Escape of large macromolecular complexes induces mechanical stress, which may threaten ER membrane integrity. How the ER responds to this threat remains unknown. Chen et al. demonstrate that the ER morphogenic protein reticulon (RTN) protects ER membrane integrity during ER escape of these complexes.
Abstract
Escape of large macromolecular complexes from the endoplasmic reticulum (ER), such as a viral particle or cellular aggregate, likely induces mechanical stress initiated on the luminal side of the ER membrane, which may threaten its integrity. How the ER responds to this threat remains unknown. Here we demonstrate that the cytosolic leaflet ER morphogenic protein reticulon (RTN) protects ER membrane integrity when polyomavirus SV40 escapes the ER to reach the cytosol en route to infection. SV40 coopts an intrinsic RTN function, as we also found that RTN prevents membrane damage during ER escape of a misfolded proinsulin aggregate destined for lysosomal degradation via ER-phagy. Our studies reveal that although ER membrane integrity may be threatened during ER escape of large macromolecular protein complexes, the action of RTN counters this, presumably by deploying its curvature-inducing activity to provide membrane flexibility and stability to limit mechanical stress imposed on the ER membrane.
Introduction
The ER supports protein biosynthesis in the secretory pathway (Rapoport, 2007, 2008; Wickner and Schekman, 2005). Upon translocation into the ER, newly synthesized proteins fold before export. However, in the event of irremediable misfolding, proteins may be retro-translocated to the cytosol for ER-associated proteasomal degradation (Brodsky, 2012; Brodsky and Skach, 2011; Olzmann et al., 2013), or, in the case of high-molecular-weight protein aggregates, may be targeted to lysosomal degradation through ER-coupled autophagy (ER-phagy; Bernales et al., 2007; Wilkinson, 2019). For instance, misfolded Akita mutant proinsulin in the ER may generate both soluble forms targeted for ER-associated proteasomal degradation and detergent-insoluble aggregates cleared by ER-phagy (Cunningham et al., 2017, 2019).
Intriguingly, elements of these ER-dependent quality control pathways can be hijacked by viruses to promote infection. This is perhaps most apparent during infection by the polyomavirus family (Bernacchi et al., 2004; Chen et al., 2019; Dupzyk and Tsai, 2016; Mayberry et al., 2017; Spooner et al., 2006; Tsai and Qian, 2010). Structurally, the archetype polyomavirus SV40 is composed of 72 pentamers of the structural protein VP1 that encloses its 5-kbp DNA genome, with each pentamer containing an internal hydrophobic protein VP2 or VP3 (Liddington et al., 1991; Stehle et al., 1996). Each properly assembled viral particle has a diameter of 45 nm and a molecular weight of ∼20,000 kD. During entry, SV40 traffics from the plasma membrane to the ER (Kartenbeck et al., 1989; Norkin et al., 2002; Tsai and Qian, 2010) and then escapes from this compartment to the cytosol (Geiger et al., 2011; Inoue and Tsai, 2011; Schelhaas et al., 2007), avoids proteasomal degradation, and enters the nucleus to cause infection (Nakanishi et al., 1996, 2007).
Due to their significant size, ER escape of either SV40 particles (to cytosol) or Akita proinsulin aggregates (to lysosomes) likely imposes considerable mechanical stress on the ER membrane. Whether the ER membrane harbors intrinsic mechanisms to protect itself is unknown. Here we demonstrate that the reticulon (RTN) preserves ER membrane integrity during ER escape of SV40 and aggregated Akita proinsulin. RTNs are a highly conserved eukaryotic protein family whose distinguishing feature is the presence of the RTN homology domain (RHD; Shemesh et al., 2014; Shibata et al., 2010; Voeltz et al., 2006; Yang and Strittmatter, 2007). This domain, located at the C terminus of the protein, is composed of two short hairpin structures. Functionally, these proteins are known for their roles in forming and stabilizing tubular ER structures. Accordingly, we propose that RTN protects the integrity of the ER membrane via its curvature-inducing activity, providing flexibility and stability to the ER membrane to withstand mechanical stress imposed by large macromolecular protein complexes.
Results
RTN4 binds to the ER membrane J proteins B12, B14, and C18
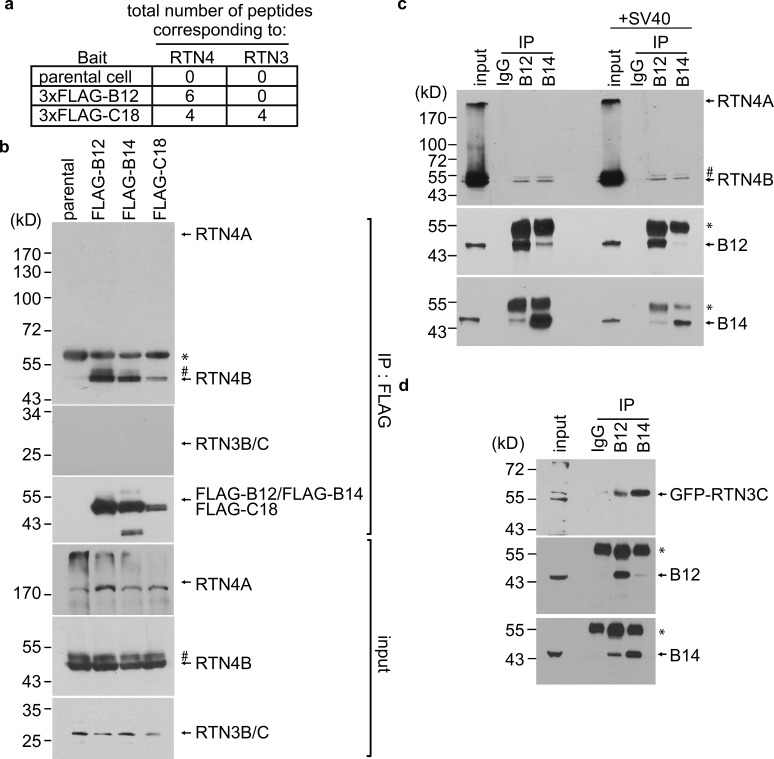
To escape from the ER to the cytosol (a decisive SV40 infection step), virions induce the formation of sub-organelle structures in the ER membrane called “foci” from which the virus emerges (Bagchi et al., 2015; Chen et al., 2019; Ravindran et al., 2015; Walczak et al., 2014). Formation of foci involves the recruitment of key ER transmembrane proteins including the J proteins B12, B14, and C18 (Goodwin et al., 2014; Inoue and Tsai, 2017; Ravindran et al., 2015; Walczak et al., 2014). Using Flp-In 293T-Rex cells stably expressing 3xFLAG-tagged B12 (3xFLAG-B12) or C18 (3xFLAG-C18), our previous immunoprecipitation (IP)–mass spectrometry (MS) analysis identified select ER membrane components (Bagchi et al., 2016, 2015; Inoue and Tsai, 2017) and a cytosolic extraction complex (Hsc70, SGTA, Hsp105, and Bag2; Dupzyk and Tsai, 2018; Dupzyk et al., 2017; Kampinga and Craig, 2010; Ravindran et al., 2015; Walczak et al., 2014) as J protein–binding partners that coordinately eject SV40 from the ER into the cytosol.
Importantly, in the same MS analysis, peptides corresponding to the ER membrane protein RTN4 were present in the 3xFLAG-B12 precipitated material, while peptides corresponding to RTN4 and RTN3 were identified in the 3xFLAG-C18 precipitated samples (Fig. 1 a; Bagchi et al., 2015; Inoue and Tsai, 2017); no peptides matching RTN3 or RTN4 were found in precipitated material derived from parental cells not expressing FLAG-tagged protein. To confirm that B12 and C18, as well as B14, associate with RTN3 or RTN4, anti-FLAG precipitations from parental cells and cells expressing 3xFLAG-B12, 3xFLAG-B14, or 3xFLAG-C18 were analyzed by SDS-PAGE and immunoblotting, with the precipitated material found to contain endogenous RTN4B (Fig. 1 b). In contrast, we could detect coprecipitation of neither RTN4A (which bears a longer N-terminal cytosolic domain than RTN4B) nor RTN3B/C forms of RTN3 (these forms lack a cytosolic region), whereas RTN3A was altogether undetected by our antibodies. Importantly, endogenous B12 or B14 coimmunoprecipitated RTN4B in uninfected cells (Fig. 1 c), indicating that these interactions require neither SV40 infection nor overexpression. Although endogenous RTN3C was not detected when the FLAG-tagged J proteins were precipitated, transfected GFP-RTN3C did coimmunoprecipitate with endogenous B12 and B14 (Fig. 1 d), suggesting that RTN3C interacts with the J proteins. Altogether, these findings support that the RTNs associate with the ER membrane J proteins B12, B14, and C18.
Figure 1.
