Abstract
The prolyl 3-hydroxylase family member 4 (P3H4) (alias SC65) is implicated in a variety of physiological and pathological processes. However, little is known about the role of P3H4 in tumors. This study aimed to investigate the role of P3H4 in bladder cancer (BC) and the regulatory mechanisms that influence its expression. P3H4 was highly expressed in BC tissues. Knockdown of P3H4 inhibited BC cell proliferation, cell cycle, migration and invasion in vitro, and inhibited BC growth in vivo. We also found that ETV4 bound directly to the promoter region of P3H4 and activated its transcription. Furthermore, overexpression of ETV4 rescued the inhibition of proliferation and invasion induced by PH4 silencing. ETV4 was significantly overexpressed in the BC tissues. In conclusion, P3H4 functioned an oncogene role in BC progression, and ETV4 bound directly to the P3H4 promoter region to regulate its transcription.
Keywords: P3H4, bladder cancer, ETV4, transcription factor
INTRODUCTION
Bladder cancer (BC) is the most common malignancy of the genitourinary system, with more than 430,000 new patients worldwide affected each year [1, 2]. BC is also the seventh leading cause of cancer-related deaths worldwide, accounting for 2.8% of these deaths [3]. Surgical resection combined with radiotherapy or chemotherapy has been applied to BC treatment [4]. However, patients receiving aggressive treatment still face the status quo of easy recurrence, poor prognosis, low survival rate and poor quality of life [5, 6]. Therefore, finding new and effective therapeutic targets is urgent and important.
The prolyl 3-hydroxylase family member 4 (P3H4) (alias SC65) is originally identified as a protein associated with the association complex (SC) [7]. However, no further evidence supports this point. Subsequent studies detect antibodies against P3H4 in cases of interstitial cystitis [8], membranous nephropathy [9], prostate cancer [10], and meningeal cancer [11]. P3H4 has also been identified as an autoantigen associated with these diseases. Furthermore, distinct functions of P3H4 are reported separately. For example, P3H4 acts as an endoplasmic reticulum (ER) protein that regulates bone mass homeostasis and skin fragility [12]. Studies have shown that P3H4 is expressed at high levels in bone, cartilage and skin. It also forms a stable complex with P3H3 in ER and interacts with lysyl hydroxylase 1 and CYPB [13, 14]. Additionally, P3H4 is an adaptor protein that links myelin protein zero (P0) and the activated C kinase 1 (RACK1) receptor [15], a gene that is up-regulated in the hippocampus during sleep [16], and is possibly a PTEN-interacting protein [17]. In general, P3H4 is involved in a variety of physiological and pathological processes. However, little is known about the role of P3H4 in BC.
In the present study, we found that P3H4 was significantly overexpressed in BC tissues, and knockdown of P3H4 inhibits BC cell proliferation, cell cycle, migration and invasion. ETV4 regulated P3H4 transcription by binding directly to its promoter region and was involved in the regulation of BC progression. Overexpression of ETV4 rescued the inhibition of proliferation and invasion induced by PH4 silencing.
RESULTS
P3H4 is highly expressed in BC
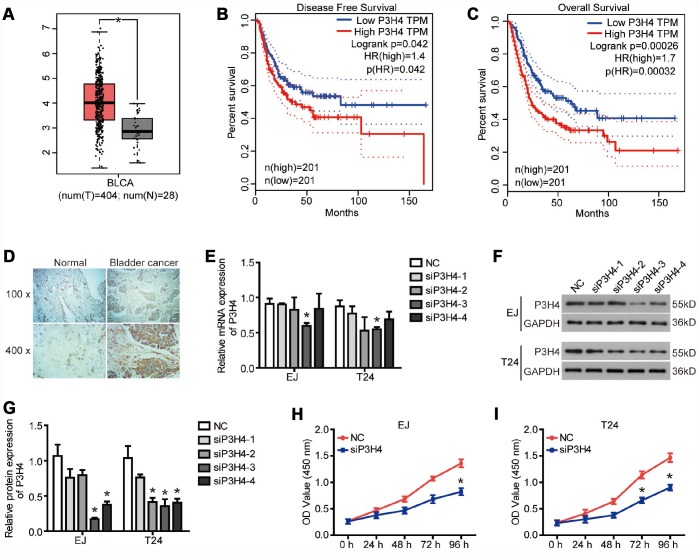
To reveal the role of P3H4 in BC, we first examined the P3H4 expression in BC tissues by database analysis and IHC analysis. The GEPIA online analysis website (http://gepia.cancer-pku.cn/) was used to analyze P3H4 mRNA levels in 404 patients with bladder urothelial carcinoma (BLCA) and 28 healthy volunteers. As shown in Figure 1A, the mRNA levels of P3H4 were significantly higher in BLCA patients compared to the healthy volunteers. The disease-free survival and overall survival of BLCA patients with low P3H4 expression was obviously higher than those of BLCA patients with high P3H4 expression (Figure 1B and 1C). In addition, we collected 32 BC tissues and 26 normal tissues to measure P3H4 expression. As shown in Figure 1D and Table 1, P3H4 was high expressed in BC tissues compared to normal tissues. Furthermore, statistical analysis demonstrated that high expression of P3H4 in BC tissues was related to gender (Table 2). Moreover, the incidence of BC in males is 3-4 times that of females. These results suggest that P3H4 may play an oncogene function in BC.
