Rahman et al. analyze the first mitotic division in C. elegans embryos via focused ion beam scanning electron microscopy. They identify novel membrane structures, including three-way sheet junctions, underlying the fusion of the two pronuclei containing the parental genomes.
Abstract
After fertilization, parental genomes are enclosed in two separate pronuclei. In Caenorhabditis elegans, and possibly other organisms, when the two pronuclei first meet, the parental genomes are separated by four pronuclear membranes. To understand how these membranes are breached to allow merging of parental genomes we used focused ion beam scanning electron microscopy (FIB-SEM) to study the architecture of the pronuclear membranes at nanometer-scale resolution. We find that at metaphase, the interface between the two pronuclei is composed of two membranes perforated by fenestrations ranging from tens of nanometers to several microns in diameter. The parental chromosomes come in contact through one of the large fenestrations. Surrounding this fenestrated, two-membrane region is a novel membrane structure, a three-way sheet junction, where the four membranes of the two pronuclei fuse and become two. In the plk-1 mutant, where parental genomes fail to merge, these junctions are absent, suggesting that three-way sheet junctions are needed for formation of a diploid genome.
Introduction
The nuclear envelope (NE) is composed of a double membrane that forms a diffusion barrier between the cytoplasm and nucleoplasm. It also includes various proteins and protein complexes, such as nuclear pore complexes (NPCs) and the nuclear lamina (for reviews, see Ungricht and Kutay, 2017; Cohen-Fix and Askjaer, 2017; De Magistris and Antonin, 2018). During mitosis in metazoans, NE breakdown (NEBD) allows microtubules nucleated by cytoplasmic centrosomes to access the chromosomes. NEBD requires the activity of various kinases, including cyclin-dependent kinase 1 and Polo-like kinase 1, which drive the disassembly of the NPCs and nuclear lamina (Heald and McKeon, 1990; Laurell et al., 2011; Rahman et al., 2015; Martino et al., 2017; Linder et al., 2017). However, in many cell types, the membranes of the NE persist and become highly fenestrated such that they exclude organelles but no longer pose a diffusion barrier (Ellenberg et al., 1997; Yang et al., 1997; Hepler and Wolniak, 1984; Schweizer et al., 2015; Luckner and Wanner, 2018).
After fertilization, the two parental genomes are initially encased within separate nuclei, called pronuclei. The two pronuclei migrate toward each other and meet, and as the cell enters mitosis, the NE begins to break down to facilitate the merging of parental chromosomes (Longo, 1973). The fate of the pronuclear membranes during fertilization is largely unknown; if they were to persist, as in somatic cell mitosis, they could interfere with the formation of the zygote’s diploid nucleus. A number of studies, mainly in the 1960s and 70s, that examined the state of the two pronuclei after fertilization with transmission EM suggested that pronuclear membranes in certain organisms can fuse (reviewed in Longo, 1973). However, owing to the inability to generate 3D reconstructions from these EM images, the extent of fusion and the membrane configuration that ultimately allowed merging of parental genomes remained unknown. The only system in which merging of parental genomes is understood at a molecular level is nuclear fusion during budding yeast mating. In this system, the two nuclei fuse in a two-step process: first, the two outer nuclear membranes fuse, followed by fusion of the two inner ones, generating a single “fusion pore” (Melloy et al., 2007; Rogers and Rose, 2014).
In Caenorhabditis elegans, after fertilization, the parental genomes merge at the first metaphase through a membrane gap that forms at the pronuclear interface (where the two pronuclei are juxtaposed) and is visible by fluorescence microscopy (Poteryaev et al., 2005; Audhya et al., 2007; see Fig. 1, A and B). Failure to form this gap, such as in plk-1ts mutants, results in a “paired nuclei” phenotype, where the daughter cells contain two nuclei each, one with the maternal chromosomes and one with the paternal ones (Rahman et al., 2015). This phenotype is observed under other conditions (Audhya et al., 2007; Galy et al., 2008; Golden et al., 2009; Gorjánácz and Mattaj, 2009), but the underlying defect is largely unknown. Here we examined the configuration of the pronuclear membranes throughout the first mitotic division in C. elegans embryos using focused ion beam scanning EM (FIB-SEM; Narayan and Subramaniam, 2015). We reasoned that uncovering the membrane configuration around the interface membrane gap would shed light on the mechanism of its formation. A priori, the membrane gap could have resulted from independent, but aligned, fenestrations in the nuclear membranes of each of the two pronuclei; such fenestrations could have resulted, for example, from the enlargement of membrane pores left behind following NPC disassembly. Alternatively, the membrane gap could have formed through interpronuclear membrane fusion, such as in nuclear fusion in budding yeast. Our data reveal that the membrane gap at the pronuclear interface forms in a process that has elements of these two mechanisms. We find that the two pronuclei become linked by different kinds of membrane structures, including a novel type of membrane junction, a three-way sheet junction. Our results also shed light on the role of PLK-1 in the process and suggest that these membrane junctions are necessary for merging of parental genomes.
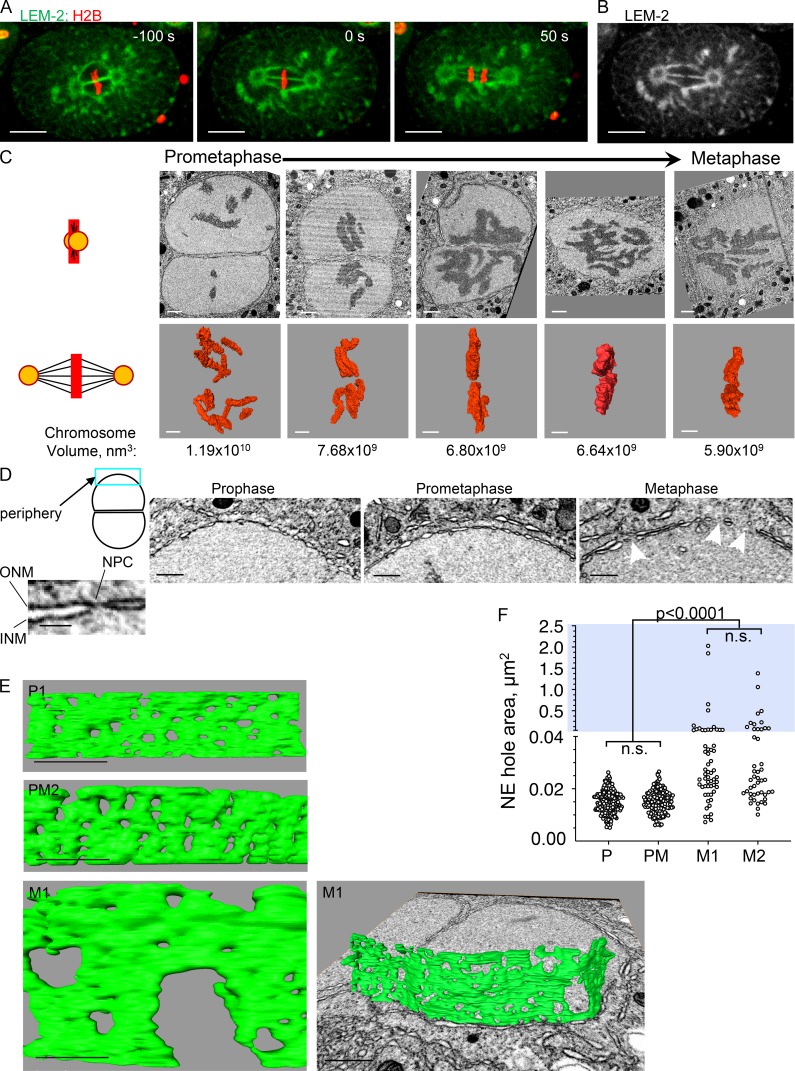
Figure 1.
Pronuclear membranes persist after prophase and become highly fenestrated in metaphase. (A and B) A one-cell embryo expressing the NE protein LEM-2 fused to GFP and histone H2B fused to mCherry was imaged at the indicated time points (metaphase = time 0). LEM-2 persists on the pronuclear membranes throughout mitosis, allowing the visualization of the pronuclear membranes between the two pronuclei. Merging of parental chromosome occurs at metaphase through a fenestration in the pronuclear interface membranes (B). Scale bars, 10 µm. (C) 2D cross sections from FIB-SEM image volumes of a subset of embryos (top row) and their reconstructed chromosomes (middle row) used in this study. The illustration on the left depicts the orientation of the centrosomes (orange circle) and spindle microtubules (black lines) relative to the images shown (chromosomes are in red). Some panels are from the plane of imaging and some are oblique 2D slices through a reconstructed FIB-SEM image volume, aligned orthogonal to the membrane interface. Embryos were ordered from prometaphase to metaphase by the degree of chromosome alignment and compaction, based on total chromosome volumes, shown below the 3D chromosome reconstructions. The pronuclei shown are from the following embryos (from left to right): PM2, PM4, M1, M2, and M3. Scale bars, 1 µm. (D) Examples of peripheral pronuclear membranes (blue rectangle in the cartoon depiction of two associated pronuclei) from prophase (P1), prometaphase (PM2), and metaphase (M1) embryos. The image on the far left is an enlargement of a membrane segment from the prometaphase embryos showing the two nuclear membranes (inner and outer) and the constriction that is seen where an NPC is embedded. Arrowheads in the “metaphase” panel point to holes that are larger than the expected size for NPC holes. Scale bars represent 500 nm, except in the inset, where the scale bar represents 100 nm. INM, inner nuclear membrane; ONM, outer nuclear membrane. (E) Segmented volumes of peripheral pronuclear membranes from the embryos shown in D. Scale bars represent 500 nm, except for the metaphase image on the right, for which the scale bar is 1 µm. P1, prophase; PM2, prometaphase; M1, metaphase. (F) Quantification of hole area in the peripheral pronuclear membranes of an embryo in prophase (P; embryo P1), prometaphase (PM; embryo PM2) and two embryos in metaphase (M1 and M2). For each embryo, data were derived from several segmented areas that were at least 1 µm2 in size. The total number of holes measured and number of areas per embryo were as follows: prophase, n = 213, nine areas; prometaphase, n = 153, nine areas; M1, n = 58, six areas; and M2, n = 46, five areas. Note that the y axis is discontinuous; blue background highlights the upper segment of the graph (hole area >0.04 µm2). Statistical analyses were done using the Kruskal-Wallis test with correction for multiple comparisons. The adjusted P values for the nonsignificant differences (n.s.) were >0.9999. See Fig. S1 for statistical analyses of the lower segment.
