Abstract
Background
MicroRNAs (miRNAs) can act as negative regulators of gene expression, and play a crucial role in cancer progression. The aim of this study was to investigate the role of miR-1294/pyruvate kinase M2 (PKM2) axis in osteosarcoma cells in vitro and in vivo.
Methods
The function of miR-1294 and its association with PKM2 in osteosarcoma cells were studied by real-time PCR, CCK-8, Western blot, scratch test, transwell assay, flow cytometry, and dual-luciferase reporter assays. The effect of miR-1294 on tumor growth in vivo was evaluated in a subcutaneous xenograft model of osteosarcoma.
Results
miR-1294 was downregulated in osteosarcoma cells. Forced overexpression of miR-1294 inhibited cell proliferation, migration, and invasion, and induced G0/G1 arrest and apoptosis. Consistently, protein expression levels of proliferating cell nuclear antigen, c-Myc, cyclin D1, active matrix metalloproteinase 2, and active matrix metalloproteinase 9 were decreased, and cleaved caspase 3 and cleaved PARP were increased following miR-1294 overexpression. Moreover, we demonstrated that PKM2 was a target of miR-1294 in osteosarcoma cells, and the effects caused by miR-1294 mimic were reversed by the overexpression of PKM2. Furthermore, we found that upregulation of miR-1294 inhibited tumorigenesis of osteosarcoma cells in vivo, which was accompanied by downregulation of PKM2.
Conclusion
Our results revealed that miR-1294/PKM2 signaling cascade exerts important roles in the regulation of tumor progression, implying that this pathway may serve as a potential therapeutic target in osteosarcoma.
Keywords: pyruvate kinase M2, miR-1294, osteosarcoma, cell proliferation, cell apoptosis, tumorigenesis
Background
Osteosarcoma is the most common malignant bone tumor, occurring predominantly in adolescents and young adults.1 There are many risk factors for osteosarcoma, such as abnormal growth hormone levels, genetic and epigenetic misregulations.2 The standard treatment of osteosarcoma is surgery, neoadjuvant, and adjuvant chemotherapy.3 The 5-year survival rate has remained at 60–70% in patients with non-metastatic disease, while it is dramatically reduced in patients with metastatic disease.4 Therefore, it is necessary to understand the pathogenesis of osteosarcoma in order to develop effective treatment strategies.
MicroRNAs (miRNAs), a class of non-coding RNAs of approximately 22–25 nucleotides in length, act as negative regulators of gene expression by repressing mRNA translation or facilitating mRNA degradation.5 Increasing evidence shows that miRNAs play important roles in regulating cancer cell development.6 Previous studies have demonstrated that miR-1294 is lowly expressed in multiple cancers, such as epithelial ovarian cancer,7 gastric cancer,8 oral squamous cell carcinoma,9 osteosarcoma,10 and glioma.11 Forced expression of miR-1294 inhibits tumor cell growth and cisplatin resistance.7,12 Moreover, circ_0005198 and circ_0004370 can sponge miR-1294 to promote glioma and esophageal cancer progression, respectively.13,14 However, the function and mechanism of miR-1294 in osteosarcoma are not fully understood and need further investigation.
Pyruvate kinase M2 (PKM2), a key enzyme in glycolysis, is found to be frequently overexpressed in cancers and stimulates cell proliferation, migration, and invasion.15,16 Previous research has shown that PKM2 is highly expressed in osteosarcoma and is associated with a poor outcome.17 However, the association between miR-1294 and PKM2 in osteosarcoma has not been studied. By prediction, we found that PKM2 is a candidate target of miR-1294, indicating that miR-1294/PKM2 pathway may play a role in osteosarcoma.
In the present study, we explored the expression and function of miR-1294 in osteosarcoma cells. Moreover, the role of PKM2 in miR-1294-mediated growth inhibition was investigated. The effect of miR-1294 on tumorigenesis of osteosarcoma cells in vivo was further studied.
Methods
Cells and Cell Culture
MG63, U2OS, and 143B were purchased from Shanghai Zhong Qiao Xin Zhou Biotechnology (Shanghai, China). Human bone marrow mesenchymal stem cells (hMSCs), Saos-2, and HOS were purchased from Procell Biological Technology (Wuhan, China). Saos-2 cells were cultured in McCoy’s 5a medium (Procell Biological Technology) supplemented with 15% fetal bovine serum (FBS; BI, Kibbutz Beit Haemek, Israel). U2OS cells were grown in Dulbecco’s modified Eagle’s medium (BD Biosciences, Franklin Lakes, NJ, USA) containing 10% FBS (BI). 143B cells were cultured in Eagle modified essential medium (Shanghai Zhong Qiao Xin Zhou Biotechnology) supplemented 10% FBS (BI). MG63 and HOS cells were grown in minimum essential medium (Gibco, Grand Island, NY, USA) containing 10% FBS (BI). hMSCs were cultured in hMSC complete medium (Procell). All cell lines were maintained in an incubator at 37°C with 5% CO2. Cisplatin (DDP) was obtained from Meilun Biotechnology (Dalian, China). In experiments using cisplatin, cells were incubated with 5 μmol/L DDP for 24 h before the detection.
Animals and Ethics Statement
Forty-eight nude mice (two months old) weighed 18–20 g were purchased from Beijing Huafukang Biological Technology Co., Ltd. (license SCXK (jing) 2014–0004; Beijing, China), and housed in a temperature-controlled room (21 ± 1°C) with a 12-h/12-h light/dark cycle. This study was approved by the Ethics Committee for Animal Experimentation of the China Medical University and conducted according to the National Institutes of Health guide for the care and use of laboratory animals.
