Ma and Starr highlight new work from Rahman et al. that reveals new insight into the mechanisms controlling embryonic pronuclei fusion.
Abstract
The mechanisms that control how the two parental pronuclei fuse in the first mitosis of the embryo are poorly understood. In this issue, Rahman et al. (2020. J. Cell Biol. https://doi.org/10.1083/jcb.201909137) found that membrane fusion between pronuclear envelopes, followed by fenestration, promotes pronuclear fusion.
The meeting of the two pronuclei before the first zygotic mitosis is a key step in development. Pronuclear fusion and the first mitotic event are complicated by the double membrane surrounding each pronucleus and the fact that in metazoans, nuclear envelope breakdown (NEBD) must occur for microtubules to access chromosomes (1, 2). The mechanisms promoting nuclear envelope fusion and the subsequent joining of two parental pronuclei in the first mitosis of the embryo are poorly understood. Pronuclear fusion occurs only once, in the middle of a large zygote. Moreover, the behavior of the four nuclear membranes between the two pronuclei are not resolvable using light microscopy, limiting studies to EM approaches. Electron tomography in budding yeast determined that, first, the perinuclear lumen fuses to resolve the four membranes into two. Then, the two remaining nuclear membranes fuse to create a single nucleoplasm (3). Similar intermediates of pronuclear fusion have been observed in algae and sea urchin embryos where four nuclear membranes are resolved to two, before nucleoplasmic fusion (4, 5, 6). Unfortunately, these EM images were thin sections, limiting the analysis. Nonetheless, nuclear membrane fusion before NEBD appears to be conserved. However, the mechanisms promoting fusion of four pronuclear membranes into two and subsequent pronuclear fusion have not yet been fully explored.
To better understand how pronuclei fuse, in this issue, Rahman et al. (7) used focused ion beam–scanning electron microscopy (FIB-SEM) in Caenorhabditis elegans zygotes, to study pronuclear fusion. The FIB-SEM approach allowed Rahman et al. (7) to generate three-dimensional reconstructions of nuclear membranes from both wild-type and mutant embryos at prophase, prometaphase, and metaphase. These images confirmed early EM findings and revealed other novel observations leading to a new, multistep model for pronuclear fusion.
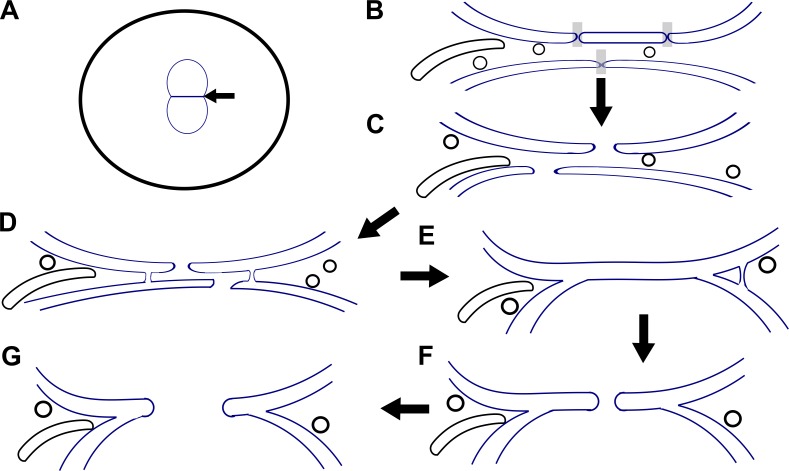
First, in prophase and prometaphase embryos, fenestrations throughout the nuclear envelope were about the size of nuclear pores (Fig. 1 B). However, two population sizes of fenestrations were observed at metaphase. Most were about the size of disassembled nuclear pores, but some were much larger (Fig. 1 C). Since they did not align at the pronuclear interface, they are likely not the key to pronuclear fusion. In contrast, during prometaphase, Rahman et al. (7) characterized several interesting structures at the pronuclear interface. They observed an abundance of ER sheet–like membrane fragments and membrane vesicles closely associated with pronuclear membranes (Fig. 1 C). These could represent cytoplasmic structures that were trapped between the two parental pronuclei. They also observed elongated amorphous densities, which could be precursors to the fused outer–outer pronuclear membranes junctions they also observed. The fused outer membranes appear to be the first step of pronuclear fusion (Fig. 1 D). Later on, by metaphase, the four membranes at prometaphase (two outer and two inner pronuclear membranes) transitioned to only two membranes at the pronuclear interface (Fig. 1 E). Once the four membranes become two, then a fenestration can form to allow the metaphase chromosomes to mix (Fig. 1, F and G). This was further supported by live fluorescent imaging, when a gap in nuclear envelope components was seen at the metaphase interface. Thus, membrane fusion events, not NEBD, drives the two pronuclei together before the first mitosis.
Figure 1.
