Ems-McClung et al. visualize a RanGTP effector gradient of association between XCTK2 and importin α/β in the spindle. The importins preferentially inhibit XCTK2-mediated antiparallel microtubule cross-linking and sliding, which allows XCTK2 to cross-link parallel microtubules and help focus spindle poles.
Abstract
High RanGTP around chromatin is important for governing spindle assembly during meiosis and mitosis by releasing the inhibitory effects of importin α/β. Here we examine how the Ran gradient regulates Kinesin-14 function to control spindle organization. We show that Xenopus Kinesin-14, XCTK2, and importin α/β form an effector gradient that is highest at the poles and diminishes toward the chromatin, which is opposite the RanGTP gradient. Importin α and β preferentially inhibit XCTK2 antiparallel microtubule cross-linking and sliding by decreasing the microtubule affinity of the XCTK2 tail domain. This change in microtubule affinity enables RanGTP to target endogenous XCTK2 to the spindle. We propose that these combined actions of the Ran pathway are critical to promote Kinesin-14 parallel microtubule cross-linking to help focus spindle poles for efficient bipolar spindle assembly. Furthermore, our work illustrates that RanGTP regulation in the spindle is not simply a switch, but rather generates effector gradients where importins α and β gradually tune the activities of spindle assembly factors.
Introduction
The small GTP-binding protein, Ran, is required for microtubule (MT) nucleation and spindle assembly (Carazo-Salas et al., 1999; Kaláb et al., 1999; Ohba et al., 1999; Wilde and Zheng, 1999). RanGTP forms a gradient around chromatin that diminishes toward the spindle poles (Kaláb et al., 2002, 2006) due to the chromatin localization of its guanine nucleotide exchange factor, RCC1 (Moore et al., 2002). RanGTP acts through the nuclear transport receptors, importin α and β, which bind to NLS-containing proteins and inhibit their activity in areas of low RanGTP. However, in areas of high RanGTP, this inhibition is released, resulting in localized activation of the NLS-containing proteins that can stimulate MT nucleation and dynamics. It is postulated that the RanGTP gradient will result in the formation of downstream effector gradients of these MT regulators (Athale et al., 2008; Caudron et al., 2005), but there has been no direct visualization of these effector gradients. It is important to note that based on this model, RanGTP interaction with importin β would happen only near the chromatin, resulting in the local release and activation of NLS-containing proteins that dissipate toward the poles (Caudron et al., 2005). However, whether all NLS-containing proteins will respond equivalently to the RanGTP gradient is unknown. In addition, the RanGTP gradient can induce feedback loops by the generation of increased numbers of MTs, which are bound by components of the Ran pathway, resulting in localized effects on the spindle (Oh et al., 2016).
Ran-regulated spindle assembly factors (SAFs) that are normally sequestered by importin α/β (Kaláb and Heald, 2008) include a number of MT-associated proteins, such as TPX2 (Gruss et al., 2001), nuclear mitotic apparatus protein (NuMA; Nachury et al., 2001; Wiese et al., 2001), hepatoma up-regulated protein (Koffa et al., 2006), and nucleolar spindle–associated protein (Ribbeck et al., 2006), as well as the molecular motor proteins Kid (Tahara et al., 2008; Trieselmann et al., 2003) and XCTK2 (Ems-McClung et al., 2004). XCTK2 is a minus end–directed Kinesin-14 motor that cross-links and slides both parallel and antiparallel MTs (Hentrich and Surrey, 2010) and contributes to proper spindle assembly, spindle length, and spindle pole formation (Cai et al., 2009; Walczak et al., 1997, 1998). XCTK2 association within the spindle is spatially controlled by the RanGTP gradient through the tail domain (Hallen et al., 2008; Weaver et al., 2015), suggesting that the RanGTP gradient may specifically regulate the MT binding of XCTK2 to the spindle. How the RanGTP gradient regulates the localization of SAFs to the spindle is unknown.
Kinesin-14 proteins play critical roles in spindle pole formation in multiple organisms (Fink et al., 2009; Goshima et al., 2005a,b; Hatsumi and Endow, 1992b; Matuliene et al., 1999; Mountain et al., 1999; Sharp et al., 1999; She and Yang, 2017) and may be especially important in cells with centrosome amplification (Kwon et al., 2008), where they use their minus end–directed motor activity to help cluster those centrosomes into a bipolar spindle. Indeed, small-molecule inhibitors that target the human Kinesin-14, HSET, may be useful as targeted therapeutics in cancers with centrosome amplification (Watts et al., 2013); therefore, understanding Kinesin-14 function in the spindle is an important avenue of pursuit. One conundrum is to understand how a molecular motor that cross-links and slides both antiparallel and parallel MTs through its motor and tail domains can be involved in MT focusing at the spindle poles, where the tail would be predicted to be inhibited from binding MTs by importin α/β. Here, we develop XCTK2 biosensors that visualize a gradient of association with importin α in the spindle that is highest at spindle poles. We show that the importins preferentially inhibit XCTK2-mediated antiparallel MT cross-linking and sliding and propose that importin α/β association with XCTK2 near the spindle poles forms an effector gradient opposite the Ran gradient that functions to facilitate parallel MT cross-linking and sliding for pole focusing. This model provides a mechanism by which XCTK2 localization and activity in the spindle are spatially controlled.
Results and discussion
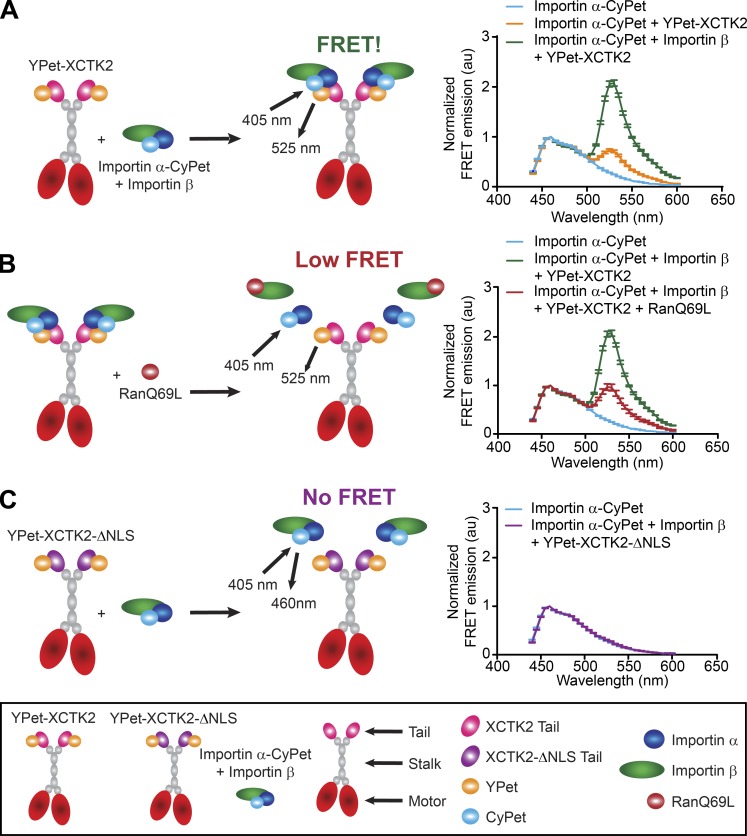
The RanGTP gradient is proposed to affect the ability of importin α/β to interact with SAFs around the chromatin where RanGTP levels are high (Kaláb et al., 2002, 2006). We hypothesized that this would create an effector gradient of importin α/β association with XCTK2 that would be highest at the poles and lowest near the chromatin. To test this idea, we developed Förster resonance energy transfer (FRET) biosensors as a readout of the Ran-regulated interaction of importin α/β with XCTK2, in which we tagged importin α with CyPet (importin α-CyPet) and full-length XCTK2 with YPet (YPet-XCTK2; Fig. 1). YPet-XCTK2 produced a strong FRET signal at 525 nm with importin α-CyPet only in the presence of importin β, indicating a strong association (Fig. 1 A). This association was disrupted by addition of the RanGTP analogue, RanQ69L (Fig. 1 B). In contrast, mutation of the NLS in XCTK2 (YPet-XCTK2-ΔNLS) did not display FRET with importin α-CyPet and importin β, demonstrating their lack of association (Fig. 1 C). These results demonstrate that we have developed FRET probes that monitor the effects of RanGTP on XCTK2 and importin α/β association and can be used to look at effector gradients in the spindle.
Figure 1.
FRET biosensors recapitulate Ran-regulated association of XCTK2 and importin α/β. (A–C) Schematic (left) and solution-based FRET assay (right) of importin α-CyPet with YPet-XCTK2 ± importin β (A) ± RanQ69L (B) or YPet-XCTK2-ΔNLS + importin β (C). The normalized FRET ratios are graphed as the mean ± SEM from 440 to 600 nm (n = 3 independent experiments).
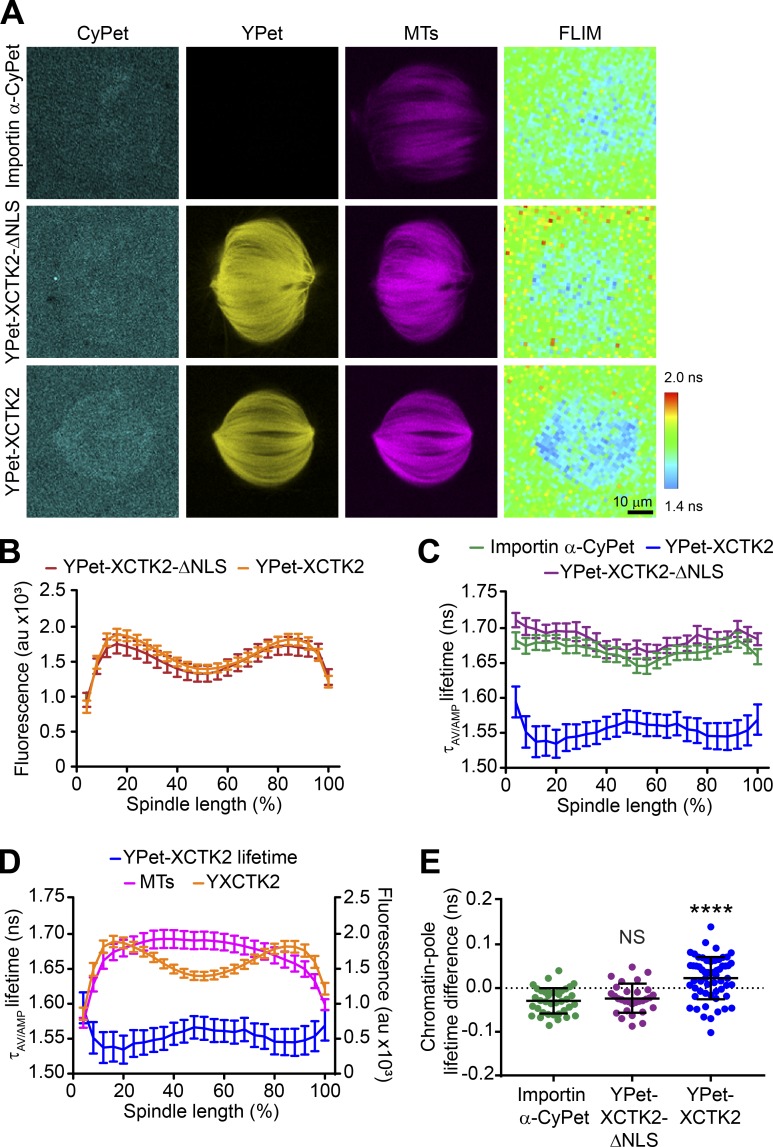
To test whether there is a gradient of importin α/β association with XCTK2 within the spindle, we added importin α-CyPet ± YPet-XCTK2 or YPet-XCTK2-ΔNLS to spindle assembly reactions in Xenopus laevis egg extracts and imaged for CyPet, YPet, and Rhodamine-MT fluorescence, followed by fluorescence lifetime imaging microscopy (FLIM; Fig. 2 A). As a control, we used YPet-XCTK2-ΔNLS that cannot bind to importin α/β and thus would be insensitive to the RanGTP gradient. Importin α-CyPet was generally diffuse in the cytoplasm, with slight accumulation on the spindle, whereas both YPet-XCTK2-ΔNLS and YPet-XCTK2 concentrated on the spindle with localization enriched toward the poles, similar to the endogenous localization of XCTK2 (Fig. 2, A and B; Ems-McClung et al., 2004; Walczak et al., 1997). The FLIM images of importin α-CyPet ± YPet-XCTK2-ΔNLS showed a diffuse pattern of lifetimes within the spindle. In contrast, FLIM images of importin α-CyPet with YPet-XCTK2 appeared to have shorter lifetimes than those of importin α-CyPet ± YPet-XCTK2-ΔNLS (Fig. 2 A), suggesting that there is an interaction between importin α-CyPet and YPet-XCTK2 on the spindle. Using line scans to look at the distribution of lifetimes across the spindle, we found that importin α-CyPet and importin α-CyPet with YPet-XCTK2-ΔNLS had relatively flat distributions, with mean lifetimes of 1.67 ± 0.06 and 1.68 ± 0.05 ns, respectively (Fig. 2 C). This result is consistent with our solution-based FRET assay, in which YPet-XCTK2-ΔNLS does not display FRET with importin α-CyPet, and demonstrates that there is no interaction of YPet-XCTK2-ΔNLS and importin α-CyPet on the spindle. In contrast, importin α-CyPet with YPet-XCTK2 showed shorter lifetimes within the spindle, demonstrated by a dramatic decrease in the mean lifetime of the line scan across the spindle (1.55 ± 0.14 ns; Fig. 2, A and C), indicating that XCTK2 interacts with importin α-CyPet on the spindle. In addition, the lifetimes for importin α-CyPet + YPet-XCTK2 were shorter toward the spindle poles and coincided with the peak localization of YPet-XCTK2 at the poles, suggesting a gradient of association of importin α with XCTK2 extending from the poles to the chromatin (Fig. 2 D). Consistent with this idea, the difference between the chromatin and pole lifetimes for YPet-XCTK2 was statistically greater than either importin α-CyPet alone or importin α-CyPet with YPet-XCTK2-ΔNLS (Fig. 2 E and Table S1). These results strongly suggest that XCTK2 and importin α form a gradient of interaction from the poles to the chromatin that is opposite the RanGTP gradient, which extends from the chromatin to the poles.
Figure 2.
