Shiga toxin (Stx)-producing Escherichia coli (STEC) is the main cause of postdiarrheal hemolytic-uremic syndrome (HUS), a life-threatening clinical complication characterized by hemolytic anemia, thrombocytopenia, and acute renal failure that mainly affects children. A relevant feature of STEC strains is the production of Stx, and all of them express Stx1 and/or Stx2 regardless of the strain serotype.
KEYWORDS: STEC, HUS, Shiga toxin, Stx2, VHH, nanobodies, ELISA
ABSTRACT
Shiga toxin (Stx)-producing Escherichia coli (STEC) is the main cause of postdiarrheal hemolytic-uremic syndrome (HUS), a life-threatening clinical complication characterized by hemolytic anemia, thrombocytopenia, and acute renal failure that mainly affects children. A relevant feature of STEC strains is the production of Stx, and all of them express Stx1 and/or Stx2 regardless of the strain serotype. Therefore, Stx detection assays are considered the most suitable methods for the early detection of STEC infections. Single-domain antibodies from camelids (VHHs) exhibit several advantages in comparison with conventional antibodies, making them promising tools for diagnosis. In this work, we have exploited VHH technology for the development of an immunocapture assay for Stx2 detection. Thirteen anti-Stx2 VHHs previously obtained from a variable-domain repertoire library were selected and evaluated in 130 capture-detection pair combinations for Stx detection. Based on this analysis, two VHHs were selected and a double VHH-based biotin-streptavidin capture enzyme-linked immunosorbent assay (ELISA) with spectrophotometric detection was developed and optimized for Stx2 detection. This assay showed an excellent analytical and clinical sensitivity in both STEC culture supernatants and stool samples even higher than the sensitivity of a commercial ELISA. Furthermore, based on the analysis of stool samples, the VHH-based ELISA showed high correlation with stx2 detection by PCR and a commercial rapid membrane-based immunoassay. The intrinsic properties of VHHs (high target affinity and specificity, stability, and ease of expression at high yields in recombinant bacteria) and their optimal performance for Stx detection make them attractive tools for the diagnosis of HUS related to STEC (STEC-HUS).
INTRODUCTION
Shiga toxin (Stx)-producing Escherichia coli (STEC) is an important foodborne pathogen that can produce sporadic cases and outbreaks of gastroenteritis with diarrhea, bloody diarrhea (BD), and hemolytic-uremic syndrome (HUS). HUS is a life-threatening clinical condition characterized by microangiopathic hemolytic anemia, thrombocytopenia, and acute renal failure (1, 2). It is estimated that STEC causes annually more than 2.5 million cases of acute illness worldwide, among which several thousand trigger HUS and hundreds of cases suffer severe kidney damage, often irreversible, or even death (3). HUS related to STEC (STEC-HUS) is the leading cause of acute renal failure in children worldwide; over 90% of HUS cases are associated with STEC infection in this population (4). The incidence of STEC-HUS varies according to the country, and Argentina shows the highest HUS incidence worldwide, with almost 10 cases a year per 100,000 children under 5 years old. In this country, HUS shows an endemic behavior and constitutes an important public health issue with high morbidity and mortality rates (5).
STEC strains are characterized by the production of Shiga toxins (Stx[s]), which constitute the main virulence factors in the pathogenesis of BD and HUS since they spread to the target organs such as kidneys, resulting in cell receptor-mediated internalization and toxicity (6, 7). Stx family members have an AB5 molecular configuration in which an enzymatically active A subunit is noncovalently linked to five B subunits. The A subunit inhibits protein synthesis in the target cells, and the B pentamer binds the cellular receptor, globotriaosylceramide (Gb3), found primarily on endothelial cells (8, 9). The STEC strains can produce two types of Stx(s), type 1 (Stx1) and type 2 (Stx2), as well as their variants. For Stx1, three variants (a, c, and d) were identified, among which Stx1a is the most prevalent. Stx1c is rare and does not cause serious complications in humans, and Stx1d has not been detected in strains isolated from humans. Within the Stx2 variants (a, b, c, d, e, f, and g), Stx2a is the most frequently found in stool samples and the Stx2 subtype with the highest clinical and epidemiological relevance. In STEC infections, severe disease is more often associated with STEC strains that produce Stx2a rather than Stx1a (10, 11).
The diagnosis of STEC infections is based on the isolation and characterization of STEC strains as well as free fecal Shiga toxin (FFStx) and/or stx gene detection. E. coli O157:H7 is the dominant STEC serotype associated with sporadic cases and outbreaks of BD and HUS in different parts of the world (2); however, a subset of non-O157:H7 STEC serotypes (O26:H11, O103:H2, O111:NM, O121:H19, O145:NM, and O45:H2, among others) can cause a similar disease (5, 12). We have recently demonstrated that serological diagnosis using serogroup-specific antigens for the most prevalent STEC serogroups is a valuable and sensitive method for diagnosing STEC infections in combination with bacteriological culture and Stx detection (13, 14). Irrespective of the strain serotype, the common virulence trait of all STEC strains is the ability to produce one or both Shiga toxins (Stx1 or Stx2). Therefore, Stx detection assays are considered the most suitable methods for the timely and accurate diagnosis of STEC infections. The gold standard test for free fecal Shiga toxin (FFStx) detection is the Vero cell cytotoxicity assay because of its picogram-level analytical sensitivity (15). In addition, a number of immunoassays for FFstx detection, such as enzyme-linked immunosorbent assays (ELISAs) (16–18), membrane-based immunoassays (19–21), and immunochromatographic systems (21–23), are available. All these tests use a combination of polyclonal and/or monoclonal antibodies against the A or B subunits of the toxins. STEC infection can also be confirmed by PCR assays targeting stx1 and/or stx2 genes that can be tested on direct fecal samples, the culture medium after enrichment of the stool samples, or samples obtained from the confluent growth zone in agar plates or from the isolated strains (24, 25). However, stx gene(s) detection does not always correlate with disease or expression of the toxin/s.
In addition to conventional heterotetrameric antibodies, camelids also produce unusual homodimeric antibodies that are composed only of heavy chains known as HCAbs for heavy-chain-only antibodies (26). The antigen-binding site of HCAbs consists of one single variable domain, and the product of its recombinant expression is called VH domain of heavy-chain antibodies (VHH) or nanobody (Ablynx). VHHs provide many advantages over conventional polyclonal and monoclonal antibodies and currently used antibody-based fragments (27). They can be easily engineered, cloned, and expressed with high yields in a variety of expression systems, including E. coli. VHHs are highly soluble and stable, even under denaturing conditions or at high temperatures, and can be efficiently functionalized. Moreover, due to their small size they can recognize unusual antigenic sites and penetrate tissues deeply. Therefore, VHHs are very valuable immunoreagents that have been extensively used in diagnostics (28–32) and therapy (33–36).
In this work, we have developed a double-VHH sandwich enzyme-linked immunosorbent assay (ELISA) for Stx2 detection. This VHH-based ELISA uses two different VHHs for capture and detection and allows a sensitive and specific detection of Stx2a in both STEC culture supernatants and stool samples. We propose that this unique, tailor-made assay could be of great value for the diagnosis of STEC infections.
MATERIALS AND METHODS
Plasmids, bacterial strains, and growth conditions.
The bacterial strains and plasmids used in this work are listed in Table 1. The E. coli TOP10 strain (Invitrogen Life Technologies) was used for cloning procedures and was grown on Luria-Bertani medium (LB; 10 g/liter tryptone, 5 g/liter yeast extract, and 10 g/liter NaCl) at 37°C. Tetracycline at 20 μg/ml, chloramphenicol at 20 μg/ml, kanamycin at 50 μg/ml, or ampicillin at 100 μg/ml was added as needed to the medium.
TABLE 1.
Plasmids and strains used in this work
| Plasmid or strain | Description/characteristicsa | Reference or source |
|---|---|---|
| Plasmids | ||
| pHEN4 | Phagemid vector for phage display; HA tag, Ampr | 47 |
| pHEN6 | Phagemid vector for phage display; 6×His tag, Ampr | 48 |
| pDONR221 | Entry vector for Gateway cloning system, Kanr | Invitrogen |
| pDEST15 | Expression vector for Gateway cloning system, Ampr | Invitrogen |
| pDEST15-Stx2B | pDEST15 containing the B subunit from Stx2a fused to GST, Ampr | This work |
| E. coli strains | ||
| TOP10 | F− mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80dlacZΔM15 ΔlacX74 endA1 recA1 Δ(ara, leu)7697 araD139 galU galK nupG rpsL λ (Strr) | Invitrogen |
| BL21-CodonPlus (DE3)-RP | E. coli B F− ompT hsdS(rB− mB−) dcm+ Tetr gal λ(DE3) endA Hte [argU proL Chlr] | Stratagene |
| WK6 | F′ lacIq Δ(lacZ) M15 proA+ B+ Δ(lac-proAB) galE rpsL | 48 |
| O157:H7 (EDL933) | Reference STEC strain, eae+/ehx+ stx1/2 O157:H7 | ANLIS-Malbrán (49) |
| O145:NM (FP 02/02) | STEC isolation strain, eae+/ehx+ stx2 O145:NM | ANLIS-Malbrán |
| O145:NM (FP 1091/10) | Non-STEC isolation strain, eae negative/ehx negative stx negative O145:NM | ANLIS-Malbrán |
| (FP 817/12) | STEC isolation strain, eae′+/ehx+ stx1a O26:H11 | ANLIS-Malbrán |
| (FP 447/11) | STEC isolation strain, eae negative/ehx+ stx1c ONT:NM | ANLIS-Malbrán |
| (FP 379/10) | STEC isolation strain, eae negative/ehx negative stx2bO118 ONT:HNT | ANLIS-Malbrán |
| (FP 550/12) | STEC isolation strain, eae+/ehx+ stx2cvh-a O157:H7 | ANLIS-Malbrán |
| (FP 862/10) | STEC isolation strain, eae negative/ehx negative stx2cvh-b ONT:H21 | ANLIS-Malbrán |
| (FP 713/09) | STEC isolation strain, eae negative/ehx+ stx2d O91:H21 | ANLIS-Malbrán |
| (GG7) | STEC isolation strain, eae+/ehx negative stx2d O145:H34 | ANLIS-Malbrán |
Abbreviations: Str, streptomycin; Chl, chloramphenicol; Amp, ampicillin; Kan, kanamycin; Tet, tetracycline; ONT, O polysaccharide not typified; HNT, flagellar H antigen not typified; NM, not motile.
