We reevaluated 20 cases of blastomycosis diagnosed in South Africa between 1967 and 2014, with Blastomyces dermatitidis considered to be the etiological agent, in light of newly described species and the use of more advanced technologies. In addition to histopathological and/or culture-based methods, all 20 isolates were phenotypically and genotypically characterized, including multilocus typing of five genes and whole-genome sequencing.
KEYWORDS: blastomycosis, mycoses, South Africa, Blastomyces, emergomycosis, histoplasmosis, tuberculosis
ABSTRACT
We reevaluated 20 cases of blastomycosis diagnosed in South Africa between 1967 and 2014, with Blastomyces dermatitidis considered to be the etiological agent, in light of newly described species and the use of more advanced technologies. In addition to histopathological and/or culture-based methods, all 20 isolates were phenotypically and genotypically characterized, including multilocus typing of five genes and whole-genome sequencing. Antifungal susceptibility testing was performed as outlined by Clinical and Laboratory Standards Institute documents M27-A3 and M38-A2. We merged laboratory and corresponding clinical case data, where available. Morphological characteristics and phylogenetic analyses of five-gene and whole-genome sequences revealed two groups, both of which were closely related to but distinct from B. dermatitidis, Blastomyces gilchristii, and Blastomyces parvus. The first group (n = 12) corresponded to the recently described species Blastomyces percursus, and the other (n = 8) is described here as Blastomyces emzantsi sp. nov. Both species exhibited incomplete conversion to the yeast phase at 37°C and were heterothallic for mating types. All eight B. emzantsi isolates belonged to the α mating type. Whole-genome sequencing confirmed distinct species identities as well as the absence of a full orthologue of the BAD-1 gene. Extrapulmonary (skin or bone) disease, probably resulting from hematogenous spread from a primary lung infection, was more common than pulmonary disease alone. Voriconazole, posaconazole, itraconazole, amphotericin B, and micafungin had the most potent in vitro activity. Over the 5 decades, South African cases of blastomycosis were caused by species that are distinct from B. dermatitidis. Increasing clinical awareness and access to simple rapid diagnostics may improve the diagnosis of blastomycosis in resource-limited countries.
INTRODUCTION
The first human case of blastomycosis was described by Gilchrist in 1894 from skin tissue (1). Blastomycosis was initially believed to be restricted to North America (2). However, since the first case was reported from Tunisia in 1952, cases of blastomycosis have also been reported throughout Africa and less commonly from India, the Middle East, and occasionally Europe (3, 4). Differences have been noted between North American and African cases of blastomycosis, both in the isolates and in the clinical presentation of disease. For instance, blastomycosis in North America primarily involves the lungs, while more African cases have involved skin and bone (3).
The genus Blastomyces belongs within the family Ajellomycetaceae and order Onygenales, which includes other thermally dimorphic fungi such as Histoplasma, Emmonsia, Emmonsiellopsis, Paracoccidioides, and a newly described genus, Emergomyces (5). Initially, the Blastomyces genus was thought to comprise a single species, Blastomyces dermatitidis (6). Three other species have since been described within Blastomyces, including B. gilchristii (a cryptic species phenotypically indistinguishable from B. dermatitidis), B. percursus, and B. silverae (5, 7, 8). In addition, Emmonsia parva (Blastomyces parvus) and Emmonsia helica (Blastomyces helicus) have been assigned to the genus (8). B. dermatitidis produces abundant large broad-based budding yeasts at 37°C; B. percursus produces large yeastlike cells from fragmented, swollen hyphal cells; and B. helicus produces variably shaped yeast cells in short chains, while thin-walled giant cells and occasional broad-based budding yeastlike cells are seen in B. parvus and B. silverae strains (5, 8, 9).
In previous studies, mycological differences were noted between North American and African Blastomyces strains (10–16). African strains were more difficult to convert from the mold to the yeast phase (10–12). While North American and African isolates shared the K antigen, North American strains additionally possessed an A antigen, which is absent from most African strains (11). When tested for sexual compatibility, African strains failed to mate with North American strains of B. dermatitidis (10). Furthermore, DNA melting-curve analyses separated the North American and African strains into groups that differed sexually, histologically, epidemiologically, and clinically (15).
In view of the recent description of new pathogenic species within the genus Blastomyces and the potential molecular epidemiological, clinical, and public health implications, we reexamined South African Blastomyces human clinical isolates archived over 5 decades.
MATERIALS AND METHODS
Case detection and isolate archive.
We conducted passive laboratory-based surveillance for thermally dimorphic fungi associated with human disease from 2008 through 2014 at nine South African diagnostic pathology laboratories in the public and private sectors. We defined a case of blastomycosis as a person with an isolate cultured from any specimen, initially identified as B. dermatitidis by standard phenotypic culture-based or histological methods. In addition, we included 17 clinical isolates from South Africa, which had been cultured from 1967 through to 1999, previously identified as B. dermatitidis by phenotypic methods and archived at the National Institute for Communicable Diseases (NICD), South Africa, or at the National Collection of Pathogenic Fungi (NCPF), United Kingdom. For 10 cases, we obtained detailed information on patient demographics; medical conditions, including HIV and coinfections; details of the patient’s clinical presentation; diagnostic investigations; management; and outcomes from previous case reports (3–5, 17–21). Limited information was available for the other 10 archived isolates. In total, we studied 20 Blastomyces isolates. Examination of isolates included morphological characterization by light and electron microscopy, determination of MICs for nine antifungal agents, and phylogenetic analyses using the sequences of five genes (22). We also performed phylogenetic analysis using whole-genome single nucleotide polymorphisms (SNPs) for 18 isolates.
Morphological description and electron microscopy.
All isolates were subcultured onto Sabouraud agar or potato dextrose agar (PDA) (Diagnostic Media Products [DMP], Sandringham, South Africa) and incubated at 25°C and 30°C for up to 4 weeks for the mycelial phase. Isolates were transferred onto brain heart infusion (BHI) agar with or without 5% sheep blood (DMP) and incubated for 1 to 3 weeks at 37°C and 40°C for conversion to the yeast phase. Mycelial growth was also inoculated onto a urea agar slope (DMP) and incubated at 37°C for 6 days. Yeast and mycelial isolates were examined using light microscopy and electron microscopy (see the supplemental material). The descriptive terminology used follows that of Jiang et al. (8).
Antifungal susceptibility testing.
Susceptibility testing was performed for both yeast- and mold-phase isolates using a reference broth microdilution (BMD) method and the commercial Etest method (bioMérieux, Marcy l’Etoile, France). The BMD method was performed according to Clinical and Laboratory Standards Institute (CLSI) documents M27-A3 and M38-A2 (23, 24), with a modified inoculum size for the latter, as described previously by Maphanga et al. (see the supplemental material) (25). The following quality control (QC) strains were included in each test run: Candida parapsilosis ATCC 22019, Candida krusei ATCC 6258, and B. dermatitidis ATCC 10225 for yeast-phase tests and Aspergillus fumigatus NCPF 7097, A. fumigatus NCPF 7100, and B. dermatitidis ATCC 10225 for mycelium-phase tests. The MICs for C. parapsilosis ATCC 22019 and C. krusei ATCC 6258 were within the CLSI-recommended ranges. We calculated a geometric mean (GM) for each MIC distribution using Stata version 14.0 (StataCorp Limited, College Station, TX, USA).
DNA extraction, PCR amplification, and sequencing.
Genomic DNA was extracted from 20 isolates using the Zymo ZR fungal/bacterial DNA miniprep kit (Zymo Research Corp., USA) according to the manufacturer’s instructions. PCR amplification of five genes (internal transcribed spacer [ITS], large subunit [LSU], actin, β-tubulin, and intein PRP8) and sequencing PCR were performed as described previously (see the supplemental material) (22). The samples were sequenced in a 3130 sequencer (Applied Biosystems, Life Technologies Corporation, USA). Sequences were subjected to BLAST analyses in GenBank (www.ncbi.nlm.nih.gov) for identification.
MLST phylogenetic analysis.
DNA sequences were aligned with MAFFT version 7, and data sets for the five-gene multilocus sequence typing (MLST) loci were trimmed with BioEdit version 7 (26, 27). The phylogenetic tree was generated by a maximum likelihood statistical method using 1,000 bootstrap replications in MEGA version 6 from the concatenated data set and individual genes (see the supplemental material) (28). A clade was defined as isolates from different patients which shared a common ancestor with >80% support values in the maximum likelihood analyses.
Whole-genome sequencing (WGS) and de novo assembly.
Paired-end libraries were prepared using the Nextera XT DNA library kit, followed by 2- by 300-bp sequencing on an Illumina MiSeq sequencer (Illumina, San Diego, CA, USA). The sequenced paired-end reads were quality trimmed and de novo assembled using Qiagen CLC Genomics Workbench version 10 (Qiagen, The Netherlands).
Phylogenetic analysis using whole-genome single nucleotide polymorphisms.
Reads were aligned to the B. percursus assembly strain BP222 (GenBank accession number GCA_001883805.1) using BWA-MEM version 0.7.12 (5, 29). Variants were then identified using GATK version 3.7 (see the supplemental material) (30). For phylogenetic analysis, the 1,712,033 sites with an unambiguous SNP in at least one isolate and with ambiguity in at most 10% of isolates were concatenated; insertions or deletions at these sites were treated as ambiguous to maintain the alignment. Maximum likelihood phylogenies were constructed using RAxML version 8.2.4 using the GTRCAT nucleotide substitution model and bootstrap analysis based on 1,000 replicates (31).
Determination of nucleotide diversity using the whole genome.
Genome-wide nucleotide diversity (π) was computed for B. emzantsi, B. percursus, and the Blastomyces genus using VCFtools v0.1.12 (32). The average nucleotide diversity (π) was computed for nonoverlapping sliding windows of 10 kb.
Identification of the mating types from WGS data.
