sleeping group size, suggesting that malaria might be a significant cost associated with larger group sizes (Davies et al., 1991; Nunn and Eckhard, 2005). However, data on primate malaria and group size are still too scarce to draw any firm conclusions. Sleeping site selection in these primates also seems to play a role in malaria infection rates by reducing the risk of attack by the vector (Nunn and Eckhard, 2005). In chimpanzees, two studies looked at malaria and sleeping sites indirectly by capturing mosquitoes at natural nest sites. While one study failed to detect any patterns of association between the choice of nest site and mosquito densities (Koops et al., 2012), the second study found patterns compatible with mosquito avoidance (Krief et al., 2012). Chimpanzees seemed to choose nest sites in drier and higher places compared to their feeding sites, and these sites were indeed characterised by lower densities of Anopheles mosquitoes. Mosquitoes were also less abundant in sites with more nests (Krief et al., 2012). Moreover, an experimental study showed that tree species preferred by chimpanzees for nest building might be selected due to their repulsive properties against flying arthropods (Samson et al., 2013).
As for intrinsic determinants of malaria infection in AGAs, based on data from human malaria, one would expect factors such as age, pregnancy status, co-infections, immune status and genetics to influence susceptibility to infection as well as levels of parasitaemia and the expression of clinical malaria.
As a matter of fact, patterns strikingly similar to those found in human populations from malaria endemic areas have been observed in wild AGAs. Faecal detection rates of Plasmodium infection were found to be higher in younger individuals of P. t. verus (De Nys et al., 2013) and G. g. gorilla (Mapua et al., 2015) as well as in pregnant females of P. t. verus (De Nys et al., 2014). This probably reflects changes in the frequency of new infections and/or in parasite densities induced by variation in susceptibility to these parasites due to age and pregnancy status. These fluctuations in susceptibility might be the result of immune protection acquired in adults through constant exposure to the parasites (Doolan et al., 2009) and, during pregnancy, a combination of immunological and hormonal changes, together with the ability of P. falciparum-infected erythrocytes to sequester in the placenta (Rogerson et al., 2007). However, variations in the exposure to the vectors and, thus, to malaria parasites might also occur and play a role in these observed patterns. It was, for instance, demonstrated in humans that pregnant women are more attractive to Anopheles mosquitoes compared to non-pregnant women, probably for physiological reasons, e.g. increased exhaled breath and abdominal temperature (Ansell et al., 2002; Lindsay et al., 2000). Additionally, possible behavioural changes, such as nesting habits, that could lead to increased exposure to the vectors should be considered.
In addition, it would be judicious to keep in mind that in a given host, malaria parasites are part of a parasite community, whereas we use the term parasite in its broad ecological sense, designating micro- and macroparasites (Faure, 2014). This implies that, just like in an ecosystem, multiple and complex interactions between these parasites and with their host (i.e. their environment) take place; this can also partly dictate the presence of parasite species or their densities in a specific host, as opposed to a single host–parasite relationship (Pedersen and Fenton, 2007; Rynkiewicz et al., 2015), and perhaps explain (at least partially) the patterns of malaria parasite detection observed in AGAs. As an illustration, Telfer et al. (2010) actually demonstrated, for the first time in a natural population of field vole (Microtus agrestis), that co-infection with several microparasites explains more of the variation in the risk of infection compared to other factors such as host condition, e.g. age, and exposure risk, e.g. season (Telfer et al., 2010). Co-infection interactions can take place via competition for shared resources, e.g. nutrients, physical site of infection, or the immune system of the host, e.g. cross-reactivity or immunosuppressant pathogens such as HIV (Graham, 2008; Pedersen and Fenton, 2007; Rynkiewicz et al., 2015). Resource-mediated parasite interactions would be likely to occur between malaria parasites and other haemoparasite species which target the same cells, like erythrocytes. For instance, Babesia microti infection in Microtus agrestis negatively affects host susceptibility to Bartonella spp. infection and vice versa (Telfer et al., 2010). Likewise, a negative association between Plasmodium spp. and Babesia spp. has been recently observed in wild Malagasy primates (Propithecus verreauxi), where high Babesia spp. infection levels in younger individuals also seem to confer protection against Plasmodium spp. infections, possibly via shared resource competition (Springer et al., 2015). Inhibition of Plasmodium cynomolgi infection by Babesia microti was also previously described in experimental rhesus macaques (van Duivenvoorde et al., 2010; Voorberg-vd Wel et al., 2008). In humans, there is evidence of a pathological interaction between the immunosuppressant HIV virus and Plasmodium, with HIV-seropositive people being at increased risk of Plasmodium infection and the development of clinical malaria (Patnaik et al., 2005; Whitworth et al., 2000). Malaria parasites and various helminth species have also been shown to interact in humans, with either an antagonistic or beneficial relationship, possibly mediated by cross-reactive immune responses (Faure, 2014). In order to fully understand the patterns of malaria infection in AGAs, it would thus be necessary to also explore and take into account co-infections with parasites that are likely to interact.
Genetic factors may additionally play a role in conferring a certain degree of resistance to infection by malaria parasites or clinical disease. Genetic regulation of the immune response by certain alleles encoding the major histocompatibility complexes I and II, several interleukins, the CD40 ligand, the nitric oxide synthase, and interferon and receptors has been associated with protection against severe malaria in humans (Hill et al., 1991; Kwiatkowski, 2005). Mutations leading to variants of erythrocyte characteristics also provide protection against parasite invasion – for example, the Duffy negativity for P. vivax; glycophorin A, B and C deficiency for P. falciparum; and haemoglobin E (-globin gene mutation) – or confer protection against clinical malaria, such as haemoglobin S (sickle haemoglobin) and C (both -globin gene mutations), glucose-6-phosphate dehydrogenase (G6PD) deficiency and -thalassemia (defective production of globin) (Kwiatkowski, 2005; Williams, 2006). To date, there is very little information available for AGAs. The diversity of the G6PD and -globin genes has been examined in chimpanzees, but so far no mutations appear to have been selected for malarial resistance (MacFie et al., 2009; Verrelli et al., 2006). Moreover, Liu et al. (2014) sequenced samples from 134 AGAs but did not find any Duffy-negative phenotypes.
