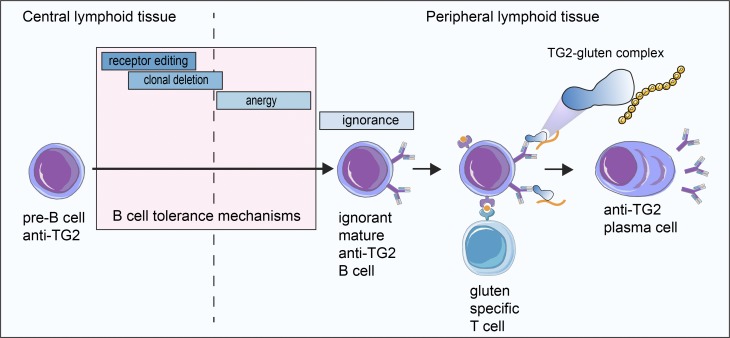
This study sheds light on the mechanism by which autoantibodies to transglutaminase 2 (TG2) are formed in celiac disease. The authors find that there is no deletion or silencing of autoreactive TG2-specific B cells and that production of anti-TG2 autoantibodies is controlled by T cell help.
Abstract
Autoantibodies to transglutaminase 2 (TG2) are hallmarks of celiac disease. To address B cell tolerance and autoantibody formation to TG2, we generated immunoglobulin knock-in (Ig KI) mice that express a prototypical celiac patient–derived anti-TG2 B cell receptor equally reactive to human and mouse TG2. We studied B cell development in the presence/absence of autoantigen by crossing the Ig KI mice to Tgm2−/− mice. Autoreactive B cells in Tgm2+/+ mice were indistinguishable from their naive counterparts in Tgm2−/− mice with no signs of clonal deletion, receptor editing, or B cell anergy. The autoreactive B cells appeared ignorant to their antigen, and they produced autoantibodies when provided T cell help. The findings lend credence to a model of celiac disease where gluten-reactive T cells provide help to autoreactive TG2-specific B cells by involvement of gluten–TG2 complexes, and they outline a general mechanism of autoimmunity with autoantibodies being produced by ignorant B cells on provision of T cell help.
Graphical Abstract
Introduction
Celiac disease, characterized by highly disease-specific antibodies to transglutaminase 2 (TG2; Dieterich et al., 1997), is a common enteropathy caused by exposure to dietary gluten proteins (Sollid, 2002). Plasma cells specific for TG2 are abundant in celiac gut lesions (Di Niro et al., 2012). Subjects with the condition exclusively carry the HLA-DQ allotypes DQ2.5, DQ2.2, or DQ8, and they have gluten-specific CD4+ T cells recognizing deamidated gluten peptides bound to these disease-associated HLA-DQ molecules (Sollid, 2002). The creation of deamidated gluten epitopes is mediated by the same enzyme to which there are autoantibodies, TG2. This dual role of TG2 in celiac disease, both creating immunogenic T cell epitopes and being the target of autoantibodies, is hardly coincidental and is likely central in the disease pathogenesis.
In general, the majority of newly generated B cells have B cell receptors (BCRs) that bind self-antigens, but most of these autoreactive B cells are removed from the repertoires of mature B cells (Wardemann et al., 2003). In the bone marrow, developing autoreactive B cells can become non–self-reactive by receptor editing (Gay et al., 1993; Tiegs et al., 1993), and if receptor editing fails, they can be deleted by apoptosis (Nemazee and Bürki, 1989). In the periphery, autoreactive B cells can be kept silent by anergy (Goodnow et al., 1988). What dictates the mode of tolerance is not fully understood, but avidity of the BCR appears central as multimeric or membrane-bound self-antigens typically promote B cell deletion, and monomeric soluble self-antigens typically result in B cell anergy (Goodnow, 1992). In some cases, autoreactive B cells termed “ignorant” can appear in the periphery as normal B cells, presumably because of nonexposure to self-antigen or because the BCR-binding signal is too weak to enforce tolerance induction (Adelstein et al., 1991; Akkaraju et al., 1997; Aplin et al., 2003; Hannum et al., 1996; Huang et al., 2006; Koenig-Marrony et al., 2001).
TG2 is constitutively expressed in most cell types and tissues (Lorand and Graham, 2003). The protein is transcribed on free ribosomes in the cytosol but can be externalized and bind to components of the extracellular matrix. Many tissues exhibit abundant TG2 staining in the extracellular matrix (Korponay-Szabó et al., 2004). For other extracellular matrix proteins, like basement membrane laminin, to which there are antibodies in lupus disease of mice and humans, transgenic BCR mouse models have revealed that there is strong B cell tolerance obtained by central deletion, L chain editing, or anergy (Foster et al., 2006).
To address how anti-TG2 autoantibodies are formed, we generated Ig knock-in (KI) mice based on a celiac patient–derived BCR with equal reactivity to human and mouse TG2 (mTG2). TG2-specific plasma cells across celiac patients have a strikingly restricted usage of VH and VL genes with a particular dominance of the IGHV5-51/IGKV1-5 pair and with a low frequency of somatic mutations (Di Niro et al., 2012; Hnida et al., 2016; Iversen et al., 2013; Roy et al., 2017). The chosen BCR is prototypic for celiac disease as it expresses IGHV5-51 in germline configuration and IGKV1-5 with two somatic mutations. By crossing the Ig KI mice to Tgm2-deficient mice, we were able to study the development and function of autoreactive B cells in the presence and in the absence of the autoantigen. We demonstrate that clonal deletion and anergy to TG2 do not develop. Instead, self-tolerance to the TG2 autoantigen appears to be regulated outside the B cell compartment and is maintained by the absence of T cell help. We also show that autoreactive anti-TG2 B cells are able to produce autoantibodies when linked T cell help is provided, suggesting that autoreactivity to TG2 in celiac disease is controlled at the T cell level.
Results
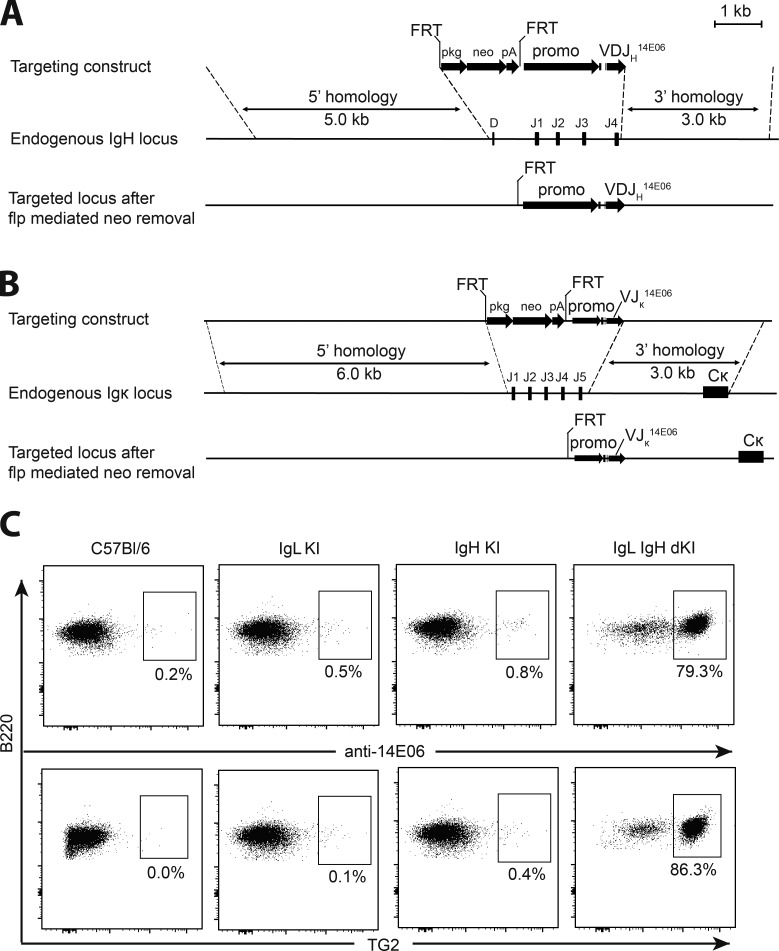
Generation of an Ig KI mouse model to study class switch–competent anti-TG2 B cells
The Ig KI mouse was based on a prototypic anti-TG2 mAb 679–14-E06 (hereafter termed 14E06) derived from a single gut plasma cell from a celiac patient (Di Niro et al., 2012). The mAb is specific for the major epitope 1 of TG2, and it binds with equal, nanomolar affinity to the human and mouse orthologue TG2 proteins (Table S1; Di Niro et al., 2012; Hnida et al., 2016; Iversen et al., 2013). The H chain KI mouse was generated with the 14E06-rearranged VDJ sequence introduced into a targeting vector with the leader-encoding sequence and promoter of a mouse H chain gene to replace the D to J4 segments of the mouse H chain locus (Fig. 1 A). Likewise, the L chain KI mouse was generated with the 14E06-rearranged VJ sequence introduced into a targeting vector with the leader-encoding sequence and promotor of mouse κ L chain gene to replace the J1 to J5 segments of the mouse κ locus (Fig. 1 B). H and L chain KI mice were crossed to obtain 14E06 double KI mice. Only B cells of the double KI mice and not the single H chain or single L chain KI mice stained with an antibody (anti-14E06; Høydahl et al., 2016) specific for the H and L chain combination of 14E06 (Fig. 1 C). Consistent with anti-14E06 staining, 14E06 B cells, but not B cells from H or L chain single KI mice, stained with mTG2 tetramerized on PE-conjugated streptavidin (Fig. 1 C). These data indicate that mature TG2-specific B cells are present in the double KI mice and that both the H and L chains of 14E06, as also observed by H and L chain shuffling (Chen et al., 2015), contribute to TG2 binding.
Table S1. Surface plasmon resonance derived binding kinetics.
| Sample | ka (107/Ms) | kd (10−2/s) | KD (nM) |
|---|---|---|---|
| hTG2 GDP | 0.60 ± 0.05 | 2.74 ± 0.29 | 4.56 ± 0.10 |
| hTG2 DP3-3/Ca2+ | 0.10 ± 0.013 | 3.01 ± 0.18 | 3.07 ± 0.20 |
| mTG2 GDP | 0.14 ± 0.00 | 0.44 ± 0.01 | 3.21 ± 0.06 |
| mTG2 DP3-3/Ca2+ | 0.10 ± 0.00 | 0.67 ± 0.01 | 6.44 ± 0.14 |
Data are presented as the means ± SD. Based on two experiments. GDP-bound TG2: TG2 bound to GDP, closed conformation, presumably representing intracellular TG2; DP3-3/Ca2+ bound TG2: TG2 with irreversible active site inhibitor in presence of Ca2+, open conformation, as expected to occur extracellularly due to the high extracellular concentration of Ca2+. ka, association rate constant; kd, dissociation rate constant.
Figure 1.
Generation of 14E06 KI mice. (A) Generation of the VDJH 14E06 KI mouse. Top: Targeting construct for the rearranged VDJH 14E06 and a FRT-flanked pkg-neo selection cassette. Middle: Mouse H chain locus with indication of the replaced D and J1-4 gene segments. Dashed lines indicate terminal points of homologous recombination. Bottom: Integrated transgene after flp mediated excision of the FRT-flanked neomycin selection gene. (B) Generation of the VJκ 14E06 KI mouse. Top: Targeting construct for the rearranged VJκ 14E06. Middle: Mouse κ L chain locus with indication of the replaced J1-5 gene segments. Bottom: integrated transgene after flp mediated excision of the FRT-flanked neomycin selection gene. (C) Detection of transgenic anti-TG2 BCR. Spleen cells from WT C57Bl/6 mice (obtained from Janvier Laboratoires), 14E06 L chain single KI and H chain single KI littermate control mice, and H and L chain double KI mice on C57Bl/6 background were stained using anti-B220 and an antibody specific for the H and L chain combination of 14E06 (anti-14E06) or mTG2 tetramers. Data are representative of two independent experiments.
