The emergence of drug resistance in Helicobacter pylori has resulted in a greater need for susceptibility-guided treatment. While the alleles associated with resistance to clarithromycin and levofloxacin have been defined, there are limited data regarding the molecular mechanisms underlying resistance to other antimicrobials. Using H. pylori isolates from 42 clinical specimens, we compared phenotypic and whole-genome sequencing (WGS)-based detection of resistance.
KEYWORDS: antimicrobial susceptibility testing, Helicobacter pylori, whole-genome sequencing, amoxicillin, clarithromycin, levofloxacin, lineage, metronidazole, rifampin, tetracyclines
ABSTRACT
The emergence of drug resistance in Helicobacter pylori has resulted in a greater need for susceptibility-guided treatment. While the alleles associated with resistance to clarithromycin and levofloxacin have been defined, there are limited data regarding the molecular mechanisms underlying resistance to other antimicrobials. Using H. pylori isolates from 42 clinical specimens, we compared phenotypic and whole-genome sequencing (WGS)-based detection of resistance. Phenotypic resistance correlated with the presence of alleles of 23S rRNA (A2142G/A2143G) for clarithromycin (kappa coefficient, 0.84; 95% confidence interval [CI], 0.67 to 1.0) and gyrA (N87I/N87K/D91Y/D91N/D91G/D99N) for levofloxacin (kappa coefficient, 0.90; 95% CI, 0.77 to 1.0). Phenotypic resistance to amoxicillin in three isolates correlated with mutations in pbp1, pbp2, and/or pbp3 within coding regions near known amoxicillin binding motifs. All isolates were phenotypically susceptible to tetracycline, although four bore a mutation in 16S rRNA (A926G). For metronidazole, nonsense mutations and R16H substitutions in rdxA correlated with phenotypic resistance (kappa coefficient, 0.76; 95% CI, 0.56 to 0.96). Previously identified mutations in the rpoB rifampin resistance-determining region (RRDR) were not present, but 14 novel mutations outside the RRDR were found in rifampin-resistant isolates. WGS also allowed for strain lineage determination, which may be important for future studies in associating precise MICs with specific resistance alleles. In summary, WGS allows for broad analyses of H. pylori isolates, and our findings support the use of WGS for the detection of clarithromycin and levofloxacin resistance. Additional studies are warranted to better define mutations conferring resistance to amoxicillin, tetracycline, and rifampin, but combinatorial analyses for rdxA gene truncations and R16H mutations have utility for determining metronidazole resistance.
INTRODUCTION
Helicobacter pylori colonizes approximately half of the population globally and causes gastritis in nearly all infected individuals (1). In some, infection progresses, resulting in dyspepsia, the formation of gastric and duodenal ulcers, mucosa-associated lymphoid tissue lymphoma, and gastric cancer. In one study conducted in Japan, 2.9% of patients with gastric ulcers, dyspepsia, or gastric hyperplasia in conjunction with H. pylori infection developed gastric cancer (2). H. pylori is the only bacterium classified as a type 1 carcinogen, with pathogen eradication resulting in a reduction of cancer risk (3–5). Anti-H. pylori therapy involves multidrug regimens that consist of at least two antibiotics and proton pump inhibitors (PPI) with or without bismuth (3, 5).
Treatment of primary H. pylori infections is largely empirical, but susceptibility-guided therapy is recommended after initial treatment failures or if the local level of resistance to clarithromycin or metronidazole is greater than 15% (5). Two meta-analyses that included 17 randomized controlled trials revealed that susceptibility-guided treatment is superior to empirical therapy with two antibiotics and a PPI (6, 7). However, phenotypic susceptibility testing for H. pylori is not offered by the majority of North American hospital laboratories due to challenges in performing the assay. H. pylori culture and susceptibility testing can take weeks because of the slow growth and fastidious nature of the organism (3).
Molecular detection of H. pylori drug resistance may provide a more rapid and amenable means for susceptibility determination. The results of real-time PCR and DNA hybridization-based assays have been shown to correlate with phenotypic results for the detection of point mutations mediating clarithromycin (93 to 97% concordance) (8–13) and levofloxacin (80 to 92% concordance) (10, 11) resistance, and the findings from targeted sequencing of the H. pylori 23S rRNA gene also show a high concordance with phenotypic results for clarithromycin (14). Molecular methods can be superior to phenotypic methods for the detection of subpopulations of resistant strains, and sequencing offers an advantage over PCR-based assays since all known mutations within a targeted gene or whole genome can be assessed (15). Recently, the agreement between whole-genome sequencing (WGS) and Etest results for clarithromycin, levofloxacin, and rifampin was found to be strong; however, the study found no agreement for metronidazole (16).
In this study, we performed WGS of H. pylori isolates originating from 42 clinical specimens to determine the strain lineage in our local population of isolates and to compare WGS-mediated resistance detection to phenotypic Etest results for all antimicrobials with Clinical and Laboratory Standards Institute (CLSI) or European Committee for Antimicrobial Susceptibility Testing (EUCAST) breakpoints. Additionally, WGS led to the identification of novel mutations that have not previously been associated with drug resistance.
MATERIALS AND METHODS
H. pylori isolates.
H. pylori isolates were recovered from gastric biopsy specimens submitted to the clinical microbiology laboratory at Montefiore Medical Center, Bronx, NY, for routine patient care between 14 May 2018 and 8 May 2019. Routine testing consisted of culture and phenotypic susceptibility testing for clarithromycin, amoxicillin, levofloxacin, and tetracycline. Isolates, which consisted of multiple colonies, were then stored in CryoSaver brucella broth with glycerol- and bead-containing tubes (Hardy Diagnostics) at −80°C. The whole genomes of isolates that remained viable after freezing were sequenced, and phenotypic susceptibility testing for rifampin and metronidazole was performed for research purposes only. Routine susceptibility data and the corresponding patient demographics were retrieved retrospectively from the electronic medical records. Primary infection was defined when no previous documentation in the notes or previous test results suggested H. pylori infection. The testing reviewed included the results of H. pylori stool antigen and rapid urease point-of-care tests and cultures, prescription history, and pathology reports. Only the first isolate for each patient recovered during the study period was included. This study was approved by the Albert Einstein College of Medicine Institutional Review Board (IRB) (approval number 2018-9702). The need for informed consent was waived by the IRB since loss to follow-up of patients was expected, given the use of remnant isolates.
Phenotypic antimicrobial susceptibility testing.
Phenotypic susceptibility testing was performed using 3- to 4-day old cultures of H. pylori when sufficient growth was obtained to prepare a no. 2 McFarland suspension (17). H. pylori clinical isolates and quality control strain ATCC 43504 were inoculated into Mueller-Hinton blood agar plates containing 5% sheep blood (BD BBL, Sparks, MD), and Etests (bioMérieux, France) were used for determination of the MIC after 72 to 96 h of incubation at 37°C under microaerophilic conditions. CLSI M45 standard (3rd edition) breakpoints (17) were used to determine susceptibility to clarithromycin. Interpretations for other antibiotics were based on EUCAST 2019 (version 9.0) guidelines, solely for the purposes of this study.
Isolation of genomic DNA and whole-genome sequencing.
