Meningococcal meningitis remains a life-threatening disease worldwide, with high prevalence in the sub-Saharan meningitis belt. A rapid diagnosis is crucial for implementing adapted antimicrobial treatment. We describe the performances of a new immunochromatographic test (MeningoSpeed, BioSpeedia, France) for detecting and grouping Neisseria meningitidis. Cerebrospinal fluids (CSFs) were collected from 5 African countries and France.
KEYWORDS: cerebrospinal fluid, Neisseria meningitidis, rapid diagnostic test, validation
ABSTRACT
Meningococcal meningitis remains a life-threatening disease worldwide, with high prevalence in the sub-Saharan meningitis belt. A rapid diagnosis is crucial for implementing adapted antimicrobial treatment. We describe the performances of a new immunochromatographic test (MeningoSpeed, BioSpeedia, France) for detecting and grouping Neisseria meningitidis. Cerebrospinal fluids (CSFs) were collected from 5 African countries and France. For the rapid diagnostic test (RDT), the CSF sample was deposited on each of the 3 cassettes for a total volume of 90 μl. The results of the RDT were compared to those of a reference multiplex PCR assay detecting the major serogroups of N. meningitidis on 560 CSF specimens. Five specimens were found uninterpretable by RDT (0.9%). The results of interpretable specimens were as follows: 305 positive and 212 negative samples by both techniques, 14 positive by PCR only, and 24 positive by RDT only (sensitivity, specificity, and positive and negative predictive values of 92.7%, 93.8%, 95.6%, and 89.8%, respectively, with an accuracy of 93.2% and a kappa test of 0.89; P < 0.05). From 319 samples positive by PCR for serogroups A, C, W, X, or Y, the grouping results were concordant for 299 specimens (sensitivity of 93.0%, 74.4%, 98.1%, 100%, and 83.3% for serogroups A, C, W, X, and Y, respectively). The MeningoSpeed RDT exhibited excellent performances for the rapid detection of N. meningitidis antigens. It can be stored at room temperature, requires a minimal amount of CSF, is performed in 15 minutes or less, and is easy to use at bedside.
INTRODUCTION
Neisseria meningitidis is a Gram-negative bacterium that infects only humans; it is one of the major agents of septic meningitis, with the potential to cause either outbreaks or sporadic cases worldwide. The “meningitis belt” of sub-Saharan Africa, involving 26 countries from Senegal in the west to Ethiopia in the east, exhibits the highest rates of the disease (1). However, sporadic and epidemic cases also occur everywhere in the world (2). Meningitis due to N. meningitidis is known to exhibit a severe prognosis, including frequent sequelae and life-threatening issues (3). There are 12 known serogroups of N. meningitidis, differentiated by the composition of the structure of their polysaccharide capsule, but only six of them (A, B, C, W, X, and Y) are commonly involved in human neurological infections. Five of these serogroups (A, B, C, W, and Y) are targets in currently licensed meningococcal vaccines (4).
Despite vaccination programs in some African countries, outbreaks due to N. meningitidis are still observed in Africa. Since the introduction of the MenAfriVac immunization program using a vaccine directed against N. meningitidis serogroup A, major epidemics involving this serogroup have nearly disappeared in Africa (5). However, other serogroups are emerging in the meningitis belt, and outbreaks involving C, W, or X serogroups (depending on the country) are now reported (2, 6). Ongoing surveillance for early detection of N. meningitidis epidemics is important, as illustrated by a recent study from the World Health Organization (WHO) Collaborating Centers for Reference Meningitis that showed that new strains of N. meningitidis can cause an epidemic at any time and everywhere (7).
WHO guidelines recommend the use of point-of-care tests, also named rapid diagnostic tests (RDTs), for the early etiological diagnosis of meningitis, notably in resource-limited countries (8). RDTs have been defined as tests able to give a result in the same visit as diagnosis (9) or that can be used in health care settings with limited infrastructure and nontrained personnel (10). Current RDTs aimed to detect N. meningitidis in resource-limited countries are mainly Gram staining, latex agglutination kits (11–13), immunochromatographic dipstick diagnostic test (14–18), and nucleic acid amplification tests (NAATs), notably those based on loop-mediated isothermal amplification methods due to high sensitivity, a rapid availability of results, and minimal specialized equipment and laboratory expertise (19–21).