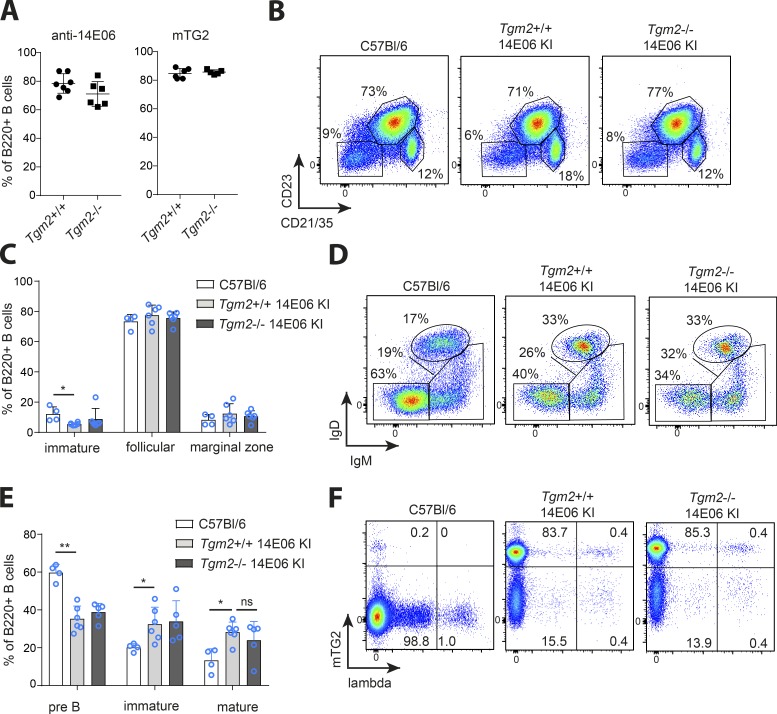
Normal B cell development in 14E06 KI mice with no signs of deletion or receptor editing
To study how the development of B cells expressing the anti-TG2 BCR is affected by recognition of the antigen in vivo, we crossed 14E06 KI mice to Tgm2-deficient mice (De Laurenzi and Melino, 2001). We found that the frequency of TG2-binding B cells in peripheral lymphoid organs was the same for 14E06 KI mice on Tgm2+/+ and Tgm2−/− genetic backgrounds (Fig. 2 A). Moreover, no differences in spleen cellularity and the total number of B cells were found between 14E06 KI mice being Tgm2+/+ or Tgm2−/− (Fig. S1, A and B). To assess if there would be spontaneous differentiation of Ig KI B cells into antibody-secreting cells, we measured basal serum total and TG2-specific IgM, IgG, and IgA levels in Tgm2+/+ and Tgm2−/− 14E06 KI mice. Both IgM and IgG anti-TG2 antibodies, but not IgA, were detected; however, titers were not increased in the presence of the self-antigen compared with control mice on Tgm2−/− background (Fig. S1 C).
Figure 2.
Analysis of B cell development in 14E06 KI mice. (A) Percentage of TG2-specific B cells as detected by anti-14E06 antibody or mTG2 tetramers in Tgm2+/+ and Tgm2−/− 14E06 KI mice. Data are pooled from six independent experiments and show mean ± SD. (B and C) Analysis of B cell subsets in spleen. (B) Representative plots of spleen B220+ B cells stained for CD23 and CD21/35 expression to identify immature/T1 (CD23−CD21−), follicular/T2 (CD23+CD21+), and marginal zone (CD23−CD21hi) subsets. (C) Pooled data from at least four independent experiments. Data are mean ± SD. (D and E) Analysis of developing B cells in bone marrow. (D) Representative plots of bone marrow B220+ B cells stained for anti-IgD and anti-IgM to identify pre-B cells (IgM−IgD−), immature and transitional B cells (IgM+IgD−/lo), and mature recirculating (IgM+IgD+) B cells in WT C57Bl/6, Tgm2+/+, and Tgm2−/− 14E06 KI mice. (E) Data are pooled from at least four independent experiments and show mean ± SD. (F) Representative plots of spleen B220+ B cells stained with mTG2 tetramers and an anti-λ L chain antibody. *, P < 0.05, **, P < 0.0005, as determined by Student’s t test (C and E). Flow plots are representative of at least four independent experiments (B, D, and F). For C57Bl/6 control mice, we used either commercially obtained C57Bl/6 from Janvier Laboratoires or nontransgenic littermates from in house breedings. ns, not significant.
Immature, mature follicular, and marginal zone B cells (as defined by CD21/CD35 and CD23 staining) in Tgm2−/− mice were present at similar proportions to those observed in Tgm2+/+ mice (Fig. 2, B and C). Peritoneal cavity lavage showed that TG2-specific B cells exclusively belonged to the B-2 cell lineage (Fig. S1 D). In the bone marrow, we detected a reduced frequency of pro/pre-B cells and a corresponding increase in immature and mature B cells compared with WT mice (Fig. 2, D and E), indicating that, as seen in other Ig transgenic mouse models (Goodnow, 1992), 14E06 KI mice had accelerated development of B cells as a result of the prearranged transgenic BCR. We next looked at the fraction of peripheral B cells expressing λ L chains as a sign of receptor editing. As the transgenic L chain is κ, these λ L chains must have been the result of expression of the endogenous λ locus. We found no increase in the percentage of λ-positive cells in TG2-expressing 14E06 KI mice (Fig. 2 F). Taken together, our data demonstrate a normal development of 14E06 B cells without evidence for receptor editing or developmental arrest at an immature stage.
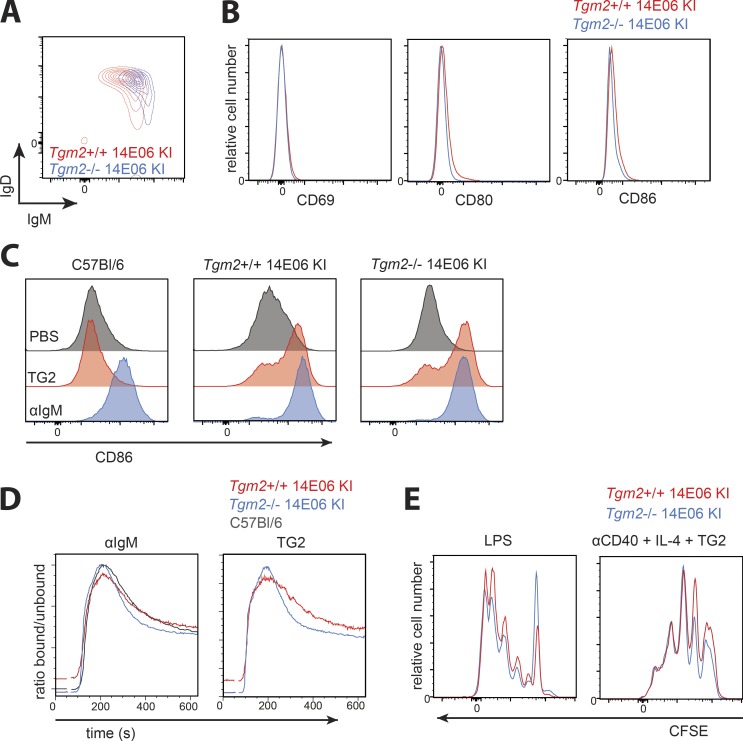
Classical features of anergy are not observed in autoreactive 14E06 KI B cells
We next assessed whether the TG2-specific B cells emerging from the bone marrow could be anergic. B cell anergy is characterized by down-regulation of surface IgM levels, impaired signaling through the BCR (shown by reduced Ca2+ influx, protein phosphorylation, and induction of activation markers), and resistance to LPS-mediated activation (Cambier et al., 2007). We analyzed these features in 14E06 KI B cells from Tgm2+/+ vs. Tgm2−/− mice comparing autoreactive and naive cells. We noted a very modest down-regulation of surface IgM levels on autoreactive cells (Fig. 3 A). Besides this minor difference in surface IgM, we found that autoreactive and naive cells were phenotypically indistinguishable with no increased expression of activation markers CD69, CD80, and CD86 on autoreactive KI B cells (Fig. 3 B). To determine whether autoreactive B cells were able to up-regulate activation markers in response to BCR ligation, we cultured isolated splenic B cells for 18 h in the presence of anti-IgM or TG2 antigen (Fig. 3 C). We observed that autoreactive KI B cells from Tgm2+/+ mice up-regulated activation markers CD86 (Fig. 3 C) and CD80 (data not shown) to the same extent as naive KI B cells. We next compared autoreactive and naive TG2-specific KI B cells for their ability to mobilize Ca2+. Both stimulation with anti-IgM and with TG2 antigen induced a clear Ca2+ flux response in freshly isolated autoreactive B cells from Tgm2+/+ KI mice, and importantly, the magnitude of this response was comparable to that seen for resting naive B cells isolated from both WT C57Bl/6 mice and 14E06 KI Tgm2−/− mice (Fig. 3 D). We also observed that anti-TG2 B cells isolated from the spleens of TG2-expressing 14E06 KI mice had slightly elevated basal Ca2+ levels when compared with resting naive B cells (Fig. 3 D). We next investigated the ability of autoreactive B cells to proliferate in response to antigen in the presence of T cell mimicking stimuli (anti-CD40 and IL-4), as well as the T cell–independent stimulus LPS. Both stimuli induced efficient proliferation of both autoreactive and naive TG2-specific B cells (Fig. 3 E).
Figure 3.
14E06 Ig KI B cells are not anergic. (A) Representative plot of spleen B220+ B cells of Tgm2+/+ (red) and Tgm2−/− 14E06 KI mice (blue) stained for anti-IgM and anti-IgD. (B) Histogram plots gated on B220+ TG2-tetramer+ B cells isolated from spleens of Tgm2+/+ (red) and Tgm2−/− (blue) 14E06 KI mice that were stained for CD69, CD80, and CD86. (C) Induction of CD86 after 18 h culture of isolated B cells from spleens of C57Bl/6, Tgm2+/+ 14E06 KI, and Tgm2−/− 14E06 KI mice in the presence of 2 µg/ml TG2 or 10 µg/ml anti-IgM. (D) Ca2+ flux of C57Bl/6, Tgm2+/+ 14E06 KI, and Tgm2−/− 14E06 KI B cells. Splenic B cells were loaded with the intracellular calcium indicator Indo-1. Baseline Ca2+ levels were acquired for 30 s before the cells were stimulated with 10 µg/ml anti-IgM or 2 µg/ml TG2. (E) CFSE dilution after 72 h of culture of isolated B cells of Tgm2+/+ (red) and Tgm2−/− (blue) 14E06 KI mice stimulated with 2 µg TG2 in the presence of 5 µg/ml anti-CD40 and 50 ng/ml IL-4 or 5 µg/ml LPS. All data are representative of at least three independent experiments (A–E). For C57Bl/6 control mice, we used either commercially obtained C57Bl/6 from Janvier Laboratoires or nontransgenic littermates from in-house breedings.