H. pylori clinical isolates were subcultured on blood agar plates for genomic DNA isolation. Three plates were used for each isolate after 3 to 4 days of incubation under microaerophilic conditions at 37°C. The isolates were resuspended in sterile saline solution and pelleted. Genomic DNA was prepared using a PureLink genomic DNA isolation kit (Invitrogen Inc.) following the manufacturer’s instructions. DNA quality was assessed using a NanoDrop One spectrophotometer (Thermo Scientific, Inc). All DNA quantifications were done with a Qubit double-stranded DNA high-sensitivity assay kit (Invitrogen Inc). A paired-end genomic library was prepared using a Nextera XT library preparation kit (Illumina, San Diego, CA) per the manufacturer’s instructions. Following cleanup using AMPure XP magnetic beads (Beckman Coulter, USA), genomic libraries were pooled in equimolar concentrations (2 nM each) with 20 pM bacteriophage phiX control, and sequencing was performed using a MiSeq reagent kit (v3; 150 cycles; 2 × 75 bp) on an Illumina MiSeq platform (Illumina, Inc.). The quality of the read files was evaluated using the FastQC (v0.11.7) program (18). The read files and adaptors were trimmed using the Trim Galore (v0.6.1) and Cutadapt (v2.2) programs. Sequence coverage for all the isolates was assessed using a MiSeq reporter by keeping H. pylori 26695 as the reference. Coverage ranged from 28 to 210 times, with an average depth of 80 times.
Genome assembly and annotation.
Short read assembly was performed using the SPAdes algorithm (19) at the Pathosystems Resource Integration Center (PATRIC) online server (https://www.patricbrc.org/) (20). The number of contigs obtained after assembly ranged from 33 to 72, with the exception of the number for one isolate (isolate MHP39), for which 188 contigs were obtained. The assembled contigs were annotated by the RASTtk algorithm (21) using the PATRIC annotation server with default parameters. Genome annotations revealed the presence of 1,573 to 1,702 open reading frames encoding proteins and RNAs among the sequenced clinical isolates. The pangenome of the sequenced isolates constituted 2,031 genes, while the core genome consisted of 1,177 genes. Quality metrics data for all sequenced genomes including coverage, genome length, and N50 and L50 values, which are detailed in Data Set S1 in the supplemental material. H. pylori isolates from 42 clinical specimens were used in this study; however, it was found upon subculture of the frozen clinical isolates that one specimen (MHP9) consisted of two H. pylori strains that were sequenced independently (MHP9C and MHP9L).
WGS and antimicrobial resistance analyses.
In order to identify drug resistance mutations, sequence data corresponding to the 23S rRNA gene for clarithromycin, gyrA and gyrB for levofloxacin, pbp1, pbp2, and pbp3 for amoxicillin, rdxA, frxA, fdxB, mdaB, omp11, and rpsU for metronidazole, the 16S rRNA gene for tetracycline, and rpoB for rifampin were retrieved from the annotated genomes and analyzed by performing a multiple-sequence alignment with the MUSCLE program using MEGA (v10.0.5) and Geneious R11 software and by keeping a sensitive strain (H. pylori 26695) as the reference (22). Defined drug resistance mutations were used to determine the correlation between phenotypic and genotypic testing results (23). The presence of novel variants associated with phenotypic resistance was evaluated separately. Isolates with discrepant phenotype and sequence results were examined for allele variations by read mapping trimmed short reads to the H. pylori 26695 sequence using the “map to reference” option with a threshold of 95 in Geneious R11 software. We observed a depth of at least 200 times to a maximum of 1,200 times for all drug resistance mutations.
Epidemiology and lineage analyses.
Multilocus sequence typing (MLST) of all isolates was performed using a web-based MLST tool hosted by the Center for Genetic Epidemiology, Denmark (http://www.genomicepidemiology.org). A phylogenetic tree was constructed using PATRIC’s Codon Tree service, which uses the amino acid and nucleotide sequences from a defined number of PATRIC’s global protein families (24). The protein families, known as PGFams, were randomly picked to build an alignment and then to generate a tree based on the differences within those selected sequences. Both the protein (amino acid) and gene (nucleotide) sequences were used for each of the selected genes from the PGFams. Protein sequences were aligned using the MUSCLE program (22), and the nucleotide sequences encoding the genes were aligned using the Codon align function of the Biopython package (25). A concatenated alignment of all proteins and nucleotides was written to a Phylip-formatted file, and then a partitions file for the RAxML program (26) describing the alignment in terms of the proteins and then the first, second, and third codon positions was generated. This Phylip file was used as an input file for FigTree (v1.4.4) software to build an unrooted phylogenetic tree. Support values were generated using 100 rounds of the Rapid bootstrapping option (27) of the RAxML program. Sequences from 512 genes (128,869 amino acids and 386,361 nucleotides) were used to generate the tree, which included 109 genomes (Data Sets S2 and S3). These included the 43 H. pylori genomes sequenced in this study and 66 additional genomes that were described by Kumar et al. (28).
Statistical analyses.
Cohen’s kappa coefficient was used to assess the agreement between phenotypic resistance and drug resistance mutations. A kappa coefficient value of <0.4 was considered low agreement, a value of 0.4 to 0.6 was considered moderate agreement, a value of 0.61 to 0.8 was considered substantial agreement, and a value of 0.81 to 1.0 was considered nearly perfect or perfect agreement. SAS (v9.4) software (SAS Institute Inc., Cary, NC) was used to estimate kappa coefficient values. Prism (v7) software was used to perform Fisher’s exact test for contingency analyses, followed by the Bonferroni-Dunn test to correct for multiple comparisons. A P value of ≤0.05 was considered statistically significant.
Data availability.
All 43 sequenced genomes of the H. pylori strains isolated from 42 clinical specimens in this study were assembled, annotated, and submitted to NCBI BioProject database under BioProject accession number PRJNA566177.
RESULTS
Patient characteristics.
H. pylori clinical isolates (n = 54) recovered from patient biopsy specimens between 14 May 2018 and 8 May 2019 were stored for this study. Isolates from 42 specimens were recoverable after storage (n = 42, 79%), and the corresponding patient demographics and clinical characteristics were retrieved for analyses. For this study population in the Bronx, NY, the majority of isolates were from adults, but 21% percent of the isolates were from pediatric patients under the age of 21 years (Table 1). Most of the isolates were from females (76%), and the most common race recorded in the medical record was “other” (38%), which likely reflects those that identify as being of Hispanic or Latino heritage in the Bronx. Review of the medical records revealed that nearly half (48%) of the isolates were from patients with a documented previous H. pylori infection. For patients with a corresponding histopathology evaluation, most exhibited active, chronic gastritis in the antrum and/or fundus (74%) without evidence of progression to intestinal metaplasia. The most common antibiotic combinations for empirical therapy or for treatment of the most recently documented H. pylori infection prior to specimen collection (n = 23) were amoxicillin in combination with clarithromycin (n = 10) followed by amoxicillin in combination with levofloxacin (n = 5) (Table 1).
TABLE 1.
Demographics and clinical characteristics of patients with sequenced H. pylori isolatesa
| Characteristic | Value |
|---|---|
| Mean age (yr) | 56 |
| No. (%) of patients ages (yr): | |
| <21 | 9 (21) |
| >21 | 33 (79) |
| No. (%) of patients by gender | |
| Female | 32 (76) |
| Male | 10 (24) |
| No. (%) of patients by race | |
| Black | 13 (31) |
| Asian | 3 (7) |
| White | 2 (5) |
| Other | 16 (38) |
| Unavailable or declined | 8 (19) |
| No. (%) of patients by infection status | |
| Primary | 22 (52) |
| Secondary or tertiary | 20 (48) |
| No. (%) of patients by histopathology finding | |
| Evaluation performed | 31 (74) |
| Helicobacter-associated inactive, chronic gastritis | 1 (3) |
| Helicobacter-associated active, chronic gastritis | 23 (74) |
| Helicobacter-associated active, chronic gastritis with focal intestinal metaplasia | 7 (23) |
| No. (%) of patients by antibiotic used in H. pylori treatment regimens prior to biopsy | 23 (55) |
| Amoxicillin and clarithromycin | 10 (44) |
| Amoxicillin and levofloxacin | 5 (22) |
| Amoxicillin and doxycycline or tetracycline | 4 (17) |
| Clarithromycin and doxycycline | 2 (9) |
| Doxycycline, levofloxacin, and metronidazole | 1 (4) |
| Moxifloxacin and rifabutin | 1 (4) |
Data are for a total of 42 patients.