XCTK2 forms a gradient of association with importin α/β from the poles to the chromatin. (A) Representative confocal fluorescence (CyPet, YPet, and MTs) and lifetime (FLIM) images of importin α-CyPet + nontagged XCTK2 (importin α-CyPet), importin α-CyPet + YPet-XCTK2-ΔNLS, and importin α-CyPet + YPet-XCTK2 spindle assembly reactions. Scale bar: 10 µm. (B) YPet fluorescence line scans of spindles assembled in A of importin α-CyPet with YPet-XCTK2-ΔNLS or YPet-XCTK2 normalized to percentage of spindle length (25 bins). YPet fluorescence percentage spindle length is graphed as the mean ± SEM (n = 30 YPet-XCTK2-ΔNLS + importin α-CyPet and 58 YPet-XCTK2 + importin α-CyPet spindles from three independent experiments). (C) Lifetime line scans of spindles imaged in A and B, where lifetimes are represented as the amplitude averaged lifetime (τAV/AMP), and lifetimes per normalized percentage spindle length are graphed as the mean ± SEM (n = 38 importin α-CyPet + XCTK2 spindles, 30 YPet-XCTK2-ΔNLS + importin α-CyPet, and 58 YPet-XCTK2 + importin α-CyPet spindles). (D) Line scans of the lifetimes from spindles with YPet-XCTK2 + importin α-CyPet relative to the fluorescence of the spindle MTs and the YPet fluorescence plotted as in B and C. (E) Chromatin and pole lifetime differences for each spindle analyzed in C with the mean ± SD indicated (D’Agostino and Pearson normality test and Brown–Forsythe’s and Welch’s with Dunnett’s multiple comparisons test: ****, P < 0.0001).
Table S1. Importin α-CyPet lifetime chromatin to pole differences.
| Condition, statistic, and number | Importin α-CyPet | YPet-XCTK2-ΔNLS | YPet-XCTK2 |
|---|---|---|---|
| Importin α-CyPet chromatin to pole lifetime difference (NS) | −0.02905 | −0.02383 | 0.02289 |
| SD | 0.02903 | 0.03273 | 0.04838 |
| D’Agostino and Pearson normality test | 0.7582 | 0.8582 | 0.5582 |
| Brown–Forsythe’s and Welch’s ANOVA with Dunnett’s multiple comparisons | 0.8689 | <0.0001 | |
| Spindle number | 38 | 30 | 58 |
| Experiment number | 3 | 3 | 3 |
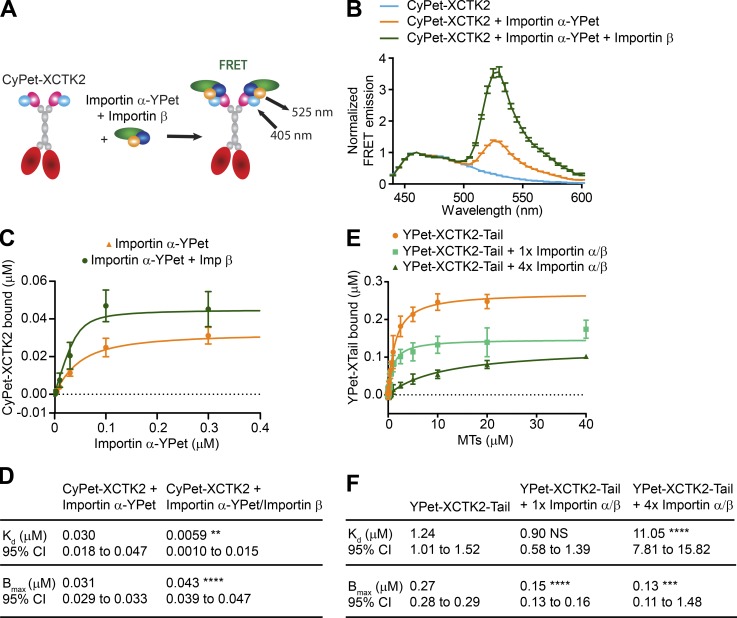
One way that RanGTP could modulate localization of SAFs within the spindle is by modulating the affinity of interactions of SAFs with importin α/β or with spindle MTs. We hypothesized that importin α/β binding could fine-tune XCTK2 function within spindles through the Ran-regulated association of the importins to the nuclear localization signal (NLS) in the XCTK2 tail domain. To test this idea, we designed additional FRET biosensors that monitored importin α/β binding to XCTK2 (Fig. S1, A and B) and used FRET as a readout to measure the binding affinity of importin α-YPet to CyPet-tagged XCTK2 (CyPet-XCTK2) in the presence and absence of importin β (Fig. S1 C). Importin β significantly increased the affinity of importin α by decreasing the Kd fivefold and increasing the total amount bound (Bmax) by 28% (Fig. S1, C and D), consistent with previous studies showing that importin β releases the autoinhibition of importin α for binding to NLS-containing proteins (Catimel et al., 2001). To test how importin α/β binding to the XCTK2 tail modulates its interaction with MTs, we generated a construct containing only the XCTK2 tail domain tagged with YPet (YPet-XCTK2-Tail) and used it to measure the affinity of MT binding in the presence and absence of an equal molar amount (1×) or fourfold molar excess (4×) of importin α/β (Fig. S1, E and F). Interestingly, addition of an equal molar amount of importin α/β did not change the Kd but decreased the amount bound by 46% (Fig. S1 F). In contrast, a fourfold excess of the importins increased the Kd by 10-fold and reduced the amount bound by 53% (Fig. S1 F). These results demonstrate that the XCTK2 tail MT affinity is tunable by importin α/β and provide a mechanism by which RanGTP could control the localization of XCTK2 within the spindle.
Figure S1.
XCTK2 tail MT binding is tunable by importin α/β association. (A) Schematic of FRET biosensors to detect XCTK2 and importin α/β association where XCTK2 is N-terminally tagged with CyPet (CyPet-XCTK2) and importin α is C-terminally tagged with YPet (importin α-YPet). (B) Solution-based FRET assay of CyPet-XCTK2 with importin α-YPet ± importin β. The normalized FRET ratios are graphed as mean ± SEM from 440 to 600 nm (n = 4–5 independent experiments). (C and D) Affinity assays of CyPet-XCTK2 with importin α-YPet ± importin β. The mean ± SD of CyPet-XCTK2 bound for each importin α-YPet concentration and the best-fit quadratic binding curves are graphed from C with the binding data summarized in D (n = 4–5 independent experiments; extra sum-of-squares F test compared with CyPet-XCTK2 + importin α-YPet: Kd, P = 0.0088; Bmax, P < 0.0001). CyPet-XCTK2 bound is graphed from 0 to 0.3 µM importin α-YPet, excluding 1 and 2.5 µM importin α-YPet data points. (E and F) MT affinity assays of YPet-XCTK2-Tail in the absence and presence of equimolar (1×) or fourfold excess (4×) of importin α/β. The amount of YPet-XCTK2-Tail bound is plotted as the mean ± SD for each MT concentration, and the best-fit quadratic binding curves are graphed (E) with the binding data summarized in F (n = 5–8 independent experiments; extra sum-of-squares F test compared with YPet-XCTK2-Tail alone: YPet-XCTK2-Tail + 1× importin α/β, Kd, NS; Bmax, P < 0.0001; YPet-XCTK2-Tail + 4× importin α/β, Kd, P < 0.0001; P = 0.0002). (D and F) **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
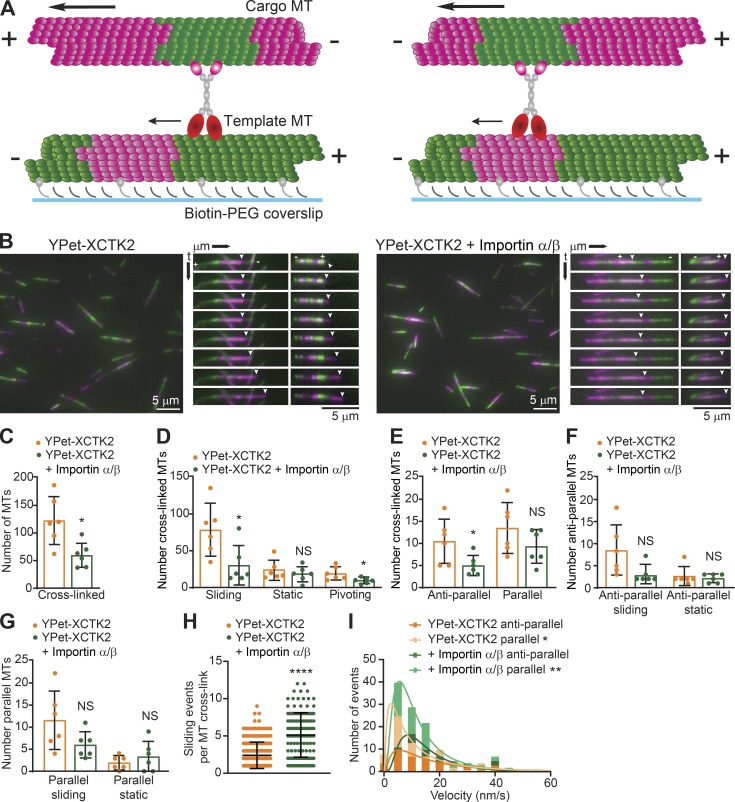
Kinesin-14s cross-link and slide both parallel and antiparallel MTs (Braun et al., 2017; Fink et al., 2009; Hentrich and Surrey, 2010), but how importins α and β regulate these activities is unknown. We devised an in vitro assay using polarity-marked MTs to assess how importin α/β modulate XCTK2 MT cross-linking and sliding (Fig. 3, A and B; and Videos 1 and 2). Addition of a molar excess of importin α/β inhibited overall XCTK2 MT cross-linking, sliding, and pivoting (Fig. 3, C and D; and Table S2), consistent with the inhibition of tail MT binding by importin α/β shown above. XCTK2 cross-linked both antiparallel and parallel MTs to similar extents (P = 0.36; Fig. 3 E and Table S2), consistent with previous studies (Fink et al., 2009; Hentrich and Surrey, 2010). Surprisingly, importin α/β preferentially inhibited antiparallel MT cross-linking by 45% with only a modest effect on parallel MT cross-links (Fig. 3 E and Table S2). The most dramatic effect of importin α/β was on the actively sliding cross-linked MTs (Fig. 3 D). In this context, the effects of importin α/β on MT sliding were much more pronounced on antiparallel MT cross-links (Fig. 3 F and Table S2) relative to parallel MT cross-links (Fig. 3 G and Table S2). These results are interesting, because they suggest that the tail of XCTK2 has two different modalities of binding MTs and that importin α/β preferentially modulates the antiparallel MT cross-linking and sliding activity.
Figure 3.
Importin α/β inhibit XCTK2 antiparallel MT cross-linking and sliding. (A) Schematic of XCTK2 antiparallel (left) and parallel (right) MT cross-linking and sliding assay with segmented MTs. (B) Representative images and kymographs of MT cross-linking and sliding with YPet-XCTK2 (left; Video 1) or YPet-XCTK2 with 4–8 molar excess of importin α/β (right; Video 2). The template MT minus (−) and plus (+) ends are indicated on the top kymograph image, and the sliding cargo MT plus end is indicated by an arrowhead. The YPet-XCTK2 kymograph images increment by 30-s intervals, and the YPet-XCTK2 + importin α/β increment by 75-s intervals to illustrate the multiple events over the course of the time lapse. Scale bars: 5 µm. (C–G) Quantification of the indicated conditions plotted as the number of cross-links per experiment, with the mean ± SD indicated as a bar graph, with the dots representing the values of individual experiments (n = 729 YPet-XCTK2 and 352 YPet-XCTK2 + importin α/β cross-linked MTs, n = 63 YPet-XCTK2 and 31 YPet-XCTK2 + importin α/β antiparallel cross-links, n = 81 YPet-XCTK2 and 56 YPet-XCTK2 + importin α/β parallel cross-links from six independent experiments; F test to compare variances and two-tailed Student’s or Welch’s t tests were performed: *, P < 0.05). (H) The number of sliding events per MT cross-link is graphed with the mean ± SD (n = 212 YPet-XCTK2 and 140 YPet-XCTK2 + importin α/β cross-links from three independent experiments; D’Agostino and Pearson normality and log-normal tests and two-tailed Mann–Whitney t test: ****, P < 0.0001). (I) Velocity of MT sliding events for antiparallel and parallel MT cross-links is graphed as a frequency histogram with the best-fit log Gaussian curve (n = 42 YPet-XCTK2 antiparallel and 71 parallel events, and n = 60 YPet-XCTK2 + importin α/β antiparallel and 116 parallel events from three independent experiments; D’Agostino and Pearson log-normal test and extra sum-of-squares F test to compare geometric means of the log Gaussian distribution of events: XCTK2 antiparallel versus parallel, *, P = 0.0362; XCTK2 + importin α/β antiparallel versus parallel, **, P = 0.0050). The frequency histogram is graphed up to 60 nm/s, excluding a single point at 120 nm/s.
Video 1.
YPet-XCTK2 slides both antiparallel (left) and parallel (right) MTs. In vitro time-lapse TIRF microscopy assay of 10 nM YPet-XCTK2 cross-linking and sliding dual-color segmented MTs shown in Fig. 3 B. Images were collected for 10 min at 15-s intervals for 41 frames and are shown at 7 frames/s. Top videos, color combined channels; middle, magenta channel; bottom, green channel. Template MTs have magenta (X-rhodamine) MT seeds with green (DyLight488) MT extensions, and cargo MTs have green MT seeds and magenta MT extensions. Template MT plus (+) and minus (−) ends are labeled, and the cargo MT plus end is labeled with an asterisk. Smaller asterisk indicates the cargo MT plus end waving in and out of frame.
Video 2.
Cargo MTs cross-linked with YPet-XCTK2 in the presence of importin α/β have more frequent sliding events. In vitro time-lapse TIRF microscopy assay of 10 nM YPet-XCTK2 + 80 nM importin α/β cross-linking and sliding dual-color segmented MTs shown in Fig. 3 B. Images were collected as for Video 1 for antiparallel (left) and parallel (right) MT cross-links. Top videos, color combined channels; middle, magenta channel; bottom, green channel. Template MT plus (+) and minus (−) ends are labeled, and the cargo MT plus end is labeled with an asterisk.