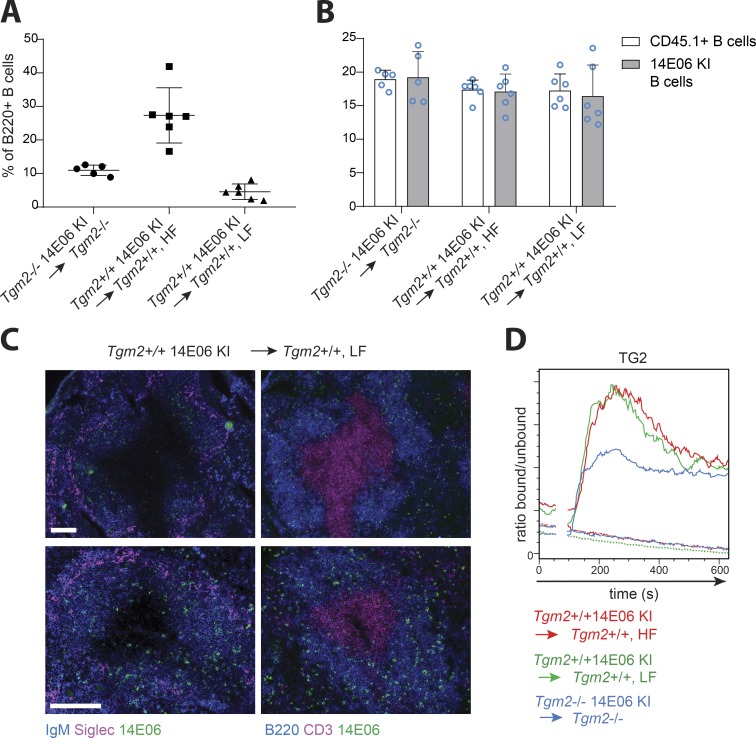
Anergic B cells have been shown to compete poorly with nonanergic B cells, resulting in exclusion from B cell follicles and a shortened life span (Cyster et al., 1994). To determine if these features were applicable to autoreactive TG2-specific B cells in our KI model, we created mixed bone marrow chimeras by mixing the bone marrow of TG2-expressing 14E06 KI mice 1:1 with that of CD45.1 WT mice followed by transfer to lethally irradiated C57Bl/6 recipient mice. As a control, we used lethally irradiated Tgm2−/− mice reconstituted with mixed bone marrow from Tgm2−/− 14E06 Ig KI mice and CD45.1 Tgm2−/− mice. The composition of the peripheral B cell pool in chimeric mice reconstituted with an equal ratio of donor cells was substantially skewed in favor of CD45.1+ (WT) cells in both Tgm2+/+ and Tgm2−/− chimeras, possibly due to competition with clones in the nontransgenic B cell population (Fig. S2 A). Anti-14E06–positive B cells in Tgm2+/+ chimeras comprised 27.3 ± 8.2% of splenic B cells and were not reduced compared with those in in Tgm2−/− chimeras (Fig. 4 A). As the frequency of antigen-specific B cells inversely correlates with the signal strength each B cells receives through its BCR (Cook et al., 1997), even lower frequencies of autoreactive B cells could be required in order to reach the threshold required to induce B cell tolerance in our model. We therefore also generated TG2-expressing chimeras reconstituted with 5% 14E06 Ig KI and 95% CD45.1 (WT) bone marrow, resulting in 4.6 ± 2.3% anti-14E06+ B cells in the spleen (Fig. 4 A). We did not observe differences in the proportions of immature, mature follicular, and marginal zone B cells in the spleens between low-frequency Tgm2+/+ chimeras, high-frequency Tgm2+/+ chimeras, and Tgm2−/− control chimeras (Fig. S2 B). To analyze the turnover of the autoreactive B cells among an excess of competitor B cells, we compared incorporation of BrdU after 8 d of continuous BrdU feeding. We found that TG2-specific B cells and WT CD45.1 B cells displayed comparable percentages of BrdU+ cells in both Tgm2+/+ and Tgm2−/− chimeras and that this was not dependent on the frequency of autoreactive TG2-specific B cells (Fig. 4 B). In keeping with this finding, analysis of the anatomical localization of autoreactive TG2-specific B cells in low-frequency chimeric mice revealed that TG2-specific B cells essentially migrated and distributed equivalently to naive control B cells and gained access to the lymphoid follicles in the presence of self-antigen (Fig. 4 C). We next analyzed whether TG2-specific B cells in mixed bone marrow chimeric mice were functional by investigating their ability to mobilize Ca2+. CD45.2 B cells, the majority of which were TG2-specific, demonstrated a clear Ca2+ flux upon stimulation with anti-IgM or TG2, whereas CD45.1 B cells in the same animal were, as expected, only responsive to anti-IgM stimulation (Fig. 4 D). These analyses demonstrate that 14E06 KI B cells are not anergic in the presence of the TG2 autoantigen.
Figure 4.
Analysis of 14E06 KI B cells in mixed bone marrow chimeric mice. To create Tgm2+/+ mixed bone marrow chimeric mice, bone marrow from CD45.2 Tgm2+/+ 14E06 KI mice was mixed with that from WT CD45.1 mice at a 1:1 ratio to obtain high frequency (HF) bone marrow chimeric mice, or mixed at 1:20 to obtain low frequency (LF) chimeric mice and used to reconstitute the hematopoietic system of lethally irradiated CD45.2 WT recipients. As a control, CD45.2 Tgm2−/− hosts were reconstituted with bone marrow from CD45.2 Tgm2−/− 14E06 KI mice and CD45.1 Tgm2−/−mice mixed 1:1. (A) The frequency of anti-TG2 B cells (as indicated by anti-14E06 staining) in spleens of bone marrow chimeric mice. Data are pooled from two independent measures and presented as mean ± SD from five or six mice in each group. (B) Mixed bone marrow chimeric mice were given BrdU in drinking water for 8 d. Splenic B220+CD45.1+ (WT) or B220+ CD45.2 α14E06+ (KI) B cells in Tgm2−/− control chimeric mice and Tgm2+/+ HF and Tgm2+/+ LF chimeric mice were analyzed for BrdU incorporation by flow cytometry. Data are pooled from two independent measures and represent mean ± SD of five or six mice in each group. The BrdU feeding experiment has been performed twice independently, once with LF bone marrow chimeric mice. (C) Immunohistochemical localization of autoreactive TG2-specific B cells in Tgm2+/+ LF bone marrow chimeric mice. Left: Spleen sections were stained for anti-14E06 (green) to identify anti-TG2 Ig KI B cells, IgM (blue) to identify the B cell zone, and CD169/SIGLEC (magenta) to identify marginal zone macrophages. Right: Spleen sections were stained for CD3 (magenta) to identify the T cell zone in combination with anti-14E06 (green) and B220 (blue). Representative images of two mice (top and bottom panels) out of five mice analyzed are shown. Scale bars, 100 µm. Top panels: 10× objective; bottom panels: 20× objective. (D) Splenic B cells were isolated from Tgm2−/− (blue), Tgm2+/+ HF (red), and Tgm2+/+ LF (green) chimeric mice, loaded with the intracellular calcium indicator Indo-1 and stained with FITC-conjugated anti-CD45.1. Baseline Ca2+ levels were acquired for 30 s before the cells were stimulated with 2 µg/ml TG2. CD45.1+ cells (WT, dotted lines) and CD45.1-negative cells (comprising mostly 14E06 KI B cells, continuous lines) were gated out and plotted for Ca2+ flux analysis. One representative plot out of three independent measures is shown.
BCR occupancy of 14E06 TG2-specific B cells
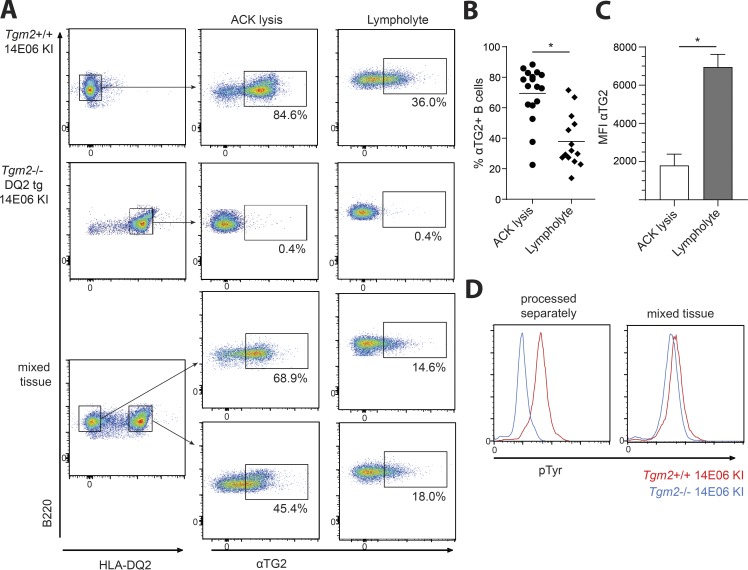
Increase in basal intracellular Ca2+ and phosphoprotein levels have been observed in models of B cell anergy (Benschop et al., 2001; Healy et al., 1997), and these findings were interpreted to reflect BCR occupancy by antigen resulting in continuous BCR signaling (Gauld et al., 2005). To assess anti-TG2 BCR occupancy in our model, we used B cells from Ig KI mice stained with a mouse TG2-reactive, non-epitope 1–specific (i.e., non-14E06-competing) mAb (679–14-D04; Iversen et al., 2013) to detect BCR-bound TG2. First, we analyzed BCR-bound TG2 on splenic TG2-specific B cells after the lysis of erythrocytes. We found that the majority of freshly isolated peripheral TG2-specific B cells in TG2-expressing 14E06 KI mice had bound TG2, whereas no BCR-bound TG2 could be detected in either WT (data not shown) or Ig KI mice on a Tgm2−/− background (Fig. 5 A). As TG2 is abundantly present intracellularly in many different cell types, including erythrocytes, we wanted to control for the possible release of TG2 during erythrocyte lysis and cell death related to tissue disruption during preparation of single cell suspensions. When erythrocytes and dead cells were removed using Lympholyte M, we detected a much lower degree of receptor occupancy (Fig. 5, A–C). To determine whether this receptor occupancy came from in vivo encountered TG2 or from release of the TG2-antigen during in vitro cell processing, we made use of Tgm2−/− 14E06 KI mice that were crossed to HLA-DQ2.5 transgenic mice (du Pre et al., 2011) and mixed the tissue with that of Tgm2+/+ 14E06 KI mice for joint tissue processing. Using surface HLA-DQ2.5 expression to discriminate between cells of different origin, we found that also splenic B cells originating from Tgm2−/− 14E06 KI mice had BCR-bound TG2 (Fig. 5 A). The same was true when lymph nodes were processed together (data not shown). Thus, TG2 thus appears to be easily released during tissue disruption and cell processing, which hinders efforts to determine whether the BCR is engaged by antigen in vivo.
Figure 5.
Receptor occupancy of 14E06 anti-TG2 B cells. (A–C) Splenocytes from Tgm2+/+ 14E06 KI and HLA-DQ2.5 Tgm2−/− 14E06 KI mice were prepared and stained for anti-B220, anti-HLA-DQ2.5, and anti-TG2 (14D04). Red blood cells were removed either by ACK lysis or using Lympholyte media. (A) Representative plots gated on B220+ splenocytes showing the percentage of B cells with surface-bound TG2. (B) Pooled data of at least 14 Tgm2+/+ 14E06 KI mice per group showing the percentage of B cells with surface-bound TG2 after red blood cell lysis (ACK lysis) or removal of red blood cells using Lympholyte media. *, P < 0.0001, as determined by Student’s t test. (C) Analysis of mean fluorescent intensity (MFI) of surface anti-TG2 staining on splenic B220+ B cells isolated from Tgm2+/+ 14E06 KI mice. Paired analysis showing representative data from n = 3 mice in one experiment. Data show mean ± SD. *, P < 0.01, as determined by Student’s t test. (D) Flow cytometric detection of phosphorylated proteins in immediately fixed splenocytes isolated from Tgm2+/+ 14E06 KI (red) and HLA-DQ2.5 Tgm2−/− 14E06 KI mice (blue) that were processed separately or together. Data are representative of two (D) or three (A and C) independent experiments.
