This study reveals a novel role of hemidesmosomes in resisting actomyosin-generated cellular tension, which is dependent on mechanical coupling of focal adhesions to hemidesmosomes and inhibition of mechanosensitive signaling. Furthermore, through their ability to influence cellular tension, hemidesmosomes also control the localization of αvβ5 in flat clathrin lattices.
Abstract
Hemidesmosomes are specialized cell-matrix adhesion structures that are associated with the keratin cytoskeleton. Although the adhesion function of hemidesmosomes has been extensively studied, their role in mechanosignaling and transduction remains largely unexplored. Here, we show that keratinocytes lacking hemidesmosomal integrin α6β4 exhibit increased focal adhesion formation, cell spreading, and traction-force generation. Moreover, disruption of the interaction between α6β4 and intermediate filaments or laminin-332 results in similar phenotypical changes. We further demonstrate that integrin α6β4 regulates the activity of the mechanosensitive transcriptional regulator YAP through inhibition of Rho–ROCK–MLC– and FAK–PI3K–dependent signaling pathways. Additionally, increased tension caused by impaired hemidesmosome assembly leads to a redistribution of integrin αVβ5 from clathrin lattices to focal adhesions. Our results reveal a novel role for hemidesmosomes as regulators of cellular mechanical forces and establish the existence of a mechanical coupling between adhesion complexes.
Introduction
The attachment of cells to the ECM is essential for the integrity and function of multiple tissues (Michelson et al., 2000). In (pseudo-)stratified epithelium, specialized structures called hemidesmosomes (HDs) stably anchor epithelial cells to the basement membrane through association of the cytoplasmic keratin intermediate filaments (IFs) with laminin-332 in the extracellular space (Walko et al., 2015). Classical type I HDs are composed of integrin α6β4, plectin, bullous pemphigoid antigen 1 isoform e (BPAG1e, also called BP230), bullous pemphigoid antigen 2 (BPAG2, also called BP180 or type XVII collagen), and the tetraspanin CD151 (Litjens et al., 2006; Walko et al., 2015; Sterk et al., 2000). Type II HDs are found in simple epithelial tissues, such as the intestine, and consist of only integrin α6β4 and plectin (Fontao et al., 1999; Uematsu et al., 1994). Integrin α6β4, the major transmembrane component of HDs, initiates HD formation by interacting with the cytoskeletal cross-linker plectin, which binds to IFs in the cytoplasm (Schaapveld et al., 1998; Geerts et al., 1999; Rezniczek et al., 1998). The importance of HDs in epithelial cell adhesion is illustrated by the fact that mutations in any of the six genes encoding the structural components of HDs cause the congenital inherited skin blistering disorder epidermolysis bullosa (McGrath, 2015; Fine et al., 2014). Beyond their adhesion function, HDs may also play an important role in signal transduction through the integrin α6β4. Signals arising from this integrin have been shown to regulate cell proliferation, survival, and migration, as well as invasion of tumor cells (Stewart and O’Connor, 2015; Cooper and Giancotti, 2019; Ramovs et al., 2017).
Other integrin-containing adhesion structures in epithelial cells include focal adhesions (FAs) and podosomes. In contrast to HDs, these adhesion structures connect to the actin cytoskeleton (Burridge and Guilluy, 2016; van den Dries et al., 2013; Geiger et al., 2001), which, along with its associated myosin II motor proteins, forms the cell’s primary force-generating apparatus (Houdusse and Sweeney, 2016; Kull and Endow, 2013). It has been demonstrated that the cellular tension created by the contractile actomyosin machinery is needed for the maturation of FAs, which originate from smaller focal complexes that are formed in a tension-independent manner at the cell’s edge (Geiger et al., 2001). FAs act as both mechanosensors and sites of force transduction. They sense and respond to both intrinsic and extracellular forces (Iskratsch et al., 2014; Oria et al., 2017; Schwartz, 2010) and play an important role in many cellular processes that are driven by mechanotransduction, including cell adhesion, polarized migration, and differentiation (Jansen et al., 2017). In contrast to the FA-anchored actomyosin cytoskeleton, the HD-associated IF system enables cells to withstand mechanical stress and tension (Sanghvi-Shah and Weber, 2017; De Pascalis et al., 2018; Goldmann, 2018). However, it is unclear whether HD-associated IFs can also reduce cellular tension generated by the actomyosin cytoskeleton.
As a protein that can link the keratin IF system with either α6β4 or F-actin (de Pereda et al., 2009; Geerts et al., 1999), plectin could play an important role in mechanotransduction events at IFs and HDs. In line with such a role, it has been shown that plectin contributes to protect cells from osmotic stress and regulates nuclear mechanics in keratinocytes (Almeida et al., 2015; Osmanagic-Myers et al., 2006). Furthermore, its absence in fibroblasts has been associated with an increased number of actin filaments and FAs, suggesting plectin is a regulator of actin filament dynamics (Andrä et al., 1998).
In the present study, we assessed the contribution of HDs to the generation of intrinsic cellular tension and traction forces applied on the ECM in immortalized keratinocytes and multiple carcinoma cell lines. Superresolution imaging and biochemical analysis revealed a close association of integrin β4/HDs and FAs. By using keratinocytes that either lack integrin β4 or stably express wild-type β4 or a β4 mutant that cannot bind plectin or laminin, we demonstrate that an intact laminin-integrin β4-plectin linkage is essential for resisting actomyosin contractility and traction force generation. Similar results were obtained by deleting plectin in integrin β4–expressing keratinocytes. In addition, increased activation of FAK, myosin light chain (MLC), and Yes-associated protein (YAP) is observed in cell lines lacking intact HDs. Moreover, we show that the presence of intact HDs affected the subcellular distribution of the integrin β5 subunit, which is controlled by the amount of cellular tension. Taken together, our data provide evidence for a role of HDs in the counterbalance of FAs and actomyosin contractility.
Results
Inverse correlation between integrin β4 expression and FA maturation
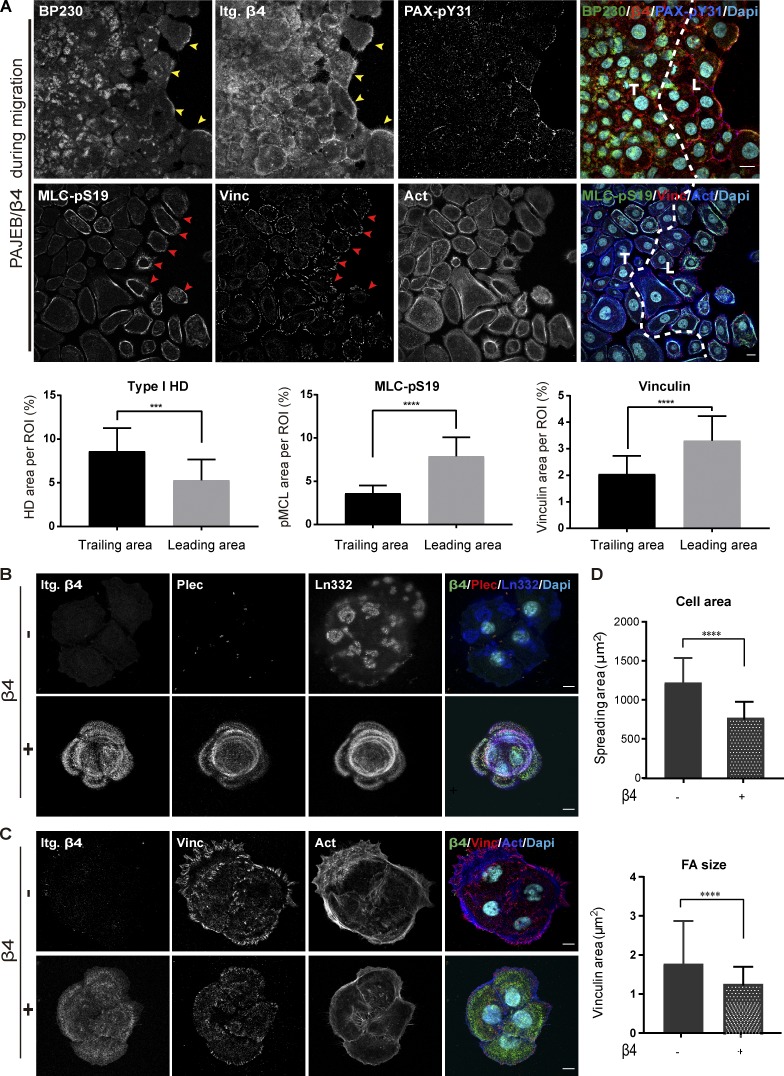
To assess whether HDs play a role in force generation via the actomyosin cytoskeleton, we examined the interplay between FA and HD adhesion complexes during cell migration in an in vitro wound-healing setting. For migration to take place, keratinocytes need to disassemble the cell-matrix adhesions that provide anchorage to the underlying basement membrane. After reepithelization of the wound area has taken place, HDs have to be reestablished, and FAs, through which mechanical forces are transmitted, are dismantled (Hopkinson et al., 2014). To study the interplay between HDs and FAs in this process, we allowed immortalized junctional epidermolysis bullosa associated with pyloric atresia (PA-JEB) keratinocytes that stably express wild-type β4 (PA-JEB/β4 keratinocytes; Sterk et al., 2000) to form a confluent cell monolayer around a cylindrical pillar with a diameter of 3 mm stuck on a glass coverslip. After careful removal of the pillar, we assessed the assembly and reorganization of FAs and HDs in keratinocytes that started migrating into the cell-free gap. Additionally, we evaluated the phosphorylation of MLC at Ser19 (MLC-pS19), which was used as a readout of cell contractility (Beningo et al., 2006; Pelham and Wang, 1999). Immunofluorescence microscopy showed increased levels of the FA markers vinculin and phosphorylated paxillin (PAX-pY31; Oakes et al., 2012), as well as of MLC-pS19 in the leading keratinocytes, while the type I HD components BP230 and integrin β4 were found to be diffusely distributed over the cell surface with some enrichment at the free border (Fig. 1 A). On the contrary, in the stationary cells located away from the wound interface, BP230 and integrin β4 concentrated in type I HD clusters along the ventral surface. These findings were confirmed by quantitative analyses (Fig. 1 A). The high level of MLC-pS19 in the leading cells, compared with the stationary cells indicates increased actomyosin contractility (Fig. 1 A) and suggests an inverse relationship between the presence of type I HDs on the one hand and FA maturation and cellular force generation on the other.
Figure 1.
Inverse correlation between integrin β4 expression and FA maturation. (A) Immunofluorescence confocal images of migrating PA-JEB/β4 keratinocytes stained for BP230 (green), integrin β4 (Itg. β4; red), phosphorylated paxillin (PAX-pY31; blue), phosphorylated MLC (MLC-pS19; green), vinculin (Vinc; red), and actin (Act; blue) 72 h after creating the gap. Nuclei were counterstained with DAPI (cyan). T means the trailing area, while L means the leading area of migrating cell monolayer. Yellow arrowheads indicate the enrichment of hemidesmosomal structure at the leading border, and red arrowheads indicate the higher level of MLC-pS19 and larger FAs in the leader cells. Scale bars: 20 µm. For quantification, type I HD (based on β4 and BP230 colocalization), MLC-pS19, and vinculin-positive areas are calculated as a percentage of the total ROI area. The values represent the mean (± SD) of three independent experiments, with ∼18 images per experiment. ***, P < 0. 001; ****, P < 0.0001. (B) Representative confocal fluorescence microscopy images of PA-JEB (β4 −) and PA-JEB/β4 (β4 +) keratinocytes cultured for 1 d in complete KGM medium and then switched to DMEM (10% FCS) for 16 h. Cells were immunostained for β4 (green), plectin (Plec; red), and laminin-332 (Ln332; blue). Colocalization of β4, plectin, and laminin-332 is visualized in the overlay images. Nuclei were counterstained with DAPI (cyan). (C) Cells were immunostained for β4 (green), vinculin (Vinc; red), and actin (Act; blue). Nuclei were counterstained with DAPI (cyan). Scale bars: 10 µm. (D) Cell area and FA size probed by vinculin were quantified with ImageJ. The values represent the mean (± SD) of three independent experiments, with ∼20 images per experiment. ****, P < 0.0001.
Building further on this notion, we compared FA assembly in PA-JEB keratinocytes that either lacked or expressed the HD component integrin β4. These keratinocytes are maintained in keratinocyte growth medium (KGM) but treated with DMEM for 24 h to induce a cell migration arrest and allow for the formation of robust HDs (Schaapveld et al., 1998). As shown by the colocalization of β4 with plectin and laminin-332, HDs were abundantly present in these cells and organize in a cauliflower-like pattern (Fig. 1 B). In contrast, in the β4-deficient PA-JEB keratinocytes, no HDs were detected, and plectin is diffusely distributed throughout the cytoplasm. Notably, these cells were also more spread and form less tight cell islands compared with the β4-proficient PA-JEB keratinocytes (Fig. 1, B and D). Furthermore, larger FAs were observed in the β4-deficient cells (Fig. 1, C and D).
Together, these data show that integrin β4 suppresses cell spreading and FA maturation, which indicates that HDs likely play a role in regulation of tensile forces in keratinocytes.
