Axon initial segments (AISs) initiate action potentials and regulate the trafficking of vesicles between somatodendritic and axonal compartments. Torii et al. show that NuMA1 is transiently located at the AIS and promotes rapid AIS assembly by inhibiting the endocytosis of neurofascin-186.
Abstract
Axon initial segments (AISs) initiate action potentials and regulate the trafficking of vesicles between somatodendritic and axonal compartments. However, the mechanisms controlling AIS assembly remain poorly defined. We performed differential proteomics and found nuclear mitotic apparatus protein 1 (NuMA1) is downregulated in AIS-deficient neonatal mouse brains and neurons. NuMA1 is transiently located at the AIS during development where it interacts with the scaffolding protein 4.1B and the dynein regulator lissencephaly 1 (Lis1). Silencing NuMA1 or protein 4.1B by shRNA disrupts AIS assembly, but not maintenance. Silencing Lis1 or overexpressing NuMA1 during AIS assembly increased the density of AIS proteins, including ankyrinG and neurofascin-186 (NF186). NuMA1 inhibits the endocytosis of AIS NF186 by impeding Lis1’s interaction with doublecortin, a potent facilitator of NF186 endocytosis. Our results indicate the transient expression and AIS localization of NuMA1 stabilizes the developing AIS by inhibiting endocytosis and removal of AIS proteins.
Introduction
Somatodendritic excitatory and inhibitory postsynaptic potentials converge on axon initial segments (AISs) to initiate and modulate axonal action potentials. Action potential initiation depends on clustered Na+ channels estimated to be at a density ∼50× higher at the AIS compared with dendrites (Kole et al., 2008; Lorincz and Nusser, 2010). Other clustered AIS proteins include the cytoskeletal and scaffolding proteins ankyrinG (AnkG) and β4 spectrin, and the cell adhesion molecule neurofascin-186 (NF186; Nelson and Jenkins, 2017). Developing neurons rapidly assemble AIS as they arrive at their final cortical destinations (Galiano et al., 2012), while in mature neurons, AIS proteins are remarkably stable with very long half-lives (Hedstrom et al., 2008; Saifetiarova et al., 2017). Both the initial clustering and stability of AIS membrane proteins is attributed mainly to their direct association with AnkG, which functions as the master scaffolding protein at the AIS; Na+ and K+ channels, NF186, and β4 spectrin are all clustered at the AIS through their AnkG-binding domains (Garrido et al., 2003; Pan et al., 2006; Yang et al., 2007). In addition, AIS membrane proteins themselves may contribute to AIS stability. For example, loss of NF186 or Na+ channels destabilizes the AIS protein complex (Leterrier et al., 2017; Xu and Shrager, 2005; Zonta et al., 2011).
Nav1.6 Na+ channels are directly trafficked to the AIS, and their insertion and retention requires AnkG binding (Akin et al., 2015). Other studies suggest that during neuronal maturation, AIS membrane proteins are first inserted into somatodendritic domains and then retained at the AIS through tethering to the AnkG-dependent AIS cytoskeleton; proteins that are not tethered to AnkG are removed by endocytosis (Fache et al., 2004). For example, NF186 is tethered to AnkG through a cytoplasmic FIGQY motif (Tuvia et al., 1997). Phosphorylation of the tyrosine inhibits the interaction between NF186 and AnkG (Garver et al., 1997) and instead promotes NF186 binding to doublecortin (DCX; Kizhatil et al., 2002). DCX facilitates the endocytosis and retrieval of NF186 from the plasma membrane (Yap et al., 2012). While tethering to the AIS may be efficient at a well-developed, AnkG-rich AIS, in developing neurons with a less well-defined AIS cytoskeleton and few AIS membrane proteins, how is the balance between endocytosis and membrane retention regulated to assemble and stabilize an AIS? Unfortunately, the mechanisms responsible for the early assembly of AIS remain poorly defined.
We used differential proteomics to search for regulators of AIS development. We found that nuclear mitotic apparatus protein 1 (NuMA1), previously reported to play important roles in cell division (Kiyomitsu and Cheeseman, 2013; Williams et al., 2011), is transiently located at the AIS in postmitotic neurons. There, it promotes the overall stability of the AIS protein complex by blocking the interaction between DCX and Lis1, thereby inhibiting endocytosis of AIS membrane proteins precisely during the time when the AIS is assembled.
Results
Endocytosis of somatodendritic NF186 contributes to its differential AIS enrichment
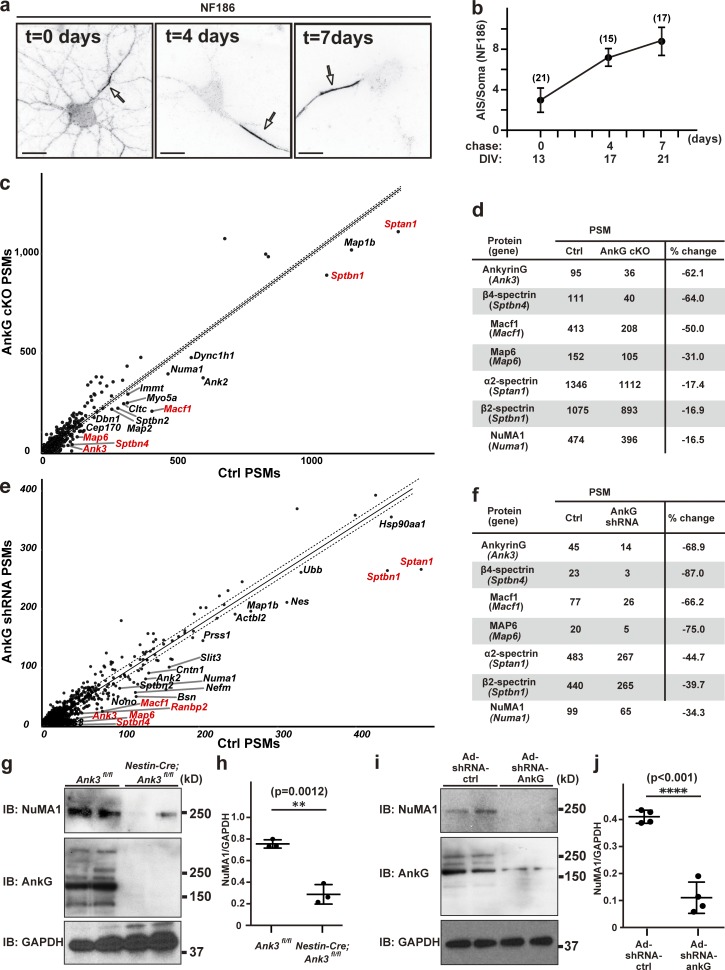
One remarkable feature of the AIS is the very high density of ion channels, cell adhesion molecules, and scaffolding proteins that can be found there. To begin to determine the temporal dynamics of AIS protein localization and stability, we live labeled NF186 for 1 h in day in vitro (DIV) 13 cultured hippocampal neurons using antibodies against NF186’s extracellular domain. Immediately after live labeling (t = 0 d), we found both AIS and somatodendritic NF186 immunoreactivity (Figs. 1, 2, 3, 4, 5, 6, 7, 8, 9, S1, and S2). However, a chase of 4 and 7 d showed that while AIS NF186 was retained, somatodendritic NF186 was efficiently removed, resulting in very strong enrichment at the AIS compared with the soma (Fig. 1, a and b).
Figure 1.
NuMA1 protein levels are reduced in AIS-deficient neurons. (a) Surface NF186 in primary hippocampal neurons. The chase time is indicated. Arrows indicate AIS in hippocampal neurons. Scale bars = 10 µm. (b) Ratio of AIS to soma NF186 immunofluorescence after live labeling and chase. Mean ± SEM is shown for three independent experiments. The total number of neurons counted is given for each data point. No statistical comparison was performed. (c) The number of PSMs for each protein identified by MS of the detergent-insoluble membrane fraction isolated from P0 control and AnkG cKO mice (Nestin-Cre; AnkGf/f). Gene names are used for simplicity, and those with protein products previously reported at the AIS are in red. The dotted lines represent a “confidence interval” based on a cutoff of 10 PSMs and the least-squares fit of the results. Proteins below the lines are candidates whose expression levels may depend on AnkG expression and may be located at the AIS. (d) Examples of proteins with reduced amounts in the AnkG cKO mice. (e) The number of PSMs for each protein identified in cultured hippocampal neurons treated with control or AnkG shRNA. (f) Examples of proteins with reduced amounts in the AnkG shRNA-treated neurons. (g) Immunoblots of NuMA1, AnkG, and GAPDH from the membrane fraction of brain homogenates from control and AnkG cKO mice at P0. (h) The NuMA1 to GAPDH ratios in immunoblots as shown in g. Mean ± SEM is shown (**, P = 0.0012; n = 3 independent experiments; unpaired two-tailed Student’s t test). (i) Immunoblots of NuMA1, AnkG, and GAPDH from cell lysates of control and AnkG shRNA-treated neurons at DIV12. (j) The NuMA1 to GAPDH ratios in immunoblots as shown in i. Mean ± SEM is shown (****, P < 0.001; n = 4 independent experiments, unpaired two-tailed Student’s t test). Ad, adenoviral delivered; IB, immunoblot.
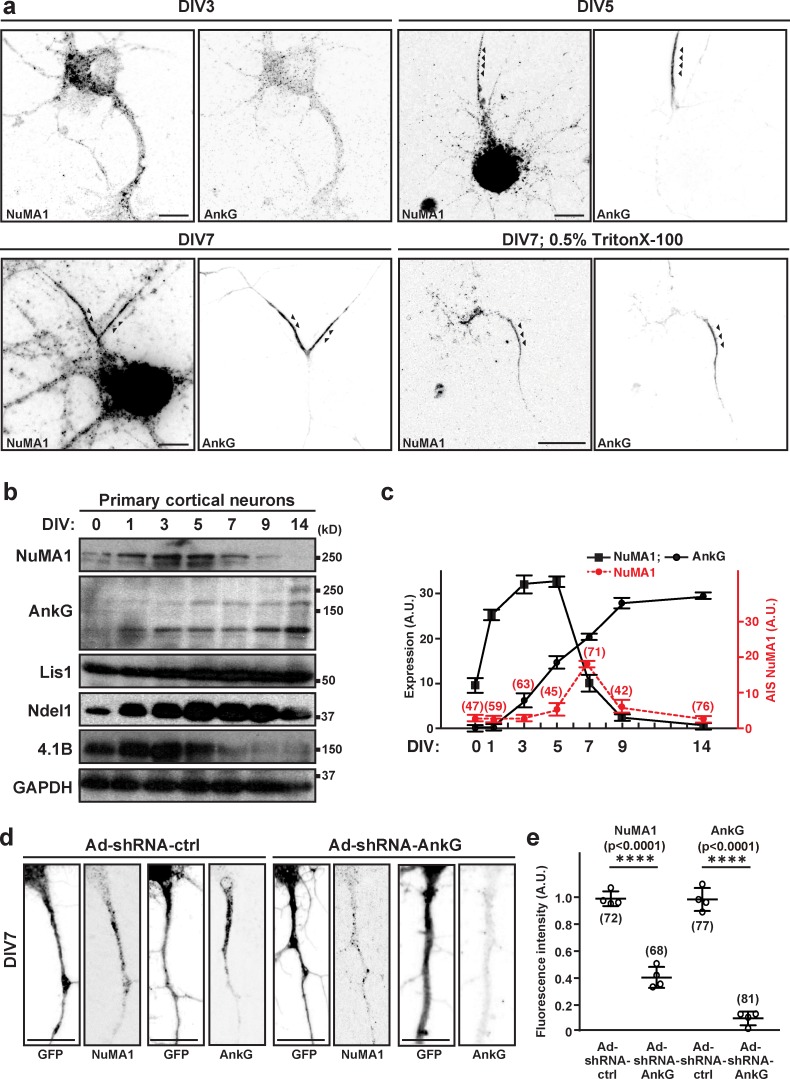
Figure 2.
NuMA1 is transiently located at the AIS during development. (a) Immunostaining of DIV3, DIV5, and DIV7 hippocampal neurons using antibodies against NuMA1 and AnkG. Arrowheads indicate AIS in hippocampal neurons. In one instance (lower right panels), neurons were treated with 0.5% Triton X-100 before fixation to reveal the detergent-insoluble pool of NuMA1 and AnkG. Scale bars = 7 µm. (b) Immunoblots of NuMA1, AnkG, Lis1, Ndel1, 4.1B, and GAPDH from lysates of developing cultured cortical neurons (DIV0, DIV1, DIV3, DIV5, DIV7, DIV9, and DIV14). (c) Quantification of protein expression for NuMA1 (black squares) and AnkG (black circles) at each developmental stage is shown. n = 3 independent immunoblotting experiments. The average immunofluorescence intensity for NuMA1 at the AIS at each developmental stage is shown (red circles); the number of neurons measured at each time is indicated. Mean ± SEM is shown. No statistical tests were performed. (d) Immunostaining of DIV7 hippocampal neurons that were transduced at DIV1 by control or AnkG shRNA-expressing adenovirus, using antibodies against GFP, NuMA1, and AnkG in DIV7 neurons. Scale bars = 5 µm. (e) The fluorescence intensity of AIS NuMA1 and AnkG after transduction with control or AnkG shRNA (****, P < 0.001; n = 4 independent experiments, Tukey’s multiple comparison test; mean ± SEM; the number of neurons analyzed for each condition is also shown). Ad, adenoviral delivered.