Tyrosine phosphorylation is rapidly induced upon BCR ligation. We therefore next analyzed tyrosine phosphorylation in 14E06 KI B cells among Tgm2+/+, Tgm2−/−, or mixed splenocytes that were immediately fixed, followed by flow cytometric analysis. When processed separately, anti-TG2 B cells from Tgm2+/+ 14E06 KI mice demonstrated high basal tyrosine phosphorylation that was not observed in B cells from Tgm2−/− 14E06 KI mice (Fig. 5 D). However, when the tissue was mixed and processed together, high tyrosine phosphorylation was also observed in TG2-specific B cells from Tgm2−/− 14E06 KI mice (Fig. 5 D), indicating that ex vivo exposure to antigen rather than in vivo BCR signaling accounts for the increase in phosphoproteins. We conclude from these studies that there is minimal BCR occupancy of 14E06 KI B cells in vivo in Tgm2+/+ mice.
Transcriptome analysis of autoreactive 14E06 KI B cells
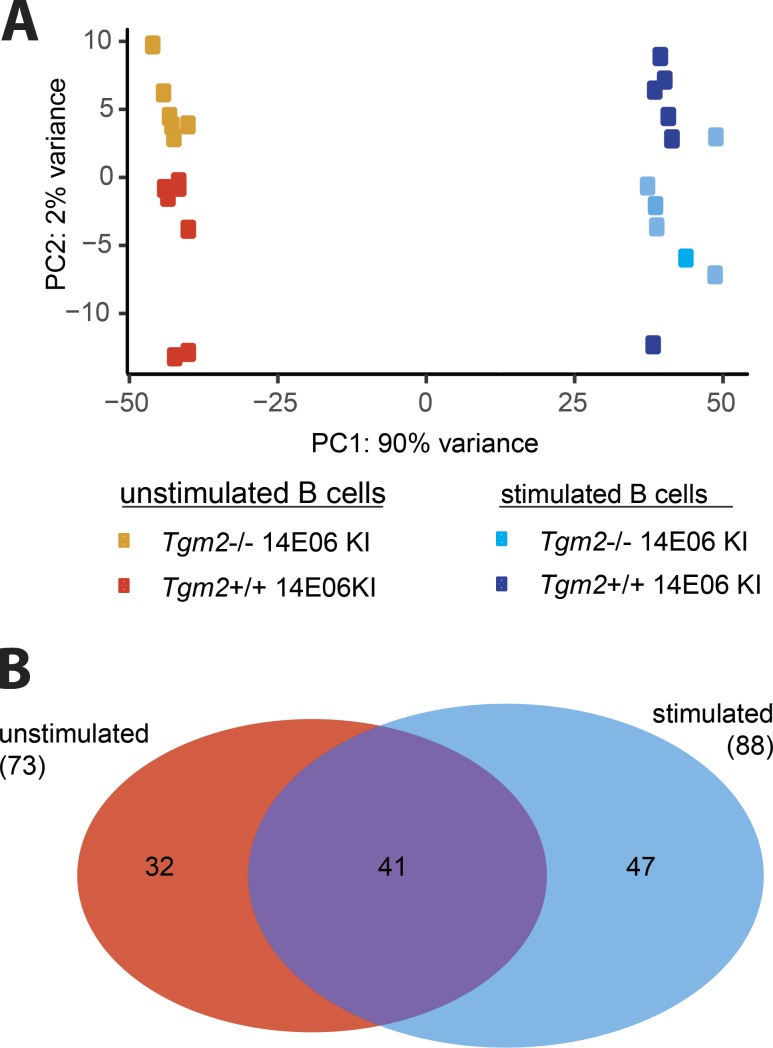
To further characterize autoreactive TG2-specific B cells, we compared their transcriptome to that of their naive counterparts under resting conditions and upon BCR stimulation with TG2. We isolated total resting B cells from spleens of Tgm2-deficient or Tgm2-expressing 14E06 KI mice by negative selection using anti-CD43 magnetic beads, resulting in a B cell purity of >95%. The analysis was done on all splenic B cells, the majority of which were TG2-specific (85.6 ± 7.4%). Of note, based on the findings reported above, this procedure would likely expose B cells to TG2 antigen briefly during the ex vivo cell processing. Principal component analysis showed distinct clustering of unstimulated vs. antigen-stimulated samples, and notably the unstimulated B cells from Tgm2+/+ vs. Tgm2−/− mice clustered closely together, as did the stimulated cells (Fig. 6 A). A total number of 120 genes were differentially expressed between samples from Tgm2−/− and Tgm2+/+ 14E06 KI mice (false discovery rate <0.05 and fold-change >0.5 in either direction; Fig. 6 B and Tables S2 and S3). These few genes pale in comparison to the 4,895 genes (out of a total of 8,305 genes) differentially expressed between stimulated vs. unstimulated B cells in Tgm2−/− mice. In unstimulated B cells, 73 genes were differentially expressed in Tgm2+/+ vs. Tgm2−/− mice. Of these 73 genes, 41 were also differentially expressed between stimulated B cells of Tgm2+/+ vs. Tgm2−/− mice; hence, these might include genes that have altered transcription as a direct result of Tgm2 deficiency (Fig. 6 B). Only 32 genes had uniquely altered expression among unstimulated B cells in Tgm2+/+ vs. Tgm2−/− mice. Early growth response 1 (Egr1) was on the top of the list of these 32 genes. Egr1 up-regulation in B cells is indicative of BCR ligation with antigen (McMahon and Monroe, 1996), and Egr1 is one of few genes identified as up-regulated in anergic hen egg lysozyme (HEL)–specific B cells compared with antigen-naive HEL-specific B cells, albeit we did not observe differential expression of other genes (Nab2, Nrgn) seen in the HEL model of anergy (Glynne et al., 2000; Merrell et al., 2006). While this is noteworthy, we cannot exclude the possibility that the signals of Egr1 and the other 31 genes with differential expression are the results of the short artifactual exposure to autoantigen during cell preparation in vitro. Antibody staining and flow cytometry did not show differences in expression of EGR1 protein (data not shown). Overall, the transcriptome analysis of autoreactive 14E06 KI B cells matured in the presence or absence of TG2 suggests minimal impact of this autoantigen.
Figure 6.
Transcriptome analysis of 14E06 KI B cells. (A and B) B cells were isolated from spleens of Tgm2+/+ and Tgm2−/− 14E06 KI mice for RNA-seq profiling before and after stimulation with 2 µg/ml TG2 for 120 min (n = 6 mice per group in a single experiment). (A) Principal component analysis (PCA). (B) Venn diagram showing differentially expressed genes between unstimulated and stimulated B cells for Tgm2+/+ vs. Tgm2−/− 14E06 KI mice.
Anti-TG2 14E06 KI B cells respond to T cell help
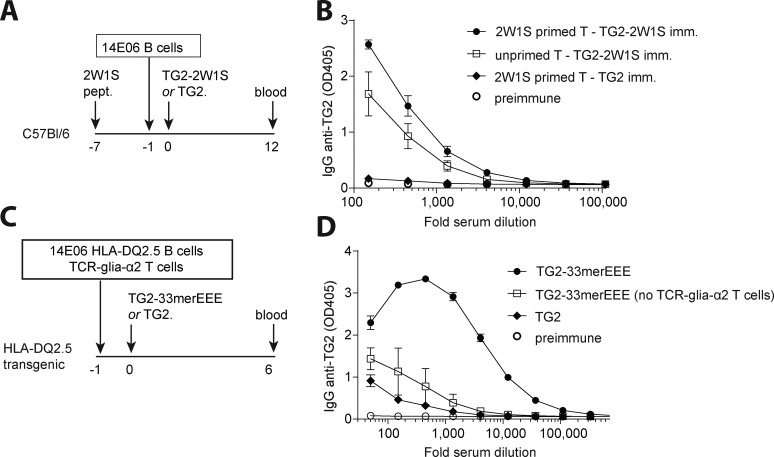
Having established that TG2-reactive B cells readily mature with no induction of tolerance, we next examined whether these B cells can isotype-switch and produce antibodies in vivo. First, we addressed whether autoreactive TG2-specific B cells would respond to endogenous T cell help. We produced in Escherichia coli a recombinant fusion protein consisting of TG2 and the 2W1S peptide (TG2-2W1S), which is highly immunogenic in C57Bl/6 mice as it stimulates alloreactive CD4+ T cells (Moon et al., 2007). We adoptively transferred autoreactive TG2-specific B cells to WT C57Bl/6 recipients that were unprimed or previously primed with the 2W1S peptide, and that were subsequently immunized with TG2 or TG2-2W1S. While mice immunized with TG2 did not develop an IgG anti-TG2 antibody response, immunization with TG2-2W1S elicited activation of TG2-specific B cells and the production of class-switched anti-TG2 IgG antibodies (Fig. 7, A and B). The anti-TG2 IgG titer was higher in mice that were primed with 2W1S peptide compared with mice that were not previously primed (Fig. 7, A and B).
Figure 7.
Self-reactive TG2-specific B cells respond to T cell help. (A) Schematic representation of the 2W1S experiment. WT C57Bl/6 mice were immunized i.p. with 50 µg 2W1S peptide (pept.) in CFA 7 d before adoptive transfer of TG2-specific B cells from Tgm2+/+ 14E06 KI mice. The next day, mice were immunized i.p. with 100 µg TG2-2W1S fusion protein or TG2 alone in IFA. (B) Serum IgG anti-TG2 antibodies were measured at day 12 after TG2/TG2-2W1S administration. Data are mean ± SEM (n = 3/group) and representative of two independent experiments. Imm., immunized. (C) Schematic representation of the TG2-gluten experiment. TG2-specific B cells obtained from Tgm2+/+ 14E06 KI HLA-DQ2.5 mice and gluten-specific T cells obtained from TCR-glia-α2 HLA-DQ2.5 mice were adoptively transferred to HLA-DQ2.5 recipient mice. The next day, mice were immunized with 100 µg TG2 fused to a deamidated gluten T cell epitope (TG2-33merEEE) or with TG2 alone in PBS. (D) Serum IgG anti-TG2 antibodies were measured at day 6 after TG2/TG2-33merEEE administration. Data are mean ± SEM (n = 3/group) and representative of at least four independent experiments.
For celiac disease, a model was outlined where TG2-specific B cells with involvement of hapten-carrier like TG2-gluten complexes can receive help from gluten-specific T cells (Mäki, 1992; Sollid et al., 1997). To address whether 14E06 KI B cells could cooperate with gluten-specific T cells in vivo, we generated a gluten-specific TCR transgenic mouse strain that recognizes the DQ2.5-glia-α2 epitope and introduced human HLA-DQ2.5 into the mice by breeding to comply with MHC restriction. Proper TCR expression (Fig. S3 A) and proliferative response to cognate gluten peptide antigen were confirmed in these TCR transgenic mice (Fig. S3 B). We next isolated naive CD4+ T cells from HLA-DQ2.5 × TCR-glia-α2 double transgenic mice and naive B cells from HLA-DQ2.5 transgenic 14E06 KI mice and adoptively transferred the T and B cells into nonirradiated HLA-DQ2.5 transgenic recipient mice. The recipient mice were administered a recombinant fusion protein of TG2 with the deamidated immunodominant gluten 33mer peptide without adjuvant on the day after the adoptive cell transfer. Importantly, the anti-TG2 antibodies were formed with a clear effect of gluten-specific T cells (Fig. 7, C and D). Using a similar fusion protein produced in insect cells, the antibody production after adoptive cell transfers was weak with a very modest effect of gluten-specific T cells (Fig. S3 C). This might possibly indicate that endotoxin contamination in the E. coli–produced protein induced the expression of costimulatory molecules on TG2-specific B cells, making them better equipped to interact with naive gluten-specific T cells. Future studies will need to address this issue. Taken together, these experiments reveal that 14E06 KI B cells can get activated when provided with T cell help.