Pathogenicity of malaria parasites in African great apes
None of the published studies have focused on the consequences of malaria infection on wild great apes' health so far, and the information collected from experimental or captive conditions is rather limited. Observations made during early experiments on the clinical course of malaria in captive chimpanzees, i.e. mainly the lack of noticeable illness behaviour during various infections with human and possibly chimpanzee strains, led Rodhain (1936, 1940) to believe that, in natural conditions, chimpanzees do not suffer significantly from malaria and that they can be subject to asymptomatic chronic infections lasting several months. On one occasion, he reported a double inoculation with P. falciparum and P. vivax in a young individual, which had induced a febrile state with loss of appetite but only mildly (Rodhain, 1936). The only indications on the duration of infections also come from early experimental studies. The infections seemed to be able to last for at least several months, with periods as long as 5 months recorded for P. reichenowi and P. schwetzi (Garnham et al., 1956) and over 1 year for P. malariae (Rodhain, 1940).
Reports from sanctuaries or other facilities hosting captive individuals are rare and variable. In Japan, two captive chimpanzees were found to be chronically infected with P. malariae, without showing any evidence of illness for 30 years (Hayakawa et al., 2009). P. falciparum infection of captive bonobos in the Democratic Republic of the Congo were also not associated with apparent clinical signs or increased body temperature (Krief et al., 2010). Moreover, a young chimpanzee newly transferred to a sanctuary after 6 years of captivity in an urban setting developed severe anaemia and hyperthermia concomitant with a high P. reichenowi parasitaemia (Herbert et al., 2015). The latter case would suggest that infection with certain malaria parasites can, under certain circumstances, be detrimental for chimpanzees' health. However, extrapolating these observations to the wild is difficult as stress related to captivity could also influence the effect of such infections. Moreover, whether individuals were initially naïve to the Plasmodium strains they were diagnosed with (whether human or chimpanzee derived) is generally undetermined (strains novel to a chimpanzee's immune system would be more likely to cause illness). One publication also reported the death of a 1-year-old captive chimpanzee infected with P. reichenowi; however, given the extremely poor and stressful conditions this individual was kept in, whether malaria effectively played a significant role in the fatal outcome is questionable (Tarello, 2005).
In contrast, pathogenicity has never been demonstrated in AGAs living in natural conditions so far. The general impression given by the above-mentioned elements, as well as by the absence or rarity of reported malaria-like clinical signs or death associated with infection in individuals observed on a sometimes daily basis (wild habituated or captive individuals) (e.g. Kaiser et al., 2010), is that wild-living chimpanzees are mostly undisturbed by malaria infections, or that at least the clinical impact and related discomfort are generally mild enough to be invisible to observers' eyes when not specifically searched for. Also, as pointed out by Rayner et al. (2011), the very high prevalence rates of malaria parasite infections observed in numerous wild chimpanzee and gorilla populations indicate that severe outcomes are probably rather rare.
As young and pregnant chimpanzees seem to be more susceptible to malaria infection, it is possible that they are also more prone to suffer from the biological consequences of infection and to develop clinical malaria, as seen in humans (Doolan et al., 2009). Healthy and non-pregnant adults probably benefit from a certain degree of acquired clinical immunity, but it would be reasonable to think that malaria infection would start having some level of detrimental effect when the host's ability to control parasite density is diminished. Higher levels of blood parasitaemia are likely to at least induce higher degrees of anaemia (Kotepui et al., 2015; Oni and Oguntibeju, 2006), which in turn could cause morbidity or even mortality. Anaemia is actually recognised as one of the major causes of morbidity and mortality related to malaria in young children and pregnant women (Ekvall et al., 2001). Moreover, in humans, malaria during pregnancy can have deleterious consequences for the pregnancy's outcome and the infant's health (Desai et al., 2007; Guyatt and Snow, 2001). Deleterious effects on pregnancy outcome and/or infant health would mean a general impact on population reproductive success and fitness, thereby shaping population dynamics.
Further investigation of the relationship between malaria parasites and chimpanzees in natural conditions is important to understand the significance of malaria infection for wild great ape populations' health, and it can serve as a model to understand the host–parasite relationship in human populations, where malaria is believed to have acted as an important driver of recent evolution (Carter and Mendis, 2002; Sabbatani et al., 2010). Wild habituated AGA communities are ideal targets to pursue investigations on illness related to malaria infection. In a more indirect way, studying the AGA malaria parasites' genes, which might play a role in pathogenesis, can also provide valuable information. For example, the very diverse var genes, found only in Laverania parasites, are unique to P. falciparum in humans and play an important role in its pathogenesis by coding for proteins which mediate the cytoadherence of infected erythrocytes (Smith et al., 2013). Analysis of var and var-like gene sequences from human and ape Laverania species showed that many characteristics of these genes are conserved across the different Laverania species and that these evolved well before the emergence of all the Laverania species (Larremore et al., 2015). In addition, the var2csa genes, known to be associated with the adherence of P. falciparum-infected erythrocytes to the placenta and placental malaria in humans, have also been identified in P. reichenowi and P. gaboni (Pacheco et al., 2013; Trimnell et al., 2006). However, the question of whether the expression of var genes is also associated with pathogenicity in AGAs remains open. Some significant differences have also been found between var genes from P. gaboni and var genes from P. falciparum and P. reichenowi, suggesting potential disparities in their biology.
Conclusions and perspectives
It is apparent that, to date, most of the knowledge on wild AGA malaria is concentrated around parasite diversity, distribution and phylogenetic relationships with malaria parasites infecting other species, humans in particular. Indeed, the majority of research studies ultimately aim at identifying potential animal reservoirs with ongoing zoonotic transmission as well as assessing the risk of emergence of new malaria strains in human populations (by, inter alia, exploring the origins of the current human malaria parasites). Many gaps, however, still need to be filled in order to achieve these objectives – e.g. further exploration of vectors, of phylogenetic relationships of ape-derived P. malariae and P. ovale strains with human strains, and of zoonotic infections in humans living close to AGA communities.
