Christophe Leterrier previews work from Lee et al. that describes a mechanism by which NKCC1 levels are regulated in GABAergic neurons.
Abstract
The GABA response switch from excitatory to inhibitory is a key event in neuronal maturation that depends on the regulated expression of chloride transporters NKCC1 and KCC2. In this issue, Lee et al. (2020. J. Cell. Biol. https://doi.org/10.1083/jcb.201903033) describe how degradation of NKCC1 by proteasomes immobilized at the axon initial segment (AIS) by the Ecm29 adaptor contributes to this regulation, driving the GABA switch and the positional maturation of the AIS.
Response of neurons to the neurotransmitter GABA switches from excitatory to inhibitory, a key event for proper maturation of the brain. This switch relies on the developmental regulation of intracellular chloride ionic concentration by two opposing cation-chloride transporters: in immature neurons, strong NKCC1 expression drives entry of chloride ions and their high intracellular concentration, before KCC2 becomes dominant and lowers this concentration by driving chloride exit. The drop in intracellular chloride concentration modifies the response profile of GABA receptors from excitatory to inhibitory around a week after birth in rodents (1). Several mechanisms have been proposed that modulate the relative NKCC1/KCC2 expression including transcriptional, post-translational, and trafficking regulations (2).
In this issue, Lee et al. demonstrate a novel subcellular mechanism contributing to the NKCC1/KCC2 expression switch: degradation of NKCC1 by proteasomes (3). Intriguingly, this involves specific localization of proteasomes and subsequent degradation of NKCC1 at the axon initial segment (AIS). The AIS is a specialized compartment located along the first 20 to 50 µm of the axon. It concentrates sodium channels that generate the action potential and plays a role in maintaining neuronal polarity by separating the axon from the cell body (4). In cultured neurons, Lee et al. first show that proteasomes are immobilized at the AIS via interaction with ankyrin G, the main AIS scaffold protein, via both the Ecm29 adaptor and direct binding of proteasomes subunits. Notably, proteasomes disperse along the whole axon in Ecm29−/− neurons. These interactions implicate two different ankyrin G domains and result in a differential nanoscale localization of adaptors and proteasomes relative to the periodic actin/spectrin scaffold that lines the AIS plasma membrane (5). At the AIS, proteasomes facilitate protein turnover, as elegantly shown by monitoring the half-life of an AIS probe fused to a destabilized GFP.
What connects this location-specific proteasome activity to the developmental GABA switch? In Ecm29 knockout mice, the expression of NKCC1 is prolonged beyond the first postnatal week. This correlates with NKCC1 accumulation at the AIS and slowed turnover in neuronal cultures. The elevated expression of NKCC1 in Ecm29−/− neurons results in a delayed GABA response switch: GABA stimulation results in chloride efflux until 9 d in culture, whereas wild-type neurons switch to chloride influx after 7 d, as monitored using a Förster resonance energy transfer–based chloride concentration probe. This is confirmed by monitoring intracellular calcium, indicating an excitatory response to GABA.
The physiological relevance of the cellular defects observed in Ecm29 knockout mice is indicated by their higher susceptibility to chemically induced seizures and higher neuronal excitability. Strikingly, inhibition of NKCC1 function by the inhibitor bumetanide is able to compensate for its prolonged expression and protect against seizure susceptibility, but only if administered perinatally, when the GABA response switch normally occurs. In a final return to their initial observation involving the AIS in the NKCC1 degradation process, Lee et al. (3) explore the relationship between elevated excitability in Ecm29−/− and the morphological plasticity of the AIS during development (6). The AIS gets closer to the cell body as neurons mature, and this proximal shift is accelerated in Ecm29−/− neurons or upon proteasome inhibition in wild-type neurons, possibly contributing to their elevated excitability (7).
This study presents a thorough and extensive range of experiments spanning from cellular details of proteasomes and chloride transporter localization to the effect on neuronal hyperactivity in knockout mice. This builds a compelling demonstration for a fundamentally novel cellular mechanism of the developmental GABA response switch, based on time-, location-, and target-selective protein degradation. Numerous studies have focused on the “input” side of chloride transporters expression, unveiling transcription, modification, and trafficking factors to the NKCC1/KCC2 transition (2). Lee et al. (3) provide a new angle, by showing the importance of the “output” side: specific protein degradation can be a powerful process to shape the availability of cellular components. Furthermore, this work links two key factors for neuronal excitability that were so far unconnected: the developmental GABA response switch and the AIS formation and morphological development (Fig. 1).