Figure 1.
P3H4 is highly expressed in BC. (A)The boxplot of P3H4 mRNA level. Red and gray boxes represent bladder urothelial carcinoma (BLCA) tissue and normal bladder tissue, respectively. The data came from the GEPIA database. Disease free survival (B) and overall survival (C) percentage of BLCA patients with high or low P3H4 expression. (D) P3H4 expression in bladder cacner (BC) tissues and adjacent normal tissues was examined by Immunohistochemical (IHC) analysis. After siRNA targeting P3H4 (siP3H4) was transfected into EJ and T24 cells, P3H4 mRNA (E) and protein (F and G) expression were detected by RT-qPCR and western blot, cell proliferation (H and I) were measured by CCK8 assays. *P<0.05.
Table 1. P3H4 expression in BC compared with normal tissue.
| Group | n | P3H4 expression | P | |
| Low (n%) | High (n%) | |||
| BC | 32 | 8 (25.0) | 24 (75.0) | 0.005* |
| Normal | 26 | 16 (61.5) | 10 (38.5) | |
Table 2. P3H4 expression associated with the clinicopathological parameters in BC.
| clinicopathological parameters | n | P3H4 expression | P | |
| Low (n%) | High (n%) | |||
| Gender | ||||
| Male | 26 | 4 (15.4) | 22 (84.6) | 0.009* |
| Female | 6 | 4 (66.7) | 2 (33.3) | |
| Age (years) | ||||
| <65 | 11 | 3 (27.3) | 8 (72.7) | 0.830 |
| ≥65 | 21 | 5 (23.8) | 16 (76.2) | |
| Tumor diameter (cm) | ||||
| <4 | 15 | 3 (20.0) | 12 (80.0) | 0.539 |
| ≥4 | 17 | 5 (29.4) | 12 (70.6) | |
| Clinical stages | ||||
| 1-2 | 9 | 2 (22.2) | 7 (77.8) | 0.820 |
| 3-4 | 23 | 6 (26.1) | 17 (73.9) | |
Knockdown of P3H4 inhibits BC cell proliferation by impeding cell cycle progression
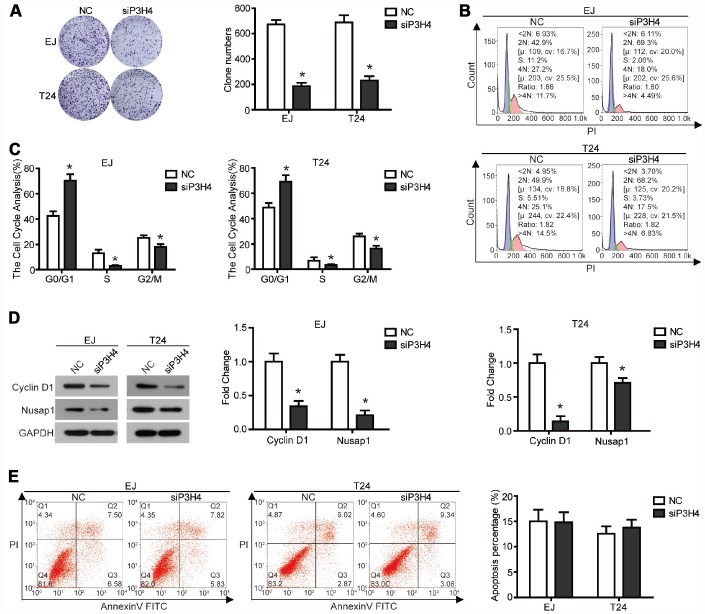
To expose the action of P3H4 in BC cells’ biological function, expression of P3H4 was down-regulated with siRNA (Figure 1E, 1F), and siRNA-4 targeting P3H4 (siP3H4) was used in subsequent experiments. As shown in Figure 1H and 1I, CCK8 assay showed that proliferation of EJ and T24 cells was inhibited when siP3H4 was transfected into cells. Similarly, colony formation assay revealed that knockdown of P3H4 inhibited the colony forming ability of EJ and T24 cells (Figure 2A). Furthermore, we detected the action of P3H4 knockdown on the cell cycle and apoptosis of BC cells. As shown in Figure 2B and 2C, the proportion of cells in G0/G1 phase increased while the proportion of cells in S and G2/M phase decreased, when siP3H4 was transfected into cells. These results indicate that knockdown of P3H4 impedes cell cycle in the G0/G1 phase. In addition, transfection of siP3H4 reduced the protein levels of Cyclin D1, Nusap1 and p70 (Figure 2D). Cyclin D1 is a key protein that regulates the G1 phase of the cell cycle [18]. Nusap1 is an indispensable microtubule and chromatin binding protein, which regulates the dynamics of kinetochore microtubules, governs chromosome oscillations, and ensures the normal progress of cell cycle [19, 20]. Finally, flow cytometry detection found that knockdown of P3H4 had no effect on the apoptosis of BC cells (Figure 2E). These results demonstrate that the knockdown of P3H4 can inhibit BC cell proliferation by arresting cell cycle progression, but that it has no effect on the apoptosis of BC cells.