RTN4 binds to the ER membrane J proteins B12, B14, and C18. (a) Total number of peptides corresponding to RTN3 and RTN4 identified by MS using the FLAG immunoprecipitated material derived from either Flp-In 293T-Rex parental cells or cells stably expressing 3xFLAG-B12 or 3xFLAG-C18. (b) 3XFLAG-B12, 3XFLAG-B14, or 3XFLAG-C18 was immunoprecipitated from induced Flp-In 293T-REx cells. Extracts from the parental cells were used as a negative control. Bound proteins were eluted by 3× FLAG peptide and subjected to SDS-PAGE, followed by immunoblotting using the indicated antibodies. The asterisks in panels b and c indicate an antibody heavy chain, and the number signs in panels b and c might be RTN4D. (c) Cell extracts from mock-infected (left panel) or SV40-infected (right panel) CV-1 cells were incubated with either anti-B12, anti-B14, or a control IgG antibody. The immunoprecipitated materials were subjected to SDS-PAGE, followed by immunoblotting using the indicated antibodies. (d) COS-7 cells were transfected with GFP-RTN3C, and the cell extracts were subjected to immunoprecipitation by anti-B12, anti-B14, or a control IgG antibody. The immunoprecipitated materials were subjected to SDS-PAGE, followed by immunoblotting using the indicated antibodies. Input represents 1% of the total sample used for the immunoprecipitation.
RTN3 and RTN4 promote SV40 infection
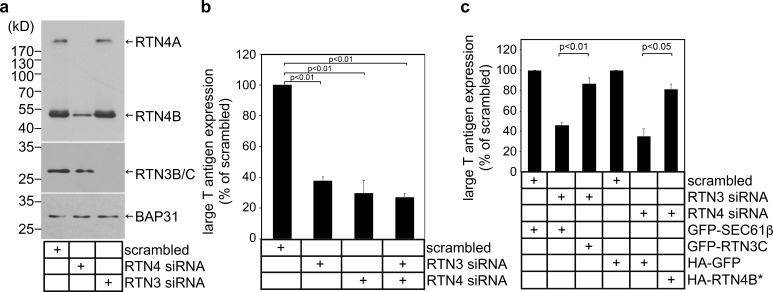
To test whether RTN4 and RTN3 might promote SV40 infection, we used simian CV-1 cells (used in classical studies of SV40) and performed siRNA knockdown to acutely deplete RTN4A and RTN4B (with a RTN4 siRNA; Fig. 2 a, lane 2) or RTN3B/C (with a RTN3 siRNA; lane 3; note that RTN3A was undetectable in these cells using two commercially available RTN3 antibodies). Under these conditions, we monitored virus infection by assessing expression of the virally encoded large T antigen, which occurs only upon successful arrival of SV40 into the host nucleus. Indeed, when compared with cells transfected with a scrambled siRNA, large T antigen expression was significantly impaired in cells with knockdown of either RTN3 or RTN4 (Fig. 2 b; simultaneous depletion of RTN3 and RTN4 led to a slightly greater impairment of virus infection when compared with single knockdown conditions). These data suggest that RTN function plays an important role during SV40 infection.
Figure 2.
RTN3 and RTN4 promote SV40 infection. (a) siRNA knockdown of RTN3 and RTN4. Cell extracts derived from CV-1 cells transfected with the indicated siRNA were subjected to SDS-PAGE and immunoblotting with the indicated antibodies. (b) CV-1 cells transfected with the indicated siRNAs were infected with SV40. 24 h after infection, cells were permeabilized, fixed, and stained for large T antigen. At least 1,000 cells were counted per condition over three biological replicates. (c) As in b, except cells were transfected with the indicated constructs 24 h before infection with SV40. Cells were then fixed, permeabilized, and stained for GFP/HA and large T antigen. Only cells expressing the GFP/HA construct were counted. At least 100 cells were counted per condition over three biological replicates. The graph represents the means ± SD. Student’s two-tailed t test was used to determine statistical significance.
To begin to verify the specificity of the RTN3 and RTN4 siRNAs, cells transfected with RTN3 siRNA were cotransfected with either a mouse GFP-RTN3C plasmid (resistant to the RTN3 siRNA) or a GFP-Sec61β control plasmid. We found that expressing GFP-RTN3C in RTN3 knockdown cells largely restored large T antigen expression, indicative of successful SV40 infection (Fig. 2 c). Further, when cells transfected with RTN4 siRNA that were cotransfected with HA-RTN4B* (resistant to the RTN4 siRNA), this too rescued large T antigen expression (Fig. 2 c). These results demonstrate that impaired SV40 infection following transfection with RTN3 or RTN4 siRNA was due to specific depletion of RTN3 or RTN4 rather than unintended off-target effects. They further support that RTN3 and RTN4 execute critical functions during SV40 infection.
RTN3 and RTN4 are recruited to viral foci to enable SV40 engagement with the cytosolic extraction machinery
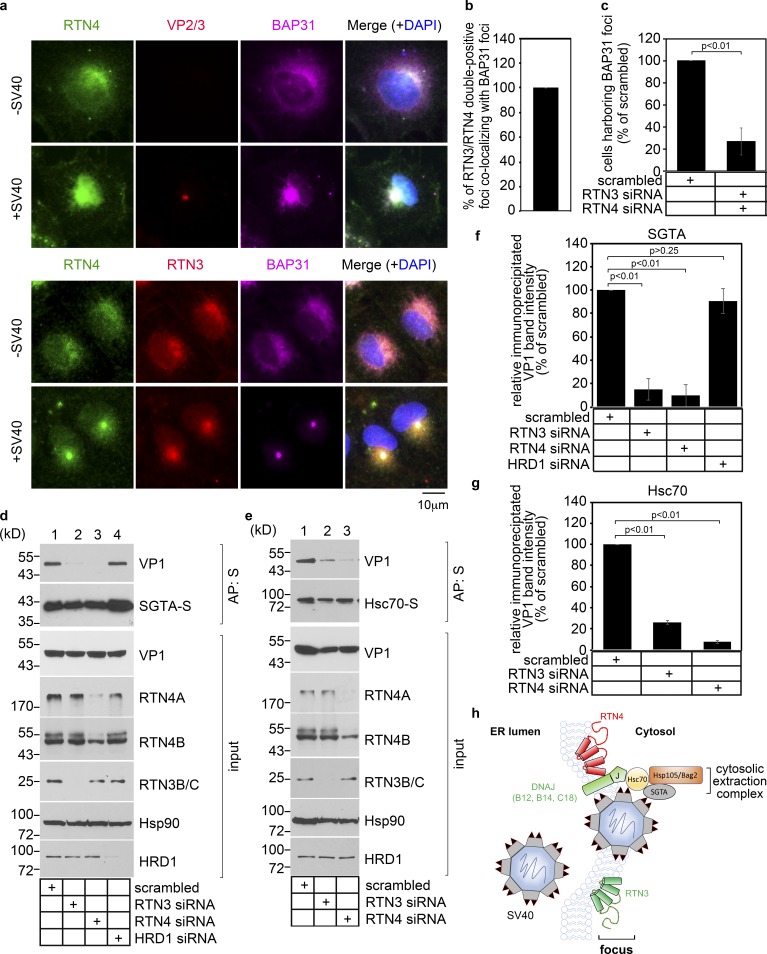
How might RTN3 and RTN4 promote SV40 infection? Formation of viral foci involves the recruitment of many ER membrane proteins, including the transmembrane protein BAP31 (Geiger et al., 2011). Not surprisingly, the membrane-penetrating form of SV40, in which the hydrophobic VP2 and VP3 of SV40 are exposed, is also concentrated at ER foci (Bagchi et al., 2015; Ravindran et al., 2017). We therefore hypothesize that RTN3 and RTN4 are similarly recruited to ER foci. To test this, CV-1 cells with or without SV40 exposure were stained for VP2/3, BAP31, RTN3, and RTN4. Strikingly, in infected cells, RTN4 was recruited to VP2/3-BAP31 double-positive foci (Fig. 3 a, second row), whereas in uninfected cells, RTN4 exhibited the typical diffuse ER pattern along with BAP31 (Fig. 3 a, first row). Likewise, staining for RTN3 revealed that it is recruited to BAP31-RTN4 double-positive foci in infected cells (Fig. 3 a, compare fourth to third row). Quantification revealed that essentially all RTN3 and RTN4 double-positive foci are also positive for the BAP31 foci (Fig. 3 b). Thus, SV40 triggers relocation of RTN3 and RTN4 to viral foci during infection. Functionally, recruitment of RTN3 and RTN4 to the foci is critical to support or stabilize these structures, as depletion of RTN3 and RTN4 robustly decreased the number of cells containing the BAP31 foci (Fig. 3 c).
Figure 3.
RTN3 and RTN4 are recruited to viral foci to enable SV40 engagement with the cytosolic extraction machinery. (a) CV-1 cells uninfected or infected with SV40 for 16 h were fixed, stained with the indicated antibodies, and processed by epifluorescence microscopy. (b) Quantification of the percentage of RTN3/RTN4 double-positive foci colocalizing with BAP31 foci. Values represent means ± SD from three independent experiments. (c) Quantification of the percentage of RTN3/RTN4 double knockdown cells harboring BAP31 foci after SV40 infection. Cells were scored positive if at least one focus was present in the cell. Values represent means ± SD from three independent experiments. (d and e) COS-7 cells were treated with the indicated siRNAs for 24 h, followed by transfection of the SGTA-S (d) or Hsc70-S (e) construct. After 24 h, cells were infected with SV40 for 16 h, and the resulting cell extracts were subjected to affinity purification (AP), SDS-PAGE, and immunoblotting with the indicated antibodies. Input represents 5% of the total sample used for the affinity purification. (f and g) The VP1 band intensities from d and e were quantified with ImageJ software, normalized relative to the precipitated SGTA-S or Hsc70-S level, and expressed as a percentage of the VP1 band intensity in the scrambled siRNA-treated sample. The graph represents the means and SDs from three independent experiments. Student’s two-tailed t test was used to determine statistical significance. (h) A model to summarize the results in this figure. During ER-to-cytosol membrane transport of SV40, RTN3 and RTN4 are recruited to the SV40-induced focus to enable virus engagement with the cytosolic extraction machinery (composed of Hsc70, Hsp105, Bag2, and SGTA).