The epidemiological determinants and the effects of malaria infection in wild AGAs have been left largely unexplored. This also leaves many questions that need to be addressed in order to further understand the basic epidemiology and biology of malaria parasites in AGAs. What are the influences of other known intrinsic and extrinsic factors affecting human malaria on infection, parasite density or, perhaps, disease in AGAs? Are there visible patterns of infection in specific populations? How frequently and for how long are individuals infected? What are the pathological consequences of malaria infection? Does infection have a significant impact on great ape health? Could the impact of malaria be sufficiently subtle to be misleading? Is this just like SIV (simian immunodeficiency virus) infection in chimpanzees, which was believed for many years to be rather trivial for their health (non-pathogenic), but whose actual deleterious impact on their health only became apparent recently when examined at the level of an entire community (Keele et al., 2009; Rudicell et al., 2010)? Does malaria play a role in sociality, demographics, population dynamics and general fitness? Are there any indications of selective pressures exerted by malaria on AGA evolution or development of resistance? These are just a fraction of the multiple questions that still need to be addressed. Using faecal samples has proven suitable and efficient, not only to address broad-scale questions such as those on the geographic distribution and genetic diversity of great ape malaria parasites, but also to address small-scale epidemiological questions which might, in comparison, require a higher (finer-scale) resolution and thus more sensitive screening (De Nys et al., 2013, 2014). As non-invasive methods to study pathogen infections and monitor health parameters (e.g. body temperature, inflammation markers and hormone levels) are now available, attempts to answer some of these questions are actually conceivable. In the meantime, further exploration, improvement and testing of non-invasive methods need to be pursued in order to enhance sensitivity, facilitate the interpretation of results and broaden research possibilities.
Acknowledgements
This research was funded by the Deutsche Forschungsgemeinschaft (LE 1813/10-1) and has profited from numerous discussions in the Sociality and Health in Primates (FOR2136) research group. Edited by: K. Mätz-Rensing Reviewed by: L. Duval and one anonymous referee
This is a review article; therefore, no data were obtained.
The authors declare that they have no conflict of interest.
References
- Adler S. Malaria in chimpanzees in Sierra Leone. Ann Trop Med Parasit. 1923;17:13–19. [Google Scholar]
- Ansell J, Hamilton KA, Pinder M, Walraven GE, Lindsay SW. Short-range attractiveness of pregnant women to Anopheles gambiae mosquitoes. T Roy Soc Trop Med H. 2002;96:113–116. doi: 10.1016/s0035-9203(02)90271-3. [DOI] [PubMed] [Google Scholar]
- Ayouba A, Mouacha F, Learn GH, Mpoudi-Ngole E, Rayner JC, Sharp PM, Hahn BH, Delaporte E, Peeters M. Ubiquitous Hepatocystis infections, but no evidence of Plasmodium falciparum-like malaria parasites in wild greater spot-nosed monkeys (Cercopithecus nictitans) Int J Parasitol. 2012;42:709–713. doi: 10.1016/j.ijpara.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blacklock B, Adler S. A parasite resembling Plasmodium falciparum in a chimpanzee. Ann Trop Med Parasit. 1922;160:99–106. [Google Scholar]
- Blanquart S, Gascuel O. Mitochondrial genes support a common origin of rodent malaria parasites and Plasmodium falciparum's relatives infecting great apes. BMC Evol Biol. 2011;11:70. doi: 10.1186/1471-2148-11-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boundenga L, Ollomo B, Rougeron V, Mouele LY, Mve-Ondo B, Delicat-Loembet LM, Moukodoum ND, Okouga AP, Arnathau C, Elguero E, Durand P, Liégeois F, Boué V, Motsch P, Le Flohic G, Ndoungouet A, Paupy C, Ba CT, Renaud F, Prugnolle F. Diversity of malaria parasites in great apes in Gabon. Malaria J. 2015;14:111. doi: 10.1186/s12936-015-0622-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray RS. Studies on malaria in chimpanzees. V. The sporogonous cycle and mosquito transmission of Plasmodium vivax shcwetzi. J Parasitol. 1958;44:46–51. a. [PubMed] [Google Scholar]
- Bray RS. Studies on malaria in chimpanzees. VI. Laverania falciparum. Am J Trop Med Hyg. 1958;7:20–24. doi: 10.4269/ajtmh.1958.7.20. b. [DOI] [PubMed] [Google Scholar]
- Bray RS. Studies on malaria in chimpanzees. VIII. The experimental transmission and pre-erythrocytic phase of Plasmodium malariae, with a note on the host-range of the parasite. Am J Trop Med Hyg. 1960;9:455–465. doi: 10.4269/ajtmh.1960.9.455. [DOI] [PubMed] [Google Scholar]
- Bray RS. The Malaria Parasites of Anthropoid Apes. J Parasitol. 1963;49:888–891. [PubMed] [Google Scholar]
- Broderick C, Nadjm B, Smith V, Blaze M, Checkley A, Chiodini PL, Whitty CJM. Clinical, geographical, and temporal risk factors associated with presentation and outcome of vivax malaria imported into the United Kingdom over 27 years: observational study. BMJ. 2015;350:h1703. doi: 10.1136/bmj.h1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumpt E. Les parasites du paludisme des chimpanzés. Comptes Rendus Soc Biol. 1939;130:834–840. [Google Scholar]
- Carter R. Speculations on the origins of Plasmodium vivax malaria. Trends Parasitol. 2003;19:214–219. doi: 10.1016/s1471-4922(03)00070-9. [DOI] [PubMed] [Google Scholar]