Table S2. YPet-XCTK2 ± importin α/β MT cross-linking and sliding.
| Condition, statistic, and number | YPet-XCTK2 | YPet-XCTK2 + importin α/β | F test | P value |
|---|---|---|---|---|
| Template MTs | 442 | 451 | 0.9565 | 0.9403 |
| SD | 189 | 194 | ||
| Total number | 2,654 | 2,705 | ||
| Cross-linked MTs | 122 | 59 | 0.1734 | 0.0109 |
| SD | 44 | 23 | ||
| Total number | 729 | 352 | ||
| Sliding cross-linked MTs | 78 | 31 | 0.5524 | 0.0230 |
| SD | 35 | 26 | ||
| Total number | 470 | 184 | ||
| Static cross-linked MTs | 24 | 19 | 0.6029 | 0.4709 |
| SD | 13 | 10 | ||
| Total number | 144 | 112 | ||
| Pivoting cross-linked MTs | 19 | 10 | 0.1145 | 0.0373 |
| SD | 9 | 4 | ||
| Total number | 116 | 58 | ||
| Antiparallel cross-linked MTs | 11 | 5 | 0.1858 | 0.0437 |
| SD | 5 | 3 | ||
| Total number | 63 | 31 | ||
| Parallel cross-linked MTs | 14 | 9 | 0.3925 | 0.1705 |
| SD | 6 | 4 | ||
| Total number | 81 | 56 | ||
| Antiparallel sliding cross-linked MTs | 8 | 3 | 0.0461 | 0.7810* |
| SD | 5 | 2 | ||
| Total number | 48 | 19 | ||
| Antiparallel static cross-linked MTs | 3 | 2 | 0.2290 | 0.6463 |
| SD | 2 | 1 | ||
| Total number | 15 | 12 | ||
| Parallel sliding cross-linked MTs | 12 | 6 | 0.1079 | 0.0899 |
| SD | 7 | 3 | ||
| Total number | 69 | 36 | ||
| Parallel static cross-linked MTs | 2 | 3 | 0.1041 | 0.4075 |
| SD | 2 | 3 | ||
| Total number | 12 | 20 | ||
| Experiment number | 6 | 6 | ||
| Sliding events per MT cross-link | 2.40 | 5.13 | <0.0001 | <0.0001** |
| SD | 1.77 | 2.97 | ||
| D’Agostino and Pearson normality test | <0.0001 | 0.0001 | ||
| D’Agostino and Pearson log-normal test | <0.0001 | <0.0001 | ||
| Total MT cross-links | 212 | 140 | ||
| Experiment number | 3 | 3 | ||
| Antiparallel velocity of sliding (nm/s) | 14.55 | 12.90 | NA | 0.4299 |
| Geometric SD | 2.71 | 1.89 | ||
| D’Agostino and Pearson log-normal test | 0.2744 | 0.7894 | ||
| Total sliding events | 42 | 60 | ||
| Parallel velocity of sliding (nm/s) | 8.55 | 9.88 | NA | 0.4020 |
| Geometric SD | 3.06 | 2.14 | ||
| D’Agostino and Pearson log-normal test | 0.3072 | 0.2937 | ||
| Total sliding events | 71 | 116 | ||
| Experiment number | 3 | 3 |
Mean number of MTs cross-linked, sliding, static, or pivoting from Fig. 3 (C–G). An F test to compare variances was determined for each sample set where applicable. Mean number of sliding events per MT cross-link and the geometric mean of MT sliding velocity for antiparallel and parallel MT cross-links from Fig. 3 (H and I). The sliding events per MT cross-link were neither normal nor log-normal; thus, a nonparametric two-tailed Mann-Whitney t test was performed for the sliding events. An extra sum-of-squares F test was performed for the geomean sliding velocities. NA, not applicable. A two-tailed Student’s t test was performed for statistical significance except where indicated (*, two-tailed Welch’s t test; **, two-tailed Mann–Whitney t test).
Cargo MTs often slid stochastically along the template MTs, being interrupted by pausing, reversing directions, and/or changing their rate of sliding. Within the 10-min time frame of the videos, the cargo MTs on average had two distinct sliding events per MT cross-link (Fig. 3 H, Video 1, and Table S2). The addition of importin α/β increased the number of sliding events per MT cross-link twofold (Fig. 3 H, Video 2, and Table S2). The increased sliding events in the presence of importin α/β are consistent with reduced tail MT affinity that could result in more frequent release of the tail domains from MTs due to competition with importin α/β. XCTK2 also slid antiparallel MTs 41% faster than parallel MT cross-links (Fig. 3 I, Video 1, and Table S2), consistent with differences in sliding observed previously (Hentrich and Surrey, 2010). Importin α/β addition did not affect the sliding velocity of either orientation of MT cross-links (Fig. 3 I, Video 2, and Table S2), suggesting that the tail domain cannot bind MTs and importin α/β simultaneously. These results, along with the increased sliding events in the presence of importin α/β, imply that importins α and β compete with MTs for binding to the tail domain (Chang et al., 2017).
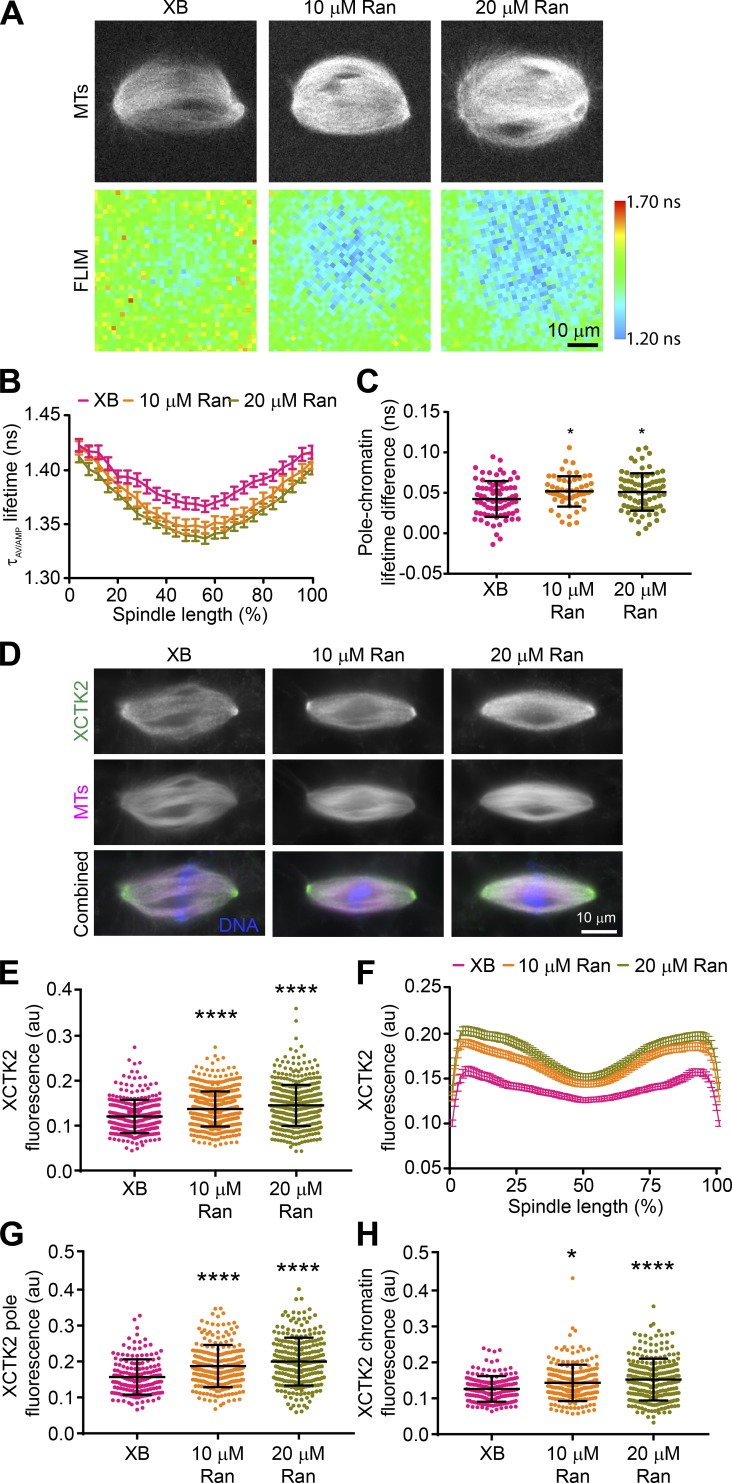
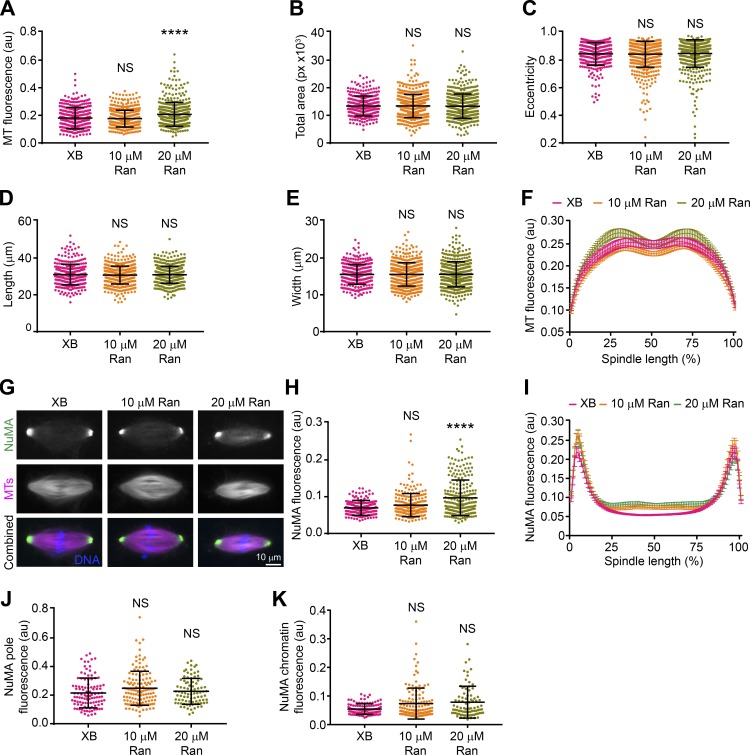
Previous work showed that the tail domains of Kinesin-14 motors are important for spindle localization (Hallen et al., 2008; Weaver et al., 2015), and thus differential interaction with importin α/β may affect XCTK2 localization. To look at the effects of the RanGTP gradient on XCTK2 localization within the spindle, we enhanced the RanGTP gradient in Xenopus egg extracts (Halpin et al., 2011) and quantified the localization of endogenous XCTK2. Adding purified Ran to spindle assembly reactions enhanced the RanGTP gradient as visualized by Rango-2, a RanGTP gradient biosensor, resulting in steeper gradients with higher RanGTP around the chromatin that decreased to control levels at the spindle poles (Fig. 4, A–C; and Table S3). Enhancing the RanGTP gradient by the addition of 10 or 20 µM Ran increased the overall localization of endogenous XCTK2 to the spindle by 12% and 17%, respectively (Fig. 4, D and E; and Table S4). Enhancing the RanGTP gradient did not change the overall morphology of the spindles, as the MT polymer level, spindle area, eccentricity, length, and width were largely unchanged, as well as the distribution of MT polymer across the spindle (Fig. S2, A–F; and Table S4). These results suggest that the RanGTP gradient promotes the global localization of Kinesin-14s to the spindle.
Figure 4.
The RanGTP gradient promotes global and local targeting of XCTK2 within the spindle. (A) Representative MT confocal and Rango-2 FLIM images of spindles assembled with XB control buffer, 10 µM Ran, or 20 µM Ran addition. Lifetimes are represented as the amplitude averaged lifetime (τAV/AMP). (B) Line scans from pole to pole of the spindle lifetime images for the indicated conditions normalized for percent spindle length (25 bins). Lifetimes per spindle length are graphed as the mean ± SEM (n = 57–78 spindles per condition from three to four independent experiments). (C) Lifetime difference between the pole and chromatin regions for each spindle in each condition from B (D’Agostino and Pearson normality test and one-way ANOVA with Tukey’s multiple comparisons test compared with XB control: *, P < 0.05). (D) Representative wide-field fluorescence microscopy images of spindle assembly reactions from parallel experiments with XB control buffer or with added Ran that were stained with α-XCTK2. (E) Spindles from D were analyzed using a Cell Profiler pipeline that measured the mean total XCTK2 spindle fluorescence based on α-XCTK2 staining in the indicated conditions and plotted with the mean ± SD (n = 314–564 spindles per condition from five independent experiments). (F) Line scans of bipolar spindles from D were performed, normalized for percentage spindle length (101 bins), and graphed as the mean ± SEM (n = 168–248 spindles per condition from four independent experiments). (G) XCTK2 peak pole fluorescence plotted as the average fluorescence from the two poles per spindle analyzed in F. (H) XCTK2 chromatin fluorescence plotted as the center spindle position for each spindle analyzed in F. In G and H, the mean ± SD is indicated. (E–H) D’Agostino and Pearson normality tests and Kruskal–Wallis with Dunn’s multiple comparisons tests compared with XB control were performed: *, P < 0.05; ****, P < 0.0001. Scale bars: 10 µm.
Table S3. Rango-2 lifetime pole-to-chromatin differences.
| Condition, statistic, and number | XB | 10 µM Ran | 20 µM Ran |
|---|---|---|---|
| Rango-2 pole-to-chromatin lifetime difference (NS) | 0.04247 | 0.05184 | 0.05115 |
| SD | 0.02217 | 0.01879 | 0.02317 |
| D’Agostino and Pearson normality test | 0.9927 | 0.4739 | 0.7530 |
| One-way ANOVA with Tukey’s multiple comparisons test | 0.0372 | 0.0351 | |
| Spindle number | 78 | 57 | 78 |
| Experiment number | 4 | 3 | 4 |
Table S4. Fluorescence and morphology of spindles assembled in enhanced RanGTP gradients in Xenopus egg extracts.