Dipsticks using immunochromatography have been recently developed at the Institut Pasteur de Paris (IPP) in France to detect meningococci of serogroups A, C, W/Y, and X (14, 16, 18) that were subsequently transferred to BioSpeedia, which developed an improved prototype for industrial use. This report describes an innovative, rapid, and easy-to-use immunochromatographic RDT called MeningoSpeed (BioSpeedia, Paris, France). This test uses monoclonal antibodies directed against capsular polysaccharides belonging to A, C, W, X, and Y antigens of N. meningitidis. This study was aimed at evaluating this new test on many cerebrospinal fluid (CSF) specimens originating, in large part, from countries of the meningitis belt.
(These results were presented, in part, as a poster at the 28th European Congress of Clinical Microbiology and Infectious Diseases, Madrid, Spain, 21 to 24 April 2018 [22].)
MATERIALS AND METHODS
Population.
The studied population included patients consulting for clinical symptoms suspect of meningitis. The study was performed on 560 CSF specimens collected between 2006 and 2017 from 7 different centers located in 5 African countries and France, including 70 samples from the Centre de Recherche Médicale et Sanitaire, Niamey, Niger; 77 from the Pasteur Institute of Abidjan, Côte d’Ivoire; 29 from the Pasteur Institute of Bangui, République Centre-Africaine; 6 from the Pasteur Institute of Casablanca, Morocco; 90 from the Agence de Médecine Préventive-Togo of Dapaong, Togo; 72 from the University-Hospital of Saint-Etienne, France; and 216 from the IPP, France.
Study design.
The diagnosis strategy to perform the initial etiological diagnosis of meningitis in each country was left to the initiative of each center and is not described herein. The CSF samples that were evaluated in this study were remaining clinical specimens that were kept frozen at −80°C. The MeningoSpeed cassettes were distributed by the IPP to the other centers, in line with the mission of these centers for the surveillance of meningococcal diseases in the corresponding countries under approvals from the internal board of the IPP. The samples were anonymized before testing. In this study, the performances of the MeningoSpeed RDT were evaluated retrospectively by comparison to the results of an in-house multiplex PCR assay described below, also performed in each center, used as a reference standard.
Microbiological cultures.
Bacterial cultures were performed for some specimens (at least 266) by plating a 10-μl volume of CSF on appropriate media; the plates were incubated at 37°C under 5% CO2.
MeningoSpeed rapid diagnostic test.
The MeningoSpeed RDT (BioSpeedia, Paris, France) is a lateral flow immunochromatographic test composed of 3 cassettes. The first one detects A and W serogroups, the second one detects Y and C serogroups, and the third one detects the X serogroup. Each cassette comprises a control line for verifying that the sample was deposited correctly. One drop (approximately 30 μl) of CSF specimen was deposited in the sample well of each cassette with the help of a disposable pipette present in the kit. The result was read as soon as the bands appeared, for a maximum time of 15 min. In the absence of positivity of the control line, the test was declared uninterpretable.
PCR assays.
An in-house real-time PCR (RT-PCR) multiplex assay developed by the IPP (referenced as IPP RT-PCR assay further in the text) was used in each center for the detection of N. meningitidis DNA targeting the meningococcal-specific gene crgA coding for a LysR-like transcriptional regulator and targeting six serogroup-specific (A, B, C, W, X, and Y) N. meningitidis capsular genes. A Qiagen column-based method was used for the optimal extraction of DNA from CSF. Internal quality controls were included to monitor extraction of DNA and PCR inhibition, as previously described (23).
Additional PCR assays were performed for the detection of Haemophilus influenzae and Streptococcus pneumoniae DNA targeting hpd and lytA genes, respectively, according to formerly described techniques (24).
Statistical analysis.
Descriptive variables and sensitivity, specificity, and predictive values were reported with their 95% confidence interval (CI). These data, together with the Cohen’s kappa concordance coefficient, were calculated by using GraphPad Prism 5 software (CA, USA). P values under 5% were considered to be statistically significant.
RESULTS
Results combining culture and PCR techniques.
Five hundred and sixty CSF samples were included in the study. By combining conventional culture (performed on at least 288 samples) and the IPP RT-PCR assay (performed on all the samples), the following results were observed: 334 samples were positive for N. meningitidis (including 44 serogroup A, 10 serogroup B, 47 serogroup C, 161 serogroup W, 60 serogroup X, and 12 serogroup Y), 83 samples were positive for Streptococcus pneumoniae, 16 samples were positive for Haemophilus influenzae, 5 samples were positive for Streptococcus suis, and 122 samples were negative.
Performances of the MeningoSpeed RDT by comparison to the IPP RT-PCR assay.