Figure 2.
Knockdown of P3H4 arrested cell cycle in the G1 phase and thus inhibited proliferation, but had no effect on apoptosis. After siP3H4 was transfected into EJ and T24 cells, (A) the clonality was evaluated by clone formation assay, (B and C) cell cycle was detected by flow cytometry, (D) the protein expression of cell cycle-related proteins Cyclin D1, Nusap1 and p70 were measured by western blot, (E) apoptosis was examined by flow cytometry. *P<0.05.
Knockdown of P3H4 inhibits BC cell migration and invasion by affecting EMT progression
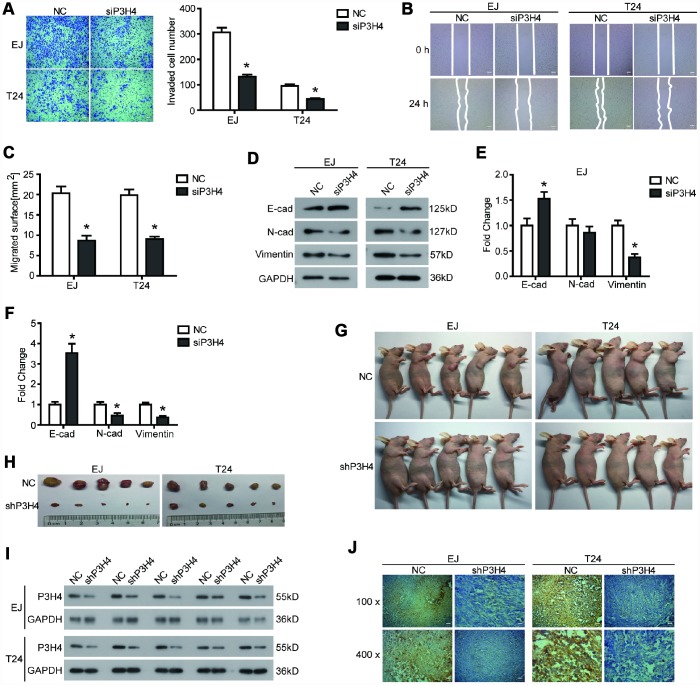
In addition to the effects on BC cell proliferation, we also examined the effects of P3H4 knockdown on BC cell motility. Transwell assay showed that compared to NC, fewer EJ and T24 cells could pass through the matrigel and invade the lower surface of the chamber membrane (Figure 3A). Additionally, wound healing assay revealed that transfection of siP3H4 caused wound healing of EJ and T24 cells to be slower (Figure 3B and 3C). Moreover, the knockdown of P3H4 reduced protein levels of N-cadherin and Vimentin, as well as increased the protein levels of E-cadherin (Figure 3D–3F). Decreased E-cadherin expression, increased N-cadherin and increased Vimentin were the most important sign change of the Epithelial-Mesenchymal transition (EMT) [21]. EMT plays a pivotal role in the primary infiltration and secondary metastasis of a variety of solid tumors [22].
Figure 3.
Knockdown of P3H4 inhibits BC cell migration and invasion in vitro and BC growth in vivo. After siP3H4 was transfected into EJ and T24 cells, (A) the invasion ability was determined by Transwell assay, the migration ability was examined (B) and quantified (C) by wound healing assay, (D–F) the expression of EMT-related proteins was detected by western blot. (G) Images of xenograft nude mice after 4 weeks of treatment. (H) Images of subcutaneous tumor. The expression of P3H4 in subcutaneous tumors was examined by western blot (I) and IHC (J). *P < 0.05.
Knockdown of P3H4 inhibits BC growth in vivo
In addition to cells in vitro, a subcutaneous xenograft model was used to further assess the biological role of P3H4 in BC progression. As shown in Figure 3G, 3H, knockdown of P3H4 in EJ and T24 cells significantly inhibited BC growth. Western blot and IHC analysis also demonstrated that the expression of P3H4 was down-regulated by the shP3H4 transfection (Figure 3I, 3J).
