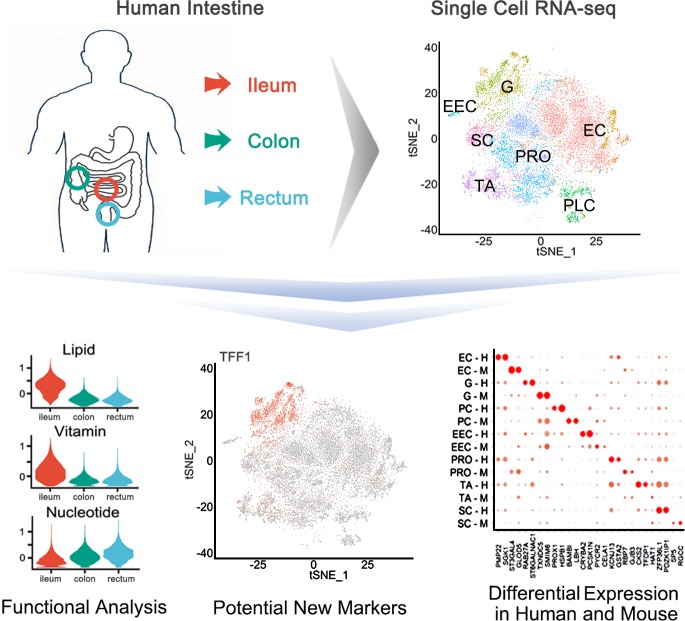
Using a single-cell RNA-seq approach, the work by Wang et al. establishes an atlas of human colon, rectum, and ileum epithelial cells. Their study reveals that different regions have specialized nutrient absorption preferences, microbe defenses, and endocrine function. They also identify new markers for a variety of cell types.
Abstract
In this issue of JEM, Wang et al. (https://doi.org/10.1084/jem.20191130), using a single-cell RNA-seq approach, establish an atlas of human colon, rectum, and ileum epithelial cells. Their study reveals that different regions have specialized nutrient absorption preferences, microbe defenses, and endocrine function. They also identify new markers for a variety of cell types.
The intestine is the largest and the most multifunctional organ in the human body and is responsible for food digestion, nutrient absorption and transportation, forming a barrier for microorganism defense, immune response to pathogens, and hormonal regulation via intestinal–brain communication (Duerkop et al., 2009; Grapin-Botton and Melton, 2000; Murphy and Bloom, 2006; Peterson and Artis, 2014). These complicated functions require intersection of the most complex biological systems. Previous studies mainly used pharmacological, pathophysiological, biochemical, and genetic approaches to characterize cell populations and types in the intestinal organ. Recent technological advancements in single-cell transcriptomic analysis have enabled more precise and comprehensive characterization of cell types and subpopulations across many organs (Tabula Muris Consortium, 2018; Han et al., 2018).
Insights from Xi C. He and Linheng Li.
To characterize cell populations of the human small and large intestines, Wang et al. (2019) isolated epithelial cells from the ileum, colon, and rectum from six donors and obtained their corresponding single-cell transcriptome of 14,537 total cells, with two for each intestinal segment as biological replicates. Using previously reported cell markers and other intestinal single-cell sequencing results (Haber et al., 2017), the researchers identified seven known cell types: enterocytes, goblets, Paneth cells, enteroendocrine cells, progenitor cells, transient amplifying (TA) cells, and stem cells. Tuft cell markers were detected in only a few cells, while the typical Tuft cell marker DCLK1 was not detected. They also identified Paneth-like cells in the large intestine, which express the genes related to microbe defense and the niche factors to support stem cells (Sato et al., 2009).
Image from Wang et al. (2019).
The data showed distinct cell compositions in the small and large intestine epithelia. There were 70% enterocytes in the ileum, but only 14% enterocytes in the large intestine; 20% were goblet cells in the large intestine, but only 5% in the ileum. Stem cells and TA cells were enriched with LGR5 and Ki67 expression, respectively. Stem cell signature genes of the three segments showed largely different functional features. For example, FABP2 and FABP6, involved in the fatty acid metabolic process, were enriched in ileum stem cells but not in large intestine stem cells, thus being consistent with the intestinal functions. Of note, the progenitor cells exhibited functional differences in different segments: progenitor cells from the ileum were marked by genes related to lipid and protein metabolism, while those from the colon and rectum were marked by genes involved in immune response.