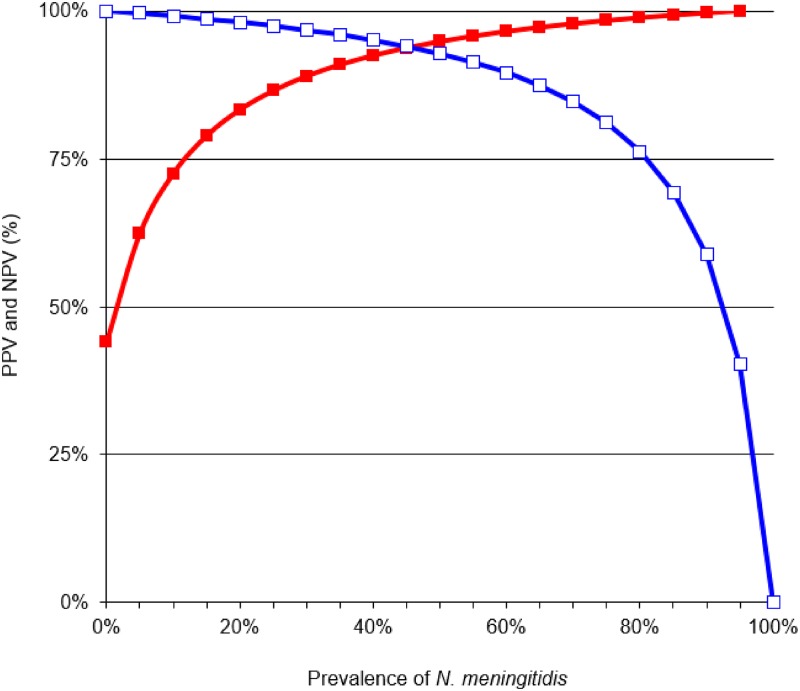
Five of the 560 specimens were found to be uninterpretable by RDT (0.9%). Table 1 depicts the comparative results between the MeningoSpeed RDT and the IPP RT-PCR assay taken as reference standard (23) on the 555 interpretable CSF specimens. The accuracy between the two tests was 93.2% (Cohen’s kappa test of 0.89, P < 0.05). The sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) of RDT were 0.927 (95% confidence interval [CI], 0.892 to 0.952), 0.938 (95% CI, 0.896 to 0.964), 0.956 (95% CI, 0.926 to 0.974), and 0.898 (95% CI, 0.851 to 0.932), respectively, by comparison to PCR. The variations of NPV and PPV for the diagnosis of N. meningitidis (serogroups A/C/W/Y/X) according to prevalence 0% to 100% are shown in Fig. 1.
TABLE 1.
Comparison of the results obtained by the rapid diagnostic test (RDT) and the IPP RT-PCR assay taken as reference standard on cerebrospinal fluids for the diagnosis of Neisseria meningitidis infection
| RDT result | No. of samples with PCR result: |
Total no. of samples | |
|---|---|---|---|
| + | − | ||
| + | 305 | 14 | 319 |
| − | 24 | 212 | 236 |
| Total | 329 | 226 | 555 |
FIG 1.
Positive (red line with filled squares) and negative predictive values (blue line with open squares) of the MeningoSpeed rapid diagnostic test for the detection of N. meningitidis in cerebrospinal fluid according to prevalence varying from 0% to 100%.
Analysis of discrepant results.
Table 2 gives the details of the 43 specimens that gave discrepant results by PCR and RDT (5 uninterpretable by RDT, 24 positive by PCR and negative by RDT, and 14 exhibiting the opposite pattern). Concerning false-positive results, they were observed in negative specimens as well as in specimens positive for another bacterium; in 8 of these 14 samples, the cross reactivity was observed with the W valence. Concerning false-negative results, it is worthwhile to note that the MeningoSpeed RDT does not include antigens targeting the B serogroup, which explains why the 10 samples found positive for this serogroup, all coming from CSF sampled in France, were all missed by the RDT.
TABLE 2.
Analysis of discrepant results between the PCR assays taken as reference standard and the MeningoSpeed rapid diagnostic test
| RDT result | Total no. of specimens | PCR assays results (no. of specimens) for: |
No. of specimens with negativea PCR result | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | W | X | Y | Streptococcus pneumoniae | Haemophilus influenzae | |||
| Uninterpretable | 5 | 1 | 0 | 0 | 2 | 2 | 0 | 0 | 0 | 0 |
| False negative | 24 | 2 | 10b | 8 | 3 | 0 | 1 | 0 | 0 | 0 |
| False positive | 14 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 3 | 9 |
Negative result by the PCR tests independently targeting Neisseria meningitidis, Streptococcus pneumoniae, and Haemophilus influenzae.