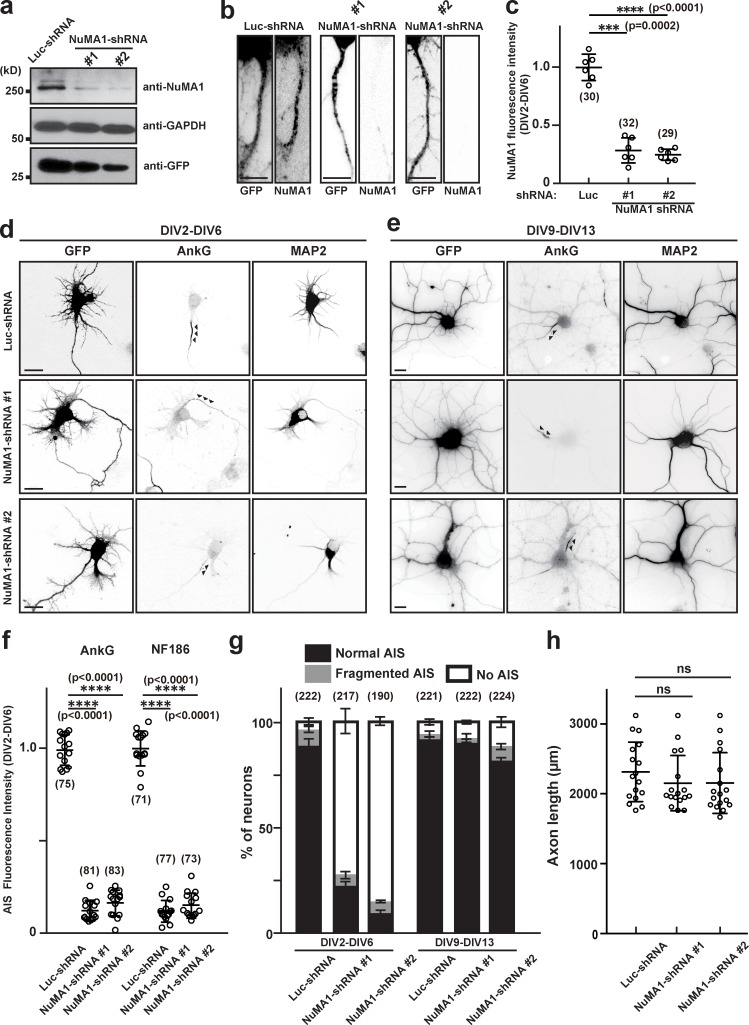
Figure 3.
Silencing expression of NuMA1 disrupts AIS assembly but not maintenance. (a) Mouse neuroblastoma Neuro-2a cells were transfected with an shRNA construct for control (Luc-shRNA), NuMA1-shRNA #1, or NuMA1-shRNA #2 to validate shRNA efficiency. The expression levels of NuMA1, GAPDH, and GFP were determined by immunoblotting. (b) Immunostaining of hippocampal neurons transfected with Luc-shRNA, NuMA1-shRNA #1, or NuMA1-shRNA #2 at DIV2 and stained at DIV6. Scale bars = 3 µm. (c) Quantification of NuMA1 fluorescence in hippocampal neurons after knockdown by shRNA. Mean ± SEM is shown (***, P = 0.0002; ****, P < 0.0001; n = 6 independent experiments, repeated measures one-way ANOVA; the number of neurons analyzed for each condition is also shown). (d and e) Hippocampal neurons were transfected with Luc-shRNA, NuMA1-shRNA #1, or NuMA1-shRNA #2 at DIV2 and stained at DIV6 (d), or transfected at DIV9 and stained at DIV13 (e) using antibodies against GFP, AnkG, and MAP2. Arrowheads indicate AIS in hippocampal neurons. Scale bars = 5 µm. (f) Fluorescence intensity of AnkG and NF186 at the AIS in neurons transfected using Luc- or NuMA1-shRNAs; neurons were transfected at DIV2 and analyzed at DIV6. Mean ± SEM is shown (****, P < 0.0001; n = 15 independent experiments, repeated measures one-way ANOVA; the number of neurons analyzed for each condition is also shown). (g) Quantification of the staining pattern for AnkG in shRNA-transfected neurons. The number of neurons counted is shown. Mean ± SEM is shown. No statistical comparison was performed. (h) Axon length in neurons transfected with control or NuMA1-shRNA. Mean ± SEM is shown (N = 3 independent experiments, n = 17 total cells measured; one neuron was measured per coverslip; repeated measures one-way ANOVA).
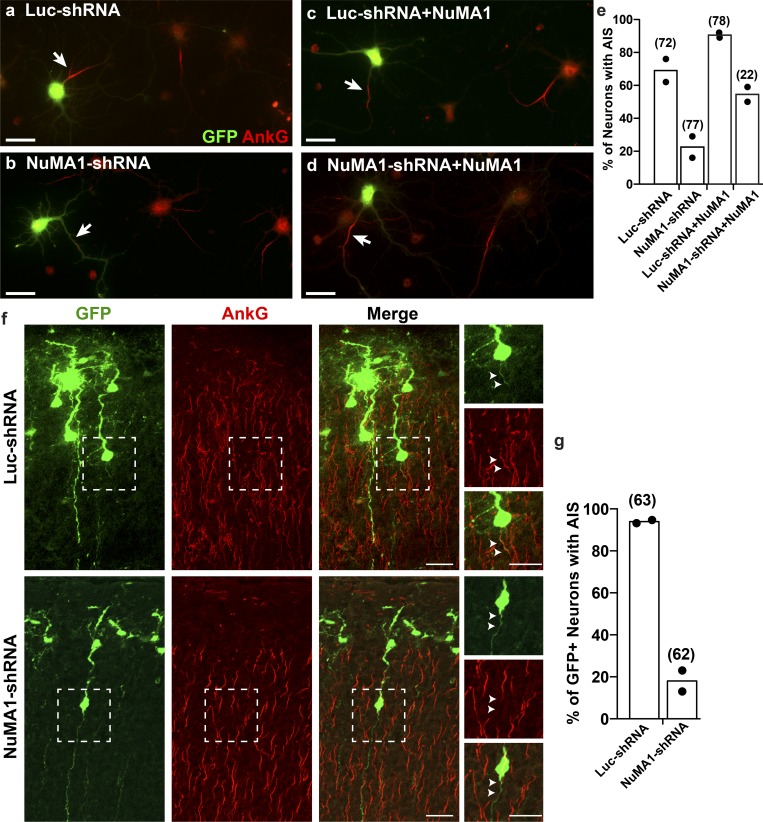
Figure 4.
Silencing expression of NuMA1 in vivo blocks AIS assembly. (a–d) An shRNA-resistant NuMA1 plasmid rescues AIS after NuMA1 knockdown. Immunostaining of cortical neurons using antibodies against GFP and AnkG. Cortical neurons were transfected with the indicated plasmids. GFP-labeled neurons express the shRNA and NuMA1 plasmids. Scale bars = 25 µm. (e) The percentage of cortical neurons in vitro with an AIS after transfection with the indicated plasmids. The mean of two independent experiments is shown, with the total number of cells analyzed shown in parentheses above each column. (f) Immunostaining of P5 cortical neurons for GFP and AnkG after in utero electroporation at E15.5. Boxed regions are shown in the panels to the right. Arrowheads indicate the proximal axon (i.e., AIS) of electroporated cortical neurons. Scale bars = 25 µm, and 20 µm for insets. (g) The percentage of cortical neurons in vivo after in utero electroporation with an AIS after transfection with the indicated plasmids. The mean of two independent experiments (mice) is shown, with the total number of cells analyzed shown in parentheses above each column.
Figure 5.
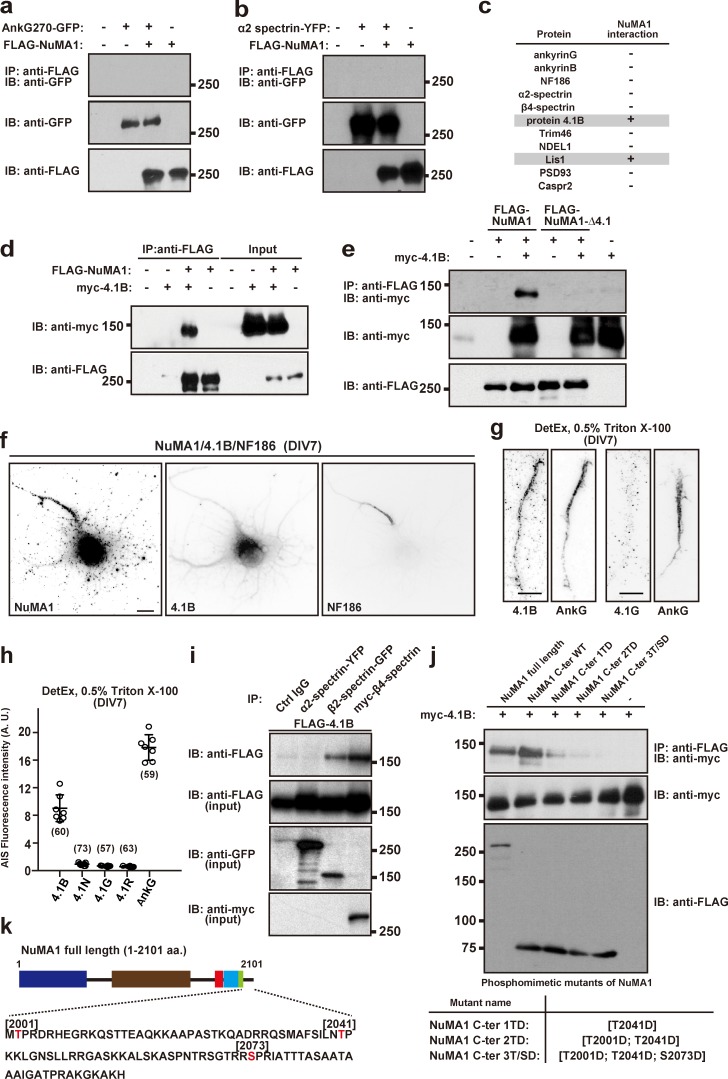
Protein 4.1B is at the AIS and interacts with NuMA1. (a and b) Coimmunoprecipitation experiments in HEK293T cells show FLAG-NuMA1 does not coimmunoprecipitate AnkG270-GFP (a) or α2 spectrin-YFP (b). The lower two panels show the starting lysates. (c) Summary of all coimmunoprecipitation experiments to test for NuMA1 interactors. (d) Coimmunoprecipitation of transfected FLAG-NuMA1 and myc-4.1B from HEK293T cell lysates. (e) FLAG-NuMA1 or FLAG-NuMA1Δ4.1 cotransfected with protein 4.1B in HEK293T cells. Cell lysates were immunoprecipitated with an anti-FLAG antibody and immunoblotted with anti-FLAG antibody and anti-myc antibody. (f) Immunostaining of DIV7 hippocampal neurons using antibodies against NuMA1, protein 4.1B, and NF186. Scale bar = 5 µm. (g) Immunostaining of proteins 4.1B, 4.1G, and AnkG in DIV7 hippocampal neurons after detergent extraction using 0.5% Triton X-100. Scale bars = 5 µm. (h) Fluorescence intensity of 4.1B, 4.1N, 4.1G, and 4.1R at AIS after detergent extraction using 0.5% Triton X-100. The mean ± SEM is shown (n = 7 independent experiments; the number of neurons analyzed for each condition is also shown). No statistical comparison was performed. (i) FLAG-4.1B and α2 spectrin-YFP, β2 spectrin-GFP, or myc-β4 spectrin constructs were cotransfected into HEK293T cells. After 48 h, cells were lysed and cell lysates were immunoprecipitated using control IgG, or antibodies against α2 spectrin, β2 spectrin, or β4 spectrin. Immunoblotting shows protein 4.1B binds to β spectrins. (j) Coimmunoprecipitation of myc-4.1B with full-length FLAG-tagged NuMA1, a C-terminal NuMA1 fragment, and C-terminal NuMA1 fragment phosphomimetics. (k) Schematic structure of NuMA1 and its C-terminal amino acids. Known Cdk1 and Cdk2 phosphorylation sites are indicated in red (Thr2001, Thr2014, and Ser2073). IB, immunoblot; IP, immunoprecipitation.
Figure 6.