ETV4 is the transcription factor of P3H4
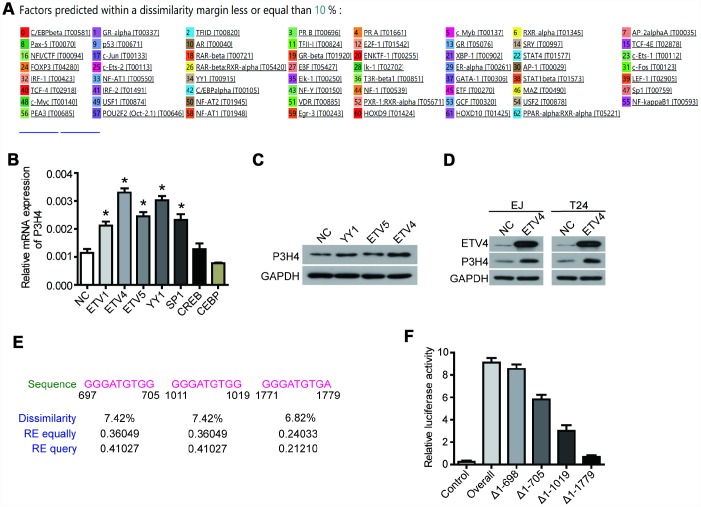
The PROMO database predicted a variety of P3H4 transcription factors (Figure 4A), of which ETV1, ETV4, ETV5, YY1, SP1, CREB, and CEBP were selected for subsequent testing. Their overexpression plasmids were transfected into EJ cells, respectively. RT-qRCP detection revealed that overexpression of the transcription factors ETV4, ETV5, and YY1 significantly enhanced the mRNA level of P3H4 (Figure 4B). Western blot results also showed that ETV4 could better enhance the expression of P3H4 (Figure 4C, 4D). Furthermore, the promoter analysis revealed the presence of three binding regions for ETV4 on the promoter of P3H4, located at 697-705, 111-1019, and 1771-1779, respectively (Figure 4E). Fragment deletion plasmid Δ1-698 (1-698 bp deletion), Δ1-705 (1-705 bp deletion), Δ1-1019 (1-1019 bp deletion), and Δ1-1779 (1-1779 bp deletion) were constructed and co-transfected into 293T cells with ETV4-overexpression plasmid. Fluorescein assay results indicated that ETV4 was able to bind to three predicted sites (Figure 4F).
Figure 4.
ETV4 is the transcription factor of P3H4. (A) The PROMO database predicted a variety of P3H4 transcription factors. After transfected with transcription factor overexpression plasmid respectively, P3H4 mRNA and protein expression in EJ cells were detected by RT-qPCR (B) and western blot (C). (D) After transfected with ETV4-overexpression plasmid, the expression of ETV4 and P3H4 was examined by western blot. (E) Three predicted ETV4 binding P3H4 promoter sites. (F) The site where ETV4 bound directly to the P3H4 promoter region was measured by luciferase activity assay. *P < 0.05.
ETV4 is essential for P3H4 to affect BC cell proliferation and motility.
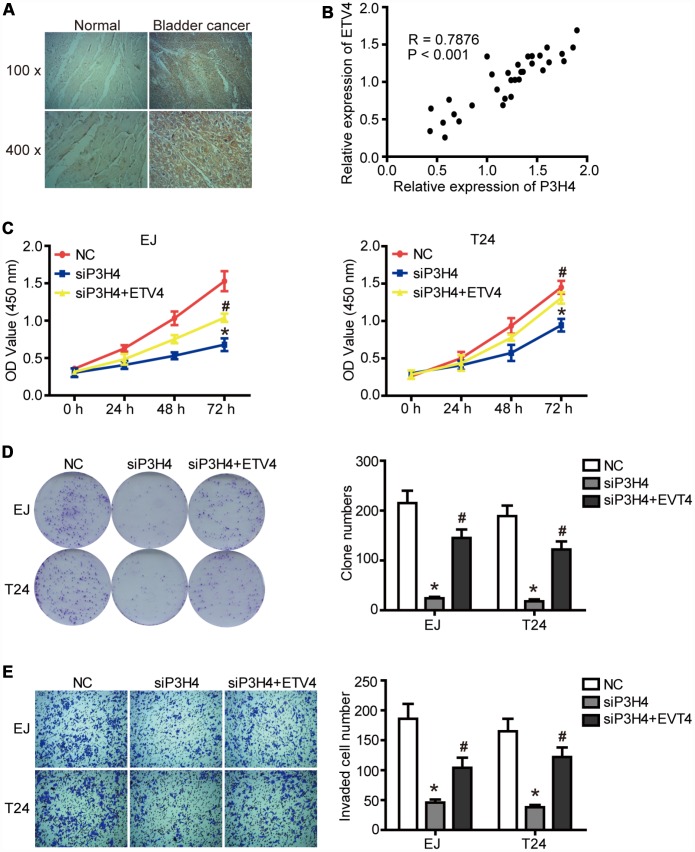
Given that ETV4 may be an important transcription factor for P3H4, we further examined the expression of ETV4 in BC tissues. As shown in Figure 5A and Tables 3, 4, ETV4 was significantly high expressed in BC tissues compared to normal tissues, and ETV4 expression was related to clinical stage of BC. Furthermore, there was a significant positive correlation between the expression of P3H4 and ETV4 (Figure 5B, P < 0.001, R = 0.7876). In addition, ETV4 overexpression partially restored the inhibitory of siP3H4 transfection on proliferation, colony formation and invasion ability of EJ and T24 cells (Figure 5C–5E).