The reverse and forward primer pairs for Blastomyces MAT1-1 and MAT1-2 obtained from Li et al. were subjected to a BLAST search with our Blastomyces contigs (29). For Blastomyces adhesion 1 (BAD-1) gene determination, the genome of a reference strain of Ajellomyces dermatitidis (ATCC 26199; GenBank accession number U37772) was mapped to those of our Blastomyces strains. The obtained sequences were visually analyzed and subjected to BLAST analyses in GenBank (www.ncbi.nlm.nih.gov) for confirmation of mating types and BAD-1 sequences.
Ethics.
Ethics clearance for the study was obtained from the ethics committees of the University of the Free State (13/2016), the University of Cape Town (704/2013 and 138/2014), and the University of the Witwatersrand (M140112), South Africa.
Data availability.
We declare that the data supporting the findings of this study are available within the paper. Contigs and sequences were submitted to GenBank (contig accession numbers QGQE00000000 [SA-NICD-12], QGQF00000000 [SA-NICD-09], QGQG00000000 [SA-NICD-10], QGQH00000000 [SA-NICD-11], QGQI00000000 [SA-NICD-15], QGQJ00000000 [SA-NICD-13], QGQK00000000 [SA-NICD-14], QGQL00000000 [SA-NICD-16], QGQM00000000 [SA-NICD-01], QGQN00000000 [SA-NICD-07], QGQO00000000 [SA-NICD-02], QGQP00000000 [SA-NICD-03], QGQQ00000000 [SA-NICD-04], QGQR00000000 [SA-NICD-08], QGQS00000000 [SA-NICD-05], QGQT00000000 [SA-NICD-06], and QKWI00000000 [NCPF 4091]; ITS accession numbers MH571851, MH571852, MH571853, MH571854, MH571855, MH571856, MH571857, MH571858, MH571859, MH571860, MH571861, MH571862, MH606201, MH606202, MH606203, MH606204, MH606205, MH606206, MH606207, and MH606208; and LSU accession numbers MH644816, MH644817, MH644818, MH644819, MH644820, MH644821, MH644822, MH644823, MH644824, MH644825, MH644826, MH644836, MH644837, MH644838, MH644839, MH644840, MH644841, MH644842, and MH644843).
RESULTS
Isolates and cases.
Isolates were available from 20 cases of blastomycosis diagnosed from 1967 through 2014. Only the year of isolation was available for nine archived Blastomyces isolates: 1975 (n = 6), 1992 (n = 2), and 1999 (n = 1). For one isolate, no information was available at all. Clinical data were available for the 10 remaining cases. Nine patients were adult males, with a median age of 37 years (interquartile range [IQR], 32.5 to 52 years). Isolates were cultured from skin biopsy specimens (n = 4), pus from a thoracic vertebral abscess (n = 2) (Fig. 1), pus from a subcutaneous abscess (n = 2), brain tissue (n = 1), and a biopsy specimen of a tongue ulcer (n = 1). For the nine patients for whom this information was available, B. dermatitidis was initially identified as the causative pathogen by histopathological examination (Table 1). All 20 isolates had previously been phenotypically identified in culture as B. dermatitidis.
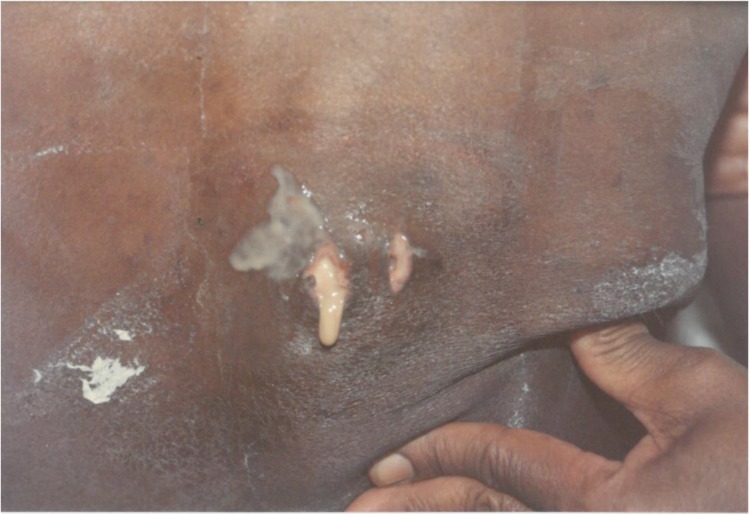
FIG 1.
A case of disseminated blastomycosis caused by Blastomyces emzantsi. Shown is purulent discharge from a cutaneous sinus tract at the T5 vertebral level (case 8). (Republished from reference 20 with permission of the publisher.)
TABLE 1.
Clinical characteristics of 10 patients with South African blastomycosis, 1967 to 2014
| Case | Yr of isolation/reference(s) (GenBank accession no.; strain) | Demographic information | Clinical presentation and radiology findings | Laboratory findingsa | Treatment(s); outcome (duration of follow-up) |
|---|---|---|---|---|---|
| 1 | 1986/5 (QKWI00000000; NCPF 4091) | Age: unknown | History: ulcerative skin lesions | Specimen type: skin | Unknown |
| Race, sex: unknown | Examination: unknown | Histopathology: yeasts morphologically resembling B. dermatitidis | |||
| Province: Gauteng | Radiology: unknown | Culture identification, B. dermatitidis | |||
| Comorbidities: unknown | ITS amplification and sequencing: B. percursus (99–100%) | ||||
| 2 | 2008/17, 18 (LGTZ00000000; BP222) | Age: 52 yrs | History: 7-mo history of headache with features suggestive of raised intracranial pressure and 2-mo history of ataxia | Specimen type: brain tissue | Amphotericin B followed by itraconazole; recovered (3 yrs) |
| Race, sex: white, male | Examination: ataxic and bilateral papilloedema; tone, power, reflexes, and sensation of extremities were normal; respiratory examination findings were not noted | Histopathology: yeasts with broad-based budding and double refractile walls suggestive of B. dermatitidis | |||
| Province: Western Cape | Radiology: computerized tomography brain scan, solid contrast-enhancing cerebellar lesion with multiple small abscesses; chest X ray, bilateral perihilar disease and pulmonary infiltrates | Culture identification: B. dermatitidis | |||
| Comorbidities: HIV seronegative | ITS amplification and sequencing: B. percursus (99–100%) | ||||
| 3 | 2014/17 (QGQT00000000; SA-NICD-06) | Age: 52 yrs | History: 6-mo history of cough with hemoptysis and wt loss, which had progressed in spite of empirical therapy for TB, and 3-mo history of fleshy skin lesions on the right lower lip and right nasolabial fold | Specimen type: skin and endobronchial tissue | Amphotericin B for 14 days followed by itraconazole; recovered (11 mos) |
| Race, sex: black, male | Examination: digital clubbing, wartlike lesions of the nasolabial fold and right lower lip, rightward deviation of the trachea, dullness of the right upper lung zone with decreased breath sounds; bronchoscopic examination visualized a fleshy endobronchial mass in the right main bronchus | Histopathology: PAS and Grocott’s stains; broad-based budding yeast and double refractile walls suggestive of B. dermatitidis; reactive squamous epithelium and stroma with sheets of foamy histiocytes and granuloma with central microabscesses, within which were scattered encapsulated yeasts resembling B. dermatitidis | |||
| Province: Western Cape | Radiology: chest X ray showing a dense right upper lobe infiltrate | Culture identification: B. dermatitidis | |||
| Comorbidities: HIV seronegative | ITS amplification and sequencing: B. percursus (99–100%) | ||||
| 4 | 2014/this study (QGQR00000000; SA-NICD-08) | Age: 34 yrs | History: 5-yr history of cutaneous lesions of the groin that resolved without intervention, followed by a 5-mo history of skin lesions in the perineum and groin that began as pustules | Specimen type: skin | Itraconazole; recovered (unknown) |
| Race, sex: white, male | Examination: partially healed cutaneous ulcer with undermined edges and cribriform scarring in the perineum | Histopathology: dermis showed dense granulomatous inflammation with neutrophilic microabscess formation; large yeasts with thick capsules were noted in giant cells and in the inflammatory material, resembling B. dermatitidis | |||
| Province: Gauteng | Radiology: not done | Culture identification, B. dermatitidis | |||
| Comorbidities: HIV seronegative | ITS amplification and sequencing: B. percursus (99–100%) | ||||
| 5 | 1975/21 (QGQS00000000; SA-NICD-05) | Age: 63 yrs | History: 7 mos of an ulcerative growth on the tongue | Specimen type: tongue biopsy specimen | None; died |
| Race, sex: white, male | Examination: lesion measured 3 cm by 4 cm and involved the right edge and lower surface about 2 cm from the tip of the tongue; enlarged, mobile submandibular lymph nodes were present in the right side; the rolled edge of the ulcer and its site suggested a malignant lesion | Histopathology: broad-based yeasts in relation to giant cell granulomas resembling B. dermatitidis; granulomas with yeasts were seen in the lung | |||
| Province: Gauteng | Radiology: not done | Culture identification: B. dermatitidis | |||
| Comorbidities: unknown | ITS amplification and sequencing: B. percursus (99–100%) | ||||