- Carter R, Mendis KN. Evolutionary and historical aspects of the burden of malaria. Clin Microbiol Rev. 2002;15:564–594. doi: 10.1128/CMR.15.4.564-594.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anopheles mosquitoes. CDC; [last access: 2015]. 2012. available at: http://www.cdc.gov/malaria/about/biology/mosquitoes/ [Google Scholar]
- Coatney GR. Simian malarias in man: facts, implications, and predictions. Am J Trop Med Hyg. 1968;17:147–155. doi: 10.4269/ajtmh.1968.17.147. [DOI] [PubMed] [Google Scholar]
- Coetzee M. Distribution of the African malaria vectors of the Anopheles gambiae complex. Am J Trop Med Hyg. 2004;70:103–104. [PubMed] [Google Scholar]
- Collins WE, Skinner JC, Pappaioanou M, Broderson JR, Mehaffey P. The sporogonic cycle of Plasmodium reichenowi. J Parasitol. 1986;72:292–298. [PubMed] [Google Scholar]
- Contacos PG, Coatney GR, Orihel TC, Collins WE, Chin W, Jeter MH. Transmission of Plasmodium schwetzi from the chimpanzee to man by mosquito bite. Am J Trop Med Hyg. 1970;19:190–195. doi: 10.4269/ajtmh.1970.19.190. [DOI] [PubMed] [Google Scholar]
- Culleton RL, Mita T, Ndounga M, Unger H., Cravo PV, Paganotti GM, Takahashi N, Kaneko A, Eto H, Tinto H, Karema C, D'Alessandro U, do Rosario V, Kobayakawa T, Ntoumi F, Carter R, Tanabe K. Failure to detect Plasmodium vivax in West and Central Africa by PCR species typing. Malaria J. 2008;7:174. doi: 10.1186/1475-2875-7-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies CR, Ayres JM, Dye C, Deane LM. Malaria infection rate of Amazonian primates increases with body weight and group size. Funct Ecol. 1991;5:655–662. [Google Scholar]
- Délicat-Loembet L, Rougeron V, Ollomo B, Arnathau C, Roche B, Elguero E, Moukodoum ND, Okougha A-P, Mve Ondo B, Boundenga L, Houzé S, Galan M, Nkoghé D, Leroy EM, Durand P, Paupy C, Renaud F, Prugnolle F. No Evidence for Ape Plasmodium Infections in Humans in Gabon. PLoS One. 2015;10:e0126933. doi: 10.1371/journal.pone.0126933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Nys HM, Calvignac-Spencer S, Thiesen U, Boesch C, Wittig RM, Mundry R, Leendertz FH. Age-related effects on malaria parasite infection in wild chimpanzees. Biol Lett. 2013;9:20121160. doi: 10.1098/rsbl.2012.1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Nys HM, Calvignac-Spencer S, Boesch C, Dorny P, Wittig RM, Mundry R, Leendertz FH. Malaria parasite detection increases during pregnancy in wild chimpanzees. Malaria J. 2014;13:413. doi: 10.1186/1475-2875-13-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai M, ter Kuile FO, Nosten F, McGready R, Asamoa K, Brabin B, Newman RD. Epidemiology and burden of malaria in pregnancy. Lancet Infect Dis. 2007;7:93–104. doi: 10.1016/s1473-3099(07)70021-x. [DOI] [PubMed] [Google Scholar]
- Doolan DL, Dobaño C, Baird JK. Acquired Immunity to Malaria. Clin Microbiol Rev. 2009;22:13–36. doi: 10.1128/CMR.00025-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duval L, Nerrienet E, Rousset D, Sadeuh Mba SA, Houze S, Fourment M, Le Bras J, Robert V, Ariey F. Chimpanzee malaria parasites related to Plasmodium ovale in Africa. PLoS One. 2009;4:e5520. doi: 10.1371/journal.pone.0005520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duval L, Fourment M, Nerrienet E, Rousset D, Sadeuh SA, Goodman SM, Andriaholinirina NV, Randrianarivelojosia M, Paul RE, Robert V, Ayala FJ, Ariey F. African apes as reservoirs of Plasmodium falciparum and the origin and diversification of the Laverania subgenus. P Natl Acad Sci USA. 2010;107:10561–10566. doi: 10.1073/pnas.1005435107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekvall H, Arese P, Turrini F, Ayi K, Mannu F, Premji Z, Bjorkman A. Acute haemolysis in childhood falciparum malaria. T Roy Soc Trop Med H. 2001;95:611–617. doi: 10.1016/s0035-9203(01)90095-1. [DOI] [PubMed] [Google Scholar]
- Escalante AA, Ayala FJ. Phylogeny of the malarial genus Plasmodium, derived from rRNA gene sequences. P Natl Acad Sci USA. 1994;91:11373–11377. doi: 10.1073/pnas.91.24.11373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escalante AA, Barrio E, Ayala FJ. Evolutionary origin of human and primate malarias: evidence from the circumsporozoite protein gene. Mol Biol Evol. 1995;12:616–626. doi: 10.1093/oxfordjournals.molbev.a040241. [DOI] [PubMed] [Google Scholar]
- Escalante AA, Cornejo OE, Freeland DE, Poe AC, Durrego E, Collins WE, Lal AA. A monkey's tale: the origin of Plasmodium vivax as a human malaria parasite. P Natl Acad Sci USA. 2005;102:1980–1985. doi: 10.1073/pnas.0409652102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure E. Malarial pathocoenosis: beneficial and deleterious interactions between malaria and other human diseases. Front Physiol. 2014;5:441. doi: 10.3389/fphys.2014.00441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnham PC, Lainson R, Gunders AE. Some observations on malaria parasites in a chimpanzee, with particular reference to the persistence of Plasmodium reichenowi and Plasmodium vivax. Ann Soc Belg Med Tr. 1956;36:811–821. [PubMed] [Google Scholar]
- Gautret P, Legros F, Koulmann P, Rodier MH, Jacquemin JL. Imported Plasmodium vivax malaria in France: Geographical origin and report of an atypical case acquired in Central or Western Africa. Acta Trop. 2001;78:177–181. doi: 10.1016/s0001-706x(00)00181-9. [DOI] [PubMed] [Google Scholar]
- Graham AL. Ecological rules governing helminth-microparasite coinfection. P Natl Acad Sci USA. 2008;105:566–570. doi: 10.1073/pnas.0707221105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyatt HL, Snow RW. Malaria in pregnancy as an indirect cause of infant mortality in sub-Saharan Africa. T Roy Soc Trop Med H. 2001;95:569–576. doi: 10.1016/s0035-9203(01)90082-3. [DOI] [PubMed] [Google Scholar]