| Condition, statistic, and number | XB | 10 µM Ran | 20 µM Ran |
|---|---|---|---|
| XCTK2 fluorescence (a.u.) | 0.1205 | 0.1369 | 0.1451 |
| SD | 0.03693 | 0.03809 | 0.04572 |
| D’Agostino and Pearson normality test | <0.0001 | 0.0001 | 0.0001 |
| Kruskal–Wallis test with Dunn’s multiple comparisons test | <0.0001 | <0.0001 | |
| Spindle number | 314 | 564 | 487 |
| Experiment number | 5 | 5 | 5 |
| MT fluorescence (a.u.) | 0.1801 | 0.1763 | 0.2069 |
| SD | 0.07748 | 0.06103 | 0.08669 |
| D’Agostino and Pearson normality test | 0.0001 | 0.0001 | 0.0001 |
| Kruskal–Wallis test with Dunn’s multiple comparisons test | >0.9999 | <0.0001 | |
| Spindle number | 328 | 509 | 453 |
| Experiment number | 4 | 4 | 4 |
| Total area (px) | 12,697 | 12,935 | 12,540 |
| SD | 3,278 | 3,924 | 3,781 |
| D’Agostino and Pearson normality test | 0.0013 | 0.0001 | 0.0001 |
| Kruskal–Wallis test with Dunn’s multiple comparisons test | >0.9999 | >0.9999 | |
| Eccentricity | 0.8712 | 0.8755 | 0.8876 |
| SD | 0.07026 | 0.05873 | 0.04387 |
| D’Agostino and Pearson normality test | 0.0001 | 0.0001 | 0.0001 |
| Kruskal–Wallis test with Dunn’s multiple comparisons test | >0.9999 | 0.4134 | |
| Length (µm) | 30.45 | 30.64 | 30.41 |
| SD | 5.368 | 4.562 | 4.331 |
| D’Agostino and Pearson normality test | 0.0001 | 0.0074 | 0.0024 |
| Kruskal–Wallis test with Dunn’s multiple comparisons test | >0.9999 | >0.9999 | |
| Width (µm) | 15.63 | 15.21 | 15.34 |
| SD | 2.854 | 3.617 | 3.423 |
| D’Agostino and Pearson normality test | 0.1077 | 0.0248 | 0.0053 |
| Kruskal–Wallis test with Dunn’s multiple comparisons test | >0.9999 | >0.9999 | |
| Spindle number | 314 | 564 | 487 |
| Experiment number | 5 | 5 | 5 |
| XCTK2 pole fluorescence (a.u.) | 0.1569 | 0.1862 | 0.1998 |
| SD | 0.04905 | 0.05667 | 0.06651 |
| D’Agostino and Pearson normality test | <0.0001 | 0.0031 | 0.0732 |
| Kruskal–Wallis test with Dunn’s multiple comparisons test | <0.0001 | <0.0001 | |
| XCTK2 chromatin fluorescence (a.u.) | 0.1262 | 0.1421 | 0.1521 |
| SD | 0.03553 | 0.04858 | 0.05817 |
| D’Agostino and Pearson normality test | 0.0005 | 0.0001 | 0.0048 |
| Kruskal–Wallis test with Dunn’s multiple comparisons test | 0.0424 | <0.0001 | |
| Spindle number | 168 | 243 | 248 |
| Experiment number | 4 | 4 | 4 |
| NuMA fluorescence (a.u.) | 0.06903 | 0.07720 | 0.09696 |
| SD | 0.01803 | 0.03147 | 0.04846 |
| D’Agostino and Pearson normality test | 0.0013 | 0.0001 | 0.0001 |
| Kruskal–Wallis test with Dunn’s multiple comparisons test | 0.1527 | <0.0001 | |
| Spindle number | 209 | 240 | 239 |
| Experiment number | 4 | 4 | 4 |
| NuMA pole fluorescence (a.u.) | 0.2153 | 0.2259 | 0.2269 |
| SD | 0.1045 | 0.09043 | 0.09038 |
| D’Agostino and Pearson normality test | 0.0067 | 0.0001 | 0.0506 |
| Kruskal–Wallis test with Dunn’s multiple comparisons test | >0.9999 | >0.9999 | |
| NuMA chromatin fluorescence (a.u.) | 0.05438 | 0.07358 | 0.07644 |
| SD | 0.01853 | 0.05421 | 0.05276 |
| D’Agostino and Pearson normality test | 0.0047 | <0.0001 | 0.0001 |
| Kruskal–Wallis test with Dunn’s multiple comparisons test | >0.9999 | >0.9999 | |
| Spindle number | 107 | 134 | 86 |
| Experiment number | 4 | 4 | 4 |
Mean fluorescence and spindle morphology parameter values graphed in Figs. 4 E and S2 (A–E and H). Mean pole and chromatin fluorescence values measured from the line scans performed in Figs. 4 F and S2 I and graphed in Fig. 4 (G and H) and Fig. S2 (J and K). Statistical tests represent P values relative to control XB addition.
Figure S2.
Enhancing the RanGTP gradient does not change spindle morphology or NuMA localization. (A–E) Spindle morphology analysis with Cell Profiler of spindles measured in Fig. 4 D and graphed with the mean ± SD (n = 314–564 spindles per condition from four to five independent experiments; D’Agostino and Pearson normality and Kruskal–Wallis with Dunn’s multiple comparisons tests were performed, and statistical significance is compared with XB control buffer: ****, P < 0.0001). (F) Line scan analysis of MT polymer distribution of spindles analyzed in Fig. 4 F graphed as the mean ± SEM. (G) Representative wide-field fluorescence microscopy images of spindle assembly reactions with XB control buffer, 10 µM Ran, or 20 µM Ran addition stained with α-NuMA that were performed in parallel to those in Fig. 4. Scale bar: 10 µm. (H) Total α-NuMA fluorescence of spindles assembled in G and plotted with the mean ± SD (n = 209–240 spindles per condition from four independent experiments; D’Agostino and Pearson normality and Kruskal–Wallis with Dunn’s multiple comparisons tests were performed compared with XB control buffer: ****, P < 0.0001). (I) Line scan analysis of bipolar spindles analyzed in G, normalized for percentage spindle length (101 bins), and graphed as the mean ± SEM (n = 86–134 spindles per condition from four independent experiments). (J and K) NuMA peak pole and chromatin fluorescence as calculated in Fig. 4 (G and H) and plotted with the mean ± SD (D’Agostino and Pearson normality and Kruskal–Wallis with Dunn’s multiple comparisons tests were performed compared with XB buffer control: NS).
Kinesin-14 localization is enriched toward the spindle poles, where it functions in bipolar spindle assembly (Hatsumi and Endow, 1992a; Walczak et al., 1997). Enhancing the RanGTP gradient showed an overall increase in XCTK2 localization across the spindle that was significantly enriched toward the spindle poles relative to the chromatin region, suggesting that the RanGTP gradient also influences the localization of XCTK2 within the spindle (Fig. 4, F–H; and Table S4). Enhancing the RanGTP gradient resulted in a small but less dramatic change in the localization of NuMA, another Ran-regulated SAF, to the spindle (Fig. S2, G and K; and Table S4). These results suggest that the RanGTP gradient differentially controls the spatial localization of SAFs within the spindle. Setting up tunable effector gradients in the spindle would provide a mechanism for cells to respond to local changes in RanGTP levels.
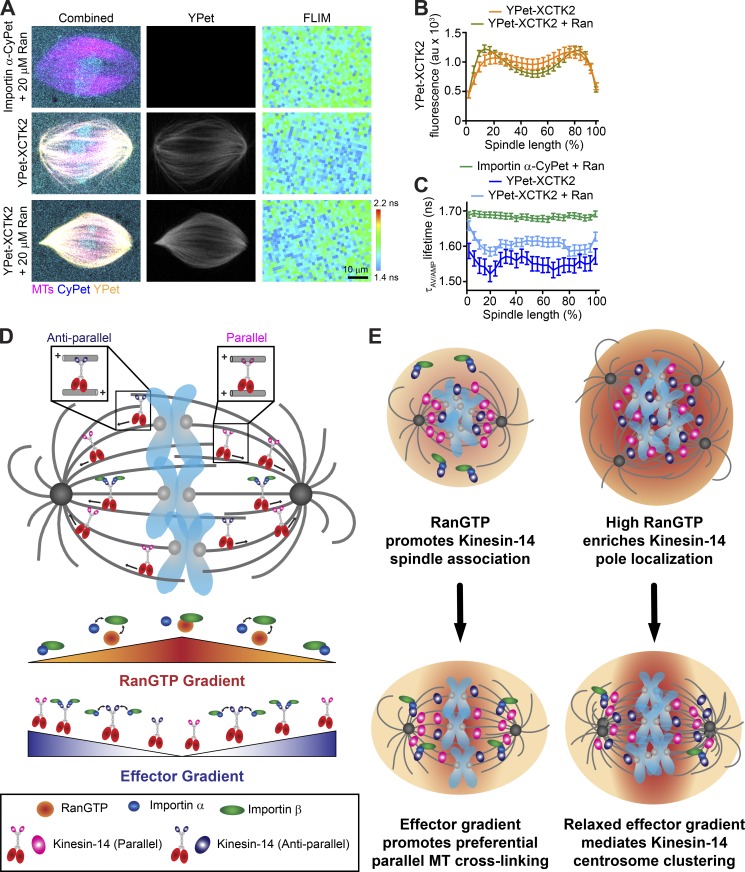
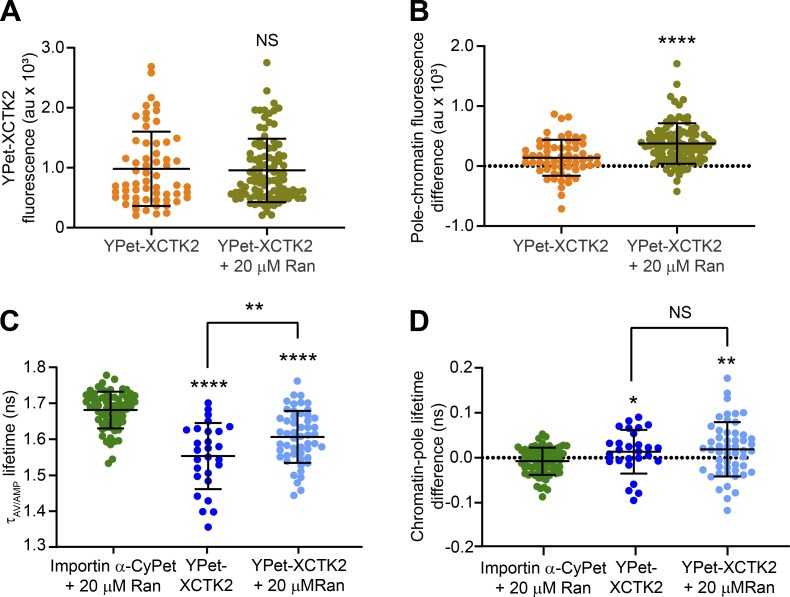
To test the idea that effector gradients within the spindle respond to different levels of RanGTP, we added 20 µM Ran to importin α-CyPet and YPet-XCTK2 spindle assembly reactions in our FLIM assay (Fig. 5, A-C). Consistent with the shift of endogenous XCTK2 to the spindle poles in enhanced RanGTP gradients shown above, YPet-XCTK2 appeared enriched at the spindle poles with added Ran (Fig. 5, A and B). While the mean YPet-XCTK2 fluorescence across the spindle did not differ with the addition of Ran, the differences between the pole and chromatin YPet-XCTK2 fluorescence with added Ran were greater than those without added Ran (Fig. S3, A and B; and Table S5), suggesting that YPet-XCTK2 had saturable binding to the spindle, but the enhanced RanGTP gradient shifted its distribution. Consistent with our FLIM results shown in Fig. 2, the lifetimes of importin α-CyPet + Ran were long and flat across the spindle, with a mean lifetime of 1.68 ± 0.05 ns (Fig. 5, A and C; Fig. S3 C; and Table S5), indicating that the added Ran did not affect the lifetime of importin α-CyPet. In addition, the lifetimes of importin α-CyPet + YPet-XCTK2 were short (1.55 ± 0.09 ns), with dips in lifetime at the spindle poles, indicative of a gradient of importin α-CyPet and YPet-XCTK2 association as seen above (Fig. 5, A and C; Fig. S3 C; and Table S5). The FLIM images of importin α-CyPet + YPet-XCTK2 + Ran showed an overall increase in lifetimes across the spindle (1.60 ± 0.07 ns) compared with YPet-XCTK2 spindles without Ran, suggesting decreased association (Fig. 5, A and C; Fig. S3 C; and Table S5). The addition of Ran did not change the chromatin-pole lifetime difference of importin α-CyPet + YPet-XCTK2 (Fig. S3 D and Table S5), suggesting that the enhanced RanGTP gradient did not affect the steepness of the gradient of YPet-XCTK2 association with importin α/β. These results suggest that the concentrations of SAFs and their affinity to importin α/β regulate the steepness of the effector gradient and not the availability of importin α/β induced by the RanGTP gradient.
Figure 5.
The RanGTP gradient generates an effector gradient of Kinesin-14s for preferential parallel MT cross-linking and sliding near the spindle poles. (A) Representative confocal fluorescence (CyPet, YPet, and MT) and lifetime (FLIM) images of spindles assembled with importin α-CyPet + nontagged XCTK2 + Ran (importin α-CyPet + 20 µM Ran), importin α-CyPet + YPet-XCTK2, or importin α-CyPet + YPet-XCTK2 + 20 µM Ran). Scale bar: 10 µm. (B) YPet fluorescence line scans of spindles assembled in A of importin α-CyPet with YPet-XCTK2 ± Ran normalized to percentage spindle length (25 bins) and graphed as the mean ± SEM (n = 60 YPet-XCTK2 and 97 YPet-XCTK2 + Ran spindles from four to five independent experiments). (C) Lifetime line scans of spindles imaged in A where lifetimes are represented as the amplitude averaged lifetime (τAV/AMP), and lifetimes per percentage of spindle length are graphed as the mean ± SEM (n = 68 importin α-CyPet + XCTK2 + Ran spindles, 27 importin α-CyPet + YPet-XCTK2, and 52 importin α-CyPet + YPet-XCTK2 + Ran spindles from four to five independent experiments). (D) XCTK2 cross-links and slides both antiparallel and parallel MTs near the chromatin where RanGTP is high and the association with importin α/β is low. Near the spindle poles where RanGTP is low, importin α/β can bind to the XCTK2 tail and can selectively inhibit XCTK2 antiparallel MT cross-linking. Thus, the RanGTP gradient sets up an opposing effector gradient of importin α/β association with XCTK2 that promotes pole focusing through preferential parallel MT cross-linking and sliding near the poles. (E) In cancer cells with centrosome amplification and high RanGTP, we propose that heightened RanGTP increases Kinesin-14 association to the spindle and that the RanGTP gradient biases Kinesin-14 localization toward the spindle poles, where reduced importin α/β association sets up an effector gradient of increased Kinesin-14 cross-linking and sliding activity that mediates centrosome clustering for cancer cell survival.
Figure S3.
Enhancing the RanGTP gradient decreases the association of XCTK2 with importin α/β without affecting the steepness of the effector gradient. (A) YPet-XCTK2 mean fluorescence across the spindle ± Ran from spindles imaged in Fig. 5 A, calculated from the line scans performed in Fig. 5 B, and graphed with the mean ± SD (n = 60 YPet-XCTK2 spindles and 97 YPet-XCTK2 + Ran spindles from four to five independent experiments; D’Agostino and Pearson normality test and two-tailed Mann–Whitney t test were performed: NS). (B) YPet-XCTK2 fluorescence difference between the pole and chromatin regions ± Ran for each spindle line scan in Fig. 5 B and graphed with the mean ± SD (D’Agostino and Pearson normality test and two-tailed Mann–Whitney t test were performed: ****, P < 0.0001). (C) Mean line scan lifetime across the spindles of importin α-CyPet ± YPet-XCTK2 ± Ran calculated from the line scans performed in Fig. 5 C and graphed with the mean ± SD (n = 68 importin α-CyPet + Ran spindles, 27 YPet-XCTK2 + importin α-CyPet spindles, and 52 YPet-XCTK2 + Ran + importin α-CyPet spindles from four to five independent experiments; D’Agostino and Pearson normality tests, F tests to compare variances, Mann–Whitney t tests compared with importin α-CyPet + Ran control and a Student’s t test between YPet-XCTK2 and YPet-XCTK2 + Ran were performed: ****, P < 0.0001; **, P < 0.01). (D) Lifetime difference between the pole and chromatin regions for each spindle from Fig. 5 C and graphed ± SD (D’Agostino and Pearson normality tests, F tests to compare variances, Welch’s t test compared with importin α-CyPet + Ran control, and Student’s t test between YPet-XCTK2 and YPet-XCTK2 + Ran were performed: *, P < 0.05; **, P < 0.01).