Close connection between HDs and FA components
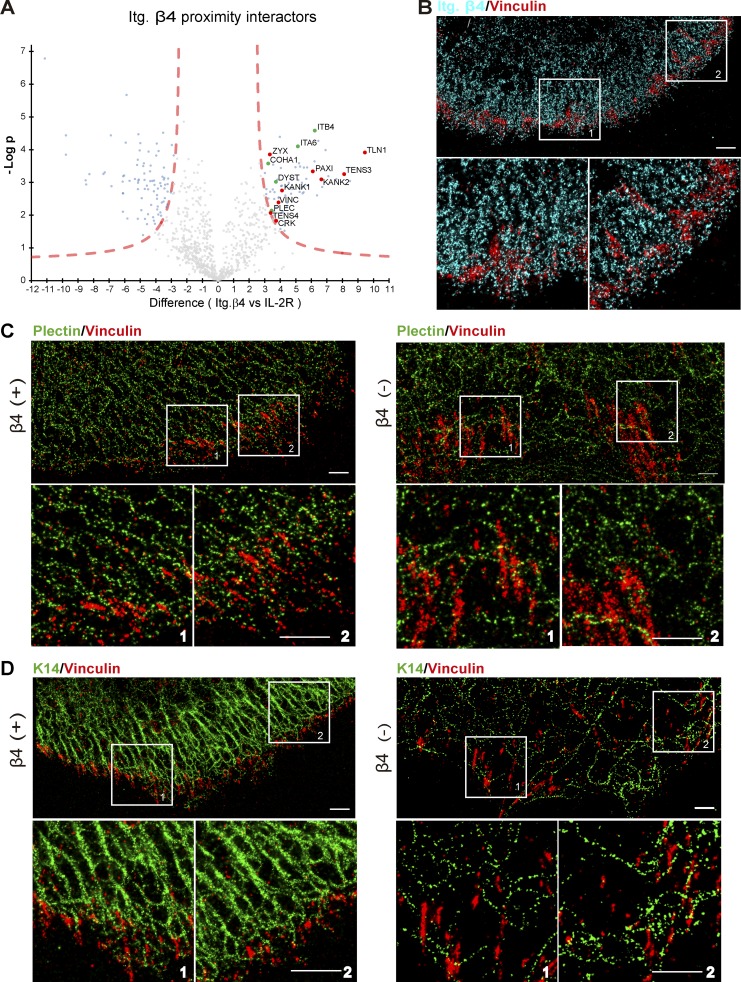
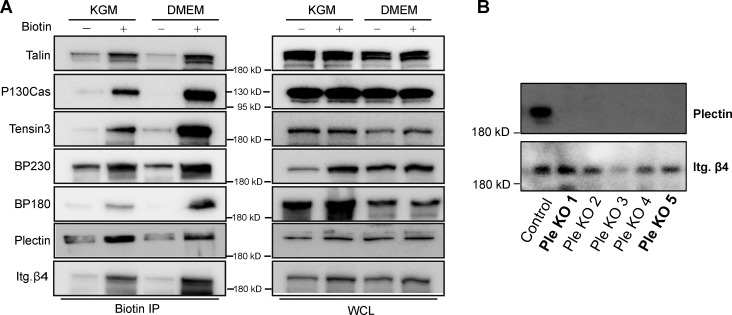
HD protein complexes localize in close proximity to FAs (Ozawa et al., 2010; Rid et al., 2005; Pora et al., 2019), which suggests that these complexes might be interconnected to regulate traction forces. To provide evidence of a physical linkage between FAs and HDs, we performed proximity-dependent biotin identification (BioID) assays using PA-JEB keratinocytes stably expressing integrin β4 or IL2R fused to a biotin ligase (BirA*). Comparison of the proximity interactors of β4 and IL2R (used as negative control; Data S1) revealed in addition to the already known interactors of α6β4, BP230 (DYST), BP180 (COHA1), and plectin, the FA components talin-1, zyxin, paxillin, tensin-3, vinculin, tensin-4, and KANK1/2 as specific proximity interactors of β4 (Fig. 2 A). The specificity of these β4 interactors was validated by additional BioID experiments using integrin β4– and IL2R-BirA*–expressing keratinocytes in the absence of biotin to exclude any false-positive interactors. These data are in line with a previous study in which some FA components, such as tensin-3, were identified as interactors of integrin β4 (Myllymäki et al., 2019). Moreover, some of these β4 proximity interactors were validated by Western blotting in the presence or absence of biotin (Fig. S1 A). Irrespective of whether the cells are grown in KGM or DMEM, we found the same FA proteins as proximal interactors of β4, suggesting the proximity interaction exists in both migratory and stationary cells.
Figure 2.
Close connection between HDs and FA components. (A) Proximity biotinylation assays were performed with PA-JEB/β4 keratinocytes expressing integrin β4 fused to the biotin ligase BirA*. An IL-2R-BirA* fusion protein, which is dispersed over the cell membrane, was used as negative control to identify the specific proximity interactors of integrin β4. The volcano plot shows the results from three independent experiments (threshold false discovery rate: 0.01 and S0: 2). Significant proximity interactors of β4 and IL-2R are indicated in light blue (IL-2R interactors, left, and integrin β4 interactors, right), red (FA components), or green (HD proteins). (B) Representative superresolution microscope image of β4 (+) cells showing vinculin (red) and integrin β4 (cyan). (C) Representative superresolution microscope image showing vinculin (red) and plectin (green) in β4 (+) and β4 (−) PA-JEB cells. (D) Representative superresolution microscope image showing vinculin (red) and keratin 14 (green) in β4 (+) and β4 (−) PA-JEB cells. Scale bars: 1 µm.
Figure S1.
Confirmation of BirA*-β4 interactions by streptavidin precipitation and Western blot. (A) PA-JEB/β4 keratinocytes (cultured in KGM or DMEM) expressing integrin β4 fused to the biotin ligase BirA* were used to perform proximity biotinylation assays. The proximity interactors of integrin β4 were pulled down by streptavidin beads and detected by Western blotting with the indicated antibodies. (B) Western blot analysis of whole-cell lysates (WCL) of plectin (Ple) knockout (KO) clones, probed with antibodies against plectin and integrin β4. β4 (+) PA-JEB cells were included as a positive control.
We also examined the association of FAs and HDs in PA-JEB/β4 keratinocytes by superresolution microscopy and found that in the cell periphery, plectin and, to a lesser extent, β4 localize in linear arrays aligned with keratin filaments (Fig. 2, B–D). The FA-associated protein vinculin was found in close association with these hemidesmosomal protein arrays at the very edge of the cells (Fig. 2, B and C). On the contrary, in β4-deficient PA-JEB keratinocytes, plectin and keratin filaments displayed a more irregular and wavy distribution (Fig. 2, C and D) and the close association between FA and HD components seemed diminished.
All in all, these findings confirm the close connection between HDs and FAs in β4-proficient keratinocytes.
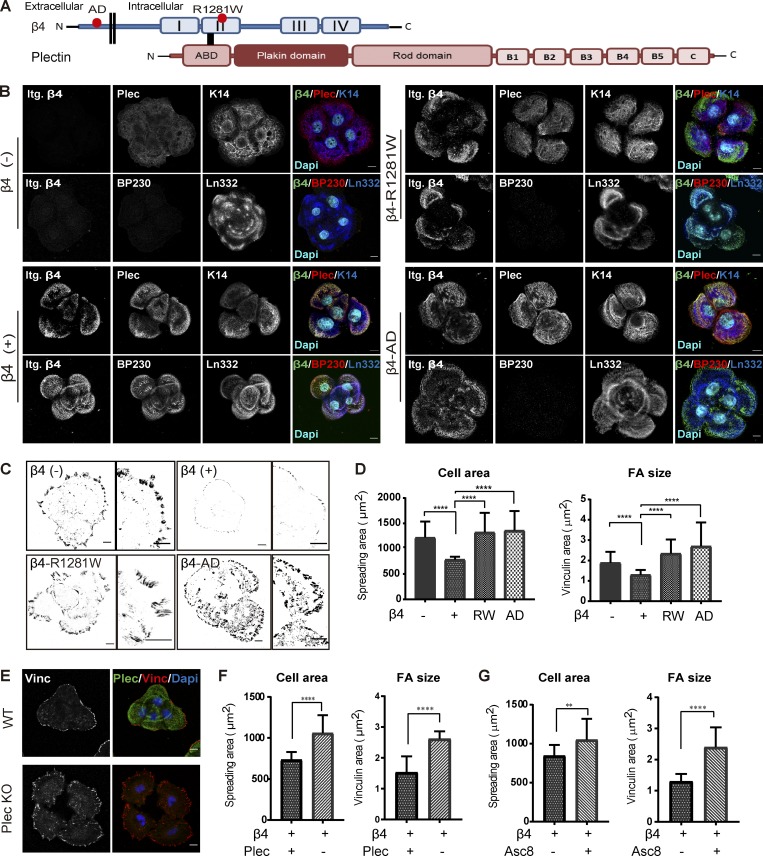
Intact laminin-integrin β4-plectin linkage reduces FA size and cell spreading
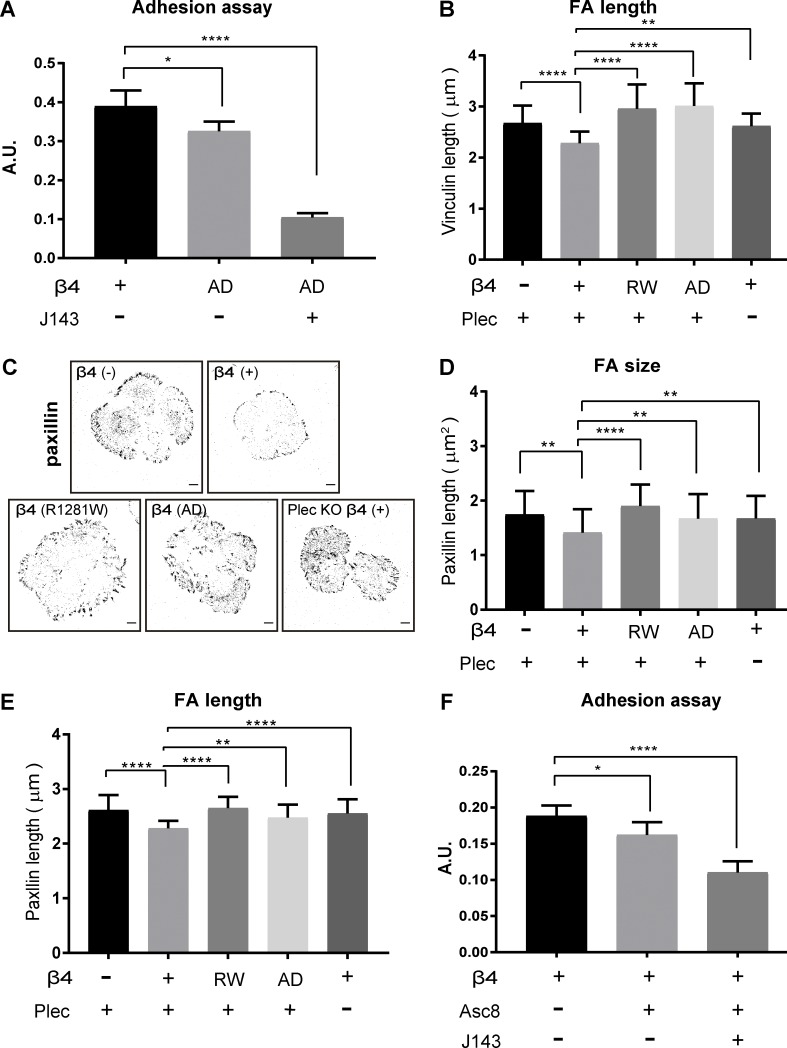
Integrin α6β4 is a transmembrane receptor for laminin-332 that associates with IF network through interactions of the large cytoplasmic domain of the β4 subunit with the cytoskeletal linker proteins plectin and BP230 (Koster et al., 2003; Manso et al., 2018 Preprint). To study whether the binding to the ECM and/or the connection with IFs via plectin is required for the observed integrin β4-mediated effects on cell area and FA size, we generated two cell lines carrying mutations in the β4 subunit. The missense mutation R1281W renders β4 unable to interact with plectin (Koster et al., 2004), while three engineered amino acid substitutions (D230A, P232A, and E233A) in β4 result in an adhesion-defective (AD) integrin (Fig. 3 A; Russell et al., 2003). We confirmed the impaired ability of PA-JEB/β4-AD to bind to laminin-332 in in vitro cell adhesion assays (Fig. S2 A) and showed that in the keratinocytes expressing β4-R1281W, the association between β4 and the keratin IF system via plectin/BP230 is abolished (Koster et al., 2004; Fig. 3 B). Previous findings have shown that the β4 cytoplasmic domain is sufficient to induce coclustering of an IL2R reporter domain and plectin at the plasma membrane (Homan et al., 1998; Nievers et al., 2000). In line with this finding, we found that the β4-AD mutant was able to induce the formation of HD-like structures, as judged from the similar distribution patterns of β4 and plectin in β4-proficient cells (Figs. 1 B and 3 B). Yet, β4 clustered more diffusely near the periphery of cell islands and showed limited colocalization with the deposited laminin-332. Similar to the PA-JEB cells lacking β4, BP230 could hardly be detected in the cells expressing β4-R1281W or β4-AD, indicating the absence of type I HDs (Fig. 3 B). Moreover, similar to β4-deficient PA-JEB keratinocytes, PA-JEB/β4-R1281W and PA-JEB/β4-AD cells exhibited larger and longer FAs probed by vinculin or paxillin and an increased mean cell area compared with PA-JEB/β4 (Fig. 3, C and D; and Fig. S2, B–E).
Figure 3.
Intact laminin-integrin β4-plectin linkage reduces FA size and cell spreading. (A) Domain structure of integrin β4 and plectin. Dots indicate the relative locations of the R1281W mutation in β4-R1281W and of the D230A, P232A and E233A mutations in β4-AD. ABD, actin-binding domain. (B) Representative confocal fluorescence microscopy images of β4 (−), β4 (+), β4-R1281W, and β4-AD PA-JEB keratinocytes. Cells were cultured for 24 h in DMEM (10% FCS) and then fixed and stained for β4 (green), plectin (Plec; red) or BP230 (red), and keratin-14 (K14; blue) or laminin-332 (Ln322; blue). Nuclei were counterstained with DAPI (cyan). Scale bars: 10 µm. (C) Inverse black-and-white images of confocal micrographs of β4 (−), β4 (+), β4-R1281W, and β4-AD PA-JEB keratinocytes showing cell morphology and vinculin-stained FAs (black). Scale bars: 10 µm. (D) Quantification of cell area and FA size with ImageJ. The data are presented as the mean (± SD) from three independent experiments, with ∼20 images per experiment. ****, P < 0.0001. (E) Confocal microscopy images of vinculin-stained FAs (red) and plectin-stained HDs (green) in β4 (+) and plectin-deficient β4 (+) keratinocytes (Plec KO). Nuclei (blue) were visualized with DAPI staining. Scale bars: 10 µm. (F) Quantification of cell area and FA size from three independent experiments, with ∼20 images per experiment. ****, P < 0.0001. (G) PA-JEB/β4 keratinocytes were cultured for 24 h in DMEM (10% FCS) with or without the β4 blocking antibody ASC-8 (supernatant diluted 1:5). Shown are quantification of cell area and FA size from three independent experiments, with ∼20 images per experiment. **, P < 0.01; ****, P < 0.0001.
Figure S2.
Focal contact area varies depending on the expression and function of integrin β4. (A) Cells were treated with or without integrin α3–blocking mAb (J143; 20 µg/ml) in suspension before a short-term (45 min) adhesion assay was performed on a laminin-332–rich matrix substrate. Data are presented as the mean (± SD) from three independent experiments. *, P < 0.05; ****, P < 0.0001. (B) Quantification of FA length probed by vinculin with ImageJ. Data are presented as the mean (± SD) from three independent experiments, with ∼20 images per experiment. **, P < 0.01; ****, P < 0.0001. (C) Inverse black-and-white images of confocal micrographs of β4 (−), β4 (+), β4-R1281W, and β4-AD PA-JEB keratinocytes showing cell morphology and paxillin-stained FAs (black). Scale bars: 10 µm. (D and E) Quantification of FA size and length probed by paxillin with ImageJ. Data are presented as the mean (± SD) from two independent experiments, with ∼20 images per experiment. **, P < 0.01; ****, P < 0.0001. (F) Integrin β4 (+) PA-JEB keratinocytes were treated with integrin β4–blocking mAb (ASC-8; supernatant diluted 1:5) alone or together with integrin α3-blocking mAb (J143; 20 µg/ml). Data are presented as the mean (± SD) from three independent experiments. *, P < 0.05; ****, P < 0.0001.