Expression and purification of BLS-Stx2B.
BLS-Stx2B was produced and purified as previously described (37). In this chimera, Stx2a B subunit is covalently linked to the enzyme lumazine synthase from Brucella spp. (BLS).
Cloning, expression, and purification of GST-Stx2B.
The fusion of Stx2a B subunit to glutathione S-transferase (GST-Stx2B) was generated using Gateway technology (38). Briefly, the gene that encodes Stx2a B subunit (Gen Bank accession no. AAG55588.1) was amplified by PCR with the oligonucleotides attB1-Stx2B-Fw (GGGGACAAGTTTGTACAAAAAAGCAGGCTTCATGGCGGCGGATTGTGCTAAA) and attB2-Stx2B-Rv (GGGGACCACTTTGTACAAGAAAGCTGGGTCTCAGTCATTATTAAACTGCAC) (underlining indicates the sequence of the attB1 or attB2 site) using Platinum Pfx DNA polymerase (Invitrogen Life Technologies) and genomic DNA from the EDL933 E. coli O157:H7 strain as the template. The amplification product was cloned into the pDONR221 plasmid using the BP Clonase to generate the entry clone pDONR221-Stx2B. The entry clone was recombined with the pDEST15 plasmid to generate the pDEST15-Stx2B using the LR Clonase. For expression of GST-Stx2B, the E. coli BL21-CodonPlus (DE3)-RP strain was transformed with the pDEST15-Stx2B plasmid. The resulting strain was grown overnight at 37°C with agitation in LB medium supplemented with 20 g/ml chloramphenicol and 100 g/ml ampicillin and reinoculated at 1:100 dilution into fresh LB. After incubation for 3 h at 37°C (optical density at 600 nm [OD600] of ∼0.5), protein expression was induced by addition of isopropyl-α-d thiogalactopyranoside (IPTG) to a final concentration of 1 mM. Following a 3-h induction period, the cells were harvested by centrifugation, and the cytoplasmic extracts were obtained by lysis of the cells with lysis buffer (50 mM Tris-HCl, pH 7.6, 150 mM NaCl, 0.5% Triton X-100, 0.5 mg/ml lysozyme, 10 mM MgCl2, 2 mM phenylmethylsulfonyl fluoride [PMSF], 2 mM EDTA, and 10 mg/ml DNase) and sonication. After centrifugation, the soluble fraction was subjected to glutathione-Sepharose affinity chromatography according to the manufacturer’s instructions (GE Healthcare Life Sciences).
Production of bacterial culture supernatants enriched in Stx1 and/or Stx2.
Culture supernatants of the STEC strains, which were used as a source of native Shiga toxins, were prepared according to the protocol described by Karmali et al. with minor modifications (39). Isolated colonies (approximately 10 to 15 colonies) of the corresponding strain (Table 1) were resuspended in 100 μl of tryptic soy broth medium (TSB; 17 g/liter tryptone, 3 g/liter peptic digest of soybean, 2.5 g/liter glucose, 5 g/liter NaCl, and 5 g/liter K2PO4) and inoculated in 5 ml of TSB medium. After 24 h of incubation at 37°C under static conditions, the cultures were diluted 1:25 in Penassay broth antibiotic medium 3 (5 g/liter peptone, 1.5 g/liter yeast extract, 1.5 g/liter beef extract, 1 g/liter dextrose, 5 g/liter NaCl, 3.68 g/liter K2HPO4, and 1.32 g/liter KH2PO4) and incubated for 24 h under the same conditions. Finally, the cultures were centrifuged at 10,000 × g for 10 min, and the resulting supernatants were filtered using an 0.2-μm membrane.
Anti-Stx2B VHHs.
Stx2B-specific VHHs were previously selected by competitive panning using the immobilized antigen BLS-Stx2B and soluble BLS as a competitor protein from phage display libraries generated using the BLS-Stx2B chimera as the immunogen (33). Thirteen clones with Stx2-binding capacity, which included 10 clones belonging to family 1 and one member each belonging to families 2, 6, and 7, were further evaluated by direct ELISA. To assess the interaction of VHHs with Stx2B, high-binding flat-bottom 96-well plates (Invitrogen) were coated with purified BLS-Stx2B, BLS, GST-Stx2B, or GST and incubated for 1 h at room temperature with periplasmic extracts (PE) enriched in the corresponding hemagglutinin (HA)-tagged VHH (HA-VHH). Horseradish peroxidase (HRP)-conjugated anti-HA antibodies (Roche) were used for detection. The absorbance was read at 450 nm.
Expression and purification of selected VHHs.
Selected VHHs were subcloned into the pHEN6 expression plasmid and introduced into the strain E. coli WK6. The resulting strains were grown in Terrific Broth (2.3 g/liter KH2PO4, 12.5 g/liter K2HPO4, 12 g/liter tryptone, 24 g/liter yeast extract, 4 ml/liter glycerol) supplemented with 100 μg/ml ampicillin and incubated at 37°C with agitation (OD600 of ∼0.6 to 0.9). The expression of VHHs was induced by adding 1 mM IPTG and incubating the mixture for 18 h at 28°C with agitation. Periplasmic fractions were obtained by the osmotic shock method (see below), and the purification was performed by metal-chelate affinity chromatography. Purified VHHs were dialyzed against phosphate-buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 2 mM KH2PO4), concentrated, and quantified by the Bradford method according to the manufacturer’s instructions (Bio-Rad).
Preparation of periplasmic fractions for VHH purification: osmotic shock method.
After induction of the expression of the corresponding VHH, the culture (600 ml) was centrifuged at 10,000 × g for 10 min. The resulting pellet was resuspended in TES buffer (200 mM Tris-HCl, pH 8.0, 0.5 mM EDTA, and 0.5 M sucrose) at a concentration factor of 100. After incubation for 1 h at 4°C with orbital agitation, 9 ml of TES/4 (TES diluted four times in water) was added, and the resulting suspension was incubated for 1 h at 4°C and centrifuged at 10,000 × g for 30 min at 4°C, producing the periplasmic fraction in the supernatant.
Formatting of HA-2vb10 VHH.
HA-2vb10 VHH was engineered and expressed as an HA-2vb10-6×His fusion protein. HA-2vb10-6×His was generated by fusion of a six-histidine tag at the carboxyl-terminal end. Nucleotide sequence was obtained from GenScript (GenScript Biotech), and the resulting molecule was cloned into pHEN6 expression vector. HA-2vb10-6×His VHH was expressed and purified as indicated above.
VHH biotinylation.
Purified 2vb10-6×His VHH was biotinylated using the EZ-Link sulfo-NHS-LC-biotinylation kit according to the manufacturer’s instructions (Thermo Fisher Scientific).
HA–anti-HA VHH-based capture ELISA.
For the development of the Stx capture ELISA with detection based on the HA–anti-HA system (HA–anti-HA capture ELISA), 96-well high-binding polystyrene microplates (Corning Inc.) were coated with purified VHHs at a concentration of 10 μg/ml in 50 mM carbonate-bicarbonate buffer (pH 9.6) and incubated for 18 h at 4°C. Blocking (2% bovine serum albumin [BSA], 20 mM Tris-HCl [pH 7.6], 150 mM NaCl, and 0.1% Tween 20) was performed by incubation for 1 h at room temperature. The recombinant proteins (BLS-Stx2B, GST-Stx2B, BLS, and GST) and the culture supernatants of the Stx-producing strains were diluted in blocking solution and incubated for 1 h at room temperature with agitation. HA-tag detection VHHs, obtained from the periplasmic extracts (PE), were diluted 1:16 in 20% blocking solution in TBS-T (200 mM Tris-HCl, pH 7.6, 150 mM NaCl, and 0.1% Tween 20) and incubated for 1 h at room temperature with agitation. HA-tag VHHs were detected using anti-HA antibodies conjugated to horseradish peroxidase (HRP; Roche) diluted in 20% blocking solution in TBS-T and incubated for 1 h at room temperature with agitation. Between each reaction step, the plates were washed three times with TBS-T. After incubation with the substrate (0.015% H2O2, 0.1% 3,3′,5,5′-tetramethylbenzidine [TMB] in 0.1 M citrate phosphate buffer, pH 5.0) for 5 min at room temperature, the reaction was stopped with 0.16 N H2SO4, and the absorbance was measured at 450 nm (A450) using a microplate reader (DTX 880 multimode detector; Beckman Coulter, Inc.).