Results and discussion
Analysis of the first zygotic mitosis in the one-cell C. elegans embryo using FIB-SEM
Analyzing the configuration of the membranes at the pronuclear interface requires imaging micron-scale structures at nanometer resolution and in 3D. To meet these requirements, we used FIB-SEM imaging, an EM approach that has been used in recent years in a variety of biological system and allows the interrogation of nanoscale architectures in 3D at near-isotropic resolution (Narayan and Subramaniam, 2015; Xu et al., 2017; Kizilyaprak et al., 2019). Briefly, a single C. elegans embryo shortly after fertilization was inserted into a cellulose capillary tube and followed by light microscopy until it reached the pronuclear migration stage, typically 7–8 min before prometaphase/metaphase (Oegema and Hyman, 2006). The tube was then sealed, and, at the desired mitotic stage, the embryo was cryofixed by high-pressure freezing, followed by freeze substitution and embedding in resin. The block was then subjected to FIB-SEM imaging, with scanning EM acquired at 3-nm lateral pixel sampling and 9-nm intervals. Image volumes typically encompassed 22 × 14 × 16 μm, sufficient to capture the two pronuclei. Image stacks, typically comprising several thousand images, were registered, binned, and inverted using in-house scripts (Murphy et al., 2011) to generate 9 × 9 × 9 nm voxel image volumes. These were then visualized, segmented, and analyzed using IMOD and Amira packages. Using this approach, we analyzed multiple one-cell embryos at various stages of prometaphase/metaphase. Embryos were staged relative to each other based on their chromosome configuration and their total chromosome volume, as obtained from 3D reconstructions (Fig. 1 C). Prometaphase and metaphase embryos differed in the spatial distribution of their chromosomes, from being dispersed to being aligned on the metaphase plate with contact between the parental chromosomes. Embryos also differed in the total volume of their chromosomes, with metaphase chromosomes that have breached the pronuclear interface membrane being the most compact (Fig. 1 C).
The peripheral pronuclear membranes become fenestrated during metaphase
As proof of principle that our experimental approach is valid, we first analyzed the configuration of the pronuclear membranes away from the interface (referred to here as the peripheral pronuclear membrane; Fig. 1 D). At prophase and prometaphase, the only fenestrations expected in the peripheral pronuclear membranes are the ones associated with NPCs. Measurements of fenestration areas from randomly chosen regions throughout the peripheral pronuclear membrane of prophase and prometaphase embryos revealed that in both cases, the average hole area was 0.015 ± 0.004 µm2 (Fig. 1, E and F). The aspect ratio of these fenestrations was nearly 1 (0.99 ± 0.19 for prophase and 1.01 ± 0.2 for prometaphase), indicating that the fenestrations are mostly round, with a diameter of roughly 68 nm. This is comparable to inner diameter of the NPC ring in other organisms (50–75 nm in Xenopus laevis [Frenkiel-Krispin et al., 2010] and 40–80 nm in human cells [Bui et al., 2013]) and in excellent agreement with previous measurements in C. elegans using transmission EM (Cohen et al., 2002). Thus, the FIB-SEM approach faithfully captures salient features of the nuclear membranes.
Our analysis of peripheral pronuclear membranes in two metaphase embryos revealed two populations of fenestrations: most fenestrations were only slightly (but significantly) larger than the prophase and prometaphase fenestrations, while a few fenestrations were much larger (>0.04 µm2, Fig. 1, E and F; and Fig. S1 A). Over 20% of the peripheral pronuclear membrane at metaphase was missing, and the degree of fenestration in the two pronuclei of a given embryo was very similar (78.04% and 71.59% of the membrane was retained in the two pronuclei of embryo M1 [total area measured: 7.06 µm2 and 7.19 µm2, respectively] and 77.62% and 77.36% of the membrane was retained in embryo M2 [total area measured: 5.16 µm2 and 7.45 µm2, respectively]). We speculate that the smaller metaphase fenestrations are the result of the enlargement of the membrane holes left behind after NPC disassembly, consistent with a model proposed by (Terasaki et al., 2001). The large fenestrations could be a result of enlargement and/or fusion of smaller fenestrations. The forces leading to the formation of larger fenestrations are currently unknown but could be a result of microtubule-dependent forces, as proposed previously (Beaudouin et al., 2002; Salina et al., 2002). Regardless, during metaphase of the one-cell C. elegans embryo, the chromosomes are enclosed within a perforated double membrane.
Figure S1.
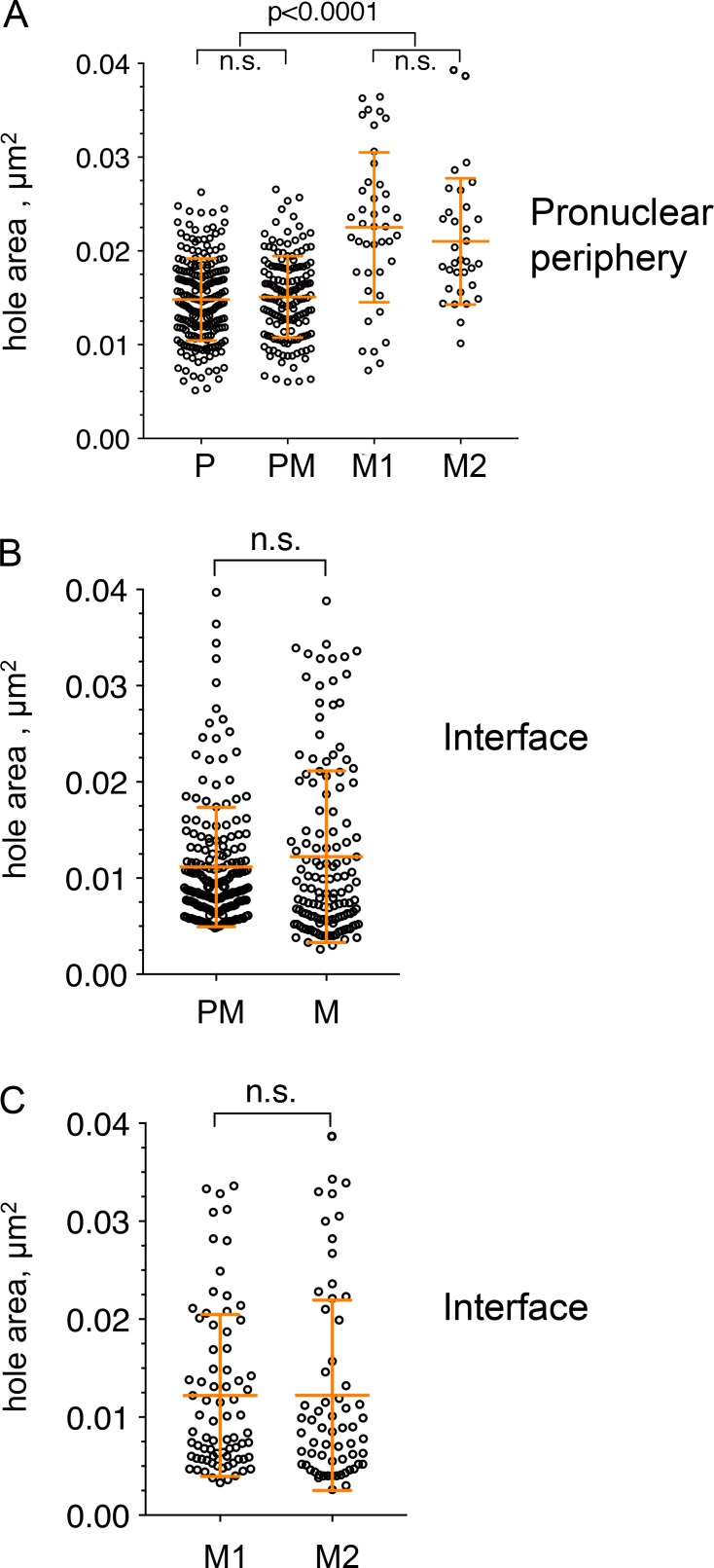
Comparison of pronuclear membrane hole sizes <0.04 µm2. (A) Analysis of hole sizes <0.04 µm2 in peripheral pronuclear membranes of an embryo in prophase (P; embryo P1) and prometaphase (PM; embryo PM2) and two embryos in metaphase (M1 and M2). The data are a subset of the data shown in Fig. 1 F. n = 213, 153, 42, and 33 for P1, PM2, M1, and M2, respectively. Error bars show SD. Statistical analyses was done using a Kruskal-Wallis test with multiple comparisons. The P values for the nonsignificant pairs were >0.9999. (B) Analysis of hole sizes <0.04 µm2 in the interface pronuclear membranes of prometaphase and metaphase embryos. The data are a subset of the data shown in Figs. 2 C and 3 D. All the values for a particular stage (i.e., prometaphase or metaphase) were combined. Statistical analysis was done using two-tailed Mann-Whitney test. The nonsignificant P value = 0.2175. (C) Analysis of hole sizes <0.04 µm2 in the interface pronuclear membranes of two metaphase embryos, M1 and M2. The data are a subset of the data shown in Fig. 3 D. Statistical analysis was done using two-tailed Mann-Whitney test. The nonsignificant P value = 0.4922.
The membranes at the interface between the two pronuclei become fenestrated during prometaphase
We next examined the configuration of the pronuclear membranes at the interface between the two pronuclei, starting at prometaphase. To do so, we reconstructed segments of the adjacent interface pronuclear membranes in two prometaphase embryos (Fig. 2, A and B). Unlike the membranes at the pronuclear periphery at prometaphase, which contained only NPC-sized holes, the membranes at the interface contained both small and larger holes (Fig. 2, B and C). There was no difference in the hole size distribution between the two surfaces (Fig. 2 C). The presence of larger holes at the interface, but not the periphery, is consistent with NEBD occurring earlier at the interphase, as was seen by others for NPC disassembly ( Schetter et al., 2006; Galy et al., 2008; Hachet et al., 2012; Martino et al., 2017). Interestingly, the holes in the interface membranes of the two pronuclei did not align (note that the membrane of the adjacent pronucleus is visible through most holes).