- Hayakawa T, Culleton R, Otani H, Horii T, Tanabe K. Big bang in the evolution of extant malaria parasites. Mol Biol Evol. 2008;25:2233–2239. doi: 10.1093/molbev/msn171. [DOI] [PubMed] [Google Scholar]
- Hayakawa T, Arisue N, Udono T, Hirai H, Sattabongkot J, Toyama T, Tsuboi T, Horii T, Tanabe K. Identification of Plasmodium malariae, a human malaria parasite, in imported chimpanzees. PLoS One. 2009;4:e7412. doi: 10.1371/journal.pone.0007412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert A, Boundenga L, Meyer A, Moukodoum DN, Okouga AP, Arnathau C, Durand P, Magnus J, Ngoubangoye B, Willaume E, Ba CT, Rougeron V, Renaud F, Ollomo B, Prugnolle F. Malaria-like symptoms associated with a natural Plasmodium reichenowi infection in a chimpanzee. Malaria J. 2015;14:220. doi: 10.1186/s12936-015-0743-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill AV, Allsopp CE, Kwiatkowski D, Anstey NM, Twumasi P, Rowe PA, Bennett S, Brewster D, McMichael AJ, Greenwood BM. Common west African HLA antigens are associated with protection from severe malaria. Nature. 1991;352:595–600. doi: 10.1038/352595a0. [DOI] [PubMed] [Google Scholar]
- Hughes AL, Verra F. Malaria parasite sequences from chimpanzee support the co-speciation hypothesis for the origin of virulent human malaria (Plasmodium falciparum) Mol Phylogenet Evol. 2010;57:135–143. doi: 10.1016/j.ympev.2010.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junglen S, Kurth A, Kuehl H, Quan PL, Ellerbrok H, Pauli G, Nitsche A, Nunn C, Rich SM, Lipkin WI, Briese T, Leendertz FH. Examining landscape factors influencing relative distribution of mosquito genera and frequency of virus infection. Ecohealth. 2009;6:239–249. doi: 10.1007/s10393-009-0260-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser M, Lowa A, Ulrich M, Ellerbrok H, Goffe AS, Blasse A, Zommers Z, Couacy-Hymann E, Babweteera F, Zuberbuhler K, Metzger S, Geidel S, Boesch C, Gillepsie T, Leendertz F. Wild Chimpanzees Infected with 5 Plasmodium Species. Emerg Infect Dis. 2010;16:1956–1959. doi: 10.3201/eid1612.100424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keele BF, Jones JH, Terio KA, Estes JD, Rudicell RS, Wilson ML, Li Y, Learn GH, Beasley TM, Schumacher-Stankey J, Wroblewski E, Mosser A, Raphael J, Kamenya S, Lonsdorf EV, Travis DA, Mlengeya T, Kinsel MJ, Else JG, Silvestri G, Goodall J, Sharp PM, Shaw GM, Pusey AE, Hahn BH. Increased mortality and AIDS-like immunopathology in wild chimpanzees infected with SIVcpz. Nature. 2009;460:515–519. doi: 10.1038/nature08200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koops K, McGrew WC, Matsuzawa T, Knapp LA. Terrestrial nest-building by wild chimpanzees (Pan troglodytes): implications for the tree-to-ground sleep transition in early hominins. Am J Phys Anthropol. 2012;148:351–361. doi: 10.1002/ajpa.22056. [DOI] [PubMed] [Google Scholar]
- Kotepui M, Piwkham D, PhunPhuech B, Phiwklam N, Chupeerach C, Duangmano S. Effects of malaria parasite density on blood cell parameters. PLoS One. 2015;10:e0121057. doi: 10.1371/journal.pone.0121057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs BL, Anderson TK, Goldberg TL, Hamer GL, Kitron UD, Newman CM, Ruiz MO, Walker ED, Brawn JD. Host group formation decreases exposure to vector-borne disease: a field experiment in a “hotspot” of West Nile virus transmission. P Roy Soc B-Biol Sci. 2014;281:20141586. doi: 10.1098/rspb.2014.1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krief S, Escalante AA, Pacheco MA, Mugisha L, Andre C, Halbwax M, Fischer A, Krief JM, Kasenene JM, Crandfield M, Cornejo OE, Chavatte JM, Lin C, Letourneur F, Gruner AC, McCutchan TF, Renia L, Snounou G. On the diversity of malaria parasites in African apes and the origin of Plasmodium falciparum from Bonobos. PLoS Pathog. 2010;6:e1000765. doi: 10.1371/journal.ppat.1000765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krief S, Levrero F, Krief J-M, Thanapongpichat S, Imwong M, Snounou G, Kasenene JM, Cibot M, Gantier J-C. Investigations on anopheline mosquitoes close to the nest sites of chimpanzees subject to malaria infection in Ugandan Highlands. Malaria J. 2012;11:116. doi: 10.1186/1475-2875-11-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski DP. How malaria has affected the human genome and what human genetics can teach us about malaria. Am J Hum Genet. 2005;77:171–192. doi: 10.1086/432519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larremore DB, Sundararaman SA, Liu W, Proto WR, Clauset A, Loy DE, Speede S, Plenderleith LJ, Sharp PM, Hahn BH, Rayner JC, Buckee CO. Ape parasite origins of human malaria virulence genes. Nat Commun. 2015;6:8368. doi: 10.1038/ncomms9368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KS, Divis PC, Zakaria SK, Matusop A, Julin RA, Conway DJ, Cox-Singh J, Singh B. Plasmodium knowlesi: reservoir hosts and tracking the emergence in humans and macaques. PLoS Pathog. 2011;7:e1002015. doi: 10.1371/journal.ppat.1002015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay S, Ansell J, Selman C, Cox V, Hamilton K, Walraven G. Effect of pregnancy on exposure to malaria mosquitoes. Lancet. 2000;355:1972. doi: 10.1016/S0140-6736(00)02334-5. [DOI] [PubMed] [Google Scholar]