| 6 | 1983/3, 4 (QGQQ00000000; SA-NICD-04) | Age: 34 yrs | History: unknown | Specimen type: skin biopsy specimen | Unknown; died |
| Race, sex: black, male | Examination: postmortem examination identified disease of lung, skin (face), kidney, liver, spleen, and thyroid | Histopathology: yeasts morphologically resembling B. dermatitidis | |||
| Province: Free State | Radiology: not done | Culture identification: B. dermatitidis | |||
| Comorbidities: unknown | ITS amplification and sequencing: B. percursus (99–100%) | ||||
| 7 | 1967/19 (SA-NICD-18) | Age: 27 yrs | History: swelling of scalp for 4 wks prior to admission; subsequently developed similar lumps over the right iliac crest, the left clavicle, and the chest wall over the left axilla; burning on micturition, pain in both thighs, feeling of lameness and low back pain for 2 wks; good appetite but lost wt for the past 4 wks | Specimen type: pus from a clavicular lesion | Kanamycin and ampicillin, later amphotericin B, nystatin, and griseofulvin; died 3.5 mos after admission |
| Race, sex: black, male | Examination: new lesion appeared in the fibula, and there was collapse of the first lumbar vertebra | Histopathology: bone showing enormous yeast cells associated with giant cells and cellular reaction with acute and chronic inflammation; hourglass-type yeast cells resembling B. dermatitidis | |||
| Province: Gauteng | Radiology: skull X ray showed large abscess cavity 6 cm in diam, from which pus discharged | Culture identification: B. dermatitidis | |||
| Comorbidities: unknown | ITS amplification and sequencing: B. percursus (99–100%) | ||||
| 8 | 1989/20 (QGQJ00000000; SA-NICD-13) | Age: 40 yrs | History: cough, wt loss, night sweats, and dyspnea; readmitted 3 mos later with chest pain and dyspnea and paravertebral abscess at the level of T4–T5 | Specimen type: paravertebral abscess | Initially given TB treatment followed by broad-spectrum antibiotics and amphotericin B (30 mg daily) intravenously; died |
| Race, sex: black, male | Examination: emaciated; clinical signs of hepatomegaly; paraplegic, with no sensation in the legs and in moderate respiratory distress; discharging sinus at the T5 level | Histopathology: chromoblastomycosis; broad-based budding yeasts resembling B. dermatitidis on potassium hydroxide stain | |||
| Province: Limpopo | Radiology: chest X ray showed a diffuse nodular infiltrate with paratracheal and hilar lymphadenopathy; spine X ray showed collapse of the T4 and T5 vertebrae | Culture identification: B. dermatitidis | |||
| Comorbidities: HIV seronegative | ITS amplification and sequencing: B. helicus or B. parvus (97%) | ||||
| 9 | Unknown/20 (QGQI00000000; SA-NICD-15) | Age: 31 yrs | History: 3-mo history of productive cough, wt loss, and dyspnea | Specimen type: subcutaneous abscess | Initially treated with antituberculous agents, amphotericin B (1.8 g over 12 wks); recovered (3 mos) |
| Race, sex: black, male | Examination: raised erythematous patches on the skin of the face, forehead, ears, and anterior chest developed 4 mos after admission; the skin was biopsied, and blastomycosis was diagnosed | Histopathology: yeasts resembling B. dermatitidis | |||
| Province: North West | Radiology: chest Xray showed a fluffy infiltrate in the left lung and bilateral nodular infiltrates, hilar lymphadenopathy, and erosion of several ribs; spine X ray showed collapse of T10 and T12 vertebrae | Culture identification: B. dermatitidis | |||
| Comorbidities: HIV seronegative | ITS amplification and sequencing: B. helicus or B. parvus (97%) | ||||
| 10 | 1975/3, 4 (QGQK00000000; SA-NICD-14) | Age: adult, unknown | History: unknown | Specimen type: subcutaneous abscess | Unknown; died |
| Race, sex: black, male | Examination: unknown | Histopathology: unknown | |||
| Province: Northern Cape | Radiology: unknown | Culture identification: B. dermatitidis | |||
| Comorbidities: severe combined immune deficiency | ITS amplification and sequencing: B. helicus or B. parvus (97%) | ||||
PAS, periodic acid-Schiff.
Mycological examination of the 20 isolates.
Based on examination of cultures grown on Sabouraud agar (at 25°C) and BHI agar (at 37°C), we distinguished two morphological groups of isolates. Twelve isolates (group 1) resembled B. percursus morphologically. The remaining 8 isolates (group 2), here named B. emzantsi, resembled B. parvus in some aspects but were morphologically distinguishable (Fig. 2 and 3). The morphological characteristics of B. emzantsi were similar to those described for B. percursus except that we observed clavate, complanate cells during conidiogenesis, with occasional helical hyphae at 25°C, while at 37°C, there were adiaspore-like cells, hyphal fragments, and infrequent broad-based, budding cells on culture (Fig. 2). No growth at 40°C was observed for any isolate from either group. Group 1 isolates were urease negative within 24 h of incubation, while group 2 isolates were urease positive within 24 h at 37°C.
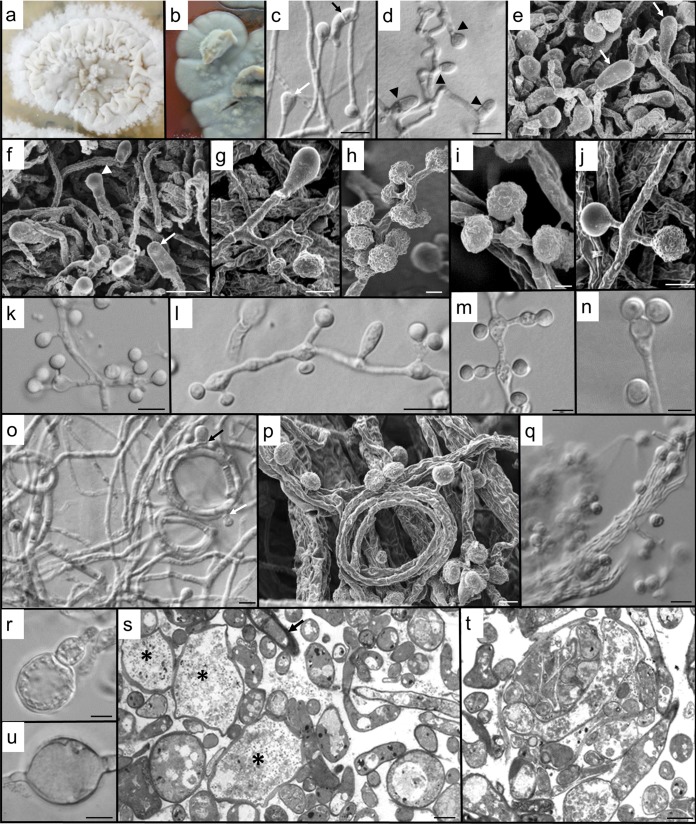
FIG 2.
Morphology of B. emzantsi. Shown are the saprophytic phase (25°C) on Sabouraud agar after 2 weeks (a, c to k, and o to q) and the intermediate phase (30°C) after 3 weeks on BHI agar with 5% horse blood (b) or Sabouraud dextrose agar (l to n and r to u). (a) Floccose, white colony, with a narrow margin peripheral to the crumpled central areas. (b) At 30°C on BHI agar with 5% horse blood, colonies were gray (darker gray on the reverse), with numerous aerial hyphal tufts and a flattened margin. (c) Two terminal conidia, a developing clavate cell (white arrow), and a clavate cell with secondary septation (black arrow). (d) Hyphal septation (arrowheads) in conidiogenesis and clavate cell formation. (e) Clavate, complanate cells (arrows) clearly distinguishable from developing conidia. (f) Complanate terminal cells (arrows) and a developing terminal conidium (arrowhead). (g) Hyphal cell subtending single, lateral conidia and a terminal, basally septate, clavate cell. (h) The apparent clusters of conidia are usually due to the short pedicels of many of the laterally positioned conidia. (i) Multiple conidia on short secondary conidiophores can be subtended from a single basal cell. (j) Conidial ornamentation is variable, from glabrous to papillate. (k) The single conidium may be lateral or clustered on vesiculate primary conidiophores. (l) Clavate cell between conidiophores, with variously arranged conidia. Some inflation of subtending cells is apparent at an intermediate temperature. (m) Increase in temperature results in conidiophores becoming increasingly ampulliform and vesiculate, with distended hyphal cells. Light microscopy creates the illusion that one of each of the laterally paired conidia is sessile. (n) Expanded primary conidiophore with two conidia on extremely short secondary conidiophores. (o) Helical hyphae, one with both developing clavate cells (black arrow) and conidia (white arrow). (p) Numerous conidia developing on hyphal gyres, with bundles of older hyphae behind them. (q) Abundant verrucose conidia and hyphae aggregated into rope-like structures. (r) Development of an adiaspore-like cell (inflated terminal conidia). (s) Sectioned material providing accurate imaging of the diversity of cell forms, including the development of giant cells (*) from enlarged, fragmented hyphal cells; increased vacuolation of enlarging cells; numerous thin hyphal profiles; and some thick-walled cells (arrow) (tangentially sectioned). (t) Section through the hyphal bundles seen by light and scanning microscopy revealing numerous endo/intrahyphal profiles. (u) An inflated intercalary cell suggestive of a chlamydospore. Bars, 5 μm (c, d, k to o, q, s, and u) and 2 μm (e to j, p, r, and t).
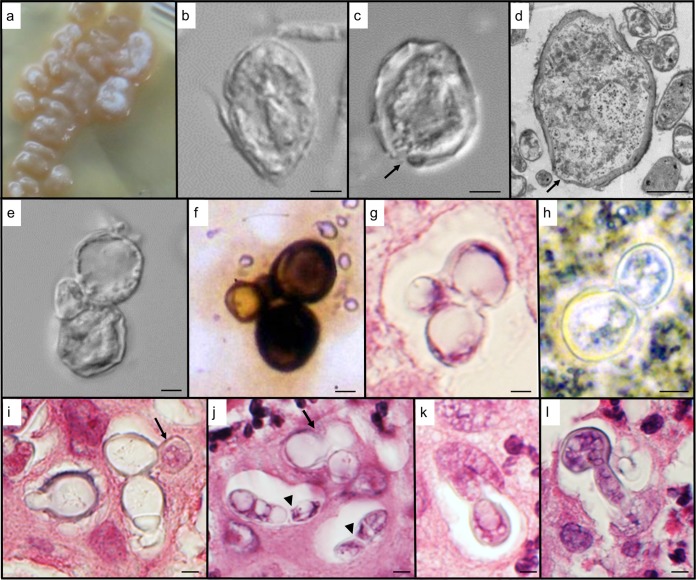
FIG 3.