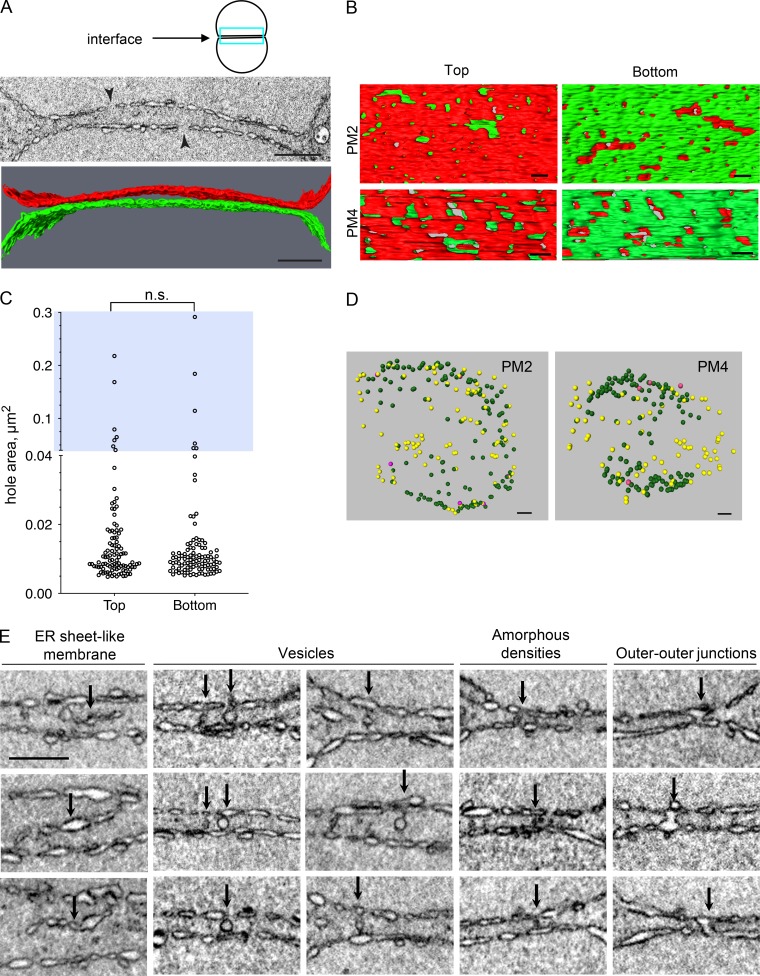
Figure 2.
The interface membranes of the two pronuclei during prometaphase have enlarged fenestration and are connected by various structures. (A) A 2D cross section from a FIB-SEM image volume (top) and segmented subvolumes (bottom) of the pronuclear membranes at the interface (blue rectangle in the cartoon above) of a prometaphase embryo (PM2). The two arrowheads in the top panel are pointing to holes that are larger than the expected size for NPC holes. Scale bars, 1 µm. The membranes of the “top” pronucleus are labeled in red and the “bottom” pronucleus in green. (B) Segmented subvolumes from the interface membranes of two embryos in prometaphase, PM2 and PM4, viewed through the plane of the membrane interface: “top down” (left panels) or “bottom up” (right panels). The membranes of the two pronuclei were labeled in red or green, as in A. Scale bars, 200 nm. (C) Quantification of the hole areas of the top and bottom pronuclear membranes at the interface (as in A and B) of the PM2 embryo. n = 100 and 102 holes for the top and bottom surfaces, respectively. Note that the y axis is discontinuous; blue background highlights the upper segment of the graph (hole area >0.04 µm2). The differences between the two sets of hole sizes are not statistically significant (P = 0.605, two-tailed Mann-Whitney test). (D) Distribution of vesicles (yellow), amorphous densities (green), and outer–outer junctions (purple) present between the two pronuclei in two prometaphase embryos (PM2 and PM4). The interface is shown from a “top” view. Scale bars, 500 nm. (E) Examples of four types of structures that are seen between the two pronuclei at prometaphase (arrows), as indicated above each column. Images are from PM2 and PM4. Scale bar, 500 nm (applies to all panels).
FIB-SEM analysis of the prometaphase pronuclear membrane interface also revealed four different types of structures between the two pronuclei (Fig. 2, D and E): (1) membrane fragments that resemble ER sheets; (2) membrane vesicles, which were associated with one, both, or neither of the two outer pronuclear membranes and distributed throughout the region between the two pronuclei; (3) amorphous densities, which often appeared as poorly demarcated dark staining features between the two pronuclei, mostly at the interface periphery; their size was roughly 89 nm wide and 105 nm long (length = between the two outer pronuclear membrane, n = 16), and the length of the amorphous structures was substantially smaller than the average distance between the two pronuclei (237.36 ± 73.62 nm, n = 16 independent locations), suggesting that the amorphous structures either tend to form where the pronuclei are closer to each other or that their formation brings the two outer pronuclear membranes into closer proximity; and (4) fusion between the two outer pronuclear membranes, referred to here as outer–outer junctions. This structure was relatively rare in prometaphase (Fig. 2 D) but prevalent in metaphase embryos (see below). The ER sheet fragments and vesicles could represent cytoplasmic structures that were trapped between the two pronuclei during pronuclear meeting. Given the distribution of the amorphous structures, they could be precursors of the structures seen between the pronuclei at metaphase, as discussed below.
At metaphase, the membrane at the pronuclear interface contains large fenestrations
The membrane interfaces of two metaphase embryos were segmented (Fig. 3 A). This allowed us to determine the sizes of the interface membrane holes (Fig. 3 B), as was done above for the prometaphase interface. It also allowed us to examine the position of the chromosomes relative to the membrane (Fig. 3 C) and determine the configuration of the interface membranes at this stage (see below). As in prometaphase, the metaphase interface had both large and small holes (Fig. 3 D). Unlike prometaphase, however, the metaphase interface membranes were perforated by holes that connected the two pronuclear lumens (Fig. 3 B). In one of the metaphase embryos (M1), the chromosomes were still separated by the interface membranes, whereas in the other (M2), the chromosomes had come in contact through one of the large holes (Fig. 3 C; see also Fig. 1 C). The two large fenestrations at the interface membrane of the M2 embryo were significantly larger than the largest holes of the M1 embryo (∼15 and 30 µm2 in M2 compared with ∼0.05–1.5 µm2 in M1; Fig. 3 D), consistent with the M2 embryo being further along in mitosis. Interestingly, the size of the small fenestrations (<0.04 µm2) was the same in prometaphase and metaphase (Fig. S1 B). At the pronuclear periphery, on the other hand, the metaphase small holes were significantly larger than the prometaphase ones (Fig. S1 A). This suggests that the regulation and/or mechanism of hole size expansion in the periphery may be distinct from that which drives hole expansion at the interface.
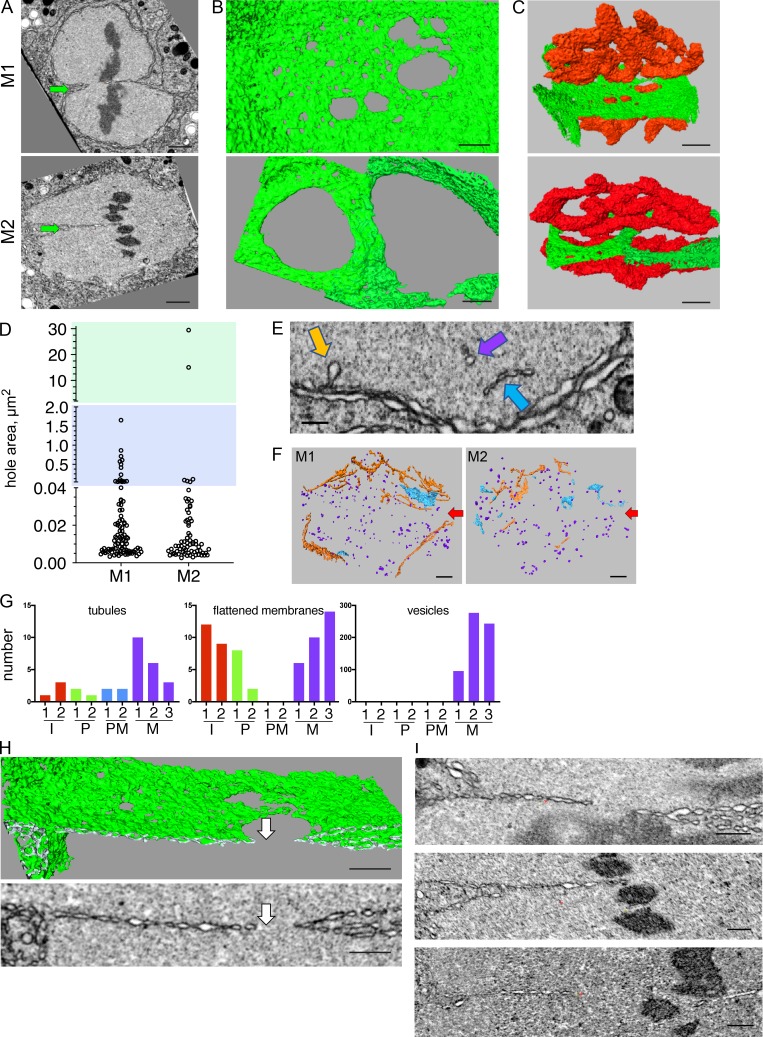
Figure 3.
The pronuclear membrane interface at metaphase contains large holes and only two membranes. (A–C) 2D image slices from a FIB-SEM reconstruction (A; arrows point to the interface), segmented volumes of part of the interface membrane (B and C, green), and segmented volumes of chromosome (C, red) of two one-cell metaphase embryos, M1 and M2. B is a top view of the interface, and C is a view of the metaphase plate from the centrosome’s viewpoint (a ∼45° rotation of the image in B around the x axis). Scale bars, 1 µm. (D) Areas of holes in the interface membranes shown in B. Note that the y axis is discontinuous; blue and green backgrounds highlight the middle and upper segment of the graph (0.04 µm2 < hole area <20 µm2 for the middle segment, and hole area >20 µm2 for the upper segment). See Fig. S1 for statistical analyses of the lower segment. n = 93 and 69 for M1 and M2, respectively, measured using the segmented areas shown in B. (E) Examples of intrapronuclear membrane structures at metaphase (M1). Colors of arrows correspond to the structures shown in F. Scale bar, 250 nm. (F) Reconstruction of all the intrapronuclear membrane structure in two metaphase embryos (M1 and M2). Orange, membrane tubules; blue, flattened membranes, or small sheet-like structures; purple, membrane vesicles. Arrows point the approximate location of the pronuclear interface. Scale bars, 500 nm. (G) Quantification of all the intrapronuclear membrane structures observed in two or three embryos at interphase (I1 and I1, red), prophase (P1 and P2, green), prometaphase (PM2 and PM4, blue), and metaphase (M1, M2, and M4, purple) using reconstructions as shown in F. (H) A 2D cross section from a FIB-SEM image volume (bottom image) and the corresponding segmented volume (top image) of a slice through the interface of M1. Note that to the left of the hole (arrows), the interface is made of only two membranes. Scale bars, 500 nm. (I) Additional images as in panel H of the two-membrane pronuclear interface from three different metaphase embryos (from top to bottom: M1, M2, and M3). Scale bars, 500 nm.
While the size of large holes at the interface of the two metaphase embryos varied widely, the size distribution of the small holes was similar (Fig. S1 C). We speculate that at the interface there is a selective expansion of only some fenestrations, either by fusion of neighboring holes or expansion of individual ones. Live imaging data (Rahman et al., 2015; Fig. 1, A and B) showed that chromosomes are aligned close to the membrane gap that is visible by confocal imaging, raising the possibility that one or more factors associated with chromosomes could affect hole expansion in the vicinity of the chromosomes. Of note, while the two parental genomes merge at metaphase of the one-cell-stage, the maternal and paternal chromosomes themselves intermingle only at the four-cell stage (Bolková and Lanctôt, 2016), similar to the situation in mouse (Mayer et al., 2000).