Table S5. YPet-XCTK2 fluorescence and lifetime means and pole-to-chromatin differences in enhanced RanGTP gradients.
| Condition, statistic, and number | Importin α/β + 20 µM Ran | YPet-XCTK2 | YPet-XCTK2 + 20 µM Ran |
|---|---|---|---|
| YPet-XCTK2 mean line scan fluorescence (a.u.) | NA | 981 | 956 |
| SD | 619 | 528 | |
| D’Agostino and Pearson normality test | 0.0137 | 0.0005 | |
| Two-tailed Mann–Whitney t test of YPet-XCTK2 ± 20 µM Ran | 0.8409 | ||
| YPet-XCTK2 pole-to-chromatin fluorescence difference (a.u.) | NA | 138 | 376 |
| SD | 300 | 338 | |
| D’Agostino and Pearson normality test | 0.4470 | <0.0001 | |
| Two-tailed Mann-Whitney t test of YPet-XCTK2 ± 20 µM Ran | <0.0001 | ||
| Spindle number | 60 | 97 | |
| Experiment number | 4 | 5 | |
| Mean line scan lifetime (NS) | 1.68 | 1.55 | 1.60 |
| SD | 0.05 | 0.09 | 0.07 |
| D’Agostino and Pearson normality test | 0.0353 | 0.4737 | 0.7387 |
| Two-tailed Mann-Whitney t test compared to importin α/β + 20 µM Ran | <0.0001 | <0.0001 | |
| F test to compare variance between YPet-XCTK2 ± 20 µM Ran | 0.1422 | ||
| Two-tailed Student’s t test of YPet-XCTK2 ± 20 µM Ran | 0.0061 | ||
| Pole to chromatin lifetime difference (NS) | −0.007662 | 0.01348 | 0.01873 |
| SD | 0.03028 | 0.04865 | 0.06032 |
| D’Agostino and Pearson normality test | 0.3706 | 0.3975 | 0.6348 |
| F test to compare variances to importin α/β + 20 µM Ran | 0.0020 | <0.0001 | |
| Two-tailed Welch’s t test compared to importin α/β + 20 µM Ran | 0.0429 | 0.0055 | |
| F test to compare variance between YPet-XCTK2 ± 20 µM Ran | 0.2374 | ||
| Two-tailed Student’s t test of YPet-XCTK2 ± 20 µM Ran | 0.6981 | ||
| Spindle number | 68 | 27 | 52 |
| Experiment number | 4 | 4 | 5 |
YPet-XCTK2 fluorescence and lifetime means and pole-to-chromatin lifetime differences of spindles measured from the line scans in Fig. 5 (B and C) and graphed in Fig. S3 (A–D).
Overall, our results uncover several important findings about how the RanGTP gradient modulates SAF activity in the spindle. First, we found that a key action of Ran and the importins is to target XCTK2 to the spindle and regulate its spatial distribution by modulating the binding affinity of the XCTK2 tail to MTs (Fig. 5 D). This is consistent with our previous work, showing that XCTK2 turnover in the spindle is spatially controlled by Ran (Weaver et al., 2015). More intriguing is the current observation that the importins preferentially inhibit XCTK2 antiparallel MT cross-linking and sliding (Fig. 5 D). Kinesin-14s can cross-link both parallel and antiparallel MTs, but how a single, small (13-kD) tail domain can bind to a structural polymer in two orientations is not known. Studies with Drosophila melanogaster Ncd suggest that there are two regions of MT binding in the tail domain (Karabay and Walker, 1999, 2003; Wendt et al., 2003). One interesting idea is that each tail domain of Kinesin-14s has two separate MT-binding domains, one that mediates parallel MT cross-links and one that mediates antiparallel MT cross-links. In this model, as cross-linked Kinesin-14s walk down the RanGTP gradient from the chromatin, competitive importin α/β binding to the NLS in the tail could inhibit antiparallel MT cross-links or could prevent nonproductive binding of the tail to MTs in the antiparallel orientation, leaving parallel MT cross-linking intact (Fig. 5 D). This idea is consistent with our biochemical analysis of XCTK2 MT sliding and tail MT binding, which is reduced but not abolished by importin α/β. This model would help explain how Kinesin-14s can focus MT minus ends at the spindle poles, which is an area of high importin α/β association. Future structural studies will thus be critical to characterize the different mechanisms of tail MT binding.
RanGTP modulates several SAFs through the action of importin α/β, suggesting a common mechanism. In contrast, our work demonstrates that not all SAFs respond equivalently to the action of RanGTP in the spindle. Most notably, high RanGTP promoted both XCTK2 association with the spindle and an increase in localization toward the poles, but high RanGTP had only minor effects on NuMA localization, which is another Ran- and importin α/β–regulated SAF (Chang et al., 2017; Nachury et al., 2001; Wiese et al., 2001). The prevailing model on how the RanGTP gradient regulates SAFs is to release the inhibitory effects of importin α/β around chromatin, essentially setting up an effector gradient parallel to the RanGTP gradient. Our combined FLIM and cross-linking results support the idea that the RanGTP gradient sets up an opposing effector gradient of importin α/β and XCTK2 association to modulate XCTK2 cross-linking, rather than turning off XCTK2 activity near the poles (Fig. 5 D, bottom). One idea is that the steep RanGTP gradient in the spindle (Athale et al., 2008) sets up importin α/β effector gradients that in turn act as rheostats to modulate activities based on local SAF concentrations and importin α/β binding affinities, rather than differential levels of free importin α/β, as previously proposed. In support of this idea, several components of the Ran pathway that influence MT behavior were shown to be controlled by their interaction with MTs rather than the RanGTP gradient per se (Oh et al., 2016). Furthermore, enhancing the RanGTP gradient decreased the level of importin α/β association with XCTK2 across the spindle in our FLIM assay but did not change the steepness of the association from the poles to chromatin. Understanding the differences in effector gradients and determining whether modulation of MT binding by importin α/β is a universal control mechanism will be important avenues for future studies.
Finally, our work brings important new insights on how cancer cells co-opt the spindle machinery for their own survival (Fig. 5 E). It is well known that many cancers have centrosome amplification, and centrosome amplification can lead to chromosome instability through clustering of centrosomes in multipolar spindles to allow for bipolar divisions and cell survival (Ganem et al., 2009; Silkworth et al., 2012). The human Kinesin-14 HSET is a key factor that promotes centrosome clustering in these cells (Chavali et al., 2016; Kwon et al., 2008; Watts et al., 2013). Various cancers also have increased Ran expression and enhanced RanGTP gradients due to increased DNA content (Hasegawa et al., 2013; Sheng et al., 2018; Xia et al., 2008). Some cancers also have increased HSET expression, which often correlates with metastasis and poor prognosis (Fu et al., 2018; Liu et al., 2016; Pannu et al., 2015; Patel et al., 2018). We propose that in cancer cells with increased HSET expression, the enhanced RanGTP gradients from higher DNA content promote HSET localization to the spindle, with a bias to the spindle poles that facilitates centrosome clustering by increased HSET-mediated cross-linking and sliding (Fig. 5 E). Thus, cancer cells have taken advantage of their enhanced RanGTP gradient for their own benefit and survival.
Materials and methods
Protein expression and purification
Plasmids for the expression of 6His-Rango-2 (pRSETA-Rango-2; Kaláb and Soderholm, 2010; Kaláb et al., 2002) and 6His-Ran (pRSETB-Ran; Wilde and Zheng, 1999) were induced in BL21(DE3)pLysS bacteria and purified using NiNTA agarose (Qiagen; Ems-McClung et al., 2004). Briefly, expression was induced with 0.1 mM isopropyl β-d-1-thiogalactopyranoside at 16°C for 24 h, cells pelleted, frozen in liquid nitrogen, and stored at −80°C. Cells were lysed in 50 mM phosphate buffer, pH 8.0, 300 mM NaCl, 10 mM imidazole, 0.1% Tween-20, 1 mM PMSF, 1 mM benzamidine, and 0.5 mg/ml lysozyme and sonicated with a Branson Sonifer two to three times at 20% output. Sodium chloride was increased to 500 mM, and β-mercaptoethanol (β-ME) was added to 5 mM. The lysate was spun at 18,000 rpm for 20 min at 4°C in a Beckman JA25.5 rotor, and the supernatant was added to equilibrated NiNTA agarose and incubated at 4°C for 1 h with rotation. Unbound proteins were separated from the proteins bound to the agarose beads by centrifugation in a clinical centrifuge for 3–4 min at setting 4. The agarose beads were then washed twice in 10 column volumes (CV) of 50 mM phosphate buffer, pH 8.0, 500 mM NaCl, 10 mM imidazole, 0.1% Tween-20, 1 mM PMSF, and 5 mM β-ME for 10 min with rotation and once in 10 CV of 50 mM phosphate buffer, pH 6.0, 500 mM NaCl, 5 mM β-ME, and 0.1 mM PMSF, and then loaded into a disposable column at 4°C. Bound protein was eluted with 10 CV of 50 mM phosphate buffer, pH 7.2, 500 mM NaCl, 400 mM imidazole, and 5 mM β-ME at 4°C in 1-ml aliquots. Aliquots containing protein were pooled and dialyzed into XB dialysis buffer (10 mM Hepes, pH 7.2, 100 mM KCl, 25 mM NaCl, 50 mM sucrose, 0.1 mM EDTA, and 0.1 mM EGTA). Rango-2 was further purified by gel filtration on a 24-ml Superose 6 10/30 column (GE Healthcare) in XB dialysis buffer. Proteins were then aliquoted and flash frozen in liquid nitrogen.
To express the 13-kD tail domain of XCTK2 fused to YPet, a parental pRSETB-YPet plasmid was created by PCR amplification of the YPET DNA from pRSETA-Rango-2 using GFPBamHIF2 and YPetSacIR2 primers and inserted into the BamHI/SacI sites of pRSETB. The cDNA for the tail domain of XCTK2, amino acids 2–120, was inserted into the SacI/HindIII sites of pRSETB-YPet for the expression of 6His-YPet-XCTK2-Tail (YPet-XCTK2-Tail). To express importin α fused to CyPet, a parental pRSETB-CyPet plasmid was created by PCR amplification of the CyPET DNA from pRSETA-Rango-2 using GFPBamHIF1 and GFP(TGA)HindIII primers and inserted into the NcoI/HindIII sites of pRSETB. The cDNA for importin α (Xenopus importin α1a) was amplified from pQE70-importin α (Ems-McClung et al., 2004) with BamHI-importin α-F and importin α-XhoI-R primers, digested with BamHI and XhoI, and cloned into the pRSETB-CyPet BamHI and XhoI sites to generate pRSETB-importin α-CyPet (importin α-CyPet). For 6His-importin α-YPet (importin α-YPet) protein expression, YPet DNA from pRSETB-YPet was digested with BamHI and HindIII and inserted into the 3′ BglII and HindIII sites of pRSETB-importin α-CyPet. YPet-XCTK2-Tail, importin α-CyPet, importin α-YPet, importin α-6His (importin α), and 6His-S-importin β (importin β) were induced, purified using NiNTA agarose, dialyzed, and aliquoted as described above for 6His-Ran except YPet-XCTK2-Tail was dialyzed into XB250 dialysis buffer (10 mM Hepes, pH 7.2, 250 mM KCl, 25 mM NaCl, 50 mM sucrose, 0.1 mM EDTA, and 0.1 mM EGTA). Primers are shown in Table 1.
Table 1. Primers used.
| Primer | Sequence (5′ to 3′) |
|---|---|
| GFPBamHIF2 | CGGGATCCGATGGTGAGCAAAGGCGAAGAG |
| YPetSacIR2 | CTAGTCGAGCTCTTATACAGTTCGTTCATGCC |
| GFPBamHIF1 | CGCGCGGGATCCATGGTGAGCAAGGGCGAGGAG |
| GFP(TGA)HindIII | CCCAAGCTTTCACTTGTACAGCTCGTC |
| BamHI–importin α-F | ATATAACGCGGATCCGATGCCGACCACAAATG |
| Importin α–XhoI-R | CTATACCCGCTCGAGCGGAAATTGAAAGACTC |
| GFPSacIR1 | CGCGAGCTCGAGATCTGAGTCC |
| YPetSacIR1 | CTAGTCGAGCTCGCTTATACAGTTCGTTCATGCC |
Restriction sites are in bold.
For expression of CyPet- or YPet-tagged full-length XCTK2, the DNA for CyPet or YPet was amplified with the GFPBamHIF1 and GFPSacIR1 primers, digested with BamHI/SacI, and inserted into pFastBac1-XCTK2 (Cai et al., 2009). For expression of YPet-XCTK2-ΔNLS, YPet DNA was amplified using GFPBamHIF1 and YPetSacIR1 primers, digested with BamHI/SacI, and inserted into pFastBac1-GFP-XCTK2-NLS2b (Cai et al., 2009), replacing the GFP DNA. Full-length XCTK2, CyPet-XCTK2, YPet-XCTK2, and YPet-XCTK2-ΔNLS proteins were expressed using the Bac-to-Bac Baculoviral System (Invitrogen) and purified using traditional ion exchange and gel filtration chromatography (Ems-McClung et al., 2004). Briefly, Sf9 insect cells were infected with baculovirus at 2–5 multiplicity of infection and incubated at 27°C for 42–48 h. Cells were pelleted, frozen in liquid nitrogen, and stored at −80°C. Cells were lysed in BRB80 (80 mM Pipes, pH 6.8, 1 mM MgCl2, and 1 mM EGTA), 100 mM KCl, 2 mM MgATP, 0.1 mM EDTA, 1 mM DTT, 1 mM PMSF, and 1 µg/ml leupeptin, pepstatin, and chymostatin (LPC) and sonicated three times at 10% output. The lysate was centrifuged at 20,000 rpm for 30 min at 4°C in a JA25.5 rotor. The supernatant was filtered through a 0.8/0.2-µm filter and subjected to ion exchange chromatography over a 2-ml HiTrap column (GE Healthcare) with a 20-CV gradient, starting with 100 mM KCl in FPLC (fast protein liquid chromatography) buffer (20 mM Pipes, pH 6.8, 1 mM MgCl2, 1 mM EGTA, 0.1 mM EDTA, 1 mM DTT, 10 µM MgATP, and 0.1 µg/ml LPC) and ending with 1 M KCl in FPLC buffer. Fractions containing protein were pooled, filtered through a 0.2-µm syringe filter, applied to a 24-ml Superose 6-gel filtration column in 1-ml aliquots, and fractionated in FPLC buffer containing 300 mM KCl. Fractions containing protein were pooled, solid sucrose added to 10%, aliquoted, and flash frozen in liquid nitrogen. All proteins were quantified by absorbance using the extinction coefficient for CyPet (35,000 M−1 cm−1) or YPet (104,000 M−1 cm−1) and/or by densitometry of proteins electrophoresed on 10% SDS-PAGE gels using BSA as a control.