It was predicted that these samples would be negative since the RDT assay lacks the B antigen.
Correlation between MeningoSpeed RDT and IPP RT-PCR assay at the serogroup level.
After exclusion of the 10 samples found positive for serogroup B that was absent from the RDT, 319 CSF samples remained positive by PCR. From these 319 samples, 299 gave concordant results at the serogroup level by both techniques with the following distribution: 40 serogroup A, 35 C, 156 W, 58 X, and 10 Y. For 6 samples, the RDT detected N. meningitidis infection but with a wrong serogroup; specifically, 3 samples were identified as A by RDT but were typed C by PCR, and 3 other samples identified as X by RDT were typed A, C, and Y, respectively, by PCR. The 14 remaining samples positive by PCR were found negative by RDT. Table 3 illustrates the performances of the RDT for each serogroup; sensitivity was excellent for serogroups A, W, and X; acceptable for serogroup Y; and weak for serogroup C. The specificity and NPV were excellent for all the 5 serogroups (Table 3).
TABLE 3.
Performances of the MeningoSpeed RDT by comparison to the IPP RT-PCR assay used as reference standard for the serogroup determination of the 319 specimens found positive for Neisseria meningitis by PCRa
| Parameterb | Values by serogroup, determined by PCR |
||||
|---|---|---|---|---|---|
| A | C | W | X | Y | |
| No. of CSF+ samples by PCR | 43 | 47 | 159 | 58 | 12 |
| Sensitivity (95% CI) | 0.930 (0.799–0.981) | 0.744 (0.594–0.856) | 0.981 (0.942–0.945) | 1 (0.923–1) | 0.883 (0.509–0.970) |
| Specificity (95% CI) | 0.990 (0.976–0.996) | 1 (0.990–1) | 0.980 (0.960–0.990) | 0.986 (0.976–0.994) | 1 (0.991–1) |
| PPV (95% CI) | 0.889 (0.751–0.958) | 1 (0.877–1) | 0.951 (0.903–0.978) | 0.892 (0.784–0.952) | 1 (0.655–1) |
| NPV (95% CI) | 0.994 (0.982–0.998) | 0.977 (0.959–0.988) | 0.992 (0.976–0.998) | 1 (0.990–1) | 0.996 (0.985–0.999) |
| Cohen’s kappa coefficient (95% CI) | 0.901 (0.834–0.969) | 0.842 (0.755–0.930) | 0.952 (0.924–0.980) | 0.936 (0.889–0.983) | 0.907 (0.780–1) |
After exclusion of the 10 samples serogrouped as NmB, as this antigen was absent from the RDT.
PPV,: positive predictive value; NPV, negative predictive value.
DISCUSSION
The identification and serogrouping of N. meningitidis infection are important for disease control and vaccination strategies (25, 26). The availability of RDTs is important for the early diagnosis of meningitis due to this bacterium, notably in the case of outbreaks that frequently occur in the meningitis belt of sub-Saharan Africa (27). The MeningoSpeed RDT evaluated in this study answers to most of the diagnostic criteria required by WHO for limited-resource areas because it is easy to store (room temperature with stability of 2 years between +4°C and 30°C), perform, and read, without needing sophisticated equipment nor electricity; small amounts of CSF are required (less than 100 μl for the 3 cassettes); the result is available within 15 minutes; 5 of the 6 most commonly identified serogroups can be detected, which permits simultaneous identification and serogrouping with a good confidence; and, finally, the overall performances of the test are very satisfactory compared to a sensitive PCR assay taken as reference standard when tested on a large panel of CSF belonging to various serogroups (Tables 1 and 3). However, the risk of missing a positive sample (as illustrated by the overall NPV of 89.8% with regard to PCR assay) confirms that a negative result with this technique does not avoid further testing and the start of an empirical antimicrobial treatment if the clinical and biological context is evocative of purulent meningitis.