Biotin-streptavidin VHH-based capture ELISA.
For the development of the Stx capture ELISA with detection based on the biotin-streptavidin system (biotin-streptavidin capture ELISA), 96-well high-binding polystyrene microplates (Corning) were coated with purified 1vb1-6×His VHH at a concentration of 2.5 μg/ml in 50 mM carbonate-bicarbonate buffer (pH 9.6) and incubated for 18 h at 4°C. The detection VHH, biotinylated 2vb10-6×His VHH (0.8 mg/ml), was diluted 1:16,000 in 50% blocking solution in TBS-T and detected with streptavidin conjugated to HRP (Thermo Fisher). All other steps were performed as indicated for the HA–anti-HA capture ELISA.
Other assays. (i) Vero cell cytotoxicity assay.
The Vero cell cytotoxicity assay was performed according to the method of Karmali et al. (15). Briefly, the cells were incubated with the culture supernatants for 48 h at 37°C. Then, the cells were washed with PBS and incubated for 15 min at room temperature with the staining solution (0.75% crystal violet, 40% methanol). The cytotoxic activity was evaluated by observing the staining of the cells with the dye.
(ii) MTT viability assay.
The MTT viability assay was performed according to the protocol described by Mosmann (40). The MTT assay is a quantitative and colorimetric assay based on the ability of living cells to reduce 4,5-dimethylthiazole-2,5-diphenyltetrazolium (MTT) bromide (Sigma-Aldrich) to formazan in a reaction catalyzed by mitochondrial dehydrogenases. Vero cell monolayers were grown as indicated in the Vero cell cytotoxicity assay and incubated for 48 h at 37°C with different dilutions of the culture supernatants obtained from the indicated strains. Then, the cells were washed with Earle’s solution (110 mM NaCl, 54 mM KCl, 1 mM NaH2PO4·H2O, and 5.5 mM glucose), and MTT (stock solution; 5 mg/ml in PBS) was added to each well at a final concentration of 0.5 mg/ml in cell culture medium without serum. After incubation for 1.5 h at 37°C, the solution was removed, and the formazan produced by living cells was solubilized in 200 μl of dimethyl sulfoxide (DMSO). The absorbance measured at 655 nm, corresponding to the substrate not converted to formazan, was subtracted from the absorbance of formazan measured at 570 nm.
(iii) Commercial ELISA (R-Biopharm).
Ridascreen Verotoxin is an enzyme immunoassay for the qualitative detection of Stx1 and Stx2 that uses specific monoclonal antibodies to Stx1 and Stx2 in a sandwich-type method (16). The assay was performed according to the manufacturer’s instructions.
(iv) Commercial membrane-based rapid immunoassay (Alere-Abbot).
The Shiga Toxin Quik Chek test is a rapid membrane enzyme immunoassay for the simultaneous qualitative detection and differentiation of Stx1 and Stx2 in a single test device (19–21). This test utilizes specific antibodies against Stx1 and Stx2 and contains a reaction window with three vertical lines of immobilized antibodies. The test lines “1” and “2” contain monoclonal antibodies against Stx1 and Stx2, respectively, and the control line “C” is a dotted line that contains anti-HRP antibodies. The conjugate consists of antibodies toward Stx1 and Stx2 coupled to HRP. The assay was performed according to the manufacturer’s instructions.
Stool samples.
The samples were obtained from patients under 6 years old with a clinical diagnosis of diarrhea, bloody diarrhea, or HUS associated or not with STEC infection. The stool samples were cultured in Gram-negative bacterial broth (GN; 20 g/liter peptone, 1 g/liter dextrose, 2 g/liter d-mannitol, 5 g/liter sodium citrate, 0.5 g/liter sodium deoxycholate, 4 g/liter K2HPO4, 1.5 g/liter KH2PO4, and 5 g/liter NaCl) for 18 h at 37°C, and the following tests were carried out: (i) FFStx detection by Vero cell cytotoxicity test, (ii) PCR for stx1/stx2/rfbO157 (41), (iii) PCR for stx1 and stx2 allelic variants as described by Scheutz et al. (42), (iv) Shiga Toxin Quik Chek membrane-based rapid immunoassay, and (v) VHH-based Stx2 capture ELISA.
Ethics statement.
All the stool samples analyzed in this study were selected from a previously characterized collection provided by the Servicio Fisiopatogenia, Instituto Nacional de Enfermedades Infecciosas (INEI)-ANLIS Dr. Carlos G. Malbrán (Argentina), the National Reference Laboratory (NRL) for HUS and diarrheal diseases associated with diarrheagenic E. coli. These samples were submitted to the NRL during the 2017–2018 period in the framework of the national surveillance protocol. To ensure anonymity, the samples were coded upon collection; therefore, no personal identification data of the patients were available during the study.
RESULTS
Selection of Stx2B-recognizing VHHs and development of a capture ELISA.
In a previous work, VHH phage display libraries were generated using the BLS-Stx2B fusion protein as an immunogen. Stx2B-specific VHHs were selected by competitive panning of the phage libraries using the immobilized antigen BLS-Stx2B and soluble BLS as a competitor protein (33).
In order to select VHHs for the development of an Stx2 capture ELISA, 17 anti-Stx2 VHHs were evaluated by direct ELISA, coating the microplates with BLS-Stx2B, BLS, GST-Stx2B, and GST. A total of 13 VHHs were selected based on their capacity to recognize the recombinant B subunit of Stx2 fused to BLS (BLS-Stx2B) and GST (GST-Stx2B) but not the carrier proteins (see Fig. S1 in the supplemental material). Previously, anti-Stx2 VHHs were classified into families based on amino acid composition and length of the CDR3 (33). VHHs selected in this work come from four different families: 10 VHHs (1vb1, 1vb19, 1vb20, 2vb1, 2vb7, vb8, 2vb10, 2vb11, 2vb21, and 2vb28) belong to family 1 and the remaining three (2vb23, 2vb20, and 2vb6) belong to families 2, 6, and 7, respectively (Fig. S2).
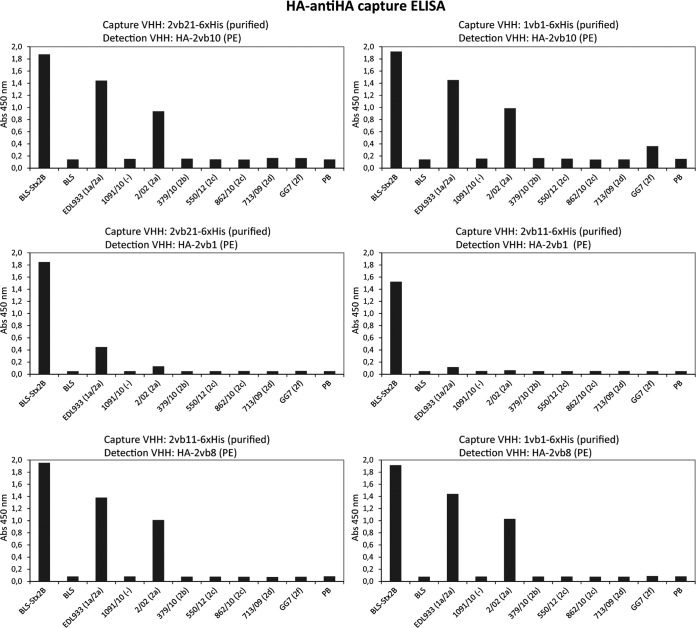
To select the best capture-detection VHH pair for the development of the capture ELISA, 10 VHHs fused to histidine tags and purified by metal-chelate affinity chromatography (2vb7, 2vb8, 2vb28, 1vb1, 2vb10, 2vb21, 1vb19, 1vb20, 2vb1, and 2vb11) were evaluated as capture antibodies. The same 10 plus three additional VHHs (2vb23, 2vb20, and 2vb6) fused to HA and obtained from periplasmic extracts (PE) enriched in the corresponding VHH were used as detection antibodies (Fig. 1A, HA–anti-HA capture ELISA). A total of 130 pairwise combinations were evaluated for the detection of BLS-Stx2B and GST-Stx2B (Fig. 2). Based on this analysis and the expression and purification yield of the corresponding VHHs, six capture-detection pairs were preliminarily selected (2vb21-2vb10, 1vb1-2vb10, 2vb21-2vb1, 2vb11-2vb1, 2vb11-2vb8, and 1vb1-2vb8). The analytical sensitivity of these pair combinations for the recombinant proteins BLS-Stx2B and GST-Stx2B was further evaluated (Fig. S3); all VHH pairs except 2vb21-2vb1 were able to detect at least 1.5 ng/well (30 ng/ml) of BLS-Stx2B. 1vb1-2vb10 and 2vb21-2vb10 were the VHH pairs with better performance for GST-Stx2B detection.