Phenotypic antimicrobial resistance of H. pylori isolates.
The most common phenotypic resistance observed was to levofloxacin (57% of isolates), metronidazole (45%), and clarithromycin (31%). In contrast, phenotypic resistance to amoxicillin was rare (7%), and none of the isolates were found to be phenotypically resistant to tetracycline. Using EUCAST guidelines for susceptibility to rifampin (in which an MIC of ≤1 μg/ml indicates susceptibility), 19% of our isolates were resistant. A recent study has suggested that an MIC of ≤4 μg/ml for susceptibility is more appropriate (29), and using this higher breakpoint, 7% of the isolates were rifampin resistant. Coresistance to both levofloxacin and clarithromycin was observed in 21% of the isolates. Resistance to clarithromycin was more common in isolates obtained from patients with a previous documented H. pylori infection (45% among isolates from patients with secondary/tertiary infections versus 18% among isolates from patients with primary infections), but the association was not statistically significant (P = 0.1). Resistance to levofloxacin was similar for isolates from patients with previous and primary infections (60% versus 50%, P = 0.6).
Genotypic determination of antimicrobial resistance.
The sequences of drug resistance-related genes were retrieved from the annotated genomes to identify potential drug resistance mutations. Mutations in genes previously reported to be associated with amoxicillin, tetracycline, clarithromycin, levofloxacin, metronidazole, and rifampin resistance were examined, and the correlation with the phenotypic results was determined (23, 30, 31). Three isolates were phenotypically resistant to amoxicillin (MHP11, MHP12, MHP22), and all exhibited point mutations in penicillin binding protein genes pbp1 (N107R, A201V, V250I, S543T), pbp2 (I259T), and/or pbp3 (D2N, A50S, F490Y, A541T, V374I) that were not present in susceptible isolates (Table 2). Although none of the amino acid substitutions were located in regions previously reported to confer amoxicillin resistance, such as SXN/SXXK/KTG in Pbp1A (32), all three isolates exhibited at least one substitution near an SXN/SXXK/KTG motif in Pbp1A, Pbp2, or Pbp3 (see Table S1 in the supplemental material). All isolates were tetracycline susceptible phenotypically, but four isolates (MHP28, MHP32, MHP38, MHP44) had point mutations in the 16S rRNA gene that are associated with resistance (A926G) (33). The 23S rRNA mutations A2142G and A2143G correlated with phenotypic Etest results for clarithromycin (kappa coefficient, 0.84; 95% confidence interval [CI], 0.67 to 1.0) (Table 3). All 13 isolates phenotypically clarithromycin resistant demonstrated point mutations in 23S rRNA (A2142G/A2143G). However, we also identified three isolates that were resistant by WGS but that were phenotypically sensitive (MHP28, MHP34, MHP50) (Table 2). None of our isolates harbored the A2142C, A2115G, G2141A, C2147G, or T2190C mutation. These mutations have previously been associated with clarithromycin resistance (12, 34). We did observe the C2195T mutation in three isolates (MHP41, MHP50, MHP52), among which MHP41 and MHP50 were sensitive phenotypically, and MHP50 and MHP52 had the A2142G mutation. We did not consider this mutation in our analyses, as previous studies have found it to be associated with resistance only when it is present in combination with other known clarithromycin resistance-associated mutations (35, 36). Twenty-three isolates were phenotypically resistant to levofloxacin, and resistance mutations in the quinolone resistance-determining region (QRDR) in gyrA (N87I/N87K/D91Y/D91N/D91G/D99N) were found in 22 isolates. Isolate MHP11 carried the R484K mutation in gyrB, in addition to a gyrA mutation. For one phenotypically resistant isolate (MHP35), mutations in gyrA or gyrB were not identified (Table 2). Agreement between methods was strong for levofloxacin (kappa coefficient, 0.9; 95% CI, 0.77 to 1.0) (Table 3). In addition, we identified heteroresistant strains for one of our isolates (MHP9), wherein one strain was resistant to clarithromycin (MHP9C) and the other was resistant to levofloxacin (MHP9L) (Table 2).
TABLE 2.
H. pylori isolate Etest MIC values and resistance mutations/genes for amoxicillin, clarithromycin, levofloxacin and tetracycline, metronidazole and rifampina
| SId no. | Isolate identifier | MIC (μg/ml) |
Mutation for resistance to: |
Infection typec | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMX | CLR | LVX | TET | MET | RIF | AMX (pbp1A, pbp2, pbp3) | CLR (23S rRNA) | LVX (gyrA) | TET (16S rRNA) | MET (rdxA) | RIF (rpoB) | |||
| 1 | MHP1 | 0.016 | 0.032 | 0.064 | 0.032 | 0.38 | 1 | P | ||||||
| 2 | MHP2 | 0.023 | 0.094 | 0.047 | 0.023 | 0.047 | 0.125 | R | ||||||
| 3 | MHP3 | 0.016 | 0.023 | 0.023 | 0.032 | 0.5 | 0.5 | P | ||||||
| 4 | MHP4 | 0.032 | 12 | <32 | 0.064 | 0.19 | 1 | A2143G | N87K | R | ||||
| 5 | MHP5 | 0.016 | 0.016 | 0.006 | 0.023 | 0.032 | 0.19 | P | ||||||
| 6 | MHP6 | 0.016 | 0.023 | 0.016 | 0.032 | 0.25 | 0.50 | P | ||||||
| 7 | MHP8 | 0.016 | 0.016 | >32 | 0.125 | >256 | 0.5 | N87K | R16H | P | ||||
| 8 | MHP9b | 0.032 | >256 | >32 | 0.064 | >256 | 1 | A2143G | N87K | 75 stop | R | |||
| 9 | MHP10 | 0.064 | 3 | >32 | 0.032 | >256 | A2143G | N87K | 50 stop | R | ||||
| 10 | MHP11 | 256 | 0.016 | >32 | 0.25 | 0.75 | 0.5 | pbp1A, I259T; pbp3, D2N, A50S, F490Y, A541T | D99N | P | ||||
| 11 | MHP12 | 0.19 | >256 | >32 | 0.032 | 0.75 | 0.5 | pbp2, T498S | A2143G | N87I | 112 stop | R | ||
| 12 | MHP13 | 0.064 | 0.016 | 0.125 | 0.5 | >256 | 0.19 | 112 stop | R | |||||
| 13 | MHP15 | 0.032 | 0.023 | 0.125 | 0.5 | 0.19 | 2 | 112 stop | R | |||||
| 14 | MHP16 | 0.064 | 24 | >32 | 0.032 | >256 | 0.38 | A2143G | D91Y | 197 stop | P | |||
| 15 | MHP17 | 0.047 | 0.023 | >32 | 0.064 | >256 | >32 | N87I | 119 stop | K2691R | R | |||
| 16 | MHP18 | 0.023 | 0.032 | 0.125 | 0.023 | >256 | 3 | 76 stop | R | |||||
| 17 | MHP19 | 0.032 | >256 | 0.19 | 0.047 | 1.0 | 0.5 | A2143G | P | |||||
| 18 | MHP20 | 0.016 | 0.016 | >32 | 0.125 | >256 | 0.5 | N87K | P | |||||