- Liu W, Li Y, Learn GH, Rudicell RS, Robertson JD, Keele BF, Ndjango JB, Sanz CM, Morgan DB, Locatelli S, Gonder MK, Kranzusch PJ, Walsh PD, Delaporte E, Mpoudi-Ngole E, Georgiev AV, Muller MN, Shaw GM, Peeters M, Sharp PM, Rayner JC, Hahn BH. Origin of the human malaria parasite Plasmodium falciparum in gorillas. Nature. 2010;467:420–425. doi: 10.1038/nature09442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Li Y, Shaw KS, Learn GH, Plenderleith LJ, Malenke JA, Sundararaman SA, Ramirez MA, Crystal PA, Smith AG, Bibollet-Ruche F, Ayouba A, Locatelli S, Esteban A, Mouacha F, Guichet E, Butel C, Ahuka-Mundeke S, Inogwabini B-I, Ndjango J-BN, Speede S, Sanz CM, Morgan DB, Gonder MK, Kranzusch PJ, Walsh PD, Georgiev AV, Muller MN, Piel AK, Stewart FA, Wilson ML, Pusey AE, Cui L, Wang Z, Färnert A, Sutherland CJ, Nolder D, Hart JA, Hart TB, Bertolani P, Gillis A, LeBreton M, Tafon B, Kiyang J, Djoko CF, Schneider BS, Wolfe ND, Mpoudi-Ngole E, Delaporte E, Carter R, Culleton RL, Shaw GM, Rayner JC, Peeters M, Hahn BH, Sharp PM. African origin of the malaria parasite Plasmodium vivax. Nat Commun. 2014;5:3346. doi: 10.1038/ncomms4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Sundararaman SA, Loy DE, Learn GH, Li Y, Plenderleith LJ, Ndjango J-BN, Speede S, Atencia R, Cox D, Shaw GM, Ayouba A, Peeters M, Rayner JC, Hahn BH, Sharp PM. Multigenomic Delineation of Plasmodium Species of the Laverania Subgenus Infecting Wild-Living Chimpanzees and Gorillas. Genome Biol Evol. 2016;8:1929–1939. doi: 10.1093/gbe/evw128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacFie TS, Nerrienet E, Bontrop RE, Mundy NI. The action of falciparum malaria on the human and chimpanzee genomes compared: absence of evidence for a genomic signature of malaria at HBB and G6PD in three subspecies of chimpanzee. Infect Genet Evol. 2009;9:1248–52. doi: 10.1016/j.meegid.2009.06.025. [DOI] [PubMed] [Google Scholar]
- Makanga B, Yangari P, Rahola N, Rougeron V, Elguero E, Boundenga L, Moukodoum ND, Okouga A.P, Arnathau C, Durand P, Willaume E, Ayala D, Fontenille D, Ayala FJ, Renaud F, Ollomo B, Prugnolle F, Paupy C. Ape malaria transmission and potential for ape-to-human transfers in Africa. P Natl Acad Sci USA. 2016;113:5329–5334. doi: 10.1073/pnas.1603008113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mapua MI, Qablan MA, Pomajbíková K, Petrželková KJ, Hůzová Z, Rádrová J, Votýpka J, Todd A, Jirků M, Leendertz FH, Lukeš J, Neel C, Modrý D. Ecology of malaria infections in western lowland gorillas inhabiting Dzanga Sangha Protected Areas, Central African Republic. Parasitology. 2015;142:890–900. doi: 10.1017/S0031182015000086. [DOI] [PubMed] [Google Scholar]
- Martin MJ, Rayner JC, Gagneux P, Barnwell JW, Varki A. Evolution of human-chimpanzee differences in malaria susceptibility: relationship to human genetic loss of N-glycolylneuraminic acid. P Natl Acad Sci USA. 2005;102:12819–12824. doi: 10.1073/pnas.0503819102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinsen ES, Perkins SL, Schall JJ. A three-genome phylogeny of malaria parasites (Plasmodium and closely related genera): Evolution of life-history traits and host switches. Mol Phylogenet Evol. 2008;47:261–273. doi: 10.1016/j.ympev.2007.11.012. [DOI] [PubMed] [Google Scholar]
- Mooring MS, Hart BL. Animal Grouping for Protection from Parasites: Selfish Herd and Encounter-Dilution Effects. Behaviour. 1992;123:173–193. doi: 10.1163/156853992X00011. [DOI] [Google Scholar]
- Mu J, Joy DA, Duan J, Huang Y, Carlton J, Walker J, Barnwell J, Beerli P, Charleston MA, Pybus OG, Su XZ. Host switch leads to emergence of Plasmodium vivax malaria in humans. Mol Biol Evol. 2005;22:1686–1693. doi: 10.1093/molbev/msi160. [DOI] [PubMed] [Google Scholar]
- Mühlberger N, Jelinek T, Gascon J, Probst M, Zoller T, Schunk M, Beran J, Gjørup I, Behrens RH, Clerinx J, Björkman A, McWhinney P, Matteelli A, Lopez-Velez R, Bisoffi Z, Hellgren U, Puente S, Schmid ML, Myrvang B, Holthoff-Stich ML, Laferl H, Hatz C, Kollaritsch H, Kapaun A, Knobloch J, Iversen J, Kotlowski A, Malvy DJM, Kern P, Fry G, Siikamaki H, Schulze MH, Soula G, Paul M, Gómez i Prat J, Lehmann V, Bouchaud O, da Cunha S, Atouguia J., Boecken G. Epidemiology and clinical features of vivax malaria imported to Europe: sentinel surveillance data from TropNetEurop. Malaria J. 2004;3:5. doi: 10.1186/1475-2875-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngoubangoye B, Boundenga L, Arnathau C, Mombo IM, Durand P, Tsoumbou T-A, Otoro BV, Sana R, Okouga A-P, Moukodoum N, Willaume E, Herbert A, Fouchet D, Rougeron V, Bâ CT, Ollomo B, Paupy C, Leroy EM, Renaud F, Pontier D, Prugnolle F. The host specificity of ape malaria parasites can be broken in confined environments. Int J Parasitol. 2016;46:737–744. doi: 10.1016/j.ijpara.2016.06.004. [DOI] [PubMed] [Google Scholar]
- Nunn C, Eckhard WH. Malaria infection and host behavior: a comparative study of Neotropical primates. Behav Ecol Sociobiol. 2005;59:30–37. doi: 10.1007/s00265-005-0005-z. [DOI] [Google Scholar]
- Oni GA, Oguntibeju OO. Relationships between the Malaria Parasite Density and Children Anaemia: Brief Communication . [last access: 28 June 2015];Int J Pathol. 2006 4 available at: http://www.researchgate.net/publication/267367309_Relationships_between_the_Malaria_Parasite_Density_and_Children_Anaemia_Brief_Communication . [Google Scholar]
- Pacheco MA, Cranfield M, Cameron K, Escalante AA. Malarial parasite diversity in chimpanzees: the value of comparative approaches to ascertain the evolution of Plasmodium falciparum antigens. Malaria J. 2013;12:328. doi: 10.1186/1475-2875-12-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patnaik P, Jere CS, Miller WC, Hoffman IF, Wirima J, Pendame R, Meshnick SR, Taylor TE, Molyneux ME, Kublin JG. Effects of HIV-1 serostatus, HIV-1 RNA concentration, and CD4 cell count on the incidence of malaria infection in a cohort of adults in rural Malawi. J Infect Dis. 2005;192:984–991. doi: 10.1086/432730. [DOI] [PubMed] [Google Scholar]