Discussion
Exploring an Ig KI mouse model system, we report that there is absence of B cell tolerance for TG2 autoreactive B cells, and that such autoreactive B cells can produce autoantibodies when provided with T cell help. The observations support a model of celiac disease autoimmunity where gluten-specific T cells by involvement of TG2–gluten complexes provide help to TG2-specific B cells. This finding of B cell clonal ignorance to TG2 antigen is surprising as transgenic B cell tolerance models of other extracellular matrix antigens have generally reported B cell deletion, κ L chain editing, or anergy (Foster et al., 2006). TG2 has been described as being expressed intracellularly and extracellularly as part of the extracellular matrix of most organs (Korponay-Szabó et al., 2004; Lorand and Graham, 2003). This notion may have to be revisited. Our findings reveal that functionally normal anti-TG2 B cells readily develop, thereby poising for an establishment of anti-TG2 antibody formation.
The observed B cell ignorance could reflect that the avidity of antigen to the BCR is too low to induce tolerance, as cells with self-reactive BCR with high affinity are more likely to be tolerized compared with cells with lower antigen affinity (Huang et al., 2006; Packard et al., 2016; Taylor et al., 2012; Wang and Shlomchik, 1997). The patient-derived, prototypic mAb 14E06 used for generation of the anti-TG2 Ig KI mice has a near-germline configuration reflecting minimal affinity maturation. Notwithstanding, 14E06 is a high-affinity antibody with similar affinity as post–germinal center BCRs observed in the anti-HEL transgenic model (Goodnow et al., 1988). Thus, lack of tolerance in our model likely cannot be ascribed to low BCR affinity, but rather points to limited antigen availability.
BCR occupancy is an important parameter for B cell tolerance induction. We therefore aimed to determine the degree of in vivo BCR occupancy in our model. This turned out not to be straightforward, as we from a series of experiments arrived at the conclusion that intracellular TG2 is released during cell isolation, from splenic tissue in particular, as well as by hemolysis during cell preparation. Thus, BCR occupancy of freshly isolated anti-TG2 B cells can easily occur. Although we are not able to say whether autoreactive TG2-specific B cells were occupied by antigen in vivo or not, our findings suggest a low degree of receptor occupancy, which is consistent with the observed lack of anergy.
Our current understanding on the biology of TG2 is incomplete. The enzyme is ubiquitously expressed in many organs (Lorand and Graham, 2003), including the thymus (Szondy et al., 1997). The transamidating/deamidating activity of TG2 is Ca2+-dependent, and hence TG2 is likely to serve an extracellular function. Staining for TG2 in frozen tissue sections gives a characteristic distribution pattern that reflects the binding of TG2 to fibronectin (Upchurch et al., 1987) and an as yet unidentified extracellular matrix component, TG2 interaction partner (Stamnaes et al., 2016). The findings of the current study call into question the asserted abundance of extracellular TG2 under normal physiological conditions, particularly in the bone marrow. Importantly, the abundant extracellular distribution of TG2 observed in frozen sections may upon tissue sectioning result from release of intracellular TG2 binding to adjacent extracellular matrix components, and as such represent an in vitro artifact (Stamnaes et al., 2016).
Tolerance induction in the T cell compartment is known to be much more efficient than tolerance induction in the B cell compartment (Adelstein et al., 1991; Chiller et al., 1971), and it is thus widely assumed that a lack of cognate T cell help prevents B cell autoimmunity. In line with this notion, there is no substantial evidence for autoreactive TG2-specific T cell responses (Jabri and Sollid, 2017) resonating with the abundant thymic expression of TG2. However, although unlikely a target for T cell autoimmunity, TG2 does efficiently target gluten peptides and generate covalent TG2–gluten complexes in vitro (Fleckenstein et al., 2004). These TG2–gluten complexes can conceivably act as hapten carrier–like complexes to allow TG2-specific B cells to receive help from gluten-reactive T cells (Sollid et al., 1997). This model neatly explains the clinical observations that TG2 autoantibodies are only seen in subjects who consume gluten (Sulkanen et al., 1998) and who express HLA-DQ2/DQ8 (Björck et al., 2010). Using a TG2–gluten fusion protein, we demonstrate that gluten-specific T cells can indeed provide help to ignorant TG2-specific B cells in vivo. Whether and under what conditions TG2–gluten complexes are formed in situ remain to be resolved.
Lack of B cell tolerance and presence of ignorant B cells that will become producers of autoantibodies once T cell help is provided are mechanisms of autoimmunity not only applying to celiac disease but also relevant to other autoimmune diseases whose autoantigens are shielded from B cells or present at very low concentrations. Our findings resemble the observations obtained for the nuclear protein La, where no evidence for deletion or anergy for high-affinity transgenic La-specific B cells was obtained (Aplin et al., 2003). In this model, ignorant La-specific B cells were stimulated to produce autoantibodies when cognate T cell help was provided (Keech et al., 2001).
The conceptual model of generating autoantibodies by means of coupling a B cell autoantigen with foreign T cell antigens is not unique to celiac disease. It has been shown that B cells producing anti-nuclear antibodies can receive help from T cells recognizing epitopes of a viral antigen that complexes with the self-nuclear antigen (Dong et al., 1994), and moreover, that BCR recognizing a self-membrane protein can capture this from membranes of virus-infected cells and simultaneously cocapture viral proteins that become recognized by virus-specific CD4+ T cells (Sanderson et al., 2017). Fundamentally, the conceptual model is reliant on weak or absent B cell tolerance, which we indeed demonstrate for TG2. Thus, in essence the results presented here support the concept where anti-TG2 autoimmunity develops when nontolerized TG2-specific B cells receive help from foreign antigen-specific T cells.
Materials and methods
Generation of 14E06 VH VL KI and glia-α2 TCR transgenic mice
The 14E06 antibody genes were originally amplified from a single TG2-reactive plasma cell from a celiac lesion (Di Niro et al., 2012). 14E06 Ig H and Ig L KI mice were prepared by Ozgene following an established protocol (Taki et al., 1993). To make the Ig H chain targeting construct, the B1-8 promotor with leader sequence (kindly provided by K. Rajewsky, Max Delbrück Center for Molecular Medicine, Berlin, Germany) and rearranged 14E06 VDJH were cloned downstream of a flippase recognition target (FRT)–flanked phosphoglycerate kinase I neomycin resistance cassette (Fig. 1 A). For the Ig L chain targeting construct, the rearranged 14E06 VJκ was used with the promotor sequence of IgKV3-12 (kindly provided by B. Bogen, University of Oslo, Oslo, Norway). The targeted constructs were linearized and introduced into C57BL/6 embryonic stem (ES) cells by electroporation. Targeted ES cells were screened by Southern blotting, and ES cell clones with correct integration were injected separately into blastocysts and then transferred into foster mothers. Chimeric offspring with germline transmission were crossed with flippase (FLP) recombinase transgenic mice to delete the selection gene.
For the generation of the TCR-glia-α2 transgenic mice, cDNA sequences representing V(D)J regions of rearranged TCRα and TCRβ chains of the T cell clone 364.1.0.14 (Qiao et al., 2011), specific for the DQ2.5-glia-α2 epitope, were synthezied (Genscript) and cloned into a pair of cassette expression vectors containing the murine TCR constant sequences as well as the natural mouse TCR promoter/enhancer elements as described (kindly provided by D. Mathis, Harvard Medical School, Boston, MA; Kouskoff et al., 1995). Transgenic mice were generated at the Norwegian Transgenic Center by microinjecting the constructs into one of the pronuclei of one-cell-stage embryos of C57Bl/6 mice.
Mice
Hemizygous 14E06 Ig KI mice on a C57Bl/6 background were generated by crossing VDJH14E06 and VJκ14E06 single KI mice. VDJH14E06 and VJκ14E06 single KI mice were crossed to Tgm2−/− mice (De Laurenzi and Melino, 2001) and crossed to generate hemizygous Tgm2−/− 14E06 Ig KI mice. HLA-DQ2.5 transgenic mice (du Pre et al., 2011) were backcrossed into C57Bl/6 for at least 10 generations and thereafter crossed to both Tgm2+/+ and Tgm2−/− 14E06 Ig KI mice. TCR-glia-γ1 transgenic mice (du Pre et al., 2011) specific for the DQ2.5-glia-γ1 epitope were backcrossed into C57Bl/6 for at least 10 generations. Mice were bred at the Department of Comparative Medicine, Oslo University Hospital, Rikshospitalet (Oslo, Norway) under specific pathogen–free conditions. CD45.1 mice (B6.SJL-Ptprca Pepcb/BoyCrl) were purchased from Charles River Laboratories, crossed to Tgm2−/− mice, and bred in house. C57Bl/6 mice were purchased from Janvier Laboratoires. All experiments were approved by the Norwegian Food Safety Authority.
Antigen production
Recombinant TG2 protein was expressed in E. coli or in Sf+ express insect cells. For insect cell expression, mouse Tgm2 or human Tgm2 was cloned into the pACAB3 vector, and protein was expressed for 3 d followed by cell lysis and purification by nickel-nitrilotriacetic acid affinity chromatography as previously described (Stamnaes et al., 2016). For E. coli expression, mouse Tgm2 was cloned into the pET28a(+) vector (Novagen) and expressed as previously described (Iversen et al., 2013). Protein was purified by nickel-nitrilotriacetic acid affinity chromatography and if required also by gel filtration chromatography using a Superdex 200 10/300 column (GE Healthcare). For TG2 T cell epitope fusion protein production in E. coli, the 2W1S peptide (EAWGALANWAVDSA) or the deamidated 33mer (LQLQPFPQPELPYPQPELPYPQPELPYPQPQPF) was cloned into the pET28a vector to the C-terminal end of the mouse Tgm2 gene spaced by a three glycine residue linker. For insect cell production, deamidated 33mer was cloned to the C-terminal end of the mouse Tgm2 gene in pACAB3. Protein purity was assessed by SDS-PAGE and Coomassie Blue staining. Protein antigen used for in vivo experiments was buffer exchanged by spin filter centrifugation to sterile PBS before use. In the 2W1S experiment, E. coli produced TG2 and TG2-2W1S were further purified by size exclusion chromatography on a Superdex 200 10/300 GL column (GE Healthcare) using PBS as running buffer. To generate biotinylated antigen, TG2 was biotinylated using BirA ligase (Avidity) according to the manufacturer’s instructions. For surface plasmon resonance analysis, insect cell–produced mouse and human TG2 with a C-terminal biotin-BirA site were used.