Figure 5.
ETV4 is essential for P3H4 to affect BC cell proliferation and motility. (A) ETV4 expression in BC tissues and adjacent normal tissues was examined by IHC. (B) The correlation analysis of P3H4 and ETV4 expression. After siP3H4 and ETV4-overexpression plasmids were co-transfected into EJ and T24 cells, cell proliferation were measured by CCK8 assays (C) and clone formation assay (D), invasion ability was determined by Transwell assay (E). * P<0.05.
Table 3. ETV4 expression in BC compared with normal tissue.
| Group | n | ETV4 expression | P | |
| Low (n%) | High (n%) | |||
| BC | 33 | 12 (36.4) | 21 (63.6) | 0.001** |
| Normal | 27 | 22 (81.5) | 5 (18.5) | |
Table 4. ETV4 expression associated with the clinicopathological parameters in BC.
| Clinicopathological parameters | n | ETV4 expression | P | |
| Low (n%) | High (n%) | |||
| Gender | ||||
| Male | 27 | 9 (33.3) | 18 (66.7) | 0.765 |
| Female | 6 | 3 (50.0) | 3 (50.0) | |
| Age (years) | ||||
| ≤65 | 15 | 5 (33.3) | 10 (66.7) | 0.974 |
| <65 | 18 | 7 (38.9) | 11 (61.1) | |
| Tumor diameter (cm) | ||||
| ≤4 | 20 | 6 (30.0) | 14 (70.0) | 0.773 |
| <4 | 12 | 5 (41.7) | 7 (58.3) | |
| Clinical staging | ||||
| 1-2 | 9 | 7 (77.8) | 2 (22.2) | 0.009* |
| 3-4 | 24 | 5 (20.8) | 19 (79.2) | |
DISCUSSION
In the present study, we discovered the role of P3H4 in BC, and found that ETV4 can act as a transcription factor to influence BC progression by regulating P3H4 transcription.
Database analysis and IHC results showed abnormally high expression of P3H4 mRNA and proteins in BC tissues. Li et al also detected P3H4 mRNA expressions in 44 pairs of BC tissues and adjacent normal tissues by qRT-PCR, as well as discovered that P3H4 mRNA were highly expressed in BC tissues [23]. Their findings are consistent with our results.
Knockdown of P3H4 arrested the EJ and T24 cell cycle in the G1 phase, thus inhibiting cell proliferation. However, this no effect on cell apoptosis. In addition, knockdown of P3H4 also impeded migration and invasion of both EJ and T24 cells by inhibiting the EMT process. The xenograft nude mouse model also showed that knockdown of P3H4 inhibited the growth of subcutaneous BC tumors. These results indicate that P3H4 may play an oncogene function in BC progression. Although studies have displayed that P3H4 may be associated with several tumors (prostate cancer [10] and meningeal cancer [11]), this study was the first to conduct experiments in vivo and in vitro to determine the role of P3H4 in BC.
Furthermore, ETV4 could regulated the P3H4 transcription in EJ and T24 cells. ETV4 (ETS Variant 4) is a transcription factor belonging to the EST family and is specifically involved in the carcinogenesis of various tissues, including pancreatic cancer, breast cancer and intestinal cancer [24–26]. However, little is known about the role of ETV4 in BC. ETV4 is frequently up-regulated in tumors and acts as an oncogenic factor via different mechanisms. IHC analysis also found a significant up-regulation of ETV4 expression in BC. By fusion with other genes, ETV4 regulates genome transcriptional pattern and carcinogenesis [27]. For example, TMPRSS2-ETV4, KLK2-ETV4 and CANTT-ETV4 [27, 28]. ETV4-related gene fusion may be an early event in prostate cancer and could define a new molecular subtype of prostate cancer [28]. On the other hand, ETV4 can directly bind to the promoter region of target genes and regulate their transcriptional activity. In pancreatic cancer and breast cancer, ETV4 directly upregulates the transcription of cyclin proteins, such as cyclin D1 [26] and cyclin D3 [29] to promote cell proliferation. MMPs, such as MMP13 and urokinase plasminogen activator (uPA) are also transcriptionally regulated by ETV4, which are involved in the ETV4-driven malignant phenotypes in prostate cancer [30] and breast cancer [25]. In the present study, our results of western blot and Luciferase activity assay demonstrated that ETV4 can directly bind to the promoter of P3H4 and activate transcription of P3H4. Moreover, three binding sites of the ETV4 and P3H4 promoter regions were found. Overexpression of ETV4 rescued the inhibition of proliferation and invasion induced by PH4 silencing. These results indicate that ETV4 can participate in the regulation of BC progression by transcriptionally regulating the expression of P3H4.