We also noticed several types of membrane structures within the pronuclei at metaphase, including tubules, flattened membranes, and vesicles (Fig. 3, E and F). The membrane tubes and flattened membrane structures (also referred to as nucleoplasmic reticulum; Drozdz and Vaux, 2017) were observed throughout the cell cycle, but the vesicles were unique to metaphase (Fig. 3 G), consistent with Waterman-Storer et al. (1993). It is tempting to speculate that the intrapronuclear vesicles originated from membrane that was lost from the NE. However, given that at this stage the peripheral pronuclear membrane is highly fenestrated (Fig. 1 E), it is also possible that these vesicles originated from the cytoplasm.
The interface membrane at metaphase is made of only two membranes and is surrounded by two types of membrane junctions: outer–outer junctions and three-way sheet junctions
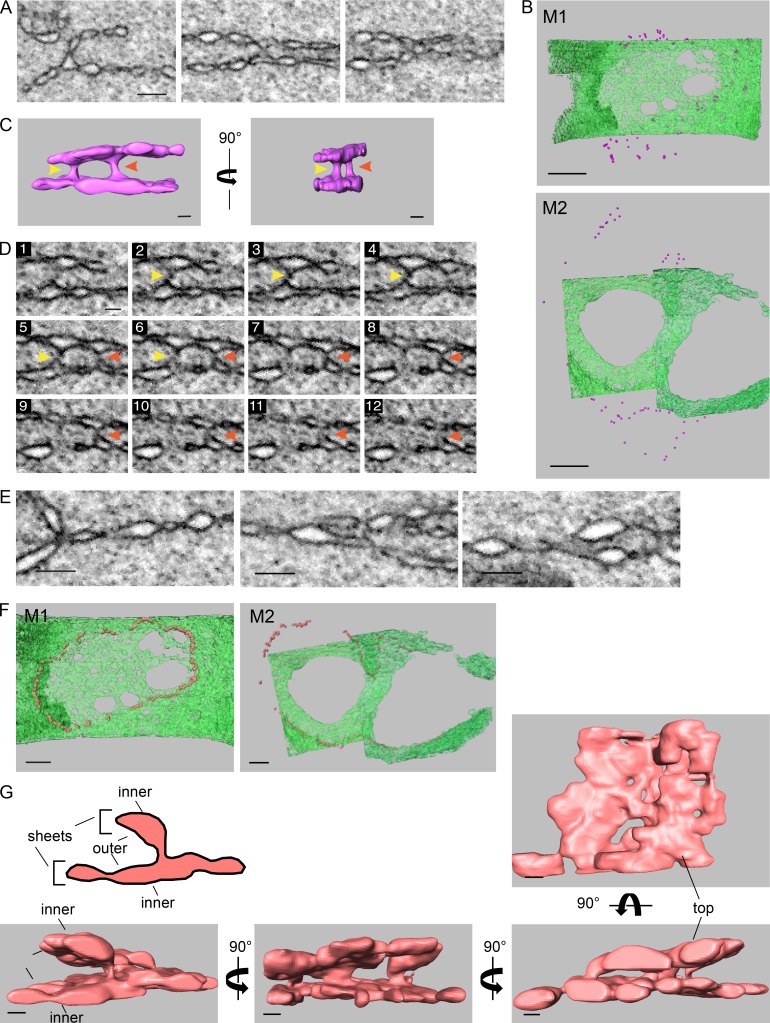
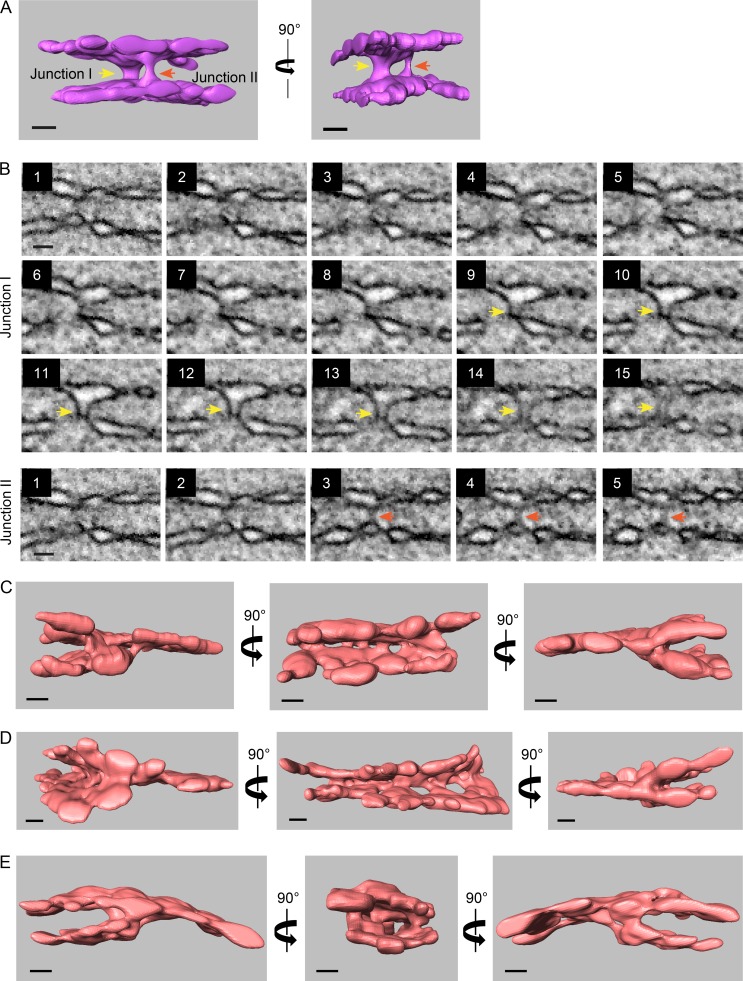
At prometaphase, the interface between the two pronuclei contained four membranes: the two inner and the two outer pronuclear membranes (Fig. 2 A). Strikingly, a cross section of the fenestrated region of the interface membrane at metaphase revealed that this region contains only two membranes (Fig. 3, A, H, and I). To understand the origin of the two remaining membranes, we analyzed the membrane structures surrounding this two-membrane interface region. Two types of structures were found. The first was termed an “outer–outer junction.” This type of junction was also seen at prometaphase, albeit to a lesser extent (Fig. 2 D). Outer–outer junctions form by fusion of the outer membranes of the two pronuclei (Fig. 4 A) and were found at the periphery of the interface where the four membranes were still present, surrounding the perforated two-membrane interface (Fig. 4 B). 3D reconstructions of these structures (Fig. 4, C and D; Fig. S2 A; and Videos 1, 2, and 3) revealed that the outer–outer junctions form membrane tubes with diameters ranging from 36 to 117 nm (average, 80.19 ± 22.74 nm; n = 16).
Figure 4.
At metaphase, the two pronuclei are connected by two types of junctions, outer–outer junctions and three-way sheet junctions, that surround the two-membrane interface. (A) 2D cross sections from FIB-SEM image volumes of outer–outer membrane junctions at the interface of metaphase pronuclei. Scale bar, 200 nm (applies to all panels). (B) Distribution of outer–outer junctions (purple spheres) relative the membrane interface (in green) of two metaphase one-cell embryos. Top panel, M1; bottom panel, M2. Scale bars, 1 µm. (C) Segmented volumes of two adjacent outer–outer junctions from the M1 metaphase embryo. The yellow and red arrowheads serve as markers for the two junctions shown in D. See also Fig. S2 A. Scale bars, 100 nm. (D) Consecutive FIB-SEM images of the outer–outer junctions shown in C. The distance between slices is 9 nm. See also Fig. S2 B. Scale bar, 100 nm (applies to all panels). (E) 2D cross sections from FIB-SEM image volumes of intersections between the four original pronuclear membranes and the two-membrane interface of the M1 embryo. Scale bars, 200 nm. (F) Distribution of the junctions (orange spheres) such as shown in E relative the membrane interface (in green) in two metaphase one-cell embryos. Left panel, M1; right panel, M2. Scale bar, 500 nm. (G) A segmented volume of three-way sheet junctions from embryo M2. Structures were rotated along the y axis as indicated, except for the right-most image, which is a top view of the segmented volume. See Fig. S2, C–E, for more examples. Scale bars, 100 nm.
Figure S2.
Additional examples of outer–outer junctions and three-way sheet junctions from metaphase one-cell embryos. (A) Segmented volumes of two adjacent outer–outer junctions from the M2 metaphase embryo. The yellow and red arrows serve as markers for each of the two junctions. Scale bars, 100 nm. See also Video 4. (B) Consecutive scanning EM images of the outer–outer junctions shown in A. The distance between slices is 9 nm. Scale bars, 100 nm. (C–E) Segmented volumes of three-way sheet junctions from embryo M1 (C and D) and M2 (E). Structures were rotated along the y axis as indicated. Scale bars, 100 nm. See also Videos 5, 6, and 7.
Video 1.
3D reconstructions of outer–outer junctions. Outer–outer junctions were reconstructed as described in the text. The video shows all sides of the junction shown in Fig. 4 C. Frames per second (FPS) = 61.
Video 2.
3D reconstructions of outer–outer junctions. Outer–outer junctions were reconstructed as described in the text. The video shows all sides of the junction shown in Fig. S1 A. FPS = 61.
Video 3.
3D reconstructions of outer–outer junctions. Outer–outer junctions were reconstructed as described in the text and shown from all sides (the junction is not in any figure). FPS = 61.
The second type of junction was termed a “three-way sheet junction” (Fig. 4 E). These junctions encircle the two-membrane interface region, internal (with respect to the interface) to the outer–outer membrane junctions (Fig. 4 F), and they form the intersections between the original four membranes of the two pronuclei and the two membranes of the interface (Fig. 4, E and G). If one considers two parallel membranes a membrane sheet, as in the case of ER sheets, then the “four membrane to two membrane” junctions are, in fact, three-way sheet junctions (Fig. 4 G; Fig. S2, C–E; and Videos 4, 5, 6, and 7). To our knowledge, this type of membrane configuration has not been demonstrated before. Pronuclear fusion is likely not the only process in which three-way sheet junctions exist; for example, the incorporation of annulate lamellae-associated NPCs into the NE in the Drosophila melanogaster embryo (and perhaps other organisms) may involve a similar membrane configuration (Hampoelz et al., 2016). The two membrane sheets join through a series of membrane tubes and cisternae of varying widths, from 37 to 179 nm (average, 89 ± 42.77 nm; n = 17; Fig. 4 G; Fig. S2, C–E; and Videos 4, 5, 6, and 7). The similar size of these tubes and the tubes in the outer–outer junctions suggests that they are structurally related.