Next, we disrupted the interaction between the F-actin and IF networks by deleting plectin in PA-JEB/β4 using CRISPR/Cas9 technology. The deletion of plectin (Fig. S1 B) resulted in increased FA size (both area and length) and cell spreading (Fig. 3, E and F; and Fig. S2, B–E), which is in agreement with the observations in β4-R1281W–expressing keratinocytes (Fig. 3, C and D). Disruption of integrin β4 binding to laminin by treatment with a blocking antibody (ASC-8; Fig. S2 F) yielded results similar to the β4-AD mutant with respect to the size of FAs and the increase in cell area (Fig. 3 G).
Collectively, these results show that the interaction of integrin β4 with laminin-332 and plectin is required for suppression of FA maturation and cell spreading.
Integrin α6β4 and plectin reduce cellular traction force and tension
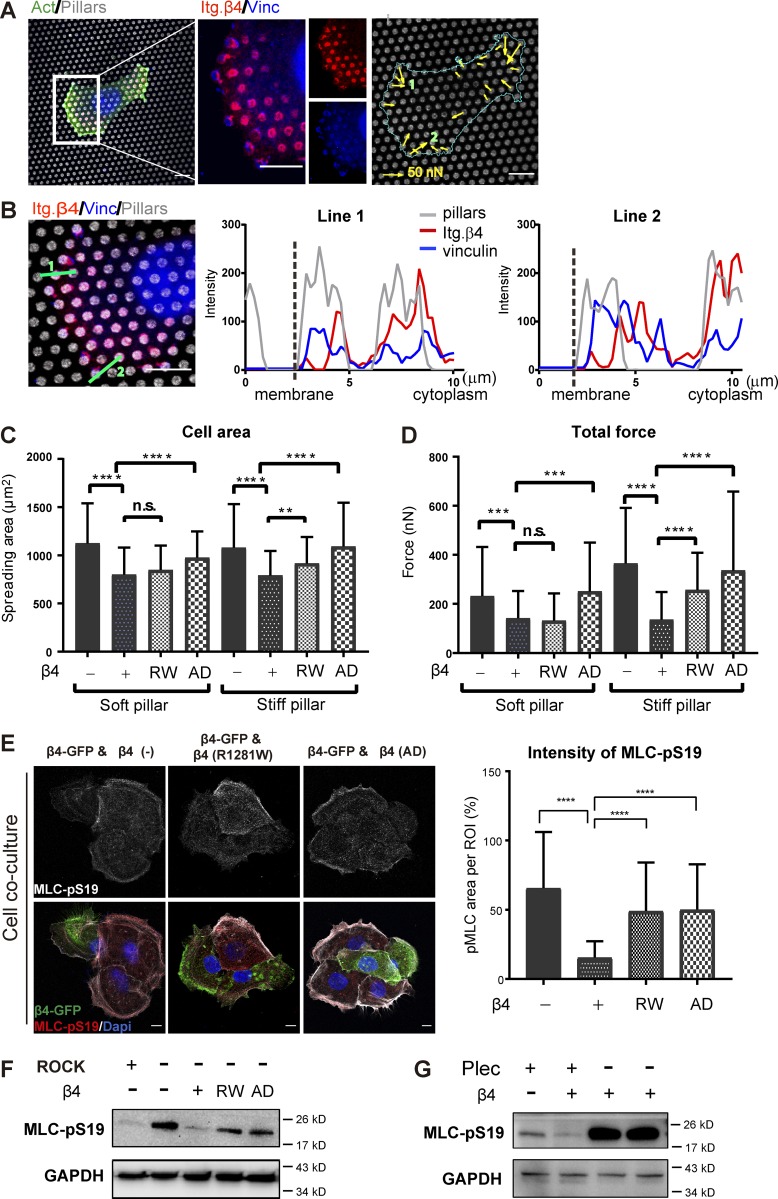
To investigate the ability of PA-JEB/β4 keratinocytes to exert traction force, we seeded the cells on fibronectin-stamped polydimethylsiloxane micropillar arrays with an effective stiffness of 137.1 kPa, roughly mimicking the stiffness of certain basement membranes (van Helvert et al., 2018). We confirmed that the keratinocytes formed both FAs and HDs when cultured on these pillars (Fig. 4 A). These adhesion structures predominantly localized on the pillar tops, which is likely due to the fibronectin coating of the pillars in combination with the efficient deposition of cellular laminin-332 at this area (Fig. S3 A). Traction forces generated by these cells were determined by visualizing the deflections of the pillar tops by confocal microscopy and calculating the forces corresponding to these deflections (Tan et al., 2003; van Hoorn et al., 2014). Deflected pillars could be observed near the cell periphery. On the deflected pillars, both vinculin and integrin β4 were present. Interestingly, vinculin mainly occupied the part of the pillar directed toward the cell periphery, while integrin β4 could be found on the pillar part directed toward the cell nucleus (Fig. 4, A and B).
Figure 4.
Integrin α6β4 and plectin reduce cellular traction force and cellular tension. (A) Representative confocal image of β4 (+) cells exerting forces on stiff pillars. The cell seeded on the pillar top was stained for actin (green), vinculin (blue), and integrin β4 (red). The secondary fluorescence antibody against vinculin has a strong nuclear background (blue). The pillar tops were coated by fibronectin tagged by fluorescence (gray). Deflection of pillars is shown by arrows, whose length is relative to force magnitude. The cyan line represents the cell periphery. Scale bars: 10 µm. (B) Intensity profile of vinculin, integrin β4 (Itg. β4), and the pillar tops from the green line drawn following the deflection of pillars in A. (C and D) Cell spreading area and total cellular force of β4 (−), β4 (+), β4-R1281W, and β4-AD keratinocytes adhering on soft (29.5 kPa) and stiff (137.1 kPa) pillars. The data are presented as the mean (± SD) from three independent experiments, with ∼70 cells in total. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. (E) Representative images from confocal microscopy comparing the phosphorylation of MLC at S19 (red; MLC-pS19) in PA-JEB cells stably expressing β4-GFP (green) with that in β4-deficient PA-JEB cells (left), PA-JEB cells expressing β4-R1281W (middle), or cells expressing β4-AD (right). Scale bars: 10 µm. Graph showing the quantification of the intensity of MLC-pS19 in different cell lines, expressed as a ratio of the total pixel intensity of MLC-pS19 staining divided by cell area. (F) Western blot analysis of whole-cell lysates from β4 (−), β4 (+), β4-R1281W, and β4-AD keratinocytes probed with antibodies against MLC-pS19 and GAPDH. PA-JEB cells treated with Y27632, a ROCK inhibitor, served as a negative control for validation of the MLC-pS19 antibody. A representative Western blot is shown (n = 3). (G) Western blot analysis of whole-cell lysates from integrin β4 (+), β4 (−) keratinocytes, and two plectin-knockout clones probed with antibodies against MLC-pS19 and GAPDH. A representative Western blot is shown (n = 2).
Figure S3.
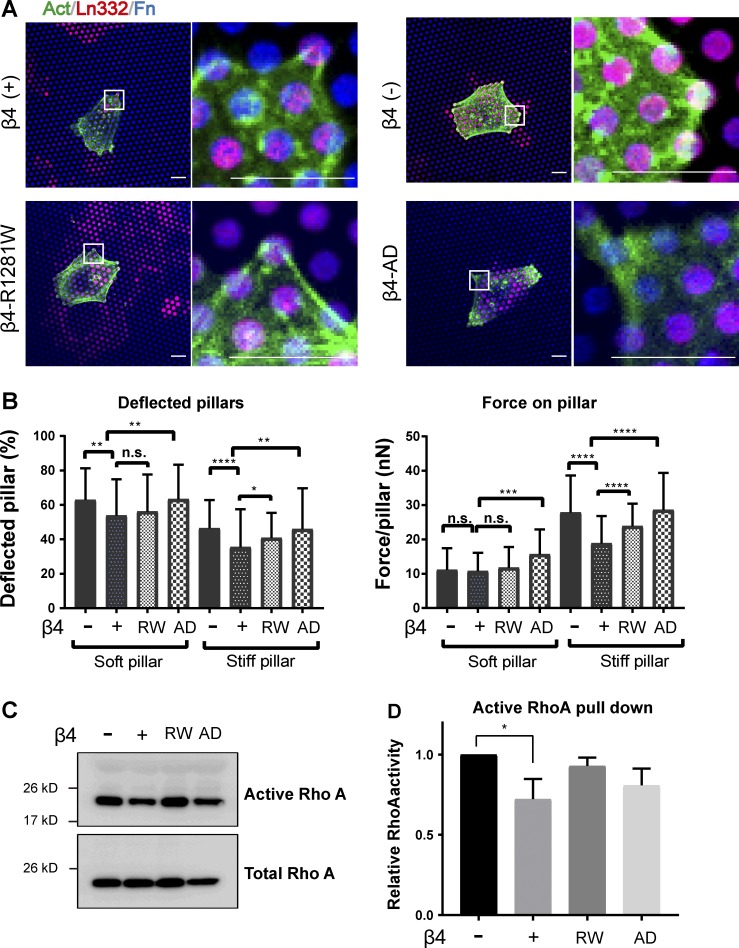
Integrin α6β4 reduces traction force generation through modulation of RhoA activity. (A) Confocal images showing the deposition of laminin-332 (red) by β4 (−) and β4 (+) PA-JEB keratinocytes seeded on micropillars. Cells were visualized by actin staining (green), and pillar tops were coated with fibronectin (blue). Scale bars: 10 µm. (B) Quantifications of percentage of deflected pillars and force per pillar of the indicated cell lines seeded on pillars. (C) Representative immunoblot of RhoA activity in integrin β4 (−), β4 (+), β4-R1281W, and β4-AD PA-JEB keratinocytes. Active RhoA was pulled down from the lysates, using GST–rhotekin–binding domain fusion proteins with the RhoA-binding region of rhotekin. (D) RhoA activity is shown as active RhoA/total RhoA, and values were normalized to the β4 (−) group. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; n.s., not significant.
Next, we investigated the effect of HDs on the ability of keratinocytes to exert traction forces and evaluated the role of substrate stiffness in this process. To this end, β4-proficient, β4-deficient, β4-R1281W, and β4-AD cells were seeded on micropillars with effective stiffnesses of 137.1 and 29.5 kPa, the latter mimicking the stiffness of granulation tissue, which forms on the surface of a wound during the healing process (van Helvert et al., 2018). The cellular spreading area was increased by ∼20% in the absence of β4, regardless of the stiffness of the pillars (Fig. 4 C). Furthermore, β4-proficient PA-JEB cells applied the lowest traction force of 137 (± 112) nN on stiff pillars, which increased to 365 (± 226) nN for β4-deficient and 337 (± 322) nN for β4-AD cells. The β4-R1281W mutant displayed an intermediate force behavior of 257 (± 152) nN (Fig. 4 D). A similar trend in total generated force could be observed on soft pillars, with the exception of β4-R1281W cells, which showed low total force generation, comparable to the force generated by β4-proficient cells (Fig. 4 D). This result shows that in contrast to β4-AD, the β4-R1281W mutation does not completely mimic the β4-null phenotype. The total force generated by each cell has two contributions: the percentage of deflected pillars with respect to the total number of pillars and the force applied to each pillar. Analysis of both yields a more subtle change of the mechanical response to HD formation and substratum stiffness (Fig. S3 B). In line with previous findings in 3T3 fibroblasts and MCF-7 cells (van Hoorn et al., 2014), we observed that the number of deflected pillars decreases with increased substratum stiffness, while the force per pillar increases. Similar to their effects on total force, cells with different β4 mutants show significant differences in the amount of force generated on single stiff pillars. The β4-proficient cell line applied the lowest force per pillar, which significantly increased for β4-deficient, β4-R1281W, and β4-AD cells.
Traction force generation is dependent on actomyosin contractility through the small GTPase RhoA–Rho kinase (ROCK)–MLC signaling axis (Amano et al., 1996; Lessey et al., 2012). To assess whether HD assembly also regulates myosin activity, we visualized the level of MLC-pS19 in a co-culture of PA-JEB cells expressing GPF-tagged integrin β4 and PA-JEB, PA-JEB/β4-R1281W, or PA-JEB/β4-AD cells. Cells that do not express GPF-tagged wild-type β4 showed an increased MLC phosphorylation (Fig. 4 E), indicating that actomyosin contractility is increased in keratinocytes that do not assemble intact HDs. This data were confirmed by Western blotting (Fig. 4 F). In line with this, MLC phosphorylation was strongly increased in plectin-deficient PA-JEB/β4 compared with plectin-knockout PA-JEB/β4 and PA-JEB keratinocytes (Fig. 4 G). Upstream of MLC phosphorylation, lower levels of GTP-bound (active) RhoA were detected in PA-JEB/β4 keratinocytes, indicating a decreased activation of ROCK and MLC phosphorylation (Fig. S3, C and D). The reduced RhoA activation in the presence of wild-type β4 indicates that HDs negatively modulate contractile forces through RhoA–ROCK–MLC signaling, even though the variation of active RhoA is not as dramatic as the level of MLC-pS19. The dramatic increase in MLC phosphorylation can also be attributed to the enhanced adhesion strength induced by larger FAs that compensate for the loss of HDs (Stricker et al., 2011).
In summary, these results demonstrate that integrin β4 and plectin reduce cellular traction force and actomyosin contractility.