SV40 foci engage a cytosolic machinery (composed of Hsc70, SGTA, Hsp105, and Bag2) that extracts the virus from the ER into the cytosol, directing it along a productive pathway to the nucleus to cause infection (Dupzyk and Tsai, 2018; Dupzyk et al., 2017; Ravindran et al., 2015). Remarkably, we observed that upon RTN3 or RTN4 knockdown, virion interaction with SGTA (transfected S-tagged SGTA-S) was blocked, whereas knockdown of the unrelated Hrd1 ER membrane protein was without effect (Fig. 3 d, top panel; the VP1 level is quantified in Fig. 3 f). Similarly, upon RTN3 or RTN4 knockdown, SV40 could not efficiently associate with Hsc70 (transfected Hsc70-S; Fig. 3 e, top panel; the VP1 level is quantified in Fig. 3 g). These findings suggest that RTN3 and RTN4 recruitment to viral foci enable successful engagement of SV40 with the cytosol extraction machinery, as depicted in Fig. 3 h.
RTN3 and RTN4 are essential to protect ER membrane integrity during SV40 infection
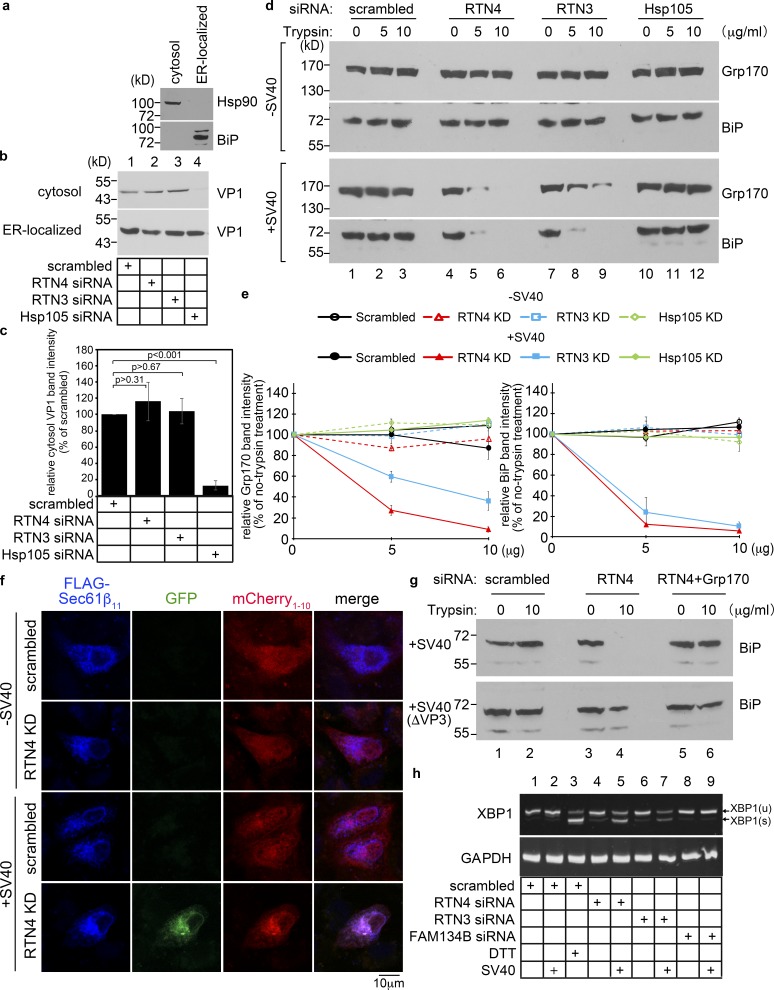
We reasoned that if RTN3 and RTN4 are required for SV40 to engage the cytosolic extraction complex (Fig. 3) in order to reach the nucleus (Fig. 2), SV40 emergence from the ER into the cytosol should be impaired under RTN3 or RTN4 knockdown. To test this, we used an established assay of semi-permeabilized cells (Dupzyk and Tsai, 2018; Dupzyk et al., 2017; Geiger et al., 2011; Inoue and Tsai, 2011). In this assay, virus-infected cells were harvested and incubated with a low concentration of digitonin to permeabilize the plasma membrane without affecting internal membranes. Cells were then subjected to centrifugation to produce a supernatant fraction (“cytosol” fraction) harboring cytosolic proteins and cytosolic virus, and a pellet fraction that contains membranes including the ER, as well as membrane-associated virus. Further fractionation of the membrane fraction isolates an “ER-localized” SV40 pool. Validity of this fractionation protocol is supported by the appearance of cytosolic Hsp90 in the cytosolic fraction and the ER-resident BiP in the ER-localized fraction (Fig. 4 a). Using this assay, we found that whereas depleting Hsp105 (a cytosol extraction machinery component) blocked appearance of SV40 VP1 in the cytosol (Fig. 4 b, lane 4; quantified in Fig. 4 c) as reported (Dupzyk and Tsai, 2018; Ravindran et al., 2015), depleting RTN3 or RTN4 did not (Fig. 4 b, lanes 2 and 3). Hence, unexpectedly, knockdown of RTN3 or RTN4 did not block SV40 arrival to the cytosol from the ER, even though SV40 infection was markedly impaired when these proteins were depleted.
Figure 4.
RTN3 and RTN4 are essential to protect ER membrane integrity during SV40 infection. (a) CV-1 cells were semi-permeabilized with digitonin and centrifuged to generate the supernatant (cytosol) and pellet (membrane) fractions; material in the membrane fraction was further extracted by Triton X-100 to isolate ER-localized SV40. The cytosol and ER-localized fractions were analyzed for the presence of the cytosolic Hsp90 and the ER-resident BiP proteins. This procedure represents the ER-to-cytosol transport assay. (b) Cells transfected with the indicated siRNA were infected with SV40 for 16 h, and subjected to the ER-to-cytosol transport assay described in a. Both the cytosol and ER-localized fractions were analyzed by SDS-PAGE and immunoblotted with the indicated antibodies. (c) The intensity of the VP1 band in b was quantified by ImageJ using scans of films after ECL. The results represent the mean of three independent experiments. A two-tailed t test was used. Error bars, ± SD. (d) Protease protection assay to monitor the integrity of the ER membrane. CV-1 cells treated with the indicated siRNAs for 48 h were infected with SV40 for 16 h. Cells were subjected to semi-permeabilization as in panel a to generate the membrane fraction. This fraction (containing the ER membrane) was resolubilized in PBS with the indicated trypsin concentration. The sample was TCA-precipitated, and the precipitated material subjected to SDS-PAGE and immunoblotted with the ER luminal Grp170 and BiP antibodies. (e) Quantification of the percentage of Grp170 and BiP band intensity in d. Values represent means ± SDs from three independent experiments. (f) Split-GFP method to probe for the integrity of the ER membrane. COS-7 cells were treated with the indicated siRNAs for 24 h followed by cotransfection with FLAG-tagged Sec61β11 and cytosolic mCherry1-10. 24 h after transfection, cells mock-infected or infected with SV40 (MOI ∼100) for 16 h were fixed, stained with anti-FLAG antibody, and analyzed by a Zeiss LSM 800 confocal microscope. Scale bars, 10 µm. (g) CV-1 cells treated with the indicated siRNAs for 48 h were infected with 10 µg of WT SV40 or SV40(ΔVP3) for 16 h. Cells were subjected to the protease protection assay as in d. Cell extracts were collected, subjected to SDS-PAGE, and immunoblotted with an antibody against BiP. (h) CV-1 cells transfected with the indicated siRNAs for 2 d were mock infected or infected with SV40 (MOI ∼50) for 16 h. RNA was extracted and reverse-transcribed to cDNA, and the cDNA was amplified using PCR to identify splicing of the XBP1 mRNA. Cells treated with DTT served as a positive control.
Because SV40 cannot engage the cytosol extraction machinery under RTN3 or RTN4 knockdown yet can reach the cytosol despite the depleted conditions, we postulated that the integrity of the ER membrane is compromised when RTN3 or RTN4 is depleted during SV40 infection. If so, membrane stress caused by the virus could lead to a localized site of ER membrane discontinuity, with viral particles diffusing into the cytosol en route to a nonproductive fate. To probe if the ER membrane is compromised during SV40 infection, we used two distinct assays. First, we used the classic protease protection assay to monitor ER membrane integrity. We envisioned that if the ER membrane is compromised, adding a general protease such as trypsin should degrade luminally localized ER factors such as Grp170 and BiP. Indeed, whereas Grp170 and BiP remained resistant to degradation in the presence of increasing concentrations of trypsin in noninfected cells depleted of either RTN4 or RTN3 (or as a control, Hsp105 [Fig. 4 d, upper two panels; quantified in Fig. 4 e]), these luminal proteins became profoundly sensitive to proteolysis in SV40-infected cells depleted of RTN4 or RTN3 (but not Hsp105 [Fig. 4 d, lower two panels]). These data suggest that the integrity of the ER membrane is compromised in SV40-infected cells deficient for RTN3 or RTN4. Although Hsp105 depletion leads to accumulation of SV40 at ER foci (Dupzyk and Tsai, 2018; Ravindran et al., 2015), it does not cause ER membrane damage. This finding indicates that the ER membrane damage is not caused merely by accumulation of SV40 at viral foci, but rather by lack of a specific activity attributed to the morphogenic proteins RTN4 and RTN3.