Video 4.
3D reconstructions three-way sheet junctions from metaphase one-cell embryos. Three-way sheet junctions were reconstructed as described in the text. The videos show a 360° rotation of the junctions shown in Fig. 4 G. FPS = 61.
Video 5.
3D reconstructions three-way sheet junctions from metaphase one-cell embryos. Three-way sheet junctions were reconstructed as described in the text. The videos show a 360° rotation of the junctions shown in Fig. S1 C. FPS = 61.
Video 6.
3D reconstructions three-way sheet junctions from metaphase one-cell embryos. Three-way sheet junctions were reconstructed as described in the text. The videos show a 360° rotation of the junctions shown in Fig. S1 D. FPS = 61.
Video 7.
3D reconstructions three-way sheet junctions from metaphase one-cell embryos. Three-way sheet junctions were reconstructed as described in the text. The videos show a 360° rotation of the junctions shown in Fig. S2 E. FPS = 61.
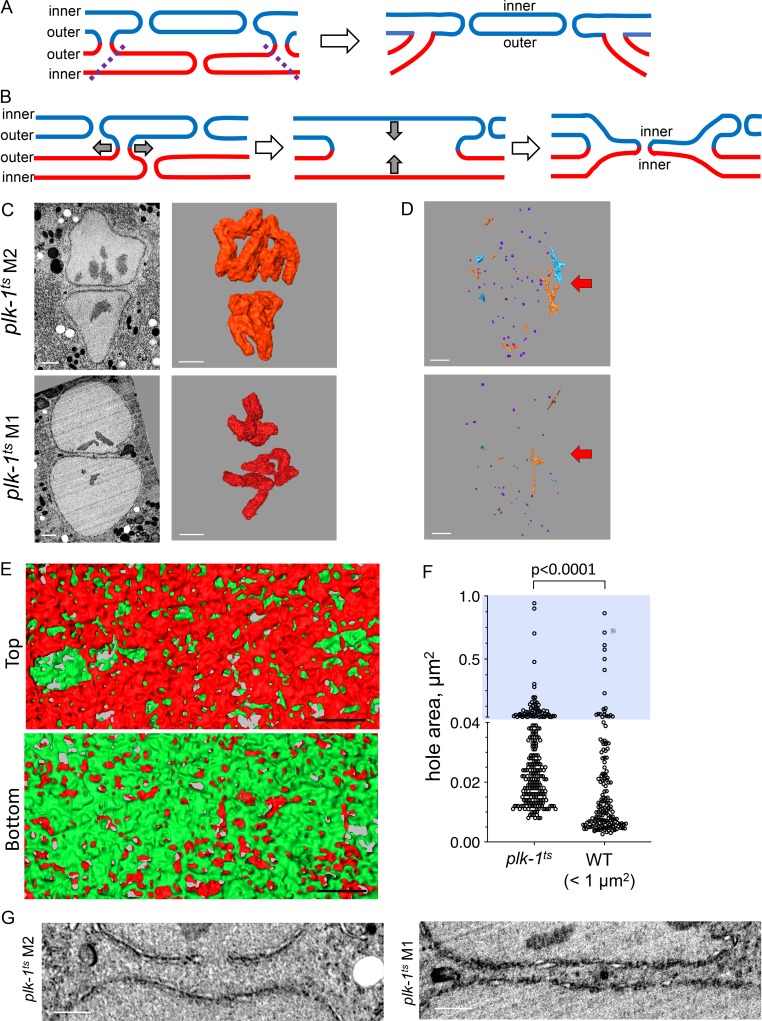
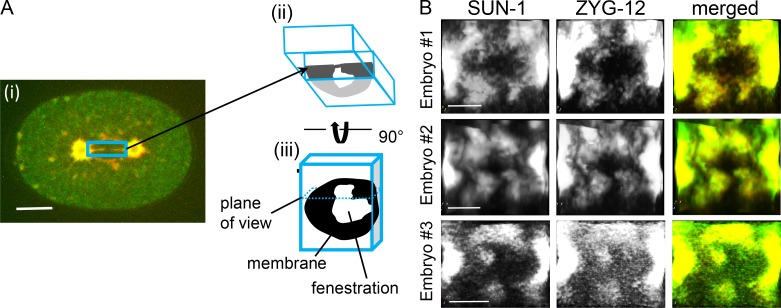
Our observations raise two related questions: how do the three-way sheet junctions form, and what is the origin of the two membranes at the metaphase interface? We propose that the first step in this process is the formation of outer–outer junctions, possibly in a process that is analogous to atlastin-driven ER tubule fusion (Wang and Rapoport, 2019). If one assumes that the small holes at the metaphase pronuclear interface originated from dissociated NPCs, then this interface must be both inner and outer pronuclear membranes. To reach this point, the three-way sheet junctions could have formed by fusion of both the inner and outer nuclear membranes of one pronucleus with the outer membrane of the second pronucleus, leaving a continuous inner and outer membrane of the second pronucleus at the interface (Fig. 5 A). Alternatively, one could imagine a mechanism that is analogous to yeast karyogamy, whereby the two outer membranes fuse and retract, leading to juxtaposition and fusion of the two inner membranes (Fig. 5 B). In this case, the interface membrane would be composed of two inner membranes. Confocal microscopy of an inner and an outer nuclear membrane-associated proteins, SUN-1 and ZYG-12, respectively (Bone and Starr, 2016), which do not disperse in mitosis, revealed that both proteins are present at the metaphase interface (Fig. S3). This, together with the relative similar size of the holes at the interface in prometaphase and metaphase, suggest that the first model is correct. However, due to the presence of membrane junctions, we cannot rule out the possibility that ZYG-12 and/or SUN-1 are no longer exclusively at the original outer and inner nuclear membranes, respectively. It is also possible that the complex does not remain intact during this stage, rendering the assignment of inner and outer pronuclear membranes ambiguous.
Figure 5.
The three-way sheet junction likely forms by loss of the membranes from one of the pronuclei and is absent in the plk-1 mutant, which fails to merge its parental genomes. (A and B) Two models to explain how three-way sheet junctions might form. (A) Removal of the membranes of the “bottom” pronucleus in the region bound by the outer–outer junctions (dashed lines) leaves behind the inner and outer membranes of the “top” pronucleus along with its holes. (B) Expansion of an outer–outer junctions (first step, gray arrows) allows juxtaposition of the inner nuclear membranes (second step, gray arrows), followed by inner nuclear membrane fusion, resulting in fenestration (final step). See text for more detail. (C) 2D cross sections from FIB-SEM image volumes (left) and segmented volumes of chromosomes (right) of pronuclei from two plk-1ts mutant one-cell embryos grown at a semipermissive condition (23°C). Scale bars, 1 µm. (D) Reconstruction of intrapronuclear membrane structures for the same two plk-1ts one-cell embryo at metaphase shown in D, as described in Fig. 3 F. Arrows show the approximate location of the interface. Scale bars, 1 µm. (E) Segmented subvolumes from the interface membranes of a one-cell plk-1ts embryo at metaphase (plk-1ts M2), viewed through the plane of the membrane interface. The membranes of the two pronuclei were labeled in red or green, as in Fig. 2 B. Scale bars, 500 nm. (F) Quantification of hole size in two pronuclear membranes at the interface of a plk-1ts embryo at metaphase (plk-1ts M2) compared with interface hole size in two wild-type metaphase embryos, excluding holes >1 µm2. n = 252 for plk-1ts and 159 for wild type. Note that the y axis is discontinuous, blue background highlights the upper segment of the graph (hole area >0.04 µm2). Statistical analyses were done using a two-tailed Mann-Whitney test. (G) Examples of the interface of two plk-1ts one-cell embryos at metaphase (plk-1ts M1 and M2). Scale bars, 1 µm. See also Video 8 (wild type) and Video 9 (plk-1ts).
Figure S3.
SUN-1 and ZYG-12 distribution at the interface is largely overlapping. (A) A diagram of how the images in B were generated; to view the interface, metaphase one-cell embryos expressing SUN-1::mRuby (red) and ZYG-12::GFP (green) were imaged by confocal microscopy (i). A volume encompassing the interface (blue rectangle) was cropped (ii) using Aivia software and rotated by 90° (iii), resulting in the images shown in B. See Materials and methods for more details about the cropping process. Scale bar, 10 µm. (B) The interface areas of three different one-cell embryos expressing SUN-1::mRuby and ZYG-12::GFP at metaphase. Scale bars, 5 µm
Pronuclear membrane junctions are absent in plk-1 mutant embryos, which fail to merge their parental genomes
Finally, we wanted to examine whether membrane junctions are needed for merging of parental genomes. We previously showed that when the activity of PLK-1 is down-regulated, parental genomes fail to merge (Rahman et al., 2015). To examine the configuration of the pronuclear membrane interface under these conditions, plk-1ts worms were grown at the semipermissive temperature, and embryos at metaphase were analyzed by FIB-SEM as described above. For reasons that are currently unknown, the plk-1 embryos were exceedingly fragile, and many did not survive the high-pressure freezing/freeze substitution steps. Nonetheless, three metaphase plk-1ts embryos were recovered. Because PLK-1 is needed for chromosome alignment, we could not ascertain the embryos’ mitotic stage by chromosome position (Fig. 5 C). However, the degree of chromosome compaction in the plk-1ts embryos was consistent with the embryos being in metaphase (4.15–5.59 × 109 nm3; compare to Fig. 1 B). Furthermore, the plk-1ts embryos contained vesicles inside the both pronuclear lumens (Fig. 5 D), which is characteristic of embryos at metaphase (Fig. 3, F and G).
The interface membranes of the metaphase plk-1ts embryos were highly fenestrated (Fig. 5, E and F), with an average hole size of 0.048 ± 0.101 µm2. However, only a fraction of openings connected the lumens of the two pronuclei, and in none of the three plk-1ts embryos did we observe fenestrations as large as the ones seen in wild-type metaphase one-cell embryos (Fig. 5, E and F; and data not shown). However, if one excludes the very large wild-type holes at the metaphase interface (>1 µm2), there was a trend toward larger holes in the plk-1ts mutant (median hole size 0.01 µm2 and 0.024 µm2 for wild type and plk-1ts, respectively; Fig. 5 F), as if there was a general expansion of membrane holes in the plk-1ts mutant as opposed to selective expansion seen in wild-type cells. Importantly, unlike in wild-type one-cell metaphase embryos, where there were membrane junctions between the two pronuclei (Video 8), in none of the three plk-1ts one-cell metaphase embryos did we observed junctions anywhere throughout the entire interface (Fig. 5 G and Video 9). As in the case of wild-type embryos in prometaphase (Fig. 2), the formation of holes larger than NPC-sized openings at the pronuclear interface was independent of junctions. However, junctions appear to be needed for efficient removal of membrane between the two pronuclei.