| 19 | MHP22 | 0.25 | 0.125 | 0.25 | 0.125 | 0.125 | 0.38 | pbp1A, N107R, A201V, V250I, S543T; pbp3, V374I | P | |||||
| 20 | MHP23 | 0.016 | >256 | >32 | 0.125 | 0.016 | 4 | A2143G | N87I | D221N, F971Y, K1229N, V2664M | P | |||
| 21 | MHP28 | 0.023 | 0.016 | 0.047 | 0.094 | >256 | 0.38 | A2143G | A926G | R16H, H99R | R | |||
| 22 | MHP29 | 0.016 | 0.047 | >32 | 0.19 | 0.064 | 0.5 | N87I | P | |||||
| 23 | MHP30 | 0.023 | 0.032 | 0.094 | 0.125 | 0.125 | 0.125 | P | ||||||
| 24 | MHP31 | 0.016 | >256 | 8 | 0.023 | 0.38 | 2 | A2143G | D91N | R | ||||
| 25 | MHP32 | 0.094 | 0.125 | >32 | 0.75 | >256 | 3 | N87I | A926G | R | ||||
| 26 | MHP33 | 0.016 | 0.064 | >32 | 0.125 | 0.032 | 1 | N87I | R | |||||
| 27 | MHP34 | 0.016 | 0.047 | >32 | 0.19 | >256 | 1.5 | A2143G | D91N | R16H | R | |||
| 28 | MHP35 | 0.016 | 0.016 | >32 | 0.094 | 0.125 | 1 | P | ||||||
| 29 | MHP36 | 0.023 | >256 | 0.047 | 0.032 | >256 | 1 | A2142G | R16H, V57A | R | ||||
| 30 | MHP37 | 0.032 | 3 | 0.094 | 0.19 | >256 | 1.5 | A2143G | N14T, R16H, W209Q, L210N | P | ||||
| 31 | MHP38 | 0.047 | <0.016 | 4 | 0.094 | 0.094 | 0.125 | D91G | A926G | 53 stop | P | |||
| 32 | MHP39 | 0.016 | 0.047 | >32 | 0.032 | 0.38 | 1.5 | N87I | P | |||||
| 33 | MHP40 | 0.016 | >256 | >32 | 0.023 | >256 | 3 | A2142G | D91Y | 143 stop | R984C, T1042A, T1965M, N2219S | R | ||
| 34 | MHP41 | 0.016 | 0.064 | 0.125 | 0.023 | 0.016 | 0.5 | P | ||||||
| 35 | MHP42 | 0.064 | 0.023 | >32 | 0.064 | 0.032 | 12 | N87I | M415V, I2021V | P | ||||
| 36 | MHP43 | 0.023 | 0.016 | >32 | 0.064 | 0.016 | >32 | N87I | I125T, T897A, Q1010R | P | ||||
| 37 | MHP44 | 0.016 | 0.047 | >32 | 0.064 | >256 | 0.38 | N87I | A926G | R16H | R | |||
| 38 | MHP47 | 0.016 | 0.50 | 0.064 | 0.023 | >256 | 1 | 75 stop | R | |||||
| 39 | MHP50 | 0.047 | 0.016 | 0.094 | 0.125 | >256 | 1 | A2142G | N87K | 72 stop | P | |||
| 40 | MHP51 | 0.023 | 0.016 | 0.125 | 0.047 | 0.19 | 0.5 | P | ||||||
| 41 | MHP52 | 0.047 | >256 | >32 | 0.047 | >256 | 2 | A2142G | N87K | 72 stop | R | |||
| 42 | MHP54 | 0.023 | >256 | >32 | 0.094 | >256 | 0.38 | A2143G | N87I | 90 stop | R | |||
Isolates with phenotypically resistant MIC values are highlighted in light gray shading. Discrepancies among WGS results compared to phenotypic resistance are highlighted in dark gray shading. Blank cells indicate that no mutations associated with resistance were found in the 23S rRNA gene for clarithromycin, gyrA and gyrB for levofloxacin, pbp1, pbp2, and pbp3 for amoxicillin, 16S rRNA for tetracycline, or rdxA for metronidazole. AMX, amoxicillin; CLR, clarithromycin; LVX, levofloxacin; TET, tetracycline; MET, metronidazole; RIF, rifampin.
MHP9 is a heteroresistant isolate comprised of strain MHP9C (resistant to clarithromycin) and MHP9L (resistant to levofloxacin).
P, primary; R, recurrent.
SI, specimen isolate.
TABLE 3.
Agreement between phenotypic MIC values and resistance genotype
| Antibiotic | MIC (μg/ml) breakpoint | Genotype | No. of isolates with the following phenotype: |
Kappa coefficient (95% CI) | P value by Fisher’s exact test | Adjusted P value | |
|---|---|---|---|---|---|---|---|
| Sensitive | Resistant | ||||||
| Amoxicillina | >0.125 | Sensitive | 39 | 0 | 1 (1.0–1.0) | <0.0001 | 0.0007 |
| Resistant | 0 | 3 | |||||
| Clarithromycin | >0.5 | Sensitive | 26 | 0 | 0.84 (0.67–1.0) | <0.0001 | 0.0007 |
| Resistant | 3 | 13 | |||||
| Levofloxacin | >1 | Sensitive | 17 | 1 | 0.9 (0.77–1.0) | <0.0001 | 0.0007 |
| Resistant | 1 | 23 | |||||
| Tetracycline | >1 | Sensitive | 38 | 0 | 1.0 | 1.0 | |
| Resistant | 4 | 0 | |||||
| Metronidazoleb | >8 | Sensitive | 20 | 2 | 0.76 (0.56–0.96) | <0.0001 | 0.0007 |
| Resistant | 3 | 17 | |||||
| Rifampinc | >1 | Sensitive | 29 | 8 | 0.46 (0.18–0.75) | 0.0015 | 0.0105 |
| Resistant | 0 | 5 | |||||
| >4 | Sensitive | 37 | 0 | 0.73 (0.37–1.0) | <0.001 | 0.0063 | |
| Resistant | 2 | 3 | |||||
Genetic resistance was defined as mutations near putative amoxicillin binding motifs.
Genetic resistance was defined as a truncation in rdxA or an R16H mutation.
Genetic resistance was defined as any mutation in rpoB.
None of the previously reported drug resistance-associated mutations within the rifampin resistance-determining region (RRDR) of rpoB were present in our isolates. However, 5 of 13 phenotypically resistant isolates with MICs of >1 μg/ml and 5 of 7 isolates with MICs of ≥3 μg/ml exhibited mutations outside the rpoB RRDR. No mutations in rpoB were present in isolates with MICs of <3 μg/ml (Table 2). The correlation between methods was poor (kappa coefficient, 0.46; 95% CI, 0.18 to 0.75) when the EUCAST breakpoint of an MIC of >1 μg/ml was used to define phenotypic resistance but improved (kappa coefficient, 0.73; 95% CI, 0.37 to 1.0) when an MIC of >4 μg/ml was used as a breakpoint (Table 3) (29).
For metronidazole resistance analyses, all sequences were assessed for the presence of intact rdxA and frxA, either of which converts the prodrug form of metronidazole into its active form. rdxA is believed to be the predominant gene responsible for mediating metronidazole resistance, with the role of frxA being discrepant among studies (37). Eleven of 19 phenotypically resistant isolates carried nonsense mutations in rdxA at amino acid positions 50, 72, 75, 76, 112, 119, 143, and 197. These mutations lead to the synthesis of a truncated/nonfunctional protein (Table S2) (38). The presence of rdxA truncations alone was present in only 58% of the resistant isolates.
Various point mutations in rdxA have been associated with metronidazole resistance, but results are not consistent between studies (23). We examined our isolates for point mutations that may be associated with metronidazole resistance and found a mutation at position R16 that was associated with six of the eight phenotypically resistant isolates that lacked a truncation in rdxA (Table 2; Table S2). This mutation has previously been associated with metronidazole phenotypic resistance (37, 39, 40). The detection of either a truncated rdxA gene or a mutation at position R16 correlated (kappa coefficient, 0.76) with 88% of phenotypic testing results (Table 2).