Cycled spindle assembly and immunofluorescence
Cytostatic factor (CSF) extract was made from Xenopus eggs (Murray, 1991), except that cytochalasin B was used in place of cytochalasin D. Briefly, eggs of excellent quality were collected, and their zona pellucida were removed with 2% cysteine in 100 mM KCl, 1 mM MgCl2, and 0.1 mM CaCl2. Eggs were then washed sequentially with 250 ml XB (10 mM Hepes, pH 7.7, 1 mM MgCl2, 0.1 mM CaCl2, 100 mM KCl, and 50 mM sucrose), 150 ml CSF-XB (XB, 1 mM MgCl2, and 5 mM EGTA), and 100 ml CSF-XB + 10 µg/ml LPC. Eggs were packed in a clinical centrifuge and then crushed at 10,000 rpm in a Beckman SW 55 Ti rotor at 16°C for 15 min. The cytoplasmic fraction was removed with an 18-gauge needle and supplemented with 1/1,000 volume of 10 mg/ml LPC, 1/1,000 volume of cytoB, 1/50 volume of 50× energy mix (150 mM creatine phosphate, 20 mM ATP, 2 mM EGTA, and 20 mM MgCl2), 1/40 volume of 2 M sucrose, and 0.3 µM X-Rhodamine tubulin. CSF extracts containing 200 demembranated sperm/µl were cycled by the addition of 25× calcium chloride solution (final: 10 mM Hepes, pH 7.7, 1 mM MgCl2, 100 mM KCl, 150 sucrose, 10 µg/ml cytochalasin D, and 10 mM CaCl2) at RT for ∼60 min followed by the addition of an equal volume CSF extract.
For immunofluorescence, 25 µl of cycled extract was mixed with equal volumes of XB dialysis buffer or a 20× stock of Ran to give 10- or 20-µM final concentrations. Reactions were incubated at RT for 45 min before mixing with 30% glycerol, 0.5% Triton X-100, and BRB80 and pelleting onto coverslips over a 40% glycerol/BRB80 cushion. Coverslips were fixed with cold methanol for 5 min and rehydrated in TBS-Tx (10 mM Tris, pH 7.6, 150 mM NaCl, and 0.1% Triton X-100). Spindle assembly structures were immunostained by blocking with AbDil-Tx (2% BSA and TBS-Tx) and then incubated with 1 µg/ml rabbit α-XCTK2 (Walczak et al., 1997) or 2.5 µg/ml rabbit α-NuMA Tail II (Walczak et al., 1998) in AbDil-Tx for 30 min, washed with TBS-Tx, incubated with 1 µg/ml donkey anti-rabbit Alexa Fluor 488 (Invitrogen) in AbDil-Tx for 30 min, washed with TBS-Tx, and then stained with 2 µg/ml Höechst (Thermo Fisher Scientific) in TBS-Tx for 5 min. Coverslips were washed in TBS-Tx, mounted onto slides with Prolong Diamond (Invitrogen), and sealed with nail polish.
Immunofluorescent spindle assembly reactions were imaged at RT on a Nikon A1 microscope mounted with a Hamamatsu Orca-Flash 4.0 sCMOS camera and a Plan Fluor 40× Oil DIC NA-1.3 objective and controlled by Elements (Nikon). Images for each condition were acquired in three different channels (DAPI, FITC, or TRITC) as a 10 × 10 or 12 × 12 array using the Scan Large Image module. Each image was acquired with equivalent exposure times per channel. Images containing spindles were manually grouped per experiment per condition for analysis with a custom-built Cell Profiler pipeline (Lamprecht et al., 2007). The MTs in the TRITC channel were enhanced for edges, smooth texture, and tubeness. Spindles were identified in the enhanced MT image by manually drawing a line inside the spindle. An outline was then created around the spindles using the IdentifySecondaryObjects module. The mean intensity of the TRITC (MTs) and FITC (XCTK2 or NuMA) channels were quantified, and the area, eccentricity, length, and width were calculated. Results were exported as a Microsoft Excel file and graphed in Prism (GraphPad). D’Agostino and Pearson normality test and Kruskal–Wallis test with Dunn’s multiple comparisons test were performed in Prism.
The localization of XCTK2 or NuMA within the bipolar spindles identified in Cell Profiler was determined using line scans generated with the XLineScan plugin for Fiji (Wilbur and Heald, 2013). Briefly, 15-pixel-wide lines were manually drawn from pole to pole of bipolar spindles on composite DAPI/FITC/TRITC images, and the FITC and TRITC channel fluorescence intensities were measured and normalized to 101 bins. The results were exported as an Excel file and analyzed in Prism. The differences between poles and chromatin fluorescence were calculated in Excel by taking the difference between the average of the three pole fluorescence values centered on the peak XCTK2 or NuMA fluorescence at both ends of the spindle (positions 6, 7, 8, 94, 95, and 96) from the average of the center three fluorescence values (positions 50, 51, and 52) for each spindle for each condition and plotted in Prism. The peak fluorescence of XCTK2 and NuMA were used because the MT fluorescence was dome shaped and did not peak at the poles. D’Agostino and Pearson normality test and Kruskal–Wallis test with Dunn’s multiple comparisons test were performed in Prism.
FLIM
Confocal and FLIM imaging of the Rango-2 RanGTP biosensor were performed at RT in parallel cycled spindle assembly reactions, except that sperm was added to the extract at a final concentration of 600 sperm/µl. Reactions were supplemented with a 12.5× stock of proteins for a final concentration of 2 µM Rango-2 with XB dialysis buffer, 10 µM Ran, or 20 µM Ran and incubated for 45–60 min at RT before squashing 4 µl under a 22 × 22-mm #1.5 coverslip and imaging sequentially for confocal and FLIM images. A Leica SP8 confocal microscope equipped with a DMi8 inverted platform and a HC PL APO CS2 63× 1.2-NA water objective controlled by LAS-X software (Leica) was used to take confocal images of X-Rhodamine fluorescence (MTs) and YPet and CyPet fluorescence (Rango-2). Images of MTs and Rango-2 were acquired with lasers at 40 MHz, as 256 × 256-pixel images at 400 MHz scanning with two-frame averaging and a zoom factor of 2. MTs and YPet were imaged with a white-light laser at 70% power using the 594-nm laser line at 25% and the HyD5 PMT (610–700 nm) with variable gain for MTs and the 514-nm laser line at 50% and the SMD2 PMT (520–580 nm) with 50% gain for YPet. CyPet was imaged with a 440-nm laser at 90% power using SMD1 PMT (460–490 nm) with 500% gain. The pinhole was set to 1 a.u. or 111.5 µm.
FLIM of Rango-2 was performed sequentially after confocal imaging using an attached Pico Harp 300 (PicoQuant) time-correlated single photon counting system controlled by SymPhoTime64 software (PicoQuant) using a pulsed 440-nm laser at 20 MHz and 90% power. Images were acquired at 128 × 128 pixels at 200 MHz scanning and a zoom factor of 2 until the photon count reached 2,000, which on average took 1.5 min. Spindle lifetimes were determined using the SymPhoTime64 software in which the decay data were binned at 2 × 2 pixels, and the overall decay of the image was used to determine the model parameters, since Rango-2 is soluble throughout the extract. Decay of CyPet in Rango-2 best fitted a three-exponential tail model based on the χ2 values and the residuals. The average amplitude-adjusted lifetimes, τAV/AMP, were then calculated per pixel by fixing the background and the individual lifetimes determined by the fitting. FLIM and photon count images were scaled identically in SymPhoTime and exported as bitmaps for color images and TIFF or ASCII files for line scan analysis.
Spindle line scans were performed on color combined images in ImageJ (National Institutes of Health) using a XiaLineScan application (https://github.com/XiaoMutt/XiaoImageJApp/blob/master/store/XiaoImageJApp.jar) based on a similar application used by Wilbur and Heald (2013). Briefly, for each FLIM acquisition in each condition and experiment, the confocal X-Rhodamine MT, YPet, and CyPet images were scaled to 64 × 64 32-bit images and color combined with the corresponding FLIM image as a ZIP file. A 3-pixel-width line was drawn from one spindle pole to the opposite pole based on the MT (X-Rhodamine) channel, and the average lifetimes from the FLIM channel were recorded and normalized to a 25-pixel line length. Results were exported into Excel and graphed in Prism. The differences between poles and chromatin lifetimes were calculated in Excel by taking the difference between the average of the three pole lifetimes centered on the peak MT fluorescence at both ends of the spindle (positions 2, 3, 4, 22, 23, and 24) from the average of the center three lifetimes (positions 12, 13, and 14) for each spindle for each condition and plotted in Prism. D’Agostino and Pearson normality test and an ANOVA with Tukey’s multiple comparisons test were performed in Prism.
For confocal and FLIM imaging of YPet-XCTK2 and importin α-CyPet, cycled egg extracts were prepared, incubated, and squashed as described above for the Rango-2 biosensor. For FLIM of importin α-CyPet ± YPet-XCTK2 or YPet-XCTK2-ΔNLS, 20× stocks of proteins in CSF-XB (10 mM Hepes, pH 7.7, 1 mM MgCl2, 100 mM KCl, 5 mM EGTA, and 50 mM sucrose) were added to 1× for final concentrations of 1 µM importin α-CyPet + 150 nM XCTK2; 1 µM importin α-CyPet + 150 nM YPet-XCTK2; or 1 µM importin α-CyPet + 100 nM YPet-XCTK2-ΔNLS. Confocal images were taken as for Rango-2 except that the X-Rhodamine channel was imaged with 100% gain and the YPet channel at 20% gain. FLIM images were acquired the same as for Rango-2. Spindles with robust YPet-XCTK2 or YPet-XCTK2-ΔNLS localization were imaged to ensure enough spindle localization to be in the dynamic range of interaction with importin α-CyPet to produce a FLIM signal. The overall importin α-CyPet decay data were binned 2 × 2 and fitted to a three-exponential reconvolution model using SymPhoTime64 software. The average amplitude-adjusted lifetimes per pixel in each image were calculated based on the fitted parameters in which the background and lifetimes were fixed. Images were scaled in SymPhoTime, processed in ImageJ, exported into Excel, and graphed in Prism for line scans as described above for Rango-2. The mean lifetimes across the spindles were determined from the lifetime line scans and graphed in Prism. The differences between the chromatin and pole lifetimes were calculated in Excel by taking the difference between the average of the three pole lifetimes centered on the peak YPet-XCTK2 and YPet-XCTK2-ΔNLS fluorescence at both ends of the spindle (positions 3, 4, 5, 21, 22, and 23) from the average of the center three lifetimes (positions 12, 13, and 14) for each spindle for each condition and plotted in Prism. D’Agostino and Pearson normality test and Brown–Forsythe’s and Welch’s ANOVA test with Dunnett’s multiple comparisons test were performed in Prism.
For FLIM of importin α-CyPet ± YPet-XCTK2 ± Ran, 12.5× stocks of proteins in CSF-XB were added to cycled egg extracts for 1× final concentrations of 1 µM importin α-CyPet + 150 nM XCTK2 + 20 µM Ran; 1 µM importin α-CyPet + 150 nM YPet-XCTK2; or 1 µM importin α-CyPet + 150 nM YPet-XCTK2 + 20 µM Ran. Confocal and FLIM images were acquired and processed as for FLIM imaging of importin α-CyPet ± YPet-XCTK2 or YPet-XCTK2-ΔNLS. Line scans were performed on the YPet fluorescence and FLIM images as described above and graphed in Prism. The YPet fluorescence or the lifetime means across the spindle and the differences in YPet fluorescence or lifetime between the poles and chromatin were calculated as described above and graphed in Prism. For the lifetime analysis of importin α-CyPet and YPet-XCTK2 ± Ran, spindles with a mean YPet-XCTK2 fluorescence >2 SD below the average of the YPet-XCTK2 fluorescence of spindles acquired in Fig. 2 were used to ensure there was sufficient YPet-XCTK2 localized to the spindle to be in the dynamic range of interaction with importin α-CyPet to produce a FLIM signal. D’Agostino and Pearson normality tests, F tests to compare variances, and two-tailed Mann-Whitney, Welch’s, or Student t tests were performed in Prism.
Reconstitution of XCTK2 interaction with importin α/β and Ran-regulation by FRET
XCTK2 interaction with importin α/β was reconstituted by FRET using CyPet-XCTK2, importin α-YPet, and importin β. A spectral scan in a Synergy H1 (Bio-Tek) was performed in a Costar #3964 half-area black plate with 100 nM CyPet-XCTK2; 100 nM CyPet-XCTK2 and 200 nM importin α-YPet; or 100 nM CyPet-XCTK2 and 200 nM importin α-YPet ± 1.6 µM importin β in 20 mM Pipes, pH 6.8, 20 mM KCl, 1 mM MgCl2, 1 mM EGTA, 0.1 mM EDTA, 1 mM DTT, and 0.2 mg/ml casein. CyPet-XCTK2 was excited at 405 nm, and emission was recorded from 440 to 600 nm with 5-nm steps and a gain of 75. Spectra were background subtracted for YPet fluorescence bleed-through and then normalized to the CyPet emission at 460 nm in Excel and plotted in Prism.
XCTK2 Ran-regulation of importin α/β binding was demonstrated by FRET using purified YPet-XCTK2 or YPet-XCTK2-ΔNLS with purified importin α-CyPet and importin β ± purified RanQ69L. A solution of 240 nM YPet-XCTK2 or YPet-XCTK2-ΔNLS ± 240 nM importin α-CyPet, 960 nM importin β, 20 mM Pipes, pH6.8, 22 mM KCl, 1 mM MgCl2, 1 mM EGTA, 0.1 mM EDTA, 0.24 mg/ml, and 1 mM DTT was incubated at RT for 10 min in a Costar #3964 half area black plate and scanned spectrally to show binding of importin α/β to XCTK2. Ran control buffer or RanQ69L was then added to 10 µM, and the reactions were incubated for 10 min and then scanned again with excitation at 405 nm. Emission was recorded from 440 to 600 nm with 5-nm steps and a gain of 75. Final protein concentrations: 200 nM YPet-XCTK2 or YPet-XCTK2-ΔNLS, 200 nM importin α-CyPet, 800 nM importin β, and 10 µM GST-RanQ69L. Spectra were background subtracted for YPet fluorescence bleed-through and then normalized to the CyPet emission at 460 nm in Excel and plotted in Prism.