Mechanotransduction-related signaling pathways are inactivated by integrin α6β4 and plectin
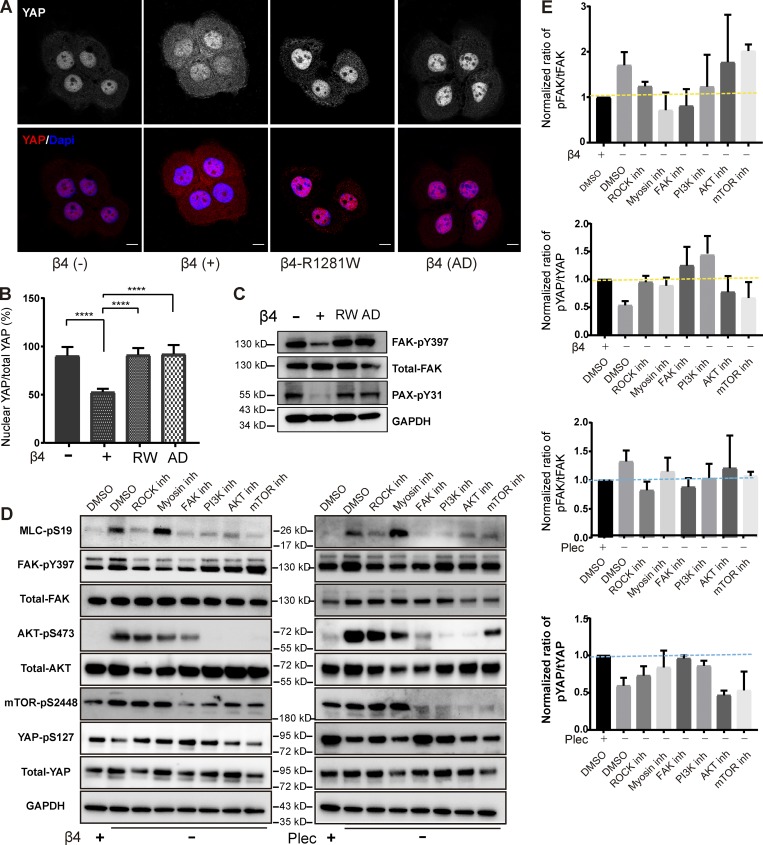
Cellular mechanoresponses not only require a rapid remolding of adhesion structures and the cytoskeleton but also involve the activation of several specific signaling pathways. As key molecules in mechanosensing, the transcriptional regulators YAP/TAZ (transcriptional coactivator with a PDZ-binding motif) control FA formation and actomyosin cytoskeleton stability in response to mechanical cues (Nardone et al., 2017; Piccolo et al., 2014). YAP and TAZ are inactivated and localized in the cytoplasm when endogenous tensile forces are inhibited (Halder et al., 2012; Dupont et al., 2011). We analyzed the effect of HDs on the subcellular distribution of YAP and found that YAP displayed a strong nuclear localization in the β4-deficient and mutant cell lines, while in PA-JEB/β4 cells, the level of nuclear YAP was reduced (Fig. 5, A and B).
Figure 5.
Mechanotransduction-related signaling pathways are inactivated by integrin α6β4 and plectin. (A) Representative images from confocal microscopy showing the distribution of YAP (red in merge) and cell nuclei (blue in merge) in β4 (−), β4 (+), β4-R1281W, and β4-AD PA-JEB keratinocytes. Scale bars: 10 µm. (B) Quantification of the percentage of nuclear YAP. The percentage of nuclear YAP was calculated by dividing the YAP staining overlapped with DAPI with the total YAP staining intensity. The data are presented as the mean (± SD) from three independent experiments, with ∼60 cells in total. ****, P < 0.0001. (C) Western blot analysis showing the levels of phosphorylated FAK (Y397) and paxillin (PAX-pY31) in β4 (−), β4 (+), β4-R1281W, and β4-AD PA-JEB keratinocytes. A representative Western blot is shown (n = 2). (D) Western blot analysis showing the levels of MLC-pS19, FAK (at Y397), AKT (at S473), mTOR (at S2448), and YAP (S127) of β4 (−) and plectin (−) keratinocytes treated with ROCK (Y27632; 10 µM), myosin (blebbistatin; 10 µM), FAK (VS-4718; 1 µM), PI3K (GDC-0941; 1 µM), AKT (MK-2206; 1 µM), or mTOR (AZD-8055; 100 nM) inhibitors. Inhibitors were added 45 min before cell lysate. β4 (+) cells are used as a control group. DMSO was used as vehicle control. (E) The relative ratios of phosphorylated FAK/total FAK (pFAK/tFAK) and phosphorylated YAP/total YAP (pYAP/tYAP) were calculated and normalized to β4 (+) group treated by DMSO. Data are presented as the mean (± SD) from two independent experiments.
It is well established that FA assembly and maturation occurs concomitantly with a change in cellular tension (Riveline et al., 2001; Oakes and Gardel, 2014; Stricker et al., 2011), which can be achieved by autophosphorylation of FAK at Y397 (Oakes and Gardel, 2014). To assess how inhibition of HD assembly influences FA maturation, we determined the extent of phosphorylation of FAK (at Y397) and paxillin (at Y31) in the four cell lines. A low level of FAK phosphorylation was detected in PA-JEB/β4 keratinocytes, while in the cells that lacked β4 or expressed β4 mutants that are unable to bind laminin-332 or plectin, the phosphorylation levels were elevated (Fig. 5 C). Similar observations were made for paxillin (Fig. 5 C).
Recent studies have shown that FAK phosphorylation positively regulates YAP activity and localization (Lachowski et al., 2018) through the Src–PI3K–PDK1 axis (Kim and Gumbiner, 2015). To investigate if β4-mediated YAP activity involves the FAK–PI3K axis, β4-deficient and plectin-knockout keratinocytes were treated with FAK (VS-4718) and PI3K (GDC-0941) inhibitors. Additionally, we treated the cells with the compounds AZD-8055 and MK-2206, which inhibit mTORC2 and AKT, respectively, two downstream effectors of PI3K. All the inhibitors effectively blocked phosphorylation of their target protein and subsequently prevented the activation of their downstream targets along the FAK–PI3K–AKT–mTOR axis. Both the FAK and PI3K inhibitors stimulated the phosphorylation of YAP (at S127), while inhibition of AKT and mTOR did not obviously alter YAP activity (Fig. 5, D and E). To directly address whether the accumulation of dephosphorylated nuclear YAP occurs due to the increased cellular tension, we reduced cellular tension in β4-deficient and plectin-knockout keratinocytes by treatment with Y27632 or blebbistatin. Y27632 inhibits ROCK and thereby blocks the phosphorylation of MLC at S19. Blebbistatin instead inhibits myosin ATPase activity and thus keeps myosin detached from actin but does not affect its phosphorylation state. Both inhibitors rescued the reduction of phosphorylated YAP induced by the loss of β4 or plectin, suggesting a RhoA–ROCK–MLC signaling axis plays a critical role in driving the altered cytoskeletal tension in the β4-deficient cells (Fig. 5 D).
In summary, these results indicate that integrin β4 and plectin decrease cell contractility and mechanotransduction through inhibition of RhoA–ROCK–MLC and FAK–PI3K signaling axes. Subsequently, decreased cellular tension prevents nuclear accumulation and dephosphorylation of YAP.
HD assembly controls the localization of integrin αVβ5
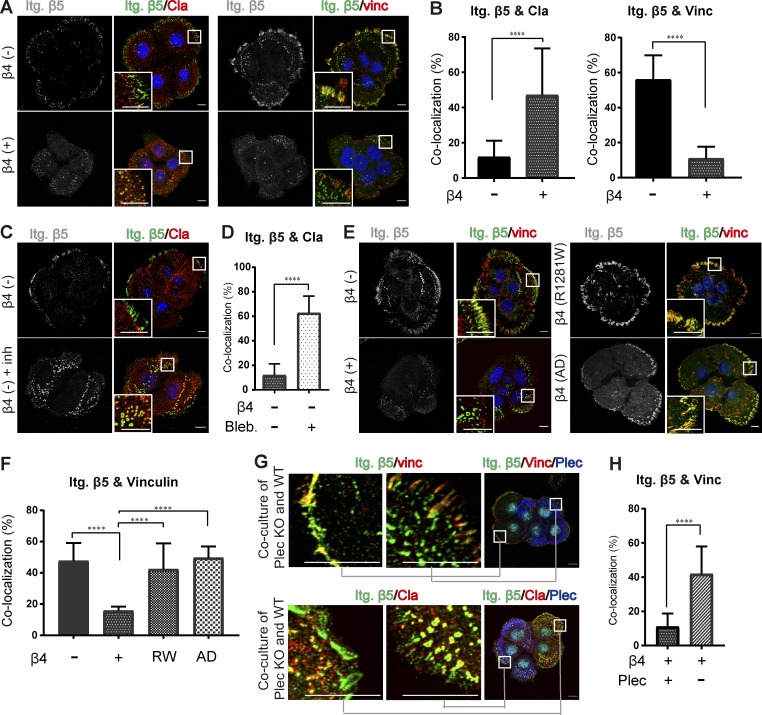
Integrin αVβ5 can reside in both flat clathrin lattices and FAs in a variety of cell lines and cell types (Lock et al., 2019). According to our recently published work, in PA-JEB/β4 keratinocytes, low levels of cytoskeletal force facilitate clustering of αVβ5 in clathrin lattices, while in highly contractile cells, αVβ5 is predominantly localized in FAs (Zuidema et al., 2018). Here, we analyzed whether increased cellular tension as a result of impaired HD assembly on stiff matrices also leads to changes in the subcellular distribution of integrin αVβ5. In β4-deficient cells, integrin β5 mainly followed the pattern of FAs, which were marked by vinculin staining. In the presence of integrin β4, integrin β5 redistributed and localized predominantly with clathrin (Fig. 6, A and B), suggesting that integrin β4 influences the localization of integrin β5. To confirm that integrin β4 alters the localization of β5 by reducing actomyosin-generated tension, we treated β4-deficient PA-JEB keratinocytes with the myosin inhibitor blebbistatin. As expected, blebbistatin treatment resulted in the redistribution of integrin β5 from FAs to clathrin lattices (Fig. 6, C and D).
Figure 6.
HD assembly controls the localization of integrin αVβ5. (A) Confocal images showing the distribution of β5 (green) together with either clathrin (Cla; red) or vinculin (Vinc; red) in β4 (−) and β4 (+) PA-JEB keratinocytes. Nuclei were counterstained with DAPI (blue). Scale bars: 10 µm. (B) Quantification of the colocalization of β5 with clathrin or vinculin. (C) Representative images showing integrin β5 clustering in clathrin lattices in β4 (−) cells, untreated or treated with the myosin inhibitor blebbistatin (20 µM) for 30 min. (D) Quantification of the colocalization of β5 with clathrin. (E) Confocal images showing the distribution and colocalization of β5 (green) and vinculin (red) in β4 (−), β4 (+), β4-R1281W, and β4-AD PA-JEB keratinocytes. Nuclei are shown in blue. Scale bars: 10 µm. (F) Quantification of the colocalization of β5 with vinculin. (G) Confocal images showing the distribution of β5 (green) together with either vinculin (red) or clathrin (red) in PA-JEB/β4 wild-type (close-up in the middle panel) and plectin knockout PA-JEB/β4 (close-up in the left panel) keratinocytes. Plectin is shown in blue, and nuclei are shown in cyan. Scale bars: 10 µm. (H) Quantification of the colocalization of β5 with vinculin. Data are presented as the mean (± SD) from three independent experiments, with ∼60 cells in total. ****, P < 0.0001.
Next, we analyzed the localization of integrin β5 in PA-JEB keratinocytes containing β4-R1281W or β4-AD and observed that similar to β4-deficient cells, integrin β5 predominantly clustered to FAs (Fig. 6, E and F). In addition, we found that integrin β5 was mostly assembled in FAs in plectin-deficient PA-JEB/β4 keratinocytes (Fig. 6, G and H). These data corroborate earlier findings (Zuidema et al., 2018) and demonstrate that impaired laminin-integrin β4-plectin linkage leads to high cellular tension, which consequently promotes integrin β5 clustering in FAs.
HDs counteract actomyosin contractility in carcinoma cell lines
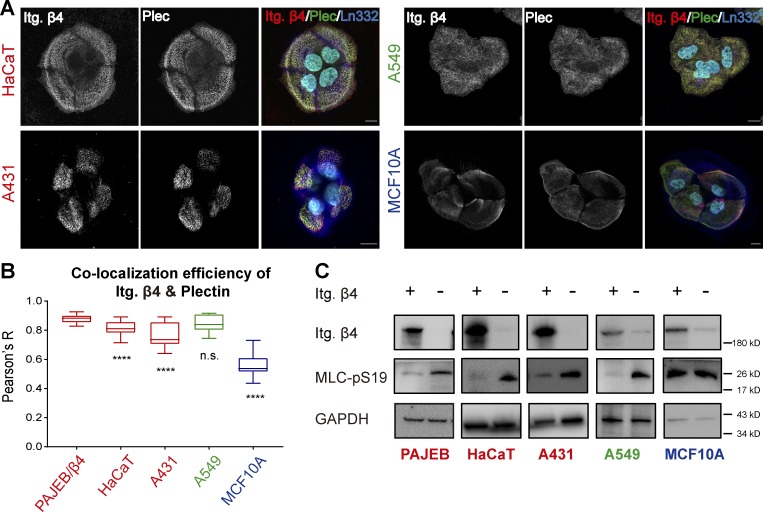
Although HDs have been most extensively studied in the epidermis, these adhesions can also be found in several other tissues, including lung (Chapman et al., 2011) and mammary gland (Klinowska et al., 2001). Furthermore, many tumor cells contain (rudimentary) HDs. We visualized HD assembly in several cancer cell lines by confocal microscopy. The results show that HaCaT keratinocytes and MCF10A mammary gland epithelial, A431 epidermoid, and A549 lung carcinoma cells expressed integrin β4 and plectin and deposited laminin-332 on the substratum. However, their ability to assemble HDs varied greatly (Fig. 7, A and B). HaCaT keratinocytes formed typical HDs in a cauliflower-like pattern, comparable to PA-JEB/β4 cells. In A431 and A549 cells, integrin β4 efficiently colocalized with plectin, although the distribution of these proteins was not completely comparable to that in normal keratinocytes. In MCF10A cells, integrin β4 and plectin showed limited colocalization (Fig. 7, A and B), suggesting that HDs are not efficiently assembled in these cells.
Figure 7.
HDs counteract actomyosin contractility in carcinoma cell lines. (A) Confocal images showing the distribution of β4 (red) together with plectin (green) and laminin (blue) in HaCaT, A431, A549, and MCF10A cells. Font in red indicates cells are skin derived, green indicates lung-derived cells, and blue indicates breast-derived cells. Scale bars: 10 µm. (B) Colocalization efficiency between the integrin β4 (Itg. β4) channel and the plectin channel, shown by Pearson’s R value. Data are presented as box-and-whisker plots in which the box extends the 25th to 75th percentiles, the middle line indicates the median, and whiskers go down to the smallest value and up to the largest (∼60 cells in total). The statistical analysis compares the PA-JEB/β4 group with each of the other groups. ****, P < 0.0001. (C) Western blot analysis of whole-cell lysates from PA-JEB, HaCaT, A431, A549, and MCF10A cells either expressing integrin β4 or lacking integrin β4 probed with antibodies against integrin β4, MLC-pS19, and GAPDH.