H. pylori lineage tree analyses.
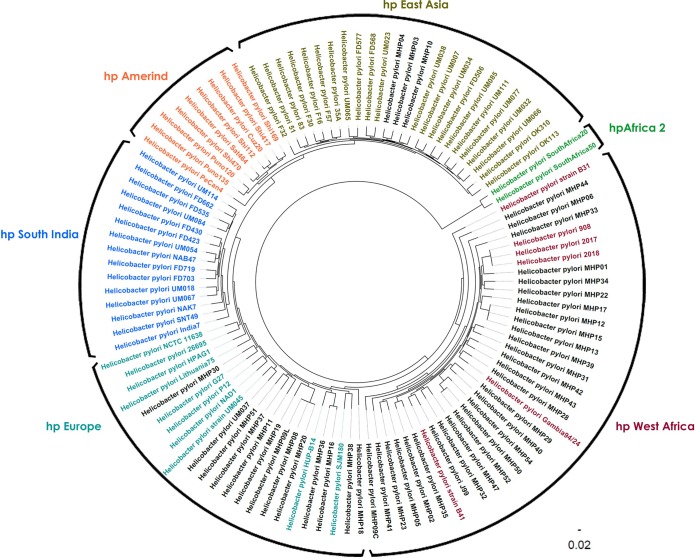
WGS data were used to construct a phylogenetic tree to determine where the 43 clinical strain genomes from the 42 clinical specimens clustered compared to previously defined H. pylori sequence types (STs) (28). The hpEastAsia, hpSouthIndia, hpWestAfrica, and hpAfrica2 sequence types are monophyletic with a common ancestor, and each has a support value of 100 at the defining branch point. The hpAmerind and hpEurope genomes were found to be paraphyletic, wherein the isolates were descended from multiple ancestors. Three of the 43 genomes sequenced here (MHP4, MHP3, and MHP10) clustered with the hspEastAsia clade, 12 clustered with the hpEurope clade, and 28 clustered with the hpWestAfrica clade. Interestingly, the heteroresistant isolate MHP9 comprised two strains from different lineages: strain MHP9C from hpWestAfrica and strain MHP9L from hpEurope (Fig. 1). None of the genomes that we sequenced were closely related to the hpAmerind, hpSouthIndia, or hpAfrica1 clade. Strains phenotypically resistant to clarithromycin, levofloxacin, and amoxicillin were found to be spread across all identified lineages. All clarithromycin-resistant strains from the West African (n = 6) and East Asian (n = 2) lineages were coresistant to levofloxacin.
FIG 1.
Phylogenetic tree based on the amino acid and nucleotide sequences of 512 genes shared across 102 Helicobacter pylori genomes. The names of some of the genomes, originally identified by Kumar et al. (28), are colored based on the phylogenetic lineages that they describe. The 43 genomes evaluated in the present study are in black.
DISCUSSION
Drug resistance among clinical isolates of H. pylori is a well-recognized issue, and susceptibility-guided treatment of H. pylori infection is crucial to attain a better treatment response among infected individuals (41). WHO has listed H. pylori as a high-priority pathogen for which there is an urgent need to develop new antimicrobials (42). We assessed the utility of WGS as a method for detecting molecular determinants of resistance by comparing the findings of WGS to phenotypic testing results. We found a strong correlation between the two methods for clarithromycin (93%) and levofloxacin (95%). Additionally, we identified mutations in our clinical isolates that correlated with phenotypic metronidazole resistance and identified mutations outside of the rpoB RRDR and novel mutations in penicillin binding proteins that may contribute to rifampin and amoxicillin resistance, respectively.
WGS allowed us to explore the putative mechanisms underlying the phenotypic resistance in our H. pylori isolates. For amoxicillin, point mutations in penicillin binding protein genes have been predicted to result in amoxicillin resistance in H. pylori, but most studies report novel mutations that require validation (23, 43). We identified 11 amino acid substitutions in Pbp1A/Pbp2/Pbp3 exclusively in three amoxicillin-resistant isolates, and 7 of these substitutions were found adjacent to SXN/SXXK/KTG motifs. In Pbp1A, mutations in these motifs have been found to confer resistance (32), and mutations adjacent to SKN402–404 and KTG555–557 have previously been identified among resistant isolates (44). Our data provide further evidence that the identification of mutations near SXN/SXXK/KTG motifs may contribute to resistance. Another observation was the detection of a D99N substitution (gyrA) in an isolate phenotypically levofloxacin resistant (MHP11). A previous study from Nepal found that a mutation at this position confers resistance, but a D99V substitution was noted (40).
WGS revealed clinically significant mutations in isolates testing phenotypically sensitive, which highlights an advantage of WGS over phenotypic testing. We noted point mutations in 23S rRNA (A2142G/A2143G) in three strains that were phenotypically sensitive to clarithromycin (MHP28, MHP34, MHP50) and a mutation in gyrA at position N87K in an isolate sensitive to levofloxacin (MHP35). The A2143G mutation is associated with clarithromycin MIC values ranging from susceptible to resistant, and patients with this mutation have a 60% increased risk for treatment failure (45). Environmental conditions and the genetic context (i.e., the strain lineage) may also influence the phenotypic susceptibility result (46). Another possible explanation for discrepancies between methods is that we may have selectively subcultured colonies with drug resistance when they were revived from stocks containing mixed populations. Nevertheless, the existence of heteroresistance is common during H. pylori infections (15).
In addition, we detected mutations in the 16S rRNA gene (A926G) in four tetracycline-sensitive isolates, but the significance of this finding is unclear, since this mutation has been associated with both low-level resistance and phenotypic susceptibility (16, 47, 48). In a recent study that examined the correlation between the detection of mutations directly from formalin-fixed, paraffin-embedded tissue and patient outcomes, only one treatment failure was noted in six patients with the A926G mutation, but studies with larger numbers of patients treated with tetracycline-containing regimens are needed to understand the clinical significance of this mutation (49).
Based on EUCAST guidelines, we categorized 13 isolates as rifampin resistant (MIC > 1 μg/ml) but did not identify any significant amino acid substitutions in the rpoB RRDR region, even for isolates with an MIC of >4 μg/ml. However, we could detect amino acid substitutions outside the rpoB RRDR exclusively in phenotypically resistant isolates with MICs of ≥3 μg/ml. Mutations outside of the RRDR have previously been associated with elevated rifampin MICs of between 2 and 4 μg/ml (29). As suggested previously, the lack of detection of any mutations in our isolates with an MIC of <3 μg/ml provides further support that the EUCAST breakpoint may need to be reevaluated (29). Further studies are needed to demonstrate that the point mutations identified outside the RRDR in our clinical isolates are responsible for the resistance phenotype observed.
Isolates from all 42 specimens were tested phenotypically for metronidazole resistance, and 19 had metronidazole MICs of >256 μg/ml. The presence of one or both intact genes (rdxA and frxA) did not always indicate low MICs, signifying that there may be other point mutations that could contribute to resistance (50). Few mutations were identified among our isolates carrying intact rdxA and frxA. R16H and V57A substitutions in rdxA and R58C, A85V, I117M, and E169K substitutions in frxA seem to be potential drug resistance mutations which were present only in drug-resistant isolates with intact rdxA and frxA genes (see Table S2 in the supplemental material). However, the R16H mutation in rdxA was the most common mutation found among our metronidazole-resistant isolates with an intact rdxA and has also been previously reported to be a significant resistance mutation (37, 39, 40, 48). We found that the combination of R16H and nonsense mutations in rdxA to be the best correlate of phenotypic resistance in our isolates. However, three isolates (MHP12, MHP15, MHP31) had nonsense mutations in rdxA but were found to be sensitive by the phenotypic Etest, supporting the fact that metronidazole resistance is multifaceted and is not yet clearly understood (50). We did not find among the metronidazole-resistant isolates any exclusive mutations in the rpsU, mdaB, omp11, or fdxB gene that have previously been associated with metronidazole resistance (data not shown). Additional studies on other possible mechanisms, such as the overexpression of efflux pumps or defects in membrane porins, might provide some novel insights into metronidazole resistance (50).