Transient and Stable Transfection
Transient transfection was performed for cell culture experiments. Briefly, miR-1294 mimic or inhibitor, PKM2 overexpressing vector, or their corresponding control was transfected into osteosarcoma cells for 48 h using the Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA). Stable transfection was used for animal studies. The pre-miR-1294 sequence was cloned into the BamHI/HindIII site of pRNA H1.1 vector. An empty vector was used as a control. The vectors were then transfected into MG63 and U2OS cells using Lipofectamine 2000 reagent (Invitrogen) and stably transfected cells were selected with G418.
Real-Time PCR
The mRNA levels of genes were measured by real-time PCR assay.18 Briefly, total RNA isolated from cells or tissues was reversely transcribed into cDNAs using Super M-MLV reverse transcriptase (Tiangen, Beijing, China). Real-time PCR was carried out with 2 × Power Taq PCR MasterMix (Tiangen) and SYBR Green (Solarbio, Beijing, China) on an ExiCycler™ 96 Real-time Quantitative Thermal Block (Bioneer Corporation, Daejeon, Korea). The sequences of the primers were as follows: PKM2 forward, 5′- CTATCCTCTGGAGGCTGTGC −3′, reverse, 5′- GTGGGGTCGCTGGTAATG −3′; miR-1294 forward, 5′- TGTGAGGTTGGCATTGTTGTCTGT −3′, reverse, 5′- GTGCAGGGTCCGAGGTATTC −3′; β-actin forward, 5′- CTTAGTTGCGTTACACCCTTTCTTG −3′, reverse, 5′- CTGTCACCTTCACCGTTCCAGTTT −3′; U6 forward, 5′- GCTTCGGCAGCACATATACT −3′, reverse, 5′- GTGCAGGGTCCGAGGTATTC −3′. The relative expression levels of miR-1294 and PKM2 were calculated using the ΔCt or 2 -ΔΔCt method.
Cell Proliferation Assay
Cell proliferation was determined by CCK-8 assay.19 In brief, cells were seeded into 96-well plates at 4×103 cells per well. After culture for 0, 24, 48, 72, or 96 h at 37°C in 5% CO2, the medium was discarded and replaced with 100 μL complete medium and 10 μL CCK-8 (Sigma, St. Louis, MO, USA). One hour later, the absorbance value of each well was determined at 450 nm using a microplate reader (BioTek, Winooski, VT, USA).
Cell Cycle Assay
Cell cycle distribution was analyzed with a flow cytometer (BD Biosciences) following propidium iodide (PI) staining.20 Briefly, cells were collected and fixed in 70% ethanol at 4 °C for 2 h. Then, the supernatant was discarded and 500 μL of dye buffer was added, followed by addition of 25 μL PI and 10 μL RNase A. After 30 min at 37°C in the dark, cells were analyzed with the flow cytometer.
Cell Apoptosis Assay
Cell apoptosis was measured using a flow cytometer (BD Biosciences) after annexin V-FITC/PI staining.21 In short, cells were gathered and resuspended in 500 μL of binding buffer and then addition of 5 μL Annexin V-FITC and 10 μL PI and mix well. After 15 min at room temperature in the dark, cells were detected by flow cytometry.
Western Blot
Protein expression levels were examined by Western blotting.22 Total protein was separated from cells or tissues using RIPA lysis buffer (Beyotime Institute of Biotechnology, Shanghai, China) supplemented with phenylmethanesulfonyl fluoride. The protein concentration was determined with the BCA assay (Beyotime Institute of Biotechnology). Then the samples were subjected to SDS–PAGE, transferred on PVDF membranes, blocked with 5% nonfat milk, and incubated with primary antibodies against proliferating cell nuclear antigen (PCNA; 1:500; Proteintech, Wuhan, China), c-Myc (1:1000; Proteintech), cyclin D1 (1:400; Boster, Wuhan, China), cleaved caspase 3 (1:500; CST, Danvers, MA, USA), cleaved PARP (1:1000; CST), matrix metalloproteinase 2 (MMP-2; 1:500; Proteintech), MMP-9 (1:1000; Proteintech), PKM2 (1:1000; Proteintech), and β-actin (1:500; KeyGEN, Nanjing, China) overnight at 4°C. The membranes were incubated with horseradish peroxidase-labeled goat-anti-rabbit/mouse secondary antibodies for 45 min at 37°C. The protein was visualized by the enhanced chemiluminescence (ECL; 7 Sea Biotech, Shanghai, China) under a gel imaging system (Liuyi, Beijing, China). The blots were analyzed by densitometry. The expression of the target gene was normalized to that of β-actin.
Assessment of Cell Migration
Cell migration was determined by a wound healing method.23 Briefly, cells seeded into six-well plates were transfected for 24 h, followed by treatment with mitomycin C (1 μg/mL) for 1 h. Then, the medium was replaced with serum-free medium, and a sterile 200-μL pipette tip was used to create a scratch at the bottom of the plate, after which cell debris was washed away and serum-free medium was added. The scratch was photographed at 0 and 24 h using an inverted phase-contrast microscope (Olympus, Tokyo, Japan) at 100 × magnification.
Assessment of Cell Invasiveness
Invasion of cancer cells was assessed by transwell Matrigel assay.24 In brief, 1.5×104 cells were seeded into each well of 24-well plates with transwell chamber (Corning, New York, USA) coated with Matrigel (BD Biosciences). After culture for 24 h, cells were fixed with paraformaldehyde (4%) for 20 min at room temperature and followed by stained using crystal violet (0.5%) for 4 min. Invasive cells were observed under an inverted microscope (Olympus) at × 200 magnification.