Surface plasmon resonance analysis
Fab fragment production of 14E06 in HEK293 cells has been described elsewhere (Hnida et al., 2016). Binding kinetics of Fab 679–14-E06 for human and mouse TG2 was assessed by surface plasmon resonance analysis (BIAcore T200 instrument; GE Healthcare) using antigen with a controlled open or closed conformation. To immobilize TG2 in an open conformation, biotinylated, insect cell–produced TG2 was incubated with the irreversible active site inhibitor DP3-3 (Ac-P(DON)LPF-NH2; Zedira) at room temperature for 15 min before addition of 5 mM CaCl2 followed by incubation on ice (TG2-DP3-3). To generate TG2 in a closed conformation (TG2-GDP), biotinylated, insect cell–produced TG2 was preincubated with GDP to a final concentration of 1 mM. NeutrAvidin-coated sensor chips were generated by coupling 10 µg/ml NeutrAvidin (Thermo Fisher Scientific) in 10 mM sodium acetate, pH 4.5, to CM5 sensor chips to reach 500 response units. Unreactive moieties were blocked with 1 M ethanolamine. Biotinylated TG2-GDP or TG2-DP3-3 was injected at 16°C at a flow rate of 10 µl/min to reach ∼200–800 resonance units. Kinetic measurements were done by injection of serial dilutions of 14E06 Fab-fragment at a flow rate of 30 µl/min for 60 s at 16°C. Binding data were automatically zero-adjusted and reference cell binding subtracted by the BIAcores T200 software. Kinetic rate constants were estimated using a Langmuir 1:1 binding model provided by the software.
Flow cytometry
Single-cell suspensions from spleens and lymph nodes were prepared by carefully grinding and filtering tissues through a 70 µm diameter nylon mesh (BD Biosciences). Bone marrow was flushed from the femur/tibia. Blood was collected using heparin. For removal of erythrocytes, cell suspensions were treated with ammonium-chloride-potassium (ACK) lysis buffer or subjected to Lympholyte M gradient (Cedarlane). Peritoneal cells were harvested by lavage with 5 ml PBS. Cells were resuspended in PBS containing 2% FCS (vol/vol) and incubated with optimal amounts of antibodies against the following molecules: B220 (RA3-6B2; BioLegend), CD19 (6D5; BioLegend), CD21/CD35 (7G6; BD), CD23 (B3B4; eBioscience), CD3 (145-2C11; BioLegend), CD45.1 (A20; BioLegend), CD45.2 (104; BioLegend), CD5 (53–7.3; BioLegend), CD69 (H1.2F3; BioLegend), CD80 (16-10A1; BioLegend), CD86 (B7-2; eBioscience), Ig λ1/λ2/λ3 (R26-46; BD), IgD (11-26c.2a; BD), IgM (II/41; eBioscience), and Vβ6.7 (OT145; Endogen). Unconjugated hIgG1 anti-14E06 and anti-TG2 (679–14-D04) were produced in our own laboratory as previously described (Di Niro et al., 2012; Høydahl et al., 2016) and detected with phycoerythrin-conjugated anti-hIgG1 (4E3; Acris Antibodies). TG2 tetramers were generated by incubating 0.4 µM of biotinylated mTG2 with 0.1 µM of phycoerythrin- or allophycocyanin-conjugated streptavidin (Thermo Fisher Scientific). Detection of BrdU-labeled cells was done using the BrdU Flow Kit (BD Biosciences). EGR1 staining was performed on fixed and permabilized cells using the Fixation/Permeabilization kit (BD Biosciences), using rabbit monoclonal antibody T.126.1 (Thermo Fisher Scientific) followed by goat-anti-rabbit IgG-alexa647 (Invitrogen). For detection of phosphotyrosine, total splenocytes were immediately fixed by incubating in RPMI containing 2% paraformaldehyde for 15 min at room temperature. Cells were briefly incubated with H2O and then permeabilized with ice-cold methanol and incubated for 30 min on ice. Cells were washed twice with PBS and incubated with anti-phosphotyrosine (PY20; BioLegend) for 30 min, combined with surface antibodies against B220 and HLA-DQ2.5 (2.12.E11; Viken et al., 1995). Cells were analyzed on a FACSCalibur, Fortessa (BD Biosciences), or Attune NxT Flow Cytometer (Thermo Fisher Scientific), and data analysis was done using FlowJo software (Tree Star).
Isolation of T and B cells, and cell culture
Splenic B cells were purified using the Dynabeads Mouse CD43 kit (Invitrogen). The purity of B cells, measured by the percentage of B220+ cells, was typically >96%. For measurement of activation markers, cells were cultured overnight with LPS (from E. coli O111:B4; Sigma-Aldrich), mTG2, or F(ab′)2 goat-anti-mouse IgM (Jackson ImmunoResearch) at a density of 1.25 × 106 cells/ml in RPMI 1640 supplemented with 10% (vol/vol) FCS, penicillin, streptomycin, and 10 µM β-mercaptoethanol at 37°C. For B cell proliferation assays, cells were labeled with CFSE (Molecular Probes) and cultured in RPMI as above with additional supplements 1 mM sodium pyruvate, 0.1 mM nonessential amino acids, and 10 mM Hepes (all from Sigma-Aldrich) for 72 h with the stimuli indicated above as well as anti-CD40 (5 µg/ml; BD) and rmIL-4 (50 ng/ml; R&D Systems). For 3H-thymidine incorporation assay, 5 × 105 splenocytes from HLA-DQ2.5 × TCR-glia-γ1 transgenic mice and HLA-DQ2.5 × TCR-glia-α2 transgenic mice were cultured in the presence of 5 µg/ml plate-bound anti-CD3 and 2 µg/ml anti-CD28 (both eBioscience) or with 10 µM synthetic gluten peptides (γ-gliadin, YQQLPQPEQPQQSFPEQERPF; α-gliadin, LQLQPFPQPELPYPQPELPYPQPELPYPQPQPF; Genscript) for 72 h. Proliferation was assessed by incorporation of 3H-thymidine. CD4+ T cells were isolated from pooled lymph nodes and spleens from gliadin TCR transgenic mice by negative selection using the EasySep Mouse CD4+ T cell isolation kit (StemCell Technologies). The purity of CD4+ T cells was typically >85%.
Intracellular Ca2+ mobilization
B cells were loaded for 60 min at 37°C with Indo-1 acetoxymethyl (final concentration, 1 µM; Molecular Probes) in RPMI containing 10% FCS. Baseline measurements were acquired for 30 s, and then cells were stimulated with 10 µg/ml F(ab′)2 goat-anti-mouse IgM (Jackson ImmunoResearch) or 2 µg/ml TG2 for 2.5 min. Mean relative [Ca2+]i was monitored over time using an LSR II flow cytometer (BD) with analysis using FlowJo software (Tree Star).
Adoptive transfer and immunization
For the 2W1S experiment, C57Bl/6 recipients were immunized i.p. with 50 µg 2W1S peptide (EAWGALANWAVDSA) in CFA (Sigma-Aldrich) 6 d before adoptive transfer of 106 TG2-binding B cells from Tgm2+/+ 14E06 KI mice in 200 µl PBS by intravenous injection. 24 h after transfer, recipient mice were i.p. injected with 100 µg mTG2 or 100 µg mTG2-2W1S in IFA (Sigma-Aldrich). For the gluten-specific T cell experiment, 106 TG2-binding B cells from Tgm2+/+HLA-DQ2 transgenic 14E06 KI mice and 106 CD4+ T cells from HLA-DQ2 × gliadin-α2 TCR transgenic mice were adoptively transferred to HLA-DQ2 tg recipient mice. The next day, recipient mice received 100 µg mTG2 or 100 µg mTG2-33merEEE in PBS by i.p. injection.
Mixed bone marrow chimera
C57Bl/6 and Tgm2−/− recipient mice were lethally irradiated (1,000 Rad) and reconstituted by i.v. injection with equal mixtures of 5 × 106 WT CD45.1 bone marrow cells and 5 × 106 Tgm2+/+ 14E06 Ig KI bone marrow cells, or 5 × 106 CD45.1 Tgm2−/− and 5 × 106 Tgm2−/− 14E06 Ig KI bone marrow cells. Bone marrow chimeric mice with low frequencies of autoreactive B cells were generated by mixing 0.5 × 106 Tgm2+/+ 14E06 Ig KI bone marrow cells with 9.5 × 106 WT CD45.1 bone marrow cells. Mice received 2 g/liter neomycin sulfate (Thermo Fisher Scientific) and 1 g/liter glucose in drinking water until day 14 after irradiation. 6 wk after bone marrow transfer, mice were i.p. injected with 1 mg BrdU (BD PharMingen) and given 1 mg/ml BrdU (Sigma-Aldrich) in their drinking water. 1 g/liter glucose was added to overcome taste aversion.
Immunohistochemistry
Spleens from bone marrow chimera mice were embedded in optimal cutting temperature compound and snap frozen in liquid nitrogen. 4-μm cryosections were briefly fixed in 4% neutral buffered formalin and stained with anti-14E06, anti-CD169/SIGLEC (clone 3D6.112; BioRad), and anti-IgM (donkey-anti-mouse-IgM-A488; Jackson ImmunoResearch) followed by donkey-anti-rat-IgG-Cy3,donkey-anti-human-IgG-biotin(both Jackson ImmunoResearch), and streptavidin-APC (ProZyme), or anti-14E06, anti-B220-biotin (clone RA3-6B2; BioLegend), and anti-CD3-PE (clone 145-2C11; BioLegend) followed by donkey-anti-human-IgG-A488 and streptavidin–aminomethylcoumarin acetate (both Jackson ImmunoResearch). Slides were mounted with ProLong Diamond Antifade (Thermo Fisher Scientific). Images were acquired on an inverted fluorescence microscope (Nikon Eclipse Ti-S; Nikon) with a Nikon 9 10/0.3 or 9 20/0.45 Plan Fluor lens (Nikon) and NIS ELEMENTS BR3.2 software (Nikon). Single channel images were merged in FIJI/ImageJ (Schindelin et al., 2012; Schneider et al., 2012). Linear adjustment of brightness and contrast was done equally across images. Anti-14E06 staining acquired in the blue channel was pseudocolored as green and anti-IgM staining acquired in the green channel was pseudocolored as blue for visualization purposes.
RNA sequencing (RNA-seq) analysis
Total RNA was isolated from B cells with an RNeasy minikit (Qiagen). RNA integrity was analyzed on an Agilent Bioanalyzer using an Agilent RNA 6000 Pico kit. RNA integrity was 9.1 ± 0.7. RNA-seq libraries were generated from purified ribosomal RNA–depleted RNA samples using the strand-specific TruSeq stranded mRNA library prep kit (Illumina), and sequenced at the Norwegian Sequencing Centre using the Illumina HiSeq 3/4000 with a read length of 150 bp and paired end reads. Salmon version 0.7.2 (Patro et al., 2017) was used for the quantification of transcript counts. In quasi-mapping mode, a variational Bayesian expectation-maximization algorithm was used for optimizing abundance estimates. With the default k-mer length of 31, the quasi-mapping index was built on the transcriptome of Ensembl genome build GRCm38 patch release 5 that includes alternative loci. Transcript-level counts were aggregated to gene level (including those on alternative loci). Models that correct sequence-specific and fragment-level biases were applied while quantifying the transcript counts.
To identify and retain the absolutely expressed genes for downstream analyses, finite normal mixture models implemented in the Mclust package (Scrucca et al., 2016) were used as suggested (Hebenstreit et al., 2011). Briefly, a model-based clustering of the log-transformed transcripts per million values of all protein-coding genes in each sample was performed to categorize each gene as either “expressed” or “not expressed.” This exercise has been performed independently for each group of mouse samples. Further, to be categorized as “expressed” in a sub-group, we imposed another criterion that the gene has to be detected in all the samples within that group. DESeq2 version 1.20 (Love et al., 2014) was used for differential expression analysis. All the samples were processed in a single batch, ruling out any batch-induced variation. Genes were categorized as differentially expressed at a false discovery rate of 10%.