Next, we deleted integrin β4 in these cell lines and evaluated its effect on cell contractility by analyzing the level of MLC phosphorylation (Fig. 7 C). HaCaT, A431, and A549 cells all exhibited a decreased level of phosphorylated MLC upon the deletion of integrin β4. In contrast, the deletion of integrin β4 in MCF10A cells had no obvious effect on MLC phosphorylation. In conclusion, these data suggest that integrin β4 can reduce cell contractility in different cell types, but this may only be applicable to cells that can form HD-like structures.
Discussion
Cells sense and respond to mechanical stimuli, which involves cooperation between adhesion structures and the cytoskeleton. In this study, we used micropillar-based and biochemical assays to demonstrate an important role for integrin α6β4 and plectin in regulating intracellular tension and traction force generation. We show that the presence of α6β4 and its binding to keratin filaments via plectin and to laminin-332 lowers the ability of keratinocytes to exert traction forces on the substratum. As a consequence, cell spreading and maturation of FAs is reduced in the presence of α6β4. In line with our previous observations (Zuidema et al., 2018), we show that HDs promote the redistribution of integrin αVβ5 from FAs to clathrin structures by reducing cellular tension. These findings together support an important role for HDs in cell mechanical features and cross-talk between integrin-containing adhesion structures in response to altered intracellular tension.
The function of integrin-mediated FAs as primary sites for cellular force transduction during cell adhesion has been widely documented. HDs represent an additional type of adhesion structure that in contrast to FAs is connected not to actomyosin but to IFs. IFs are not associated with motor proteins and, therefore, cannot by themselves mediate traction forces. However, because IFs form an integrated network with F-actin through cytoskeletal linker proteins such as plectin and fimbrin (Jiu et al., 2015; Tang and Gerlach, 2017; Correia et al., 1999), they may influence traction forces exerted by actomyosin contractility.
We confirm the close association between FA and HD components and the IF network by BioID assays and superresolution imaging. Moreover, we demonstrate for the first time that HDs counteract cellular tension and traction-force generation. These findings build on previous studies that reported increased cellular tension in U2OS osteosarcoma cells in the absence of vimentin IFs (Jiu et al., 2017) and in plectin-deficient keratinocytes and endothelial cells (Bonakdar et al., 2015; Osmanagic-Myers et al., 2015).
However, these phenotypical changes observed after deletion and/or mutation of HD components might be very cell type specific. We validated the role of HDs in the regulation of cellular tension in HaCaT keratinocytes and A431 and A549 carcinoma cells. In contrast, deletion of integrin β4 in MCF10A cells had no effect on myosin activity, which could be attributed to the limited colocalization of integrin β4, plectin, and laminin-332. In line with this, cells that do not assemble HDs (i.e., myoblast and fibroblasts) can show opposite phenotypical changes compared with those observed in keratinocytes (Bonakdar et al., 2015; Na et al., 2009; Gregor et al., 2014). The reasons for the cell type–specific mechanical response on HD and IFs are not yet clear. Possible explanations include differences in how IFs are mechanically anchored to the plasma membrane, i.e., only via FAs or via FAs and HDs. Similarly, individual HD components might also have distinct functions during keratinocyte migration, in which type I HDs are disassembled and cannot be efficiently linked to the IF network to counteract force generation. Under these conditions, it was reported that the HD component BP180 was directly binding to and stabilizing actin bundles and, subsequently, its depletion resulted in a minor decrease in traction force (Hiroyasu et al., 2016).
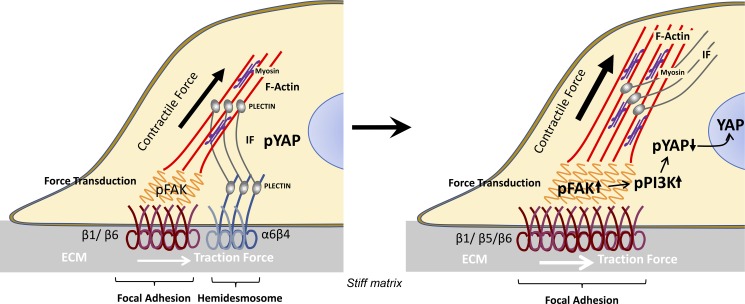
Here, we propose that HDs are mechanically coupled to FAs in keratinocytes and reduce cellular traction forces through a mechanism that involves binding of IF-anchored HDs to the actomyosin cytoskeleton in FAs (Fig. 8). Indeed, in β4-proficient cells, keratin IFs could be found closely associated with FAs (Fig. 2 D). These IFs appear to be more linear than IFs that do not seem to be associated with FAs, and we hypothesize that this might be the result of IF stretching by actomyosin-generated tension.
Figure 8.
Proposed model for the role of HDs in cellular tension regulation. Contractile forces generated by the actomyosin cytoskeleton can prestress IFs, which can counterbalance a further increase in force generation. Binding of the prestressed IFs to β4 in HDs and F-actin–anchored FAs is mediated by plectin (situation on the left). When the HD constraint on actomyosin generated force is eliminated, force transduction across integrins will increase and FAs can further grow and reorganize into larger and more complex structures (situation on the right). Upon increased cellular tension, more FAK can be phosphorylated and subsequently trigger the activation of the PI3K–YAP signaling pathway.
Furthermore, our data indicate that the regulation of cellular force by HDs further affects activation of YAP through the RhoA–ROCK–MLC and FAK–PI3K signaling axes. Since IFs are elastic and can be stretched several times their original lengths without breaking (Charrier and Janmey, 2016), we speculated that the effects of HD perturbation on traction forces should be most dramatic on stiff substrates at which cells can generate high traction forces (Zhao et al., 2018). Consistent with this notion, we observed a more dramatic effect of HD pertubation on stiff compared with soft pillars. Moreover, on soft pillars, we detected an effect of β4-AD, but not the plectin-binding deficient mutant (Fig. 4, C and D). Although not statistically significant, we observed a trend suggesting that on stiff pillars, the effect of β4-AD on force generation was larger than that of the β4-R1281W mutant. It thus seems that α6β4 can regulate force generation in a somewhat IF-binding–independent but adhesion-dependent manner. It is also important to note that in the absence of α6β4-mediated cell adhesion, adhesion by the interaction of α3β1 or α6β1 with the available vacant laminin-332 might be promoted, and therefore, it is not clear whether the effects of the β4-AD on force generation are actually due to a loss of α6β4-mediated adhesion or a gain of α3β1-mediated cell adhesion. Adding to complexity of the issue is the notion that α3β1 may regulate the actin cytoskeleton through its interaction with the tetraspanin CD151 (Hemler, 2005; te Molder et al., 2019).
During wound healing, cellular traction forces and cell contractility are the main drivers for epithelial restoration through regulating cell contraction at wound edges and cell crawling toward the wound bed (Brugués et al., 2014; Fenteany et al., 2000). At the early stage of wound healing, dissolved HDs can be found in keratinocytes at the wound edge, in which cells exhibit elongated and flattened morphology, with long lamellipodia extending into the matrix (Koivisto et al., 2014). Previous studies showed that type I HDs are only reformed after completion of reepithelialization, since cells at wound margin partially disassemble HDs in order to detach from the basement membrane (Hopkinson et al., 2014). Even though both integrin β4 and BP230 were found at the leading front of keratinocytes in a scratch-wound assay (Hamill et al., 2009) and an in vivo incisional wound (Underwood et al., 2009), they were much less clustered in the leading area than in the trailing area of the wound. Furthermore, integrin α6β4 was shown to move into assembling lamellipodium at the leading edge of the wound and interact with actin fibers, but not to associate with keratin filaments (Tsuruta et al., 2011; Svitkina et al., 1997), indicating that typical type I HDs were not efficiently formed. Consistent with this notion, our own data indicate that type I HDs are less abundantly present in keratinocytes at the wound edge. Although different mechanisms may underlie the high cytoskeletal tension in the front wound edge, the loss of HDs in leading cells may be one of the factors that have contributed to this increased tension.
Interestingly, in many tumors, the distribution and expression of integrin α6β4 have been altered as well (Davis et al., 2001; Stewart and O’Connor, 2015; Ramovs et al., 2017). It is therefore feasible that alterations and/or loss of HDs may change the mechanical properties of these cells and contribute to tumor dissemination. In agreement with this hypothesis, we demonstrated that the regulatory role of HDs in cellular tension correlates with efficient HD assembly in tumor cells.
In summary, we demonstrate the crucial role of integrin α6β4 and plectin in force modulation. Impaired HD assembly leads to increased cellular tension and traction forces and subsequently promotes the maturation of FAs. Our results reveal a novel role for HDs as regulators of cellular mechanical forces and establish the existence of a mechanical coupling between adhesion complexes.
Materials and methods
Construction of expression plasmids
Full-length human β4A cDNA was cloned as a 5.6-kb EcoRI fragment into the EcoRI site of retroviral expression vector LZRS-IRES-zeo, a modified LZRS retroviral vector conferring resistance to zeocin (Kinsella and Nolan, 1996). β4-R1281W was made through mutating the CGG to TGG in codon 1,281 of β4, which was found from a human patient (Pulkkinen and Uitto, 1998). The R1281W mutation was introduced into human β4 cDNA by sequence overhang extension PCR (Geerts et al., 1999). The mutagenesis primers were 5′-GACAACCCTAAGAACTGGATGCTGCTTATTG-3′ and 5′ CAATAAGCAGCATCCAGTTCTTAGGGTTGTC-3′. The β4-AD construct (a kind gift from M.P. Marinkovich, Stanford University School of Medicine, Stanford, CA) was produced through cloning the EcoRI β4 cDNA insert into the EcoRI site of pSK and performing mutagenesis using the GeneEditor in vitro site-directed mutagenesis system (Promega). The primer used for the point mutation of β4 sequence was β4-AD (D230A, P232A, and E233A, incorporating with a novel NaeI site): 5′-GGCAACCTGGCTGCTGCTGCCGGCGGCTTCG-3′ (Russell et al., 2003).
The coding sequence of the BirA* (R118G) enzyme was amplified by PCR from the pcDNA3.1 MCS-BirA*(R118G)-HA plasmid (a kind gift from K. Roux, University of South Dakota, Sioux Falls, SD; plasmid 30647; Addgene; Roux et al., 2012) with the oligonucleotides 5′-CCCAAGCTTGAATTCGGATCCAAGGACAAC-3′ and 5′-GCTCTAGATCAGCGGTTTAAACTTAAGGC-3′. The amplified fragment was cut with HindIII and Xbal and ligated into pCMV-IL-2R (a kind gift from K.M. Yamada, National Institutes of Health, Bethesda, MD; LaFlamme et al., 1992). Subsequently, the pcDNA3-IL-2R-BirA*(R118G) was cut with PmeI and NheI, blunt ended with Klenow fragment, and finally cloned into the SnaBI site of the LZRS-IRES-zeo retroviral expression vector.
The β4-BirA* construct was created by a three-point ligation. The full-length β4 cDNA was cut from pUC18-β4A (Delwel et al., 1993) using EcoRI and EcoRV, generating a 5-kb EcoRI–EcoRV and 0.6-kb EcoRV–EcoRI fragment. After removal of the stop codon and the introduction of a new EcoRI site in the 0.6-kb fragment, the two fragments were ligated in the pcDNA3.1MCS-BirA*(R118G)-HA plasmid digested with EcoRI. Subsequently, the β4-BirA* cDNA was released from pcDNA3.1 by digestion with HpaI and PmeI, and the resulting fragment was ligated into LZRS-IRES-zeo cut with SnaBI. Primers used to introduce restriction enzyme site and remove the stop codon of β4 were 5′-CGGAATTCAGTTTGGAAGAACTGTTG-3′ and 5′-CGGGATCCCACCCGCAGAGCCCA-3′.
Cell culture and generation of stable cell lines
Immortalized PA-JEB keratinocytes were isolated from a patient with JEB associated with pyloric atresia (Schaapveld et al., 1998). Since the derivation of this cell line was done for diagnostic purposes, the study using these cells was exempt from the requirement for ethical approval. PA-JEB keratinocytes were cultured in KGM (Invitrogen) supplemented with bovine pituitary gland extract (50 µg ml−1), EGF (5 ng ml−1), and streptomycin/penicillin (100 U ml−1 for each; Sigma-Aldrich). The human carcinoma cell lines HT29, SW620, Caco-2, A431, and A549 (obtained from American Type Culture Collection), HaCaT keratinocytes (Boukamp et al., 1988), and UM-SCC 22B cells (Brenner et al., 2010) were grown in DMEM (Gibco) containing 10% fetal bovine serum (Serana Europe) and 100 U ml−1 each of streptomycin and penicillin (Sigma-Aldrich). Human mammary gland–derived MCF 10A cells (American Type Culture Collection) were cultured in DMEM/F12 Ham’s mixture supplemented with 5% horse serum (Sigma-Aldrich), EGF (20 ng ml−1; Sigma-Aldrich), insulin (10 µg ml−1; Sigma-Aldrich), hydrocortisone (0.5 mg ml−1; Sigma-Aldrich), and cholera toxin (100 ng ml−1; Sigma-Aldrich). MDA-MB-231 cells were maintained in RPMI medium 1640 (Thermo Fisher Scientific) containing 10% fetal bovine serum and streptomycin/penicillin.
Retroviral vectors were introduced into the Phoenix packaging cells by the calcium phosphate precipitation method, and virus-containing supernatant was collected (Kinsella and Nolan, 1996). PA-JEB cells were infected with the recombinant virus by the 1,2-dioleoyl-3-trimethylammonio-propane (DOTAP) method (Boehringer). After infection overnight at 37°C, infected cells were selected with 0.2 mg ml−1 zeocin (Invitrogen). Cells expressing α6β4 or IL-2R at their surface were isolated by using a Beckman Coulter Moflo Astrios cell sorter.
To generate plectin-deficient PA-JEB/β4 keratinocytes (te Molder et al., 2019) and integrin β4-deficient cell lines, target sgRNAs against PLEC (exon2; 5′-GAGGTGCTTGTTGACCCACTTGG-3′) and ITGB4 (exon1; 5′-AGGCGCGGAGGGAGCGAGTC-3′) were cloned into pX330-U6-Chimeric_BB-CBh-hSpCas9 (plasmid 42230; Addgene; deposited by Feng Zhang; Cong et al., 2013). The plasmids containing sgRNA against PLEC or ITGB4, together with a blasticidin cassette (Blomen et al., 2015), were transfected into cells using Lipofectamine 2000 (Invitrogen) in accordance with the manufacturer’s instructions. Cells were incubated overnight with the transfection mixture, after which the solution was replaced by medium supplemented with Blasticidin (4 µg ml−1; Sigma-Aldrich). 4 d after selection, cells were trypsinized and washed twice with PBS containing 2% FCS and incubated with PE-conjugated anti-human integrin β4 antibody (439-9B; dilution 1:200; BD Biosciences), and the PE-negative populations were sorted using a Beckman Coulter Moflo Astrios cell sorter.