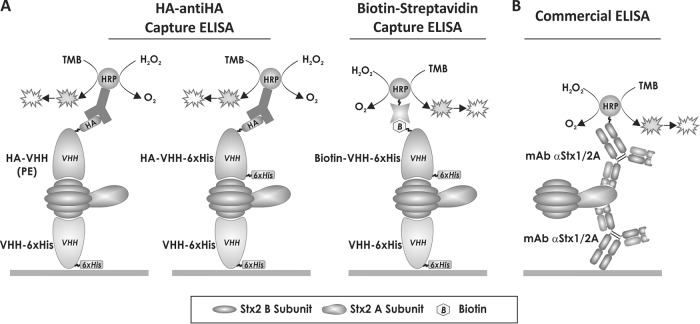
FIG 1.
Schematic representation of the capture ELISAs developed and used in this work for Stx2 detection. (A) VHH-based capture ELISAs developed in this work for Stx2 detection using specific nanobodies. Purified VHHs fused to a histidine tag (VHH-6×His) were used as capture antibodies. As detection antibodies, three types of VHHs were used: VHHs fused to the HA tag and obtained from the periplasmic extracts (PE) enriched in the corresponding HA-VHH, a VHH fused to the 6×His and HA tags and purified by metal-chelate affinity chromatography (HA-VHH-6×His), and an in vitro biotinylated VHH (biotin-VHH-6×His). In the last case, detection was performed by either spectrophotometry or chemiluminescence. (B) Commercial capture ELISA. This ELISA is based on a pair of monoclonal antibodies directed to the A subunit of Stx1 and Stx2. TMB, 3,3′,5,5′-tetramethylbenzidine.
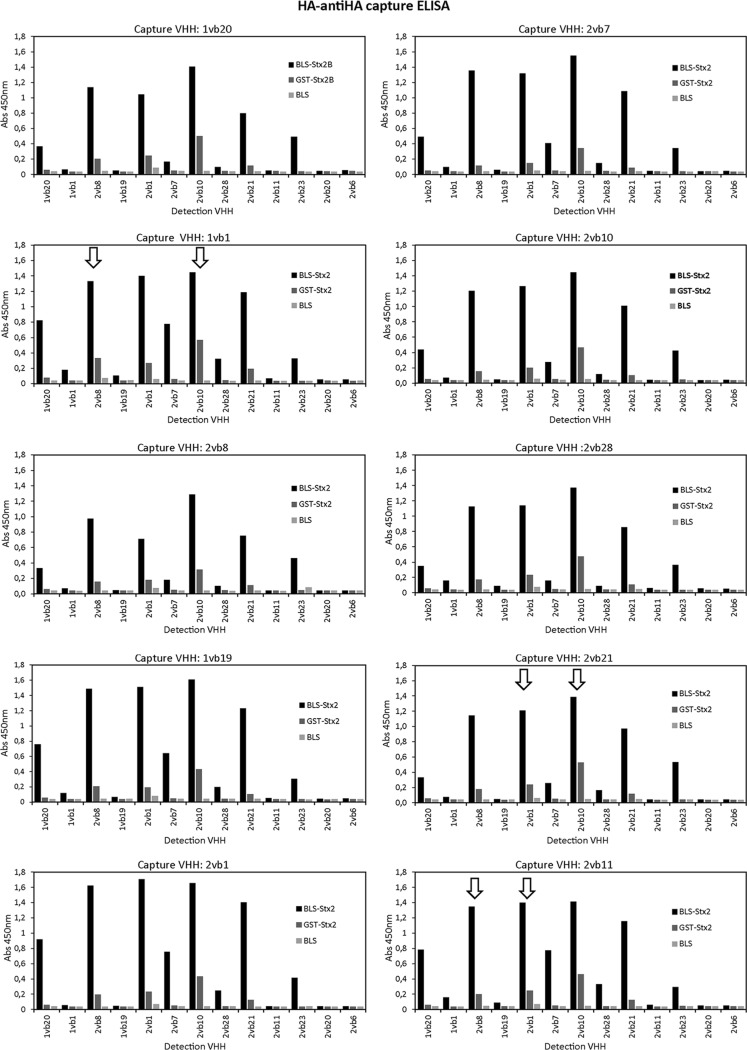
FIG 2.
Analysis of 130 VHH capture-detection combinations by the HA–anti-HA capture ELISA. Capture antibodies (VHH-6×His) were purified by metal-chelate affinity chromatography, and detection antibodies (HA-VHHs) were obtained from the periplasmic extracts (PE) enriched in the corresponding VHH (Fig. 1A). Each panel corresponds to one capture VHH in combination with each of the 13 detection VHHs. The recombinant proteins BLS-Stx2B, GST-Stx2B, and BLS (control) were assayed at 1 μg/ml (50 ng/well). The arrows indicate the six VHH pairs selected for further analysis. Reactivity values with GST were similar to those obtained with BLS (data not shown).
In order to determine if the six selected VHH pairs were capable of recognizing native Stx2 and its variants, and considering that Stx2 is secreted into the culture medium by producing bacteria (43), culture supernatants from different STEC strains were obtained as toxin sources and analyzed by the HA–anti-HA capture ELISA. Bacterial culture supernatants were obtained as described by Karmali et al. with minor modifications, and Stx release was evaluated by the Vero cell cytotoxicity assay (39) (Fig. S4). As sources of Stx2a, the prototype variant of Stx2, the reference strains EDL933 (1a/2a) and 02/02 (2a) were used; strains 379/10 (2b), 550/10 (2c), 862/10 (2c), 713/09 (2d), and GG7 (2f) were used as sources of the other Stx2 variants (Table 1). Strain 1091/10, which does not produce Stx2 or Stx1, was used as a negative control. No VHH pair showed reactivity with the culture supernatants obtained from strains 817/12 and 447/11 (Table 1) used as sources of Stx1a and Stx1c, respectively (data not shown). As shown in Fig. 3, the VHH pairs 2vb11-2vb1 and 2vb21-2vb1 were not able to detect or only poorly detected Stx2a; in contrast, the remaining pairs (1vb1-2vb10, 2vb21-2vb10, 2vb11-2vb10, and 2vb11-2vb8) showed high levels of reactivity toward Stx2a (EDL933 and 02/02 strains), which is the Stx2 subtype with the highest clinical and epidemiological relevance. For the other Stx2 variants, only some reactivity against Stx2f (strain GG7) was observed with 1vb1-2vb10.
FIG 3.
Analysis of culture supernatants of the indicated strains by the HA–anti-HA capture ELISA using the six selected VHH pairs. Capture VHHs (VHH-6×His) were purified by metal-chelate affinity chromatography, and detection antibodies (HA-VHHs) were obtained from the periplasmic extracts (PE) enriched in the corresponding VHH. The recombinant proteins BLS-Stx2B and BLS and also PB (Penassay broth antibiotic medium 3; culture medium used to grow the different strains) were used as controls. The Shiga toxin type and variant expressed by each strain are indicated in parentheses.
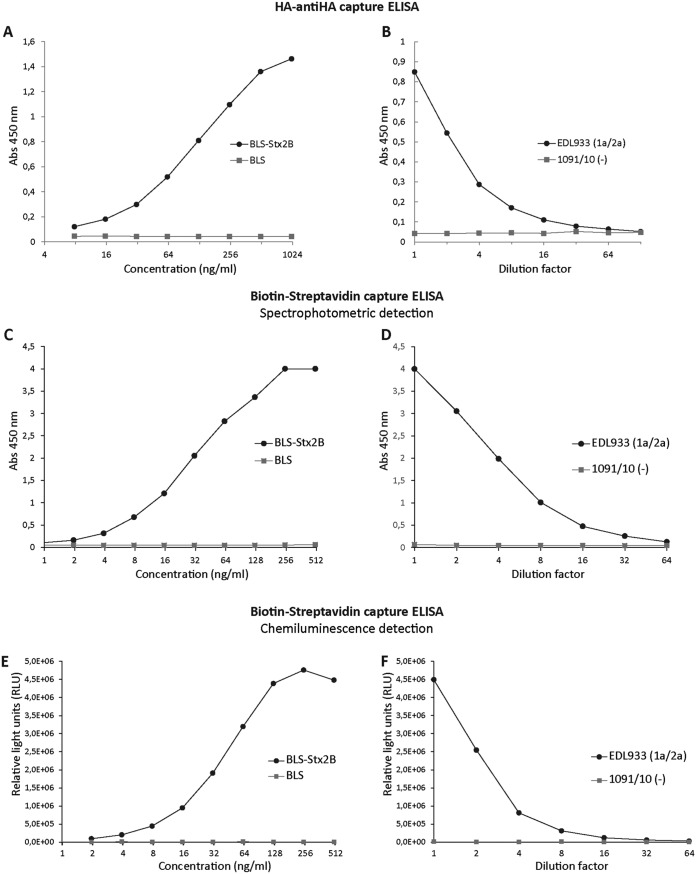
Based on these results, the 1vb1-2vb10 VHH pair was selected to continue with the development and optimization of the capture ELISA for Stx2 detection. To determine the limit of detection (LOD) of the HA–anti-HA capture ELISA using the 1vb1-2vb10 VHH pair, different concentrations of the recombinant protein BLS-Stx2B and serial dilutions of the EDL933 culture supernatant were analyzed (Fig. 4A and B). The LOD for BLS-Stx2B ranged between 16 ng/ml and 32 ng/ml, and the maximum dilution of STEC culture supernatant for which native Stx2a could be detected was 1/16.