Video 8.
The interface of a wild-type one-cell embryo in metaphase. An image stack encompassing ∼700 nm of the interface of a one-cell wild-type embryo in metaphase (M1). FPS = 6.
Video 9.
The interface of a plk-1ts one-cell embryo in metaphase. An image stack encompassing ∼700 nm of the interface of a one-cell plk-1ts embryo in metaphase (plk-1ts M2). FPS = 6.
We also noted that distance between the two pronuclei was greater in the plk-1ts metaphase embryos than in wild-type embryos at prometaphase (389 ± 146 nm vs. 237 ± 73 nm, respectively; distances were measured at 16 [wild type] and 21 [plk-1ts] locations). Thus, PLK-1 is needed for junction formation either directly, by activating one or more proteins involved in junction formation, or indirectly, for example by promoting pronuclear juxtaposition and/or NPC disassembly (Rahman et al., 2015; Martino et al., 2017; Linder et al., 2017) that may be a prerequisite for junction formation. Taken together, our data suggest that membrane junction formation is needed for merging of parental genomes in the zygote.
This study examines the process by which parental genomes merge after fertilization at a resolution that allowed the visualization of both pronuclear membranes up to the point when parental chromosomes come in contact. It is largely assumed that NEBD at mitosis removes the barrier between the two genomes. However, our data show that pronuclear membranes persist all the way to metaphase and that membrane junctions, in the form of outer–outer junctions and three-way sheet junctions, are involved in configuring the pronuclear membranes to facilitate formation of a diploid genome. If the two pronuclei independently form enlarged fenestration at the interface between them, then why go through a membrane fusion process? First, in the plk-1ts embryos, where the interface membranes are highly fenestrated, there is still a “web” of membrane that likely precludes parental chromosome merging (Fig. 5 E). Second, our analysis revealed that trapped between the two pronuclei are membranous structures, in particular flattened membranes that resemble ER sheets that also fuses with the pronuclear membranes (Fig. 2 E and Video 8). We propose that membrane junctions serve to remove all membranes except for a pair of inner and outer membranes and that the fenestrations through which the two genomes ultimately merge originate from enlarged and/or fused holes left behind after NPC disassembly. It is likely that parental chromosome merging requires the activity of a membrane fusion machinery, the nature of which remains to be determined. An obvious candidate for this process is the protein Kar5/Brambleberry, which is involved in nuclear and karyomere fusion in yeast and zebrafish, respectively (Abrams et al., 2012; Rogers and Rose, 2014). However, we and others (Ning et al., 2013) have not been able to find a Kar5/Brambleberry homologue in C. elegans.
It was recently shown that in mice, the first zygotic mitosis occurs on two separate, and often parallel, spindles (Reichmann et al., 2018). If the mechanism of pronuclear membrane remodeling in C. elegans applies to vertebrates, then these two spindles may be separated by remnants of the pronuclear membranes. In humans, the sperm brings with it centrosomes, similar to the situation in C. elegans and unlike the situation in mice, where the first zygotic division is acentrosomal (Sathananthan et al., 1991). Thus, after fertilization, the human embryo is likely to use a single spindle for its first mitosis and may encounter similar challenges as the worm embryo in terms of ensuring that the nuclear membranes are removed in a timely fashion to allow merging of parental chromosome. We suspect that three-way sheet junctions also exist in vertebrates, and understanding the mechanism of junction formation will shed light on early steps in human fertilization.
Materials and methods
C. elegans strains and feeding RNAi
The following C. elegans strains were used in this study: OCF3 (unc-119(ed3); jjIs1092[pNUT1:npp-1::GFP + unc-119(+)] ltIs37[pAA64: pie-1p::mCherry::his-58 + unc-119(+)]; Golden et al., 2009); OCF46 (unc-119(ed3); qaIs3507[unc-119(+) + pie-1::GFP::lem-2] III; ltIs37 [pAA64: pie-1p::mCherry::his-58 + unc-119 (+)]; Rahman et al., 2015); OCF85 (ojIs9 [zyg-12::GFP + unc-119(+)]; and ieSi21 [sun-1p::sun-1::mRuby::sun-1 3′UTR + Cbr-unc-119(+)] IV). FIB-SEM experiments were done with N2 (Bristol; Brenner, 1974) and OCF64 (plk-1(or683)III/hT2 [qIs48] (I:III); ltIs37 [pAA64: pie-1p::mCherry::his-58 + unc-119(+)]). Strains were maintained at 20°C using standard methods unless noted otherwise (Brenner, 1974).
Confocal microscopy and image processing
The images in Fig. 1, A and B, were taken using a Nikon Eclipse TE2000U spinning-disk confocal microscope with IPLab4.0.8 (BioVision Technologies) or Metamorph7.8.8.0 (Molecular Devices) software. The microscope was equipped with a 60× 1.4-NA Apo objective, an LMM5 laser merge module with four diode lasers (excitation at 405, 491, 561, and 655 nm) from Applied Research, a Yokogawa CSU10 spinning disk, and a Hamamatsu C9100-13 EM-CCD camera. Embryos were placed in standard M9 media (IPM scientific) on 2% agarose (Invitrogen) pads for imaging at z = 1-µm intervals. Images were processed with ImageJ (release 1.50i; http://imagej.nih.gov/ij) or Adobe Photoshop CC (release 19.0). Images in Fig. S3 were taken on a Nikon confocal Ti2 with Yokagawa CSU-X1 spinning disk and a photometrix Prime 95B camera using a Nikon water 60× 1.2-NA Apo Plan objective. Images were captured and deconvolved using Elements version 5.20.00 software, after which they were cropped with Aivia software (DRVision Technologies); four cropping planes were applied to each dataset to remove fluorescence signals from outside of the interface. The angles of the cropping planes were adjusted manually to align best with the interface while keeping them close to 90° to each other. The cropped volume was then rotated in 3D to show the membrane structure en face. Linear histogram adjustments (brightness) were applied to both the GFP (488 nm) and RFP (568 nm) channels.
High-pressure freezing of C. elegans embryos
Samples for FIB-SEM were prepared as described previously (Woog et al., 2012); gravid adults grown on plates with OP50 bacteria at 20°C (23°C for plk-1 worms) were dissected in 20% BSA. Fertilized embryos were selected under a stereo microscope (SMZ645, 50× total magnification; Nikon) and collected into cellulose capillary tubes (16706869; Leica Microsystems) with an inner diameter of 200 µm and a wall thickness of 8 µm (dry). The embryos inside a capillary were followed under the stereo microscope until they reached the appropriate cell cycle stage, at which point they were transferred into metal carriers (16770152, Cu-Au 3.0 × 0.5 mm; Leica Microsystems) for immediate cryoimmobilization using a high-pressure freezer (model EM ICE; Leica Microsystems). We typically used a combination of A and B carriers that resulted in a cavity of 200 µm.
Freeze substitution and resin embedding of embryos
High-pressure frozen (HPF) samples were freeze-substituted using a quick freeze substitution (QFS) method as published previously (McDonald and Webb, 2011) with slight modifications: 0.2 g OsO4 (Electron Microscopy Sciences) was dissolved in in 9 ml acetone and mixed with 0.5 ml of 2% uranyl acetate (Electron Microscopy Sciences) in methanol and 0.5 ml double-deionized water. 1 ml of the QFS cocktail was transferred into each of up to eight cryo vials and frozen in a metal block placed inside liquid N2 in an ice bucket. Metal carrier sandwiches of HPF embryos were opened under liquid N2 and transferred into individual cryo vials. The cold metal block was quickly transferred and placed sideways in a second ice bucket containing dry ice and then further buried under dry ice, covered, and placed on a rotary shaker at 60 rpm for 3 h. After decanting the dry ice and removing the lid, the bucket was returned to the shaker until the metal block reached room temperature (typically 1 h), as determined by an infrared thermometer (model IRT207; General Tools & Instruments). After removal of the QFS cocktail, the freeze-substituted samples were subjected to three gentle 10-min washes with acetone and then infiltrated with Polybed 812 resin (08791; Polysciences) of medium hardness (14.6 g Polybed, 8.4 g DDSA, 7.0 g NMA, and 0.42 ml DMP-30) in increasing resin/acetone ratios of 1:2, 1:1, and 2:1 (1 h each). The samples were subsequently incubated in 100% resin overnight, transferred to fresh 100% resin for 6 h before they were embedded in beem capsules, cavity side facing inward, and cured in a 55°C oven for ∼64 h.
FIB-SEM sample preparation
Excess resin around the metal carriers was removed using a jeweler’s saw and razor blades, and the exposed carriers themselves were removed carefully by repeated dipping in liquid N2 and heating with a heat gun (Master Appliance Corp). A microtome (Ultracut UC7; Leica Microsystems) was used to smoothen the face of the sample block, and resin was further removed in 200-nm increments until the top surface of the embryo was exposed, as confirmed by toluidine blue staining of the sections. The sample block was cut off from the beem capsule, cleaned in a series of distilled water baths with an ultrasonicator, mounted on a standard scanning EM stub (16111-9; Ted Pella) using super glue, and painted with colloidal silver solution (Electron Microscopy Sciences). The sample was left to dry overnight in a chemical hood and then coated with gold using a gold coater (EMITECH) to provide a conductive pathway from the resin-embedded embryo to ground.
FIB-SEM data acquisition and postprocessing
The sample stub was placed in a Zeiss FIB-SEM instrument (Crossbeam 540; Carl Zeiss); using the partially exposed oblong embryo on the surface, the sample was oriented for milling such that the long axis of the embryo was perpendicular to the FIB sectioning and scanning EM imaging plane. For some samples, the long axis was oriented parallel to this plane to reduce the total run length; however, the relative orientation of the embryo had little impact on the quality of the data. After a protective carbon pad, 300 nm thick, was deposited over the embryo and a trench well in front of the expected front edge of the embryo was milled, with FIB beam current set at 65 nA. We then reduced the FIB current to 3 nA and executed a “sneak and peek” protocol, where 1-µm-thick sections were milled until the embryo was detected by the scanning EM at the cliff face. At this point, a 1.5-µm-thick patterned platinum pad was deposited over the carbon pad, followed by a further carbon pad to protect the notches (Narayan et al., 2014). We then continued FIB milling across the entire embryo and more with a current of 1.5 nA until a recognizable feature such as the hazy pericentriolar material was observed, at which point pronuclei could be reliably expected proximally. Now, an imaging run was initiated using ATLAS 5 (Fibics). A high-resolution region of interest was drawn over the area thought to contain the pronuclei, and the following milling and imaging parameters were chosen: the scanning electron microscope was operated at 1.5 kV accelerating voltage and 1.0-nA current and FIB parameters were 30 kV with a 1.5-nA or 700-pA current. The in-column energy selective backscatter detector was used with a grid voltage of 1,000 V to record high-resolution, high-m/z contrast signals. 8-bit grayscale images were collected at total dwell time of 3 µs at 3 × 3-nm pixel size and a FIB slice thickness of 9 nm. Focus and stigmation of the beams were executed on the fly every ∼0.5 µm, and key frames were usually not collected. A typical run generated several thousand high-resolution images, collected over 40–60 h continuously and automatically.