In conclusion, P3H4 regulated proliferation, cell cycle, migration and invasion, playing an oncogene role in BC progression. In BC cells, ETV4 bound directly to the P3H4 promoter region to regulate its transcription.
MATERIALS AND METHODS
Tissues specimens
32 fresh BC tissues and 26 normal adjacent tissues were obtained from BC patients who underwent a urethral cystectomy (TURB) at Xuzhou Central Hospital in 2018. Normal tissues were excised from 5 to 7 cm away from the tumor. The specimens were immediately frozen in liquid nitrogen and stored at -80 °C. The pathological samples were reassessed by an experienced urinary tract division (ALB) to confirm the diagnosis. The hospital ethics committee approved the study and all patients signed written informed consent.
Immunohistochemistry (IHC)
Collected clinical samples were immersed in formalin for 24 h at 4 °C, followed by paraffin embedding. Paraffin blocks were cut into 4 μm sections by a cryo-cutting machine and IHC was performed to evaluate P3H4 or ETV4 protein expression. IHC was performed using non-biotinylated rabbit polyclonal anti-human P3H4 (Proteintech, Manchester, UK, 15288-1-AP, 1:100) and rabbit polyclonal anti-human ETV4 (Proteintech, Manchester, UK, 10684-1-AP, 1:200). Van-Clear (Hongci., Shanghai, China) and concentration gradient ethanol were used for the dewaxing of paraffin sections. Antigen retrieval was performed by microwave pretreatment in 0.01 M citrate buffer for 10 min. After being blocked with 5% sheep serum at room temperature for 1 h, the sections were incubated with the primary antibody, and incubated with Enzyme-labeled goat anti-mouse/rabbit IgG polymer (160101405L, Maixin., Shanghai, China). The immune response was visualized by a Reinforced DAB Chromogenic Kit (1705252031, Maixin., Shanghai, China), and hematoxylin was used for counterstaining. Finally, images were obtained using an upright microscope system (Nikon, JAPAN).
The total P3H4 or ETV4 immunostaining score was calculated by multiplying the positive staining cell ratio ® and staining intensity score (S) which were given by two pathologists blinded to the clinical parameters. R was divided into four grades: 0 (<5%, negative), 1 (5-25%, sporadic), 2 (25-50%, focal), 3 (> 51%, diffuse). S was also divided into four grades: 0 (negative), 1 (weak), 2 (moderate), 3 (strong). Finally, a total of 0-3 was considered to be low staining, while 4-9 was considered to be high staining.
Cell culture
BC cell lines EJ and T24, as well as 293T cells were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). All cells were cultured in DMEM medium (Gibco, NY, USA) containing 10% fetal bovine serum (FBS; Gibco, Australia) and 1% penicillin–streptomycin with 5% CO2 at 37 °C in a humidified incubator.
Plasmid, construction of the lentivirus and transfection
Four small interfering RNA (siRNA) sequences (Ruibo, Guangzhou, China) were synthesized to target P3H4 mRNA. Their sequence was as follows: siRNA-1: GGGCUGUGAAGCUCUACAATT, siRNA-2: CCAA GUAUCUCAACUACUATT, siRNA-3: CGGCCAUA GCAGAUCUCUUTT, siRNA-4: GGAGCCUGAGGA UGCCCUATT.
SiRNA targeting P3H4 sequence was transformed into short hairpin RNA (shRNA), which was cloned into a pLVX-shRNA2 green fluorescent protein (GFP) vector. The recombinant plasmid was transfected into the 293T cells together with two lentiviral packaging plasmids to generate lentivirus. After incubation for 18 h and 42 h, the lentivirus from the culture medium was collected and concentrated by Lentiviral Packaging Kits (TR30037, Origene, Billerica, MA, USA). For cell infection, EJ and T24 cells at a concentration of 5 × 104/well in a six-well plate were incubated with shRNA-P3H4 lentivirus (P3H4 knockdown group) or negative control lentivirus (NC), with a multiplicity of infection (MOI) of 50, lasting 24 h. Pools of stable transductions were generated by selection using puromycin (4 μg/ml) for 2 weeks. Transduction efficiency was determined using fluorescence microscopy (MicroPublisher 3.3 RTV, Olympus, Tokyo, Japan), western blot and qRT-PCR analysis of GFP expression.