A recent study comparing WGS to phenotypic testing using H. pylori isolates from a collection in Switzerland demonstrated an agreement between methods of 99% for clarithromycin and levofloxacin, 100% agreement for rifampin, but no agreement for tetracycline (the A926 mutation) or metronidazole (16). Taken together, our data and those from the previous study (16) support the use of WGS for determination of susceptibility to clarithromycin and levofloxacin but do not support a role for the A926G mutation in conferring tetracycline resistance. In contrast to the findings of the previously published study (16), we found that mutations outside the rpoB RRDR were associated with elevated MICs of rifampin, suggesting that a complete catalogue of resistance determinants for H. pylori must be established. Our study also adds novel mutations associated with amoxicillin resistance and provides molecular correlates of metronidazole resistance that best predict phenotypic testing results. Continued WGS-based characterization of H. pylori isolates will support future studies to determine which genetic determinants contribute to eradication failure.
MLST analyses revealed the presence of unique alleles among all the sequenced isolates, which is not uncommon in H. pylori, and none of the isolates belonged to known sequence types (STs) (51). Phylogenetic analysis with reference genomes from known lineages revealed the existence of East Asian, European, and West African (Africa1) lineages in the Bronx. This observation is in agreement with the observations from the earlier studies from North America, wherein the West African lineage is predominant and may be due to the migration of people of different ethnic backgrounds (52). H. pylori infection usually occurs during childhood and results from transmission through the oral-oral or oral-fecal route, with several studies demonstrating transmission between members of the same family or between individuals sharing the same house (53–57). The presence of two different lineages (MHP9C and MHP9L) with different resistance patterns in one patient sample is not surprising, since coinfection with multiple strains in one host can occur and in-host evolution can occur due to recombination events between H. pylori strains (54, 58). Resistant strains among all three lineages were identified; however, clarithromycin- and levofloxacin-coresistant strains of the East Asian and West African lineages were largely observed. Analyses of a larger cohort of isolates is needed to confirm if any association between strain lineage and resistance exists.
In summary, WGS provides a means for the more detailed analysis of H. pylori resistance than is possible by phenotypic testing. The strengths of our study are the inclusion of H. pylori isolates from pediatric patients and adults; patients with recurrent H. pylori infections, in which phenotypic resistance was the greatest; and patients with diverse racial backgrounds from our community in the Bronx. The limitations of our study are the low number of isolates that were phenotypically resistant to amoxicillin or tetracycline, and because of the limited availability of patient follow-up data, we could not determine whether the detection of resistance by WGS was associated with patient outcomes. Because of the use of remnant specimens and retrospective chart review, we cannot confirm that all strains originated from unrelated individuals. Strains transmitted within families would be expected to be similar, which would reduce the significance of our findings. However, when MLST was performed, we found unique alleles among all strains, suggesting that transmission among individuals in our cohort would be unlikely. Our findings support the utilization of WGS for the determination of H. pylori resistance to clarithromycin and levofloxacin. Our data suggest that the single nucleotide mutation A926G is not associated with phenotypic tetracycline resistance and provide potential genetic determinants of amoxicillin, rifampin, and metronidazole resistance for further investigation.
Supplementary Material
ACKNOWLEDGMENT
We thank Yungtai Lo, associate professor of epidemiology and population health, Albert Einstein College of Medicine, for his assistance with statistical analyses.
This work was supported by National Institutes of Health grant P30CA013330 (W.R.J.).
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.McColl KE. 2010. Clinical practice. Helicobacter pylori infection. N Engl J Med 362:1597–1604. doi: 10.1056/NEJMcp1001110. [DOI] [PubMed] [Google Scholar]
- 2.Uemura N, Okamoto S, Yamamoto S, Matsumura N, Yamaguchi S, Yamakido M, Taniyama K, Sasaki N, Schlemper RJ. 2001. Helicobacter pylori infection and the development of gastric cancer. N Engl J Med 345:784–789. doi: 10.1056/NEJMoa001999. [DOI] [PubMed] [Google Scholar]
- 3.Chey WD, Leontiadis GI, Howden CW, Moss SF. 2017. ACG clinical guideline: treatment of Helicobacter pylori infection. Am J Gastroenterol 112:212–239. doi: 10.1038/ajg.2016.563. [DOI] [PubMed] [Google Scholar]
- 4.Pan KF, Zhang L, Gerhard M, Ma JL, Liu WD, Ulm K, Wang JX, Zhang L, Zhang Y, Bajbouj M, Zhang LF, Li M, Vieth M, Liu RY, Quante M, Wang LH, Suchanek S, Zhou T, Guan WX, Schmid R, Classen M, You WC. 2016. A large randomised controlled intervention trial to prevent gastric cancer by eradication of Helicobacter pylori in Linqu County, China: baseline results and factors affecting the eradication. Gut 65:9–18. doi: 10.1136/gutjnl-2015-309197. [DOI] [PubMed] [Google Scholar]
- 5.Malfertheiner P, Megraud F, O’Morain CA, Gisbert JP, Kuipers EJ, Axon AT, Bazzoli F, Gasbarrini A, Atherton J, Graham DY, Hunt R, Moayyedi P, Rokkas T, Rugge M, Selgrad M, Suerbaum S, Sugano K, El-Omar EM, European Helicobacter and Microbiota Study Group and Consensus Panel. 2017. Management of Helicobacter pylori infection—the Maastricht V/Florence consensus report. Gut 66:6–30. doi: 10.1136/gutjnl-2016-312288. [DOI] [PubMed] [Google Scholar]
- 6.López-Góngora S, Puig I, Calvet X, Villoria A, Baylina M, Muñoz N, Sanchez-Delgado J, Suarez D, García-Hernando V, Gisbert JP. 2015. Systematic review and meta-analysis: susceptibility-guided versus empirical antibiotic treatment for Helicobacter pylori infection. J Antimicrob Chemother 70:2447–2455. doi: 10.1093/jac/dkv155. [DOI] [PubMed] [Google Scholar]
- 7.Wenzhen Y, Yumin L, Quanlin G, Kehu Y, Lei J, Donghai W, Lijuan Y. 2010. Is antimicrobial susceptibility testing necessary before first-line treatment for Helicobacter pylori infection? Meta-analysis of randomized controlled trials. Intern Med 49:1103–1109. doi: 10.2169/internalmedicine.49.3031. [DOI] [PubMed] [Google Scholar]
- 8.Lehours P, Siffre E, Megraud F. 2011. DPO multiplex PCR as an alternative to culture and susceptibility testing to detect Helicobacter pylori and its resistance to clarithromycin. BMC Gastroenterol 11:112. doi: 10.1186/1471-230X-11-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ducournau A, Benejat L, Sifre E, Bessede E, Lehours P, Megraud F. 2016. Helicobacter pylori resistance to antibiotics in 2014 in France detected by phenotypic and genotypic methods. Clin Microbiol Infect 22:715–718. doi: 10.1016/j.cmi.2016.06.003. [DOI] [PubMed] [Google Scholar]