Morphology of B. emzantsi at 37°C. (a to e) Cultured isolates on BHI agar at 4 weeks; (f) Grocott’s stain of a paravertebral pus smear; (g and i to l) hematoxylin-and-eosin-stained section through the sinus tissue of a vertebral abscess; (h) potassium hydroxide stain of pus. (a) A beige, butyrous, cerebriform colony still with some white, aerial hyphal tufts. (b) Incomplete transformation in culture typified by large, thick-walled, yeastlike cells juxtaposed to hyphal fragments. (c) Thick-walled giant cell filled with cytoplasm (indicative of an active, viable cell). The slight disruption in the cell wall (arrow) may be the beginning of a budding process. (d) Section through one of the large, thick-walled cells filled with cytoplasm. A number of thin areas in the cell wall (arrows) are suggestive of the initiation of multiple budding sites. Note the many vegetative hyphal profiles surrounding the giant cell. (e) Budding yeastlike cells with a smaller lateral cell, from a cultured isolate. (f) Similar configuration of cells as in panel e but photographed from the clinical specimen. (g) Section through infected sinus tissue, again illustrating yeastlike budding cells with a smaller laterally positioned cell. (h) Image showing a typical broad-based budding yeast cell. (i) A number of budding, thick-walled cells, sectioned through various planes. It appears that one of the daughter cells (arrow) has penetrated the tissue around the phagocytic vacuole. (j) Although budding cells are apparent in the section (arrow), other budding-cell profiles appear to contain hyphal cells (arrowheads), as the cytoplasm is delimited by a cell wall on the inside of the host vacuole. (k) Hyphal invasion of surrounding tissue from a daughter cell within the phagocytic vacuole. (l) Hypha-like extrusion of a thicker-walled cell into surrounding sinus tissue. Bars, 2 μm (b, d to i, and k) and 3 μm (c, j, and l).
Antifungal susceptibility testing of isolates.
All isolates grew for MIC determination after 5 to 6 days of incubation. Tables 2 and 3 summarize the distribution, range, and GM MIC/minimum effective concentration (MEC) for nine antifungal agents for the yeast and mold phases of 17 isolates. There was no difference between the BMD and Etest GM MICs for voriconazole, posaconazole, and itraconazole for the yeast and mold phases of B. percursus and B. emzantsi (Tables 2 and 3). The yeast- and mold-phase GM MICs of fluconazole were higher for both B. percursus and B. emzantsi (BMD GM MIC range, 1.41 mg/liter to 2.18 mg/liter) than those of the other tested azoles. The amphotericin B Etest GM MIC for the B. percursus mold phase was lower (0.01 mg/liter) than the MIC for the B. emzantsi mold phase (0.26 mg/liter). The BMD GM MICs for anidulafungin were higher (range, 0.27 mg/liter to 1 mg/liter) than those for micafungin (range, 0.03 mg/liter to 0.13 mg/liter) for both yeast and mold phases of B. percursus and B. emzantsi.
TABLE 2.
Yeast-phase MIC distribution of Blastomyces isolates (9 B. percursus isolates, 8 B. emzantsi isolates, and 1 B. dermatitidis isolate [ATCC 10225])a
| Case (GenBank accession no.; isolate) | Reference(s) | Etest MIC (mg/liter) |
BMD MIC (mg/liter) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMB | VRC | POS | ITC | FLC | VRC | POS | ITC | 5FC | CAS | MFG | ANG | ||
| Group 1 (Blastomyces percursus) | |||||||||||||
| 1 (QKWI00000000; NCPF 4091) | 5 | 0.004 | 0.002 | 0.002 | 0.002 | 4 | 0.008 | 0.008 | 0.008 | 64 | 0.06 | 0.008 | 0.12 |
| 2 (LGTZ00000000; BP222) | 17, 18 | 0.064 | 0.002 | 0.002 | 0.002 | 8 | 0.03 | 0.25 | 0.25 | 256 | 0.5 | 0.12 | 0.12 |
| 3 (QGQT00000000; SA-NICD-06) | 17 | 0.012 | 0.002 | 0.002 | 0.002 | 1 | 0.008 | 0.03 | 0.06 | 128 | 0.5 | 0.06 | 0.5 |
| 4 (QGQR00000000; SA-NICD-08) | This study | 0.047 | 0.002 | 0.002 | 0.002 | 4 | 0.015 | 0.008 | 0.015 | 256 | 1 | 0.03 | 2 |
| 5 (QGQS00000000; SA-NICD-05) | 21 | 0.094 | 0.012 | 0.002 | 0.003 | 8 | 0.015 | 0.008 | 0.008 | 32 | 4 | 0.12 | 1 |
| 6 (QGQQ00000000; SA-NICD-04) | 3, 4 | 0.003 | 0.002 | 0.002 | 0.002 | 0.25 | 0.008 | 0.008 | 0.008 | 32 | 0.5 | 0.03 | 4 |
| 7 | |||||||||||||
| SA-NICD-18 | 19 | 0.003 | 0.002 | 0.002 | 0.002 | 0.5 | 0.008 | 0.008 | 0.008 | 32 | 1 | 0.03 | 0.5 |
| QGQP00000000; SA-NICD-03 | This study | 0.004 | 0.002 | 0.002 | 0.002 | 2 | 0.015 | 0.008 | 0.008 | 32 | 0.5 | 0.12 | 0.5 |
| QGQN01000000; SA-NICD-07 | This study | 0.004 | 0.002 | 0.002 | 0.002 | 2 | 0.015 | 0.015 | 0.03 | 16 | 0.5 | 0.015 | 0.25 |
| MIC range | 0.003–0.094 | 0.002–0.012 | 0.002–0.002 | 0.002–0.003 | 0.25–8 | 0.008–0.03 | 0.008–0.25 | 0.008–0.25 | 32–256 | 0.06–4 | 0.008–0.12 | 0.12–4 | |
| GM | 0.02 | 0.002 | 0.002 | 0.002 | 2 | 0.01 | 0.02 | 0.02 | 59.26 | 0.59 | 0.03 | 0.54 | |
| ATCC | 0.002 | 0.002 | 0.002 | 0.002 | 0.5 | 0.015 | 0.12 | 0.06 | 256 | 2 | 8 | 2 | |
| Group 2 (Blastomyces emzantsi) | |||||||||||||
| 8 (QGQJ00000000; SA-NICD-13) | 20 | 0.25 | 0.002 | 0.002 | 0.006 | 2 | 0.015 | 0.008 | 0.03 | 128 | 2 | 0.06 | 1 |
| 9 (QGQI00000000; SA-NICD-15) | 20 | 0.25 | 0.002 | 0.002 | 0.003 | 2 | 0.008 | 0.008 | 0.015 | 32 | 1 | 0.06 | 1 |
| 10 | |||||||||||||
| QGQK00000000; SA-NICD-14 | 3, 4 | 0.023 | 0.003 | 0.002 | 0.002 | 1 | 0.008 | 0.008 | 0.008 | 16 | 2 | 0.06 | 1 |
| QGQL00000000; SA-NICD-16 | This study | 0.002 | 0.002 | 0.002 | 0.002 | 1 | 0.008 | 0.008 | 0.008 | 64 | 1 | 0.03 | 0.5 |
| QGQE00000000; SA-NICD-12 | This study | 0.032 | 0.002 | 0.002 | 0.002 | 2 | 0.015 | 0.008 | 0.008 | 128 | 4 | 0.06 | 2 |
| QGQF00000000; SA-NICD-09 | This study | 0.047 | 0.002 | 0.004 | 0.008 | 4 | 0.03 | 0.008 | 0.12 | 64 | 0.5 | 0.008 | 1 |
| QGQG00000000; SA-NICD-10 | This study | 0.047 | 0.002 | 0.003 | 0.016 | 2 | 0.008 | 0.008 | 0.008 | 64 | 4 | 0.03 | 2 |
| QGQH00000000; SA-NICD-11 | This study | 0.64 | 0.002 | 0.002 | 0.002 | 0.25 | 0.008 | 0.008 | 0.008 | 16 | 1 | 0.03 | 0.5 |
| MIC range | 0.002–0.64 | 0.002–0.003 | 0.002–0.004 | 0.002–0.016 | 0.25–4 | 0.008–0.03 | 0.008–0.008 | 0.008–0.12 | 16–128 | 0.5–4 | 0.008–0.3 | 0.5–2 | |
| GM | 0.06 | 0.002 | 0.002 | 0.004 | 1.41 | 0.01 | 0.01 | 0.01 | 49.35 | 1.54 | 0.03 | 1 | |
BMD, broth microdilution; GM, geometric mean; ATCC, B. dermatitidis ATCC 10225; AMB, amphotericin B; VRC, voriconazole; POS, posaconazole; ITC, itraconazole; FLC, fluconazole; 5FC, 5-flucytosine; CAS, caspofungin; MFG, micafungin; ANG, anidulafungin.
TABLE 3.