At the serogroup level, the analytical performances of the MeningoSpeed RDT were in good concordance with those of other rapid antigenic tests, as illustrated in Table 4 for sensitivity and specificity. Even if the comparison of results is not totally pertinent given the size and origin of the panels that were evaluated in these different studies, it is worthwhile to note that the performances of the MeningoSpeed RDT were at least equivalent to and often higher than those of former tests. The only exception concerned serogroup C for which a few samples were missed; the same moderate sensitivity for this serogroup was noticed in the study of Uadiale et al. conducted in Nigeria (13). These observations stressed once more that a negative result for N. meningitidis antigens in a CSF sample from a patient suspect with purulent meningitis does not discard the diagnosis of meningococcal infection and must be confirmed by additional microbiological investigations. In the future, an improvement in the quality of the antibodies used in the MeningoSpeed test may contribute to reduce the number of cross-reactions and of false-negative results.
TABLE 4.
Respective sensitivity and specificity of RDT by comparison to PCR as referenced in different studies of the literaturea
| Reference and yr | Type of RDT | No. of CSF samples | Origin of CSF samples | Serogroup | % Sensitivity (95%CI) | % Specificity (95% CI) |
|---|---|---|---|---|---|---|
| (11) 2006 | LA | 484 | Niger 2003 | A | 88 (85–91) | 93 (90–95) |
| (12) 2009 | LA | 126 | Niger 2006 | A | 69 (57–79) | 81 (68–91) |
| Dipstick | 89 (80–95) | 62 (48–75) | ||||
| (15) 2010 | Dipstick | 265 | Burkina Faso 2007 | A | 70.0 (55.4–82.1) | 97.0 (93.4–98.7) |
| (16) 2013 | Dipstick | 401 | France, Burkina Faso, Niger | A | 88 (81–92) | 99 (98–100) |
| (17) 2014 | Dipstick | 1,113b | Niger 2008–2012 | 87.6 (85.7–89.3) | 85.7 (83.1–87.2) | |
| 1,162c | Niger 2008–2012 | A/W | 92.5 (89.8–92.9) | 84.6 (82.0–86.9) | ||
| (18) 2015 | Dipstick | 369 | Cameroon, Côte d’Ivoire, Niger, France | X | 94 (86–98) | 100 (99–100) |
| (13) 2016 | LA | 63 | Nigeria 2013–2015 | C | 80.0 (65.4–90.4) | 94.4 (72.7–99.9) |
| This study 2019 | Dipstick | 560 | Niger, Côte d’Ivoire, République Centre-Africaine, Togo, France | A | 93.0 (79.9–98.1) | 99.0 (97.6–99.6) |
| C | 74.4 (59.4–85.6) | 100 (99.0–100) | ||||
| W | 98.1 (94.2–99.5) | 98.0 (96.0–99.0) | ||||
| X | 100 (92.3–100) | 98.6 (97.6–99.4) | ||||
| Y | 83.3 (50.9–97.0) | 100 (99.1–100) |
RDT, rapid diagnostic test; CSF, cerebrospinal fluid; LA, latex agglutination.
Tested in point of care.
Tested in reference laboratory.
We are aware that the present study has some limitations. First, its retrospective design is not compatible with a correct evaluation of the feasibility of the RDT in field condition. However, the technology of the MeningoSpeed RDT is very close to that already evaluated in points of care by other studies; the study of Collard et al. (17) performed in Niger on a large panel of samples is illustrative of the good concordance between results obtained with a dipstick system in the field and in a reference laboratory (see Table 4). A prospective in-field study is currently being conducted in Niger and Burkina Faso to validate the MeningoSpeed assay. Second, despite the very low proportion of uninterpretable results (0.9%), it is important to consider that all of them were found positive for N. meningitidis DNA by the IPP RT-PCR assay (Table 2), which means that such samples need imperatively to be tested by an alternative technique. Third, absence of the B serogroup in the MeningoSpeed panel led us to miss 10 samples found positive for this serogroup by the IPP RT-PCR assay. Although the NmB serogroup is less frequently involved than others in meningitis outbreaks occurring in Africa (all the B strains of this study were detected in CSF sampled in France), it was shown to be responsible for 29% of all the outbreaks observed in other continents (25). In order to fill the gap, we have the objective to add this valence to a future version of the MeningoSpeed RDT. Fourth, a few methodological aspects were neglected, including an intersite quality control in order to assess that the results of the different tests were reproducible from one site to another, or a blind reading of each RDT by two independent observers in order to minimize the misinterpretations due to eye reading. These limitations are linked to the difficulty of organizing a ground trial in 7 different centers, including 5 of them in African countries. It is obvious that these limitations may have underestimated the performances of the tested RDT. Finally, the eye reading of the result of the present RDT may introduce subjectivity in the interpretation of the test, notably when nontrained personnel is in charge of it; for the detection of other agents, and notably viruses, automatic readers are available, which improves substantially the traceability and the performances of such “digital immunoassays” (28). It could be interesting to propose this improvement for a future version of the test.