Dual-Luciferase Reporter Assay
The wild type PKM2 3′UTR containing the putative miR-1294 target site or mutant PKM2 3′UTR sequence was inserted into the pmirGLO vector containing both the firefly luciferase gene and Renilla luciferase gene. 293T cells were co-transfected with miR-1294 mimic or control and pmirGLO vector containing wild- or mutant type PKM2 3′UTR using Lipofectamine 2000 reagent (Invitrogen) for 24 h. The firefly and Renilla luciferase activities were measured using the dual-luciferase assay system (Promega, Wisconsin, WI, USA). The activity of firefly luciferase was normalized to that of Renilla luciferase.
Determination of Tumorigenicity in vivo
Tumorigenicity assays were performed in nude mice as previously described.25 The mice were randomly divided into 4 groups (n = 12 each group): MG63 cells stably transfected with pre-miR-1294, MG63 cells stably transfected with empty vector, U2OS cells stably transfected with pre-miR-1294, and U2OS cells stably transfected with empty vector. 1.5 × 106 cells were subcutaneously injected into nude mice. Tumor volume was recorded every three days and mice were sacrificed on day 28. Then the tumor tissues were isolated from the mice. Each group of tumor tissue was divided into two parts. One part was fixed in neutral formalin for immunohistochemical study. The other was frozen in liquid nitrogen for protein and mRNA isolation.
Immunohistochemistry Analysis
Immunohistochemistry was performed as previously described.26 Formalin-fixed tissue samples were sectioned to 5-μm thickness using a microtome (Leica, Bensheim, Germany). After dewaxing with xylene and rehydration with gradient ethanol, the sections were subjected to antigen retrieval and incubation with one of the primary antibodies against c-Myc (1:200; Proteintech), cyclin D1 (1:200; Boster), MMP-2; (1:200; Proteintech), MMP-9 (1:200; Proteintech), and PKM2 (1:200; Proteintech) overnight at 4°C. Horseradish peroxidase-labeled goat anti-rabbit secondary antibody was used to probe the primary antibody for 60 min at 37°C. Following DAB and hematoxylin staining, the sections were observed with a microscope (Olympus) at × 400 magnification.
Statistical Analysis
Results are expressed as mean ± standard deviation (SD). Each in vitro experiment was repeated three times. Statistical significance was analyzed using Student’s t-test or variance analysis with the GraphPad Prism 7.0 software (GraphPad Software, La Jolla, CA, USA). A P value of less than 0.05 was accepted as statistically significant.
Results
miR-1294 Was Downregulated in Osteosarcoma Cells
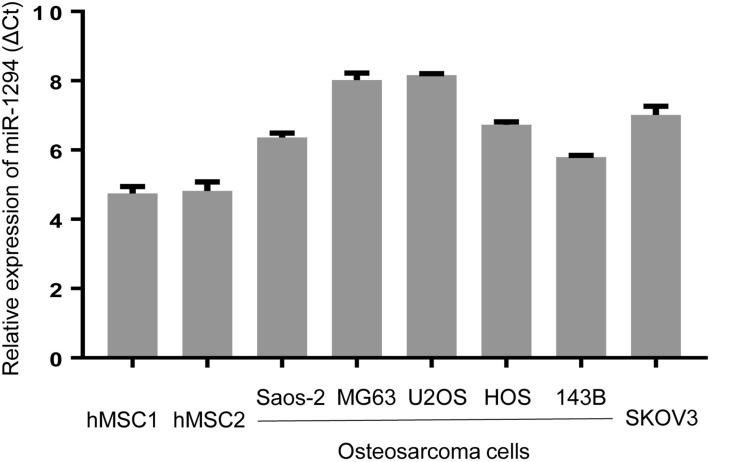
We first evaluated the expression level of miR-1294 in five osteosarcoma cell lines (Saos-2, MG63, U2OS, HOS, and 143B) and human mesenchymal stem cells (hMSC1 and hMSC2). Moreover, an ovarian cancer cell line SKOV3, which has a relatively low expression of miR-1294,7 was used as a reference. The results from real-time PCR showed a decreased expression of miR-1294 in osteosarcoma cells compared with in hMSCs (Figure 1). Interestingly, a similar result was obtained from Gene Expression Omnibus (GSE70415) (Additional file 1).
Figure 1.
Expression levels of miR-1294 in osteosarcoma cells. Real-time PCR was employed to determine miR-1294 expression in five osteosarcoma cell lines (Saos-2, MG63, U2OS, HOS, and 143B) and human mesenchymal stem cells (hMSCs).
Overexpression of miR-1294 Restrained Proliferation and Induced Apoptosis of Osteosarcoma Cells
The influence of miR-1294 upregulation on osteosarcoma cell proliferation and apoptosis was determined. The efficiency of miR-1294 mimic transfection was validated in MG63 and U2OS cells by real-time PCR (Figure 2A). Forced overexpression of miR-1294 significantly inhibited cell proliferation, as measured by CCK-8 assay (Figure 2B). Flow cytometry with propidium iodide (PI) staining showed that transfection of miR-1294 mimic caused distinct G0/G1 cell cycle arrest (Figure 2C). The percent of apoptotic cells was higher in miR-1294 mimic-transfected osteosarcoma cells compared to control cells (Figure 2D). Furthermore, proliferation/apoptosis-related genes were evaluated by Western blot analysis. As shown in Figure 2E, we demonstrated that upregulation of miR-1294 suppressed the expression of PCNA, c-Myc and cyclin D1, and promoted the expression of cleaved caspase 3 and cleaved PARP in MG63 and U2OS cells. Moreover, the expression of miR-1294 was downregulated in cells treated with DDP (Additional file 2A). Upregulation of miR-1294 enhanced the chemosensitivity of osteosarcoma cells to DDP, as evidenced by increased cell apoptosis (Additional file 2C).