ELISA
Serum was collected from recipient mice serially bled through the saphenous vein by incubating blood for 20 min at room temperature followed by centrifugation for 15 min at 2,000 g, 4°C. Supernatant was collected and stored at −20°C until assayed. Total IgM, IgG, and IgA serum antibodies were measured using the ELISA Ready-SET-Go! kits (eBioscience) according to the manufacturer’s instructions. Sera were diluted 1:10,000. For detection of TG2-specific antibodies, wells were coated with 5 µg/ml mTG2 in PBS overnight at 4°C. After washing and blocking, serial dilutions of serum were incubated for 2.5 h, washed, and detected with alkaline-phosphatase-conjugated goat anti-mouse IgG (Sigma-Aldrich), or biotinylated mouse anti-mouse IgM (BD) or goat anti-mouse IgA (Southern Biotech) followed by alkaline-phosphatase-conjugated streptavidin (Southern Biotech). OD was determined at 405 nm. Monoclonal mouse IgM, IgG2c, and IgA 14E06 antibodies were purified from 14E06 B cell hybridomas (see below) using HiTrap protein L columns (BD) and used as standard curves.
Generation of B cell hybridomas
Single-cell suspensions were prepared from spleens, lymph nodes, Peyer’s patches, and bone marrow of Tgm2+/+ 14E06 KI mice and fused with murine plasmacytoma cells (OURI cells, an X63-Ag8.653 variant, kindly provided by B. Bogen) using polyethylene glycol 1500 (Roche) according to the manufacturer’s instructions. After fusion, the cells were cultured in RPMI 1640 containing 10% (vol/vol) FCS, penicillin, streptomycin, 10 µM β-mercaptoethanol, 1 mM sodium pyruvate, 0.1 mM nonessential amino acids, and hypoxanthine-aminopterin-thymidine supplement (all Sigma-Aldrich) in 96-well plates. Aminopterin was diluted from the culture after 6 d by switching to hypoxanthine-thymidine supplement (Sigma-Aldrich). After 14 d, culture supernatants were assessed for anti-TG2 reactivity by ELISA, and limiting dilution culture was performed at least twice on positive cultures. The chosen clones were cultured in RPMI-based medium containing 10% Ultralow IgG FBS (Gibco).
Data availability
RNA-seq data have been deposited to the EBI ArrayExpress database (accession no. E-MTAB-7567).
Online supplemental material
Fig. S1 shows that there is no difference in total splenocyte count or in splenic B cell numbers between Tgm2+/+ and Tgm2−/− 14E06 KI mice and that there are no differences in TG2-specific serum IgM, IgG, or IgA levels. It is also shown that anti-TG2 B cells are not diverted to the B-1 compartment. Fig. S2 shows that there is a skewed CD45.1/CD45.2 ratio among B cells in mixed bone marrow chimeric mice. Fig. S2 also shows that there are no differences in the percentages of TG2-specific B cells that belong to the immature, follicular, or marginal zone B cell subsets between Tgm2−/− control chimeras and Tgm2+/+ chimeric mice with a high or low frequency of TG2-specific B cells. Fig. S3 shows the characterization of TCR-glia-α2 transgenic mice as well as the response of 14E06 KI B cells to insect cell–produced TG2-33mer fusion protein. Table S1 shows that the 14E06 antibody has an equal nanomolar affinity to both human and mouse TG2 protein. Tables S2 and S3 list differentially expressed genes related to the RNA-seq experiment in Fig. 6.
Acknowledgments
We are grateful to Bjørg Simonsen, Nicolay Rustad Nilssen, and Liv Kleppa for technical assistance, and the staff of the Department of Comparative Medicine, Oslo University Hospital, for animal husbandry. We thank Johanne Jacobsen and Bjarne Bogen for sharing knowledge and Ig-gene plasmids during the process of generating the Ig KI mice, Diane Mathis for sharing TCR-expression plasmids, Klaus Rajewsky for sharing Ig-gene plasmid, Rasmus Iversen for advice, and Mark Shlomchik for helpful discussions.
This work was supported by grants from the European Commission (project ERC-2010-Ad-268541), the Research Council of Norway (project 179573/V40, Centre of Excellence funding scheme and project 275053), the South-Eastern Norway Regional Health Authority (projects 2010048, 2011050, and 2016113), and Stiftelsen KG Jebsen (project SKGJ-MED-017).
The authors declare no competing financial interests.
Author contributions: M.F. du Pré designed and performed experiments, interpreted data, and wrote the manuscript; J. Blazevski and A.E. Dewan designed and performed experiments and interpreted data; J. Stamnaes designed the recombinant TG2 fusion proteins, performed immunohistochemistry, and edited the manuscript; C. Kanduri and G.K. Sandve analyzed RNA-seq data; M.K. Johannesen prepared critical reagents and expression vectors; C.B. Lindstad made 14E06 B-cell hybridomas; K. Hnida performed surface plasmon resonance analysis; G. Melino provided Tgm2−/− mice; L. Fugger provided HLA-DQ2.5 and TCR-glia-γ1 transgenic mice; S.W. Qiao designed TCR-glia-α2 transgenic mice; L.M. Sollid conceptualized and supervised the study, interpreted data, and wrote the manuscript. All authors read and approved the final version of the manuscript.
References
- Adelstein S., Pritchard-Briscoe H., Anderson T.A., Crosbie J., Gammon G., Loblay R.H., Basten A., and Goodnow C.C.. 1991. Induction of self-tolerance in T cells but not B cells of transgenic mice expressing little self antigen. Science. 251:1223–1225. 10.1126/science.1900950 [DOI] [PubMed] [Google Scholar]
- Akkaraju S., Canaan K., and Goodnow C.C.. 1997. Self-reactive B cells are not eliminated or inactivated by autoantigen expressed on thyroid epithelial cells. J. Exp. Med. 186:2005–2012. 10.1084/jem.186.12.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aplin B.D., Keech C.L., de Kauwe A.L., Gordon T.P., Cavill D., and McCluskey J.. 2003. Tolerance through indifference: autoreactive B cells to the nuclear antigen La show no evidence of tolerance in a transgenic model. J. Immunol. 171:5890–5900. 10.4049/jimmunol.171.11.5890 [DOI] [PubMed] [Google Scholar]
- Benschop R.J., Aviszus K., Zhang X., Manser T., Cambier J.C., and Wysocki L.J.. 2001. Activation and anergy in bone marrow B cells of a novel immunoglobulin transgenic mouse that is both hapten specific and autoreactive. Immunity. 14:33–43. 10.1016/S1074-7613(01)00087-5 [DOI] [PubMed] [Google Scholar]
- Björck S., Brundin C., Lörinc E., Lynch K.F., and Agardh D.. 2010. Screening detects a high proportion of celiac disease in young HLA-genotyped children. J. Pediatr. Gastroenterol. Nutr. 50:49–53. 10.1097/MPG.0b013e3181b477a6 [DOI] [PubMed] [Google Scholar]
- Cambier J.C., Gauld S.B., Merrell K.T., and Vilen B.J.. 2007. B-cell anergy: from transgenic models to naturally occurring anergic B cells? Nat. Rev. Immunol. 7:633–643. 10.1038/nri2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Hnida K., Graewert M.A., Andersen J.T., Iversen R., Tuukkanen A., Svergun D., and Sollid L.M.. 2015. Structural basis for antigen recognition by transglutaminase 2-specific autoantibodies in celiac disease. J. Biol. Chem. 290:21365–21375. 10.1074/jbc.M115.669895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiller J.M., Habicht G.S., and Weigle W.O.. 1971. Kinetic differences in unresponsiveness of thymus and bone marrow cells. Science. 171:813–815. 10.1126/science.171.3973.813 [DOI] [PubMed] [Google Scholar]
- Cook M.C., Basten A., and Fazekas de St Groth B.. 1997. Outer periarteriolar lymphoid sheath arrest and subsequent differentiation of both naive and tolerant immunoglobulin transgenic B cells is determined by B cell receptor occupancy. J. Exp. Med. 186:631–643. 10.1084/jem.186.5.631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyster J.G., Hartley S.B., and Goodnow C.C.. 1994. Competition for follicular niches excludes self-reactive cells from the recirculating B-cell repertoire. Nature. 371:389–395. 10.1038/371389a0 [DOI] [PubMed] [Google Scholar]
- De Laurenzi V., and Melino G.. 2001. Gene disruption of tissue transglutaminase. Mol. Cell. Biol. 21:148–155. 10.1128/MCB.21.1.148-155.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Niro R., Mesin L., Zheng N.Y., Stamnaes J., Morrissey M., Lee J.H., Huang M., Iversen R., du Pré M.F., Qiao S.W., et al. . 2012. High abundance of plasma cells secreting transglutaminase 2-specific IgA autoantibodies with limited somatic hypermutation in celiac disease intestinal lesions. Nat. Med. 18:441–445. 10.1038/nm.2656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterich W., Ehnis T., Bauer M., Donner P., Volta U., Riecken E.O., and Schuppan D.. 1997. Identification of tissue transglutaminase as the autoantigen of celiac disease. Nat. Med. 3:797–801. 10.1038/nm0797-797 [DOI] [PubMed] [Google Scholar]
- Dong X., Hamilton K.J., Satoh M., Wang J., and Reeves W.H.. 1994. Initiation of autoimmunity to the p53 tumor suppressor protein by complexes of p53 and SV40 large T antigen. J. Exp. Med. 179:1243–1252. 10.1084/jem.179.4.1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Pré M.F., Kozijn A.E., van Berkel L.A., ter Borg M.N., Lindenbergh-Kortleve D., Jensen L.T., Kooy-Winkelaar Y., Koning F., Boon L., Nieuwenhuis E.E., et al. . 2011. Tolerance to ingested deamidated gliadin in mice is maintained by splenic, type 1 regulatory T cells. Gastroenterology. 141:610–620: 620.e1–620.e2. 10.1053/j.gastro.2011.04.048 [DOI] [PubMed] [Google Scholar]
- Fleckenstein B., Qiao S.W., Larsen M.R., Jung G., Roepstorff P., and Sollid L.M.. 2004. Molecular characterization of covalent complexes between tissue transglutaminase and gliadin peptides. J. Biol. Chem. 279:17607–17616. 10.1074/jbc.M310198200 [DOI] [PubMed] [Google Scholar]
- Foster M.H., Zhang Y., and Clark A.G.. 2006. Deconstructing B cell tolerance to basement membranes. Arch. Immunol. Ther. Exp. (Warsz.). 54:227–237. 10.1007/s00005-006-0027-x [DOI] [PubMed] [Google Scholar]
- Gauld S.B., Benschop R.J., Merrell K.T., and Cambier J.C.. 2005. Maintenance of B cell anergy requires constant antigen receptor occupancy and signaling. Nat. Immunol. 6:1160–1167. 10.1038/ni1256 [DOI] [PubMed] [Google Scholar]
- Gay D., Saunders T., Camper S., and Weigert M.. 1993. Receptor editing: an approach by autoreactive B cells to escape tolerance. J. Exp. Med. 177:999–1008. 10.1084/jem.177.4.999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynne R., Ghandour G., Rayner J., Mack D.H., and Goodnow C.C.. 2000. B-lymphocyte quiescence, tolerance and activation as viewed by global gene expression profiling on microarrays. Immunol. Rev. 176:216–246. 10.1034/j.1600-065X.2000.00614.x [DOI] [PubMed] [Google Scholar]