Second, we used a split-GFP method to probe for the integrity of the ER membrane. In this assay, we generated an ER membrane protein Sec61β construct that contains a FLAG tag at its N terminus and β-strand 11 of GFP at its C terminus (FLAG-Sec61β11); when expressed, GFP11 of FLAG-Sec61β11 faces the ER lumen. We also engineered the cytosolic protein mCherry construct harboring β-strands 1–10 of GFP positioned at its C terminus (mCherry1-10). We reasoned that if any experimental condition damages the ER membrane in cells expressing FLAG-Sec61β11 and mCherry1-10, GFP fluorescence should be reconstituted because mCherry1-10 can now diffuse across the compromised ER membrane and reach the luminal side to bind to GFP11. Using this approach, we found that RTN4 knockdown did not generate any GFP fluorescence signal in uninfected cells, but GFP fluorescence was clearly detected in SV40-infected cells deficient of RTN4 (Fig. 4 f). In agreement with the protease protection assay, these results strongly suggest that the ER membrane was compromised in SV40-infected cells deficient for RTN4.
Is the simple presence of SV40 in the ER sufficient to induce ER membrane damage, or is the act of viral emergence from ER to cytosol required? We used two different approaches to address this. We previously showed that the ER luminal chaperone Grp170 is necessary to generate hydrophobic SV40 that binds the ER membrane at sites of viral foci, priming the virus for emergence into the cytosol (Inoue and Tsai, 2015). Knockdown of Grp170 blocks SV40 emergence from ER to cytosol without disrupting virus arrival to the ER from the cell surface (Inoue and Tsai, 2015). Using the protease protection assay, we found that in SV40-infected cells, whereas RTN4 knockdown impaired ER membrane integrity as in Fig. 4 d, additional depletion of Grp170 in RTN4 knockdown cells actually preserved ER membrane integrity (Fig. 4 g, top panel). Hence, Grp170-dependent emergence of SV40 viral particles from ER to cytosol is necessary to induce focal ER membrane discontinuity. To strengthen this idea, we used a SV40 mutant lacking VP3 (SV40[ΔVP3]); this mutant can successfully reach the ER from the cell surface but cannot induce viral foci, ER-to-cytosol viral delivery, or viral infection (Bagchi et al., 2015). Importantly, we find that the ER membrane integrity is largely preserved when cells depleted of RTN4 (or both RTN4 and Grp170) were infected with this mutant virus (Fig. 4 g, lower panel). These data support the notion that the mere arrival of SV40 virus to the ER is not sufficient to cause ER membrane damage in RTN-depleted cells, but rather the act of ER-to-cytosol membrane penetration is required.
Our data thus far demonstrate that RTN3 and RTN4 protect against membrane damage during SV40 emergence from the ER. Whether this protective mechanism subserves a normal homeostatic ER function is unknown. One major ER function is an exquisite protein quality control machinery designed to facilitate proper protein folding and to detect protein misfolding, which can trigger an ER stress response. It is likely that if ER membrane damage perturbs ER protein folding, then an ER stress response may be triggered. One sensitive assay of ER stress signaling involves Ire1-dependent XBP1 splicing (Yoshida et al., 2001). Importantly, infection of cells with SV40 did not itself trigger XBP1 splicing (Fig. 4 h, lane 2; DTT addition to cells served as a positive control in lane 3). Additionally, RTN4 and RTN3 knockdown in uninfected cells triggered only minimal XBP1 splicing (Fig. 4 h, lanes 4 and 6). However, RTN3 or RTN4 knockdown in SV40-infected cells triggered a dramatic increase of XBP1 splicing (Fig. 4 h, lanes 5 and 7). (Knockdown of another ER membrane protein, FAM134B, did not induce XBP1 splicing even in SV40 infected cells [Fig. 4 h, lane 9].) These findings strongly suggest that RTN-dependent protection of the ER membrane preserves proper ER function that can help to limit ER stress.
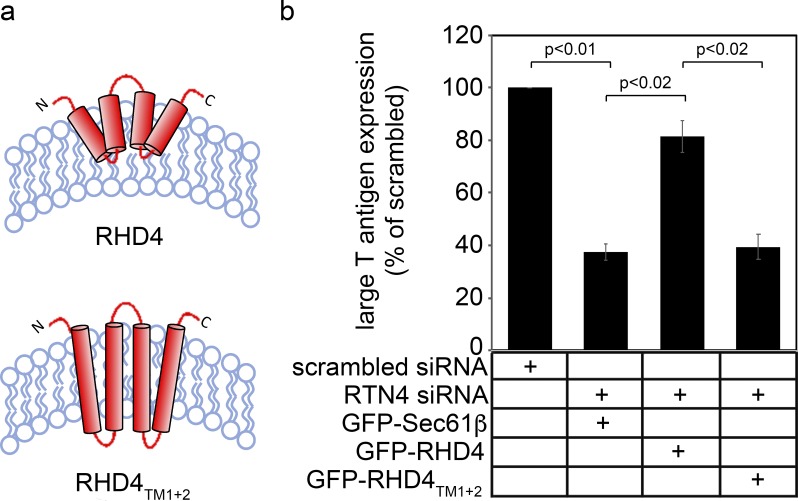
Because RTNs are well-known for their ability to generate and maintain ER membrane curvature, we posit that this activity is essential to support ER membrane integrity during SV40 infection. To test this, we used a GFP-tagged RTN4 construct that contains only its RHD and lacks the cytosolic domain (GFP-RHD4; Fig. S1 a). This construct is sufficient to constrict ER tubules and convert peripheral ER cisternae into tubules (Zurek et al., 2011). In RTN4-depleted cells, we found that expression of GFP-RHD4 restored SV40 infection (Fig. S1 b), suggesting that RHD4’s ability to induce membrane curvature is sufficient to support SV40 ER membrane penetration leading to infection. We then used a mutant construct derived from GFP-RHD4 that cannot constrict ER tubules and change peripheral ER cisternae into tubules (Zurek et al., 2011). This construct was generated by conversion of its two short hairpin RHDs into bona fide two-pass transmembrane domains that span both leaflets of the bilayer via insertion of additional amino acid residues (GFP-RHD4TM1+2, Fig. S1 a). Strikingly, in RTN4 knockdown cells, expressing GFP-RHD4TM1+2 failed to restore SV40 infection (Fig. S1 b). These results are consistent with the notion that the ability of RTNs to induce ER membrane curvature is critical to support ER escape of SV40, resulting in successful infection.
Figure S1.
A RHD4 mutant cannot support SV40 infection (related to Fig. 4). (a) Predicted membrane topology of human RHD4 and RHD4-lengthened transmembrane (TM) domain mutant RHD4TM1+2 (Zurek et al., 2011). (b) CV-1 cells transfected with the indicated siRNAs and constructs were infected with SV40. 24 h after infection, cells were permeabilized, fixed, and stained for large T antigen. At least 100 cells were counted per condition over three biological replicates. The graph represents the means ± SDs. Student’s two-tailed t test was used to determine statistical significance.
RTN3 protects ER membrane integrity during ER exit of misfolded Akita proinsulin aggregates to the lysosome for degradation
We hypothesize that the membrane-protective function of RTNs uncovered during emergence of viral particles from the ER reflects an intrinsic activity of these membrane proteins that may operate in a physiological context. To test this, we asked if RTN function preserves the integrity of the ER membrane during ER exit of another large macromolecular complex in the form of a misfolded protein aggregate. We previously demonstrated that when mutant Akita proinsulin is translocated into the ER, it can form a massive (>40,000 kD) aggregated protein complex that is delivered to lysosomes via a RTN3 (but not RTN4)-dependent ER-phagy pathway (Cunningham et al., 2019). We therefore assessed whether RTN3 protects the integrity of the ER membrane during ER-phagy of aggregated Akita proinsulin.