Figure 2.
miR-1294 upregulation inhibited proliferation and promoted apoptosis of osteosarcoma cells. (A) Overexpression of miR-1294 in MG63 and U2OS cells by transfection of miR-1294 mimic. (B) Cell proliferation was assessed using the CCK-8 assay. (C) Cell cycle distribution was analyzed by flow cytometry using propidium iodide (PI) staining. (D) Cell apoptosis was determined by flow cytometry using Annexin V and PI staining. (E) Expression levels of PCNA, c-Myc, cyclin D1, cleaved caspase 3, and cleaved PARP were examined by Western blotting. Data are shown as mean ± SD. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
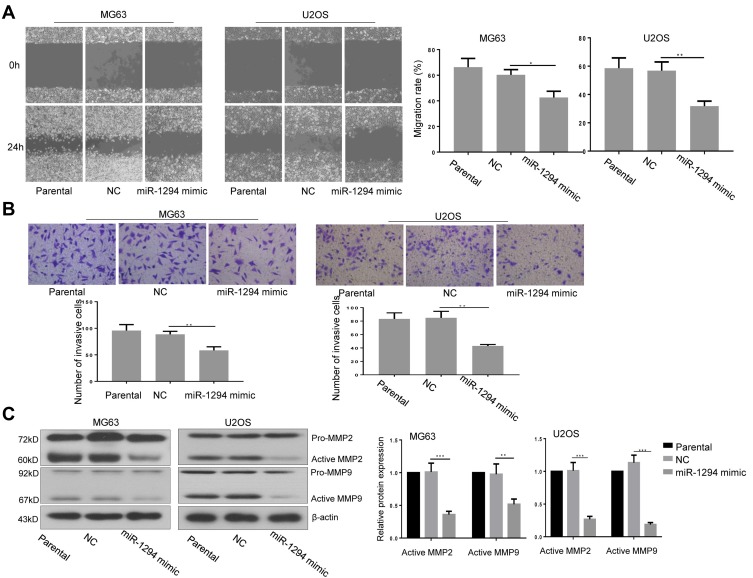
Overexpression of miR-1294 Inhibited Migration and Invasiveness of Osteosarcoma Cells
The effect of miR-1294 upregulation on osteosarcoma cell migration and invasion was assessed. The results from wound healing assay showed that overexpression of miR-1294 dramatically impeded the ability of cell migration (Figure 3A). Cell invasive capacity in osteosarcoma cells transfected with miR-1294 mimic was lower than that in control cells (Figure 3B). Moreover, Western blot analysis revealed that the protein expression levels of active MMP-2 and active MMP-9 were significantly downregulated following transfection of osteosarcoma cells with miR-1294 mimic (Figure 3C).
Figure 3.
miR-1294 upregulation impaired migration and invasiveness of osteosarcoma cells. (A) Cell migration was determined by a wound-healing test in MG63 and U2OS cells with or without miR-1294 overexpression. (B) Cell invasiveness was evaluated by transwell assay in MG63 and U2OS cells following miR-1294 mimic treatment. (C) Expression levels of MMP-2 and MMP-9 in miR-1294 overexpressing MG63 and U2OS cells were detected by Western blotting. Data are shown as mean ± SD. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
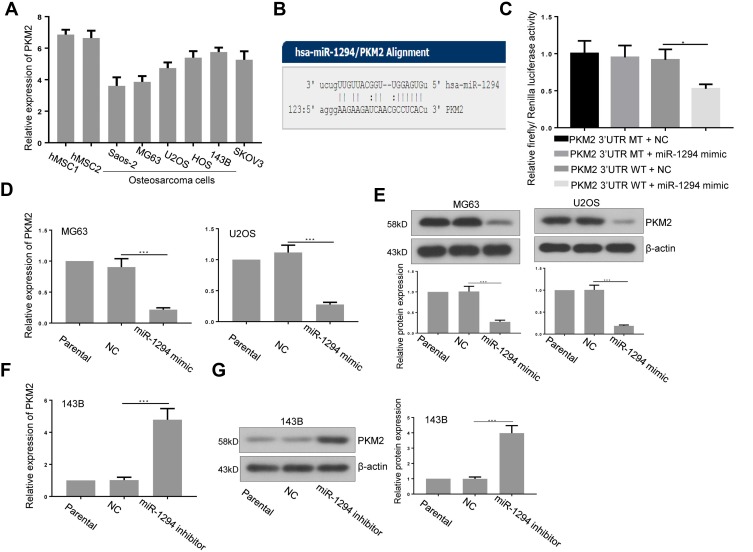
PKM2 Was a Target of miR-1294 in Osteosarcoma Cells
Furthermore, we predicted that PKM2 is a potential target of miR-1294 by using miRanda (http://www.microrna.org/microrna/home.do) (Figure 4B). The expression of PKM2 in osteosarcoma cells used in Figure 1 was relatively more abundant than that in hMSCs (Figure 4A). The predicted combination was validated in 293T cells by dual luciferase reporter assay. As shown in Figure 4C, relative firefly/Renilla luciferase activity was decreased in cells co-transfected with wild-type PKM2 3ʹUTR containing the putative miR-1294 target site and miR-1294 mimic, while no obvious change was observed in other groups. Additionally, the results from real-time PCR and Western blotting demonstrated that the expression of PKM2 was significantly inhibited in both mRNA and protein levels in MG63 and U2OS cells after miR-1294 mimic treatment (Figure 4D and E). On the contrary, transfection of 143B cells with miR-1294 inhibitor upregulated PKM2 protein and mRNA expression (Figure 4F and G).
Figure 4.