The ETV1, ETV4, ETV5, YY1, SP1, CREB and CEBP cDNA sequences were cloned into the pcDNA3.1 vector, respectively, and were transfected into EJ and T24 cells to up-regulate the expression of target gene. The P3H4 promoter sequence 697-1779 (1-698 bp deletion), 706-1779 (1-705 bp deletion), and 1019-1779 (1-1019 bp deletion) were cloned into the pGL3.0 vector, respectively, and were transfected into EJ and T24 cells together with pcDNA3.1-ETV4 plasmid. Transfection of all siRNA and plasmids was performed by Lipofectamine 2000 (Invitrogen, USA).
qRT-PCR
Total mRNA from cells was extracted using a TRizol reagent (Invitrogen, Carlsbad, CA, USA), and reverse transcribed to cDNAs by the PrimeScript Reverse Transcription Reagent Kit (TaKaRa, Dalian, China). P3H4 mRNA expression was quantitatively analyzed using SYBR Premix Ex Taq™ (TaKaRa, Dalian, China) with the 2−ΔΔCt method. β-actin as an internal reference. The primers used were as follows: P3H4 forward: 5′- CGGGCTGTGAAGCTCTACAA-3′, P3H4 reverse: 5′- GCGTCAGATAGGGCATCCTC-3′; β-actin forward: 5′- CCCGAGCCGTGTTTCCT-3′, β-actin reverse: 5′- GTCCCAGTTGGTGACGATGC-3’. ′
Western blot
Total proteins from cells and tissues were dissolved with RIPA buffer and quantified with the BCA Protein Assay Kit (Tiangen, Beijing, China). 15 μg of proteins were separated through 10% SDS-PAGE gel and then transferred onto PVDF membranes. After being incubated with 5% fat-free skim milk for 1 h at room temperature, PVDF membranes were incubated with primary antibodies at 4 °C overnight and subsequently incubated with secondary antibody at room temperature for 2 h. Immunoreactive bands were developed using Enhanced chemiluminescence kit (Amersham, Little Chalfont, UK).
Anti-P3H4 (1:1000, 15288-1-AP), anti-E-cadherin (1:1000, 29874-1-AP) and anti-ETV4 (1:1000, 10684-1-AP) were purchased from Proteintech (Manchester, UK, USA). Anti-Cyclin D1 (1:1000, ab16663), anti-Nusap1 (1:1000, ab169083), anti-p70 (1:1000, ab131526), anti-N-cadherin (1:1000, ab18203), anti-Vimentin (1:1000, ab8978), anti-GAPDH (1:1000, ab181602) were purchased from Abcam (Cambridge, MA, USA).
CCK8 assay
5×103 cells were seeded in each well of a 96-well plate and cultured overnight. After 0 h, 24 h, 48 h and 72 h of transfection, 100 μl of fresh medium containing 10 μl CCK8 solution (Beyotime Biotechnology, Shanghai, China) was added to each well and incubated at 37 °C for 2 h. The OD value at a wavelength of 450 nm was measured with a microplate reader.
Colony forming assay
200 transfected cells were normally cultured in each well of a six-well plate. After two weeks, cells were fixed with methanol and stained with 0.1% crystal violet. Visible colonies were counted and pictured.
Flow cytometry detection for cell cycle
After 48 h of transfection, cells were trypsinized (without EDTA), and resuspended in cold PBS. Subsequently, cells were fixed in cold 70% ethanol at -20 °C for 24 h. Fixed cells were incubated with RNase A for 30 min, and stained with propidium oxide for another 30 min in the dark by a Cell Cycle Analysis Kit (Beyotime Biotechnology, Beijing, China). Cell cycle progression was immediately detected using a flow cytometer, and analyzed using FCS Express 4 software (De NovoSoftware, Los Angeles, CA).
Flow cytometry detection for cell apoptosis
After 48 h of transfection, cells were trypsinized and collected by the Annexin V binding buffer. Subsequently, cells were double stained with V-FITC and propidium iodide (BD Biosciences, Franklin Lakes, NJ, USA), then measured by BD FACS Canto II (BD Biosciences). Data was analyzed using FlowJo software.
Transwell assay
Transwell chambers (BD Biosciences) were used to evaluate the invasion ability of EJ and T24 cells. The upper surface of chambers was coated with Matrigel. 200 μl of cell suspension with a concentration of 1x106 cells/ml was added into the upper chamber. The lower chambers were filled with 600 μl of DMEM containing 10% FBS. After incubation at 37 °C for 24 h, cells on the upper chambers were removed, and invaded cells on the lower surface of the chambers were fixed and stained with 0.1% crystal violet. Invaded cells were counted and photographed under a microscope (200x magnification, Nikon TE2000).