- Paupy C, Makanga B, Ollomo B, Rahola N, Durand P, Magnus J, Willaume E, Renaud F, Fontenille D, Prugnolle F. Anopheles moucheti and Anopheles vinckei Are Candidate Vectors of Ape Plasmodium Parasites, Including Plasmodium praefalciparum in Gabon. PLoS One. 2013;8:e57294. doi: 10.1371/journal.pone.0057294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen AB, Fenton A. Emphasizing the ecology in parasite community ecology. Trends Ecol Evol. 2007;22:133–139. doi: 10.1016/j.tree.2006.11.005. [DOI] [PubMed] [Google Scholar]
- Prugnolle F, Durand P, Neel C, Ollomo B, Ayala FJ, Arnathau C, Etienne L, Mpoudi-Ngole E, Nkoghe D, Leroy E, Delaporte E, Peeters M, Renaud F. African great apes are natural hosts of multiple related malaria species, including Plasmodium falciparum. P Natl Acad Sci USA. 2010;107:1458–1463. doi: 10.1073/pnas.0914440107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prugnolle F, Ollomo B, Durand P, Yalcindag E, Arnathau C, Elguero E, Berry A, Pourrut X, Gonzalez JP, Nkoghe D, Akiana J, Verrier D, Leroy E, Ayala FJ, Renaud F. African monkeys are infected by Plasmodium falciparum nonhuman primate-specific strains. P Natl Acad Sci USA. 2011;108:11948–11953. doi: 10.1073/pnas.1109368108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prugnolle F, Rougeron V, Becquart P, Berry A, Makanga B, Rahola N, Arnathau C, Ngoubangoye B, Menard S, Willaume E, Ayala FJ, Fontenille D, Ollomo B, Durand P, Paupy C, Renaud F. Diversity, host switching and evolution of Plasmodium vivax infecting African great apes. P Natl Acad Sci USA. 2013;110:8123–8128. doi: 10.1073/pnas.1306004110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayner JC, Liu W, Peeters M, Sharp PM, Hahn BH. A plethora of Plasmodium species in wild apes: a source of human infection? Trends Parasitol. 2011;27:222–229. doi: 10.1016/j.pt.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichenow E. Über das Vorkommen der Malariaparasiten des Menschen bei den Afrikanischen Menschenaffen. Cent f Bakt I Abt Orig. 1920;85:207–216. [Google Scholar]
- Rich SM, Leendertz FH, Xu G, LeBreton M, Djoko CF, Aminake MN, Takang EE, Diffo JL, Pike BL, Rosenthal BM, Formenty P, Boesch C, Ayala FJ, Wolfe ND. The origin of malignant malaria. P Natl Acad Sci USA. 2009;106:14902–14907. doi: 10.1073/pnas.0907740106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodhain J. Contribution a l'étude des plasmodiums des singes africains. Ann Soc Belg Med Tr. 1936;16:237–253. [PubMed] [Google Scholar]
- Rodhain J. Les plasmodiums des anthropoïdes de l'Afrique centrale et leurs relations avec les plasmodiums humains. Ann Soc Belge Med Tr. 1939;19:563–572. [Google Scholar]
- Rodhain J. Les plasmodium des anthropoids de l'Afriqe centrale et leurs relations avec les plasmodiums humains. Ann Soc Belg Med Tr. 1940;35:69–73. [Google Scholar]
- Rodhain J, Dellaert R. L'infection a Plasmodium malariae du chimpanze chez l'homme. Etude d'une premiere souche isolee de l'anthropoide Pan satyrus verus. Ann Soc Belg Med Tr. 1943;23:19–46. [Google Scholar]
- Rodhain J, Dellaert R. Contribution a l'etude du Plasmodium schwetzi E. Brumpt (2me note). Transmission du Plasmodium schwetzi a l'homme. Ann Soc Belg Med Tr. 1955;35:73–76. [PubMed] [Google Scholar]
- Rogerson SJ, Hviid L, Duffy PE, Leke RFG, Taylor DW. Malaria in pregnancy: pathogenesis and immunity. Lancet Infect Dis. 2007;7:105–117. doi: 10.1016/s1473-3099(07)70022-1. [DOI] [PubMed] [Google Scholar]
- Rudicell RS, Holland Jones J, Wroblewski EE, Learn GH, Li Y, Robertson JD, Greengrass E, Grossmann F, Kamenya S, Pintea L, Mjungu DC, Lonsdorf EV, Mosser A, Lehman C, Collins DA, Keele BF, Goodall J, Hahn BH, Pusey AE, Wilson ML. Impact of simian immunodeficiency virus infection on chimpanzee population dynamics. PLoS Pathog. 2010;6:e1001116. doi: 10.1371/journal.ppat.1001116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rynkiewicz EC, Pedersen AB, Fenton A. An ecosystem approach to understanding and managing within-host parasite community dynamics. Trends Parasitol. 2015;31:212–221. doi: 10.1016/j.pt.2015.02.005. [DOI] [PubMed] [Google Scholar]
- Sabbatani S, Manfredi R, Fiorino S. Malaria infection and human evolution. Infez Med. 2010;18:56–74. [PubMed] [Google Scholar]
- Samson DR, Muehlenbein MP, Hunt KD. Do chimpanzees (Pan troglodytes schweinfurthii) exhibit sleep related behaviors that minimize exposure to parasitic arthropods? A preliminary report on the possible anti-vector function of chimpanzee sleeping platforms . Primates. 2013;54:73–80. doi: 10.1007/s10329-012-0329-z. [DOI] [PubMed] [Google Scholar]
- Schaer J, Perkins SL, Decher J, Leendertz FH, Fahr J, Weber N, Matuschewski K. High diversity of West African bat malaria parasites and a tight link with rodent Plasmodium taxa. P Natl Acad Sci USA. 2013;110:17415–17419. doi: 10.1073/pnas.1311016110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwetz J. Contribution à l'étude des parasites malariens des singes supérieurs africains. Rev di Malariol. 1934;13:143–147. [Google Scholar]
- Sharp PM, Rayner JC, Hahn BH. Evolution. Great apes and zoonoses. Science. 2013;340:284–286. doi: 10.1126/science.1236958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinka ME, Bangs MJ, Manguin S, Coetzee M, Mbogo CM, Hemingway J, Patil AP, Temperley WH, Gething PW, Kabaria CW, Okara RM, Van Boeckel T, Godfray HC, Harbach RE, Hay SI. The dominant Anopheles vectors of human malaria in Africa, Europe and the Middle East: occurrence data, distribution maps and bionomic precis. Parasite Vector. 2010;3:117. doi: 10.1186/1756-3305-3-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skarbinski J, James EM, Causer LM, Barber AM, Mali S, Nguyen-Dinh P, Roberts JM, Parise ME, Slutsker L, Newman RD. Malaria surveillance – United States. 2004, MMWR (Morbidity and Mortality weekly report) Vol. 55. Washington, D.C., USA: Surveillance summaries, 2002; 2006. pp. 23–37. [PubMed] [Google Scholar]