- Goodnow C.C. 1992. Transgenic mice and analysis of B-cell tolerance. Annu. Rev. Immunol. 10:489–518. 10.1146/annurev.iy.10.040192.002421 [DOI] [PubMed] [Google Scholar]
- Goodnow C.C., Crosbie J., Adelstein S., Lavoie T.B., Smith-Gill S.J., Brink R.A., Pritchard-Briscoe H., Wotherspoon J.S., Loblay R.H., Raphael K., et al. . 1988. Altered immunoglobulin expression and functional silencing of self-reactive B lymphocytes in transgenic mice. Nature. 334:676–682. 10.1038/334676a0 [DOI] [PubMed] [Google Scholar]
- Hannum L.G., Ni D., Haberman A.M., Weigert M.G., and Shlomchik M.J.. 1996. A disease-related rheumatoid factor autoantibody is not tolerized in a normal mouse: implications for the origins of autoantibodies in autoimmune disease. J. Exp. Med. 184:1269–1278. 10.1084/jem.184.4.1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Healy J.I., Dolmetsch R.E., Timmerman L.A., Cyster J.G., Thomas M.L., Crabtree G.R., Lewis R.S., and Goodnow C.C.. 1997. Different nuclear signals are activated by the B cell receptor during positive versus negative signaling. Immunity. 6:419–428. 10.1016/S1074-7613(00)80285-X [DOI] [PubMed] [Google Scholar]
- Hebenstreit D., Fang M., Gu M., Charoensawan V., van Oudenaarden A., and Teichmann S.A.. 2011. RNA sequencing reveals two major classes of gene expression levels in metazoan cells. Mol. Syst. Biol. 7:497 10.1038/msb.2011.28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnida K., Stamnaes J., du Pré M.F., Mysling S., Jørgensen T.J., Sollid L.M., and Iversen R.. 2016. Epitope-dependent functional effects of celiac disease autoantibodies on transglutaminase 2. J. Biol. Chem. 291:25542–25552. 10.1074/jbc.M116.738161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Høydahl L.S., Nilssen N.R., Gunnarsen K.S., Pré M.F., Iversen R., Roos N., Chen X., Michaelsen T.E., Sollid L.M., Sandlie I., and Løset G.A.. 2016. Multivalent pIX phage display selects for distinct and improved antibody properties. Sci. Rep. 6:39066 10.1038/srep39066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Kearney J.F., Grusby M.J., Benoist C., and Mathis D.. 2006. Induction of tolerance in arthritogenic B cells with receptors of differing affinity for self-antigen. Proc. Natl. Acad. Sci. USA. 103:3734–3739. 10.1073/pnas.0600214103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iversen R., Di Niro R., Stamnaes J., Lundin K.E., Wilson P.C., and Sollid L.M.. 2013. Transglutaminase 2-specific autoantibodies in celiac disease target clustered, N-terminal epitopes not displayed on the surface of cells. J. Immunol. 190:5981–5991. 10.4049/jimmunol.1300183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabri B., and Sollid L.M.. 2017. T cells in celiac disease. J. Immunol. 198:3005–3014. 10.4049/jimmunol.1601693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keech C.L., Farris A.D., Beroukas D., Gordon T.P., and McCluskey J.. 2001. Cognate T cell help is sufficient to trigger anti-nuclear autoantibodies in naive mice. J. Immunol. 166:5826–5834. 10.4049/jimmunol.166.9.5826 [DOI] [PubMed] [Google Scholar]
- Koenig-Marrony S., Soulas P., Julien S., Knapp A.M., Garaud J.C., Martin T., and Pasquali J.L.. 2001. Natural autoreactive B cells in transgenic mice reproduce an apparent paradox to the clonal tolerance theory. J. Immunol. 166:1463–1470. 10.4049/jimmunol.166.3.1463 [DOI] [PubMed] [Google Scholar]
- Korponay-Szabó I.R., Halttunen T., Szalai Z., Laurila K., Király R., Kovács J.B., Fésüs L., and Mäki M.. 2004. In vivo targeting of intestinal and extraintestinal transglutaminase 2 by coeliac autoantibodies. Gut. 53:641–648. 10.1136/gut.2003.024836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouskoff V., Signorelli K., Benoist C., and Mathis D.. 1995. Cassette vectors directing expression of T cell receptor genes in transgenic mice. J. Immunol. Methods. 180:273–280. 10.1016/0022-1759(95)00002-R [DOI] [PubMed] [Google Scholar]
- Lorand L., and Graham R.M.. 2003. Transglutaminases: crosslinking enzymes with pleiotropic functions. Nat. Rev. Mol. Cell Biol. 4:140–156. 10.1038/nrm1014 [DOI] [PubMed] [Google Scholar]
- Love M.I., Huber W., and Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäki M. 1992. Seventh International Symposium on Coeliac Disease. C.L. Feighery and C. O'Farelly, editors. Oak Tree Press, Dublin. 246–252. [Google Scholar]
- McMahon S.B., and Monroe J.G.. 1996. The role of early growth response gene 1 (egr-1) in regulation of the immune response. J. Leukoc. Biol. 60:159–166. 10.1002/jlb.60.2.159 [DOI] [PubMed] [Google Scholar]
- Merrell K.T., Benschop R.J., Gauld S.B., Aviszus K., Decote-Ricardo D., Wysocki L.J., and Cambier J.C.. 2006. Identification of anergic B cells within a wild-type repertoire. Immunity. 25:953–962. 10.1016/j.immuni.2006.10.017 [DOI] [PubMed] [Google Scholar]
- Moon J.J., Chu H.H., Pepper M., McSorley S.J., Jameson S.C., Kedl R.M., and Jenkins M.K.. 2007. Naive CD4(+) T cell frequency varies for different epitopes and predicts repertoire diversity and response magnitude. Immunity. 27:203–213. 10.1016/j.immuni.2007.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemazee D.A., and Bürki K.. 1989. Clonal deletion of B lymphocytes in a transgenic mouse bearing anti-MHC class I antibody genes. Nature. 337:562–566. 10.1038/337562a0 [DOI] [PubMed] [Google Scholar]
- Packard T.A., Smith M.J., Conrad F.J., Johnson S.A., Getahun A., Lindsay R.S., Hinman R.M., Friedman R.S., Thomas J.W., and Cambier J.C.. 2016. B cell receptor affinity for insulin dictates autoantigen acquisition and B cell functionality in autoimmune diabetes. J. Clin. Med. 5:E98 10.3390/jcm5110098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro R., Duggal G., Love M.I., Irizarry R.A., and Kingsford C.. 2017. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods. 14:417–419. 10.1038/nmeth.4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao S.W., Ráki M., Gunnarsen K.S., Løset G.A., Lundin K.E., Sandlie I., and Sollid L.M.. 2011. Posttranslational modification of gluten shapes TCR usage in celiac disease. J. Immunol. 187:3064–3071. 10.4049/jimmunol.1101526 [DOI] [PubMed] [Google Scholar]
- Roy B., Neumann R.S., Snir O., Iversen R., Sandve G.K., Lundin K.E.A., and Sollid L.M.. 2017. High-Throughput Single-Cell Analysis of B Cell Receptor Usage among Autoantigen-Specific Plasma Cells in Celiac Disease. J. Immunol. 199:782–791. 10.4049/jimmunol.1700169 [DOI] [PubMed] [Google Scholar]
- Sanderson N.S., Zimmermann M., Eilinger L., Gubser C., Schaeren-Wiemers N., Lindberg R.L., Dougan S.K., Ploegh H.L., Kappos L., and Derfuss T.. 2017. Cocapture of cognate and bystander antigens can activate autoreactive B cells. Proc. Natl. Acad. Sci. USA. 114:734–739. 10.1073/pnas.1614472114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. . 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., and Eliceiri K.W.. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 9:671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scrucca L., Fop M., Murphy T.B., and Raftery A.E.. 2016. mclust 5: Clustering, classification and density estimation using Gaussian finite mixture models. R J. 8:289–317. 10.32614/RJ-2016-021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollid L.M. 2002. Coeliac disease: dissecting a complex inflammatory disorder. Nat. Rev. Immunol. 2:647–655. 10.1038/nri885 [DOI] [PubMed] [Google Scholar]
- Sollid L.M., Molberg O., McAdam S., and Lundin K.E.. 1997. Autoantibodies in coeliac disease: tissue transglutaminase--guilt by association? Gut. 41:851–852. 10.1136/gut.41.6.851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamnaes J., Cardoso I., Iversen R., and Sollid L.M.. 2016. Transglutaminase 2 strongly binds to an extracellular matrix component other than fibronectin via its second C-terminal beta-barrel domain. FEBS J. 283:3994–4010. 10.1111/febs.13907 [DOI] [PubMed] [Google Scholar]
- Sulkanen S., Halttunen T., Laurila K., Kolho K.L., Korponay-Szabó I.R., Sarnesto A., Savilahti E., Collin P., and Mäki M.. 1998. Tissue transglutaminase autoantibody enzyme-linked immunosorbent assay in detecting celiac disease. Gastroenterology. 115:1322–1328. 10.1016/S0016-5085(98)70008-3 [DOI] [PubMed] [Google Scholar]
- Szondy Z., Molnar P., Nemes Z., Boyiadzis M., Kedei N., Tóth R., and Fésüs L.. 1997. Differential expression of tissue transglutaminase during in vivo apoptosis of thymocytes induced via distinct signalling pathways. FEBS Lett. 404:307–313. 10.1016/S0014-5793(97)00140-3 [DOI] [PubMed] [Google Scholar]
- Taki S., Meiering M., and Rajewsky K.. 1993. Targeted insertion of a variable region gene into the immunoglobulin heavy chain locus. Science. 262:1268–1271. 10.1126/science.8235657 [DOI] [PubMed] [Google Scholar]
- Taylor J.J., Martinez R.J., Titcombe P.J., Barsness L.O., Thomas S.R., Zhang N., Katzman S.D., Jenkins M.K., and Mueller D.L.. 2012. Deletion and anergy of polyclonal B cells specific for ubiquitous membrane-bound self-antigen. J. Exp. Med. 209:2065–2077. 10.1084/jem.20112272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiegs S.L., Russell D.M., and Nemazee D.. 1993. Receptor editing in self-reactive bone marrow B cells. J. Exp. Med. 177:1009–1020. 10.1084/jem.177.4.1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upchurch H.F., Conway E., Patterson M.K. Jr., Birckbichler P.J., and Maxwell M.D.. 1987. Cellular transglutaminase has affinity for extracellular matrix. In Vitro Cell. Dev. Biol. 23:795–800. 10.1007/BF02623682 [DOI] [PubMed] [Google Scholar]
- Viken H.D., Paulsen G., Sollid L.M., Lundin K.E., Tjønnfjord G.E., Thorsby E., and Gaudernack G.. 1995. Characterization of an HLA-DQ2-specific monoclonal antibody. Influence of amino acid substitutions in DQ β 1*0202. Hum. Immunol. 42:319–327. 10.1016/0198-8859(94)00110-C [DOI] [PubMed] [Google Scholar]
- Wang H., and Shlomchik M.J.. 1997. High affinity rheumatoid factor transgenic B cells are eliminated in normal mice. J. Immunol. 159:1125–1134. [PubMed] [Google Scholar]
- Wardemann H., Yurasov S., Schaefer A., Young J.W., Meffre E., and Nussenzweig M.C.. 2003. Predominant autoantibody production by early human B cell precursors. Science. 301:1374–1377. 10.1126/science.1086907 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
RNA-seq data have been deposited to the EBI ArrayExpress database (accession no. E-MTAB-7567).