- 10.Miendje Deyi VY, Burette A, Bentatou Z, Maaroufi Y, Bontems P, Lepage P, Reynders M. 2011. Practical use of GenoType(R) HelicoDR, a molecular test for Helicobacter pylori detection and susceptibility testing. Diagn Microbiol Infect Dis 70:557–560. doi: 10.1016/j.diagmicrobio.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 11.Cambau E, Allerheiligen V, Coulon C, Corbel C, Lascols C, Deforges L, Soussy CJ, Delchier JC, Megraud F. 2009. Evaluation of a new test, genotype HelicoDR, for molecular detection of antibiotic resistance in Helicobacter pylori. J Clin Microbiol 47:3600–3607. doi: 10.1128/JCM.00744-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Agudo S, Alarcon T, Urruzuno P, Martinez MJ, Lopez-Brea M. 2010. Detection of Helicobacter pylori and clarithromycin resistance in gastric biopsies of pediatric patients by using a commercially available real-time polymerase chain reaction after NucliSens semiautomated DNA extraction. Diagn Microbiol Infect Dis 67:213–219. doi: 10.1016/j.diagmicrobio.2010.02.021. [DOI] [PubMed] [Google Scholar]
- 13.Chisholm SA, Owen RJ, Teare EL, Saverymuttu S. 2001. PCR-based diagnosis of Helicobacter pylori infection and real-time determination of clarithromycin resistance directly from human gastric biopsy samples. J Clin Microbiol 39:1217–1220. doi: 10.1128/JCM.39.4.1217-1220.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen D, Cunningham SA, Cole NC, Kohner PC, Mandrekar JN, Patel R. 2017. Phenotypic and molecular antimicrobial susceptibility of Helicobacter pylori. Antimicrob Agents Chemother 61:e02530-16. doi: 10.1128/AAC.02530-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun L, Talarico S, Yao L, He L, Self S, You Y, Zhang H, Zhang Y, Guo Y, Liu G, Salama NR, Zhang J. 2018. Droplet digital PCR-based detection of clarithromycin resistance in Helicobacter pylori isolates reveals frequent heteroresistance. J Clin Microbiol 56:e00019-18. doi: 10.1128/JCM.00019-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lauener FN, Imkamp F, Lehours P, Buissonniere A, Benejat L, Zbinden R, Keller PM, Wagner K. 2019. Genetic determinants and prediction of antibiotic resistance phenotypes in Helicobacter pylori. J Clin Med 8:E53. doi: 10.3390/jcm8010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clinical and Laboratory Standards Institute. 2016. Methods for antimicrobial dilution and disk susceptibility testing of infrequently isolated or fastidious bacteria, 3rd ed. CLSI guideline M45 Clinical and Laboratory Standards Institute, Wayne, PA. [DOI] [PubMed] [Google Scholar]
- 18.Wingett SW, Andrews S. 2018. FastQ screen: a tool for multi-genome mapping and quality control. F1000Res 7:1338. doi: 10.12688/f1000research.15931.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wattam AR, Davis JJ, Assaf R, Boisvert S, Brettin T, Bun C, Conrad N, Dietrich EM, Disz T, Gabbard JL, Gerdes S, Henry CS, Kenyon RW, Machi D, Mao C, Nordberg EK, Olsen GJ, Murphy-Olson DE, Olson R, Overbeek R, Parrello B, Pusch GD, Shukla M, Vonstein V, Warren A, Xia F, Yoo H, Stevens RL. 2017. Improvements to PATRIC, the all-bacterial Bioinformatics Database and Analysis Resource Center. Nucleic Acids Res 45:D535–D542. doi: 10.1093/nar/gkw1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brettin T, Davis JJ, Disz T, Edwards RA, Gerdes S, Olsen GJ, Olson R, Overbeek R, Parrello B, Pusch GD, Shukla M, Thomason JA III, Stevens R, Vonstein V, Wattam AR, Xia F. 2015. RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci Rep 5:8365. doi: 10.1038/srep08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gong Y, Yuan Y. 2018. Resistance mechanisms of Helicobacter pylori and its dual target precise therapy. Crit Rev Microbiol 44:371–392. doi: 10.1080/1040841X.2017.1418285. [DOI] [PubMed] [Google Scholar]
- 24.Davis JJ, Gerdes S, Olsen GJ, Olson R, Pusch GD, Shukla M, Vonstein V, Wattam AR, Yoo H. 2016. PATtyFams: protein families for the microbial genomes in the PATRIC database. Front Microbiol 7:118. doi: 10.3389/fmicb.2016.00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cock PJ, Antao T, Chang JT, Chapman BA, Cox CJ, Dalke A, Friedberg I, Hamelryck T, Kauff F, Wilczynski B, de Hoon MJ. 2009. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 25:1422–1423. doi: 10.1093/bioinformatics/btp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML web servers. Syst Biol 57:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- 28.Kumar N, Mariappan V, Baddam R, Lankapalli AK, Shaik S, Goh KL, Loke MF, Perkins T, Benghezal M, Hasnain SE, Vadivelu J, Marshall BJ, Ahmed N. 2015. Comparative genomic analysis of Helicobacter pylori from Malaysia identifies three distinct lineages suggestive of differential evolution. Nucleic Acids Res 43:324–335. doi: 10.1093/nar/gku1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hays C, Burucoa C, Lehours P, Tran CT, Leleu A, Raymond J, Hays C, Burucoa C, Lehours P, Tran CT, Leleu A, Raymond J. 2018. Molecular characterization of Helicobacter pylori resistance to rifamycins. Helicobacter 23:e12451. doi: 10.1111/hel.12451. [DOI] [PubMed] [Google Scholar]
- 30.Rimbara E, Noguchi N, Kawai T, Sasatsu M. 2008. Mutations in penicillin-binding proteins 1, 2 and 3 are responsible for amoxicillin resistance in Helicobacter pylori. J Antimicrob Chemother 61:995–998. doi: 10.1093/jac/dkn051. [DOI] [PubMed] [Google Scholar]
- 31.Okamoto T, Yoshiyama H, Nakazawa T, Park ID, Chang MW, Yanai H, Okita K, Shirai M. 2002. A change in PBP1 is involved in amoxicillin resistance of clinical isolates of Helicobacter pylori. J Antimicrob Chemother 50:849–856. doi: 10.1093/jac/dkf140. [DOI] [PubMed] [Google Scholar]
- 32.Harris AG, Hazell SL, Netting AG. 2000. Use of digoxigenin-labelled ampicillin in the identification of penicillin-binding proteins in Helicobacter pylori. J Antimicrob Chemother 45:591–598. doi: 10.1093/jac/45.5.591. [DOI] [PubMed] [Google Scholar]
- 33.Bachir M, Allem R, Benejat L, Tifrit A, Medjekane M, Drici AE, Megraud F, Douidi KT. 2018. Molecular detection of mutations involved in Helicobacter pylori antibiotic resistance in Algeria. J Antimicrob Chemother 73:2034–2038. doi: 10.1093/jac/dky167. [DOI] [PubMed] [Google Scholar]
- 34.De Francesco V, Zullo A, Fiorini G, Saracino IM, Pavoni M, Vaira D. 2019. Role of MIC levels of resistance to clarithromycin and metronidazole in Helicobacter pylori eradication. J Antimicrob Chemother 74:772–774. doi: 10.1093/jac/dky469. [DOI] [PubMed] [Google Scholar]
- 35.Kim JM, Kim JS, Kim N, Kim YJ, Kim IY, Chee YJ, Lee CH, Jung HC. 2008. Gene mutations of 23S rRNA associated with clarithromycin resistance in Helicobacter pylori strains isolated from Korean patients. J Microbiol Biotechnol 18:1584–1589. [PubMed] [Google Scholar]
- 36.Posteraro P, Branca G, Sanguinetti M, Ranno S, Cammarota G, Rahimi S, De Carlo M, Posteraro B, Fadda G. 2006. Rapid detection of clarithromycin resistance in Helicobacter pylori using a PCR-based denaturing HPLC assay. J Antimicrob Chemother 57:71–78. doi: 10.1093/jac/dki406. [DOI] [PubMed] [Google Scholar]