FIG 4.
Detection of recombinant and native Stx2 by the capture ELISAs using the 1vb1-2vb10 VHH capture-detection pair. (A and B) HA–anti-HA capture ELISA using the 1vb1-6×His (purified) and HA-2vb10 VHHs (PE) as capture and detection antibodies, respectively. (C and D) Biotin-streptavidin capture ELISA using the 1vb1-6×His (purified) and biotin-2vb10-6×His VHHs as capture and detection antibodies, respectively, and spectrophotometric detection. (E and F) Biotin-streptavidin capture ELISA using the 1vb1-6×His (purified) and biotin-2vb10-6×His VHHs as capture and detection antibodies, respectively, and detection by chemiluminescence. The culture supernatants of the indicated strains were evaluated in several dilutions. The Shiga toxin type and variant expressed by each strain are indicated in parentheses. The 1091/10 strain does not express any Shiga toxin (control).
Optimization and improvement of the capture ELISA.
In the assays mentioned above, detection VHHs were expressed fused to the HA tag and obtained from the periplasmic extracts (PE) enriched in the corresponding VHH. To be able to purify the HA-2vb10 VHH used as the detection antibody, it was engineered and expressed as an HA-2vb10-6×His fusion protein (see Materials and Methods). The HA-2vb10-6×His was purified by metal-chelate affinity chromatography, and the ELISA conditions were optimized. Using the formatted HA-2vb10-6×His VHH and the HA–anti-HA capture ELISA format (Fig. 1A), high reactivity levels for the recombinant protein BLS-Stx2B were observed. However, no reactivity for the native Stx2a or the other variants was detected (Fig. S5). For this reason, and to improve the sensitivity of the capture ELISA, the purified 2vb10-6×His VHH was in vitro biotinylated and a new biotin-streptavidin capture ELISA was developed and optimized (Fig. 1A, biotin-streptavidin capture ELISA).
To determine the detection limit of the biotin-streptavidin capture ELISA, different concentrations of the recombinant protein BLS-Stx2B and serial dilutions of the EDL933 culture supernatant were analyzed (Fig. 4C and D). The LOD for BLS-Stx2B ranged between 4 and 8 ng/ml, 4-fold lower than the LOD for the HA–anti-HA capture ELISA. The maximum dilution of EDL933 culture supernatant for which native Stx2a could be detected was 1/32 versus 1/16 for the HA–anti-HA capture ELISA. Additionally, in order to improve the analytical sensitivity of the biotin-streptavidin capture ELISA, the detection was performed by chemiluminescence instead of spectrophotometry (see Materials and Methods), but no improvement in the sensitivity of the assay was observed (Fig. 4E and F).
Taken together, these results demonstrate that the biotin-streptavidin capture ELISA with spectrophotometric detection is more sensitive than the HA–anti-HA capture ELISA for the detection of the recombinant as well as the native Stx2a toxins.
Detection of Stx in stool samples.
To test the feasibility of the biotin-streptavidin capture ELISA for the detection of Stx2 in stool, the assay was applied to determine Stx2 in spiked stool samples. For this, a pool of 10 stool samples obtained from patients with diarrhea not associated with STEC (determined by stool culture, FFStx, and/or stx detection by PCR) was spiked with different amounts of culture supernatants obtained from the EDL933, 02/02, or 1091/10 strain and analyzed by the biotin-streptavidin capture ELISA with spectrophotometric detection (Fig. 5A). The obtained results indicate that the stool matrix does not interfere with Stx2a detection and does not affect the sensitivity of the capture ELISA; the maximum dilution of STEC culture supernatant in which the native Stx2a could be detected was 1/32 (Fig. 5A), similar to the one obtained when diluting the samples in the sample diluent (Fig. 4D). The same spiked samples were analyzed in parallel with the commercial ELISA kit Ridascreen Verotoxin (see Materials and Methods). As shown in Fig. 5B, the maximum dilution of EDL933 and 02/02 culture supernatants in which native Stx2a could be detected was 1/8 and 1/2, respectively, compared to 1/32 and 1/8 for the biotin-streptavidin capture ELISA, respectively, indicating that the biotin-streptavidin capture ELISA is more sensitive than the commercial kit for Stx2a detection.
FIG 5.
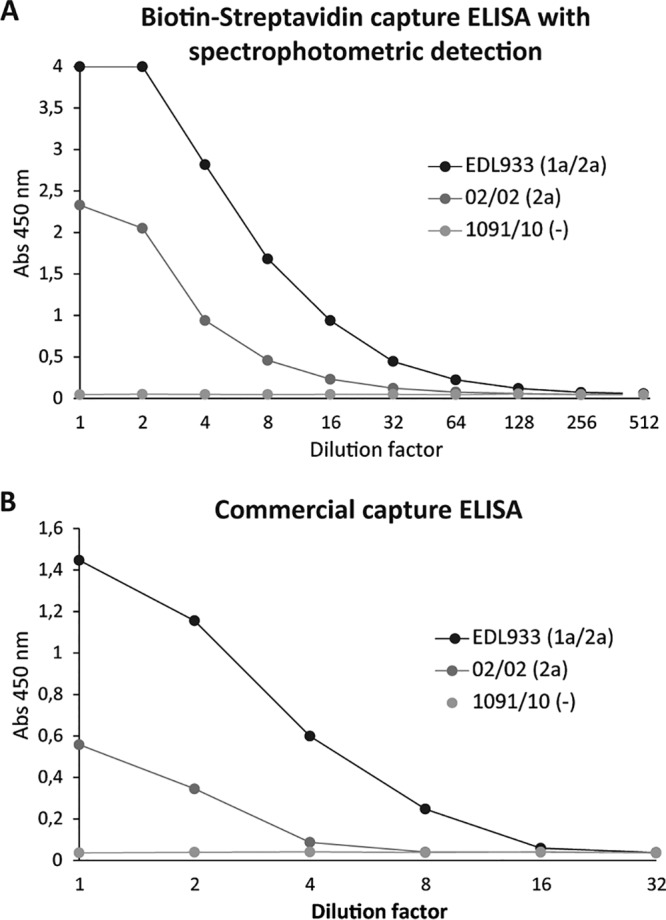
Detection of Stx2 in spiked stool samples. A pool of 10 stool samples was spiked with different amounts of the culture supernatants obtained from the indicated strains. (A) Biotin-streptavidin capture ELISA with detection by spectrophotometry. (B) Commercial capture ELISA (R-Biopharm). The EDL933 strain expresses Stx1a and Stx2a, 02/02 expresses only Stx2a, and 1091/10 does not express Shiga toxins (control).
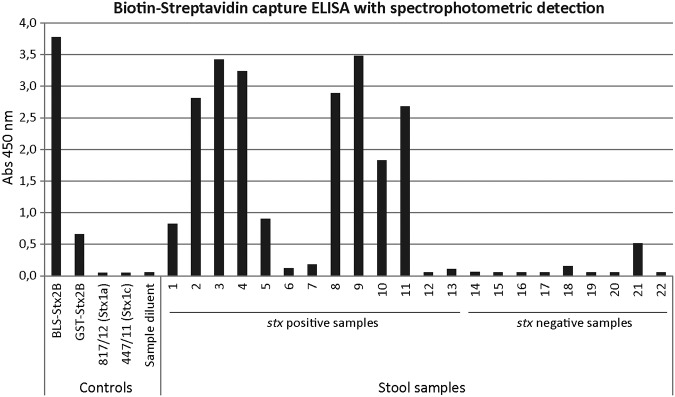
Finally, as a proof of concept, stool samples obtained from patients with diarrhea, BD, or HUS were analyzed for the presence of Stx2 (Table 2 and Fig. 6). In these samples, STEC association was confirmed by PCR for stx1 and/or stx2 gene detection from the confluent growth zone in MacConkey sorbitol agar plates or from the isolated strains. Based on this analysis, 13 samples were positive for stx2 or stx1 and 9 were negative (Table 2). By the biotin-streptavidin capture ELISA, high reactivity values were observed with 9 of 11 stx2-positive samples; as expected, stx1-positive samples (samples 6 and 7) were negative, and all the stx-negative samples except one (sample 21) showed negative results (Fig. 6). In parallel, all the samples were also analyzed using the membrane-based rapid immunoassay Shiga Toxin Quik Chek designed for the detection of Stx1 and Stx2. The results obtained with the rapid test were similar to those observed with the VHH-based capture ELISA, except for sample 21, which was negative by the rapid assay (Table 2). Interestingly, sample 21, obtained from a 2-year-old patient with clinical diagnosis of HUS, resulted positive for Stx2 by the Vero cell cytotoxicity assay and the biotin-streptavidin VHH-based capture ELISA developed in this work but negative for the rest of the laboratory tests, including the PCR techniques (Table 2). Taken together, these data indicate a high correlation and agreement of the biotin-streptavidin capture ELISA results with those obtained with stx2 gene detection by PCR and the commercial kit Shiga Toxin Quik Chek.