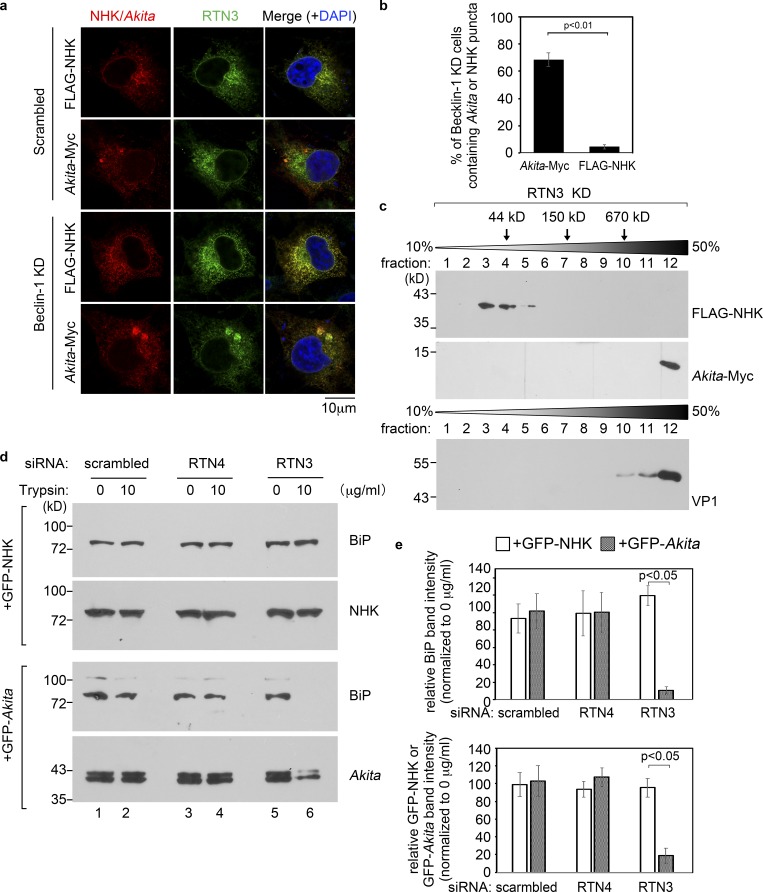
To this end, scrambled siRNA-treated cells were transfected with Myc-tagged Akita proinsulin (“Akita-Myc”) or the control FLAG-tagged null Hong Kong mutant of α-1-antitrypsin (“FLAG-NHK”), which is another protein that misfolds in the ER but does not form a massive protein complex therein (see below). In cells transfected with FLAG-NHK, immunostaining for NHK revealed a diffuse pattern that colocalizes with RTN3, indicative of ER localization (Fig. 5 a, top row), and in cells expressing Akita-Myc, immunostaining for Akita showed a similar ER localization pattern other than the occasional presence of small puncta (Fig. 5 a, second row). However, whereas the diffuse ER pattern of FLAG-NHK did not change in cells depleted of the macro-autophagy component Beclin-1 (Fig. 5 a, third row; and quantified in Fig. 5 b), Beclin-1 knockdown resulted in the formation of large Akita-positive puncta that colocalize with concentrated RTN3 (Fig. 5 a, fourth row; and quantified in Fig. 5 b); the large puncta represent Akita aggregates that can also be generated by RTN3 knockdown (Cunningham et al., 2019). These findings support the idea that, in contrast to NHK, Akita forms large aggregated complexes that are disposed of by the autophagy pathway (Cunningham et al., 2019). This notion is further supported by sucrose gradient analysis, demonstrating that in cells depleted of RTN3, Akita exists as a massive protein complex similar to purified SV40, whereas NHK fractionates as a significantly smaller protein (Fig. 5 c).
Figure 5.
RTN3 protects ER membrane integrity during ER exit of misfolded Akita proinsulin aggregates to the lysosome for degradation. (a) COS-7 cells expressing FLAG-NHK or Akita-Myc were transfected with either scrambled or Beclin-1 siRNA, fixed, permeabilized, and stained with FLAG/Myc and RTN3 antibody. Images were taken on a Zeiss LSM 800 confocal microscope and Airyscan processed. Scale bars, 10 µm. (b) Quantification of the percentage of Beclin-1 knockdown cells containing Akita-Myc or FLAG-NHK puncta. Values represent means ± SD from three independent experiments. (c) The detergent-insoluble fraction derived from RTN3-depleted COS-7 cells expressing FLAG-NHK (first panel) or Akita-Myc (second panel) were layered over a 10% to 50% discontinuous sucrose gradient and centrifuged. Each fraction was collected, subjected to SDS-PAGE, and immunoblotted with antibody against FLAG or Myc. Purified SV40 was also layered over a 10–50% sucrose gradient and centrifuged. Each fraction was collected, subjected to SDS-PAGE, and immunoblotted with the antibody against VP1 (third panel). (d) COS-7 cells expressing GFP-NHK or GFP-Akita were transfected with scrambled, RTN3, or RTN4 siRNA. Cells were then subjected to the protease protection assay as in Fig. 4 d. (e) The graph represents the intensity of the BiP and GFP-NHK/GFP-Akita band in c. The band intensity was quantified by ImageJ using scans of films after ECL. Results represent the mean of three independent experiments. A two-tailed t test was used. Error bars, ± SD.
Strikingly, using the same protease protection assay as in Fig. 4, we found that under RTN3 (but not RTN4) knockdown, BiP (and GFP-Akita) became markedly sensitive to trypsin digestion in cells expressing GFP-tagged Akita, whereas BiP (and GFP-NHK) remained completely resistant to proteolysis in cells expressing GFP-tagged NHK (Fig. 5 d, last lane; and quantified in Fig. 5 e). These results demonstrate that the integrity of the ER membrane is compromised in Akita-expressing cells when RTN3 is depleted. Because RTN3 (but not RTN4) is required during ER-phagy of aggregated Akita (Cunningham et al., 2019), our data demonstrate that in addition to physically linking to cytosolically recruited autophagy machinery, another critical role of RTN3 is to preserve ER membrane integrity during ER-phagy. This observation, coupled with the finding that RTNs are used to prevent ER membrane damage during ER escape of a virus, suggests a general function of RTNs in preserving the stability of the ER membrane during escape of macromolecular protein complexes from this organelle.
Discussion
Misfolded proteins in the ER can polymerize to form large protein aggregates. At least some of these aggregates can exit the ER for delivery to lysosomes for degradation via a recently described ER-phagy pathway (Bernales et al., 2006, 2007; Bhaskara et al., 2019; Forrester et al., 2019; Fregno et al., 2018). This process has some parallels to the emergence of polyomavirus SV40 viral particles from the ER to cytosol en route to productive infection (Bagchi et al., 2015; Dupzyk and Tsai, 2018; Dupzyk et al., 2017; Inoue and Tsai, 2015; Ravindran et al., 2015, 2017, 2018; Walczak et al., 2014). In both cases, the need to retain unusually large cargo likely imposes significant mechanical stress initiated upon the luminal leaflet of the ER membrane that may damage ER membrane continuity during the process of cargo transport. In this study, we have sought to determine whether cells may defend against such ER discontinuity by protection on the cytosolic leaflet of the ER membrane.
We have uncovered a novel mechanism of such protection. Specifically, we found that during emergence of SV40 virus from ER to cytosol, both RTN3 and RTN4 relocate to the site of viral foci (Fig. 6, top). Using the classic protease protection assay and a split-GFP strategy, we found that loss of these RTNs compromised the ER membrane integrity during the cytosolic emergence of SV40. The mere presence of SV40 in the ER is not sufficient to trigger membrane damage under RTN knockdown; the additional act of ER membrane penetration by the virus is required. Not surprisingly, under RTN knockdown conditions, SV40 cannot engage the cytosolic chaperone machinery that extracts the virus into the cytosol en route to successful infection. Instead, the viral particle nonspecifically leaks into the cytosol as a result of ER membrane discontinuity, thus leading the virus to a nonproductive fate.
Figure 6.
A model describing how RTN3 and RTN4 protect ER membrane integrity during ER escape of macromolecular protein complexes. Top: During ER exit of SV40 into the cytosol, a decisive infection step, both RTN3 and RTN4 are recruited to the ER focus. The RTN proteins are thought to provide flexibility to the ER membrane by inducing ER membrane curvature at the ER focus. This activity protects the fidelity of the ER membrane when the viral particles penetrate this barrier. Bottom: During ER exit of aggregated Akita (A), RTN3 is recruited to a site that initiates ER-phagy. Similar to SV40, we propose that RTN3 imparts flexibility to the ER membrane by triggering ER membrane curvature at the ER exit site. Once the Akita aggregates exit the ER, it is targeted to the lysosome for degradation.
Our analysis indicates that SV40 exploits an intrinsic protective function of the RTNs, as demonstrated by RTN3 protection from ER membrane damage during ER-phagy of aggregated Akita proinsulin (Fig. 6, bottom). We previously reported that when Akita aggregates are formed, a RTN3-dependent ER-phagy pathway is implemented to remove the aggregates from the ER (Cunningham et al., 2019). This agrees with our present finding that Akita forms large puncta colocalizing with concentrated RTN3 when the autophagy machinery is impaired. These puncta potentially represent the ER exit sites during ER-phagy of Akita aggregates. Using the same protease protection assay, we again observed that upon RTN3 knockdown, the integrity of the ER membrane is compromised in the presence of Akita aggregates, but this is not the case for nonaggregated misfolded ER proteins. Thus, for both SV40 virus and Akita aggregates, we propose that RTNs are recruited to the site of ER membrane stress to support its existing curved structure (a well-established function of the RTN morphogenic proteins; Shibata et al., 2010; Voeltz et al., 2006; Weller et al., 2014; Yang and Strittmatter, 2007) and protect from bilayer discontinuity. This reinforcement likely provides the ER membrane with improved stability to accommodate the mechanical stress imposed by these macromolecular complexes, thereby preserving its integrity. Not surprisingly, ER stress can induce RTN expression (Wan et al., 2007), while RTN-dependent protection against ER membrane damage preserves basic ER homeostatic function to help limit ER stress.
How might mechanical stress be generated during ER escape of these two macromolecular cargos that recruit the RTNs? SV40 exits the ER by creating membrane penetration sites called foci. Numerous viral particles, each with a diameter of 45 nm and a molecular weight of ∼20,000 kD, are recruited to the foci, where they bind to and insert into the luminal side of the ER membrane, priming them for ER membrane penetration (Bagchi et al., 2015, 2016; Dupzyk and Tsai, 2018; Dupzyk et al., 2017; Inoue and Tsai, 2011; Ravindran et al., 2015). In this scenario, accumulation of multiple SV40 particles at the ER focus likely imposes mechanical stress on the ER membrane. We hypothesize that a change in the biophysical property of the membrane recruits or entraps RTN3 and RTN4 at the site. The precise nature of these biophysical changes requires further investigation. Because both RTN3 and RTN4 are inserted only into the cytosolic leaflet of the ER membrane, we posit that their interaction with SV40 viral particles in the ER lumen is likely to be indirect and bridged by one or more other factors.