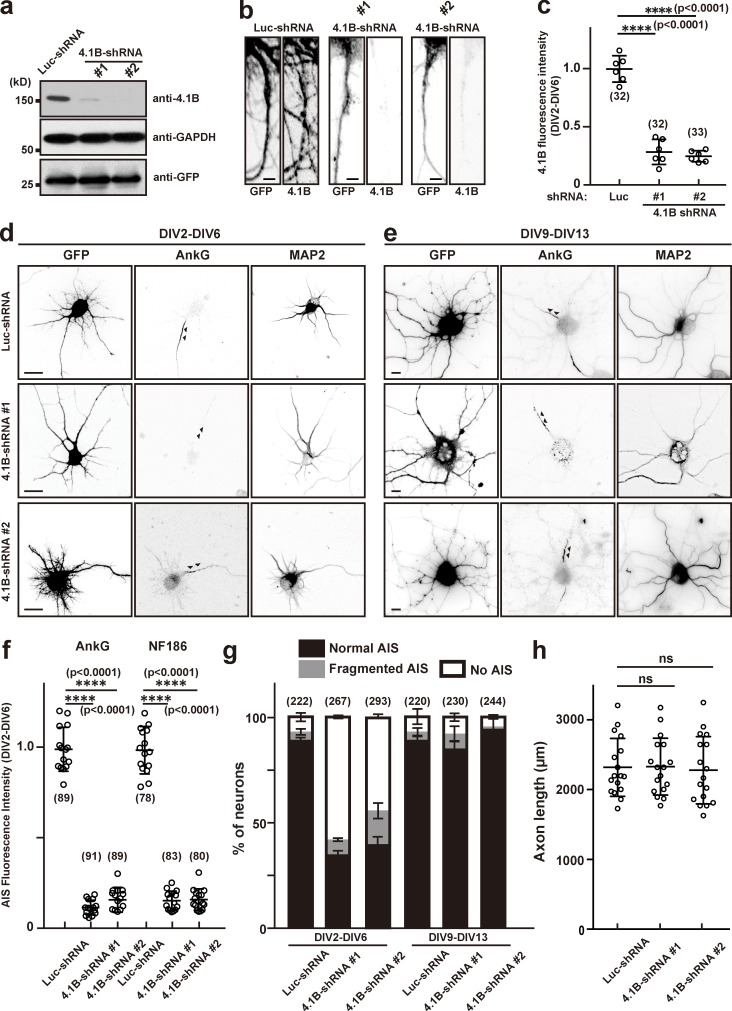
Silencing expression of protein 4.1B disrupts AIS assembly but not maintenance. (a) Mouse neuroblastoma Neuro-2a cells were transfected with an shRNA construct for control, (Luc-shRNA), 4.1B-shRNA #1, or 4.1B-shRNA #2 to validate shRNA efficiency. The expression levels of 4.1B, GAPDH, and GFP were determined by immunoblotting. (b) Immunostaining of hippocampal neurons transfected with Luc-shRNA, 4.1B-shRNA #1, or 4.1B-shRNA #2 at DIV2 and stained at DIV6. Scale bars = 3 µm. (c) Quantification of 4.1B immunofluorescence in hippocampal neurons after knockdown by shRNA. Mean ± SEM is shown (****, P < 0.0001; n = 6 independent experiments, repeated measures one-way ANOVA; the number of neurons analyzed for each condition is also shown). (d and e) Hippocampal neurons were transfected with Luc-shRNA, 4.1B-shRNA #1, or 4.1B-shRNA #2 at DIV2 and stained at DIV6 (d), or transfected at DIV9 and stained at DIV13 (e) using antibodies against GFP, AnkG, and MAP2. Arrowheads indicate AIS in hippocampal neurons. Scale bars = 5 µm. (f) Fluorescence intensity of AnkG and NF186 at the AIS in neurons transfected using Luc- or 4.1B-shRNAs; neurons were transfected at DIV2 and analyzed at DIV6. Mean ± SEM is shown (****, P < 0.0001; n = 15 independent experiments, repeated measures one-way ANOVA; the number of neurons analyzed for each condition is also shown). (g) Quantification of the staining pattern for AnkG in shRNA-transfected neurons. The number of neurons counted is shown. Mean ± SEM is shown. No statistical comparison was performed. (h) Axon length in neurons transfected with control or 4.1B-shRNAs. Mean ± SEM is shown (N = 3 independent experiments, n = 17 total cells measured; one neuron was measured per coverslip; repeated measures one-way ANOVA).
Figure 7.
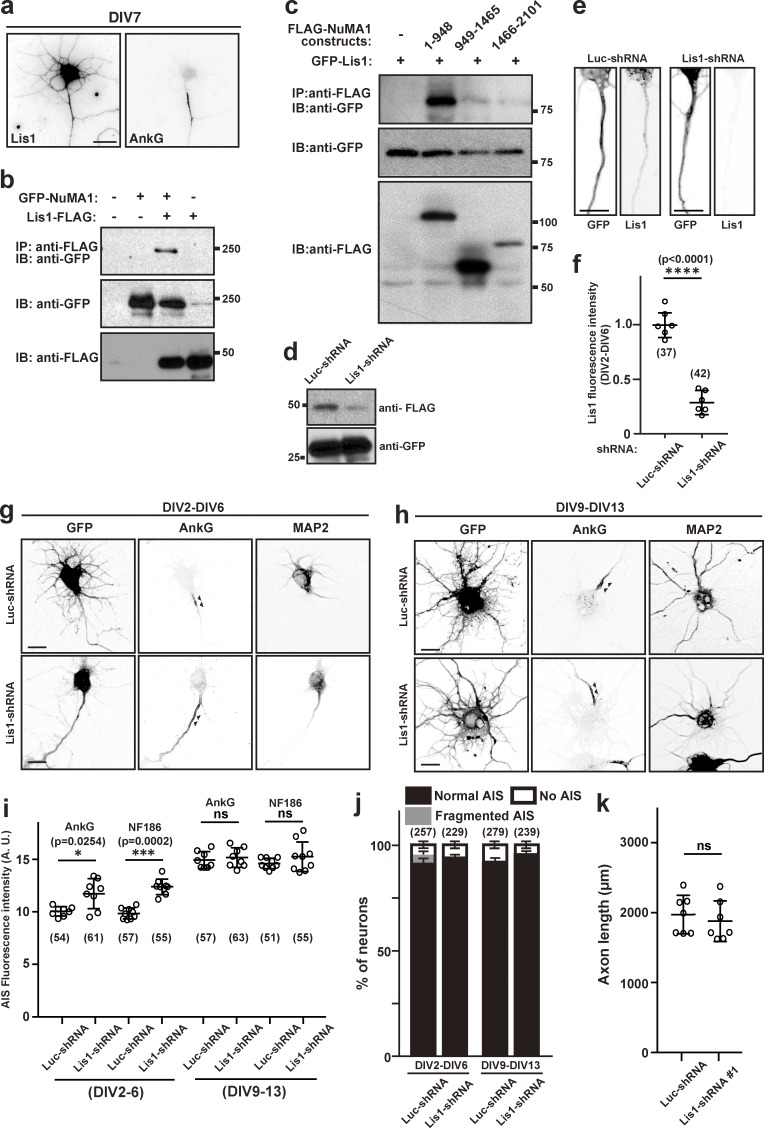
Lis1 interacts with NuMA1 and regulates AIS structure. (a) DIV7 hippocampal neurons immunostained using antibodies against Lis1 and AnkG. Scale bar = 10 µm. (b) Coimmunoprecipitation of transfected GFP-NuMA1 and FLAG-Lis1 from HEK293T cell lysates. (c) FLAG-tagged fragments of NuMA1 were cotransfected with GFP-Lis1 into HEK293T cells and lysed after 48 h. Immunoprecipitations were performed using FLAG antibodies, and immunoprecipitates were immunoblotted using both GFP and FLAG antibodies. (d) Immunoblot of HEK293T cells transfected with Lis1-FLAG and Luc-shRNA or Lis1-FLAG and Lis1-shRNA. (e) Immunostaining of hippocampal neurons transfected with Luc-shRNA or Lis1-shRNA at DIV2 and stained at DIV6. Scale bars = 10 µm. (f) Quantification of Lis1 immunofluorescence in hippocampal neurons after knockdown by shRNA. Mean ± SEM is shown (****, P < 0.0001; n = 6 independent experiments, unpaired two-tailed Student’s t test; the number of neurons analyzed for each condition is also shown). (g and h) Hippocampal neurons were transfected with Luc-shRNA or Lis1-shRNA at DIV2 and stained at DIV6 (g), or transfected at DIV9 and stained at DIV13 (h) using antibodies against GFP, AnkG, and MAP2. Arrowheads indicate AIS in hippocampal neurons. Scale bars = 5 µm. (i) Fluorescence intensity of AnkG and NF186 at the AIS in neurons transfected using Luc- or Lis1-shRNA; neurons were transfected at DIV2 and analyzed at DIV6, or transfected at DIV9 and analyzed at DIV13. Mean ± SEM is shown (***, P = 0.0002; *, P = 0.0254; n = 9 independent experiments for each time point, one-way repeated measures ANOVA; the number of neurons analyzed for each condition is also shown). (j) Quantification of the staining pattern for AnkG in shRNA-transfected neurons. The number of neurons counted is shown. Mean ± SEM is shown. No statistical comparison was performed. (k) Axon length in neurons transfected with control or Lis1-shRNA. Mean ± SEM is shown (n = 7 independent experiments, unpaired two-tailed Student’s t test; one neuron was measured per coverslip). IB, immunoblot; IP, immunoprecipitation.
Figure 8.
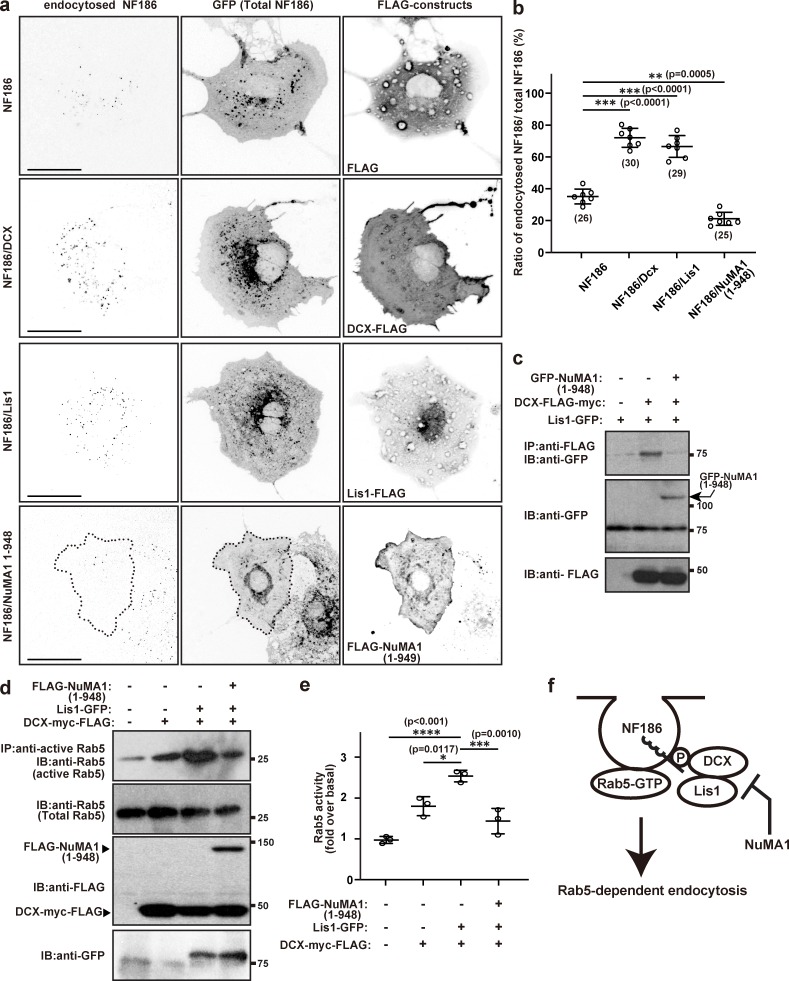
NuMA1 inhibits endocytosis of NF186 by disrupting the interaction between Lis1 and DCX. (a) Endocytosis assay in COS-7 cells for NF186 cotransfected with 3XFLAG vector, Dcx-FLAG, Lis1-FLAG, or FLAG-tagged NuMA1 (1–948). Scale bars = 10 µm. (b) Quantification of NF186 endocytosis in COS-7 cells. Mean ± SEM is shown (***, P < 0.001; **, P = 0.0005; n = 7 independent experiments, ordinary one-way ANOVA; the number of neurons analyzed for each condition is also shown). (c) GFP-NuMA1 (1–948), Lis1-GFP, and/or DCX-FLAG constructs were transfected into HEK293T cells and lysed after 48 h. Cell lysates were immunoprecipitated with an anti-FLAG antibody and immunoblotted with FLAG and GFP antibodies. (d) Rab5 activity assay in COS-7 cells. NF186-GFP, DCX-myc-FLAG, Lis1-myc-FLAG, and/or FLAG-NuMA1 1–948 mutant were transfected into COS-7 cells and lysed after 48 h of transfection. Cell lysates were immunoprecipitated using anti-active Rab5 antibody and immunoblotted with antibodies against Rab5, FLAG, and GFP. (e) Quantification of Rab5 activity over basal levels. Mean ± SEM is shown (****, P < 0.001; ***, P = 0.0010; *, P = 0.0117; n = 3 independent experiments, unpaired two-tailed Student’s t test). (f) Model for NuMA1-dependent inhibition of DCX–Lis1-mediated endocytosis of NF186. IB, immunoblot; IP, immunoprecipitation; P, phosphorylation.