PKM2 was a target of miR-1294 in osteosarcoma cells. (A) PKM2 expression in human mesenchymal stem cells (hMSCs) and five osteosarcoma cell lines (Saos-2, MG63, U2OS, HOS, and 143B) was assessed by real-time PCR. (B) The miR-1294 binding site predictions for PKM2 3ʹUTR by microRNA.org. (C) A dual luciferase reporter assay was performed to confirm the binding in 293T cells. (D and E) PKM2 expression in U2OS and HOS cells transfected with miR-1294 mimic by real-time PCR and Western blot assays. (F and G) PKM2 expression in 143B cells transfected with miR-1294 inhibitor by real-time PCR and Western blot assays. Data are shown as mean ± SD. ∗P < 0.05, ∗∗∗P < 0.001.
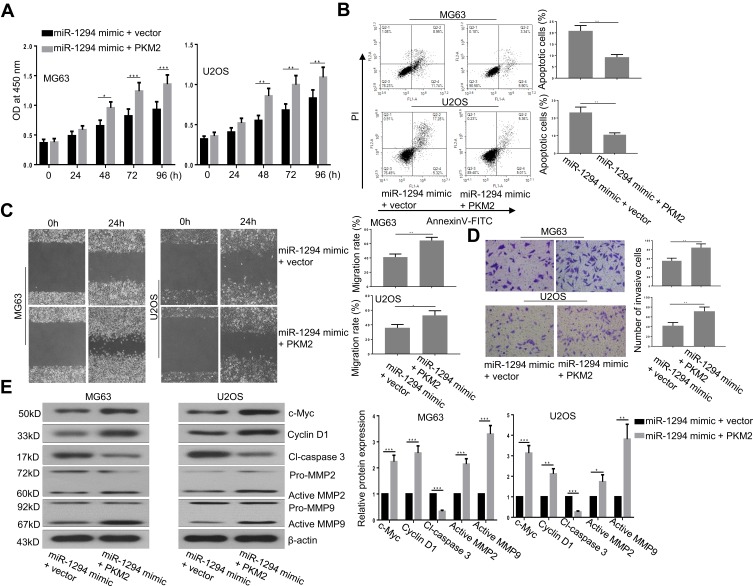
Re-Expression of PKM2 Reversed the Effects of miR-1294 Mimic on Osteosarcoma Cells
To further confirm the role of PKM2 in miR-1294 overexpression-mediated actions in osteosarcoma cells, we examined the effect of forced upregulation of PKM2 on miR-1294 mimic-transfected MG63 and U2OS cells. As shown in Figure 5A, C, and D, cell proliferation, migration, and invasion mediated by miR-1294 mimic were markedly enhanced following overexpression of PKM2. Co-upregulation of miR-1294 and PKM2 inhibited osteosarcoma cell apoptosis compared to that of miR-1294 mimic transfected cells (Figure 5B). Furthermore, cleaved caspase 3 expression levels were decreased, and the expression levels of c-Myc, cyclin D1, active MMP-2, and active MMP-9 were increased in osteosarcoma cells with co-transfection of the PKM2 expression vector and miR-1294 mimic compared with that in miR-1294 mimic-transfected cells (Figure 5E). Moreover, PKM2 expression was increased in osteosarcoma cells treated with DDP (Additional file 2B). Overexpression of PKM2 reversed the effect of miR-1294 mimic on DDP-treated cells (Additional file 2C).
Figure 5.
Overexpression of PKM2 reversed the actions of miR-1294 upregulation on osteosarcoma cells. (A) Cell proliferation was evaluated by CCK-8 assay. (B) Cell apoptosis was examined by flow cytometry using Annexin V and PI staining. (C) Cell migration was assessed by a wound-healing assay. (D) Cell invasiveness was determined by transwell assay. (E) Expression levels of c-Myc, cyclin D1, cleaved-caspase 3, MMP-2, and MMP-9 were texted by Western blotting. Data are shown as mean ± SD. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
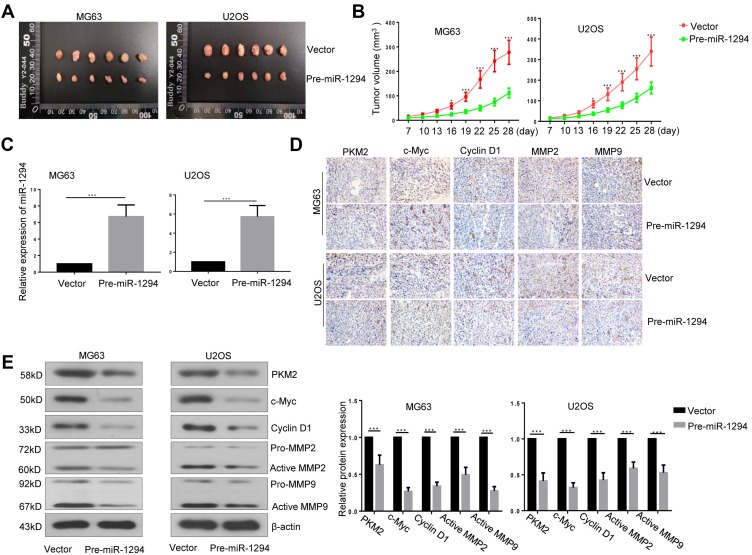
Upregulation of miR-1294 Inhibited Tumorigenicity of Osteosarcoma Cells
Lastly, we assessed the effect of miR-1294 overexpression on the tumorigenicity of osteosarcoma cells in vivo. As shown in Figure 6A and B, The tumor volume was significantly reduced in mice inoculated with miR-1294 overexpressing osteosarcoma cells, as compared to those inoculated with control cells. Confirmation of the expression miR-1294 in cancer tissues with or without miR-1294 upregulation by real-time PCR (Figure 6C). The results of immunohistochemistry and Western blot analysis showed that the expression levels of PKM2, c-Myc, cyclin D1, active MMP-2, and active MMP-9 were reduced in tumor tissues from mice injected with miR-1294 overexpressing osteosarcoma cells (Figure 6D and E).