Mold-phase MIC distribution of Blastomyces isolates (9 B. percursus isolates, 8 B. emzantsi isolates, and 1 B. dermatitidis isolate [ATCC 10225])a
| Case (GenBank accession no.; isolate | Reference(s) | Etest MIC (mg/liter) |
BMD MIC and MEC (mg/liter) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMB | VRC | POS | ITC | FLC | VRC | POS | ITC | 5FC | CAS | MFG | ANG | ||
| Group 1 (Blastomyces percursus) | |||||||||||||
| 1 (QKWI00000000; NCPF 4091) | 5 | 0.008 | 0.002 | 0.002 | 0.002 | 0.5 | 0.008 | 0.06 | 0.12 | 256 | 0.12 | 0.06 | 0.25 |
| 2 (LGTZ00000000; BP222) | 17, 18 | 0.004 | 0.002 | 0.002 | 0.012 | 4 | 0.008 | 0.06 | 0.03 | 64 | 0.06 | 0.008 | 0.008 |
| 3 (QGQT00000000; SA-NICD-06) | 17 | 0.008 | 0.002 | 0.002 | 0.012 | 2 | 0.008 | 0.015 | 0.03 | 256 | 0.5 | 0.25 | 0.5 |
| 4 (QGQR00000000; SA-NICD-08) | This study | 0.012 | 0.002 | 0.012 | 0.006 | 2 | 0.008 | 0.06 | 0.06 | 32 | 0.5 | 0.03 | 4 |
| 5 (QGQS00000000; SA-NICD-05) | 21 | 0.002 | 0.002 | 0.002 | 0.003 | 2 | 0.008 | 0.008 | 0.015 | 256 | 1 | 0.03 | 0.25 |
| 6 (QGQQ00000000; SA-NICD-04) | 3, 4 | 0.002 | 0.002 | 0.002 | 0.003 | 2 | 0.008 | 0.015 | 0.03 | 64 | 0.5 | 0.03 | 0.25 |
| 7 | |||||||||||||
| SA-NICD-18 | 19 | 0.064 | 0.002 | 0.002 | 0.004 | 2 | 0.008 | 0.008 | 0.06 | 128 | 2 | 0.06 | 1 |
| QGQP00000000; SA-NICD-03 | This study | 0.002 | 0.002 | 0.002 | 0.003 | 0.12 | 0.008 | 0.008 | 0.008 | 8 | 0.25 | 0.008 | 0.12 |
| QGQN01000000; SA-NICD-07 | This study | 0.002 | 0.002 | 0.002 | 0.002 | 4 | 0.008 | 0.06 | 0.12 | 256 | 0.5 | 0.03 | 0.25 |
| MIC range | 0.002–0.064 | 0.002–0.002 | 0.002–0.012 | 0.002–0.012 | 0.12–4 | 0.008–0.008 | 0.008–0.06 | 0.015–0.12 | 8–256 | 0.06–2 | 0.008–0.25 | 0.008–4 | |
| GM | 0.01 | 0.002 | 0.002 | 0.004 | 1.46 | 0.01 | 0.02 | 0.04 | 94.06 | 0.39 | 0.03 | 0.27 | |
| ATCC | 0.002 | 0.002 | 0.002 | 0.012 | 0.5 | 0.008 | 0.06 | 0.03 | 256 | 1 | 8 | 2 | |
| Group 2 (Blastomyces emzantsi) | |||||||||||||
| 8 (QGQJ00000000; SA-NICD-13) | 20 | 0.25 | 0.004 | 0.002 | 0.012 | 4 | 0.008 | 0.06 | 0.12 | 256 | 4 | 0.25 | 1 |
| 9 (QGQI00000000; SA-NICD-15) | 20 | 0.094 | 0.006 | 0.002 | 0.016 | 4 | 0.008 | 0.015 | 0.06 | 128 | 1 | 0.25 | 0.5 |
| 10 | |||||||||||||
| QGQK00000000; SA-NICD-14 | 3, 4 | 0.032 | 0.002 | 0.003 | 0.03 | 1 | 0.008 | 0.008 | 0.008 | 128 | 0.5 | 0.06 | 0.5 |
| QGQL00000000; SA-NICD-16 | This study | 0.47 | 0.002 | 0.002 | 0.003 | 1 | 0.008 | 0.008 | 0.008 | 128 | 4 | 0.25 | 1 |
| QGQE00000000); SA-NICD-12 | This study | 1 | 0.002 | 0.002 | 0.012 | 1 | 0.015 | 0.008 | 0.008 | 128 | 1 | 0.12 | 2 |
| QGQF00000000; SA-NICD-09 | This study | 0.19 | 0.002 | 0.002 | 0.006 | 8 | 0.015 | 0.008 | 0.06 | 128 | 4 | 0.12 | 2 |
| QGQG00000000; SA-NICD-10 | This study | 0.64 | 0.002 | 0.002 | 0.012 | 2 | 0.008 | 0.03 | 0.06 | 128 | 2 | 0.12 | 1 |
| QGQH00000000; SA-NICD-11 | This study | 0.5 | 0.002 | 0.002 | 0.006 | 2 | 0.008 | 0.015 | 0.06 | 128 | 2 | 0.06 | 0.5 |
| MIC range | 0.032–1 | 0.002–0.006 | 0.002–0.003 | 0.003–0.03 | 1–8 | 0.008–0.015 | 0.008–0.06 | 0.008–0.12 | 128–256 | 0.5–4 | 0.06–0.25 | 0.5–2 | |
| GM | 0.26 | 0.002 | 0.002 | 0.01 | 2.18 | 0.01 | 0.01 | 0.03 | 139.59 | 1.83 | 0.13 | 0.92 | |
BMD, broth microdilution; GM, geometric mean; ATCC, B. dermatitidis ATCC 10225; AMB, amphotericin B; VRC, voriconazole; POS, posaconazole; ITC, itraconazole; FLC, fluconazole; 5FC, 5-flucytosine; CAS, caspofungin; MFG, micafungin; ANG, anidulafungin.
Multilocus and whole-genome phylogenetic analyses.
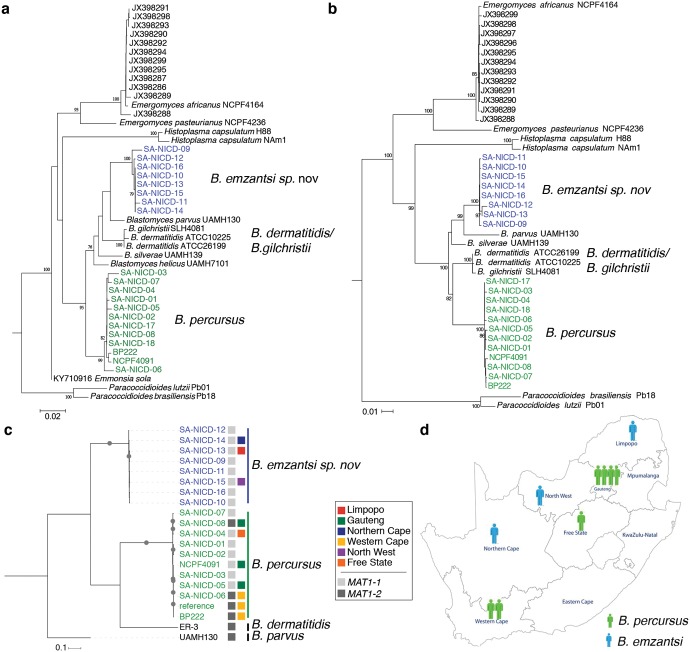
Based on ITS sequences only, the 12 strains of group 1 were confirmed as B. percursus, while the 8 strains of group 2 were very similar to B. parvus (97%) and B. helicus (97%). A phylogenetic tree of the ITS sequences separated the two groups from B. dermatitidis/B. gilchristii, B. parvus, B. silverae, B. helicus, and Emmonsia sola (Fig. 4a). Multilocus phylogenetic analysis of the concatenated sequences of ITS–LSU–PRP8–β-tubulin–actin confirmed the separation and clearly demonstrated that group 1 (B. percursus) and group 2 (B. emzantsi) isolates were distinct species (Fig. 4b). Phylogenetic analysis of each gene (ITS, LSU, intein PRP8, beta tubulin, and actin) sequence also revealed 2 distinct groups (see Fig. S2 in the supplemental material). B. dermatitidis and B. gilchristii did not separate into two different clades. Whole-genome phylogenetic analysis of 1,712,033 variant sites identified from aligning whole-genome reads to B. percursus strain BP222 strongly supported these phylogenetic relationships, also confirming that isolates from group 1 clustered with B. percursus (100% bootstrap support) and that isolates from group 2 conformed to an intermediate monophyletic clade between B. parvus and B. percursus/B. dermatitidis that represents a novel Blastomyces species (100% bootstrap support) (Fig. 4c). Large genetic diversity was found in sympatric B. emzantsi and B. percursus strains, with an average of 900,000 SNPs between species.
FIG 4.
Multigene phylogenies of B. percursus (group 1) (SA-NICD-01 to SA-NICD-08, SA-NICD-17, and SA-NICD-18) and B. emzantsi (group 2) (SA-NICD-09 to SA-NICD-16). (a and b) Maximum likelihood tree inferred from the ITS gene and concatenated gene alignment of ITS2–LSU–PRP8–β-tubulin–actin based on 1,000 replicates. (a) Sequences of Paracoccidioides lutzii, Paracoccidioides brasiliensis, Blastomyces dermatitidis, Histoplasma capsulatum, and Emergomyces africanus from Kenyon et al. (22) and of Blastomyces silverae, Blastomyces helicus, Blastomyces parvus, and Emmonsia sola from Jiang et al. (8). (c) Maximum likelihood tree inferred from SNPs, including three annotated genome assemblies of Blastomyces species (B. dermatitidis ER3, B. parvus UAMH 130, and B. percursus BP222). All nodes were supported by 100% of bootstrap replicates. The boxes indicate the mating type for each Blastomyces species. (d) Geographic distribution of B. percursus and B. emzantsi cases in South African provinces.
Clinical presentation and treatment of 10 cases.
(i) B. percursus. Four patients (cases 2 to 5) presented with a 2- to 7-month history of symptoms, including weight loss; productive cough with hemoptysis; cutaneous lesions, including chronic ulcers; and headache. Most of these patients had multisystem involvement. All seven patients (cases 1 to 7) had extrapulmonary disease, and three additional patients (cases 2, 3, and 6) had pulmonary disease. Four patients (cases 1, 3, 4, and 6) had cutaneous involvement. One patient presented with vertebral disease (case 7). B. percursus was isolated in the Gauteng (cases 1, 4, 5, and 7), Western Cape (cases 1 and 2), and Free State (case 6) provinces of South Africa (Fig. 4d).