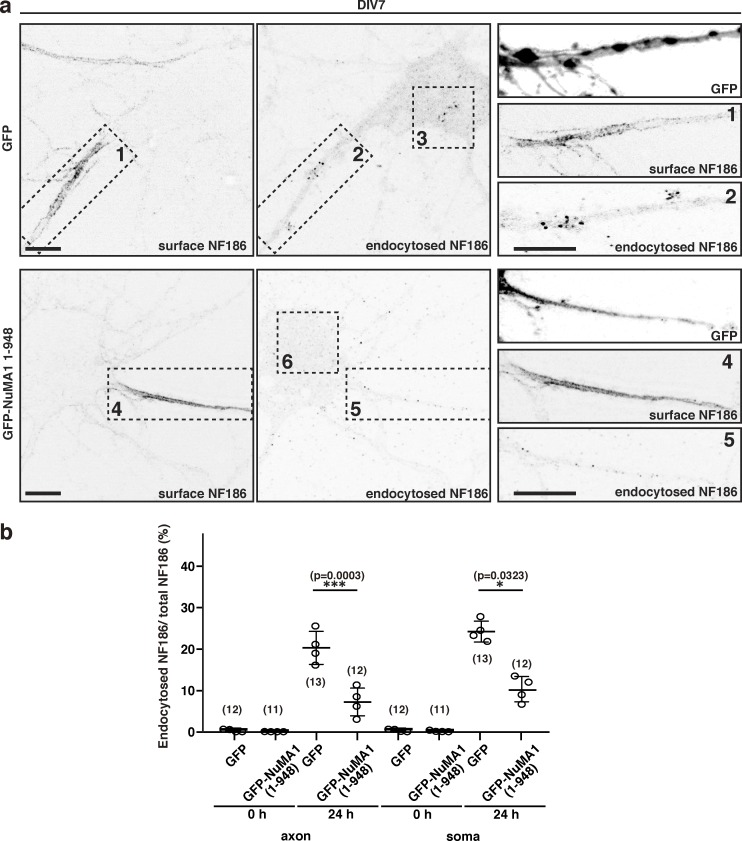
Figure 9.
Overexpression of NuMA1 suppresses endocytosis of NF186 in hippocampal neurons. (a) Endocytosis assay for endogenous NF186 in hippocampal neurons. GFP or GFP-tagged NuMA1 (1–948) were transfected into hippocampal neurons at DIV5. Surface and endocytosed NF186 are shown. Higher magnification of insets a1, a2, a4, and a5 are shown in the right panels. Scale bars = 10 µm. (b) The ratio of endocytosed NF186 to total NF186 in primary hippocampal neurons. Mean ± SEM is shown (***, P = 0.0003; *, P = 0.0323; n = 4 independent experiments, repeated measures one-way ANOVA; the number of cells counted is also indicated).
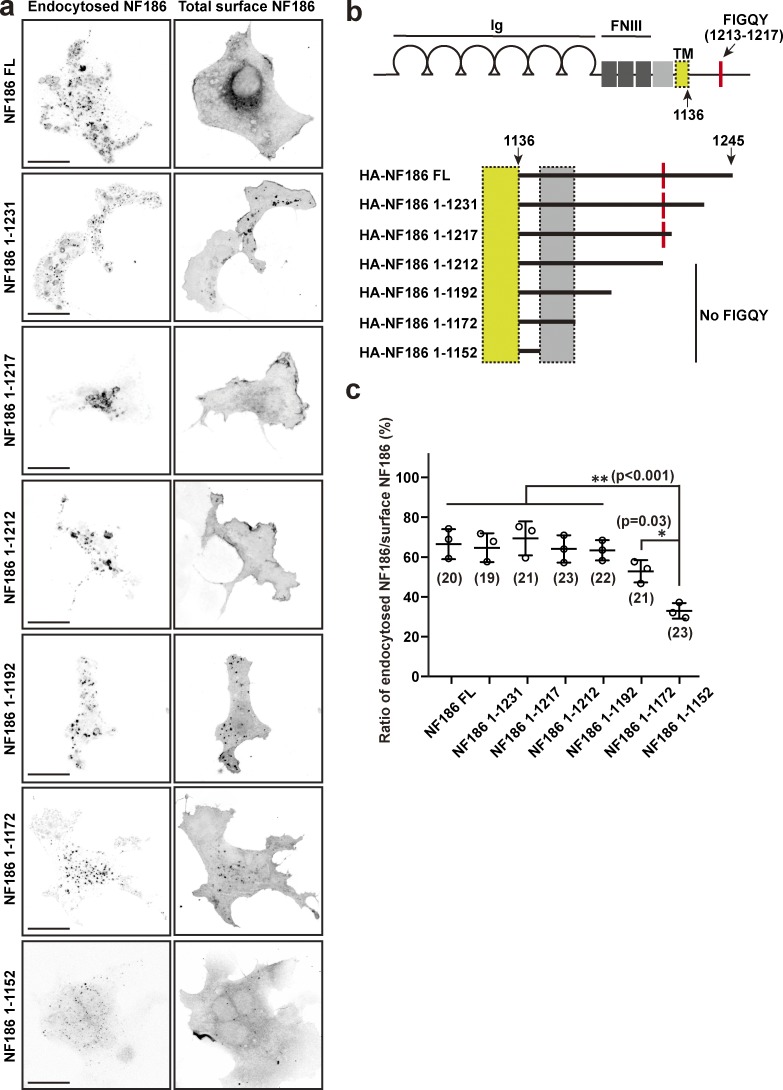
To begin to determine how NF186 is removed from the cell membrane, we performed endocytosis assays in COS-7 cells transfected with NF186 harboring C-terminal serial truncations; we measured the ratio of endocytosed NF186 to surface NF186 (Fig. S1). Whereas full-length NF186 was efficiently endocytosed, serial truncations gradually impaired its endocytosis (Fig. S1). However, the critical region mediating endocytosis did not include the FIGQY DCX–AnkG interacting motif (Tuvia et al., 1997), suggesting that interactions with additional cytoplasmic partners can efficiently remove somatodendritic NF186 and contribute to the enrichment of NF186 at AIS compared with the soma. Thus, stabilization of NF186 at the AIS may involve proteins that inhibit NF186 endocytosis and retrieval.
Figure S1.
NF186 deletion constructs reveal domains important for its endocytosis. (a) Endocytosis assay in COS-7 cells using transfected HA-tagged NF186 full-length and truncated mutants (HA-NF186; 1–1231 aa, 1–1217 aa, 1–1212 aa, 1–1192 aa, 1–1172 aa, and 1–1152 aa). Cells were immunostained with anti-HA antibody (endocytosed NF186) and anti-NF186 antibody (total surface NF186). Scale bars = 10 µm. (b) NF186 domain structure and truncation mutants. The transmembrane domain is located in the yellow boxes, while the critical region regulating endocytosis is contained in the gray boxes. FNIII, fibronectin type III domain; Ig, immunoglobulin domain, TM, transmembrane domain. (c) Ratio of endocytosed NF186/surface NF186 for full-length and deletion constructs. Mean ± SEM is shown (NF186 FL vs. NF186 1–1152, P = 0.0003; NF186 1–1231 vs. NF186 1–1152, P = 0.0006; NF186 1–1217 vs. NF186 1–1152, P = 0.0001; NF186 1212 vs. NF186 1–1152, P = 0.0007; NF186 1192 vs. NF186 1–1152, P = 0.0009; NF186 1–1172 vs. NF186 1–1152, P = 0.03; **, P < 0.001; *, P = 0.03; n = 3 independent experiments, ordinary one-way ANOVA).
Differential proteomics reveals potential AIS proteins
To identify proteins involved in assembly, retention, or retrieval of AIS proteins, we performed proteomics on control (Ank3f/f) and AnkG-deficient (Nestin-Cre;Ank3f/f or AnkG conditional knockout [cKO]) mouse brains. We prepared brain homogenates, then performed mass spectrometry (MS) on the detergent-insoluble fraction of the homogenate, since AISs are resistant to solubilization in Triton X-100 and are therefore enriched in the insoluble fraction (Boiko et al., 2007). We reasoned that because AISs are extremely stable, AIS-deficient neurons might have reduced amounts of proteins important for AIS assembly or maintenance in the detergent-insoluble fraction. We used postnatal day 0 (P0) control and AnkG cKO mice, since the latter lack AIS and die at P0 (Ho et al., 2014). We compared the number of peptide spectral matches (PSMs) for each protein identified between control and AnkG cKO mouse brains as a semi-quantitative measure of each protein’s relative levels in AIS-deficient brains (Fig. 1 c and Dataset S1; for simplicity, each protein is identified by its corresponding gene name). In other words, proteins whose amounts depend on AnkG or an AIS should have a lower number of PSMs in AnkG cKO than control. Proteins that are unchanged should have approximately equal numbers of PSMs in both control and AnkG cKO brains. These proteins reside along the diagonal least-squares fit of all purified proteins (Fig. 1 c, solid line); proteins that may be important for the AIS should lie below this line. We also used a stringent “cutoff” and only investigated proteins with at least 10 PSMs and that were below a line parallel to the least-squares fit (Fig. 1 c, lower dashed line). Thus, proteins found below the dashed line in Fig. 1 c are candidate AIS proteins, or proteins whose amounts in the detergent-insoluble fraction depend on AnkG. Importantly, we found several previously described AIS proteins were significantly reduced including AnkG (Ank3), β4 spectrin (Sptbn4), and α2 spectrin (Sptan1; red values in Fig. 1, c and d). We found that β2 spectrin (Sptbn1), recently reported at the AIS of cultured hippocampal neurons (Lazarov et al., 2018), was also reduced. Among the proteins with the largest reduction in spectral matches between AnkG cKO and control (i.e., potential AIS proteins), we identified the microtubule-binding protein NuMA1 (Fig. 1, c and d). Immunoblots from control and AnkG cKO mouse brains also showed significantly reduced NuMA1 in the AnkG cKO samples (Fig. 1, g and h).
In a complementary experiment, we silenced AnkG expression in DIV2 cultured hippocampal neurons using an adenovirus to transduce neurons with a control shRNA or a highly efficient AnkG shRNA (Hedstrom et al., 2008). At DIV12, we collected neurons and performed MS. As with the differential proteomics described above using brain homogenates (Fig. 1, c and d), we found very similar reductions in known AIS proteins (Fig. 1, e and f; and Dataset S1). For example, we found a 68.9% reduction in AnkG PSMs. Furthermore, we found a 34.3% reduction in NuMA1 PSMs (Fig. 1 h). Immunoblots of neurons transduced with control or AnkG shRNA showed a significant reduction in both AnkG and NuMA1 protein levels in the AnkG shRNA–treated samples (Fig. 1, i and j). Together, these complementary proteomic experiments show that NuMA1 is reduced after loss of AnkG, suggesting that it may be an AIS protein.
NuMA1 is transiently expressed and located at the AIS
NuMA1 is a microtubule-binding protein that was previously reported to play important roles in cell division (Kiyomitsu and Cheeseman, 2013; Kotak et al., 2013). However, the function of NuMA1 in postmitotic neurons is unknown. To determine the location of NuMA1 in neurons, we immunostained developing hippocampal neurons in culture (Fig. 2 a). We found that at DIV3, before AIS assembly and AnkG clustering, NuMA1 was distributed throughout the neuronal cytoplasm. By DIV5, NuMA1 was highly enriched in the somatodendritic compartment and in the proximal axon where it began to colocalize with AnkG at the nascent AIS. By DIV7, robust NuMA1 immunoreactivity was detected at the AIS, and that overlapped with AnkG (Fig. 2 a). This AIS pool of NuMA1 was also resistant to detergent extraction (Fig. 2 a), a common characteristic of AIS proteins due to their tight association with the AIS cytoskeleton (Boiko et al., 2007). Immunoblotting of primary cortical neurons also showed the transient expression of NuMA1; in contrast, AnkG protein levels increased from DIV0 to DIV14 (Fig. 2, b and c).
To determine if NuMA1’s AIS localization depends on AnkG, we silenced AnkG using adenoviral-delivered AnkG shRNA (Ad-shRNA-AnkG) and compared it to control shRNA (Ad-shRNA-ctrl). We found that control shRNA did not affect AIS NuMA1 or AnkG, but silencing of AnkG dramatically reduced the AIS localization of NuMA1 (Fig. 2, d and e), a result consistent with the reduced NuMA1 PSMs measured in AnkG-deficient neurons (Fig. 1, c–j). Together, these results demonstrate that NuMA1 is transiently expressed and located at the AIS, and NuMA1’s localization depends on AnkG.