Despite these limitations, the test evaluated in this study brings together most of the qualities of a point-of-care RDT. In most cases, its performances can guarantee an accurate detection of N. meningitidis antigens and a correct serogrouping of the agent. The test is now in evaluation in large-scale studies performed in the African meningitis belt.
ACKNOWLEDGMENTS
BioSpeedia delivered the MeningoSpeed RDT kits for free. This company had no role in study design, data collection, interpretation of results, or the decision to submit the work for publication.
C.H.H. is a Ph.D. student of the University of Saint-Etienne whose thesis was cofunded by the CIFRE French Ministry of Higher Education and Research (CIFRE fellowship number 1283/2014) and BioSpeedia.
E.B. and Y.G. are working at BioSpeedia, a spin-off the Paris Pasteur Institute. The other authors have no conflict of interest to declare in relation with the matter of this study.
C.H.H. did some experiments, analyzed the results, and contributed to the writing of the paper. A.T., P.V., B.-M.N.-L., M.D., F.S., A.E.M., J.-P.L., A.R., E.H., and A.A. provided the results of the different centers involved in the trial and approved the final manuscript. E.B. and Y.G. designed the MeningoSpeed RDT and approved the final manuscript. B.P. contributed to the analysis of the results and wrote the manuscript. M.-K.T. conceived the design of the study, contributed to the analysis of results, and contributed to the writing of the manuscript. All the authors agreed with the revised manuscript.
REFERENCES
- 1.Harrison LH, Pelton SI, Wilder-Smith A, Holst J, Safadi MAP, Vazquez JA, Taha M-K, LaForce FM, von Gottberg A, Borrow R, Plotkin SA. 2011. The Global Meningococcal Initiative: recommendations for reducing the global burden of meningococcal disease. Vaccine 29:3363–3371. doi: 10.1016/j.vaccine.2011.02.058. [DOI] [PubMed] [Google Scholar]
- 2.Acevedo R, Bai X, Borrow R, Caugant DA, Carlos J, Ceyhan M, Christensen H, Climent Y, De Wals P, Dinleyici EC, Echaniz-Aviles G, Hakawi A, Kamiya H, Karachaliou A, Lucidarme J, Meiring S, Mironov K, Sáfadi MAP, Shao Z, Smith V, Steffen R, Stenmark B, Taha M-K, Trotter C, Vázquez JA, Zhu B. 2019. The Global Meningococcal Initiative meeting on prevention of meningococcal disease worldwide: epidemiology, surveillance, hypervirulent strains, antibiotic resistance and high-risk populations. Expert Rev Vaccines 18:15–30. doi: 10.1080/14760584.2019.1557520. [DOI] [PubMed] [Google Scholar]
- 3.Rosenstein NE, Perkins BA, Stephens DS, Popovic T, Hughes JM. 2001. Meningococcal disease. N Engl J Med 344:1378–1388. doi: 10.1056/NEJM200105033441807. [DOI] [PubMed] [Google Scholar]
- 4.McCarthy PC, Sharyan A, Sheikhi Moghaddam L. 2018. Meningococcal vaccines: current status and emerging strategies. Vaccines 6:E12. doi: 10.3390/vaccines6010012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daugla DM, Gami JP, Gamougam K, Naibei N, Mbainadji L, Narbé M, Toralta J, Kodbesse B, Ngadoua C, Coldiron ME, Fermon F, Page AL, Djingarey MH, Hugonnet S, Harrison OB, Rebbetts LS, Tekletsion Y, Watkins ER, Hill D, Caugant DA, Chandramohan D, Hassan-King M, Manigart O, Nascimento M, Woukeu A, Trotter C, Stuart JM, Maiden M, Greenwood BM. 2014. Effect of a serogroup A meningococcal conjugate vaccine (PsA-TT) on serogroup A meningococcal meningitis and carriage in Chad: a community study. Lancet 383:40–47. doi: 10.1016/S0140-6736(13)61612-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Soumahoro M-K, Kouamé-Elogne C, Anné J-C, Noufé S, N'Guessan KC, Kacou-N'Douba A, Hanslik T, Dosso M. 2018. Emergence of Neisseria meningitidis W135 in Côte d’Ivoire: laboratory based-surveillance. Epidemiol Health 40:e2018058. doi: 10.4178/epih.e2018058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Topaz N, Caugant DA, Taha MK, Brynildsrud OB, Debech N, Hong E, Deghmane AE, Ouédraogo R, Ousmane S, Gamougame K, Njanpop-Lafourcade BM, Diarra S, Fox LM, Wang X. 2019. Phylogenetic relationships and regional spread of meningococcal strains in the meningitis belt, 2011–2016. EBioMedicine 41:488–496. doi: 10.1016/j.ebiom.2019.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.World Health Organization. 2014. Revised guidance on meningitis outbreak response in sub-Saharan Africa. Wkly Epidemiol Rec 89:580–586. [PubMed] [Google Scholar]