TABLE 2.
Stool samples obtained from patients with diarrhea, bloody diarrhea, or HUS
| Sample ID | Clinical diagnosisa | Age | Serotypeb | Genotypic characterizationc
|
FFStxd
|
|||
|---|---|---|---|---|---|---|---|---|
| eae/ehx | stx | Vero cell cytotoxicity assay | Stx Quik Chek assay | VHH-based ELISA | ||||
| 1 | HUS | 3 mo | O145:NM | +/+ | stx2a | ND | Pos (Stx2) | Pos |
| 2 | BD | 6 yr | O145:NM | +/+ | stx2a | ND | Pos (Stx2) | Pos |
| 3 | BD | 13 mo | O157:H7 | +/+ | stx2a/2c | ND | Pos (Stx2) | Pos |
| 4 | BD | 1 yr | O145:NM | +/+ | stx2a | ND | Pos (Stx2) | Pos |
| 5 | HUS | 41 mo | O59:H19 | −/+ | stx2a | Pos | Pos (Stx2) | Pos |
| 6 | UFS | 10 mo | O26:H11 | +/+ | stx1a | ND | Pos (Stx1) | Neg |
| 7 | UFS | 1 mo | ONT:HNT | +/+ | stx1a | ND | Neg | Neg |
| 8 | HUS | 20 mo | O157:H7 | +/+ | stx2a/2c | ND | Pos (Stx2) | Pos |
| 9 | D | 7 yr | O121:HNT | +/+ | stx2a | ND | Pos (Stx2) | Pos |
| 10 | D | 11 yr | O145:NM | +/+ | stx2a | ND | Pos (Stx2) | Pos |
| 11 | D | 10 mo | O145:NM | +/+ | stx2a | ND | Pos (Stx2) | Pos |
| 12 | HUS | 18 mo | O145:NM | +/+ | stx2a | Neg | Neg | Neg |
| 13 | HUS | 2 yr | O157:H7 | +/+ | stx2a/2c | Neg | Neg | Neg |
| 14 | HUS | 11 mo | O157:H7 | +/+ | Neg | Neg | Neg | Neg |
| 15 | HUS | 21 mo | ONT:HNT | ND | Neg | Neg | Neg | Neg |
| 16 | HUS | 60 mo | ONT:HNT | ND | Neg | Neg | Neg | Neg |
| 17 | D | 3 yr | ONT:HNT | ND | Neg | ND | Neg | Neg |
| 18 | BD | 2 yr | ONT:HNT | ND | Neg | ND | Neg | Neg |
| 19 | D | 2 yr | ONT:HNT | ND | Neg | ND | Neg | Neg |
| 20 | BD | 11 mo | ONT:HNT | ND | Neg | ND | Neg | Neg |
| 21e | HUS | 2 yr | ONT:HNT | ND | Neg | Pos | Neg | Pos |
| 22 | BD | 3 mo | ONT:HNT | ND | Neg | ND | Neg | Neg |
HUS, hemolytic-uremic syndrome; BD, bloody diarrhea; D, diarrhea; UFS, unspecific febrile syndrome.
Serotype was determined by seroagglutination using specific antisera. ONT, O polysaccharide not typified; HNT, flagellar H antigen not typified; NM, not motile.
eae, ehxA, and stx gene(s) detection by PCR. ND, not determined.
Free fecal Shiga toxin (FFStx) detection in stool samples. Pos, positive; Neg, negative; ND, not determined; VHH-based ELISA, biotin-streptavidin VHH-based capture ELISA with spectrophotometric detection.
Bacterial isolation, negative; immunomagnetic separation for O157, negative; multiple PCR for rfbO157/stx1/stx2, negative; PCR for diarrheagenic Escherichia coli including enteropathogenic E. coli (eae), enteroaggregative E. coli (aggR), enteroinvasive E. coli (ipaH), and enterotoxigenic E. coli (heat-labile enterotoxin/heat-stable enterotoxin), negative; Chemlis E. coli O157, O145, and O121 (Chemtest Argentina S.A., detection of specific IgM and IgG antibodies in serum sample), negative.
FIG 6.
Analysis of stool samples obtained from patients with diarrhea, bloody diarrhea, or HUS by the biotin-streptavidin capture ELISA with spectrophotometric detection. The analysis was performed after enrichment of the samples in Gram-negative broth (Hajna) (see Materials and Methods). The recombinant proteins BLS-Stx2B and GST-Stx2B, the culture supernatants obtained from STEC strains that produce Stx1a or Stx1c, and the sample diluent were included as controls. A sample was considered positive when the A450 value was 3 times higher than the value obtained for the sample diluent (A450 > 0.18).
DISCUSSION
Shiga toxins are the distinctive feature and most important virulence factors of STEC strains irrespective of the E. coli serotype. Therefore, free Stx detection assays and stx gene detection by PCR are considered the most suitable methods for the early diagnosis of STEC infections. In addition, timely and accurate diagnosis of STEC infection is crucial for ensuring proper care of patients as well as avoiding delays in starting a supportive or, in the case it becomes available, a specific treatment for STEC-HUS. For these reasons and since VHHs provide many advantages over conventional polyclonal and monoclonal antibodies or currently used antibody-based fragments, we decided to exploit VHH single-domain antibodies (nanobodies) derived from camelids to develop an immunocapture assay for the detection of Stx2.
In the present work, 13 anti-Stx2 VHHs previously obtained from a variable-domain repertoire library isolated from a llama immunized with the recombinant protein BLS-Stx2B were selected and evaluated in 130 capture-detection pair combinations for the detection of the recombinant proteins BLS-Stx2B and GST-Stx2B. Based on this analysis and the purification yield of the corresponding VHHs, six capture-detection pairs were selected and further evaluated for their ability to detect the recombinant proteins and native Stx2 including its variants. In all the VHH capture-detection combinations, we systematically observed a higher reactivity against BLS-Stx2B than against GST-Stx2B. This result could be explained by the fact that the llama from which the library was generated to obtain the VHHs was immunized with the BLS-Stx2B chimera, and it was previously demonstrated that the BLS scaffold promotes the pentamerization of the toxin, maintaining its Gb3Cer-binding capacity (37). Among the six VHH capture-detection pairs preliminarily selected, the 1vb1-2vb10 VHH capture-detection pair displayed the best performance for native Stx2a detection, even showing some level of reactivity for Stx2f. Therefore, this VHH combination was selected to develop and optimize a high-performance capture ELISA. To improve the sensitivity of the assay, the 2vb10-6×His detection VHH was purified, in vitro biotinylated, and used to develop a biotin-streptavidin capture ELISA. This capture ELISA was evaluated using spectrophotometric and chemiluminescence detection methods, showing a significant improvement in the LOD for the recombinant and native Stx2a, but no difference in the performance of the assay was observed between the two detection methods. Additionally, to test the performance of the VHH-based capture ELISA for Stx2 detection in stool samples, stools spiked with culture supernatants of Stx2-producing E. coli strains and fecal samples obtained from patients with diarrhea, bloody diarrhea, or HUS were evaluated. Stool-spiked experiments showed that the stool matrix does not interfere with Stx2 detection since the LOD was similar to the one obtained by diluting native Stx2 in sample buffer. Furthermore, the analytical sensitivity was 4-fold higher than that of a commercial capture ELISA. Finally, the analysis of diarrheal stool samples obtained from patients demonstrated a high correlation and agreement of the biotin-streptavidin Stx2 capture ELISA results with those obtained with the stx2 detection method by PCR and the commercial rapid Shiga Toxin Quik Chek immunoassay.
Although anti-Stx VHHs with therapeutic potential have been developed (33, 35), until now they have not been applied for the development of new Stx detection assays for the diagnosis of STEC infections. Here, we have demonstrated that VHHs could be used as new valuable tailor-made diagnostic tools for the detection of Stx in both STEC culture supernatants and stool samples. The double VHH-based biotin-streptavidin capture ELISA showed an excellent performance for the detection of Stx2a subtype, the variant of greater clinical and epidemiological relevance, and to a lesser extent Stx2f. In addition, VHHs provide many advantages over conventional polyclonal and monoclonal antibodies and currently used antibody-based fragments. These advantages are given by the intrinsic properties of VHHs, including their solubility and structural stability even under denaturing or high-temperature conditions (44). Furthermore, they can be efficiently engineered, cloned, and expressed with high yields in economical expression systems, such as bacteria and yeast, and with high batch-to-batch reproducibility, which allows their production on a large scale and in a standardized manner in a short period (45). In summary, these characteristics may facilitate the development, production, and standardization of these immunoreagents, resulting in less expensive and better diagnostics for STEC-HUS that could be easily transferred to clinical practice. Additionally, we are applying the VHHs selected in this work for the development of a rapid (10-min) immunochromatographic assay that would improve access to diagnostics, especially for health care units with minimal infrastructure and untrained staff. Furthermore, the potential of the VHH technology should allow us to easily broaden the detection spectrum to Stx1 and its variants as well as the other subtypes of Stx2. All these aspects will further facilitate the translation of our findings to clinical practice.