- Smith JD, Rowe JA, Higgins MK, Lavstsen T. Malaria's deadly grip: cytoadhesion of Plasmodium falciparum-infected erythrocytes. Cell Microbiol. 2013;15:1976–1983. doi: 10.1111/cmi.12183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer A, Fichtel C, Calvignac-Spencer S, Leendertz FH, Kappeler PM. Hemoparasites in a wild primate: Infection patterns suggest interaction of Plasmodium and Babesia in a lemur species. Int J Parasitol-Parasites Wildl. 2015;4:385–395. doi: 10.1016/j.ijppaw.2015.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundararaman SA, Liu W, Keele BF, Learn GH, Bittinger K, Mouacha F, Ahuka-Mundeke S, Manske M, Sherrill-Mix S, Li Y, Malenke JA, Delaporte E, Laurent C, Mpoudi Ngole E, Kwiatkowski DP, Shaw GM, Rayner JC, Peeters M, Sharp PM, Bushman FD, Hahn BH. Plasmodium falciparum-like parasites infecting wild apes in southern Cameroon do not represent a recurrent source of human malaria. P Natl Acad Sci USA. 2013;110:7020–7025. doi: 10.1073/pnas.1305201110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe K, Mita T, Jombart T, Eriksson A, Horibe S, Palacpac N, Ranford-Cartwright L, Sawai H, Sakihama N, Ohmae H, Nakamura M, Ferreira MU, Escalante AA, Prugnolle F, Bjorkman A, Farnert A, Kaneko A, Horii T, Manica A, Kishino H, Balloux F. Plasmodium falciparum accompanied the human expansion out of Africa. Curr Biol. 2010;20:1283–1289. doi: 10.1016/j.cub.2010.05.053. [DOI] [PubMed] [Google Scholar]
- Tarello W. A fatal Plasmodium reichenowi infection in a chimpanzee? Rev Méd Vét. 2005;156:503–505. [Google Scholar]
- Taylor DW, Wells RA, Vernes A, Rosenberg YJ, Vogel S, Diggs CL. Parasitologic and immunologic studies of experimental Plasmodium falciparum infection in nonsplenectomized chimpanzees (Pan troglodytes) Am J Trop Med Hyg. 1985;34:36–44. doi: 10.4269/ajtmh.1985.34.36. [DOI] [PubMed] [Google Scholar]
- Tazi L, Ayala FJ. Unresolved direction of host transfer of Plasmodium vivax v. P. simium and P. malariae v. P. brasilianum. Infect Genet Evol. 2011;11:209–221. doi: 10.1016/j.meegid.2010.08.007. [DOI] [PubMed] [Google Scholar]
- Telfer S, Lambin X, Birtles R, Beldomenico P, Burthe S, Paterson S, Begon M. Species interactions in a parasite community drive infection risk in a wildlife population. Science. 2010;330:243–246. doi: 10.1126/science.1190333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurber MI, Ghai RR, Hyeroba D, Weny G, Tumukunde A, Chapman CA, Wiseman RW, Dinis J, Steeil J, Greiner EC, Friedrich TC, O'Connor DH, Goldberg TL. Co-infection and cross-species transmission of divergent Hepatocystis lineages in a wild African primate community. Int J Parasitol. 2013;43:613–619. doi: 10.1016/j.ijpara.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimnell AR, Kraemer SM, Mukherjee S, Phippard DJ, Janes JH, Flamoe E, Su XZ, Awadalla P, Smith JD. Global genetic diversity and evolution of var genes associated with placental and severe childhood malaria. Mol Biochem Parasit. 2006;148:169–180. doi: 10.1016/j.molbiopara.2006.03.012. [DOI] [PubMed] [Google Scholar]
- van Duivenvoorde LM, Voorberg-van der Wel A, van der Werff NM, Braskamp G, Remarque EJ, Kondova I, Kocken CH, Thomas AW. Suppression of Plasmodium cynomolgi in rhesus macaques by coinfection with Babesia microti. Infect Immun. 2010;78:1032–1039. doi: 10.1128/IAI.00921-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhulst NO, Smallegange RC, Takken W. Mosquitoes as potential bridge vectors of malaria parasites from non-human primates to humans. Front Physiol. 2012;3:197. doi: 10.3389/fphys.2012.00197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verrelli BC, Tishkoff SA, Stone AC, Touchman JW. Contrasting histories of G6PD molecular evolution and malarial resistance in humans and chimpanzees. Mol Biol Evol. 2006;23:1592–1601. doi: 10.1093/molbev/msl024. [DOI] [PubMed] [Google Scholar]
- Volkman SK, Barry AE, Lyons EJ, Nielsen KM, Thomas SM, Choi M, Thakore SS, Day KP, Wirth DF, Hartl DL. Recent origin of Plasmodium falciparum from a single progenitor. Science. 2001;293:482–484. doi: 10.1126/science.1059878. [DOI] [PubMed] [Google Scholar]
- Voorberg-vd Wel A, Kocken CH, Zeeman AM, Thomas AW. Detection of new Babesia microti-like parasites in a rhesus monkey (Macaca mulatta) with a suppressed Plasmodium cynomolgi infection. Am J Trop Med Hyg. 2008;78:643–645. [PubMed] [Google Scholar]
- White NJ. In: Malaria, Manson's tropical diseases. 21st ed. Cook GC, Zumla A, editors. Saunders; London, UK: 2003. pp. 1205–1295. [Google Scholar]
- Whitworth J, Morgan D, Quigley M, Smith A, Mayanja B, Eotu H, Omoding N, Okongo M, Malamba S, Ojwiya A. Effect of HIV-1 and increasing immunosuppression on malaria parasitaemia and clinical episodes in adults in rural Uganda: a cohort study. Lancet. 2000;356:1051–1056. doi: 10.1016/s0140-6736(00)02727-6. [DOI] [PubMed] [Google Scholar]
- WHO [llast access: 2016];Malaria – Fact sheet #94 . 2015 available at: http://www.who.int/mediacentre/factsheets/fs094/en/index.html .
- Williams TN. Red blood cell defects and malaria. Mol Biochem Parasit. 2006;149:121–127. doi: 10.1016/j.molbiopara.2006.05.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This is a review article; therefore, no data were obtained.