Figure 6.
Overexpression of miR-1294 inhibited tumorigenicity of osteosarcoma cells. (A, B) Tumor volume in nude mice after subcutaneous injection of ME63 and U2O2 cells with or without overexpression of miR-1294. (C) The expression of miR-1294 in tumor tissues was determined by real-time PCR. (D, E) The expression of PKM2, c-Myc, cyclin D1, MMP-2, and MMP-9 in tumor tissues was assessed using immunohistochemistry and Western blot analysis. Data are shown as mean ± SD from 6 mice/group. ∗P < 0.05, ∗∗∗P < 0.001.
Discussion
The important function of miRNAs such as miR-26a,7 miR-29b,12 miR-155-5p,27 and miR-148a-3p27 in regulating tumor progression of osteosarcoma has been widely reported. Nevertheless, there are still many miRNAs need to be investigated. Very recently, the study from Zhang et al showed that decreased miR-1294 was associated with shorter survival in patients with osteosarcoma, and that upregulation of miR-1294 inhibited proliferation and invasion of osteosarcoma cells by targeting Homeobox A9.10 In the present study, we further demonstrated that forced overexpression of miR-1294 inhibited migration and tumorigenicity of osteosarcoma cells, induced cell apoptosis, and enhanced the chemosensitivity of osteosarcoma cells to DDP. More importantly, we identified that PKM2, a new target gene of miR-1294, played key roles in miR-1294-mediated actions in osteosarcoma cells. Moreover, the tumor-suppressing role of miR-1294 has been reported in other of tumors, such as gastric cancer,8 esophageal squamous cell carcinoma,19 epithelial ovarian cancer,7 oral squamous cell carcinoma,9 and glioma.11
Uncontrolled growth, invasion, and metastasis are the main characteristics of tumor cells.23 A variety of molecules are involved in these processes. For instance, PCNA, a key protein of DNA replication in eukaryotes, is upregulated and reported as a useful biomarker for poor outcome in patients with osteosarcoma.24 Overexpression of c-Myc activates MEK-ERK pathway leading to the increase in cell invasion.26 Cyclin D1 is the main regulator for G1 phase progression, and its knockdown induces apoptosis and cell cycle arrest in osteosarcoma cells.25 Cleavage of caspase 3 and PARP are two major fundamental proteins of programmed cell death.28 MMP-2 and MMP-9 are related to tumor metastasis and are highly expressed in various malignant tumors.29 In this study, the change of above-mentioned factors was in accordance with impaired cell proliferation, migration, and invasion, and increased cell apoptosis in miR-1294-upregulated osteosarcoma cells, which further confirm the anti-cancer function of miR-1294 in osteosarcoma.
miRNAs mainly function as negative regulators of gene expression. Our research revealed that miR-1294 regulated PKM2 expression by direct interaction with its 3ʹUTR. Re-induction of PKM2 reversed the inhibitory actions of miR-1294 mimic on osteosarcoma cells. It has been reported that PKM2 is increased and its knockdown inhibits survival and invasion of osteosarcoma cells.17,23 Other miRNAs including miR-133b,30 miR-214,31 and miR-139-5p32 have been implicated in inhibiting cancer cell growth by downregulation of PKM2. Moreover, it is also known that PKM2 contributes to tumor metabolism.33,34 In consideration of the association between PKM2 and miR-1294, it is possible that miR-1294 plays a role in the metabolism of osteosarcoma. In addition, multiple pathways are involved in the regulation of PKM2 in cancer cells, such as β-catenin,35 mechanistic target of rapamycin (mTOR),36 epidermal growth factor receptor (EGFR),37 and p2738 pathways. Whether these signaling pathways participate in the regulation of miR-1294-mediated anti-tumor effects in osteosarcoma need to be further investigated. Moreover, we found that miR-1294/PKM2 axis enhanced DDP chemosensitivity in osteosarcoma cells. Shikonin, a PKM2 inhibitor, has been proved to overcome the DDP resistance in bladder cancer.27 The study from Zhang et al showed that miR-1294 prevented ovarian cancer DDP resistance by targeting IGF1R.12 Our data showed that PKM2 upregulation only partly reversed the effect of miR-1294 on DDP-treated cells, indicating that other targets of miR-1294, apart from PKM2 may contribute to miR-1294-mediated anti-tumor activity, such as IGF1R. Previous studies showed that silencing of PKM2 enhanced radiosensitivity of cervical cancer and non-small cell lung cancer,28,39 suggesting that miR-1294/PKM2 axis may play a role in radiosensitivity of osteosarcoma.
Conclusions
This study demonstrated that upregulation of miR-1294 is an effective strategy to inhibit cell growth and tumorigenicity and enhanced DDP chemosensitivity. Moreover, PKM2 plays an important role in miR-1294-mediated anti-tumor actions, indicating that miR-1294/PKM2 axis may act as a potential therapeutic target for the treatment of osteosarcoma.
Funding Statement
This study was supported by grants from the Natural Science Foundation of China (No. 81870505), the Guiding Program of Liaoning Natural Science Foundation in China (No. 20180550610), and the Natural Science and Medical Health Foundation of Liaoning Province (No. 20180530088).
Data Sharing Statement
The datasets supporting the conclusions of the current study are available from the corresponding author on reasonable request. Please contact corresponding author, if you want to request the dataset.