Antibodies for immunofluorescence
Primary antibodies used are listed in Table S1. Secondary antibodies used were goat anti-mouse Alexa Fluor 561, goat anti-guinea pig Alexa Fluor 488, goat anti-rat Texas Red, goat anti-rabbit Alexa Fluor 647, goat anti-human Alexa Fluor 488, goat anti-rabbit Alexa Fluor 488 (Invitrogen), and goat anti-mouse Dylight 405 (BioLegend).
Immunofluorescence imaging and image analysis
For immunofluorescence microscopy, integrin β4–deficient, β4-proficient, β4-R1281W, and β4-AD PA-JEB keratinocytes were seeded on glass coverslips and cultured for 24 h in complete KGM and then cultured for a further 24 h in DMEM with 10% FCS. Additionally, β4-deficient cells were treated with Blebbistatin (20 µM; Sigma-Aldrich) for 30 min before fixation or β4-proficient cells were treated with β4-blocking antibody mAb ASC-8 (supernatant diluted 1:5; gift form A. Skubitz, University of Minnesota, Minneapolis, MN) for 24 h. Cells were fixed in 2% paraformaldehyde and permeabilized with 0.2% Triton X-100 for 5 min, blocked with PBS containing 2% BSA (Sigma-Aldrich) for 45 min, and subsequently incubated with primary and secondary antibodies for 1 h. Between antibody steps, the coverslips were washed three times with PBS. Actin fibers were visualized by staining with Flash Phalloidin Green 488 (BioLegend) or Phalloidin Alexa Fluor 647 (Cell Signaling Technology). Nuclei were stained with DAPI. Samples were mounted on glass slides in Mowiol after three washing steps with PBS. Images usually were acquired with a Leica TCS SP5 confocal microscope with a 63× (NA 1.4) oil objective, except images for cell migration, which were acquired with a 40× (NA 1.3) oil objective. The software used for image acquisition was Leica LAS AF 3.0.2.
Image analysis was performed using ImageJ (Schindelin et al., 2012). For wound-healing assays, the FAs (based on vinculin staining), type I HDs and MLC-pS19 were quantified in the keratinocytes at the leading wound edge (two or three rows of keratinocytes near the cell-free area [region of interest {ROI} L]) versus in stationary cells at the trailing area (ROI T). Type I HD was defined by the colocalization of integrin β4 and BP230 using the multiply function (Image calculator) to merge these two channels. Background staining of vinculin was subtracted (command “Subtract Background…”; rolling ball radius: 10.0 pixels), and the Analyze Particle function was used to calculate the FA area, MLC-pS19 area, and HD area per ROI as a percentage of the total ROI area. Cell area was measured by drawing an ROI at the cell periphery, and FA size was calculated using the Particle Analyze function. The percentage of nuclear YAP was calculated by dividing the YAP staining overlapped with DAPI with the total YAP staining intensity. Colocalization of integrin β5 with FAs or clathrin lattices was determined by calculating the area of β5 clusters overlapping with FAs or clathrin lattices out of the total integrin β5 area per cell, as described previously (Zuidema et al., 2018). The colocalization efficiency of β4 and plectin was performed using Pearson’s coefficient analysis in the JACoP module of ImageJ (Bolte and Cordelières, 2006).
Superresolution microscopy
According to an established method (Nahidiazar et al., 2015), cells for superresolution microscopy were fixed in 4% paraformaldehyde and permeabilized with 0.2% Triton X-100, blocked with PBS containing 5% BSA (Sigma-Aldrich), and subsequently incubated with primary and secondary antibodies with washing steps in between. Imaging was performed with a Leica SR-GSD microscope (Leica Microsystems) equipped with 405-nm/30-mW, 488-nm/300-mW, and 642-nm/500-mW lasers, with samples immersed in OxEA buffer (Nahidiazar et al., 2016). A 160× oil-immersion objective (NA 1.47) and an EMCCD iXon camera (Andor Technology) were used to collect images. Between 10,000 and 50,000 frames were collected, with a frame rate of 100 Hz. All the datasets were analyzed with the ThunderSTORM module of ImageJ software (Ovesný et al., 2014).
Micropillar-based traction force microscopy
Cellular traction force measurements were performed using elastic micropillar arrays produced in our laboratories. A hexagonal array of polydimethylsiloxane (Sylgard 184; Dow Corning) micropillars of 2 µm diameter, 4 µm center-to-center distance, and 6.1 µm (Young’s modulus 29.5 kPa effective stiffness) or 3.2 µm (137.1 kPa effective stiffness) height were produced using replica molding from a silicon wafer (van Hoorn et al., 2014). The pillar arrays were flanked by integrated 50-µm-high spacers to allow the inversion onto glass coverslips, without compromising the limited working distance of a high-NA objective on an inverted microscope. The mounting buffer is PBS and imaging was performed in room temperature. The tops of the micropillars were coated with a mixture of unlabeled and Alexa Fluor 647–labeled fibronectin (1:5; Life Technologies) using microcontact printing. The position of the pillar tops was observed by confocal fluorescence microscopy with same setting mentioned in the Immunofluorescence imaging and image analysis section and determined down to subwavelength accuracy using custom software (MATLAB; MathWorks). Forces were obtained by multiplying the pillar deflections by the array’s characteristic spring constant (41 nN μm−1 and 191 nN μm−1, respectively, determined by finite element modeling; van Hoorn et al., 2014). The pillars’ spring constants were converted to an equivalent Young’s modulus for continuous substrates (Ghibaudo et al., 2008) of 30 kPa (soft pillars) and 137 kPa (stiff pillars), respectively. Only pillars closer to cell perimeter (<3 µm) and with a deflection (<65 nm for soft pillars and <75 nm for stiff pillar) were considered for the calculation of the average force per pillar (per cell), total cellular forces, and percentage of deflected pillars (per cell). The deflection thresholds, which reflect the positional accuracy by which individual pillars could be localized, were determined for each confocal image as the 75th percentile of the displacements of pillars outside the cell area (i.e., not bent by the cells). The cell area was determined by thresholding the fluorescence signal of FlashPhalloidin Green 488–labeled actin filaments using a triangulation method (Zack et al., 1977).
BioID assay
PA-JEB expressing β4-BirA* cells cultured in KGM or DMEM were treated with 50 µM biotin (Sigma #B4501) overnight. After washing with cold PBS, cells were lysed at 4°C in lysis buffer (1% Nonidet P-40, 20 mM Tris-HCl, pH 7.5, 100 mM NaCl, and 4 mM EDTA) supplemented with Na3VO4 (1.5 mM), NaF (15 mM), and protease inhibitor cocktail (1:1,000; Sigma-Aldrich) and cleared by centrifugation at 14,000 ×g for 1 h at 4°C. Cell lysates were incubated together with Streptavidin Sepharose High Performance beads (GE Healthcare) overnight at 4°C. After washing the beads with lysis buffer and cold PBS, bound proteins were eluted from the beads at 95°C for 5 min in SDS-loading buffer (50 mM Tris-HCl, pH 6.8, 2% SDS, 10% glycerol, 12.5 mM EDTA, 0.02% Bromophenol Blue, and 2% β-mercaptoethanol) and analyzed by gel electrophoresis. Biotinylated proteins were detected by Western blotting or mass spectrometry
Mass spectrometry and data analysis
Samples from BioID experiment were separated on a 4–12% SDS-PAGE gel. The gel was stained with Coomassie Blue, and lanes were excised and then reduced by treating with dithiothreitol and alkylated with iodoacetamide. After digestion with trypsin (mass spectrometry grade; Promega), peptides were extracted with acetonitrile. A vacuum centrifuge was used to dry the digests, which were resuspended in 10% formic acid for mass spectrometry analysis. Peptides were analyzed by nano-liquid chromatography–tandem mass spectrometry on an Orbitrap Fusion Tribrid mass spectrometer equipped with a Proxeon nLC1000 system (Thermo Fisher Scientific). Next, samples were eluted from the analytical column at a constant flow of 250 nl min−1 in a 140-min gradient, containing a 124-min linear increase from 6% to 30% solvent A (0.1% formic acid/water), followed by a 16-min wash at 100% solvent B (0.1% formic acid/80% acetonitrile).
We applied MaxQuant (version 1.6.5.0; Cox et al., 2014) with standard settings to our raw data for label-free quantitation. The human Swiss-Prot database (20,432 entries, release 2019_09) was used as data source. Tandem mass spectrometry data were concatenated with the reversed version of all sequences from the database. Trypsin/P was selected as cleavage specificity allowing two missed cleavages; Carbamidomethylation (C) was set as a fixed modification, while oxidation (M) was used as variable modification. Label-free quantitation intensities were log2 transformed in Perseus (version 1.6.7.0; Tyanova et al., 2016), after which proteins were filtered for at least two valid values (out of three total). Missing values were replaced by imputation based a normal distribution using a width of 0.3 and a downshift of 1.8. Differentially expressed proteins were determined using a t test (threshold false discovery rate: 0.01 and S0: 2).
Western blotting
Integrin β4–deficient, β4-proficient, β4-R1281W, and β4-AD PA-JEB keratinocytes were grown to 50–60% confluence and treated with inhibitors of ROCK (Y27632, 10 µM; Sigma-Aldrich), myosin (blebbistatin, 10 µM; Abcam), FAK (VS-4718, 1 µM; Selleck Chemicals), PI3K (GDC-0941, 1 µM; Selleck Chemicals), AKT (MK-2206, 1 µM; Selleck Chemicals), or mTOR (AZD-8055, 100 nM; Selleck Chemicals) for 45 min before cell lysis. Cells were lysed in radioimmunoprecipitation assay buffer supplemented with Na3VO4 (1.5 mM), NaF (15 mM), protease inhibitor cocktail (1:1,000; Sigma-Aldrich), and Phosphatase Inhibitor Cocktail 3 (1:100; Sigma-Aldrich) and cleared by centrifugation at 14,000 ×g for 1 h at 4°C. Samples for gel electrophoresis were prepared in SDS-loading buffer supplemented with 2% β-mercaptoethanol and heated for 5 min at 95°C. After separation of the proteins on Novex 4–12% or 3–12% gradient Bis-Tris gel (Invitrogen) or 15% homemade SDS-PAGE, proteins were transferred onto Immobilon-P transfer membranes (Millipore). The membranes containing the blotted proteins were incubated in TBS buffer containing 2% BSA and 0.1% Tween-20 for 1 h and then treated with primary antibodies. Antibodies used are listed in Table S1. After several washing steps with TBS containing 0.1% Tween-20 and TBS, membranes were incubated with secondary antibodies (goat anti-mouse IgG conjugated with horseradish peroxidase [Bio-Rad] or goat anti-rabbit HRP-conjugated Ab [Dako]). Finally, proteins were visualized using Clarity Western ECL Substrate (Bio-Rad) or SuperSignal West Dura Extended Duration Substrate (Thermo Fisher Scientific). Signal intensities were quantified using ImageJ.
Adhesion assay
Microtiter plates (96-well; Greiner Bio-one) coated with a laminin-332–rich matrix were prepared as previously described (Delwel et al., 1993). Briefly, mouse RAC-11P cells grown to confluence were incubated overnight at 4°C in PBS containing 20 mM EDTA. Subsequently, the cells were removed from the plate as a single continuous sheet by brief but forceful pipetting and washed three times with PBS. Integrin β4–proficient PA-JEB cells were resuspended in serum-free KGM in the presence or absence of ASC-8 antibody (supernatant diluted 1:5) and/or anti-α3-blocking mAb (J143; 20 µg ml−1). The cells, together with β4-AD PA-JEB cells, were seeded in the precoated plates at a density of 105 cells per well and incubated at 37°C for 45 min. After removing the nonadherent cells by washing, the adherent cells were fixed with 4% paraformaldehyde for 10 min, washed twice with water, and stained for 10 min with 0.2% crystal violet. Next, the plates were washed with water and air-dried overnight. The violet-stained cells were lysed by 2% SDS and absorbance was measured at 490 nm using a microplate reader (Bio-Rad). Data were exported with MPM6 software and statistically processed.
Cell migration assay
Coverslips were coated by RAC-11P–secreted matrix in advance. PA-JEB/β4 keratinocytes were plated on coverslips whose central region was occupied by a round metal pillar with a diameter of 3 mm and allowed to grow to confluence surrounding the pillar. The metal pillar was removed to create a gap in the cell monolayer and induce cell migration. After 72-h culture in KGM medium, the cell monolayer with the gap was fixed and subjected to immunofluorescence staining with primary and secondary antibodies.
RhoA activity assay
Integrin β4–deficient, β4-proficient, β4-R1281W, and β4-AD PA-JEB keratinocytes at 50–60% confluency were treated overnight with DMEM containing 10% FCS. Cells were lysed in NP-40 buffer (1% Nonidet P-40, 150 mM NaCl, and 50 mM Tris, pH 8) supplemented with protease inhibitor cocktail and cleared at 14,000 ×g for 60 min. Lysates were incubated with 50 µg glutathione-Sepharose–bound (ab193267; Abcam) or GST–rhotekin–binding domain for 30 min at 4°C. Beads were then washed three times in NP-40 lysis buffer. Proteins bound to beads were released by adding SDS-loading buffer supplemented with a final concentration of 2% β-mercaptoethanol. Subsequently, released proteins and whole-cell lysates as input control were subjected to Western blotting analysis and probed with an antibody against RhoA.
Statistical analysis
The Mann–Whitney U test was used to calculate significance between two groups using GraphPad Prism 7.2. Graphs were made in GraphPad Prism, and values represent the mean (± SD) of three independent experiments. Statistically significant values are indicated as *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001.
Online supplemental material
Fig. S1 validates the integrin β4 interactors belonging to FA and HD proteins and the successful knockout of plectin from β4 (+) PA-JEB cells. Fig. S2 shows the impaired adhesion ability of β4-AD PA-JEB keratinocytes and the effect of the β4 blocking antibody ASC-8 on cell adhesion; FA size and length probed by paxillin staining are also quantified. Fig. S3 shows the deposition of laminin-332 by cells seeded on fibronectin-coated micropillar; extra data for the micropillar assay comparing the percentage of deflected pillars and traction force applied on each pillar among β4-deficient, β4-proficient, β4-R1281W, and β4-AD PA-JEB keratinocytes; and comparison of the level of active RhoA in these cells. Table S1 lists detailed information regarding the primary antibodies used in our work. Data S1 is an Excel file with the mass spectrometry data for the proximity interactors of the integrin β4 analysis.
Supplementary Material
lists primary antibodies.
shows the mass spectrometry data for the proximity interactors of the integrin β4 analysis.
Acknowledgments
We thank Dr. H. Herrmann, Dr. M. Aumailley, Dr. M. Glukhova, Dr. T. Hashimoto, K. Roux, K.M. Yamada, and P.M. Marinkovich for sharing reagents and Veronika Ramovs, Kevin Wilhemsen, and Metello Innocenti for critical reading of the manuscript.