(ii) B. emzantsi. Patient 10 had an underlying severe combined immune deficiency, with a subcutaneous abscess, but other clinical data were lacking. Patients 8 and 9 presented with cough, weight loss, night sweats, and dyspnea. Both were found to have pulmonary disease with collapsed thoracic vertebrae, and patient 8 had a purulent discharge from a sinus tract at the T5 vertebral level (Fig. 1). Both patients were treated empirically for tuberculosis (TB). Patient 8 died soon after starting amphotericin B, while patient 9 recovered after receiving a cumulative total of 1.8 g of amphotericin B (Table 1). B. emzantsi was isolated in the Limpopo (case 8), North West (case 9), and Northern Cape (case 10) provinces of South Africa (Fig. 4d).
Genome assembly sizes.
The genomic sizes of B. percursus (32.1 to 34.2 Mb) and B. emzantsi (27.4 Mb to 32.5 Mb) were similar to those of other closely related dimorphic fungi; however, they differed from those reported for B. dermatitidis (66.6 Mb) and B. gilchristii (75.4 Mb) (5).
Nucleotide diversity.
The estimated genome-wide nucleotide diversity (π) for B. emzantsi is 0.00029, which is almost twice as large as that for B. percursus, at 0.00052. Relative to the Blastomyces genus (π = 0.00905), this represents ∼31-fold- and ∼17-fold-lower levels of diversity for B. emzantsi and B. percursus populations than observed at the genus level, respectively. Compared to other dimorphic fungal pathogens from the Ajellomycetaceae causing endemic mycoses, the level of diversity in B. emzantsi is lower than that reported for Paracoccidioides brasiliensis S1a and PS2 (π = 0.00053 and 0.00066, respectively) but higher than the clonal diversity reported for P. brasiliensis PS3, which is mainly found in regions of endemicity in Colombia (π = 0.00008) (33). The degree of genetic variation supports that B. emzantsi is a single species, well separated within the Blastomyces genus.
Mating-type analysis.
Of the 18 Blastomyces strains analyzed for mating type, only the 3 recently diagnosed cases of B. percursus contained the MAT1-2 gene. Furthermore, all B. emzantsi strains contained the MAT1-1 gene. The population ratio for each mating type is 1:1 in other dimorphic pathogens from the Ajellomycetaceae, such as Paracoccidioides. Either MAT1-1 or MAT1-2 has been reported in other dimorphic fungal pathogens, demonstrating that these strains are heterothallic (7, 29).
BAD-1 gene identification from WGS.
The full orthologue of the BAD-1 gene was absent in all 18 Blastomyces strains; only a partial paralogue region was found.
Taxonomy.
Blastomyces emzantsi sp. nov.: Maphanga, Govender, Birkhead, Frean; MycoBank accession number MB828102. Etymology: referring to South Africa, the country of origin of the type species. Emzantsi means south in the isiXhosa language. Morphologically, B. emzantsi can be differentiated based on the combination of saprobe-phase helical hyphae, abundant conidiogenesis, and clavate, complanate cells extending terminally and laterally from hyphal cells, together with an incomplete transition to the yeast phase at 37°C (hyphae, adiaspore-like/giant cells, and broad-based budding yeasts are all present in the thermotolerant phase). Holotype: South Africa, National Institute for Communicable Diseases, dried herbarium specimen culture from a subcutaneous abscess of an HIV-seronegative black African man with blastomycosis, described by Frean et al. in 1993 (20); living strain, NICD (SA-NICD-15; case 9). Physiology: minimum growth temperature of 8°C, optimum temperature of 25°C with growth reaching 55 mm on Sabouraud agar in 29 days, and maximum temperature of 37°C; urease positive. For differential molecular diagnosis, B. emzantsi can be diagnosed by the following nucleotide characters, which are fixed in B. percursus, B. dermatitidis, B. gilchristii, B. parvus, B. helicus, and B. silverae: internal transcribed spacer 2 of the ribosomal DNA (rDNA) at positions 140 (T:C) and 169 (A:T).
DISCUSSION
We reexamined clinical isolates from 20 human cases of blastomycosis from South Africa from 1967 to 2014 and found that these were not B. dermatitidis, as initially believed (3–5, 17–21), but rather were different species: 12 isolates belonged to the recently described species B. percursus (5), and 8 isolates belonged to a novel species, described here as B. emzantsi. None of the 20 isolates clustered with B. dermatitidis, B. gilchristii, or B. parvus.
Blastomycosis is an infrequently diagnosed endemic mycosis in South Africa compared to emergomycosis and histoplasmosis (22, 34). This more likely reflects a diagnostic bias rather than a true difference in prevalence owing to a lack of awareness among clinicians and a lack of access to simple non-culture-based diagnostic methods. The very closely related fungi B. dermatitidis and B. gilchristii cause infections with varying clinical phenotypes (16, 35–37). On the other hand, investigators have previously hypothesized that African blastomycosis is caused by different Blastomyces strains that result in disseminated (cutaneous and bone) disease rather than localized pulmonary disease (3, 4, 35). Similar to Emergomyces and Histoplasma, the lungs are the primary site of infection, and acquisition of infection occurs following inhalation of conidia. Dissemination of the fungus from the lungs via the bloodstream is presumed to lead to skin and bone involvement. Our study supports the latter hypothesis, mainly because we found that South African cases of blastomycosis were caused by species other than B. dermatitidis. However, our sample size was too small to draw any firm conclusions about any differences in the clinical presentations of South African versus North American cases of blastomycosis. Nevertheless, 4 of the 10 cases with detailed clinical information in our study had pulmonary infiltrates on chest X ray, while all 10 cases had extrapulmonary disease. This is consistent with previous reports showing that cutaneous and bone diseases are more common than pulmonary disease in African blastomycosis, in contradistinction to the pattern in North America (3, 38). Six of 10 cases had cutaneous lesions. It is possible that extrapulmonary disease could have resulted because of delayed or inaccurate diagnosis and/or a lack of pulmonary clinical assessment (35). Both B. percursus and B. emzantsi caused vertebral and bone collapse, which is a classical feature noted in patients with African blastomycosis (3, 4, 35). The clinical picture of vertebral blastomycosis mimics that of vertebral TB, thus making clinical diagnosis challenging, particularly in a high-prevalence setting for TB (39).
The treatments of choice for North American blastomycosis are itraconazole for mild infections and amphotericin B followed by itraconazole for severe infections (39, 40). Four people with B. percursus infection for whom treatment and outcome data were available recovered after receiving azole or amphotericin B treatment. Although there are no clinical trials to compare the efficacies of these regimens for the treatment of blastomycosis, in vitro MIC data for B. dermatitidis, B. percursus, and B. helicus suggest that the azoles (excluding fluconazole) and amphotericin B are active (9, 41–44). There were some important differences between the susceptibility profiles of B. percursus and B. emzantsi. For instance, the amphotericin B Etest GM MIC for the mold phase was 0.01 mg/liter for B. percursus, versus 0.26 mg/liter for B. emzantsi. Also, both B. percursus and B. emzantsi isolates had high anidulafungin BMD GM MICs. According to previous reports, anidulafungin is weakly active against the mold phase of B. percursus, B. parvus, and B. silverae (41). Zhanel et al. reported an MIC range of 4.0 μg/ml to 64.0 μg/ml for the mold phase of B. dermatitidis with this antifungal agent (45). Similarly, we observed high anidulafungin activity against the two phases of B. percursus and B. emzantsi as well as the ATCC 10225 strain of B. dermatitidis in our study.
Macroscopically, there was an incomplete transition from the mold to the yeast phase for all 20 isolates, with most of the strains producing hyphal elements at 37°C. Similar results were reported previously by Kaufman et al. and Lombardi et al. (11, 12). The observation of hyphal elements in tissue warrants special mention, as histopathologists should be aware of these findings. Microscopically, the maximum size of the yeast cells of B. percursus (9 μm by 14 μm) was larger than that previously reported (6.5 μm by 12.2 μm) (5, 8). B. emzantsi produced larger broad-based budding yeast cells (9.0 to 10.2 by 13.8 to 20.9 μm) that were more similar to those of B. parvus, which until recently were thought to produce adiaspores (8). Helical hyphal gyres bearing conidia were present in the mycelial phase, similar to those previously described for B. parvus and Emmonsia sola (at 21°C) (8, 46). A urease test was positive within 24 h of incubation for B. emzantsi; however, similar results have been reported for other dimorphic pathogens, such as Emergomyces africanus, Emergomyces europaeus, Emergomyces pasteurianus, Emmonsia sola, and Emmonsia crescens (8).
The ITS region is a formal fungal barcode, which cannot always distinguish closely related cryptic species (47). In this study, all eight B. emzantsi isolates were erroneously identified as B. helicus or B. parvus based on ITS sequencing and a BLAST search; however, phylogenetic analysis of these ITS sequences separated the species into different clades. Of the 20 presumptive B. dermatitidis South African strains analyzed, none clustered with B. dermatitidis, B. gilchristii, or B. parvus. In 2013, Brown et al. reported the existence of a clade distinct from North American B. dermatitidis strains; this cryptic species was named B. gilchristii (7). Dukik et al. recently reported a novel species, B. percursus, which was isolated from cases in South Africa and Israel (5). Thereafter, using MLST, other Blastomyces species were identified, which cause disease in mammalian hosts (8). Our concatenated sequences of the ITS–LSU–PRP8–β-tubulin–actin genes could not separate B. dermatitidis from B. gilchristii but clearly differentiated B. percursus, B. parvus, and B. emzantsi from the former two species, forming four clades and confirming the existence of Blastomyces species different from B. dermatitidis/B. gilchristii. Furthermore, based on whole-genome sequence analysis, the degree of genetic variation supported that B. emzantsi is a single species well separated within the Blastomyces genus. Although none of the 20 Blastomyces isolates currently in the NICD’s collection belong to B. dermatitidis/B. gilchristii, other studies have reported B. gilchristii (CDC B1566 [UAMH 10245]) from South Africa (7).