- 37.Tanih NF, Ndip LM, Ndip RN. 2011. Characterisation of the genes encoding resistance to metronidazole (rdxA and frxA) and clarithromycin (the 23S-rRNA genes) in South African isolates of Helicobacter pylori. Ann Trop Med Parasitol 105:251–259. doi: 10.1179/136485911X12899838683485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matteo MJ, Perez CV, Domingo MR, Olmos M, Sanchez C, Catalano M. 2006. DNA sequence analysis of rdxA and frxA from paired metronidazole-sensitive and -resistant Helicobacter pylori isolates obtained from patients with heteroresistance. Int J Antimicrob Agents 27:152–158. doi: 10.1016/j.ijantimicag.2005.09.019. [DOI] [PubMed] [Google Scholar]
- 39.Rasheed F, Campbell BJ, Alfizah H, Varro A, Zahra R, Yamaoka Y, Pritchard DM. 2014. Analysis of clinical isolates of Helicobacter pylori in Pakistan reveals high degrees of pathogenicity and high frequencies of antibiotic resistance. Helicobacter 19:387–399. doi: 10.1111/hel.12142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miftahussurur M, Shrestha PK, Subsomwong P, Sharma RP, Yamaoka Y. 2016. Emerging Helicobacter pylori levofloxacin resistance and novel genetic mutation in Nepal. BMC Microbiol 16:256. doi: 10.1186/s12866-016-0873-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blumel B, Goelz H, Kist M, Glocker EO. 2018. Retrospective study on outcome of salvage Helicobacter pylori eradication therapies based on molecular genetic susceptibility testing. Helicobacter 23:e12494. doi: 10.1111/hel.12494. [DOI] [PubMed] [Google Scholar]
- 42.World Health Organization. 2017. Prioritization of pathogens to guide discovery, research and development of new antibiotics for drug-resistant bacterial infections, including tuberculosis. Report WHO/EMP/IAU/2017.12 World Health Organization, Geneva, Switzerland. [Google Scholar]
- 43.Chua EG, Debowski AW, Webberley KM, Peters F, Lamichhane B, Loke MF, Vadivelu J, Tay CY, Marshall BJ, Wise MJ. 2019. Analysis of core protein clusters identifies candidate variable sites conferring metronidazole resistance in Helicobacter pylori. Gastroenterol Rep (Oxf) 7:42–49. doi: 10.1093/gastro/goy048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gerrits MM, Godoy AP, Kuipers EJ, Ribeiro ML, Stoof J, Mendonca S, van Vliet AH, Pedrazzoli J Jr, Kusters JG. 2006. Multiple mutations in or adjacent to the conserved penicillin-binding protein motifs of the penicillin-binding protein 1A confer amoxicillin resistance to Helicobacter pylori. Helicobacter 11:181–187. doi: 10.1111/j.1523-5378.2006.00398.x. [DOI] [PubMed] [Google Scholar]
- 45.Park CG, Kim S, Lee EJ, Jeon HS, Han S. 2018. Clinical relevance of point mutations in the 23S rRNA gene in Helicobacter pylori eradication: a prospective, observational study. Medicine (Baltimore) 97:e11835. doi: 10.1097/MD.0000000000011835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hughes D, Andersson DI. 2017. Environmental and genetic modulation of the phenotypic expression of antibiotic resistance. FEMS Microbiol Rev 41:374–391. doi: 10.1093/femsre/fux004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Toledo H, López-Solís R. 2010. Tetracycline resistance in Chilean clinical isolates of Helicobacter pylori. J Antimicrob Chemother 65:470–473. doi: 10.1093/jac/dkp457. [DOI] [PubMed] [Google Scholar]
- 48.Tuan VP, Narith D, Tshibangu-Kabamba E, Dung HDQ, Viet PT, Sokomoth S, Binh TT, Sokhem S, Tri TD, Ngov S, Tung PH, Thuan NPM, Truc TC, Phuc BH, Matsumoto T, Fauzia KA, Akada J, Trang TTH, Yamaoka Y. 2019. A next-generation sequencing-based approach to identify genetic determinants of antibiotic resistance in Cambodian Helicobacter pylori clinical isolates. J Clin Med 8:E858. doi: 10.3390/jcm8060858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nezami BG, Jani M, Alouani D, Rhoads DD, Sadri N, Nezami BG, Jani M, Alouani D, Rhoads DD, Sadri N. 2019. Helicobacter pylori mutations detected by next-generation sequencing in formalin-fixed paraffin embedded gastric biopsies are associated with treatment failure. J Clin Microbiol 57:e01834-18. doi: 10.1128/JCM.01834-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marques B, Donato MM, Cardoso O, Luxo C, Martinho A, Almeida N. 2019. Study of rdxA and frxA genes mutations in metronidazole-resistant and -susceptible Helicobacter pylori clinical isolates from the central region of Portugal. J Glob Antimicrob Resist 17:300–304. doi: 10.1016/j.jgar.2019.01.008. [DOI] [PubMed] [Google Scholar]
- 51.Secka O, Moodley Y, Antonio M, Berg DE, Tapgun M, Walton R, Worwui A, Thomas V, Corrah T, Thomas JE, Adegbola RA. 2014. Population genetic analyses of Helicobacter pylori isolates from Gambian adults and children. PLoS One 9:e109466. doi: 10.1371/journal.pone.0109466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thorell K, Yahara K, Berthenet E, Lawson DJ, Mikhail J, Kato I, Mendez A, Rizzato C, Bravo MM, Suzuki R, Yamaoka Y, Torres J, Sheppard SK, Falush D. 2017. Rapid evolution of distinct Helicobacter pylori subpopulations in the Americas. PLoS Genet 13:e1006546. doi: 10.1371/journal.pgen.1006546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Krebes J, Didelot X, Kennemann L, Suerbaum S. 2014. Bidirectional genomic exchange between Helicobacter pylori strains from a family in Coventry, United Kingdom. Int J Med Microbiol 304:1135–1146. doi: 10.1016/j.ijmm.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 54.Didelot X, Nell S, Yang I, Woltemate S, van der Merwe S, Suerbaum S. 2013. Genomic evolution and transmission of Helicobacter pylori in two South African families. Proc Natl Acad Sci U S A 110:13880–13885. doi: 10.1073/pnas.1304681110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Furuta Y, Konno M, Osaki T, Yonezawa H, Ishige T, Imai M, Shiwa Y, Shibata-Hatta M, Kanesaki Y, Yoshikawa H, Kamiya S, Kobayashi I. 2015. Microevolution of virulence-related genes in Helicobacter pylori familial infection. PLoS One 10:e0127197. doi: 10.1371/journal.pone.0127197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Osaki T, Konno M, Yonezawa H, Hojo F, Zaman C, Takahashi M, Fujiwara S, Kamiya S. 2015. Analysis of intra-familial transmission of Helicobacter pylori in Japanese families. J Med Microbiol 64:67–73. doi: 10.1099/jmm.0.080507-0. [DOI] [PubMed] [Google Scholar]
- 57.Kurosawa M, Kikuchi S, Inaba Y, Ishibashi T, Kobayashi F. 2000. Helicobacter pylori infection among Japanese children. J Gastroenterol Hepatol 15:1382–1385. doi: 10.1046/j.1440-1746.2000.02360.x. [DOI] [PubMed] [Google Scholar]
- 58.Kennemann L, Didelot X, Aebischer T, Kuhn S, Drescher B, Droege M, Reinhardt R, Correa P, Meyer TF, Josenhans C, Falush D, Suerbaum S. 2011. Helicobacter pylori genome evolution during human infection. Proc Natl Acad Sci U S A 108:5033–5038. doi: 10.1073/pnas.1018444108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All 43 sequenced genomes of the H. pylori strains isolated from 42 clinical specimens in this study were assembled, annotated, and submitted to NCBI BioProject database under BioProject accession number PRJNA566177.