Although the events triggering ER-phagy of an aggregated cargo (including Akita proinsulin) are unclear, the present model suggests that the cargo associates with the luminal surface of the ER membrane, representing an ER-phagy exit site (Bhaskara et al., 2019; Forrester et al., 2019; Fregno et al., 2018; Grumati et al., 2017). This site also functions to recruit the phagophore membrane essential for initiating autophagy. As the phagophore membrane encases that portion of the ER harboring the aggregated cargo, the ER membrane must fragment to separate aggregated cargo from the rest of the ER. Although the phagophore membrane will seal to generate an autophagosome for ultimate lysosomal delivery and digestion, we envision that mechanical stress caused by the Akita proinsulin protein aggregate imposes a similar tension to the ER membrane as that created by the SV40 virus. For this reason, we hypothesize that this change in the biophysical property of the ER membrane may itself be a sufficient signal for RTN recruitment to the site, with RTN function needed to preserve ER membrane integrity, which can limit ER stress (Fig. 4 h) and help to support cell survival (Kanekura et al., 2015).
Whether RTNs are deployed to protect against ER membrane damage during ER escape of other large cargos is a fascinating question that deserves future investigation. For instance, ER exit of massive secretory proteins such as procollagen via distended COPII-coated membrane carriers might also require RTN function to preserve ER membrane integrity. Intriguingly, RTN3 may play an important role during ER exit of the large lipoprotein very-low-density lipoprotein before secretion (Siddiqi et al., 2018), raising the possibility that RTN3 might protect the ER membrane in this process as well. Regardless, the present findings suggest that the RTN proteins, which provide curved membrane support, are not only fundamental to the morphology of the normal ER, but also needed to functionally preserve the integrity of this organelle under conditions of membrane stress.
Materials and methods
Reagents
CV-1 (CCL-70) and COS-7 (CRL-1651) cells were purchased from ATCC. Cells were grown in complete DMEM (10% fetal bovine serum, 10 U/ml penicillin, and 10 mg/ml streptomycin; Gibco BRL). Opti-MEM and 0.25% trypsin-EDTA were purchased from Gibco. We note that all SV40 infection experiments were performed in the CV-1 cells (Fig. 2, b and c; and Fig. S1) since these cells are permissive for SV40 infection and are used classically to study SV40 entry. COS-7 cells are derived from CV-1 cells, and we use COS-7 cells because they have a much higher transfection efficiency than CV-1 cells. Hence, experiments in which high transfection efficiency is required use COS-7 cells (Fig. 1 d; Fig. 3, d and e; Fig. 4, d and f; and Fig. 5, a, c, and d). Because the stable 3xFLAG-B12–, -B14–, and -C18–expressing cells were generated in the Flp-In 293T-REx cell line, we performed the initial IP-MS analysis and FLAG-tagged B12, B14, and C18 IP experiments in these 293T cells (Fig. 1, a and b).
Chemicals and antibodies
Triton X-100, PMSF, N-ethymaleimide, and anti-FLAG M2 antibody–conjugated agarose beads were purchased from Sigma-Aldrich, S protein–conjugated beads and digitonin from EMD Millipore Chemicals, and Protein A/G–conjugated agarose beads from Thermo Fisher Scientific. The antibodies used were FLAG (Millipore Sigma-Aldrich, F1365), RTN4 (Santa Cruz Biotechnology, sc-271878), RTN3 (Boster Biologival Technology, PA2256; Bethyl Laboratories, A302-860A), B12 (Proteintech Group, 16780–1-AP), B14 (Proteintech, PTG11019-2-AP), BAP31 (Pierce, MA3-002), VP1 (W. Scott, University of Miami, Coral Gables, FL), VP2/3 (Abcam, ab53983), S (Abcam, ab18588), Hsp90 (Santa Cruz Biotechnology, sc7947), HRD1 (Protein Group, 13473-1-AP), Hsp105 (Santa Cruz Biotechnology, sc6241), BiP (Abcam, ab21685), Grp170 (Abcam, ab124884), Myc (Immunology Consultants Laboratory, RMYC-45A), and GFP (Proteintech Group, 66002).
DNA plasmid and siRNA transfection
The pcDNA3.1-GFP-Sec61β, pAc-GFP-RTN3C, and HA-RTN4B were gifts from T. Rapoport, Harvard University, Cambridge, MA. The pAc-GFP-RHD4 and pAc-GFP-RHD4TM1+2 were kindly provided by G. Voeltz, University of Colorado, Boulder, CO. The mCherry-GFP1-10 was a gift from Y. Ye, National Institutes of Health, Bethesda, MD (Addgene plasmid 78591). To generate an siRNA-resistant RTN4 construct, overlapping PCR was performed using the following primers: forward, 5′-ATCTGAGGAGTTGGTTCAGAAGTACAGTAATTCTGCTCTTGGTCATGTG-3′, and reverse, 5′-CACATGACCAAGAGCAGAATTACTGTACTTCTGAACCAACTCCTCAGAT-3′. The bold nucleotide sequences indicate the specific sequences that were altered in order to generate the siRNA-resistant construct. To generate the FLAG-Sec61β11 construct, PCR-amplified FLAG-Sec61β sequence was inserted into the KIF5C(1–560)-GFP11 plasmid (Norris et al., 2014) in which KIF5C(1–560) was removed by standard cloning methods. The RTN3 siRNA sequence is 5′-UCAGUGUCAUCAGUGUGGUUUCUUADTDT-3′ (Millipore Sigma-Aldrich), RTN4 siRNA sequence is 5′-GUUCAGAAGUACAGUAAUUDTDT-3′, and the scrambled siRNA sequence is the “all-star negative” siRNA from QIAGEN. All DNA and siRNA transfections were incubated for at least 24 h.
DNA transfection
50% confluent CV-1 cells were transfected with the indicated plasmid using the FuGENE HD (Promega) transfection reagent at a ratio of 1:4 (plasmid to transfection reagent; wt/vol). For COS-7 cells, polyethylenimine (PEI; Polysciences) was used as the transfection reagent. Cells were allowed to express the protein for 24 to 48 h before the experiments were conducted.
IP and affinity purification
For IP of FLAG-tagged proteins, HEK 293T cells stably expressing 3xFLAG-B12, 3xFLAG-B14, or 3xFLAG-C18 were seeded in 15-cm plates and induced overnight with freshly prepared 0.1–0.5 µg/ml tetracycline (Sigma-Aldrich) to express FLAG-tagged protein at a near endogenous level. The cells were lysed in 1% Deoxy Big CHAP (DBC) in a physiological buffer at 4°C for 10 min and centrifuged at 16,000 rpm at 4°C for 10 min. The supernatant was collected and rotated at 4°C for 2 h with M2 FLAG-conjugated beads. The beads were washed three times and were eluted by 3X FLAG peptide, and subjected to SDS-PAGE, followed by immunoblotting using the indicated antibodies. For IP of endogenous B12 or B14, cells were lysed, and the resulting lysate was incubated with an antibody against B12 or B14 (or a nonspecific IgG control antibody) for 2 h. The lysates were then incubated with protein A agarose beads for 1 h. The precipitated materials were processed as described above. To examine the interaction between SV40 and SGTA or Hsc70, COS-7 cells were seeded in 10-cm plates and treated with either 50 nM RTN3, RTN4, Hsp105, or a scrambled siRNA. After 24 h, cells were transfected with SGTA-S or Hsc70-S for 24 h. The cells were then infected with SV40 (MOI ∼10) for 16 h before being lysed in 1% Triton X-100 in a physiological buffer at 4°C for 10 min. The cells were then centrifuged at 16,000 rpm for 10 min at 4°C. The supernatant was collected and rotated with S protein–conjugated beads for 2 h at 4°C. After 2 h, the beads were washed three times in lysis buffer. The beads were then treated with 5× SDS sample buffer and subjected to SDS-PAGE and immunoblotting. The graph in Fig. 3, d and e, represents three biological replicates, and the VP1 band was quantified using ImageJ software (National Institutes of Health) and normalized to the precipitated SGTA-S or Hsc70-S level.
SV40 infection
CV-1 cells (2 × 105) were seeded and transfected with 50 nM RTN3 siRNA, RTN4 siRNA, or scrambled siRNA (along with Opti-MEM and RNAiMax) on glass coverslips in six-well plates. The cells were incubated at 37°C for 48 h. For knockdown-rescue experiments, 1.5 × 105 CV-1 cells were seeded and transfected using siRNA as described above. After 24 h of siRNA transfection, the cells were washed and transfected with the indicated GFP- or HA-tagged constructs using Fugene and Opti-MEM. After 48 h of siRNA transfection, cells were infected with purified SV40 (MOI ∼0.3) for 24 h. Cells were then fixed in 1% PFA for 15 min, permeabilized in 0.2% Triton X-100 for 5 min, and incubated with rabbit anti-HA and mouse anti-large T antigen antibodies for 1 h at 25°C. Cells were then washed and incubated with anti-rabbit (Alexa Fluor 488) and anti-mouse (Alexa Fluor 594) antibodies in the dark at 25°C for 30 min. Cells were washed, dried, and mounted on slides using Prolong Diamond antifade mount with DAPI (Invitrogen). For evaluating SV40 infection in cells without DNA transfection, at least 1,000 cells were counted per condition. To assess SV40 infection in cells transfected with DNA, at least 100 cells were counted per condition. The graphs in Fig. 2, b and c, represent the mean and SD of at least three biological replicates, with paired Student’s two-tailed t tests used to determine the P values.