BAD-1 is an important conserved adhesion-promoting protein and virulence factor in B. dermatitidis strains (48). We could not identify the entire BAD-1 gene in our strains. Previous studies have also reported the absence of the BAD-1 gene from African Blastomyces strains (14, 49). A study of African Blastomyces strains that lacked the BAD-1 gene showed that they exhibited an attenuated disease profile (35). Furthermore, in a mouse model, mice infected with a B. dermatitidis strain containing the BAD-1 gene died within 4 weeks, while those that lacked the BAD-1 gene survived (50). The BAD-1 gene seems to be an important virulence factor in North American Blastomyces strains; however, the impact of the absence of this gene in African strains remains unclear. More studies are required to determine virulence genes that may contribute to differences in clinical manifestations noted between cases of African and North American blastomycoses.
Sexual reproduction greatly increases genetic diversity within Blastomyces and may contribute to the emergence of new genotypes with different virulence profiles (29). Of the 18 Blastomyces strains analyzed for mating-type distribution, all B. emzantsi strains contained MAT1-1, while the B. percursus isolates contained both the MAT1-1 and MAT1-2 genes. The fact that two alleles are not evenly distributed in B. emzantsi and B. percursus suggests a clonal expansion of MAT1-1 and selection favoring MAT1-1; however, wider sampling of clinical and environmental strains and examination of additional loci are needed to confirm these trends (29).
Our study has several limitations. Only cases reported to a reference laboratory were included, which precludes estimation of the prevalence of blastomycosis in South Africa. Our sample size was small (20 isolates over almost 5 decades), which could be due to cases being missed because of limited resources and the lack of clinical awareness in our setting. Furthermore, if B. percursus and/or B. emzantsi is more likely to cause cutaneous disease than other species, then given that skin lesions (or draining pus) may be more accessible for culture, there may be a bias toward this species being represented in the NICD’s culture collection. This is plausible because we suspect that cases of pulmonary blastomycosis are misdiagnosed as smear- or Xpert MTB/RIF-negative pulmonary TB in South Africa. Our analysis included only clinical isolates from humans. In fact, neither veterinary blastomycosis nor environmental isolation of Blastomyces species has been reported from South Africa. Small numbers and incomplete clinical details limited our comparison of the diseases caused by these species. With this in mind, the two persons infected with B. emzantsi for whom clinical data were available had systemic pyogranulomatous pulmonary and vertebral blastomycosis consistent with disease observed for B. percursus and not unusual for North American blastomycosis (14). Whether B. emzantsi is more likely to be associated with this syndrome will require characterization of additional cases.
Conclusion.
South African cases of blastomycosis reported to a national mycology reference laboratory over 5 decades were caused by two species, B. percursus and B. emzantsi, and not B. dermatitidis, as had been presumed. Both taxa caused disease with overlapping clinical presentations, with a high proportion of extrapulmonary disease. Although histology and culture are useful methods in the diagnosis of blastomycosis, our analysis highlights the importance of molecular methods to identify emerging species correctly as causes of old diseases. Increasing awareness of African blastomycosis among clinicians and clinical microbiologists and access to simple, rapid, non-culture-based diagnostics in resource-limited settings may improve the early diagnosis of this illness.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge Sydney Mogokotleng (National Institute for Communicable Diseases) for his assistance with retrieving case records from NICD archives and Rudzani Mathebula for constructing the map in Fig. 4d.
T.G.M., I.S.S., J.A.F., and N.P.G. collected data. M.B. performed electron microscopy. T.G.M., M.B., T.G.Z., J.A.F., and N.P.G. performed phenotypic identification. T.G.M., T.G.Z., and R.S.M. performed antifungal susceptibility testing. T.G.M., J.F.M., M.A., and A.I. performed whole-genome sequencing. T.G.M., I.S.S., M.B., J.F.M., and N.P.G. analyzed the data and wrote the manuscript. T.G.M., M.B., J.F.M., C.A.C., I.S.S., C.C., M.A., A.I., S.D.N., C.K., A.M.B., S.D.H., J.A.F., and N.P.G. critically reviewed the manuscript.
This work was supported by the National Institute for Communicable Diseases, a division of the National Health Laboratory Service, South Africa.
We have no conflicts of interest to declare.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Gilchrist TC. 1894. Protozoan dermatitis. J Cutan Gen Dis 12:496–499. [Google Scholar]
- 2.Jerray M, Hayouni A, Benzarti M, Klabi N, Garrouche A. 1992. Blastomycosis in Africa: a new case from Tunisia. Eur Respir J 5:365–367. [PubMed] [Google Scholar]
- 3.Carman WF, Frean JA, Crewe-Brown HH, Culligan GA, Young CN. 1989. Blastomycosis in Africa. A review of known cases diagnosed between 1951 and 1987. Mycopathologia 107:25–32. doi: 10.1007/bf00437587. [DOI] [PubMed] [Google Scholar]
- 4.Frean JA, Carman WF, Crewe-Brown HH, Culligan GA, Young CN. 1989. Blastomyces dermatitidis infections in the RSA. S Afr Med J 76:13–16. [PubMed] [Google Scholar]
- 5.Dukik K, Muñoz JF, Jiang Y, Feng P, Sigler L, Stielow JB, Freeke J, Jamalian A, van den Ende BG, McEwen JG, Clay OK, Schwartz IS, Govender NP, Maphanga TG, Cuomo CA, Moreno LF, Kenyon C, Borman AM, de Hoog S. 2017. Novel taxa of thermally dimorphic systemic pathogens in the Ajellomycetaceae (Onygenales). Mycoses 60:296–309. doi: 10.1111/myc.12601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gilchrist TC, Stokes WR. 1898. A case of pseudo-lupus vulgaris caused by a Blastomyces. J Exp Med 3:53–78. doi: 10.1084/jem.3.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown EM, McTaggart LR, Zhang SX, Low DE, Stevens DA, Richardson SE. 2013. Phylogenetic analysis reveals a cryptic species Blastomyces gilchristii, sp. nov. within the human pathogenic fungus Blastomyces dermatitidis. PLoS One 11:e0059237. doi: 10.1371/journal.pone.0059237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang Y, Dukik K, Muñoz JF, Sigler L, Schwartz IS, Govender NP, Kenyon C, Feng P, van den Ende BG, Stielow JB, Stchigel AM, Lu H, de Hoog S. 2018. Phylogeny, ecology and taxonomy of systemic pathogens and their relatives in Ajellomycetaceae (Onygenales): Blastomyces, Emergomyces, Emmonsia, Emmonsiellopsis. Fungal Divers 90:245–291. doi: 10.1007/s13225-018-0403-y. [DOI] [Google Scholar]
- 9.Schwartz IS, Wiederhold NP, Patterson TF, Sigler L. 2019. Blastomyces helicus, a new dimorphic fungus causing fatal pulmonary and systemic disease in humans and animals in western Canada and United States. Clin Infect Dis 68:188–195. doi: 10.1093/cid/ciy483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kwon-Chung KJ. 1971. Genetic analysis on the incompatibility system of Ajellomyces dermatitidis. Sabouraudia 9:231–238. doi: 10.1080/00362177185190461. [DOI] [PubMed] [Google Scholar]
- 11.Kaufman L, Standard PG, Weeks RJ, Padhye AA. 1983. Detection of two Blastomyces dermatitidis serotypes by exoantigen analysis. J Clin Microbiol 18:110–114. doi: 10.1128/JCM.18.1.110-114.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lombardi G, Padhye AA, Ajello L. 1988. In vitro conversion of African isolates of Blastomyces dermatitidis to their yeast form. Mycoses 31:447–450. [PubMed] [Google Scholar]
- 13.Mercantini R, Marsella R, Moretto D, Mercantini P, Balus L, Mastroianni A, Ferraro C. 1995. Macroscopic and microscopic characteristics of an African Blastomyces dermatitidis strain. Mycoses 38:477–480. doi: 10.1111/j.1439-0507.1995.tb00023.x. [DOI] [PubMed] [Google Scholar]
- 14.Klein BS, Aizenstein BD, Hogan LH. 1997. African strains of Blastomyces dermatitidis that do not express surface adhesin WI-1. Infect Immun 65:1505–1509. doi: 10.1128/IAI.65.4.1505-1509.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guého E, Leclerc M, de Hoog GS, Dupont B. 1997. Molecular taxonomy and epidemiology of Blastomyces and Histoplasma species. Mycoses 40:69–81. doi: 10.1111/j.1439-0507.1997.tb00191.x. [DOI] [PubMed] [Google Scholar]
- 16.Meece JK, Anderson JL, Gruszka S, Sloss BL, Sullivan B, Reed KD. 2013. Variation in clinical phenotype of human infection among genetic groups of Blastomyces dermatitidis. J Infect Dis 207:814–822. doi: 10.1093/infdis/jis756. [DOI] [PubMed] [Google Scholar]
- 17.Schwartz IS, Govender NP, Corcoran C, Dlamini S, Prozesky H, Burton R, Mendelson M, Taljaard J, Lehloenya R, Calligaro G, Colebunders R, Kenyon C. 2015. Clinical characteristics, diagnosis, management, and outcomes of disseminated emmonsiosis: a retrospective case series. Clin Infect Dis 61:1004–1012. doi: 10.1093/cid/civ439. [DOI] [PubMed] [Google Scholar]