The domain structure of VHHs consists in four conserved framework regions (FR1 to FR4) and three hypervariable complementarity-determining regions (CDR1 to CDR3). FRs maintain the tertiary structure of the paratope, while the CDRs form the hypervariable loops that directly interact with the antigen epitope (46). The typically long and flexible CDR3 of VHHs makes them particularly capable of binding concave and hidden epitopes (cryptic epitopes, enzyme active sites, etc.) that are not accessible to conventional antibodies (44). However, the six VHHs selected in this work which displayed the highest performance (2vb21, 2vb10, 1vb1, 2vb1, 2vb11, and 2vb8) belong to the same VHH family (family 1) and have a CDR3 significantly shorter (7 amino acid residues) than the CDR3 length (12, 17, and 20 amino acid residues) of the VHHs of the other families (see Fig. S2 in the supplemental material). Interestingly, these six VHHs belong to the same family (family 1) of a previously developed VHH (2vb27 VHH) with therapeutic potential, which showed an excellent in vitro and in vivo Stx2-neutralizing capacity (33). Further work is needed to characterize and understand how these VHHs with a short CDR3 are able to efficiently interact with the B subunit of Stx2.
In summary, we have developed and optimized a double (capture-detection) VHH-based biotin-streptavidin capture ELISA with spectrophotometric detection for Stx2 detection. This assay showed an analytical sensitivity higher than a commercial ELISA and an excellent clinical sensitivity comparable to the one obtained with the PCR-based stx2 gene detection method and a commercial rapid immunoassay. This excellent performance coupled with the intrinsic properties of VHHs (high target affinity and specificity, stability, and ease of expression at high yields in recombinant bacteria) makes it an attractive tool for Stx detection and could be of great value for STEC-HUS diagnosis.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants PICT Start Up 2014/3755 (préstamo BID) and FONARSEC FITS SALUD 2011 Proyecto no. 0002, from the Agencia Nacional de Promoción Científica y Tecnológica (ANPCyT), Argentina. L.J.M. is a research fellow of the Consejo Nacional de Investigaciones Científicas y Técnicas, CONICET, Buenos Aires, Argentina. A.E.C., J.E.U., and D.J.C. are career investigators of CONICET.
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Noris M, Mescia F, Remuzzi G. 2012. STEC-HUS, atypical HUS and TTP are all diseases of complement activation. Nat Rev Nephrol 8:622–633. doi: 10.1038/nrneph.2012.195. [DOI] [PubMed] [Google Scholar]
- 2.Tarr PI, Gordon CA, Chandler WL. 2005. Shiga-toxin-producing Escherichia coli and haemolytic uraemic syndrome. Lancet 365:1073–1086. doi: 10.1016/S0140-6736(05)71144-2. [DOI] [PubMed] [Google Scholar]
- 3.Majowicz SE, Scallan E, Jones-Bitton A, Sargeant JM, Stapleton J, Angulo FJ, Yeung DH, Kirk MD. 2014. Global incidence of human Shiga toxin-producing Escherichia coli infections and deaths: a systematic review and knowledge synthesis. Foodborne Pathog Dis 11:447–455. doi: 10.1089/fpd.2013.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mayer CL, Leibowitz CS, Kurosawa S, Stearns-Kurosawa DJ. 2012. Shiga toxins and the pathophysiology of hemolytic uremic syndrome in humans and animals. Toxins (Basel) 4:1261–1287. doi: 10.3390/toxins4111261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rivas M, Miliwebsky E, Chinen I, Deza N, Leotta GA. 2006. The epidemiology of hemolytic uremic syndrome in Argentina. Diagnosis of the etiologic agent, reservoirs and routes of transmission. Medicina (B Aires) 66(Suppl 3):27–32. (In Spanish.) [PubMed] [Google Scholar]
- 6.Niu S, Paluszynski J, Bian Z, Shi L, Kidder K, Liu Y. 2018. LPS-primed CD11b(+) leukocytes serve as an effective carrier of Shiga toxin 2 to cause hemolytic uremic syndrome in mice. Sci Rep 8:3994. doi: 10.1038/s41598-018-22327-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Palermo MS, Exeni RA, Fernandez GC. 2009. Hemolytic uremic syndrome: pathogenesis and update of interventions. Expert Rev Anti Infect Ther 7:697–707. doi: 10.1586/eri.09.49. [DOI] [PubMed] [Google Scholar]
- 8.Johannes L, Romer W. 2010. Shiga toxins—from cell biology to biomedical applications. Nat Rev Microbiol 8:105–116. doi: 10.1038/nrmicro2279. [DOI] [PubMed] [Google Scholar]
- 9.Melton-Celsa AR. 2014. Shiga toxin (Stx) classification, structure, and function. Microbiol Spectr 2:EHEC-0024-2013. doi: 10.1128/microbiolspec.EHEC-0024-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nishikawa Y, Zhou Z, Hase A, Ogasawara J, Cheasty T, Haruki K. 2000. Relationship of genetic type of Shiga toxin to manifestation of bloody diarrhea due to enterohemorrhagic Escherichia coli serogroup O157 isolates in Osaka City, Japan. J Clin Microbiol 38:2440–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Orth D, Grif K, Khan AB, Naim A, Dierich MP, Wurzner R. 2007. The Shiga toxin genotype rather than the amount of Shiga toxin or the cytotoxicity of Shiga toxin in vitro correlates with the appearance of the hemolytic uremic syndrome. Diagn Microbiol Infect Dis 59:235–242. doi: 10.1016/j.diagmicrobio.2007.04.013. [DOI] [PubMed] [Google Scholar]
- 12.Brooks JT, Sowers EG, Wells JG, Greene KD, Griffin PM, Hoekstra RM, Strockbine NA. 2005. Non-O157 Shiga toxin-producing Escherichia coli infections in the United States, 1983–2002. J Infect Dis 192:1422–1429. doi: 10.1086/466536. [DOI] [PubMed] [Google Scholar]
- 13.Melli LJ, Ciocchini AE, Caillava AJ, Vozza N, Chinen I, Rivas M, Feldman MF, Ugalde JE, Comerci DJ. 2015. Serogroup-specific bacterial engineered glycoproteins as novel antigenic targets for diagnosis of Shiga toxin-producing-Escherichia coli-associated hemolytic-uremic syndrome. J Clin Microbiol 53:528–538. doi: 10.1128/JCM.02262-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wijnsma KL, Veissi ST, van Bommel SAM, Heuver R, Volokhina EB, Comerci DJ, Ugalde JE, van de Kar N, van den Heuvel L. 2019. Glyco-iELISA: a highly sensitive and unambiguous serological method to diagnose STEC-HUS caused by serotype O157. Pediatr Nephrol 34:631–639. doi: 10.1007/s00467-018-4118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karmali MA, Petric M, Lim C, Fleming PC, Arbus GS, Lior H. 1985. The association between idiopathic hemolytic uremic syndrome and infection by verotoxin-producing Escherichia coli. J Infect Dis 151:775–782. doi: 10.1093/infdis/151.5.775. [DOI] [PubMed] [Google Scholar]
- 16.Beutin L, Steinruck H, Krause G, Steege K, Haby S, Hultsch G, Appel B. 2007. Comparative evaluation of the Ridascreen Verotoxin enzyme immunoassay for detection of Shiga-toxin producing strains of Escherichia coli (STEC) from food and other sources. J Appl Microbiol 102:630–639. doi: 10.1111/j.1365-2672.2006.03139.x. [DOI] [PubMed] [Google Scholar]
- 17.Gavin PJ, Peterson LR, Pasquariello AC, Blackburn J, Hamming MG, Kuo KJ, Thomson RB Jr. 2004. Evaluation of performance and potential clinical impact of ProSpecT Shiga toxin Escherichia coli microplate assay for detection of Shiga toxin-producing E. coli in stool samples. J Clin Microbiol 42:1652–1656. doi: 10.1128/jcm.42.4.1652-1656.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kehl KS, Havens P, Behnke CE, Acheson DW. 1997. Evaluation of the premier EHEC assay for detection of Shiga toxin-producing Escherichia coli. J Clin Microbiol 35:2051–2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boone JT, Campbell DE, Dandro AS, Chen L, Herbein JF. 2016. A rapid immunoassay for detection of Shiga toxin-producing Escherichia coli directly from human fecal samples and its performance in detection of toxin subtypes. J Clin Microbiol 54:3056–3063. doi: 10.1128/JCM.01785-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Rauw K, Breynaert J, Piérard D. 2016. Evaluation of the Alere SHIGA TOXIN QUIK CHEK in comparison to multiplex Shiga toxin PCR. Diagn Microbiol Infect Dis 86:35–39. doi: 10.1016/j.diagmicrobio.2016.05.016. [DOI] [PubMed] [Google Scholar]
- 21.Staples M, Fang NX, Graham RM, Smith HV, Jennison AV. 2017. Evaluation of the SHIGA TOXIN QUIK CHEK and ImmunoCard STAT! EHEC as screening tools for the detection of Shiga toxin in fecal specimens. Diagn Microbiol Infect Dis 87:95–99. doi: 10.1016/j.diagmicrobio.2016.03.011. [DOI] [PubMed] [Google Scholar]