ER-to-cytosol transport assay
The protocol for the ER-to-cytosol transport assay has been described previously (Dupzyk and Tsai, 2018; Dupzyk et al., 2017; Inoue and Tsai, 2011). Briefly, CV-1 cells were seeded in 6-cm plates and transfected with 50 nM RTN3, 50 nM RTN4, 25 nM Hsp105, or 50 nM scrambled siRNA for 48 h. Cells were then infected with purified SV40 (MOI ∼5–10) for 16 h, treated with 0.1% digitonin, and centrifuged. The resulting supernatant fraction represents the cytosol fraction containing SV40 that reached the cytosol, while the pellet fraction represents the membrane fraction. The membrane fraction was subsequently treated with 1% Triton X-100, and the extracted material was isolated; this fraction contains ER-localized SV40. The VP1 band intensity was quantified using ImageJ software.
Protease protection assay
CV-1 cells were seeded in 6-cm plates and transfected with 50 nM RTN3, 50 nM RTN4, 25 nM Hsp105, or 50 nM scrambled siRNA for 48 h. Cells were then infected with purified SV40 (MOI ∼30) for 16 h, treated with 0.1% digitonin, and centrifuged. The membrane fraction, as described above, was resuspended in PBS containing 0, 5, or 10 µg/ml trypsin. After 15 min treatment on ice, the proteolysis reaction was stopped by addition of 10% trichloroacetic acid (Acros, 42145) and the sample was precipitated. The pellets were then resolubilized with 5× SDS sample buffer and subjected to SDS-PAGE and immunoblotting. The graph in Fig. 4 e represents three biological replicates, and the Grp170 and BiP band were quantified using ImageJ software. To examine ER membrane integrity in GFP-NHK– or GFP-Akita–expressing cells, 3 × 105 COS-7 cells were seeded and transfected using siRNA as described above. After 24 h of siRNA transfection, cells were washed and transfected with the GFP-NHK or GFP-Akita constructs using PEI and Opti-MEM. After 48 h of siRNA transfection, cells were harvested, treated with 0.02% digitonin, and centrifuged. The pellet materials were processed as described above.
Split-GFP method
COS-7 cells were plated on coverslips in a six-well plate at a density of ∼2 × 105 cells/well and transfected with 50 nM RTN4 or scrambled siRNA. After 24 h transfection, cells were cotransfected with the FLAG-Sec61β11 and mCherry1-10 constructs using PEI. 24 h after DNA transfection, cells were infected with SV40 (MOI ∼100) for 16 h. 16 h after infection, cells were fixed and subjected to an immunofluorescence method using a mouse monoclonal FLAG antibody as described above. Microscopy was performed using a Zeiss LSM 800 confocal laser scanning microscope with a Plan-Apochromat 40×/1.4 oil differential interference contrast M27 objective. Images were Airyscan processed in Zen 2.3 software.
XBP1 splicing assay
CV-1 cells were transfected with either scrambled, RTN3, RTN4, or FAM134B siRNA. XBP1 splicing assay was performed as described previously (Uemura et al., 2009), using the following primers: 5′-CGCGGATCCGAATGTGAGGCCAGTGG-3′ and 5′-GGGGCTTGGTATATATGTGG-3′.
Sucrose gradient fractionation assay
This assay was previously described (Cunningham et al., 2017, 2019). Briefly, cells were lysed in RIPA buffer supplemented with 10 mM N-ethylmaleimide and 1 mM PMSF, incubated on ice for 10 min, and then centrifuged. The insoluble material in the resulting pellet was further extracted by 2% SDS RIPA buffer supplemented with N-ethylmaleimide and PMSF. The extract was cleared using an ultracentrifuge at 50,000 rpm for 20 min. The resulting cleared extract was layered on top of a 10–50% discontinuous sucrose gradient and centrifuged in a Beckman SW50.1 rotor at 29,000 rpm for 24 h at 4°C. After centrifugation, 12 50-µl fractions were collected and analyzed by SDS-PAGE followed by immunoblotting.
Online supplemental material
Fig. S1 shows a RHD4 mutant that cannot support SV40 infection (related to Fig. 4).
Supplementary Material
Acknowledgments
We thank members of the Tsai laboratory for their helpful suggestions and feedback.
B. Tsai is funded by the National Institutes of Health (RO1 AI064296). B. Tsai and P. Arvan are also funded by the National Institutes of Health (RO1 DK111174).
The authors declare no competing financial interests.
Author contributions: Y.J. Chen designed and performed experiments in Figs. 1; 2; 3; 4; 5, a, b, d, and e; and S1. J. Williams contributed to Fig. 5 c. Y.J. Chen, P. Arvan, and B. Tsai conceived the project, designed the experiments, and wrote the manuscript.
References
- Bagchi P., Walczak C.P., and Tsai B.. 2015. The endoplasmic reticulum membrane J protein C18 executes a distinct role in promoting simian virus 40 membrane penetration. J. Virol. 89:4058–4068. 10.1128/JVI.03574-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagchi P., Inoue T., and Tsai B.. 2016. EMC1-dependent stabilization drives membrane penetration of a partially destabilized non-enveloped virus. eLife. 5:e21470 10.7554/eLife.21470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernacchi S., Mueller G., Langowski J., and Waldeck W.. 2004. Characterization of simian virus 40 on its infectious entry pathway in cells using fluorescence correlation spectroscopy. Biochem. Soc. Trans. 32:746–749. 10.1042/BST0320746 [DOI] [PubMed] [Google Scholar]
- Bernales S., McDonald K.L., and Walter P.. 2006. Autophagy counterbalances endoplasmic reticulum expansion during the unfolded protein response. PLoS Biol. 4:e423 10.1371/journal.pbio.0040423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernales S., Schuck S., and Walter P.. 2007. ER-phagy: selective autophagy of the endoplasmic reticulum. Autophagy. 3:285–287. 10.4161/auto.3930 [DOI] [PubMed] [Google Scholar]
- Bhaskara R.M., Grumati P., Garcia-Pardo J., Kalayil S., Covarrubias-Pinto A., Chen W., Kudryashev M., Dikic I., and Hummer G.. 2019. Curvature induction and membrane remodeling by FAM134B reticulon homology domain assist selective ER-phagy. Nat. Commun. 10:2370 10.1038/s41467-019-10345-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky J.L. 2012. Cleaning up: ER-associated degradation to the rescue. Cell. 151:1163–1167. 10.1016/j.cell.2012.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky J.L., and Skach W.R.. 2011. Protein folding and quality control in the endoplasmic reticulum: Recent lessons from yeast and mammalian cell systems. Curr. Opin. Cell Biol. 23:464–475. 10.1016/j.ceb.2011.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.J., Liu X., and Tsai B.. 2019. SV40 Hijacks Cellular Transport, Membrane Penetration, and Disassembly Machineries to Promote Infection. Viruses. 11:917 10.3390/v11100917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham C.N., He K., Arunagiri A., Paton A.W., Paton J.C., Arvan P., and Tsai B.. 2017. Chaperone-Driven Degradation of a Misfolded Proinsulin Mutant in Parallel With Restoration of Wild-Type Insulin Secretion. Diabetes. 66:741–753. 10.2337/db16-1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham C.N., Williams J.M., Knupp J., Arunagiri A., Arvan P., and Tsai B.. 2019. Cells Deploy a Two-Pronged Strategy to Rectify Misfolded Proinsulin Aggregates. Mol. Cell. 75:442–456.e4. 10.1016/j.molcel.2019.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupzyk A., and Tsai B.. 2016. How Polyomaviruses Exploit the ERAD Machinery to Cause Infection. Viruses. 8:242 10.3390/v8090242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupzyk A., and Tsai B.. 2018. Bag2 Is a Component of a Cytosolic Extraction Machinery That Promotes Membrane Penetration of a Nonenveloped Virus. J. Virol. 92:e00607-18 10.1128/JVI.00607-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupzyk A., Williams J.M., Bagchi P., Inoue T., and Tsai B.. 2017. SGTA-Dependent Regulation of Hsc70 Promotes Cytosol Entry of Simian Virus 40 from the Endoplasmic Reticulum. J. Virol. 91:e00232-17 10.1128/JVI.00232-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrester A., De Leonibus C., Grumati P., Fasana E., Piemontese M., Staiano L., Fregno I., Raimondi A., Marazza A., Bruno G., et al. 2019. A selective ER-phagy exerts procollagen quality control via a Calnexin-FAM134B complex. EMBO J. 38:e99847 10.15252/embj.201899847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fregno I., Fasana E., Bergmann T.J., Raimondi A., Loi M., Soldà T., Galli C., D’Antuono R., Morone D., Danieli A., et al. 2018. ER-to-lysosome-associated degradation of proteasome-resistant ATZ polymers occurs via receptor-mediated vesicular transport. EMBO J. 37:e99259 10.15252/embj.201899259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger R., Andritschke D., Friebe S., Herzog F., Luisoni S., Heger T., and Helenius A.. 2011. BAP31 and BiP are essential for dislocation of SV40 from the endoplasmic reticulum to the cytosol. Nat. Cell Biol. 13:1305–1314. 10.1038/ncb2339 [DOI] [PubMed] [Google Scholar]