Ethics Approval
This study was approved by the Ethics Committee for Animal Experimentation of the China Medical University and conducted according to the National Institutes of Health guide for the care and use of laboratory animals.
Disclosure
The authors declare that they have no competing interests in this work.
References
- 1.Kansara M, Teng MW, Smyth MJ, Thomas DM. Translational biology of osteosarcoma. Nat Rev Cancer. 2014;14(11):722–735. doi: 10.1038/nrc3838 [DOI] [PubMed] [Google Scholar]
- 2.Chen D, Zhao Z, Huang Z, et al. Super enhancer inhibitors suppress MYC driven transcriptional amplification and tumor progression in osteosarcoma. Bone Res. 2018;6:11. doi: 10.1038/s41413-018-0009-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tiram G, Segal E, Krivitsky A, et al. Identification of dormancy-associated MicroRNAs for the design of osteosarcoma-targeted dendritic polyglycerol nanopolyplexes. ACS Nano. 2016;10(2):2028–2045. doi: 10.1021/acsnano.5b06189 [DOI] [PubMed] [Google Scholar]
- 4.Gianferante DM, Mirabello L, Savage SA. Germline and somatic genetics of osteosarcoma - connecting aetiology, biology and therapy. Nat Rev Endocrinol. 2017;13(8):480–491. doi: 10.1038/nrendo.2017.16 [DOI] [PubMed] [Google Scholar]
- 5.Kato M, Putta S, Wang M, et al. TGF-beta activates Akt kinase through a microRNA-dependent amplifying circuit targeting PTEN. Nat Cell Biol. 2009;11(7):881–889. doi: 10.1038/ncb1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16(3):203–222. doi: 10.1038/nrd.2016.246 [DOI] [PubMed] [Google Scholar]
- 7.Guo TY, Xu HY, Chen WJ, Wu MX, Dai X. Downregulation of miR-1294 associates with prognosis and tumor progression in epithelial ovarian cancer. Eur Rev Med Pharmacol Sci. 2018;22(22):7646–7652. doi: 10.26355/eurrev_201811_16381 [DOI] [PubMed] [Google Scholar]
- 8.Shi YX, Ye BL, Hu BR, Ruan XJ. Expression of miR-1294 is downregulated and predicts a poor prognosis in gastric cancer. Eur Rev Med Pharmacol Sci. 2018;22(17):5525–5530. doi: 10.26355/eurrev_201809_15813 [DOI] [PubMed] [Google Scholar]
- 9.Wang Z, Yan J, Zou T, Gao H. MicroRNA-1294 inhibited oral squamous cell carcinoma growth by targeting c-Myc. Oncol Lett. 2018;16(2):2243–2250. doi: 10.3892/ol.2018.8967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang ZF, Li GR, Cao CN, Xu Q, Wang GD, Jiang XF. MicroRNA-1294 targets HOXA9 and has a tumor suppressive role in osteosarcoma. Eur Rev Med Pharmacol Sci. 2018;22(24):8582–8588. [DOI] [PubMed] [Google Scholar]
- 11.Chen H, Liu L, Li X, Shi Y, Liu N. MicroRNA-1294 inhibits the proliferation and enhances the chemosensitivity of glioma to temozolomide via the direct targeting of TPX2. Am J Cancer Res. 2018;8(2):291–301. [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y, Huang S, Guo Y, Li L. MiR-1294 confers cisplatin resistance in ovarian cancer cells by targeting IGF1R. Biomed Pharmacother. 2018;106:1357–1363. doi: 10.1016/j.biopha.2018.07.059 [DOI] [PubMed] [Google Scholar]
- 13.Wang J, Li J, Wang H, Lv L, Sun J. Overexpression of circ_0005198 sponges miR-1294 to regulate cell proliferation, apoptosis, migration, and invasion in glioma. J Cell Biochem. 2019;120(9):15538–15545. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Z, Lin W, Gao L, et al. Hsa_circ_0004370 promotes esophageal cancer progression through miR-1294/LASP1 pathway. Biosci Rep. 2019;39(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luo W, Semenza GL. Emerging roles of PKM2 in cell metabolism and cancer progression. Trends Endocrinol Metab. 2012;23(11):560–566. doi: 10.1016/j.tem.2012.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chattopadhyay E, Roy B. Altered mitochondrial signalling and metabolism in cancer. Front Oncol. 2017;7:43. doi: 10.3389/fonc.2017.00043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu ZX, Hong L, Fang SQ, et al. Overexpression of pyruvate kinase M2 predicts a poor prognosis for patients with osteosarcoma. Tumour Biol. 2016;37(11):14923–14928. doi: 10.1007/s13277-016-5401-7 [DOI] [PubMed] [Google Scholar]
- 18.Xin H, Wang C, Liu Z. miR-196a-5p promotes metastasis of colorectal cancer via targeting IkappaBalpha. BMC Cancer. 2019;19(1):30. doi: 10.1186/s12885-018-5245-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yao J, Zhang H, Li H, Qian R, Liu P, Huang J. P53-regulated lncRNA uc061hsf.1 inhibits cell proliferation and metastasis in human esophageal squamous cell cancer. IUBMB Life. 2019. doi: 10.1002/iub.2196 [DOI] [PubMed] [Google Scholar]
- 20.Ma J, Zeng S, Zhang Y, et al. BMP4 enhances hepatocellular carcinoma proliferation by promoting cell cycle progression via ID2/CDKN1B signaling. Mol Carcinog. 2017;56(10):2279–2289. doi: 10.1002/mc.v56.10 [DOI] [PubMed] [Google Scholar]