Loss of NuMA1 disrupts AIS assembly but not maintenance
To determine the role of NuMA1, we performed loss-of-function experiments using shRNA to silence expression of NuMA1 (NuMA1-shRNA). We generated two shRNAs against NuMA1 and confirmed that, compared with a control luciferase shRNA (Luc-shRNA), they efficiently silence NuMA1 expression (Fig. 3 a). Transfection of NuMA1-shRNAs into DIV2 hippocampal neurons revealed that by DIV6, AIS NuMA1 was no longer detectable (Fig. 3 b), and the overall NuMA1 immunoreactivity was dramatically reduced (Fig. 3 c). Transfection of DIV2 hippocampal neurons using control or NuMA1-shRNAs showed that NuMA1 is required for the accumulation of AIS AnkG by DIV6 (arrowheads in Fig. 3, d and g). Furthermore, silencing expression of NuMA1 from DIV2 to DIV6 significantly reduced the intensity of AIS AnkG and NF186 immunoreactivity (Fig. 3 f), but loss of NuMA1 did not affect axon length (Fig. 3 h). In contrast, and consistent with its transient expression, silencing NuMA1 from DIV9 to DIV13 had no effect on the AIS (arrowheads in Fig. 3, e and g). Thus, NuMA1 plays important roles during assembly, but not maintenance, of the AIS.
To confirm the specificity of the NuMA1-shRNAs, we cotransfected neurons with NuMA1-shRNA and a NuMA1-shRNA–resistant plasmid. Silencing NuMA1 expression at DIV1 dramatically reduced the number of neurons with an AIS compared with a Luc-shRNA (Fig. 4, a, b, and e) at DIV6. In contrast, cotransfecting the Luc-shRNA or NuMA1-shRNA with the NuMA1-shRNA–resistant plasmid rescued the AIS (NuMA1-shRNA+NuMA1) and even promoted the assembly of AIS (Luc-shRNA+NuMA1) compared with the Luc-shRNA control (Fig. 4, c–e).
Previous studies showed that constitutive deletion of NuMA1 results in embryonic lethality at embryonic day (E) 9.5 (Silk et al., 2009). To investigate the in vivo function of NuMA1 in postmitotic neurons, we performed in utero electroporation at E15.5 using NuMA1-shRNA or Luc-shRNA. Remarkably, and consistent with our in vitro results, we found that at P5, far fewer cortical neurons had an AIS when expressing NuMA1-shRNA compared with the control Luc-shRNA (Fig. 4, f and g). Together, these results suggest that NuMA1 contributes to AIS assembly.
NuMA1 interacts with protein 4.1B
Since the amount of NuMA1 protein decreased in AnkG-deficient neurons (Fig. 1), NuMA1 was retained at AIS after detergent extraction (Fig. 2 a), and NuMA1 was lost from the AIS after silencing AnkG (Fig. 2, d and e), we reasoned NuMA1 might associate with AnkG or some other previously described AIS protein. Therefore, we performed coimmunoprecipitation experiments using human embryonic kidney (HEK) 293T cells cotransfected with an epitope-tagged NuMA1 (FLAG or GFP) and previously reported AIS proteins. We found that NuMA1 does not coimmunoprecipitate with AnkG (Fig. 5 a), α2 spectrin (Fig. 5 b), or a number of other previously reported AIS proteins (Fig. 5 c). However, NuMA1 is a well-known binding partner for the cytoskeletal proteins 4.1N, 4.1R, and 4.1G (Kiyomitsu and Cheeseman, 2013; Mattagajasingh et al., 1999; Ye et al., 1999), and their paralog protein 4.1B is a binding partner for Caspr2, which is also found at the AIS (Pinatel et al., 2017). Therefore, we considered whether NuMA1 binds 4.1B. We found that NuMA1 can coimmunoprecipitate protein 4.1B (Fig. 5 d), and deletion of NuMA1’s 4.1-binding domain abolishes their interaction (Fig. 5 e). Immunostaining of DIV7 hippocampal neurons showed diffuse 4.1B labeling (Fig. 5 f), but a pool of this 4.1B was retained at the AIS after detergent extraction (Fig. 5 g). In contrast, although other 4.1 proteins have been reported to bind to NuMA1, none were retained at the AIS after detergent extraction (Fig. 5, g and h). Although 4.1B PSMs were found in control, but not AIS-deficient neurons, the amount was below our cutoff of 10 PSMs (data not shown). Protein 4.1B interacts with β spectrins, and we found that both β2 and β4 spectrin coimmunoprecipitated protein 4.1B (Fig. 5 i).
NuMA1 may be phosphorylated by cyclin-dependent kinases 1 and 2 (Cdk1 and Cdk2, respectively) at T2041, and NuMA1’s membrane association is reduced by T2041 phosphomimetics (Kotak et al., 2013; Zheng et al., 2014). Cdk2 has been reported to regulate the AIS targeting of Kv1 channels (Vacher et al., 2011), and the AIS is a well-recognized hotspot for protein phosphorylation (Berger et al., 2018; Buffington et al., 2012). To determine if NuMA1’s interaction with 4.1B depends on phosphorylation, we constructed three phosphomimetics of NuMA1’s C-terminus, including amino acids 1466–2101 (Fig. 5, j and k). Immunoprecipitation of full-length NuMA1, or the C-terminal fragment of NuMA1, efficiently coimmunoprecipitated 4.1B. However, NuMA1 phosphomimetics dramatically reduced the binding of 4.1B (Fig. 5 j), suggesting that phosphorylation of NuMA1 may inhibit its interaction with the membrane-associated scaffolding protein 4.1B.
Loss of protein 4.1B disrupts AIS assembly but not maintenance
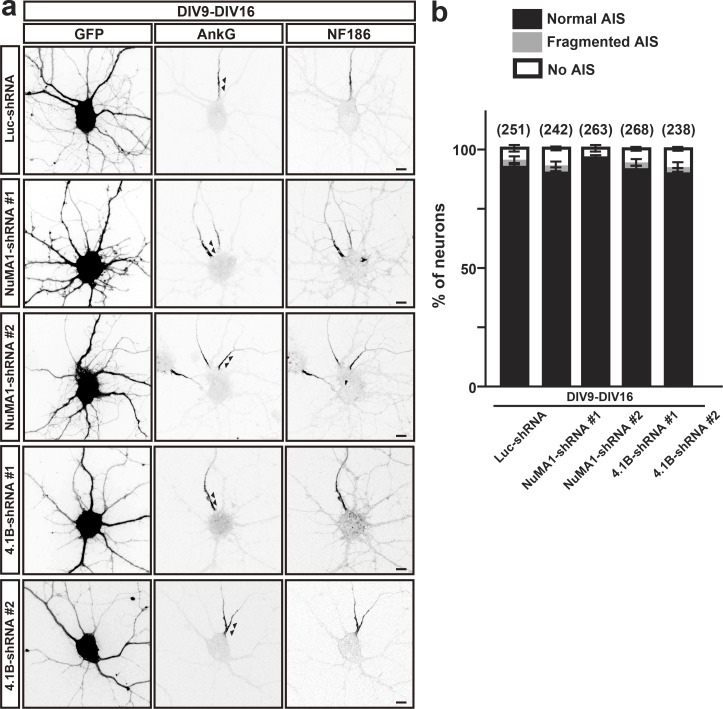
Like NuMA1, protein 4.1B is transiently expressed in cultured hippocampal neurons (Fig. 2 b). To determine the role of protein 4.1B at the AIS, we performed loss-of-function experiments using two highly efficient shRNA to silence 4.1B’s expression (Fig. 6 a). Transfection of 4.1B shRNAs into DIV2 hippocampal neurons revealed that by DIV6, 4.1B immunoreactivity was dramatically reduced (Fig. 6, b and c). Transfection of DIV2 hippocampal neurons using control or 4.1B shRNAs showed that 4.1B is required for the clustering of AIS AnkG by DIV6 (arrowheads in Fig. 6, d and g). Furthermore, silencing expression of 4.1B from DIV2 to DIV6 significantly reduced the intensity of AIS AnkG and NF186 immunoreactivity (Fig. 6 f), but loss of 4.1B did not affect axon length (Fig. 6 h). In contrast, and consistent with its transient expression, silencing protein 4.1B from DIV9 to DIV13 had no effect on the AIS (arrowheads in Fig. 6, e and g). We also considered whether even longer periods (DIV9–DIV16) of NuMA1 or protein 4.1B silencing might affect AIS maintenance. However, silencing of these proteins for this duration had no effect on AIS (Fig. S2). Thus, both NuMA1 and protein 4.1B play important roles during assembly, but not maintenance, of the AIS.
Figure S2.
Silencing expression of NuMA1 or 4.1B does not affect AIS maintenance in hippocampal neurons. (a) Hippocampal neurons were transfected with Luc-shRNA, NuMA1-shRNA #1, NuMA1-shRNA #2, 4.1B-shRNA #1, or 4.1B-shRNA #2 at DIV9 and immunostained at DIV16 for GFP, AnkG, and NF186. Arrowheads in the AnkG-labeled panels indicate AIS. Scale bars = 5 µm. (b) Quantification of the staining pattern for AnkG in shRNA-transfected neurons. Mean ± SEM is shown (the number of neurons analyzed is indicated). No statistical comparison was performed.
NuMA1 interacts with Lis1
Previous studies suggest that NuMA1 interacts with the motor protein dynein–dynactin complex to regulate the pulling forces necessary for mitotic spindle positioning (Williams et al., 2011). Furthermore, Lis1 facilitates the interaction between dynein and dynactin (Wang et al., 2013). Lis1 is located at the AIS, interacts with Ndel1, and participates in trafficking of cargoes through the AIS (Kuijpers et al., 2016). We confirmed that a pool of Lis1 may be found at the AIS (Fig. 7 a) and that NuMA1 can coimmunoprecipitate Lis1 (Fig. 7 b) through interaction between Lis1 and NuMA1’s N-terminus (amino acids 1–948; Fig. 7 c). To determine the role of Lis1 at the AIS, we generated shRNA to silence its expression (Fig. 7, d–f). In stark contrast to loss of NuMA1 or protein 4.1B, we found that silencing Lis1 expression in DIV2–DIV6 neurons did not disrupt AIS assembly but instead increased the amount of AIS AnkG and NF186 (Fig. 7, e–h). Consistent with our observations, Kuijpers et al. (2016) previously showed that silencing Lis1 increased AIS size. In contrast, silencing Lis1 had no effect on axon length (Fig. 7 i). Similar to NuMA1 and 4.1B shRNAs, Lis1-shRNA had no effect on DIV9–DIV13 neurons (Fig. 7, h–j).
NuMA1 inhibits NF186 endocytosis by blocking DCX–Lis1 interactions
Since silencing of Lis1 and NuMA1 had opposite effects on AIS NF186, we considered whether these proteins modulate the endocytosis of NF186 through DCX (Kizhatil et al., 2002; Yap et al., 2012). To test this, we performed endocytosis assays in COS-7 cells transfected with NF186 and an empty FLAG vector, or COS-7 cells transfected with NF186 and FLAG-tagged DCX, Lis1, or NuMA1. We measured the ratio of endocytosed NF186 to total NF186. Compared with NF186 alone, we found that cotransfection of NF186 with DCX or Lis1 promotes NF186 endocytosis (Fig. 8, a and b). In contrast, cotransfection with NuMA1 significantly inhibits NF186 endocytosis (Fig. 8, a and b). Previous studies showed that DCX and Lis1 physically interact (Caspi et al., 2000), and we confirmed this interaction in vitro (Fig. 8 c). The interaction between DCX and Lis1 suggests these proteins function in the same pathway to regulate NF186 endocytosis. To determine if NuMA1 also participates in a complex with DCX and Lis1, we cotransfected all three proteins and immunoprecipitated DCX. Remarkably, we found that NuMA1 inhibits Lis1 from binding to DCX (Fig. 8 c).
To determine if NuMA1 regulates DCX–Lis1-dependent endocytosis, we measured the levels of active Rab5 in COS-7 cells cotransfected with combinations of DCX, Lis1, and NuMA1. Rab5 is a key regulator of endocytosis and interacts with the dynein regulator Ndel1 (Chansard et al., 2011; Stenmark et al., 1994); Ndel1 binds Lis1 and is also located at the AIS (Kuijpers et al., 2016). Although we observed no change in the total Rab5 protein, we found that DCX promoted Rab5 activity above baseline, and cotransfection of DCX and Lis1 further potentiated Rab5 activation (Fig. 8, d and e). However, cotransfection of NuMA1 with DCX and Lis1 inhibited Rab5 activation, compared with transfection with DCX alone or cotransfection of DCX and Lis1 (Fig. 8, d and e). Together, these results suggest that NuMA1 inhibits DCX–Lis1-dependent endocytosis of NF186 (Fig. 8 f).
NuMA1 inhibits endocytosis of NF186 in neurons
To determine if NuMA1 regulates the endocytosis of endogenous neuronal NF186, we performed endocytosis assays in cultured hippocampal neurons; we compared the amount of endocytosed NF186 to the remaining surface NF186. We found that transfection with GFP alone did not inhibit NF186 endocytosis at the soma or AIS (Fig. 9, a2, a3, and b). However, transfection with the N-terminal domain of NuMA1 (the domain that inhibits Lis1’s interaction with DCX; Fig. 8 c) was sufficient to significantly increase surface NF186 (Fig. 9, a4 and b). This also significantly inhibited the amount of endocytosed NF186 in the soma and axon (Fig. 9, a5, a6, and b). Thus, during development, NuMA1 at the AIS promotes the surface and AIS accumulation of NF186 by inhibiting NF186 endocytosis mediated by DCX and Lis1.