This work was supported by the Netherlands Organization for Scientific Research as part of the National Roadmap Large-scale Research Facilities of the Netherlands Proteins@Work (grant 184.032.201). W. Wang is funded by the China Scholarship Council (2016-2020). S. Coppola is beneficiary of an AXA Research Fund postdoctoral grant (16-AXA-PDOC-004).
The authors declare no competing financial interests.
Author contributions: W. Wang, A. Zuidema, and A. Sonnenberg conceived the study, designed experiments, interpreted data, and wrote the manuscript. W. Wang, A. Zuidema, L. te Molder, and L. Nahidiazar performed and analyzed the cell biological experiments. L. Hoekman conducted the mass spectrometry analysis. T. Schmidt and S. Coppola provided the micropillar arrays and expertise required to conduct and interpret the measurements of cellular force.
References
- Almeida F.V., Walko G., McMillan J.R., McGrath J.A., Wiche G., Barber A.H., and Connelly J.T.. 2015. The cytolinker plectin regulates nuclear mechanotransduction in keratinocytes. J. Cell Sci. 128:4475–4486. 10.1242/jcs.173435 [DOI] [PubMed] [Google Scholar]
- Amano M., Ito M., Kimura K., Fukata Y., Chihara K., Nakano T., Matsuura Y., and Kaibuchi K.. 1996. Phosphorylation and activation of myosin by Rho-associated kinase (Rho-kinase). J. Biol. Chem. 271:20246–20249. 10.1074/jbc.271.34.20246 [DOI] [PubMed] [Google Scholar]
- Andrä K., Nikolic B., Stöcher M., Drenckhahn D., and Wiche G.. 1998. Not just scaffolding: plectin regulates actin dynamics in cultured cells. Genes Dev. 12:3442–3451. 10.1101/gad.12.21.3442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beningo K.A., Hamao K., Dembo M., Wang Y.L., and Hosoya H.. 2006. Traction forces of fibroblasts are regulated by the Rho-dependent kinase but not by the myosin light chain kinase. Arch. Biochem. Biophys. 456:224–231. 10.1016/j.abb.2006.09.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomen V.A., Májek P., Jae L.T., Bigenzahn J.W., Nieuwenhuis J., Staring J., Sacco R., van Diemen F.R., Olk N., Stukalov A., et al. . 2015. Gene essentiality and synthetic lethality in haploid human cells. Science. 350:1092–1096. 10.1126/science.aac7557 [DOI] [PubMed] [Google Scholar]
- Bolte S., and Cordelières F.P.. 2006. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 224:213–232. 10.1111/j.1365-2818.2006.01706.x [DOI] [PubMed] [Google Scholar]
- Bonakdar N., Schilling A., Spörrer M., Lennert P., Mainka A., Winter L., Walko G., Wiche G., Fabry B., and Goldmann W.H.. 2015. Determining the mechanical properties of plectin in mouse myoblasts and keratinocytes. Exp. Cell Res. 331:331–337. 10.1016/j.yexcr.2014.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boukamp P., Petrussevska R.T., Breitkreutz D., Hornung J., Markham A., and Fusenig N.E.. 1988. Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J. Cell Biol. 106:761–771. 10.1083/jcb.106.3.761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner J.C., Graham M.P., Kumar B., Saunders L.M., Kupfer R., Lyons R.H., Bradford C.R., and Carey T.E.. 2010. Genotyping of 73 UM-SCC head and neck squamous cell carcinoma cell lines. Head Neck. 32:417–426. 10.1002/hed.21198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugués A., Anon E., Conte V., Veldhuis J.H., Gupta M., Colombelli J., Muñoz J.J., Brodland G.W., Ladoux B., and Trepat X.. 2014. Forces driving epithelial wound healing. Nat. Phys. 10:683–690. 10.1038/nphys3040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burridge K., and Guilluy C.. 2016. Focal adhesions, stress fibers and mechanical tension. Exp. Cell Res. 343:14–20. 10.1016/j.yexcr.2015.10.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman H.A., Li X., Alexander J.P., Brumwell A., Lorizio W., Tan K., Sonnenberg A., Wei Y., and Vu T.H.. 2011. Integrin α6β4 identifies an adult distal lung epithelial population with regenerative potential in mice. J. Clin. Invest. 121:2855–2862. 10.1172/JCI57673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrier E.E., and Janmey P.A.. 2016. Mechanical properties of intermediate filament proteins. Methods Enzymol. 568:35–57. 10.1016/bs.mie.2015.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., and Zhang F.. 2013. Multiplex genome engineering using CRISPR/Cas systems. Science. 339:819–823. 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper J., and Giancotti F.G.. 2019. Integrin Signaling in Cancer: Mechanotransduction, Stemness, Epithelial Plasticity, and Therapeutic Resistance. Cancer Cell. 35:347–367. 10.1016/j.ccell.2019.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correia I., Chu D., Chou Y.-H., Goldman R.D., and Matsudaira P.. 1999. Integrating the actin and vimentin cytoskeletons. adhesion-dependent formation of fimbrin-vimentin complexes in macrophages. J. Cell Biol. 146:831–842. 10.1083/jcb.146.4.831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J., Hein M.Y., Luber C.A., Paron I., Nagaraj N., and Mann M.. 2014. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics. 13:2513–2526. 10.1074/mcp.M113.031591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis T.L., Cress A.E., Dalkin B.L., and Nagle R.B.. 2001. Unique expression pattern of the α6β4 integrin and laminin-5 in human prostate carcinoma. Prostate. 46:240–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Pascalis C., Pérez-González C., Seetharaman S., Boëda B., Vianay B., Burute M., Leduc C., Borghi N., Trepat X., and Etienne-Manneville S.. 2018. Intermediate filaments control collective migration by restricting traction forces and sustaining cell-cell contacts. J. Cell Biol. 217:3031–3044. 10.1083/jcb.201801162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Pereda J.M., Lillo M.P., and Sonnenberg A.. 2009. Structural basis of the interaction between integrin α6β4 and plectin at the hemidesmosomes. EMBO J. 28:1180–1190. 10.1038/emboj.2009.48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delwel G.O., Hogervorst F., Kuikman I., Paulsson M., Timpl R., and Sonnenberg A.. 1993. Expression and function of the cytoplasmic variants of the integrin alpha 6 subunit in transfected K562 cells. Activation-dependent adhesion and interaction with isoforms of laminin. J. Biol. Chem. 268:25865–25875. [PubMed] [Google Scholar]
- Dupont S., Morsut L., Aragona M., Enzo E., Giulitti S., Cordenonsi M., Zanconato F., Le Digabel J., Forcato M., Bicciato S., et al. . 2011. Role of YAP/TAZ in mechanotransduction. Nature. 474:179–183. 10.1038/nature10137 [DOI] [PubMed] [Google Scholar]
- Fenteany G., Janmey P.A., and Stossel T.P.. 2000. Signaling pathways and cell mechanics involved in wound closure by epithelial cell sheets. Curr. Biol. 10:831–838. 10.1016/S0960-9822(00)00579-0 [DOI] [PubMed] [Google Scholar]
- Fine J.-D., Bruckner-Tuderman L., Eady R.A.J., Bauer E.A., Bauer J.W., Has C., Heagerty A., Hintner H., Hovnanian A., Jonkman M.F., et al. . 2014. Inherited epidermolysis bullosa: updated recommendations on diagnosis and classification. J. Am. Acad. Dermatol. 70:1103–1126. 10.1016/j.jaad.2014.01.903 [DOI] [PubMed] [Google Scholar]
- Fontao L., Stutzmann J., Gendry P., and Launay J.F.. 1999. Regulation of the type II hemidesmosomal plaque assembly in intestinal epithelial cells. Exp. Cell Res. 250:298–312. 10.1006/excr.1999.4549 [DOI] [PubMed] [Google Scholar]
- Geerts D., Fontao L., Nievers M.G., Schaapveld R.Q.J., Purkis P.E., Wheeler G.N., Lane E.B., Leigh I.M., and Sonnenberg A.. 1999. Binding of integrin α6β4 to plectin prevents plectin association with F-actin but does not interfere with intermediate filament binding. J. Cell Biol. 147:417–434. 10.1083/jcb.147.2.417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger B., Bershadsky A., Pankov R., and Yamada K.M.. 2001. Transmembrane crosstalk between the extracellular matrix--cytoskeleton crosstalk. Nat. Rev. Mol. Cell Biol. 2:793–805. 10.1038/35099066 [DOI] [PubMed] [Google Scholar]
- Ghibaudo M., Saez A., Trichet L., Xayaphoummine A., Browaeys J., Silberzan P., Buguin A., and Ladoux B.. 2008. Traction forces and rigidity sensing regulate cell functions. Soft Matter. 4:1836–1843. 10.1039/b804103b [DOI] [Google Scholar]
- Goldmann W.H. 2018. Intermediate filaments and cellular mechanics. Cell Biol. Int. 42:132–138. 10.1002/cbin.10879 [DOI] [PubMed] [Google Scholar]
- Gregor M., Osmanagic-Myers S., Burgstaller G., Wolfram M., Fischer I., Walko G., Resch G.P., Jörgl A., Herrmann H., and Wiche G.. 2014. Mechanosensing through focal adhesion-anchored intermediate filaments. FASEB J. 28:715–729. 10.1096/fj.13-231829 [DOI] [PubMed] [Google Scholar]
- Halder G., Dupont S., and Piccolo S.. 2012. Transduction of mechanical and cytoskeletal cues by YAP and TAZ. Nat. Rev. Mol. Cell Biol. 13:591–600. 10.1038/nrm3416 [DOI] [PubMed] [Google Scholar]
- Hamill K.J., Hopkinson S.B., DeBiase P., and Jones J.C.R.. 2009. BPAG1e maintains keratinocyte polarity through β4 integrin-mediated modulation of Rac1 and cofilin activities. Mol. Biol. Cell. 20:2954–2962. 10.1091/mbc.e09-01-0051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemler M.E. 2005. Tetraspanin functions and associated microdomains. Nat. Rev. Mol. Cell Biol. 6:801–811. 10.1038/nrm1736 [DOI] [PubMed] [Google Scholar]
- Hiroyasu S., Colburn Z.T., and Jones J.C.R.. 2016. A hemidesmosomal protein regulates actin dynamics and traction forces in motile keratinocytes. FASEB J. 30:2298–2310. 10.1096/fj.201500160R [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homan S.M., Mercurio A.M., and LaFlamme S.E.. 1998. Endothelial cells assemble two distinct alpha6beta4-containing vimentin-associated structures: roles for ligand binding and the beta4 cytoplasmic tail. J. Cell Sci. 111:2717–2728. [DOI] [PubMed] [Google Scholar]
- Hopkinson S.B., Hamill K.J., Wu Y., Eisenberg J.L., Hiroyasu S., and Jones J.C.R.. 2014. Focal Contact and Hemidesmosomal Proteins in Keratinocyte Migration and Wound Repair. Adv. Wound Care (New Rochelle). 3:247–263. 10.1089/wound.2013.0489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houdusse A., and Sweeney H.L.. 2016. How Myosin Generates Force on Actin Filaments. Trends Biochem. Sci. 41:989–997. 10.1016/j.tibs.2016.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iskratsch T., Wolfenson H., and Sheetz M.P.. 2014. Appreciating force and shape—the rise of mechanotransduction in cell biology. Nat. Rev. Mol. Cell Biol. 15:825–833. 10.1038/nrm3903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen K.A., Atherton P., and Ballestrem C.. 2017. Mechanotransduction at the cell-matrix interface. Semin. Cell Dev. Biol. 71:75–83. 10.1016/j.semcdb.2017.07.027 [DOI] [PubMed] [Google Scholar]
- Jiu Y., Lehtimäki J., Tojkander S., Cheng F., Jäälinoja H., Liu X., Varjosalo M., Eriksson J.E., and Lappalainen P.. 2015. Bidirectional Interplay between Vimentin Intermediate Filaments and Contractile Actin Stress Fibers. Cell Reports. 11:1511–1518. 10.1016/j.celrep.2015.05.008 [DOI] [PubMed] [Google Scholar]
- Jiu Y., Peränen J., Schaible N., Cheng F., Eriksson J.E., Krishnan R., and Lappalainen P.. 2017. Vimentin intermediate filaments control actin stress fiber assembly through GEF-H1 and RhoA. J. Cell Sci. 130:892–902. 10.1242/jcs.196881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N.-G., and Gumbiner B.M.. 2015. Adhesion to fibronectin regulates Hippo signaling via the FAK-Src-PI3K pathway. J. Cell Biol. 210:503–515. 10.1083/jcb.201501025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinsella T.M., and Nolan G.P.. 1996. Episomal vectors rapidly and stably produce high-titer recombinant retrovirus. Hum. Gene Ther. 7:1405–1413. 10.1089/hum.1996.7.12-1405 [DOI] [PubMed] [Google Scholar]
- Klinowska T.C., Alexander C.M., Georges-Labouesse E., Van der Neut R., Kreidberg J.A., Jones C.J., Sonnenberg A., and Streuli C.H.. 2001. Epithelial development and differentiation in the mammary gland is not dependent on alpha 3 or alpha 6 integrin subunits. Dev. Biol. 233:449–467. 10.1006/dbio.2001.0204 [DOI] [PubMed] [Google Scholar]
- Koivisto L., Heino J., Häkkinen L., and Larjava H.. 2014. Integrins in Wound Healing. Adv. Wound Care (New Rochelle). 3:762–783. 10.1089/wound.2013.0436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster J., Geerts D., Favre B., Borradori L., and Sonnenberg A.. 2003. Analysis of the interactions between BP180, BP230, plectin and the integrin α6β4 important for hemidesmosome assembly. J. Cell Sci. 116:387–399. 10.1242/jcs.00241 [DOI] [PubMed] [Google Scholar]
- Koster J., van Wilpe S., Kuikman I., Litjens S.H., and Sonnenberg A.. 2004. Role of binding of plectin to the integrin β4 subunit in the assembly of hemidesmosomes. Mol. Biol. Cell. 15:1211–1223. 10.1091/mbc.e03-09-0697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kull F.J., and Endow S.A.. 2013. Force generation by kinesin and myosin cytoskeletal motor proteins. J. Cell Sci. 126:9–19. 10.1242/jcs.103911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachowski D., Cortes E., Robinson B., Rice A., Rombouts K., and Del Río Hernández A.E.. 2018. FAK controls the mechanical activation of YAP, a transcriptional regulator required for durotaxis. FASEB J. 32:1099–1107. 10.1096/fj.201700721R [DOI] [PubMed] [Google Scholar]