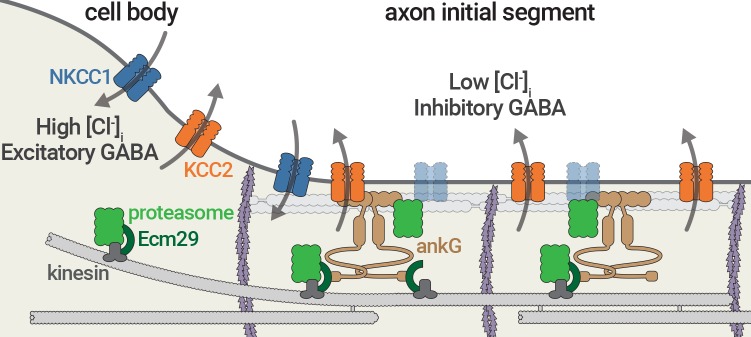
Figure 1.
AIS-located proteasomes degrade chloride transporters and drive GABA response reversal. Proteasomes associated with the adaptor Ecm29 (green) are transported to the AIS by kinesins along microtubules (gray). Here, they interact with ankyrin G (ankG; brown) and degrade NKCC1 chloride transporters (blue). This shifts the NKCC1 import/KCC2 export (orange) equilibrium, reducing the intracellular chloride concentration and shifting the GABA response from excitatory to inhibitory.
By revealing the existence of an AIS-located proteasomal activity, this study opens new perspectives and raises exciting new questions. The AIS is the location of specific GABA synapses that can exert a powerful effect on intrinsic excitability by directly acting on the site of action potential generation (8). Does the degradation of NKCC1 ensure robust reversal for these particular synapses? Another intriguing question is how protein degradation can specifically target NKCC1 for down-regulation. It may be that NCC1 is simply more abundant, hence preferentially targeted, as suggested by the authors. Degradation at the AIS could also be coupled to selective trafficking of NKCC1 toward the axon, providing a dynamic way to progressively clear the cell body and dendrites from NKCC1. Interestingly, the absence of axonal targeting for KCC2 (9) would then preserve it from degradation. Finally, proteasomal activity at the AIS is likely to have consequences beyond the regulation of the NKCC1/KCC2 balance. Lee et al. (3) have found that perturbing this activity does not impact the AIS scaffold itself or sodium channel accumulation, but it would be interesting to test if this affects how it maintains neuronal polarity and regulates trafficking from and to the axon (4).
Acknowledgments
The author declares no competing financial interests.
References
- 1.Ben-Ari Y., et al. Neuroscientist. 2012 doi: 10.1177/1073858412438697. [DOI] [Google Scholar]
- 2.Watanabe M., and Fukuda A. Front. Cell. Neurosci. 2015 doi: 10.3389/fncel.2015.00371. [DOI] [Google Scholar]
- 3.Lee M., et al. J. Cell Biol. 2020 doi: 10.1083/jcb.201903033. [DOI] [Google Scholar]
- 4.Leterrier C. J. Neurosci. 2018 doi: 10.1523/JNEUROSCI.1922-17.2018. [DOI] [Google Scholar]
- 5.Leterrier C., et al. Cell Reports. 2015 doi: 10.1016/j.celrep.2015.11.051. [DOI] [Google Scholar]
- 6.Jamann N., et al. Neuroscience. 2018 doi: 10.1016/j.neuroscience.2017.07.035. [DOI] [PubMed] [Google Scholar]
- 7.Gulledge A.T., and Bravo J.J. eNeuro. 2016 doi: 10.1523/ENEURO.0085-15.2016. [DOI] [Google Scholar]
- 8.Nathanson A.J., et al. Front. Mol. Neurosci. 2019 doi: 10.3389/fnmol.2019.00266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Côme E., et al. Front. Cell. Neurosci. 2019 doi: 10.3389/fncel.2019.00048. [DOI] [Google Scholar]