Once acquisition of the dataset was complete, the image stack was locally cropped as required and then processed on the National Institutes of Health Biowulf cluster using IMOD-based scripts to produce aligned, inverted, and binned .mrc files (Kremer et al., 1996). The resulting image volumes were made of 9-nm isotropic voxels and also benefitted from denoising by averaging in x and y, allowing for easy visualization and accurate segmentation from any angle with minimal warping. These files were subsequently used for model visualization, segmentation, and rendering. Occasionally, beam instabilities caused a “wash effect” in some datasets; this is a known artifact of multifactorial provenance, where FIB milling progresses through the insulating resin trench in jumps and spurts rather than in a smooth linear fashion. This results in contrast gradation, especially toward the bottom of the high-resolution regions of interest, resulting from a combination of actions such as redeposition and beam-induced baking. The slight brightening in background grayscale value along the y axis in the raw image has a periodicity over several slices; thus, the effect is most evident as stripes of repeating contrast variation when the image volume is viewed from oblique angles. Importantly, the few datasets with the “wash effect” showed the artifact on the order of a tens of nanometers and did not result in major physical artifacts such as shelves in the cliff face. Postprocessing routines to remove the wash did result in some improvement but also introduced some variation in signal from biological features. This approach was therefore abandoned. For segmentation of features that occurred in areas with these grayscale variations in the images, in practical terms, this meant that automated or threshold-based segmentation routines were supplanted with significant manual cleanup and cross-checked for accuracy, as detailed below.
Analysis of FIB-SEM datasets to display nuclear architecture with 3D models
Amira 6.5.0 (release 2018-03-07; Thermo Fisher Scientific) software with XIMagePAQ, XMesh, and XSkeleton extension packages were used for a threshold-based automated segmentation. Segmentation using Amira was done following a general scheme unless specified elsewhere. A threshold-based selection of membranes or chromosomes was followed by three rounds of slice-by-slice visual inspection (through xy, yz, and xz planes separately). Upon careful visual inspection, all undetected areas were manually traced/added to the segmented volume. Any unrelated structures that were automatically segmented along with the NE due to similar threshold levels (e.g., ER membranes surrounding NE; lipid droplets, mitochondria, and other organelles in the proximity of the NE) were removed manually. In total, FIB-SEM data from seven wild-type embryos were included in this study: one prophase embryo (P), two prometaphase embryos (PM2 and PM4), and four metaphase embryos (M1, M2, M3, and M4). In addition, the study includes FIB-SEM data from three plk-1ts embryos.
Segmentation of pronuclear membrane junctions
Pronuclear membrane junctions were segmented as above on separate subvolumes created with Amira. A standard Gaussian filter (interpretation: xy planes; kernel type: separable, SD x:1, SD y:1) was applied before segmentation unless otherwise specified. Typically, a threshold range between 110 and 165 was chosen for automated segmentation/selection. Selected voxels were added to a new material in a specific label field. A surface was generated from selected voxels using a variable smoothing range between 2.5 and 5 (range, 0–10). No filter was used for segmentation of PM2 interface membrane or M1 three-way junctions. For PM4, we used a Gaussian kernel, and for M2 segmentation, we used 2, 2 (other parameters unchanged) for both outer–outer and three-way sheet junctions.
DNA volume measurements
The regions containing chromosomes in each FIB-SEM dataset was volume cropped to generate a separate Amira file. After applying a Gaussian filter with SD range between 2 and 3 and leaving other general scheme parameters unchanged, a surface was generated as described in the general scheme. We used the “Materials Statistics” module in Amira to measure the volume of chromosomes from its 3D surface generated from selected voxels based on a threshold-based segmentation/selection method as per the general scheme.
NE openings/hole size measurements
We used the slicer module in 3dmod to rotate and capture tiff images of NE regions with openings/holes captured in-plane. All tiff images were imported to ImageJ, and then the ImageJ 2D measure tool was used to determine size/area of NE openings/hole by measuring two perpendicular diameters of each hole. These diameters were used to calculate aspect ratio and hole area assuming circularity. The native 2D measure tool in Amira was used to analyze NE openings/hole size from segmented 3D reconstructions/models.
Measurements of membrane tube width in outer–outer and three-way sheet junctions
The width of the membrane tubes that form at prometaphase or metaphase between parallel membrane sheets in either the outer–outer junctions or three-way sheet junctions were measured using the Amira 2D scale tool using 3D segmented volumes. Each measured tube was reconfirmed by manually scoring through the scanning EM image slices. At least 20 tubes for each junction types were measured. Average widths were determined using Excel.
Measurements of distances between pronuclear membranes
The distances between pronuclear membranes were measured based on tiff images from scanning EM image stacks, using 3dmod software package and the ImageJ measurement tool. The distance between pronuclear membrane was defined as the distance between the outer nuclear membranes of opposing maternal and paternal pronuclei.
Data
Raw FIB-SEM data files (mrc and/or am files) used for segmentation and 3D surface reconstructions (surf files) can be obtained at https://cssi-dcc.nci.nih.gov/cssiportal/view/5dea900b0c4bb14f0da5f027/.
Statistical analyses
Statistical analyses were done using Prism 8 version 8.2.0 (GraphPad Software). For each dataset, normalcy was determined and the appropriate test (parametric vs. nonparametric) was applied as indicated in the figure legends.
Online supplemental material
Fig. S1 provides additional statistical analyses of membrane hole sizes <0.04 µm2 (related to Figs. 1, 2, and 3). Fig. S2 provides additional examples of outer–outer junctions and three-way sheet junctions (related to Fig. 4). Fig. S3 provides fluorescent images to test the models shown in Fig. 5, A and B. Videos 1, 2, and 3 show 3D reconstructions of outer–outer junctions (related to Fig. 4). Videos 4, 5, 6, and 7 show 3D reconstructions of three-way sheet junctions (related to Fig. 4). Video 8 shows the interface of wild-type one-cell embryos at metaphase (related to Fig. 5). Video 9 shows the interface of plk-1ts one-cell embryos at metaphase (related to Fig. 5).
Supplementary Material
Acknowledgments
We thank Drs. Thomas Mueller-Reichert for advice throughout this project, Michael Kozlov for inspiring the model shown in Fig. 5 B, and Mark Terasaki for input on membrane junctions and coining the term “three-way sheet junction.” We are grateful to Trevor Lancon from DRVision Technologies for his invaluable assistance in processing images with Aivia. We also thank Will Prinz, Kevin O’Connell, and Madhuri Arya for critical reading of the manuscript.
M. Rahman, R. Maheshwari, K. Amoateng, and O. Cohen-Fix were supported by the National Institute of Diabetes and Digestive and Kidney Disease (intramural grant DK069012). A. Harned, I.Y. Chang, and K. Narayan were supported by the National Cancer Institute (contract HHSN261200800001E). This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
The authors declare no competing financial interests.
Author contributions: M. Rahman, K. Narayan, and O. Cohen-Fix conceived the experiments. M. Rahman and R. Maheshwari carried out the C. elegans experiments, and I.Y. Chang and M. Rahman carried out freezing and freeze substitution. A. Harned, I.Y. Chang, and K. Narayan carried out the FIB-SEM, and M. Rahman, K. Amoateng, and K. Narayan analyzed FIB-SEM data. M. Rahman and O. Cohen-Fix wrote the first draft, and M. Rahman, A. Harned, I.Y. Chang, R. Maheshwari, K. Amoateng, K. Narayan, and O. Cohen-Fix contributed to the revised version.