- LaFlamme S.E., Akiyama S.K., and Yamada K.M.. 1992. Regulation of fibronectin receptor distribution. J. Cell Biol. 117:437–447. 10.1083/jcb.117.2.437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessey E.C., Guilluy C., and Burridge K.. 2012. From mechanical force to RhoA activation. Biochemistry. 51:7420–7432. 10.1021/bi300758e [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litjens S.H.M., de Pereda J.M., and Sonnenberg A.. 2006. Current insights into the formation and breakdown of hemidesmosomes. Trends Cell Biol. 16:376–383. 10.1016/j.tcb.2006.05.004 [DOI] [PubMed] [Google Scholar]
- Lock J.G., Baschieri F., Jones M.C., Humphries J.D., Montagnac G., Strömblad S., and Humphries M.J.. 2019. Clathrin-containing adhesion complexes. J. Cell Biol. 218:2086–2095. 10.1083/jcb.201811160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manso J.A., Gómez-Hernández M., Carabias A., Alonso-García N., García-Rubio I., Kreft M., Sonnenberg A., and de Pereda J.M.. 2018. Integrin α6β4 recognition of a linear motif of bullous pemphigoid antigen BP230 controls its recruitment to hemidesmosomes. bioRxiv. doi: (Preprint posted December 20, 2018) 10.1101/402123 [DOI] [PubMed] [Google Scholar]
- McGrath J.A. 2015. Recently Identified Forms of Epidermolysis Bullosa. Ann. Dermatol. 27:658–666. 10.5021/ad.2015.27.6.658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelson P.H., Tigue M., and Jones J.C.. 2000. Human bronchial epithelial cells secrete laminin 5, express hemidesmosomal proteins, and assemble hemidesmosomes. J. Histochem. Cytochem. 48:535–544. 10.1177/002215540004800411 [DOI] [PubMed] [Google Scholar]
- Myllymäki S.-M., Kämäräinen U.-R., Liu X., Cruz S.P., Miettinen S., Vuorela M., Varjosalo M., and Manninen A.. 2019. Assembly of the β4-Integrin Interactome Based on Proximal Biotinylation in the Presence and Absence of Heterodimerization. Mol. Cell. Proteomics. 18:277–293. 10.1074/mcp.RA118.001095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na S., Chowdhury F., Tay B., Ouyang M., Gregor M., Wang Y., Wiche G., and Wang N.. 2009. Plectin contributes to mechanical properties of living cells. Am. J. Physiol. Cell Physiol. 296:C868–C877. 10.1152/ajpcell.00604.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahidiazar L., Kreft M., van den Broek B., Secades P., Manders E.M.M., Sonnenberg A., and Jalink K.. 2015. The molecular architecture of hemidesmosomes, as revealed with super-resolution microscopy. J. Cell Sci. 128:3714–3719. 10.1242/jcs.171892 [DOI] [PubMed] [Google Scholar]
- Nahidiazar L., Agronskaia A.V., Broertjes J., van den Broek B., and Jalink K.. 2016. Optimizing Imaging Conditions for Demanding Multi-Color Super Resolution Localization Microscopy. PLoS One. 11:e0158884 10.1371/journal.pone.0158884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardone G., Oliver-De La Cruz J., Vrbsky J., Martini C., Pribyl J., Skládal P., Pešl M., Caluori G., Pagliari S., Martino F., et al. . 2017. YAP regulates cell mechanics by controlling focal adhesion assembly. Nat. Commun. 8:15321 10.1038/ncomms15321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nievers M.G., Kuikman I., Geerts D., Leigh I.M., and Sonnenberg A.. 2000. Formation of hemidesmosome-like structures in the absence of ligand binding by the (alpha)6(beta)4 integrin requires binding of HD1/plectin to the cytoplasmic domain of the (beta)4 integrin subunit. J. Cell Sci. 113:963–973. [DOI] [PubMed] [Google Scholar]
- Oakes P.W., and Gardel M.L.. 2014. Stressing the limits of focal adhesion mechanosensitivity. Curr. Opin. Cell Biol. 30:68–73. 10.1016/j.ceb.2014.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakes P.W., Beckham Y., Stricker J., and Gardel M.L.. 2012. Tension is required but not sufficient for focal adhesion maturation without a stress fiber template. J. Cell Biol. 196:363–374. 10.1083/jcb.201107042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oria R., Wiegand T., Escribano J., Elosegui-Artola A., Uriarte J.J., Moreno-Pulido C., Platzman I., Delcanale P., Albertazzi L., Navajas D., et al. . 2017. Force loading explains spatial sensing of ligands by cells. Nature. 552:219–224. 10.1038/nature24662 [DOI] [PubMed] [Google Scholar]
- Osmanagic-Myers S., Gregor M., Walko G., Burgstaller G., Reipert S., and Wiche G.. 2006. Plectin-controlled keratin cytoarchitecture affects MAP kinases involved in cellular stress response and migration. J. Cell Biol. 174:557–568. 10.1083/jcb.200605172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmanagic-Myers S., Rus S., Wolfram M., Brunner D., Goldmann W.H., Bonakdar N., Fischer I., Reipert S., Zuzuarregui A., Walko G., and Wiche G.. 2015. Plectin reinforces vascular integrity by mediating crosstalk between the vimentin and the actin networks. J. Cell Sci. 128:4138–4150. 10.1242/jcs.172056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovesný M., Křížek P., Borkovec J., Švindrych Z., and Hagen G.M.. 2014. ThunderSTORM: a comprehensive ImageJ plug-in for PALM and STORM data analysis and super-resolution imaging. Bioinformatics. 30:2389–2390. 10.1093/bioinformatics/btu202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa T., Tsuruta D., Jones J.C.R., Ishii M., Ikeda K., Harada T., Aoyama Y., Kawada A., and Kobayashi H.. 2010. Dynamic relationship of focal contacts and hemidesmosome protein complexes in live cells. J. Invest. Dermatol. 130:1624–1635. 10.1038/jid.2009.439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham R.J. Jr., and Wang Y.. 1999. High resolution detection of mechanical forces exerted by locomoting fibroblasts on the substrate. Mol. Biol. Cell. 10:935–945. 10.1091/mbc.10.4.935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccolo S., Dupont S., and Cordenonsi M.. 2014. The biology of YAP/TAZ: hippo signaling and beyond. Physiol. Rev. 94:1287–1312. 10.1152/physrev.00005.2014 [DOI] [PubMed] [Google Scholar]
- Pora A., Yoon S., Windoffer R., and Leube R.E.. 2019. Hemidesmosomes and Focal Adhesions Treadmill as Separate but Linked Entities during Keratinocyte Migration. J. Invest. Dermatol. 139:1876–1888.e4. 10.1016/j.jid.2019.03.1139 [DOI] [PubMed] [Google Scholar]
- Pulkkinen L., and Uitto J.. 1998. Hemidesmosomal variants of epidermolysis bullosa. Mutations in the alpha6beta4 integrin and the 180-kD bullous pemphigoid antigen/type XVII collagen genes. Exp. Dermatol. 7:46–64. 10.1111/j.1600-0625.1998.tb00304.x [DOI] [PubMed] [Google Scholar]
- Ramovs V., Te Molder L., and Sonnenberg A.. 2017. The opposing roles of laminin-binding integrins in cancer. Matrix Biol. 57-58:213–243. 10.1016/j.matbio.2016.08.007 [DOI] [PubMed] [Google Scholar]
- Rezniczek G.A., de Pereda J.M., Reipert S., and Wiche G.. 1998. Linking integrin α6β4-based cell adhesion to the intermediate filament cytoskeleton: direct interaction between the β4 subunit and plectin at multiple molecular sites. J. Cell Biol. 141:209–225. 10.1083/jcb.141.1.209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rid R., Schiefermeier N., Grigoriev I., Small J.V., and Kaverina I.. 2005. The last but not the least: the origin and significance of trailing adhesions in fibroblastic cells. Cell Motil. Cytoskeleton. 61:161–171. 10.1002/cm.20076 [DOI] [PubMed] [Google Scholar]
- Riveline D., Zamir E., Balaban N.Q., Schwarz U.S., Ishizaki T., Narumiya S., Kam Z., Geiger B., and Bershadsky A.D.. 2001. Focal contacts as mechanosensors: externally applied local mechanical force induces growth of focal contacts by an mDia1-dependent and ROCK-independent mechanism. J. Cell Biol. 153:1175–1186. 10.1083/jcb.153.6.1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux K.J., Kim D.I., Raida M., and Burke B.. 2012. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 196:801–810. 10.1083/jcb.201112098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell A.J., Fincher E.F., Millman L., Smith R., Vela V., Waterman E.A., Dey C.N., Guide S., Weaver V.M., and Marinkovich M.P.. 2003. α6β4 integrin regulates keratinocyte chemotaxis through differential GTPase activation and antagonism of α3β1 integrin. J. Cell Sci. 116:3543–3556. 10.1242/jcs.00663 [DOI] [PubMed] [Google Scholar]
- Sanghvi-Shah R., and Weber G.F.. 2017. Intermediate Filaments at the Junction of Mechanotransduction, Migration, and Development. Front. Cell Dev. Biol. 5:81 10.3389/fcell.2017.00081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaapveld R.Q., Borradori L., Geerts D., van Leusden M.R., Kuikman I., Nievers M.G., Niessen C.M., Steenbergen R.D., Snijders P.J., and Sonnenberg A.. 1998. Hemidesmosome formation is initiated by the beta4 integrin subunit, requires complex formation of beta4 and HD1/plectin, and involves a direct interaction between beta4 and the bullous pemphigoid antigen 180. J. Cell Biol. 142:271–284. 10.1083/jcb.142.1.271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. . 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz M.A. 2010. Integrins and extracellular matrix in mechanotransduction. Cold Spring Harb. Perspect. Biol. 2:a005066 10.1101/cshperspect.a005066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterk L.M.T., Geuijen C.A.W., Oomen L.C.J.M., Calafat J., Janssen H., and Sonnenberg A.. 2000. The tetraspan molecule CD151, a novel constituent of hemidesmosomes, associates with the integrin α6β4 and may regulate the spatial organization of hemidesmosomes. J. Cell Biol. 149:969–982. 10.1083/jcb.149.4.969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart R.L., and O’Connor K.L.. 2015. Clinical significance of the integrin α6β4 in human malignancies. Lab. Invest. 95:976–986. 10.1038/labinvest.2015.82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stricker J., Aratyn-Schaus Y., Oakes P.W., and Gardel M.L.. 2011. Spatiotemporal constraints on the force-dependent growth of focal adhesions. Biophys. J. 100:2883–2893. 10.1016/j.bpj.2011.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkina T.M., Verkhovsky A.B., McQuade K.M., and Borisy G.G.. 1997. Analysis of the actin-myosin II system in fish epidermal keratocytes: mechanism of cell body translocation. J. Cell Biol. 139:397–415. 10.1083/jcb.139.2.397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J.L., Tien J., Pirone D.M., Gray D.S., Bhadriraju K., and Chen C.S.. 2003. Cells lying on a bed of microneedles: an approach to isolate mechanical force. Proc. Natl. Acad. Sci. USA. 100:1484–1489. 10.1073/pnas.0235407100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D.D., and Gerlach B.D.. 2017. The roles and regulation of the actin cytoskeleton, intermediate filaments and microtubules in smooth muscle cell migration. Respir. Res. 18:54 10.1186/s12931-017-0544-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- te Molder L., Juksar J., Harkes R., Wang W., Kreft M., and Sonnenberg A.. 2019. Tetraspanin CD151 and integrin α3β1 contribute to the stabilization of integrin α6β4-containing cell-matrix adhesions. J. Cell Sci. 132:jcs235366 10.1242/jcs.235366 [DOI] [PubMed] [Google Scholar]
- Tsuruta D., Hashimoto T., Hamill K.J., and Jones J.C.R.. 2011. Hemidesmosomes and focal contact proteins: functions and cross-talk in keratinocytes, bullous diseases and wound healing. J. Dermatol. Sci. 62:1–7. 10.1016/j.jdermsci.2011.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M.Y., Geiger T., Mann M., and Cox J.. 2016. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods. 13:731–740. 10.1038/nmeth.3901 [DOI] [PubMed] [Google Scholar]
- Uematsu J., Nishizawa Y., Sonnenberg A., and Owaribe K.. 1994. Demonstration of type II hemidesmosomes in a mammary gland epithelial cell line, BMGE-H. J. Biochem. 115:469–476. 10.1093/oxfordjournals.jbchem.a124361 [DOI] [PubMed] [Google Scholar]
- Underwood R.A., Carter W.G., Usui M.L., and Olerud J.E.. 2009. Ultrastructural localization of integrin subunits β4 and α3 within the migrating epithelial tongue of in vivo human wounds. J. Histochem. Cytochem. 57:123–142. 10.1369/jhc.2008.952176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Dries K., Meddens M.B.M., de Keijzer S., Shekhar S., Subramaniam V., Figdor C.G., and Cambi A.. 2013. Interplay between myosin IIA-mediated contractility and actin network integrity orchestrates podosome composition and oscillations. Nat. Commun. 4:1412 10.1038/ncomms2402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Helvert S., Storm C., and Friedl P.. 2018. Mechanoreciprocity in cell migration. Nat. Cell Biol. 20:8–20. 10.1038/s41556-017-0012-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoorn H., Harkes R., Spiesz E.M., Storm C., van Noort D., Ladoux B., and Schmidt T.. 2014. The nanoscale architecture of force-bearing focal adhesions. Nano Lett. 14:4257–4262. 10.1021/nl5008773 [DOI] [PubMed] [Google Scholar]
- Walko G., Castañón M.J., and Wiche G.. 2015. Molecular architecture and function of the hemidesmosome. Cell Tissue Res. 360:529–544. 10.1007/s00441-015-2216-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zack G.W., Rogers W.E., and Latt S.A.. 1977. Automatic measurement of sister chromatid exchange frequency. J. Histochem. Cytochem. 25:741–753. 10.1177/25.7.70454 [DOI] [PubMed] [Google Scholar]
- Zhao T., Zhang Y., Wei Q., Shi X., Zhao P., Chen L.-Q., and Zhang S.. 2018. Active cell-matrix coupling regulates cellular force landscapes of cohesive epithelial monolayers. NPJ Computational Materials. 4:10 10.1038/s41524-018-0069-8 [DOI] [Google Scholar]
- Zuidema A., Wang W., Kreft M., te Molder L., Hoekman L., Bleijerveld O.B., Nahidiazar L., Janssen H., and Sonnenberg A.. 2018. Mechanisms of integrin αVβ5 clustering in flat clathrin lattices. J. Cell Sci. 131:jcs221317 10.1242/jcs.221317 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists primary antibodies.
shows the mass spectrometry data for the proximity interactors of the integrin β4 analysis.