- 9.Pai NP, Vadnais C, Denkinger C, Engel N, Pai M. 2012. Point-of-care testing for infectious diseases: diversity, complexity, and barriers in low- and middle-income countries. PLoS Med 9:e1001306. doi: 10.1371/journal.pmed.1001306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yansouni CP, Bottieau E, Lutumba P, Winkler AS, Lynen L, Büscher P, Jacobs J, Gillet P, Lejon V, Alirol E, Polman K, Utzinger J, Miles MA, Peeling RW, Muyembe JJ, Chappuis F, Boelaert M. 2013. Rapid diagnostic tests for neurological infections in central Africa. Lancet Infect Dis 13:546–558. doi: 10.1016/S1473-3099(13)70004-5. [DOI] [PubMed] [Google Scholar]
- 11.Borel T, Rose AM, Guillerm M, Sidikou F, Gerstl S, Djibo A, Nathan N, Chanteau S, Guerin PJ. 2006. High sensitivity and specificity of the Pastorex latex agglutination test for Neisseria meningitidis serogroup A during a clinical trial in Niger. Trans R Soc Trop Med Hyg 100:964–969. doi: 10.1016/j.trstmh.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 12.Rose AM, Gerstl S, Mahamane AE, Sidikou F, Djibo S, Bonte L, Caugant DA, Guerin PJ, Chanteau S. 2009. Field evaluation of two rapid diagnostic tests for Neisseria meningitidis serogroup A during the 2006 outbreak in Niger. PLoS One 4:e7326. doi: 10.1371/journal.pone.0007326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Uadiale K, Bestman A, Kamau C, Caugant DA, Greig J. 2016. Evaluation of Pastorex meningitis kit performance for the rapid identification of Neisseria meningitidis serogroup C in Nigeria. Trans R Soc Trop Med Hyg 110:381–385. doi: 10.1093/trstmh/trw046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chanteau S, Dartevelle S, Mahamane AE, Djibo S, Boisier P, Nato F. 2006. New rapid diagnostic tests for Neisseria meningitidis serogroups A, W135, C, and Y. PLoS Med 3:e337. doi: 10.1371/journal.pmed.0030337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rose AM, Mueller JE, Gerstl S, Njanpop-Lafourcade BM, Page AL, Nicolas P, Traoré RO, Caugant DA, Guerin PJ. 2010. Meningitis dipstick rapid test: evaluating diagnostic performance during an urban Neisseria meningitidis serogroup A outbreak, Burkina Faso, 2007. PLoS One 5:e11086. doi: 10.1371/journal.pone.0011086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Terrade A, Collard J-M, Nato F, Taha M-K. 2013. Laboratory evaluation of a rapid diagnostic test for Neisseria meningitidis serogroup A. Trans R Soc Trop Med Hyg 107:460–461. doi: 10.1093/trstmh/trt041. [DOI] [PubMed] [Google Scholar]
- 17.Collard J-M, Wang X, Mahamane AE, Idi I, Issaka B, Ousseni M, Mayer LW, Nato F, Moulia-Pelat J-P. 2014. A five-year field assessment of rapid diagnostic tests for meningococcal meningitis in Niger by using the combination of conventional and real-time PCR assays as a gold standard. Trans R Soc Trop Med Hyg 108:6–12. doi: 10.1093/trstmh/trt104. [DOI] [PubMed] [Google Scholar]