A model for pronuclear fusion in the one-cell C. elegans embryo. (A) A cartoon of two pronuclei in the post-fertilized C. elegans embryo (arrow indicates the pronuclear interface). (B) The pronuclear interface at prometaphase, with gray boxes indicating nuclear pore complex–associated pores. ER-like sheet membrane fragments and circular vesicles are also drawn. (C) The pronuclear interface at metaphase, with the enlarged fenestrations represented. (D) The pronuclear interface further along metaphase, with outer–outer junctions forming. (E) The formation of a three-way junction, facilitating the transition from four membranes into two. (F) After the formation of a three-way junction, an enlarged fenestration forms to facilitate interaction between the two parental chromosomes. (G) The fenestration is enlarged, and mitosis can proceed.
Once it was established that the membranes of the two pronuclei fuse, Rahman et al. (7) aimed to determine how four pronuclear membranes were resolved into two. The group found two types of structures that are likely intermediates—outer–outer junctions and three-way sheet junctions. The outer–outer junctions, formed by the two outer-membranes fusing, were found on the periphery of the pronuclear interface of metaphase embryos at places close to where four membranes transition into two (Fig. 1, D and E). This location is consistent with a hypothesis that the outer–outer nuclear membrane fusions represent an initial step of pronuclear fusion. In contrast, three-way sheet junctions encapsulate the two-membrane pronuclear interface at the intersections between two and four membrane regions. Rahman et al. (7) propose two theories for the formation of these three-way sheet junctions. In the first, outer–outer junctions grow and are resolved into three-way sheet junctions formed by the inner and outer nuclear membranes of one pronucleus with the outer membrane of the other pronucleus. The second theory is similar to yeast karyogamy, where the interface membrane is made by the fusion of the two outer membranes. They tested these theories by observing the localization of the inner and outer nuclear membrane proteins, SUN-1 and ZYG-12 (8). They found that both were present at the metaphase interface, suggesting that the interface is formed by outer and inner nuclear membranes, as predicted in the first model. Given the presence of membrane junctions, it was uncertain whether these nuclear membrane proteins were exclusively localized at their original inner and outer nuclear membranes. Perhaps the distinction of whether the two membranes represent an inner and outer membrane of a single pronucleus or the inner membranes of two pronuclei is moot. Given the number of fenestrations, outer–outer fusions, and three-way junctions, the membrane networks are likely mixed and no longer maintain their identity as inner or outer nuclear membranes.
Rahman et al. (7) also examined pronuclear membranes in mutant embryos. It was previously shown that plk-1 is necessary for the mixture of parental genomes. In plk-1 mutants, pronuclei fail to fuse, but still undergo mitosis. In the resulting two-cell embryo, each cell has two paired nuclei, one with the maternal and one with the paternal genome (9). Here, Rahman et al. examined plk-1 mutants at metaphase using FIB-SEM. Despite these embryos being highly fenestrated, only a few of the holes connected the two pronuclei, and these fenestrations were significantly smaller than those found in wild-type embryos. Most significantly, no membrane junctions were detected in the plk-1 mutants at metaphase. Thus, plk-1 is necessary for pronuclear fusion events. Future studies are required to identify the targets of plk-1 that mediate pronuclear membrane fusion.
Overall, Rahman et al. (7) have successfully used FIB-SEM to uncover novel three-way sheet junctions between the membranes of pronuclear envelopes. This implies a novel mechanism in which membrane fusion, and not NEBD, drives pronuclear fusion before the first mitosis. Since similar two-membrane interfaces between pronuclei have been hinted at in budding yeast, sea urchin, and algal systems (4, 5, 6), the findings described here from C. elegans are likely conserved. Thus, Rahman et al. (7) have made a significant advance in delineating the mechanisms of pronuclear fusion in the presence of persisting membranes. Further mechanistic understanding of this new paradigm may inform studies on aneuploidy and fertilization.
Acknowledgments
Studies in the Starr lab and L. Ma are supported by the National Institutes of Health/National Institute of General Medical Sciences (grant R01 GM073874 to D.A. Starr and T32 GM007377 to L. Ma).
The authors declare no competing financial interests.
References
- 1.Bajer A., and Molè-Bajer J. Chromosoma. 1969 doi: 10.1007/BF00325682. [DOI] [Google Scholar]
- 2.Ungricht R., and Kutay U. Nat. Rev. Mol. Cell Biol. 2017 doi: 10.1038/nrm.2016.153. [DOI] [PubMed] [Google Scholar]
- 3.Melloy P., et al. J. Cell Biol. 2007 doi: 10.1083/jcb.200706151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Longo F.J. Biol. Reprod. 1973 doi: 10.1093/biolreprod/9.2.149. [DOI] [Google Scholar]
- 5.Urban P. J. Cell Biol. 1969 doi: 10.1083/jcb.42.2.606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Longo F.J., and Anderson E. J. Cell Biol. 1968 doi: 10.1083/jcb.39.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rahman M., et al. J. Cell Biol. 2020 doi: 10.1083/jcb.201909137. [DOI] [Google Scholar]
- 8.Bone C.R., and Starr D.A. J. Cell Sci. 2016 doi: 10.1242/jcs.179788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rahman M.M., et al. Mol. Biol. Cell. 2015 doi: 10.1091/mbc.E15-04-0244. [DOI] [PMC free article] [PubMed] [Google Scholar]