- 22.Park CH, Kim HJ, Hixon DL, Bubert A. 2003. Evaluation of the duopath verotoxin test for detection of Shiga toxins in cultures of human stools. J Clin Microbiol 41:2650–2653. doi: 10.1128/jcm.41.6.2650-2653.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burgos Y, Beutin L. 2012. Evaluation of an immuno-chromatographic detection system for Shiga toxins and the E. coli O157 antigen, p 29–40. In Abuelzein DE. (ed), Trends in immunolabelled and related techniques. InTech, Rijeka, Croatia. doi: 10.5772/35160. [DOI] [Google Scholar]
- 24.Margot H, Cernela N, Iversen C, Zweifel C, Stephan R. 2013. Evaluation of seven different commercially available real-time PCR assays for detection of shiga toxin 1 and 2 gene subtypes. J Food Prot 76:871–873. doi: 10.4315/0362-028X.JFP-12-365. [DOI] [PubMed] [Google Scholar]
- 25.Qin X, Klein EJ, Galanakis E, Thomas AA, Stapp JR, Rich S, Buccat AM, Tarr PI. 2015. Real-time PCR assay for detection and differentiation of Shiga toxin-producing Escherichia coli from clinical samples. J Clin Microbiol 53:2148–2153. doi: 10.1128/JCM.00115-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hamers-Casterman C, Atarhouch T, Muyldermans S, Robinson G, Hamers C, Songa EB, Bendahman N, Hamers R. 1993. Naturally occurring antibodies devoid of light chains. Nature 363:446–448. doi: 10.1038/363446a0. [DOI] [PubMed] [Google Scholar]
- 27.Lafaye P, Li T. 2018. Use of camel single-domain antibodies for the diagnosis and treatment of zoonotic diseases. Comp Immunol Microbiol Infect Dis 60:17–22. doi: 10.1016/j.cimid.2018.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Odongo S, Sterckx YG, Stijlemans B, Pillay D, Baltz T, Muyldermans S, Magez S. 2016. An anti-proteome nanobody library approach yields a specific immunoassay for Trypanosoma congolense diagnosis targeting glycosomal aldolase. PLoS Negl Trop Dis 10:e0004420. doi: 10.1371/journal.pntd.0004420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tu Z, Chen Q, Li Y, Xiong Y, Xu Y, Hu N, Tao Y. 2016. Identification and characterization of species-specific nanobodies for the detection of Listeria monocytogenes in milk. Anal Biochem 493:1–7. doi: 10.1016/j.ab.2015.09.023. [DOI] [PubMed] [Google Scholar]
- 30.Yang S, Yin S, Shang Y, Wang D, Ma W, He J, Guo J, Cai J, Liu X. 2015. Specific detection of foot-and-mouth disease serotype Asia 1 virus by carboxyl-magnetic beads conjugated with single-domain antibody. BMC Biotechnol 15:83. doi: 10.1186/s12896-015-0201-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gelkop S, Sobarzo A, Brangel P, Vincke C, Romao E, Fedida-Metula S, Strom N, Ataliba I, Mwiine FN, Ochwo S, Velazquez-Salinas L, McKendry RA, Muyldermans S, Lutwama JJ, Rieder E, Yavelsky V, Lobel L. 2018. The development and validation of a novel nanobody-based competitive ELISA for the detection of foot and mouth disease 3ABC antibodies in cattle. Front Vet Sci 5:250. doi: 10.3389/fvets.2018.00250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murad H, Assaad JM, Al-Shemali R, Abbady AQ. 2017. Exploiting nanobodies in the detection and quantification of human growth hormone via phage-sandwich enzyme-linked immunosorbent assay. Front Endocrinol (Lausanne) 8:115. doi: 10.3389/fendo.2017.00115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mejias MP, Hiriart Y, Lauche C, Fernandez-Brando RJ, Pardo R, Bruballa A, Ramos MV, Goldbaum FA, Palermo MS, Zylberman V. 2016. Development of camelid single chain antibodies against Shiga toxin type 2 (Stx2) with therapeutic potential against hemolytic uremic syndrome (HUS). Sci Rep 6:24913. doi: 10.1038/srep24913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schutze K, Petry K, Hambach J, Schuster N, Fumey W, Schriewer L, Rockendorf J, Menzel S, Albrecht B, Haag F, Stortelers C, Bannas P, Koch-Nolte F. 2018. CD38-specific biparatopic heavy chain antibodies display potent complement-dependent cytotoxicity against multiple myeloma cells. Front Immunol 9:2553. doi: 10.3389/fimmu.2018.02553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tremblay JM, Mukherjee J, Leysath CE, Debatis M, Ofori K, Baldwin K, Boucher C, Peters R, Beamer G, Sheoran A, Bedenice D, Tzipori S, Shoemaker CB. 2013. A single VHH-based toxin-neutralizing agent and an effector antibody protect mice against challenge with Shiga toxins 1 and 2. Infect Immun 81:4592–4603. doi: 10.1128/IAI.01033-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lo AW, Moonens K, De Kerpel M, Brys L, Pardon E, Remaut H, De Greve H. 2014. The molecular mechanism of Shiga toxin Stx2e neutralization by a single-domain antibody targeting the cell receptor-binding domain. J Biol Chem 289:25374–25381. doi: 10.1074/jbc.M114.566257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mejias MP, Ghersi G, Craig PO, Panek CA, Bentancor LV, Baschkier A, Goldbaum FA, Zylberman V, Palermo MS. 2013. Immunization with a chimera consisting of the B subunit of Shiga toxin type 2 and brucella lumazine synthase confers total protection against Shiga toxins in mice. J Immunol 191:2403–2411. doi: 10.4049/jimmunol.1300999. [DOI] [PubMed] [Google Scholar]
- 38.Katzen F. 2007. Gateway recombinational cloning: a biological operating system. Expert Opin Drug Discov 2:571–589. doi: 10.1517/17460441.2.4.571. [DOI] [PubMed] [Google Scholar]
- 39.Karmali MA, Petric M, Lim C, Cheung R, Arbus GS. 1985. Sensitive method for detecting low numbers of verotoxin-producing Escherichia coli in mixed cultures by use of colony sweeps and polymyxin extraction of verotoxin. J Clin Microbiol 22:614–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mosmann T. 1983. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods 65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 41.Leotta GA, Chinen I, Epszteyn S, Miliwebsky E, Melamed IC, Motter M, Ferrer M, Marey E, Rivas M. 2005. Validation of a multiplex PCR for detection of Shiga toxin-producing Escherichia coli. Rev Argent Microbiol 37:1–10. (In Spanish.) [PubMed] [Google Scholar]
- 42.Scheutz F, Teel LD, Beutin L, Piérard D, Buvens G, Karch H, Mellmann A, Caprioli A, Tozzoli R, Morabito S, Strockbine NA, Melton-Celsa AR, Sanchez M, Persson S, O’Brien AD. 2012. Multicenter evaluation of a sequence-based protocol for subtyping Shiga toxins and standardizing Stx nomenclature. J Clin Microbiol 50:2951–2963. doi: 10.1128/JCM.00860-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karmali MA. 1989. Infection by verocytotoxin-producing Escherichia coli. Clin Microbiol Rev 2:15–38. doi: 10.1128/cmr.2.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gonzalez-Sapienza G, Rossotti MA, Tabares-da Rosa S. 2017. Single-domain antibodies as versatile affinity reagents for analytical and diagnostic applications. Front Immunol 8:977. doi: 10.3389/fimmu.2017.00977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pant N, Hultberg A, Zhao Y, Svensson L, Pan-Hammarstrom Q, Johansen K, Pouwels PH, Ruggeri FM, Hermans P, Frenken L, Boren T, Marcotte H, Hammarstrom L. 2006. Lactobacilli expressing variable domain of llama heavy-chain antibody fragments (lactobodies) confer protection against rotavirus-induced diarrhea. J Infect Dis 194:1580–1588. doi: 10.1086/508747. [DOI] [PubMed] [Google Scholar]
- 46.Goldman ER, Liu JL, Zabetakis D, Anderson GP. 2017. Enhancing stability of camelid and shark single domain antibodies: an overview. Front Immunol 8:865. doi: 10.3389/fimmu.2017.00865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arbabi Ghahroudi M, Desmyter A, Wyns L, Hamers R, Muyldermans S. 1997. Selection and identification of single domain antibody fragments from camel heavy-chain antibodies. FEBS Lett 414:521–526. doi: 10.1016/s0014-5793(97)01062-4. [DOI] [PubMed] [Google Scholar]
- 48.Conrath KE, Lauwereys M, Galleni M, Matagne A, Frere JM, Kinne J, Wyns L, Muyldermans S. 2001. Beta-lactamase inhibitors derived from single-domain antibody fragments elicited in the camelidae. Antimicrob Agents Chemother 45:2807–2812. doi: 10.1128/AAC.45.10.2807-2812.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Riley LW, Remis RS, Helgerson SD, McGee HB, Wells JG, Davis BR, Hebert RJ, Olcott ES, Johnson LM, Hargrett NT, Blake PA, Cohen ML. 1983. Hemorrhagic colitis associated with a rare Escherichia coli serotype. N Engl J Med 308:681–685. doi: 10.1056/NEJM198303243081203. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.