- 18.Agnememel A, Traincard F, Dartevelle S, Mulard L, Mahamane AE, Oukem-Boyer OOM, Denizon M, Kacou-N’douba A, Dosso M, Gake B, Lombart J-P, Taha M-K. 2015. Development and evaluation of a dipstick diagnostic kit for Neisseria meningitidis serogroup X. J Clin Microbiol 53:449–454. doi: 10.1128/JCM.02444-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McKenna JP, Fairley DJ, Shields MD, Cosby SL, Wyatt DE, McCaughey C, Coyle PV. 2011. Development and clinical validation of a loop-mediated isothermal amplification method for the rapid detection of Neisseria meningitidis. Diagn Microbiol Infect Dis 69:137–144. doi: 10.1016/j.diagmicrobio.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 20.Lee D, Kim EJ, Kilgore PE, Kim SA, Takahashi H, Ohnishi M, Anh DD, Dong BQ, Kim JS, Tomono J, Miyamoto S, Notomi T, Kim DW, Seki M. 2015. Clinical evaluation of a loop-mediated isothermal amplification (LAMP) assay for rapid detection of Neisseria meningitidis in cerebrospinal fluid. PLoS One 10:e0122922. doi: 10.1371/journal.pone.0122922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seki M, Kilgore PE, Kim EJ, Ohnishi M, Hayakawa S, Kim DW. 2018. Loop-mediated isothermal amplification methods for diagnosis of bacterial meningitis. Front Pediatr 6:57. doi: 10.3389/fped.2018.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haddar C, Terrade A, Verhoeven P, Begaud E, Hamon S, Rakoto Andrianarivelo M, Grattard F, Paranhos-Baccalà G, Bizet C, Pozzetto B, Germani Y, Taha M. 2018. Sensitive rapid detection and typing of Neisseria meningitidis in cerebrospinal fluid (CSF) using an innovative rapid diagnostic test (RDT), poster 0849. Abstr 28th Eur Congr Clin Microbiol Infect Dis, Madrid, Spain, 21 to 24 April 2018. [Google Scholar]
- 23.Deghmane A-E, Hong E, Taha M-K. 2019. Diagnosis of meningococcal infection using internally controlled multiplex real-time PCR. Methods Mol Biol 1969:17–31. doi: 10.1007/978-1-4939-9202-7_2. [DOI] [PubMed] [Google Scholar]
- 24.World Health Organization. 2011. PCR for detection and characterization of bacterial meningitidis pathogens: Neisseria meningitidis, Haemophilus influenza, and Streptococcus pneumoniae, p 105–156. In World Health Organization (ed), Laboratory methods for the diagnosis of meningitis caused by Neisseria meningitidis, Streptococcus pneumoniae, and Haemophilus influenza, 2nd ed, WHO/IVB.11.09 World Health Organization, Geneva, Switzerland: https://www.cdc.gov/meningitis/lab-manual/full-manual.pdf. [Google Scholar]
- 25.van Kessel F, van den Ende C, Oordt-Speets AM, Kyaw MH. 2019. Outbreaks of meningococcal meningitis in non-African countries over the last 50 years: a systematic review. J Glob Health 9:010411. doi: 10.7189/jogh.09.010411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bai X, Borrow R, Bukovski S, Caugant DA, Culic D, Delic S, Dinleyici EC, Eloshvili M, Erdősi T, Galajeva J, Křížová P, Lucidarme J, Mironov K, Nurmatov Z, Pana M, Rahimov E, Savrasova L, Skoczyńska A, Smith V, Taha MK, Titov L, Vázquez J, Yeraliyeva L. 2019. Prevention and control of meningococcal disease: updates from the Global Meningococcal Initiative in Eastern Europe. J Infect 79:528–541. doi: 10.1016/j.jinf.2019.10.018. [DOI] [PubMed] [Google Scholar]
- 27.Patel JC, Soeters HM, Diallo AO, Bicaba BW, Kadadé G, Dembélé AY, Acyl MA, Nikiema C, Lingani C, Hatcher C, Acosta AM, Thomas JD, Diomande F, Martin S, Clark TA, Mihigo R, Hajjeh RA, Zilber CH, Aké F, Mbaeyi SA, Wang X, Moisi JC, Ronveaux O, Mwenda JM, Novak RT, MenAfriNet Consortium. 2019. MenAfriNet: a network supporting case-based meningitis surveillance and vaccine evaluation in the meningitis belt of Africa. J Infect Dis 220:S148–S154. doi: 10.1093/infdis/jiz308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Merckx J, Wali R, Schiller I, Caya C, Gore GC, Chartrand C, Dendukuri N, Papenburg J. 2017. Diagnostic accuracy of novel and traditional rapid tests for influenza infection compared with reverse transcriptase polymerase chain reaction: a systematic review and meta-analysis. Ann Intern Med 167:394–409. doi: 10.7326/M17-0848. [DOI] [PubMed] [Google Scholar]