XCTK2 importin α/β and MT affinity assays
For importin α/β affinity, 100 nM CyPet-XCTK2 (monomer concentration) was mixed with equal volumes of 60 nM to 36 µM importin α-YPet or 2 nM to 5 µM importin α-YPet with 16 nM to 40 µM importin β in 20 mM Pipes, pH 6.8, 125 mM KCl, 1 mM MgCl2, 1 mM EGTA, 0.1 mM EDTA, and 0.2 mg/ml casein in a Nunc 384-well nontreated black plate (#264556), incubated for 10 min, and scanned in a Synergy H1 (BioTek). A spectral scan was performed in which CyPet was excited at 405 nm, and emission was recorded from 440 to 600 nm in 5-nm steps with a gain of 150. The spectra were background subtracted for YPet fluorescence and normalized to CyPet emission at 460 nm. Maximum FRET attained with 50 nM CyPet-XCTK2 and 2.5 µM importin α-YPet and 20 µM importin β was a value of 5, which was set as 100% CyPet-XCTK2 bound for fitting to the quadratic equation for one-site binding with ligand depletion in Prism. An extra sum-of-squares F test was performed to compare the Kd and capacity of importin α/β binding.
To determine the affinity of the tail domain for MTs in the presence and absence of different levels of importin α and/or importin β, purified YPet-Tail was used to circumvent the complexity of the motor domain binding to MTs in addition to the tail domain in the full-length protein. Tubulin was polymerized into MTs at 10 µM tubulin with 0.5 mM guanylyl-(α,β)-methylene-diphosphonate (GMPCPP; Jena Bioscience) in BRB80/DTT (80 mM Pipes, pH 6.8, 1 mM MgCl2, 1 mM EGTA, and 1 mM DTT) for 20 min at 37°C, paclitaxel was added to 10 µM, and microtubules were incubated an additional 10 min. The MTs were then sedimented at 45,000 rpm in a TLA100 rotor (Beckman) for 10 min at 35°C and resuspended in BRB80/DTT and 10 µM paclitaxel at RT. A solution of 1 µM YPet-Tail with or without 4 or 16 µM importin α/β in 20 mM Pipes, pH 6.8, 100 mM KCl, 1 mM MgCl2, 1 mM EGTA, 0.1 mM EDTA, and 0.4 mg/ml casein was mixed with an equal volume of 0–20 µM MTs in BRB80/DTT and 10 µM paclitaxel to start the reaction. Reactions were incubated for 15 min at RT and sedimented at 45,000 rpm in a TLA100 in an Optima-TLX at 22°C for 10 min. Supernatants were removed, and the pellet was resuspended in an equal volume of resuspension buffer (50 mM Pipes, pH 6.8, 50 mM, 1 mM MgCl2, 1 mM EGTA, 0.05 mM EDTA, 1 mM DTT, 0.2 mg/ml casein, and 5 µM paclitaxel). Equal volumes (15 µl) of the supernatants and pellets were moved to a Nunc 384-well nontreated black plate, and the YPet fluorescence was measured in a Synergy HI plate reader with 500-nm excitation and emission measured at 530 nm with a gain of 100. The fraction bound was determined from the amount of YPet fluorescence in the pellet divided by the sum of the supernatant and pellet fluorescence and then normalized to the concentration bound (fraction bound × 0.5 µM), plotted in Prism, and fitted to the quadratic equation for one-site binding with ligand depletion in Prism. An extra sum-of-squares F test was performed to compare the Kd and capacity of MT binding.
Visualization of XCTK2 MT cross-linking and sliding by total internal reflection fluorescence (TIRF) microscopy
To visually characterize XCTK2 parallel and antiparallel MT cross-linking and sliding, segmented red/green template MTs and green/red cargo MTs were generated by MT extension from GMPCPP MT seeds. Seeds were polymerized from 2 µM tubulin and 1 mM GMPCPP mixes for 30 min and then added 1:10 to 0.5 µM tubulin and 0.5 mM GMPCPP extension mix for 30 min before being sedimented and resuspended in BRB80, 1 mM DTT, and 10 µM paclitaxel. The red/green template MTs were generated from 12% X-Rhodamine-tubulin, 10% biotin-tubulin GMPCPP seeds with 10% Alexa Fluor 488– or Dylight488-tubulin and 10% biotin-tubulin extensions. The green/red cargo MTs were generated from 1% Alexa Fluor 488– or Dylight488-tubulin GMPCPP seeds with 23% X-Rhodamine-tubulin extensions. MT concentration was determined in terms of tubulin dimer concentration by absorbance using the extinction coefficient for tubulin (115,000 M−1 cm−1).
To visualize sliding, flow chambers were made from 24 × 30- and 22 × 22-mm biotin-polyethylene glycol (PEG)–coated #1.5 coverslips and double-sided tape (Ems-McClung et al., 2013). Biotin-PEG–coated coverslips were made by washing coverslips in 2% Micro-90 (Cole-Parmer), MilliQ water, and 0.1 M KOH, and then drying with nitrogen gas. The coverslips were activated with 1% Vectabond (Vector Laboratories) in acetone, rinsed with MilliQ water, dried with nitrogen gas, and then coated with a 3% biotin-PEG and 25% PEG solution for ≥3 h. Coverslips were washed with MilliQ water, dried with nitrogen gas, and stored in a vacuum desiccator at RT. For each reaction condition, a chamber was rinsed with BRB80 and 1 mM DTT and then incubated in 5% Pluronic F-127 in BRB80 for 3 min. The chamber was rinsed with Block (120 mM KCl, 1 µM MgATP, 20 mM Pipes, pH 6.8, 1 mM MgCl2, 1 mM EGTA, 0.1 mM EDTA, 1 mg/ml casein, 10 µM paclitaxel, and 1 mM DTT) and incubated with 0.05 mg/ml NeutrAvidin (Life Technologies), diluted in Block for 3 min, and then rinsed with Block. Template MTs in Block (0.1 µM) were introduced and incubated for 3 min and then rinsed with Block. YPet-XCTK2 (10 nM) ± importin α/β (40–80 nM) in 120 mM KCl, 20 mM Pipes, pH 6.8, 1 mM MgCl2, 1 mM EGTA, 0.1 mM EDTA, 1 mg/ml casein, and 5 µM paclitaxel was flowed in and incubated for 3 min. The chamber was rinsed with Block and then incubated with 0.1 µM cargo MTs in Block for 5 min. To activate sliding before imaging, the chamber was washed with Block containing 5 mM MgATP, 0.32 mg/ml glucose oxidase, 0.055 mg/ml catalase, and 25 mM sucrose. Prepared chambers were imaged on a Nikon A1 microscope equipped with a Hamamatsu ORCA Flash 4.0 sCMOS camera controlled by Nikon Elements and an Apo TIRF 100× oil DIC N2 NA-1.49 (working distance 120 µm) objective for 10 min. The 488-nm argon laser was set to 30% power, and the sapphire 561-nm laser was set to 15% power. Videos were taken at 15-s intervals (41 time points) as a 2 × 2 grid using the Large Image (λ) setting in Elements with both the red and green channels being acquired for 150 ms before moving to the adjacent field. One to two videos were taken per chamber. Alternatively, videos were imaged on a Nikon 90i microscope with a Nikon Plan Fluor 100× oil, 1.3-NA objective and captured with a Photometrics CoolSNAP HQ CCD camera (Roper Scientific) controlled by Molecular Devices MetaMorph. Four individual videos were acquired as a multidimensional time-lapse image for 5 min with 30-s intervals for 500 ms with FITC and Texas Red filter sets.
Each λ video acquired on the Nikon A1 was split into four separate videos before sliding analysis. MT cross-linking, sliding, and velocity were scored by hand in Fiji (ImageJ/Fiji, National Institutes of Health). MT cross-links were scored for orientation (parallel, antiparallel, or not determined) and type of cross-link (sliding, static, or pivoting). Polarity was determined by the lengths of extensions under the assumption that the plus ends had longer extensions than the minus ends due to the differences in dynamic instability for plus and minus ends of MTs (Kristofferson et al., 1986). MT cross-link orientation was determined by switching between the magenta and green channels of the video and comparing the lengths of the extensions to determine the polarity of the template and cargo MTs (see Videos 1 and 2). Only cross-links that could have their orientation determined unequivocally were included in the quantifications. An MT cross-link was scored as sliding if a cargo MT moved in one direction for at least two consecutive frames in the video in reference to a coverslip-bound template MT. Many MTs moved both directions relative to the template MT. Static cross-linked MTs were identified based on their higher fluorescence intensities relative to nearby template MTs and the presence of a green fluorescent MT seed with red fluorescent MT extensions. Pivoting cross-links were MTs swiveling or waving in and out of the focal plane of the video, typically at the ends of the MTs. Some MT cross-links displayed multiple types of movement, e.g., sliding to the MT end and then pivoting, or pivoting and then cross-linking along the length of the template MT, and either sliding in different directions or speeds or remaining static. The first sliding or pivoting event observed in the video was used to characterize the type of cross-link, whereas static cross-links were static throughout the duration of the video. For each experiment, similar numbers of template MTs (difference <30) were scored for each condition. The types of MT cross-links were tabulated in Excel as the sum of MTs from two to four separate videos per condition per experiment from six independent experiments and plotted in Prism. F tests to compare variances and two-tailed Student’s t tests or two-tailed Welch’s t tests were performed in Prism.
The velocity of MT sliding was measured from the two-color composite videos acquired on the Nikon A1 in Fiji using the MTrackJ plugin. Each sliding and static template-cargo MT cross-link was tracked for the entire length of the video (10 min) as long as the cargo MT remained cross-linked. Three template MTs in each video were also tracked to register each video due to the stage not returning precisely to the same position after acquisition of the four videos per time interval. The tracks were then measured and exported into Excel. To register the measurements of each video, the average change in x and y for each time interval was determined from the three template MT tracks and then subtracted from x and y positions in each cross-link time interval in Excel. The distance the MT moved was then calculated using the Pythagorean theorem. The distances for each video were plotted, the sliding events were identified as described above, and the velocities were calculated by dividing by the time interval that the MT slid. Some MT cross-links had multiple sliding events, which were measured and included as individual sliding events in the velocity results. Velocities for each orientation were then tabulated per condition and graphed as a frequency histogram based on the number of values with the best-fit log Gaussian curve. D’Agostino and Pearson log-normal test and the extra sum-of-squares F test for the geometric mean of the log Gaussian distribution of velocities were performed in Prism. The number of sliding events per MT cross-link was determined per condition, tabulated, and graphed in Prism with the mean ± SD. D’Agostino and Pearson normality and log-normal tests and the two-tailed Mann–Whitney t test were performed in Prism.
Online supplemental material
Fig. S1 shows the affinity of XCTK2 to importin α and importin α/β as well as the MT affinity of the XCTK2 tail domain in the presence and absence of excess importin α/β. Fig. S2 shows the morphology of the spindles with enhanced RanGTP gradients that were stained with α-XCTK2, imaged, and analyzed in Fig. 4 (D–H) as well as the localization of NuMA in enhanced RanGTP gradients that were imaged and analyzed similar to those stained for XCTK2 localization in Fig. 4. Fig. S3 shows the mean YPet-XCTK2 fluorescence and lifetimes of spindles with enhanced Ran gradients imaged by FLIM in Fig. 5 (A–C). Tables S1, S2, S3, S4, and S5 document the means and SD or SEM for each analysis in Figs. 2, 3, 4, and 5, the statistical analysis used, the number of samples, and the number of independent experiments. Videos 1 and 2 show the in vitro cross-linking and sliding assay performed in Fig. 3.
Supplementary Material
Acknowledgments
We thank Ben Walker, Kelly Hartsough, and Rebecca Heald for critical comments on the manuscript, and all members of the Walczak laboratory for helpful discussion. We thank Petr Kaláb and Sid Shaw for help on the FLIM analysis, Tarun Kapoor for suggestions on the MT sliding assay, and Jim Powers for his help with multiple types of imaging.
This work was supported by National Institutes of Health grants GM059618 and GM122482 to C.E. Walczak and GM099309 to L.N. Weaver. The Light Microscopy Imaging Center is supported in part by the Office of the Vice Provost for Research, College of Arts and Sciences, School of Medicine, Indiana University, Bloomington, and the School of Optometry at Indiana University.
The authors declare no competing financial interests.
Author contributions: S.C. Ems-McClung designed the study, performed most of the experiments, and analyzed the data. S. Zhang performed the immunofluorescence assays of spindle assembly experiments with Ran addition. M. Emch, S. Zhang, and S. Mahnoor contributed to data collection and image analysis. L.N. Weaver contributed reagents and design advice to the study. C.E. Walczak contributed to the design and interpretation of the study, as well as writing and editing of the manuscript.