- Goodwin E.C., Motamedi N., Lipovsky A., Fernández-Busnadiego R., and DiMaio D.. 2014. Expression of DNAJB12 or DNAJB14 causes coordinate invasion of the nucleus by membranes associated with a novel nuclear pore structure. PLoS One. 9:e94322 10.1371/journal.pone.0094322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grumati P., Morozzi G., Hölper S., Mari M., Harwardt M.I., Yan R., Müller S., Reggiori F., Heilemann M., and Dikic I.. 2017. Full length RTN3 regulates turnover of tubular endoplasmic reticulum via selective autophagy. eLife. 6:e25555 10.7554/eLife.25555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T., and Tsai B.. 2011. A large and intact viral particle penetrates the endoplasmic reticulum membrane to reach the cytosol. PLoS Pathog. 7:e1002037 10.1371/journal.ppat.1002037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T., and Tsai B.. 2015. A nucleotide exchange factor promotes endoplasmic reticulum-to-cytosol membrane penetration of the nonenveloped virus simian virus 40. J. Virol. 89:4069–4079. 10.1128/JVI.03552-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T., and Tsai B.. 2017. Regulated Erlin-dependent release of the B12 transmembrane J-protein promotes ER membrane penetration of a non-enveloped virus. PLoS Pathog. 13:e1006439 10.1371/journal.ppat.1006439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampinga H.H., and Craig E.A.. 2010. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol. 11:579–592. 10.1038/nrm2941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanekura K., Ma X., Murphy J.T., Zhu L.J., Diwan A., and Urano F.. 2015. IRE1 prevents endoplasmic reticulum membrane permeabilization and cell death under pathological conditions. Sci. Signal. 8:ra62 10.1126/scisignal.aaa0341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kartenbeck J., Stukenbrok H., and Helenius A.. 1989. Endocytosis of simian virus 40 into the endoplasmic reticulum. J. Cell Biol. 109:2721–2729. 10.1083/jcb.109.6.2721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liddington R.C., Yan Y., Moulai J., Sahli R., Benjamin T.L., and Harrison S.C.. 1991. Structure of simian virus 40 at 3.8-A resolution. Nature. 354:278–284. 10.1038/354278a0 [DOI] [PubMed] [Google Scholar]
- Mayberry C.L., Nelson C.D.S., and Maginnis M.S.. 2017. JC Polyomavirus Attachment and Entry: Potential Sites for PML Therapeutics. Curr. Clin. Microbiol. Rep. 4:132–141. 10.1007/s40588-017-0069-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi A., Clever J., Yamada M., Li P.P., and Kasamatsu H.. 1996. Association with capsid proteins promotes nuclear targeting of simian virus 40 DNA. Proc. Natl. Acad. Sci. USA. 93:96–100. 10.1073/pnas.93.1.96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi A., Li P.P., Qu Q., Jafri Q.H., and Kasamatsu H.. 2007. Molecular dissection of nuclear entry-competent SV40 during infection. Virus Res. 124:226–230. 10.1016/j.virusres.2006.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norkin L.C., Anderson H.A., Wolfrom S.A., and Oppenheim A.. 2002. Caveolar endocytosis of simian virus 40 is followed by brefeldin A-sensitive transport to the endoplasmic reticulum, where the virus disassembles. J. Virol. 76:5156–5166. 10.1128/JVI.76.10.5156-5166.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris S.R., Soppina V., Dizaji A.S., Schimert K.I., Sept D., Cai D., Sivaramakrishnan S., and Verhey K.J.. 2014. A method for multiprotein assembly in cells reveals independent action of kinesins in complex. J. Cell Biol. 207:393–406. 10.1083/jcb.201407086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olzmann J.A., Kopito R.R., and Christianson J.C.. 2013. The mammalian endoplasmic reticulum-associated degradation system. Cold Spring Harb. Perspect. Biol. 5:a013185 10.1101/cshperspect.a013185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapoport T.A. 2007. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature. 450:663–669. 10.1038/nature06384 [DOI] [PubMed] [Google Scholar]
- Rapoport T.A. 2008. Protein transport across the endoplasmic reticulum membrane. FEBS J. 275:4471–4478. 10.1111/j.1742-4658.2008.06588.x [DOI] [PubMed] [Google Scholar]
- Ravindran M.S., Bagchi P., Inoue T., and Tsai B.. 2015. A Non-enveloped Virus Hijacks Host Disaggregation Machinery to Translocate across the Endoplasmic Reticulum Membrane. PLoS Pathog. 11:e1005086 10.1371/journal.ppat.1005086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravindran M.S., Engelke M.F., Verhey K.J., and Tsai B.. 2017. Exploiting the kinesin-1 molecular motor to generate a virus membrane penetration site. Nat. Commun. 8:15496 10.1038/ncomms15496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravindran M.S., Spriggs C.C., Verhey K.J., and Tsai B.. 2018. Dynein Engages and Disassembles Cytosol-Localized Simian Virus 40 To Promote Infection. J. Virol. 92:e00353-18 10.1128/JVI.00353-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schelhaas M., Malmström J., Pelkmans L., Haugstetter J., Ellgaard L., Grünewald K., and Helenius A.. 2007. Simian Virus 40 depends on ER protein folding and quality control factors for entry into host cells. Cell. 131:516–529. 10.1016/j.cell.2007.09.038 [DOI] [PubMed] [Google Scholar]
- Shemesh T., Klemm R.W., Romano F.B., Wang S., Vaughan J., Zhuang X., Tukachinsky H., Kozlov M.M., and Rapoport T.A.. 2014. A model for the generation and interconversion of ER morphologies. Proc. Natl. Acad. Sci. USA. 111:E5243–E5251. 10.1073/pnas.1419997111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata Y., Shemesh T., Prinz W.A., Palazzo A.F., Kozlov M.M., and Rapoport T.A.. 2010. Mechanisms determining the morphology of the peripheral ER. Cell. 143:774–788. 10.1016/j.cell.2010.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqi S., Zhelyabovska O., and Siddiqi S.A.. 2018. Reticulon 3 regulates very low density lipoprotein secretion by controlling very low density lipoprotein transport vesicle biogenesis. Can. J. Physiol. Pharmacol. 96:668–675. 10.1139/cjpp-2018-0077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spooner R.A., Smith D.C., Easton A.J., Roberts L.M., and Lord J.M.. 2006. Retrograde transport pathways utilised by viruses and protein toxins. Virol. J. 3:26 10.1186/1743-422X-3-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehle T., Gamblin S.J., Yan Y., and Harrison S.C.. 1996. The structure of simian virus 40 refined at 3.1 A resolution. Structure. 4:165–182. 10.1016/S0969-2126(96)00020-2 [DOI] [PubMed] [Google Scholar]
- Tsai B., and Qian M.. 2010. Cellular entry of polyomaviruses. Curr. Top. Microbiol. Immunol. 343:177–194. [DOI] [PubMed] [Google Scholar]
- Uemura A., Oku M., Mori K., and Yoshida H.. 2009. Unconventional splicing of XBP1 mRNA occurs in the cytoplasm during the mammalian unfolded protein response. J. Cell Sci. 122:2877–2886. 10.1242/jcs.040584 [DOI] [PubMed] [Google Scholar]
- Voeltz G.K., Prinz W.A., Shibata Y., Rist J.M., and Rapoport T.A.. 2006. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell. 124:573–586. 10.1016/j.cell.2005.11.047 [DOI] [PubMed] [Google Scholar]
- Walczak C.P., Ravindran M.S., Inoue T., and Tsai B.. 2014. A cytosolic chaperone complexes with dynamic membrane J-proteins and mobilizes a nonenveloped virus out of the endoplasmic reticulum. PLoS Pathog. 10:e1004007 10.1371/journal.ppat.1004007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Q., Kuang E., Dong W., Zhou S., Xu H., Qi Y., and Liu Y.. 2007. Reticulon 3 mediates Bcl-2 accumulation in mitochondria in response to endoplasmic reticulum stress. Apoptosis. 12:319–328. 10.1007/s10495-006-0574-y [DOI] [PubMed] [Google Scholar]
- Weller J.I., Glick G., Shirak A., Ezra E., Seroussi E., Shemesh M., Zeron Y., and Ron M.. 2014. Predictive ability of selected subsets of single nucleotide polymorphisms (SNPs) in a moderately sized dairy cattle population. Animal. 8:208–216. 10.1017/S1751731113002188 [DOI] [PubMed] [Google Scholar]
- Wickner W., and Schekman R.. 2005. Protein translocation across biological membranes. Science. 310:1452–1456. 10.1126/science.1113752 [DOI] [PubMed] [Google Scholar]
- Wilkinson S. 2019. Emerging Principles of Selective ER Autophagy. J. Mol. Biol. 10.1016/j.jmb.2019.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.S., and Strittmatter S.M.. 2007. The reticulons: a family of proteins with diverse functions. Genome Biol. 8:234 10.1186/gb-2007-8-12-234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H., Matsui T., Yamamoto A., Okada T., and Mori K.. 2001. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 107:881–891. 10.1016/S0092-8674(01)00611-0 [DOI] [PubMed] [Google Scholar]
- Zurek N., Sparks L., and Voeltz G.. 2011. Reticulon short hairpin transmembrane domains are used to shape ER tubules. Traffic. 12:28–41. 10.1111/j.1600-0854.2010.01134.x [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.