Cell cycle wise, the study data showed that TA cells were mainly in the S and G2/M phase, while differentiated cells were mainly in the G1 phase. Researchers identified some TA-associated genes such as NUSAP1 (nucleolar and spindle associated protein) in the TA cluster in the ileum, colon, and rectum. Interestingly, unlike PCNA, expression of NUSAP1 revealed no overlaps with LGR5 in either human or mouse intestines. Thus, NUSAP1 may serve as a potential specific marker of a subset of TA cells.
Another finding from the study was distinct expression patterns of genes related to nutrient absorption in the small and large intestine. In general, the genes involved in protein digestion and absorption and mineral and organic substance transport were enriched in all three segments. The genes involved in lipid metabolism and drug metabolic process were predominantly expressed in the ileum, whereas the genes related to small molecule transport were enriched in the large intestine. The transporter genes, including the solute carrier (SLC) family genes involved in lipid, bile salt, vitamin, and water absorption, were enriched in the ileum, and the genes associated with metal ion and nucleotide absorption were highly expressed in the large intestine. While there were no significant differences in the expression levels of transporters for amino acid, sugar, inorganic, and organic solutes among these segments, the small intestine predominantly expressed SLC genes pertaining to transportation of monosaccharides such as glucose, fructose, galactose, and xylose. The expression of Na, K, and Ca channels was consistent with the role of the large intestine in metal ion absorption.
Wang et al. (2019) also compared expression of signaling molecules in the small and large intestine. For instance, higher expression of TGF-β/BMP signaling (He et al., 2004) was found in the large intestine, especially in the rectum. FZD5 and DVL3, involved in Wnt signaling (Gregorieff et al., 2005), were upregulated in the rectum. The high expression of both the pro-proliferative and pro-death genes suggests that the epithelium of the large intestine, particularly the rectal epithelium, has a fast turnover rate.
Researchers revealed that expressions of some hormone-related genes in enteroendocrine cells were enriched in the small intestine, such as secretin (SCT), neurotensin (NTS), and cholecystokinin (CCK), while some were higher in the large intestine, such as peptidyl glycine α-amidating monooxygenase (PAM), peptide tyrosine-tyrosine (PYY), and insulin-like peptide 5 (INSL5).
Mouse models have been widely used to elucidate the mechanisms and to understand the etiology of human diseases, as well as to test drug toxicity and efficacy. However, lack of assessment of the similarities and differences between humans and mice may affect the proper application of mouse models and the related translation into humans. Indeed, Wang et al. (2019) found different signature gene expression patterns in mouse and human ileum. For instance, ITLN1 and Reg4 were enriched in human goblet cells, but not in Paneth and enteroendocrine cells as reported in the mouse ileum (Haber et al., 2017). Future studies to compare these findings with human diseases will provide critical insight for human digestive, infectious, immune/autoimmune, and hormone-related diseases.
References
- Tabula Muris Consortium Nature. 2018 doi: 10.1038/s41586-018-0590-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duerkop B.A., et al. Immunity. 2009 doi: 10.1016/j.immuni.2009.08.009. [DOI] [Google Scholar]
- Grapin-Botton A., and Melton D.A. Trends Genet. 2000 doi: 10.1016/S0168-9525(99)01957-5. [DOI] [PubMed] [Google Scholar]
- Gregorieff A., et al. Gastroenterology. 2005 doi: 10.1016/j.gastro.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Haber A.L., et al. Nature. 2017 doi: 10.1038/nature24489. [DOI] [Google Scholar]
- Han X., et al. Cell. 2018 doi: 10.1016/j.cell.2018.05.012. [DOI] [Google Scholar]
- He X.C., et al. Nat. Genet. 2004 doi: 10.1038/ng1430. [DOI] [Google Scholar]
- Murphy K.G., and Bloom S.R. Nature. 2006 doi: 10.1038/nature05484. [DOI] [Google Scholar]
- Peterson L.W., and Artis D. Nat. Rev. Immunol. 2014 doi: 10.1038/nri3608. [DOI] [PubMed] [Google Scholar]
- Sato T., et al. Nature. 2009 doi: 10.1038/nature07935. [DOI] [Google Scholar]
- Wang Y., et al. J. Exp. Med. 2019 doi: 10.1084/jem.20191130. [DOI] [Google Scholar]