References
- Athale C.A., Dinarina A., Mora-Coral M., Pugieux C., Nedelec F., and Karsenti E.. 2008. Regulation of microtubule dynamics by reaction cascades around chromosomes. Science. 322:1243–1247. 10.1126/science.1161820 [DOI] [PubMed] [Google Scholar]
- Braun M., Lansky Z., Szuba A., Schwarz F.W., Mitra A., Gao M., Lüdecke A., Ten Wolde P.R., and Diez S.. 2017. Changes in microtubule overlap length regulate kinesin-14-driven microtubule sliding. Nat. Chem. Biol. 13:1245–1252. 10.1038/nchembio.2495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S., Weaver L.N., Ems-McClung S.C., and Walczak C.E.. 2009. Kinesin-14 family proteins HSET/XCTK2 control spindle length by cross-linking and sliding microtubules. Mol. Biol. Cell. 20:1348–1359. 10.1091/mbc.e08-09-0971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carazo-Salas R.E., Guarguaglini G., Gruss O.J., Segref A., Karsenti E., and Mattaj I.W.. 1999. Generation of GTP-bound Ran by RCC1 is required for chromatin-induced mitotic spindle formation. Nature. 400:178–181. 10.1038/22133 [DOI] [PubMed] [Google Scholar]
- Catimel B., Teh T., Fontes M.R., Jennings I.G., Jans D.A., Howlett G.J., Nice E.C., and Kobe B.. 2001. Biophysical characterization of interactions involving importin-alpha during nuclear import. J. Biol. Chem. 276:34189–34198. 10.1074/jbc.M103531200 [DOI] [PubMed] [Google Scholar]
- Caudron M., Bunt G., Bastiaens P., and Karsenti E.. 2005. Spatial coordination of spindle assembly by chromosome-mediated signaling gradients. Science. 309:1373–1376. 10.1126/science.1115964 [DOI] [PubMed] [Google Scholar]
- Chang C.C., Huang T.L., Shimamoto Y., Tsai S.Y., and Hsia K.C.. 2017. Regulation of mitotic spindle assembly factor NuMA by Importin-β. J. Cell Biol. 216:3453–3462. 10.1083/jcb.201705168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavali P.L., Chandrasekaran G., Barr A.R., Tátrai P., Taylor C., Papachristou E.K., Woods C.G., Chavali S., and Gergely F.. 2016. A CEP215-HSET complex links centrosomes with spindle poles and drives centrosome clustering in cancer. Nat. Commun. 7:11005 10.1038/ncomms11005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ems-McClung S.C., Hainline S.G., Devare J., Zong H., Cai S., Carnes S.K., Shaw S.L., and Walczak C.E.. 2013. Aurora B inhibits MCAK activity through a phosphoconformational switch that reduces microtubule association. Curr. Biol. 23:2491–2499. 10.1016/j.cub.2013.10.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ems-McClung S.C., Zheng Y., and Walczak C.E.. 2004. Importin α/β and Ran-GTP regulate XCTK2 microtubule binding through a bipartite nuclear localization signal. Mol. Biol. Cell. 15:46–57. 10.1091/mbc.e03-07-0454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink G., Hajdo L., Skowronek K.J., Reuther C., Kasprzak A.A., and Diez S.. 2009. The mitotic kinesin-14 Ncd drives directional microtubule-microtubule sliding. Nat. Cell Biol. 11:717–723. 10.1038/ncb1877 [DOI] [PubMed] [Google Scholar]
- Fu X., Zhu Y., Zheng B., Zou Y., Wang C., Wu P., Wang J., Chen H., Du P., Liang B., and Fang L.. 2018. KIFC1, a novel potential prognostic factor and therapeutic target in hepatocellular carcinoma. Int. J. Oncol. 52:1912–1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem N.J., Godinho S.A., and Pellman D.. 2009. A mechanism linking extra centrosomes to chromosomal instability. Nature. 460:278–282. 10.1038/nature08136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Nédélec F., and Vale R.D.. 2005a Mechanisms for focusing mitotic spindle poles by minus end-directed motor proteins. J. Cell Biol. 171:229–240. 10.1083/jcb.200505107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Wollman R., Stuurman N., Scholey J.M., and Vale R.D.. 2005b Length control of the metaphase spindle. Curr. Biol. 15:1979–1988. 10.1016/j.cub.2005.09.054 [DOI] [PubMed] [Google Scholar]
- Gruss O.J., Carazo-Salas R.E., Schatz C.A., Guarguaglini G., Kast J., Wilm M., Le Bot N., Vernos I., Karsenti E., and Mattaj I.W.. 2001. Ran induces spindle assembly by reversing the inhibitory effect of importin alpha on TPX2 activity. Cell. 104:83–93. 10.1016/S0092-8674(01)00193-3 [DOI] [PubMed] [Google Scholar]
- Hallen M.A., Liang Z.Y., and Endow S.A.. 2008. Ncd motor binding and transport in the spindle. J. Cell Sci. 121:3834–3841. 10.1242/jcs.038497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halpin D., Kalab P., Wang J., Weis K., and Heald R.. 2011. Mitotic spindle assembly around RCC1-coated beads in Xenopus egg extracts. PLoS Biol. 9:e1001225 10.1371/journal.pbio.1001225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa K., Ryu S.J., and Kaláb P.. 2013. Chromosomal gain promotes formation of a steep RanGTP gradient that drives mitosis in aneuploid cells. J. Cell Biol. 200:151–161. 10.1083/jcb.201206142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatsumi M., and Endow S.A.. 1992a The Drosophila ncd microtubule motor protein is spindle-associated in meiotic and mitotic cells. J. Cell Sci. 103:1013–1020. [DOI] [PubMed] [Google Scholar]
- Hatsumi M., and Endow S.A.. 1992b Mutants of the microtubule motor protein, nonclaret disjunctional, affect spindle structure and chromosome movement in meiosis and mitosis. J. Cell Sci. 101:547–559. [DOI] [PubMed] [Google Scholar]
- Hentrich C., and Surrey T.. 2010. Microtubule organization by the antagonistic mitotic motors kinesin-5 and kinesin-14. J. Cell Biol. 189:465–480. 10.1083/jcb.200910125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaláb P., and Heald R.. 2008. The RanGTP gradient - a GPS for the mitotic spindle. J. Cell Sci. 121:1577–1586. 10.1242/jcs.005959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaláb P., and Soderholm J.. 2010. The design of Förster (fluorescence) resonance energy transfer (FRET)-based molecular sensors for Ran GTPase. Methods. 51:220–232. 10.1016/j.ymeth.2010.01.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaláb P., Pralle A., Isacoff E.Y., Heald R., and Weis K.. 2006. Analysis of a RanGTP-regulated gradient in mitotic somatic cells. Nature. 440:697–701. 10.1038/nature04589 [DOI] [PubMed] [Google Scholar]
- Kaláb P., Pu R.T., and Dasso M.. 1999. The ran GTPase regulates mitotic spindle assembly. Curr. Biol. 9:481–484. 10.1016/S0960-9822(99)80213-9 [DOI] [PubMed] [Google Scholar]
- Kaláb P., Weis K., and Heald R.. 2002. Visualization of a Ran-GTP gradient in interphase and mitotic Xenopus egg extracts. Science. 295:2452–2456. 10.1126/science.1068798 [DOI] [PubMed] [Google Scholar]
- Karabay A., and Walker R.A.. 1999. Identification of microtubule binding sites in the Ncd tail domain. Biochemistry. 38:1838–1849. 10.1021/bi981850i [DOI] [PubMed] [Google Scholar]
- Karabay A., and Walker R.A.. 2003. Identification of Ncd tail domain-binding sites on the tubulin dimer. Biochem. Biophys. Res. Commun. 305:523–528. 10.1016/S0006-291X(03)00827-1 [DOI] [PubMed] [Google Scholar]
- Koffa M.D., Casanova C.M., Santarella R., Köcher T., Wilm M., and Mattaj I.W.. 2006. HURP is part of a Ran-dependent complex involved in spindle formation. Curr. Biol. 16:743–754. 10.1016/j.cub.2006.03.056 [DOI] [PubMed] [Google Scholar]
- Kristofferson D., Mitchison T., and Kirschner M.. 1986. Direct observation of steady-state microtubule dynamics. J. Cell Biol. 102:1007–1019. 10.1083/jcb.102.3.1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon M., Godinho S.A., Chandhok N.S., Ganem N.J., Azioune A., Thery M., and Pellman D.. 2008. Mechanisms to suppress multipolar divisions in cancer cells with extra centrosomes. Genes Dev. 22:2189–2203. 10.1101/gad.1700908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamprecht M.R., Sabatini D.M., and Carpenter A.E.. 2007. CellProfiler: free, versatile software for automated biological image analysis. Biotechniques. 42:71–75. 10.2144/000112257 [DOI] [PubMed] [Google Scholar]
- Liu Y., Zhan P., Zhou Z., Xing Z., Zhu S., Ma C., Li Q., Zhu Q., Miao Y., Zhang J., et al. . 2016. The overexpression of KIFC1 was associated with the proliferation and prognosis of non-small cell lung cancer. J. Thorac. Dis. 8:2911–2923. 10.21037/jtd.2016.10.67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matuliene J., Essner R., Ryu J., Hamaguchi Y., Baas P.W., Haraguchi T., Hiraoka Y., and Kuriyama R.. 1999. Function of a minus-end-directed kinesin-like motor protein in mammalian cells. J. Cell Sci. 112:4041–4050. [DOI] [PubMed] [Google Scholar]
- Moore W., Zhang C., and Clarke P.R.. 2002. Targeting of RCC1 to chromosomes is required for proper mitotic spindle assembly in human cells. Curr. Biol. 12:1442–1447. 10.1016/S0960-9822(02)01076-X [DOI] [PubMed] [Google Scholar]
- Mountain V., Simerly C., Howard L., Ando A., Schatten G., and Compton D.A.. 1999. The kinesin-related protein, HSET, opposes the activity of Eg5 and cross-links microtubules in the mammalian mitotic spindle. J. Cell Biol. 147:351–366. 10.1083/jcb.147.2.351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray A.W. 1991. Cell cycle extracts. In Methods Cell Biol. Vol. 36 Kay B.K., and Peng H.B., editors. Academic Press, San Diego: 581–605. [PubMed] [Google Scholar]
- Nachury M.V., Maresca T.J., Salmon W.C., Waterman-Storer C.M., Heald R., and Weis K.. 2001. Importin β is a mitotic target of the small GTPase Ran in spindle assembly. Cell. 104:95–106. 10.1016/S0092-8674(01)00194-5 [DOI] [PubMed] [Google Scholar]
- Oh D., Yu C.H., and Needleman D.J.. 2016. Spatial organization of the Ran pathway by microtubules in mitosis. Proc. Natl. Acad. Sci. USA. 113:8729–8734. 10.1073/pnas.1607498113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohba T., Nakamura M., Nishitani H., and Nishimoto T.. 1999. Self-organization of microtubule asters induced in Xenopus egg extracts by GTP-bound Ran. Science. 284:1356–1358. 10.1126/science.284.5418.1356 [DOI] [PubMed] [Google Scholar]
- Pannu V., Rida P.C., Ogden A., Turaga R.C., Donthamsetty S., Bowen N.J., Rudd K., Gupta M.V., Reid M.D., Cantuaria G., et al. . 2015. HSET overexpression fuels tumor progression via centrosome clustering-independent mechanisms in breast cancer patients. Oncotarget. 6:6076–6091. 10.18632/oncotarget.3475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel N., Weekes D., Drosopoulos K., Gazinska P., Noel E., Rashid M., Mirza H., Quist J., Brasó-Maristany F., Mathew S., et al. . 2018. Integrated genomics and functional validation identifies malignant cell specific dependencies in triple negative breast cancer. Nat. Commun. 9:1044 10.1038/s41467-018-03283-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribbeck K., Groen A.C., Santarella R., Bohnsack M.T., Raemaekers T., Köcher T., Gentzel M., Görlich D., Wilm M., Carmeliet G., et al. . 2006. NuSAP, a mitotic RanGTP target that stabilizes and cross-links microtubules. Mol. Biol. Cell. 17:2646–2660. 10.1091/mbc.e05-12-1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp D.J., Yu K.R., Sisson J.C., Sullivan W., and Scholey J.M.. 1999. Antagonistic microtubule-sliding motors position mitotic centrosomes in Drosophila early embryos. Nat. Cell Biol. 1:51–54. 10.1038/9025 [DOI] [PubMed] [Google Scholar]
- She Z.Y., and Yang W.X.. 2017. Molecular mechanisms of kinesin-14 motors in spindle assembly and chromosome segregation. J. Cell Sci. 130:2097–2110. 10.1242/jcs.200261 [DOI] [PubMed] [Google Scholar]
- Sheng C., Qiu J., Wang Y., He Z., Wang H., Wang Q., Huang Y., Zhu L., Shi F., Chen Y., et al. . 2018. Knockdown of Ran GTPase expression inhibits the proliferation and migration of breast cancer cells. Mol. Med. Rep. 18:157–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silkworth W.T., Nardi I.K., Paul R., Mogilner A., and Cimini D.. 2012. Timing of centrosome separation is important for accurate chromosome segregation. Mol. Biol. Cell. 23:401–411. 10.1091/mbc.e11-02-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahara K., Takagi M., Ohsugi M., Sone T., Nishiumi F., Maeshima K., Horiuchi Y., Tokai-Nishizumi N., Imamoto F., Yamamoto T., et al. . 2008. Importin-beta and the small guanosine triphosphatase Ran mediate chromosome loading of the human chromokinesin Kid. J. Cell Biol. 180:493–506. 10.1083/jcb.200708003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trieselmann N., Armstrong S., Rauw J., and Wilde A.. 2003. Ran modulates spindle assembly by regulating a subset of TPX2 and Kid activities including Aurora A activation. J. Cell Sci. 116:4791–4798. 10.1242/jcs.00798 [DOI] [PubMed] [Google Scholar]
- Walczak C.E., Verma S., and Mitchison T.J.. 1997. XCTK2: a kinesin-related protein that promotes mitotic spindle assembly in Xenopus laevis egg extracts. J. Cell Biol. 136:859–870. 10.1083/jcb.136.4.859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak C.E., Vernos I., Mitchison T.J., Karsenti E., and Heald R.. 1998. A model for the proposed roles of different microtubule-based motor proteins in establishing spindle bipolarity. Curr. Biol. 8:903–913. 10.1016/S0960-9822(07)00370-3 [DOI] [PubMed] [Google Scholar]
- Watts C.A., Richards F.M., Bender A., Bond P.J., Korb O., Kern O., Riddick M., Owen P., Myers R.M., Raff J., et al. . 2013. Design, synthesis, and biological evaluation of an allosteric inhibitor of HSET that targets cancer cells with supernumerary centrosomes. Chem. Biol. 20:1399–1410. 10.1016/j.chembiol.2013.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver L.N., Ems-McClung S.C., Chen S.H., Yang G., Shaw S.L., and Walczak C.E.. 2015. The Ran-GTP gradient spatially regulates XCTK2 in the spindle. Curr. Biol. 25:1509–1514. 10.1016/j.cub.2015.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt T., Karabay A., Krebs A., Gross H., Walker R., and Hoenger A.. 2003. A structural analysis of the interaction between ncd tail and tubulin protofilaments. J. Mol. Biol. 333:541–552. 10.1016/j.jmb.2003.08.051 [DOI] [PubMed] [Google Scholar]
- Wiese C., Wilde A., Moore M.S., Adam S.A., Merdes A., and Zheng Y.. 2001. Role of importin-beta in coupling Ran to downstream targets in microtubule assembly. Science. 291:653–656. 10.1126/science.1057661 [DOI] [PubMed] [Google Scholar]
- Wilbur J.D., and Heald R.. 2013. Mitotic spindle scaling during Xenopus development by kif2a and importin α. eLife. 2:e00290 10.7554/eLife.00290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilde A., and Zheng Y.. 1999. Stimulation of microtubule aster formation and spindle assembly by the small GTPase Ran. Science. 284:1359–1362. 10.1126/science.284.5418.1359 [DOI] [PubMed] [Google Scholar]
- Xia F., Lee C.W., and Altieri D.C.. 2008. Tumor cell dependence on Ran-GTP-directed mitosis. Cancer Res. 68:1826–1833. 10.1158/0008-5472.CAN-07-5279 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.