Discussion
One defining feature of the AIS is the high density of AnkG, β4 spectrin, Na+ channels, and NF186 restricted to the proximal axon. Thus, the AIS is a remarkable example of how precise protein localization can control neuronal excitability. In addition, the AIS functions to regulate neuronal polarity, and this also depends on a stable AIS protein complex. Thus, loss of AnkG dismantles the AIS and axons acquire dendritic features including the presence of MAP2, spines, and postsynaptic structures (Hedstrom et al., 2008; Sobotzik et al., 2009). Despite these important functions, the mechanisms controlling neuronal polarity and enrichment of AIS proteins remain poorly understood. Our discovery that NuMA1 inhibits membrane protein endocytosis at the developing AIS helps to explain how such a high density of AIS proteins is achieved.
Experiments to define the AIS proteome have been much less successful than similar efforts to identify the synapse proteome (Husi and Grant, 2001). This is primarily because AIS proteins are refractory to conventional immunoaffinity purification and no biochemical methods exist to purify AIS. Instead, our unbiased approach to identify proteins whose amounts are reduced in AIS-deficient brains and neurons overcomes these limitations. However, our studies also begin with a much more heterogenous pool of proteins, suggesting that high background levels of proteins may make it difficult to identify small changes. Nevertheless, our two complementary strategies using AnkG cKO brains (Fig. 1, c and d) and AnkG-deficient neurons (Fig. 1, e and f) yielded remarkably concordant results, with many of the same proteins reduced. Our strategy also required us to perform these proteomic experiments on young neurons with developing and immature AIS since AnkG cKO mice die at P0. This experimental limitation was actually fortuitous since NuMA1 is transiently expressed and localized at the AIS; proteomic studies using mature neurons or adult brains may not have revealed NuMA1. These observations highlight the dynamic nature of the AIS and emphasize the need for future longitudinal studies of the AIS proteome. Furthermore, our studies were restricted to hippocampal neurons, and future studies of other types of neurons may reveal cell type–specific differences in the molecular organization or assembly of AIS.
NuMA1 has been studied extensively in the context of cell division. It functions together with dynein to generate pulling forces that correctly position the mitotic spindle (Greenberg et al., 2018). Like the mitotic spindle, AIS are highly enriched in microtubules and the dynein regulator Ndel1 (Kuijpers et al., 2016). Our experiments reveal new functions for NuMA1 in postmitotic and highly polarized neurons. What mechanisms control NuMA1’s AIS localization? We found that NuMA1’s clustering at the AIS depends on AnkG, although NuMA1 does not bind directly to AnkG. Instead, our results suggest that NuMA1 is recruited to the AIS through binding to protein 4.1B, which in turn interacts with AIS β spectrins. Remarkably, like NuMA1, protein 4.1B is transiently located at the AIS, and silencing its expression phenocopied the loss of NuMA1; silencing of protein 4.1B had no effect in mature neurons after the AIS was fully established.
The phosphorylation of many AIS proteins modulates their localization and retention at the AIS. For example, phosphorylation of Na+ channels by the protein kinase CK2 increases their affinity for AnkG 1,000-fold (Bréchet et al., 2008). On the other hand, phosphorylation of other AIS proteins inhibits their localization at the AIS. For example, phosphorylation of NF186 blocks its interaction with AnkG and instead promotes its interaction with DCX and subsequent endocytosis (Kizhatil et al., 2002; Yap et al., 2012). Cdk-mediated phosphorylation of the Kv1 channel’s Kvβ2 subunit inhibits its localization at the AIS (Vacher et al., 2011). NuMA1’s protein 4.1B-binding domain is also phosphorylated by Cdk’s (Kotak et al., 2013; Zheng et al., 2014), and NuMA1 phosphomimetics block its association with protein 4.1B. Thus, our studies suggest that Cdk-dependent phosphorylation of NuMA1 negatively regulates its association with 4.1B and localization to the AIS.
Besides protein 4.1B, we found that the N-terminus of NuMA1 can interact with Lis1. Lis1 can be located at the AIS (Kuijpers et al., 2016), and Lis1 facilitates the interaction between dynein and dynactin, key regulators of early endosome dynamics (Wang et al., 2013). DCX was previously reported to control the endocytosis and retrieval of NF186 from the plasma membrane in a phosphorylation-dependent manner (Yap et al., 2012). Cotransfection of NF186 with either Lis1 or DCX promoted NF186 endocytosis, but cotransfection of NF186 with NuMA1 had the opposite effect and inhibited endocytosis. These differences can be explained by our discovery that NuMA1 inhibits the interaction between Lis1 and DCX.
What does NuMA1 do at the AIS? We used both gain- and loss-of-function experiments to investigate the role of NuMA1 at the AIS. Silencing NuMA1 reduced the density of AIS NF186 and AnkG, but only during early AIS development and not after the AIS is fully developed. These contrasting effects are consistent with NuMA1’s transient expression. Conversely, overexpression of NuMA1 reduced endocytosis of NF186 and promoted AIS assembly. Silencing Lis1 also reduced endocytosis of NF186. Our experiments support a model where NuMA1 inhibits NF186 endocytosis by blocking the activity of Lis1–DCX. This shifts the balance to a stable membrane pool of NF186, prolonging the time where NF186 may interact with AIS AnkG, further stabilizing and establishing the AIS. Inhibiting endocytosis of AIS NF186 may also promote the formation of a larger AIS protein complex including other AnkG-binding partners like Na+ channels. Inhibition of NF186 endocytosis may also facilitate interactions with the AIS ECM that includes versican and brevican, both of which bind directly to NF186 (Susuki et al., 2013). This AIS ECM is proposed to function as an “exoskeleton” that, through NF186, can stabilize the AIS cytoskeleton and its associated proteins (Hedstrom et al., 2007). Thus, stabilizing AIS NF186 may facilitate the efficient assembly of both AIS cytoskeleton and exoskeleton.
In summary, our results suggest a new role for NuMA1: to promote the enrichment of AIS proteins in time and space by inhibiting AIS endocytosis, precisely at the time when AIS protein complexes are targeted to the proximal axon, assembled, and stabilized.
Materials and methods
Animals
AnkG-deficient mice were generated by cell type–specific Cre-mediated deletion of the Ank3 gene. Ank3fl/fl mice were generated as previously described (Ho et al., 2014). Nestin-Cre transgenic mice (C57BL/6) were obtained from The Jackson Laboratory (Cat# 003771). Timed pregnant ICR mice were ordered from Charles River for in utero electroporation. All animal work and procedures were approved by the Animal Care and Use Committee at Baylor College of Medicine and performed in accordance with the National Institutes of Health (NIH) guide for the humane care and use of animals.
Antibodies
The following antibodies were used: mouse monoclonal antibodies to NuMA1 (Santa Cruz Biotechnology, Cat# sc-365532; RRID: AB_10846197), Lis1 (Millipore Sigma, Cat# L7391; RRID: AB_260418), HA (BioLegend, Cat# 901503; RRID: AB_2565005), FLAG (Millipore Sigma, Cat# F1804; RRIB:AB_262044), c-myc (BioLegend, Cat# 626802; RRID: AB_2148451), βII-spectrin (BioLegend, Cat# 803201; RRID: AB_2564660), βII-spectrin (R&D Systems, Cat# 612563; RRID: AB_399854), active Rab5-GTP (NewEast Biosciences, Cat# 26911; RRID: AB_2617182), Rab5 (NewEast Biosciences, Cat# 26016), and 4.1N (R&D Systems, Cat# 611836; RRID: AB_2098366). AnkG (N106/36; RRID: AB_10673030, and N106/65; RRID: AB_10675130) and neurofascin (A12/18; AB_2282826) antibodies were purchased from the University of California, Davis/NIH NeuroMab Facility. Rabbit polyclonal antibodies against proteins 4.1B, 4.1G, and 4.1R were gifts from Dr. Elior Peles (Weizmann Institute of Science, Rehovot, Israel). We purchased the following rabbit polyclonal antibodies: GFP (Thermo Fisher Scientific, Cat# A-11122; RRID: AB_221569), NDEL1 (Thermo Fisher Scientific, Cat# PA5-53669; RRID: AB_2644530), GAPDH (Millipore Sigma, Cat# G9545; RRID: AB_796208), and NuMA1 (Santa Cruz Biotechnology, Cat# sc-48773; RRID: AB_2154276). We purchased the following chicken antibodies: MAP2 (EnCor, Cat# CACP-MAP2; RRID: AB_2138173) and neurofascin (R&D Systems, Cat# AF3235; RRID: AB_10890736). Guinea pig polyclonal antibody for AnkG was purchased from Synaptic Systems (Cat# 386 004; RRID: AB_2725774). Species-appropriate aminomethylcoumarin (AMCA), Alexa Fluor-488, and Alexa Fluor-594 secondary antibodies were purchased from Thermo Fisher Scientific. Anti-mouse HRP-labeled secondary antibody was purchased from GE Healthcare (Cat# NA9310V; RRID: AB_7721983), and anti-rabbit HRP antibody was purchased from The Jackson Laboratory (Cat# 111-035-003; RRID: AB_2313567).
Plasmids
pEGFPC1-NuMA1 human cDNA was purchased from Addgene (Cat# 81029); we substituted a 3XFLAG/V5-tag for the GFP to create p3XFLAG/V5-NuMA1. The NuMA1 protein 4.1 deletion mutant (deleted amino acids 1785–1810) was generated with the Q5 site-directed mutagenesis kit (New England BioLabs). The full-length human 4.1B (also known as EPB41L3) with N-terminal myc tag was a gift from Dr. Elior Peles. Human 4.1B was inserted into p3XFLAG-CMV-7.1 (Millipore Sigma) vector to create p3XFLAG-4.1B construct. AnkG270-GFP was a gift from Dr. Vann Bennett (Duke University, Durham, NC). αII-spectrin–GFP and βII-spectrin–GFP were gifts from Dr. Michael Stankewich (Yale University, New Haven, CT). The full-length β4Σ1-spectrin with an N-terminal myc tag was subcloned and inserted into pCS3+MT expression vector. Mouse Lis1 (also known as Pafah1b1)-myc-FLAG (Cat# BC026141), mouse NDE1-myc-FLAG (Cat# MR228536), and mouse DCX-myc-FLAG (Cat# MR205614) constructs were purchased from Origene. Lis1-myc-FLAG was inserted into the pEGFP-N1 vector to create pEGFPN1-Lis1. Phosphomimetic mutants of NuMA1 (NuMA1 C-ter 1TD, NuMA1 C-ter 2TD, and NuMA1 C-ter 3T/SD) were generated with the Q5 site-directed mutagenesis kit (New England BioLabs). NuMA1 1–948 (1–948 amino acids), NuMA1 949–1465 (949–1465 amino acids), and NuMA1 1466–2101 (1466–2101 amino acids) were inserted into p3XFLAG-CMV-7.1 vector or pEGFPC1 vector (Clonetech). The NuMA1 1–948 fragment was used as the shRNA-resistant version of NuMA1 since this fragment was sufficient to inhibit endocytosis. HA-tagged truncation mutants of rat NF186 (HA-NF186 1–1231, HA-NF186 1–1217, HA-NF186 1–1212, HA-NF186 1–1192, HA-NF186 1–1172, and HA-NF186 1–1152) were created using the Q5 site-directed mutagenesis kit. The 21-nucleotide shRNA were synthesized by Millipore Sigma. ShRNA oligos were annealed and inserted into the BglII and HidIII sites of GFP-expressing pSuper vector. The specific target sequences of shRNA for mouse and rat NuMA1 were 5′-GAGGGGCAACAAATCCTACAG-3′ (NuMA1-shRNA#1) and 5′-GACTAGACTTTGTTTGCAGTT-3′ (NuMA1-shRNA#2). The specific target sequences of 4.1B shRNA for mouse and rat 4.1B were 5′-GCATGCAGTGCAAAGTGACGC-3′ (4.1B-shRNA#1) and 5′-GCAGGGGCACAGGCTGCACCT-3′ (4.1B-shRNA#2). The specific target sequences of Lis1-shRNA for mouse and rat Lis1 was 5′-GGAAGCTGAACTAGATATGAA-3′. The specific target sequence of Luc-shRNA for Photinus pyralis luciferase was 5′-CGCTGAGTACTTCGAAATGTC-3′. DNA and shRNA constructs were verified by sequencing (Genewiz or GenScript).
Cell cultures for HEK293T cells and Cos-7 Cells
HEK293T cells (ATCC, Cat# CRL-3216; RRID: CVCL_0063), COS-7 cells (ATCC, Cat# CRL-1651; RRID: CVCL_0224), and mouse neuroblastoma Neuro2a cells (ATCC, Cat# CCL-131; RRID: CVCL_0470) were purchased and cultured on cell culture dishes at 37°C in DMEM containing 10% heat-inactivated FBS (GE Healthcare), 50 units/ml penicillin, and 50 µg/ml streptomycin.