- 18.Heys I, Taljaard J, Orth H. 2014. An Emmonsia species causing disseminated infection in South Africa. N Engl J Med 370:283–284. doi: 10.1056/NEJMc1314277. [DOI] [PubMed] [Google Scholar]
- 19.Martin PM, Berson SD. 1973. Fungus diseases in southern Africa. Mycopathol Mycol Appl 50:1–84. doi: 10.1007/bf02050005. [DOI] [PubMed] [Google Scholar]
- 20.Frean J, Blumberg L, Woolf M. 1993. Disseminated blastomycosis masquerading as tuberculosis. J Infect 26:203–206. doi: 10.1016/0163-4453(93)93031-x. [DOI] [PubMed] [Google Scholar]
- 21.Simon GB, Berson SD, Young CN. 1977. Blastomycosis of the tongue: a case report. S Afr Med J 52:82–83. [PubMed] [Google Scholar]
- 22.Kenyon C, Bonorchis K, Corcoran C, Meintjes G, Locketz M, Lehloenya R, Vismer HF, Naicker P, Prozesky H, van Wyk M, Bamford C, du Plooy M, Imrie G, Dlamini S, Borman AM, Colebunders R, Yansouni CP, Mendelson M, Govender NP. 2013. A dimorphic fungus causing disseminated infection in South Africa. N Engl J Med 369:1416–1424. doi: 10.1056/NEJMoa1215460. [DOI] [PubMed] [Google Scholar]
- 23.Clinical and Laboratory Standards Institute. 2008. Reference method for broth dilution antifungal susceptibility testing of filamentous fungi, 2nd ed Document M38-A2 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 24.Clinical and Laboratory Standards Institute. 2012. Reference method for broth dilution antifungal susceptibility testing of yeasts, 4th informational supplement. Document M27-A3 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 25.Maphanga TG, Britz E, Zulu TG, Mpembe RS, Naicker SD, Schwartz IS, Govender NP. 2017. In vitro antifungal susceptibility of yeast and mold phases of isolates of dimorphic fungal pathogen Emergomyces africanus (formerly Emmonsia sp.) from HIV-infected South African patients. J Clin Microbiol 55:1812–1820. doi: 10.1128/JCM.02524-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Katoh K, Rozewicki J, Yamada KD. 2019. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform 20:1160–1166. doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98. [Google Scholar]
- 28.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li W, Sullivan TD, Walton E, Averette AF, Sakthikumar S, Cuomo CA, Klein BS, Heitman J. 2013. Identification of the mating-type (MAT) locus that controls sexual reproduction of Blastomyces dermatitidis. Eukaryot Cell 12:109–117. doi: 10.1128/EC.00249-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, McVean G, Durbin R, 1000 Genome Project Analysis Group . 2011. The variant call format and VCFtools. Bioinformatics 27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muñoz JF, Gauthier GM, Desjardins CA, Gallo JE, Holder J, Sullivan TD, Marty AJ, Carmen JC, Chen Z, Ding L, Gujja S, Magrini V, Misas E, Mitreva M, Priest M, Saif S, Whiston EA, Young S, Zeng Q, Goldman WE, Mardis ER, Taylor JW, McEwen JG, Clay OK, Klein BS, Cuomo CA. 2015. The dynamic genome and transcriptome of the human fungal pathogen Blastomyces and close relative Emmonsia. PLoS Genet 11:e1005493. doi: 10.1371/journal.pgen.1005493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwartz IS, Kenyon C, Lehloenya R, Claasens S, Spengane Z, Prozesky H, Burton R, Parker A, Wasserman S, Meintjes G, Mendelson M, Taljaard J, Schneider JW, Beylis N, Maloba B, Govender NP, Colebunders R, Dlamini S. 2017. AIDS-related endemic mycoses in Western Cape, South Africa, and clinical mimics: a cross-sectional study of adults with advanced HIV and recent-onset, widespread skin lesions. Open Forum Infect Dis 4:ofx186. doi: 10.1093/ofid/ofx186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baily GG, Robertson VJ, Neill P, Garrido P, Levy LF. 1991. Blastomycosis in Africa: clinical features, diagnosis and treatment. Rev Infect Dis 13:1005–1008. doi: 10.1093/clinids/13.5.1005. [DOI] [PubMed] [Google Scholar]
- 36.Anderson JL, Meece JK, Hall MC, Frost HM. 2016. Evidence of delayed disseminated or re-infection with Blastomyces in two immunocompetent hosts. Med Mycol Case Rep 13:9–11. doi: 10.1016/j.mmcr.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frost HM, Anderson J, Ivacic L, Meece J. 2017. Blastomycosis in children: an analysis of clinical, epidemiologic and genetic features. J Pediatr Infect Dis Soc 6:49–56. doi: 10.1093/jpids/piv081. [DOI] [PubMed] [Google Scholar]
- 38.Gatti F, de Broe M, Ajello L. 1968. Blastomyces dermatitidis infection in the Congo. Report of a second autochthonous case. Am J Trop Med Hyg 17:96–101. doi: 10.4269/ajtmh.1968.17.96. [DOI] [PubMed] [Google Scholar]
- 39.Chapman SW, Dismukes WE, Proia LA, Bradsher RW, Pappas PG, Threlkeld MG, Kauffman CA, Infectious Diseases Society of America . 2008. Clinical practice guidelines for the management of blastomycosis: 2008 update by the Infectious Diseases Society of America. Clin Infect Dis 46:1801–1812. doi: 10.1086/588300. [DOI] [PubMed] [Google Scholar]
- 40.Saccente M, Woods GL. 2010. Clinical and laboratory update on blastomycosis. Clin Microbiol Rev 23:367–381. doi: 10.1128/CMR.00056-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dukik K, Al-Hatmi AMS, Curfs-Breuker I, Faro D, de Hoog S, Meis JF. 2018. Antifungal susceptibility of emerging dimorphic pathogens in the family Ajellomycetaceae. Antimicrob Agents Chemother 62:e01886-17. doi: 10.1128/AAC.01886-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Espinel-Ingroff A. 1998. In vitro activity of the new triazole voriconazole (UK-109,496) against opportunistic filamentous and dimorphic fungi and common and emerging yeast pathogens. J Clin Microbiol 36:198–202. doi: 10.1128/JCM.36.1.198-202.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Espinel-Ingroff A. 1998. Comparison of in vitro activities of the new triazole SCH56592 and the echinocandins MK-0991 (L-743,872) and LY303366 against opportunistic filamentous and dimorphic fungi and yeasts. J Clin Microbiol 36:2950–2956. doi: 10.1128/JCM.36.10.2950-2956.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chapman SW, Rogers PD, Rinaldi MG, Sullivan DC. 1998. Susceptibilities of clinical and laboratory isolates of Blastomyces dermatitidis to ketoconazole, itraconazole, and fluconazole. Antimicrob Agents Chemother 42:978–980. doi: 10.1128/AAC.42.4.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhanel GG, Karlowsky JA, Harding GA, Balko TV, Zelenitsky SA, Friesen M, Kabani A, Turik M, Hoban DJ. 1997. In vitro activity of a new semisynthetic echinocandin, LY-303366, against systemic isolates of Candida species, Cryptococcus neoformans, Blastomyces dermatitidis, and Aspergillus species. Antimicrob Agents Chemother 41:863–865. doi: 10.1128/AAC.41.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sigler L. 1996. Ajellomyces crescens sp. nov., taxonomy of Emmonsia spp., and relatedness with Blastomyces dermatitidis (teleomorph Ajellomyces dermatitidis). J Med Vet Mycol 34:303–314. doi: 10.1080/02681219680000531. [DOI] [PubMed] [Google Scholar]
- 47.Nguyen HDT, Jančič S, Meijer M, Tanney JB, Zalar P, Gunde-Cimerman N, Seifert KA. 2015. Application of the phylogenetic species concept to Wallemia sebi from house dust and indoor air revealed by multi-locus genealogical concordance. PLoS One 10:e0120894. doi: 10.1371/journal.pone.0120894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Finkel-Jimenez B, Wüthrich M, Klein BS. 2002. BAD1, an essential virulence factor of Blastomyces dermatitidis, suppresses host TNF-alpha production through TGF-beta-dependent and -independent mechanisms. J Immunol 168:5746–5755. doi: 10.4049/jimmunol.168.11.5746. [DOI] [PubMed] [Google Scholar]
- 49.Meece JK, Anderson JL, Klein BS, Sullivan TD, Foley SL, Baumgardner DJ, Brummitt CF, Reed KD. 2010. Genetic diversity in Blastomyces dermatitidis: implications for PCR detection in clinical and environmental samples. Med Mycol 48:285–290. doi: 10.1080/13693780903103952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brandhorst TT, Wüthrich M, Warner T, Klein B. 1999. Targeted gene disruption reveals an adhesin indispensable for pathogenicity of Blastomyces dermatitidis. J Exp Med 189:1207–1216. doi: 10.1084/jem.189.8.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
We declare that the data supporting the findings of this study are available within the paper. Contigs and sequences were submitted to GenBank (contig accession numbers QGQE00000000 [SA-NICD-12], QGQF00000000 [SA-NICD-09], QGQG00000000 [SA-NICD-10], QGQH00000000 [SA-NICD-11], QGQI00000000 [SA-NICD-15], QGQJ00000000 [SA-NICD-13], QGQK00000000 [SA-NICD-14], QGQL00000000 [SA-NICD-16], QGQM00000000 [SA-NICD-01], QGQN00000000 [SA-NICD-07], QGQO00000000 [SA-NICD-02], QGQP00000000 [SA-NICD-03], QGQQ00000000 [SA-NICD-04], QGQR00000000 [SA-NICD-08], QGQS00000000 [SA-NICD-05], QGQT00000000 [SA-NICD-06], and QKWI00000000 [NCPF 4091]; ITS accession numbers MH571851, MH571852, MH571853, MH571854, MH571855, MH571856, MH571857, MH571858, MH571859, MH571860, MH571861, MH571862, MH606201, MH606202, MH606203, MH606204, MH606205, MH606206, MH606207, and MH606208; and LSU accession numbers MH644816, MH644817, MH644818, MH644819, MH644820, MH644821, MH644822, MH644823, MH644824, MH644825, MH644826, MH644836, MH644837, MH644838, MH644839, MH644840, MH644841, MH644842, and MH644843).