- 21.Zhang M, Yang F Jr, Yang F, Chen J, Zheng CY, Liang Y. Cytotoxic aggregates of alpha-lactalbumin induced by unsaturated fatty acid induce apoptosis in tumor cells. Chem Biol Interact. 2009;180(2):131–142. doi: 10.1016/j.cbi.2009.03.019 [DOI] [PubMed] [Google Scholar]
- 22.Tao Y, Sun C, Zhang T, Song Y. SMURF1 promotes the proliferation, migration and invasion of gastric cancer cells. Oncol Rep. 2017;38(3):1806–1814. doi: 10.3892/or.2017.5825 [DOI] [PubMed] [Google Scholar]
- 23.Yuan Q, Yu H, Chen J, Song X, Sun L. Knockdown of pyruvate kinase type M2 suppresses tumor survival and invasion in osteosarcoma cells both in vitro and in vivo. Exp Cell Res. 2018;362(1):209–216. doi: 10.1016/j.yexcr.2017.11.020 [DOI] [PubMed] [Google Scholar]
- 24.Nam EH, Lee Y, Moon B, Lee JW, Kim S. Twist1 and AP-1 cooperatively upregulate integrin alpha5 expression to induce invasion and the epithelial-mesenchymal transition. Carcinogenesis. 2015;36(3):327–337. doi: 10.1093/carcin/bgv005 [DOI] [PubMed] [Google Scholar]
- 25.Wu Z, Li S, Liu J, et al. RNAi-mediated silencing of AQP1 expression inhibited the proliferation, invasion and tumorigenesis of osteosarcoma cells. Cancer Biol Ther. 2015;16(9):1332–1340. doi: 10.1080/15384047.2015.1070983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Toyonaga T, Nakano K, Nagano M, et al. Blockade of constitutively activated janus kinase/signal transducer and activator of transcription-3 pathway inhibits growth of human pancreatic cancer. Cancer Lett. 2003;201(1):107–116. doi: 10.1016/S0304-3835(03)00482-8 [DOI] [PubMed] [Google Scholar]
- 27.Wang Y, Hao F, Nan Y, et al. PKM2 inhibitor shikonin overcomes the cisplatin resistance in bladder cancer by inducing necroptosis. Int J Biol Sci. 2018;14(13):1883–1891. doi: 10.7150/ijbs.27854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang S, Ma Y, Wang P, et al. Knockdown of PKM2 enhances radiosensitivity of non-small cell lung cancer. Cell Biochem Biophys. 2015;73(1):21–26. doi: 10.1007/s12013-015-0567-y [DOI] [PubMed] [Google Scholar]
- 29.Klein G, Vellenga E, Fraaije MW, Kamps WA, de Bont ES. The possible role of matrix metalloproteinase (MMP)-2 and MMP-9 in cancer, e.g. acute leukemia. Crit Rev Oncol Hematol. 2004;50(2):87–100. doi: 10.1016/j.critrevonc.2003.09.001 [DOI] [PubMed] [Google Scholar]
- 30.Sugiyama T, Taniguchi K, Matsuhashi N, et al. MiR-133b inhibits growth of human gastric cancer cells by silencing pyruvate kinase muscle-splicer polypyrimidine tract-binding protein 1. Cancer Sci. 2016;107(12):1767–1775. doi: 10.1111/cas.2016.107.issue-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang K, Zhang M, Jiang H, Liu F, Liu H, Li Y. Down-regulation of miR-214 inhibits proliferation and glycolysis in non-small-cell lung cancer cells via down-regulating the expression of hexokinase 2 and pyruvate kinase isozyme M2. Biomed Pharmacother. 2018;105:545–552. doi: 10.1016/j.biopha.2018.06.009 [DOI] [PubMed] [Google Scholar]
- 32.Chen J, Yu Y, Chen X, et al. MiR-139-5p is associated with poor prognosis and regulates glycolysis by repressing PKM2 in gallbladder carcinoma. Cell Prolif. 2018;51(6):e12510. doi: 10.1111/cpr.2018.51.issue-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dayton TL, Jacks T, Vander Heiden MG. PKM2, cancer metabolism, and the road ahead. EMBO Rep. 2016;17(12):1721–1730. doi: 10.15252/embr.201643300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wong N, Ojo D, Yan J, Tang D. PKM2 contributes to cancer metabolism. Cancer Lett. 2015;356(2Pt A):184–191. doi: 10.1016/j.canlet.2014.01.031 [DOI] [PubMed] [Google Scholar]
- 35.Yang W, Xia Y, Ji H, et al. Nuclear PKM2 regulates beta-catenin transactivation upon EGFR activation. Nature. 2011;480(7375):118–122. doi: 10.1038/nature10598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu H, Wu J, Zhang W, et al. PKM2 enhances chemosensitivity to cisplatin through interaction with the mTOR pathway in cervical cancer. Sci Rep. 2016;6:30788. doi: 10.1038/srep30788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hsu MC, Hung WC, Yamaguchi H, et al. Extracellular PKM2 induces cancer proliferation by activating the EGFR signaling pathway. Am J Cancer Res. 2016;6(3):628–638. [PMC free article] [PubMed] [Google Scholar]
- 38.Mukherjee J, Ohba S, See WL, Phillips JJ, Molinaro AM, Pieper RO. PKM2 uses control of HuR localization to regulate p27 and cell cycle progression in human glioblastoma cells. Int J Cancer. 2016;139(1):99–111. doi: 10.1002/ijc.30041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin Y, Zhai H, Ouyang Y, et al. Knockdown of PKM2 enhances radiosensitivity of cervical cancer cells. Cancer Cell Int. 2019;19:129. doi: 10.1186/s12935-019-0845-7 [DOI] [PMC free article] [PubMed] [Google Scholar]