Culture for primary hippocampal neurons
Primary cultures of hippocampal or cortical neurons were dissected from E18 rat embryos and plated on poly-D-lysine- (Millipore Sigma) and laminin- (Life Technologies) coated glass coverslips (Ogawa et al., 2006). The media were replaced with the maintaining medium (97% neurobasal media, 2% B-27 supplement, and 1% GlutaMAX [all from Life Technologies]) after 4 h of plating.
Plasmid transfection
HEK293T cells and COS-7 cells were transfected with plasmid DNA using Lipofectamine 3000 (Invitrogen) according to the manufacturer’s instructions. The media were replaced after 24 h of transfection. Primary hippocampal neurons were transfected with Lipofectamine 2000 (Invitrogen). After neurons were washed with neurobasal media three times, the medium was replaced after 4 h of transfection.
Immunofluorescence microscopy
Cultured rat primary neurons, HEK293T cells, COS-7 cells, and brain sections were fixed in 4% PFA (pH 7.2) and immunostained with the appropriate antibodies in phosphate buffer with 0.3% Triton X-100 and 10% goat serum. Images of immunofluorescence were captured using an Axio-imager Z1 microscope (Carl Zeiss MicroImaging) or Axio-observer Z1 microscope (Carl Zeiss MicroImaging) fitted with Apotome and an AxioCam digital camera (Carl Zeiss MicroImaging). Zeiss 20× (0.8 NA) Plan Apochromat, 40× (0.75 NA) Plan Neofluar, and 63× (1.4 NA) Plan Apochromat objectives were used to acquire images. Images were collected at room temperature using Zen acquisition software (Carl Zeiss MicroImaging). Fluorophores included Alexa Fluor-488, Alexa Fluor-594, and AMCA conjugated to secondary antibodies. To measure fluorescence intensity, we used ImageJ analysis software version 2.0.0-rc-68/1.52h. Briefly, the cell or region of interest was outlined and the fluorescence intensity in the selected region was measured. Within an experiment, exposure times for all cells measured were kept constant. Only linear adjustments were made to the lookup table to maximize the range of fluorescence.
In utero electroporation
Timed pregnant ICR mice were electroporated using Luc-shRNAs or NuMA1-shRNAs at E15.5 (Saito, 2006). Pregnant mice were anesthetized and the uterine horns exposed. A small needle electrode filled with plasmid DNA (0.3–0.6 μg/μl) and 0.1% Fast Green in PBS was used to inject ∼1 µl into the lateral ventricles of embryos. Plasmid DNA was then electroporated using electric paddles. Neonatal mice were euthanized at P5. Brains were dissected, fixed, and immunolabeled as described.
Detergent extraction study
Hippocampal neurons were washed with PBS and incubated with ice-cold PBS containing 0.5% Triton X-100 at 4°C for 30 min (Huang et al., 2017). Neurons were then washed with PBS three times and fixed. Immunostaining procedure was followed as above.
Extraction and fractionation of tissue
Mouse forebrains were dissected and homogenized in ice-cold homogenization buffer (0.32 M sucrose, 5 mM sodium phosphate, pH 7.2, 1 mM sodium fluoride, 1 mM sodium orthovanadate, and protease inhibitors). The homogenates were centrifuged at 700 g for 10 min at 4°C to remove nuclei and tissue debris. The supernatant was centrifuged at 27,200 g for 90 min at 4°C and the pellet was resuspended in homogenization buffer as membrane fraction. Protein concentrations were measured using bicinchoninic acid protein assay kit (Thermo Fisher Scientific). The membrane fraction was then extracted at a concentration of 1 mg/ml for 1 h using lysis buffer containing 1% Triton X-100, 20 mM Tris-HCl, pH 8.0, 10 mM EDTA, 0.15 M NaCl, 10 mM iodoacetamide, protease inhibitors, and 0.5 mM PMSF. Soluble and insoluble materials were separated by centrifugation at 13,000 g for 30 min at 4°C. The insoluble pellet material was used for MS. In some instances, the membrane fraction was also used for immunoblotting.
In-gel digestion and MS analysis
The insoluble pellets were solubilized, reducing sample buffer, boiled, and then loaded and size fractionated on SDS-polyacrylamide gels. Polyacrylamide gels were stained using the SilverSnap silver staining kit (Pierce). Gels were cut (30 slices/lane for the AnkG shRNA vs. control experiment and 26 slices/lane for the AnkG cKO vs. control experiment), and proteins contained in the slices were digested in-gel with trypsin as described previously (Rosenfeld et al., 1992). The extracted digests were vacuum evaporated and resuspended in 10 µl of 0.1% formic acid in water. Peptides were analyzed on hybrid linear ion trap-Orbitrap mass spectrometer (LTQ-Velos, Thermo Fisher Scientific), connected to a nanoACQUITY Ultra Performance LC system (Waters). A 15-cm EasySpray C18 column (Thermo Fisher Scientific) was used to resolve peptides using 60-min gradients (mobile phase A, 0.1% formic acid in water; mobile phase B, 0.1% formic acid in acetonitrile). Following equilibration of the column in 2% solvent B, approximately one-half of each digest (5 µl) was injected and loaded, then B was increased linearly to 25% over 18 min, then to 31% B in 6 min, 40% B in 3 min, and 80% B in 3 min. Peptides were analyzed in positive-ion mode and in information-dependent acquisition mode to automatically switch between MS and MS/MS acquisition. MS spectra were acquired in profile mode using the Orbitrap analyzer in the m/z range between 350 and 1,400. For each MS spectrum, the six most intense multiple-charged ions over a threshold of 1,000 counts were selected to perform collision-induced dissociation (CID) experiments (used when analyzing the AnkG shRNA KO and corresponding control samples). A threshold of 2,000 was required for higher-energy collisional dissociation (HCD) experiments (done when analyzing the Cre-lox AnkG cKO and corresponding control samples). Product ions were analyzed on the linear ion trap in centroid mode for CID, and in the Orbitrap analyzer for HCD. The CID collision energy was automatically set to 25%. Normalized collision energy for HCD was set at 30%. A dynamic exclusion window of 0.5 D was applied, which prevented the same m/z from being selected for 60 s after its acquisition. Peak lists were generated using PAVA in-house software (Guan et al., 2011). The peak lists were searched against the mouse subset of the UniProtKB database as of June 17, 2013 (containing 73,955 entries) for the AnkG cKO and control experiment, and against the UniProtKB rat subset as of November 1, 2017 (containing 36,081 entries) for the shRNA AnkG KO vs. control experiment, using in-house ProteinProspector version 5.8.0. Peptide tolerance in searches was 20 ppm for precursor and 0.6 D (CID data) or 30 ppm (HCD data) for product ions, respectively. Cleavage specificity was selected to trypsin. Peptides containing two miscleavages were allowed. Carbamidomethylation of cysteine, acetylation of the N-terminus of the protein, pyroglutamate formation from N-terminal glutamine, and oxidation of methionine were allowed as variable modifications. The number of modifications was limited to two per peptide. Hits were considered significant when peptide sequences matched a protein entry and the Prospector score was above the significance level. False discovery rate was limited to 1% in the sets.
Immunoblotting and immunoprecipitation
HEK293T cells were transfected with plasmid DNA using Lipofectamine 3000 transfection reagent. After 48 h of transfection, cells were lysed in RIPA buffer (50 mM Tris-HCl, 150 mM NaCl, 0.5% sodium deoxycholate, 0.1% SDS, and 1% NP-40) or lysis buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.1% Triton X-100, and 1 mM EDTA), and the lysates were centrifuged at 14,000 rpm for 10 min at 4°C. For immunoblotting, the proteins in supernatants were denatured in Laemmli sample buffer and then subjected to SDS-PAGE. The electrophoretically separated proteins were transferred to a polyvinylidene difluoride (GE Healthcare) membrane, blocked with 5% skim milk, and immunoblotted with each of the primary antibodies and in turn with peroxidase-conjugated secondary antibodies. For immunoprecipitation, cell lysates were mixed with protein G resin (GE Healthcare) preabsorbed with each of the primary antibodies. The immune complexes were precipitated by centrifugation and washed three times with RIPA buffer. The immunoprecipitates were boiled in sample buffer and then separated by SDS-PAGE. The protein complexes were detected by immunoblotting using each antibody, respectively.
Activity assay for Rab5
Rat NF186-GFP, Dcx-myc-FLAG, Lis1-GFP, and 3XFLAG-NuMA1 1–948 were transfected into COS-7 cells and incubated for 48 h. After incubation, cells were collected using lysis buffer and immunoprecipitated using anti-active Rab5 antibody. Activated Rab5 and total Rab5 were detected by immunoblotting using anti-Rab5 antibody.
Endocytosis assay for NF186 in COS-7 cells
NF186-GFP, NF186-deletion constructs, Dcx-myc-FLAG, Lis1-myc-FLAG, or FLAG-NuMA1 1–948 constructs were transfected into COS-7 cells. After 48 h of transfection, the media was changed and chicken NF186 primary antibody (1:1,000 dilution) was added to the media. Cells were incubated for 30 min at 4°C and then cells were washed with ice-cold PBS three times. After the media was changed again, the cells were moved to 37°C for 1 h to allow endocytosis. The cells were then washed with ice-cold 100 mM glycine-HCl, pH 2.0, and HBSS (Life Technologies) to remove surface-bound antibodies. Finally, cells were fixed and immunostained with anti-GFP antibody and anti-FLAG antibody. Next, the cells were incubated with secondary antibodies AMCA, Alexa Fluor-488, and Alexa Fluor-594 to detect endocytosed NF186, NF186-GFP (total NF186), and FLAG-tagged constructs, respectively. Fluorescence intensities were measured using Zen acquisition software or ImageJ 1.50i software (NIH; RRID: SCR_003070).
Endocytosis assay for endogenous NF186 in hippocampal neurons
GFP and GFP-NuMA1 1–948 were transfected into DIV5 rat hippocampal neurons and then incubated for 24 h. Next, mouse monoclonal NF186 primary antibody (A12/18), which specifically recognizes the extracellular domain of rat NF186, was added into culture medium (1:1,000 dilution) and incubated for 1 h at 4°C. After incubation, neurons were washed with PBS three times. After the media was changed, the cells were moved to 37°C for 24 h to allow endocytosis. The neurons were then washed with ice-cold 100 mM glycine-HCl (pH 2.0) and HBSS (Gibco) to remove surface-bound antibodies and fixed using 4% PFA. Neurons were immunostained with chicken NF186 (also against NF186 ectodomain) to detect surface NF186. Finally, neurons were processed by the regular immunostaining procedures.
Statistical analyses
All statistical analyses were performed on Microsoft Excel software (Microsoft; RRID: SCR_016137) or GraphPad Prism 8 software (GraphPad Software; RRID: SCR_002798). Data were presented as mean ± SEM of independent experiments (the number of independent experiments is given in each figure legend). Tests of statistical significance included unpaired two-tailed Student’s t test, Tukey’s multiple comparison test, and one-way ANOVA; specific tests are indicated in each figure legend. Data distribution was assumed to be normal, but this was not formally tested.
Online supplemental materials
Fig. S1 shows that NF186-deletion constructs reveal domains important for its endocytosis. Fig. S2 shows that silencing expression of NuMA1 or 4.1B does not affect AIS maintenance in hippocampal neurons. All peptide spectral mass counts are included in Dataset S1.
Acknowledgments
We thank Cameron Smith for assistance with data presentation.
The work reported here was supported by the following research grants: National Institutes of Health NS044916 and NS069688 (to M.N. Rasband); and by the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation (to A.L. Burlingame and M.N. Rasband).
The authors declare no competing financial interests.
Author contributions: Methodology, validation, investigation, and writing – original draft and editing, T. Torii. Validation and investigation, Y. Ogawa, C-H. Liu, T.S-Y. Ho, H. Hamdan, and C-c. Wang. Investigation, data curation, resources, and writing – review and editing, J.A. Oses-Prieto. Resources, supervision, and funding acquisition, A.L. Burlingame. Conceptualization, methodology, investigation, data curation, writing – original draft and editing, project administration, and funding acquisition, M.N. Rasband.