References
- Abrams E.W., Zhang H., Marlow F.L., Kapp L., Lu S., and Mullins M.C.. 2012. Dynamic assembly of brambleberry mediates nuclear envelope fusion during early development. Cell. 150:521–532. 10.1016/j.cell.2012.05.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audhya A., Desai A., and Oegema K.. 2007. A role for Rab5 in structuring the endoplasmic reticulum. J. Cell Biol. 178:43–56. 10.1083/jcb.200701139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudouin J., Gerlich D., Daigle N., Eils R., and Ellenberg J.. 2002. Nuclear envelope breakdown proceeds by microtubule-induced tearing of the lamina. Cell. 108:83–96. 10.1016/S0092-8674(01)00627-4 [DOI] [PubMed] [Google Scholar]
- Bolková J., and Lanctôt C.. 2016. Live imaging reveals spatial separation of parental chromatin until the four-cell stage in Caenorhabditis elegans embryos. Int. J. Dev. Biol. 60:5–12. 10.1387/ijdb.150222cl [DOI] [PubMed] [Google Scholar]
- Bone C.R., and Starr D.A.. 2016. Nuclear migration events throughout development. J. Cell Sci. 129:1951–1961. 10.1242/jcs.179788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. 1974. The genetics of Caenorhabditis elegans. Genetics. 77:71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui K.H., von Appen A., DiGuilio A.L., Ori A., Sparks L., Mackmull M.-T., Bock T., Hagen W., Andrés-Pons A., Glavy J.S., and Beck M.. 2013. Integrated structural analysis of the human nuclear pore complex scaffold. Cell. 155:1233–1243. 10.1016/j.cell.2013.10.055 [DOI] [PubMed] [Google Scholar]
- Cohen M., Tzur Y.B., Neufeld E., Feinstein N., Delannoy M.R., Wilson K.L., and Gruenbaum Y.. 2002. Transmission electron microscope studies of the nuclear envelope in Caenorhabditis elegans embryos. J. Struct. Biol. 140:232–240. 10.1016/S1047-8477(02)00516-6 [DOI] [PubMed] [Google Scholar]
- Cohen-Fix O., and Askjaer P.. 2017. Cell Biology of the Caenorhabditis elegans Nucleus. Genetics. 205:25–59. 10.1534/genetics.116.197160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Magistris P., and Antonin W.. 2018. The Dynamic Nature of the Nuclear Envelope. Curr. Biol. 28:R487–R497. 10.1016/j.cub.2018.01.073 [DOI] [PubMed] [Google Scholar]
- Drozdz M.M., and Vaux D.J.. 2017. Shared mechanisms in physiological and pathological nucleoplasmic reticulum formation. Nucleus. 8:34–45. 10.1080/19491034.2016.1252893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellenberg J., Siggia E.D., Moreira J.E., Smith C.L., Presley J.F., Worman H.J., and Lippincott-Schwartz J.. 1997. Nuclear membrane dynamics and reassembly in living cells: targeting of an inner nuclear membrane protein in interphase and mitosis. J. Cell Biol. 138:1193–1206. 10.1083/jcb.138.6.1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenkiel-Krispin D., Maco B., Aebi U., and Medalia O.. 2010. Structural analysis of a metazoan nuclear pore complex reveals a fused concentric ring architecture. J. Mol. Biol. 395:578–586. 10.1016/j.jmb.2009.11.010 [DOI] [PubMed] [Google Scholar]
- Galy V., Antonin W., Jaedicke A., Sachse M., Santarella R., Haselmann U., and Mattaj I.. 2008. A role for gp210 in mitotic nuclear-envelope breakdown. J. Cell Sci. 121:317–328. 10.1242/jcs.022525 [DOI] [PubMed] [Google Scholar]
- Golden A., Liu J., and Cohen-Fix O.. 2009. Inactivation of the C. elegans lipin homolog leads to ER disorganization and to defects in the breakdown and reassembly of the nuclear envelope. J. Cell Sci. 122:1970–1978. 10.1242/jcs.044743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorjánácz M., and Mattaj I.W.. 2009. Lipin is required for efficient breakdown of the nuclear envelope in Caenorhabditis elegans. J. Cell Sci. 122:1963–1969. 10.1242/jcs.044750 [DOI] [PubMed] [Google Scholar]
- Hachet V., Busso C., Toya M., Sugimoto A., Askjaer P., and Gönczy P.. 2012. The nucleoporin Nup205/NPP-3 is lost near centrosomes at mitotic onset and can modulate the timing of this process in Caenorhabditis elegans embryos. Mol. Biol. Cell. 23:3111–3121. 10.1091/mbc.e12-03-0204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampoelz B., Mackmull M.-T., Machado P., Ronchi P., Bui K.H., Schieber N., Santarella-Mellwig R., Necakov A., Andrés-Pons A., Philippe J.M., et al. . 2016. Pre-assembled Nuclear Pores Insert into the Nuclear Envelope during Early Development. Cell. 166:664–678. 10.1016/j.cell.2016.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heald R., and McKeon F.. 1990. Mutations of phosphorylation sites in lamin A that prevent nuclear lamina disassembly in mitosis. Cell. 61:579–589. 10.1016/0092-8674(90)90470-Y [DOI] [PubMed] [Google Scholar]
- Hepler P.K., and Wolniak S.M.. 1984. Membranes in the mitotic apparatus: their structure and function. Int. Rev. Cytol. 90:169–238. 10.1016/S0074-7696(08)61490-4 [DOI] [PubMed] [Google Scholar]
- Kizilyaprak C., Stierhof Y.-D., and Humbel B.M.. 2019. Volume microscopy in biology: FIB-SEM tomography. Tissue Cell. 57:123–128. 10.1016/j.tice.2018.09.006 [DOI] [PubMed] [Google Scholar]
- Kremer J.R., Mastronarde D.N., and McIntosh J.R.. 1996. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116:71–76. 10.1006/jsbi.1996.0013 [DOI] [PubMed] [Google Scholar]
- Laurell E., Beck K., Krupina K., Theerthagiri G., Bodenmiller B., Horvath P., Aebersold R., Antonin W., and Kutay U.. 2011. Phosphorylation of Nup98 by multiple kinases is crucial for NPC disassembly during mitotic entry. Cell. 144:539–550. 10.1016/j.cell.2011.01.012 [DOI] [PubMed] [Google Scholar]
- Linder M.I., Köhler M., Boersema P., Weberruss M., Wandke C., Marino J., Ashiono C., Picotti P., Antonin W., and Kutay U.. 2017. Mitotic Disassembly of Nuclear Pore Complexes Involves CDK1- and PLK1-Mediated Phosphorylation of Key Interconnecting Nucleoporins. Dev. Cell. 43:141–156.e7. 10.1016/j.devcel.2017.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo F.J. 1973. Fertilization: a comparative ultrastructural review. Biol. Reprod. 9:149–215. 10.1093/biolreprod/9.2.149 [DOI] [PubMed] [Google Scholar]
- Luckner M., and Wanner G.. 2018. Precise and economic FIB/SEM for CLEM: with 2 nm voxels through mitosis. Histochem. Cell Biol. 150:149–170. 10.1007/s00418-018-1681-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martino L., Morchoisne-Bolhy S., Cheerambathur D.K., Van Hove L., Dumont J., Joly N., Desai A., Doye V., and Pintard L.. 2017. Channel Nucleoporins Recruit PLK-1 to Nuclear Pore Complexes to Direct Nuclear Envelope Breakdown in C. elegans. Dev. Cell. 43:157–171.e7. 10.1016/j.devcel.2017.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer W., Smith A., Fundele R., and Haaf T.. 2000. Spatial separation of parental genomes in preimplantation mouse embryos. J. Cell Biol. 148:629–634. 10.1083/jcb.148.4.629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald K.L., and Webb R.I.. 2011. Freeze substitution in 3 hours or less. J. Microsc. 243:227–233. 10.1111/j.1365-2818.2011.03526.x [DOI] [PubMed] [Google Scholar]
- Melloy P., Shen S., White E., McIntosh J.R., and Rose M.D.. 2007. Nuclear fusion during yeast mating occurs by a three-step pathway. J. Cell Biol. 179:659–670. 10.1083/jcb.200706151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy G.E., Narayan K., Lowekamp B.C., Hartnell L.M., Heymann J.A.W., Fu J., and Subramaniam S.. 2011. Correlative 3D imaging of whole mammalian cells with light and electron microscopy. J. Struct. Biol. 176:268–278. 10.1016/j.jsb.2011.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayan K., and Subramaniam S.. 2015. Focused ion beams in biology. Nat. Methods. 12:1021–1031. 10.1038/nmeth.3623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayan K., Danielson C.M., Lagarec K., Lowekamp B.C., Coffman P., Laquerre A., Phaneuf M.W., Hope T.J., and Subramaniam S.. 2014. Multi-resolution correlative focused ion beam scanning electron microscopy: applications to cell biology. J. Struct. Biol. 185:278–284. 10.1016/j.jsb.2013.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning J., Otto T.D., Pfander C., Schwach F., Brochet M., Bushell E., Goulding D., Sanders M., Lefebvre P.A., Pei J., et al. . 2013. Comparative genomics in Chlamydomonas and Plasmodium identifies an ancient nuclear envelope protein family essential for sexual reproduction in protists, fungi, plants, and vertebrates. Genes Dev. 27:1198–1215. 10.1101/gad.212746.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oegema K., and Hyman A.A.. 2006. Cell division. WormBook. 1–40. 10.1895/wormbook.1.72.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poteryaev D., Squirrell J.M., Campbell J.M., White J.G., and Spang A.. 2005. Involvement of the actin cytoskeleton and homotypic membrane fusion in ER dynamics in Caenorhabditis elegans. Mol. Biol. Cell. 16:2139–2153. 10.1091/mbc.e04-08-0726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman M.M., Munzig M., Kaneshiro K., Lee B., Strome S., Müller-Reichert T., and Cohen-Fix O.. 2015. Caenorhabditis elegans polo-like kinase PLK-1 is required for merging parental genomes into a single nucleus. Mol. Biol. Cell. 26:4718–4735. 10.1091/mbc.E15-04-0244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichmann J., Nijmeijer B., Hossain M.J., Eguren M., Schneider I., Politi A.Z., Roberti M.J., Hufnagel L., Hiiragi T., and Ellenberg J.. 2018. Dual-spindle formation in zygotes keeps parental genomes apart in early mammalian embryos. Science. 361:189–193. 10.1126/science.aar7462 [DOI] [PubMed] [Google Scholar]
- Rogers J.V., and Rose M.D.. 2014. Kar5p is required for multiple functions in both inner and outer nuclear envelope fusion in Saccharomyces cerevisiae. G3 (Bethesda). 5:111–121. 10.1534/g3.114.015800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salina D., Bodoor K., Eckley D.M., Schroer T.A., Rattner J.B., and Burke B.. 2002. Cytoplasmic dynein as a facilitator of nuclear envelope breakdown. Cell. 108:97–107. 10.1016/S0092-8674(01)00628-6 [DOI] [PubMed] [Google Scholar]
- Sathananthan A.H., Kola I., Osborne J., Trounson A., Ng S.C., Bongso A., and Ratnam S.S.. 1991. Centrioles in the beginning of human development. Proc. Natl. Acad. Sci. USA. 88:4806–4810. 10.1073/pnas.88.11.4806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schetter A., Askjaer P., Piano F., Mattaj I., and Kemphues K.. 2006. Nucleoporins NPP-1, NPP-3, NPP-4, NPP-11 and NPP-13 are required for proper spindle orientation in C. elegans. Dev. Biol. 289:360–371. 10.1016/j.ydbio.2005.10.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer N., Pawar N., Weiss M., and Maiato H.. 2015. An organelle-exclusion envelope assists mitosis and underlies distinct molecular crowding in the spindle region. J. Cell Biol. 210:695–704. 10.1083/jcb.201506107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terasaki M., Campagnola P., Rolls M.M., Stein P.A., Ellenberg J., Hinkle B., and Slepchenko B.. 2001. A new model for nuclear envelope breakdown. Mol. Biol. Cell. 12:503–510. 10.1091/mbc.12.2.503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungricht R., and Kutay U.. 2017. Mechanisms and functions of nuclear envelope remodelling. Nat. Rev. Mol. Cell Biol. 18:229–245. 10.1038/nrm.2016.153 [DOI] [PubMed] [Google Scholar]
- Wang N., and Rapoport T.A.. 2019. Reconstituting the reticular ER network - mechanistic implications and open questions. J. Cell Sci. 132:jcs227611 10.1242/jcs.227611 [DOI] [PubMed] [Google Scholar]
- Waterman-Storer C.M., Sanger J.W., and Sanger J.M.. 1993. Dynamics of organelles in the mitotic spindles of living cells: membrane and microtubule interactions. Cell Motil. Cytoskeleton. 26:19–39. 10.1002/cm.970260104 [DOI] [PubMed] [Google Scholar]
- Woog I., White S., Büchner M., Srayko M., and Müller-Reichert T.. 2012. Correlative Light and Electron Microscopy of Intermediate Stages of Meiotic Spindle Assembly in the Early Caenorhabditis elegans Embryo. Methods Cell Biol. 111:223–234. [DOI] [PubMed] [Google Scholar]
- Xu C.S., Hayworth K.J., Lu Z., Grob P., Hassan A.M., García-Cerdán J.G., Niyogi K.K., Nogales E., Weinberg R.J., and Hess H.F.. 2017. Enhanced FIB-SEM systems for large-volume 3D imaging. eLife. 6:e25916 10.7554/eLife.25916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Guan T., and Gerace L.. 1997. Integral membrane proteins of the nuclear envelope are dispersed throughout the endoplasmic reticulum during mitosis. J. Cell Biol. 137:1199–1210. 10.1083/jcb.137.6.1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.