References
- Akin E.J., Solé L., Dib-Hajj S.D., Waxman S.G., and Tamkun M.M.. 2015. Preferential targeting of Nav1.6 voltage-gated Na+ Channels to the axon initial segment during development. PLoS One. 10:e0124397 10.1371/journal.pone.0124397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger S.L., Leo-Macias A., Yuen S., Khatri L., Pfennig S., Zhang Y., Agullo-Pascual E., Caillol G., Zhu M.S., Rothenberg E., et al. . 2018. Localized Myosin II Activity Regulates Assembly and Plasticity of the Axon Initial Segment. Neuron. 97:555–570.e6. 10.1016/j.neuron.2017.12.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boiko T., Vakulenko M., Ewers H., Yap C.C., Norden C., and Winckler B.. 2007. Ankyrin-dependent and -independent mechanisms orchestrate axonal compartmentalization of L1 family members neurofascin and L1/neuron-glia cell adhesion molecule. J. Neurosci. 27:590–603. 10.1523/JNEUROSCI.4302-06.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bréchet A., Fache M.P., Brachet A., Ferracci G., Baude A., Irondelle M., Pereira S., Leterrier C., and Dargent B.. 2008. Protein kinase CK2 contributes to the organization of sodium channels in axonal membranes by regulating their interactions with ankyrin G. J. Cell Biol. 183:1101–1114. 10.1083/jcb.200805169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffington S.A., Sobotzik J.M., Schultz C., and Rasband M.N.. 2012. IκBα is not required for axon initial segment assembly. Mol. Cell. Neurosci. 50:1–9. 10.1016/j.mcn.2012.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi M., Atlas R., Kantor A., Sapir T., and Reiner O.. 2000. Interaction between LIS1 and doublecortin, two lissencephaly gene products. Hum. Mol. Genet. 9:2205–2213. 10.1093/oxfordjournals.hmg.a018911 [DOI] [PubMed] [Google Scholar]
- Chansard M., Hong J.H., Park Y.U., Park S.K., and Nguyen M.D.. 2011. Ndel1, Nudel (Noodle): flexible in the cell? Cytoskeleton (Hoboken). 68:540–554. 10.1002/cm.20532 [DOI] [PubMed] [Google Scholar]
- Fache M.P., Moussif A., Fernandes F., Giraud P., Garrido J.J., and Dargent B.. 2004. Endocytotic elimination and domain-selective tethering constitute a potential mechanism of protein segregation at the axonal initial segment. J. Cell Biol. 166:571–578. 10.1083/jcb.200312155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galiano M.R., Jha S., Ho T.S., Zhang C., Ogawa Y., Chang K.J., Stankewich M.C., Mohler P.J., and Rasband M.N.. 2012. A distal axonal cytoskeleton forms an intra-axonal boundary that controls axon initial segment assembly. Cell. 149:1125–1139. 10.1016/j.cell.2012.03.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido J.J., Giraud P., Carlier E., Fernandes F., Moussif A., Fache M.P., Debanne D., and Dargent B.. 2003. A targeting motif involved in sodium channel clustering at the axonal initial segment. Science. 300:2091–2094. 10.1126/science.1085167 [DOI] [PubMed] [Google Scholar]
- Garver T.D., Ren Q., Tuvia S., and Bennett V.. 1997. Tyrosine phosphorylation at a site highly conserved in the L1 family of cell adhesion molecules abolishes ankyrin binding and increases lateral mobility of neurofascin. J. Cell Biol. 137:703–714. 10.1083/jcb.137.3.703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg S.R., Tan W., and Lee W.L.. 2018. Num1 versus NuMA: insights from two functionally homologous proteins. Biophys. Rev. 10:1631–1636. 10.1007/s12551-018-0472-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan S., Price J.C., Prusiner S.B., Ghaemmaghami S., and Burlingame A.L.. 2011. A data processing pipeline for mammalian proteome dynamics studies using stable isotope metabolic labeling. Mol. Cell. Proteomics. 10:M111.010728 10.1074/mcp.M111.010728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedstrom K.L., Xu X., Ogawa Y., Frischknecht R., Seidenbecher C.I., Shrager P., and Rasband M.N.. 2007. Neurofascin assembles a specialized extracellular matrix at the axon initial segment. J. Cell Biol. 178:875–886. 10.1083/jcb.200705119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedstrom K.L., Ogawa Y., and Rasband M.N.. 2008. AnkyrinG is required for maintenance of the axon initial segment and neuronal polarity. J. Cell Biol. 183:635–640. 10.1083/jcb.200806112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho T.S., Zollinger D.R., Chang K.J., Xu M., Cooper E.C., Stankewich M.C., Bennett V., and Rasband M.N.. 2014. A hierarchy of ankyrin-spectrin complexes clusters sodium channels at nodes of Ranvier. Nat. Neurosci. 17:1664–1672. 10.1038/nn.3859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C.Y., Zhang C., Ho T.S., Oses-Prieto J., Burlingame A.L., Lalonde J., Noebels J.L., Leterrier C., and Rasband M.N.. 2017. αII Spectrin Forms a Periodic Cytoskeleton at the Axon Initial Segment and Is Required for Nervous System Function. J. Neurosci. 37:11311–11322. 10.1523/JNEUROSCI.2112-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husi H., and Grant S.G.. 2001. Proteomics of the nervous system. Trends Neurosci. 24:259–266. 10.1016/S0166-2236(00)01792-6 [DOI] [PubMed] [Google Scholar]
- Kiyomitsu T., and Cheeseman I.M.. 2013. Cortical dynein and asymmetric membrane elongation coordinately position the spindle in anaphase. Cell. 154:391–402. 10.1016/j.cell.2013.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kizhatil K., Wu Y.X., Sen A., and Bennett V.. 2002. A new activity of doublecortin in recognition of the phospho-FIGQY tyrosine in the cytoplasmic domain of neurofascin. J. Neurosci. 22:7948–7958. 10.1523/JNEUROSCI.22-18-07948.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kole M.H., Ilschner S.U., Kampa B.M., Williams S.R., Ruben P.C., and Stuart G.J.. 2008. Action potential generation requires a high sodium channel density in the axon initial segment. Nat. Neurosci. 11:178–186. 10.1038/nn2040 [DOI] [PubMed] [Google Scholar]
- Kotak S., Busso C., and Gönczy P.. 2013. NuMA phosphorylation by CDK1 couples mitotic progression with cortical dynein function. EMBO J. 32:2517–2529. 10.1038/emboj.2013.172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuijpers M., van de Willige D., Freal A., Chazeau A., Franker M.A., Hofenk J., Rodrigues R.J., Kapitein L.C., Akhmanova A., Jaarsma D., and Hoogenraad C.C.. 2016. Dynein Regulator NDEL1 Controls Polarized Cargo Transport at the Axon Initial Segment. Neuron. 89:461–471. 10.1016/j.neuron.2016.01.022 [DOI] [PubMed] [Google Scholar]
- Lazarov E., Dannemeyer M., Feulner B., Enderlein J., Gutnick M.J., Wolf F., and Neef A.. 2018. An axon initial segment is required for temporal precision in action potential encoding by neuronal populations. Sci. Adv. 4:eaau8621 10.1126/sciadv.aau8621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leterrier C., Clerc N., Rueda-Boroni F., Montersino A., Dargent B., and Castets F.. 2017. Ankyrin G Membrane Partners Drive the Establishment and Maintenance of the Axon Initial Segment. Front. Cell. Neurosci. 11:6 10.3389/fncel.2017.00006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorincz A., and Nusser Z.. 2010. Molecular identity of dendritic voltage-gated sodium channels. Science. 328:906–909. 10.1126/science.1187958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattagajasingh S.N., Huang S.C., Hartenstein J.S., Snyder M., Marchesi V.T., and Benz E.J.. 1999. A nonerythroid isoform of protein 4.1R interacts with the nuclear mitotic apparatus (NuMA) protein. J. Cell Biol. 145:29–43. 10.1083/jcb.145.1.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson A.D., and Jenkins P.M.. 2017. Axonal Membranes and Their Domains: Assembly and Function of the Axon Initial Segment and Node of Ranvier. Front. Cell. Neurosci. 11:136 10.3389/fncel.2017.00136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa Y., Schafer D.P., Horresh I., Bar V., Hales K., Yang Y., Susuki K., Peles E., Stankewich M.C., and Rasband M.N.. 2006. Spectrins and ankyrinB constitute a specialized paranodal cytoskeleton. J. Neurosci. 26:5230–5239. 10.1523/JNEUROSCI.0425-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Z., Kao T., Horvath Z., Lemos J., Sul J.-Y., Cranstoun S.D., Bennett V., Scherer S.S., and Cooper E.C.. 2006. A common ankyrin-G-based mechanism retains KCNQ and NaV channels at electrically active domains of the axon. J. Neurosci. 26:2599–2613. 10.1523/JNEUROSCI.4314-05.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinatel D., Hivert B., Saint-Martin M., Noraz N., Savvaki M., Karagogeos D., and Faivre-Sarrailh C.. 2017. The Kv1-associated molecules TAG-1 and Caspr2 are selectively targeted to the axon initial segment in hippocampal neurons. J. Cell Sci. 130:2209–2220. 10.1242/jcs.202267 [DOI] [PubMed] [Google Scholar]
- Rosenfeld J., Capdevielle J., Guillemot J.C., and Ferrara P.. 1992. In-gel digestion of proteins for internal sequence analysis after one- or two-dimensional gel electrophoresis. Anal. Biochem. 203:173–179. 10.1016/0003-2697(92)90061-B [DOI] [PubMed] [Google Scholar]
- Saifetiarova J., Taylor A.M., and Bhat M.A.. 2017. Early and Late Loss of the Cytoskeletal Scaffolding Protein, Ankyrin G Reveals Its Role in Maturation and Maintenance of Nodes of Ranvier in Myelinated Axons. J. Neurosci. 37:2524–2538. 10.1523/JNEUROSCI.2661-16.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T. 2006. In vivo electroporation in the embryonic mouse central nervous system. Nat. Protoc. 1:1552–1558. 10.1038/nprot.2006.276 [DOI] [PubMed] [Google Scholar]
- Silk A.D., Holland A.J., and Cleveland D.W.. 2009. Requirements for NuMA in maintenance and establishment of mammalian spindle poles. J. Cell Biol. 184:677–690. 10.1083/jcb.200810091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobotzik J.M., Sie J.M., Politi C., Del Turco D., Bennett V., Deller T., and Schultz C.. 2009. AnkyrinG is required to maintain axo-dendritic polarity in vivo. Proc. Natl. Acad. Sci. USA. 106:17564–17569. 10.1073/pnas.0909267106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark H., Parton R.G., Steele-Mortimer O., Lütcke A., Gruenberg J., and Zerial M.. 1994. Inhibition of rab5 GTPase activity stimulates membrane fusion in endocytosis. EMBO J. 13:1287–1296. 10.1002/j.1460-2075.1994.tb06381.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susuki K., Chang K.J., Zollinger D.R., Liu Y., Ogawa Y., Eshed-Eisenbach Y., Dours-Zimmermann M.T., Oses-Prieto J.A., Burlingame A.L., Seidenbecher C.I., et al. . 2013. Three mechanisms assemble central nervous system nodes of Ranvier. Neuron. 78:469–482. 10.1016/j.neuron.2013.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuvia S., Garver T.D., and Bennett V.. 1997. The phosphorylation state of the FIGQY tyrosine of neurofascin determines ankyrin-binding activity and patterns of cell segregation. Proc. Natl. Acad. Sci. USA. 94:12957–12962. 10.1073/pnas.94.24.12957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacher H., Yang J.W., Cerda O., Autillo-Touati A., Dargent B., and Trimmer J.S.. 2011. Cdk-mediated phosphorylation of the Kvβ2 auxiliary subunit regulates Kv1 channel axonal targeting. J. Cell Biol. 192:813–824. 10.1083/jcb.201007113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Ketcham S.A., Schön A., Goodman B., Wang Y., Yates J. III, Freire E., Schroer T.A., and Zheng Y.. 2013. Nudel/NudE and Lis1 promote dynein and dynactin interaction in the context of spindle morphogenesis. Mol. Biol. Cell. 24:3522–3533. 10.1091/mbc.e13-05-0283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams S.E., Beronja S., Pasolli H.A., and Fuchs E.. 2011. Asymmetric cell divisions promote Notch-dependent epidermal differentiation. Nature. 470:353–358. 10.1038/nature09793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., and Shrager P.. 2005. Dependence of axon initial segment formation on Na+ channel expression. J. Neurosci. Res. 79:428–441. 10.1002/jnr.20378 [DOI] [PubMed] [Google Scholar]
- Yang Y., Ogawa Y., Hedstrom K.L., and Rasband M.N.. 2007. betaIV spectrin is recruited to axon initial segments and nodes of Ranvier by ankyrinG. J. Cell Biol. 176:509–519. 10.1083/jcb.200610128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap C.C., Vakulenko M., Kruczek K., Motamedi B., Digilio L., Liu J.S., and Winckler B.. 2012. Doublecortin (DCX) mediates endocytosis of neurofascin independently of microtubule binding. J. Neurosci. 32:7439–7453. 10.1523/JNEUROSCI.5318-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye K., Compton D.A., Lai M.M., Walensky L.D., and Snyder S.H.. 1999. Protein 4.1N binding to nuclear mitotic apparatus protein in PC12 cells mediates the antiproliferative actions of nerve growth factor. J. Neurosci. 19:10747–10756. 10.1523/JNEUROSCI.19-24-10747.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z., Wan Q., Meixiong G., and Du Q.. 2014. Cell cycle-regulated membrane binding of NuMA contributes to efficient anaphase chromosome separation. Mol. Biol. Cell. 25:606–619. 10.1091/mbc.e13-08-0474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zonta B., Desmazieres A., Rinaldi A., Tait S., Sherman D.L., Nolan M.F., and Brophy P.J.. 2011. A critical role for Neurofascin in regulating action potential initiation through maintenance of the axon initial segment. Neuron. 69:945–956. 10.1016/j.neuron.2011.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]