Wound healing assay
Wound healing assay was performed to measure the migration ability of GC cells. Cells were grown to confluence in 6-well plates. A sterile plastic tip was used to create wounds. After washing with PBS, cells were cultured in serum-free medium for 48 h. Images were taken using a microscope. The average of five random widths of each wound was measured for quantification.
Xenografts in mice
All experimental procedures were approved by the Ethics Committee. Approximately 1x107 cells were injected subcutaneously into the armpits of female athymic BALB/C nude mice (4-6 weeks old, 18-22 g, 5 mice per group). Tumor growth was monitored weekly. After 4 weeks, the mice were euthanized and photographed. The subcutaneous tumor mass removed, and a picture was taken. A portion of the tumor mass was fixed and embedded for IHC, and a portion of the tumor mass was used for western blot.
Luciferase activity assay
The pGL3.0 recombinant plasmid and pcDNA3.1-ETV4 plasmid were co-transfected into EJ and T24 cells using Lipofectamine 2000. After 48 h, cells were harvested and luciferase activity was detected using a luciferase assay system (Promega, Madison, Wisconsin, USA).
Statistical analysis
Statistical analysis was performed as the mean ± SD and conducted using SPSS 22.0 (SPSS, IBM, Beijing, China) and GraphPad Prism 6 (GraphPad, San Diego, CA). Differences were calculated with the Student's t-test between two groups or with one-way ANOVA among multiple groups. P < 0.05 was considered statistically significant.
Footnotes
AUTHOR CONTRIBUTIONS: Lin Hao, Kun Pang and Hui Pang generated the hypothesis, designed and performed the experiments, and analyzed the data. All author performed the experiments, analyzed the data, provided conceptual advice and technical expertise, and edited the manuscript. Zhenduo Shi and Conghui Han conceived and supervised the study. All authors have reviewed and approved the final version of the manuscript.
CONFLICTS OF INTEREST: All author declare that they have no conflicts of interest.
FUNDING: This work was supported by the Jiangsu Province Key Research and Development Program (BE2017635, BE2015623); Jiangsu Province Young Medical Talents (QNRC2016386, QNRC2016383, QNRC2016389); Jiangsu Medical Innovation Team (CXTD-2016-48); Xuzhou Health Innovation Team (XWCX201603); National Natural Science Foundation of China (81774089); Xuzhou Science and Technology Program (KC18036); Jiangsu Province Natural Science Research Projects (17KJB360001, 18KJB180007); Xuzhou Medical Outstanding Talents (Xuzhou Health Education Research [2017] No. 22).
REFERENCES
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015; 65:87–108. 10.3322/caac.21262 [DOI] [PubMed] [Google Scholar]
- 2.Burger M, Catto JW, Dalbagni G, Grossman HB, Herr H, Karakiewicz P, Kassouf W, Kiemeney LA, La Vecchia C, Shariat S, Lotan Y. Epidemiology and risk factors of urothelial bladder cancer. Eur Urol. 2013; 63:234–41. 10.1016/j.eururo.2012.07.033 [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017; 67:7–30. 10.3322/caac.21387 [DOI] [PubMed] [Google Scholar]
- 4.Joshi M, Warrick JI, Yin M, Holder SL, Drabick JJ. Need for a personalized approach for muscle invasive bladder cancer: role of tumor biology in response to neoadjuvant chemotherapy. Transl Androl Urol. 2019. (Suppl 1); 8:S99–103. 10.21037/tau.2018.12.04 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bladder cancer: diagnosis and management of bladder cancer: © NICE (2015) Bladder cancer: diagnosis and management of bladder cancer. BJU Int. 2017; 120:755–65. 10.1111/bju.14045 [DOI] [PubMed] [Google Scholar]
- 6.Smith AB, Jaeger B, Pinheiro LC, Edwards LJ, Tan HJ, Nielsen ME, Reeve BB. Impact of bladder cancer on health-related quality of life. BJU Int. 2018; 121:549–57. 10.1111/bju.14047 [DOI] [PubMed] [Google Scholar]
- 7.Chen Q, Pearlman RE, Moens PB. Isolation and characterization of a cDNA encoding a synaptonemal complex protein. Biochem Cell Biol. 1992; 70:1030–8. 10.1139/o92-147 [DOI] [PubMed] [Google Scholar]
- 8.Ochs RL, Stein TW Jr, Chan EK, Ruutu M, Tan EM. cDNA cloning and characterization of a novel nucleolar protein. Mol Biol Cell. 1996; 7:1015–24. 10.1091/mbc.7.7.1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhai Y, Zhao L, Wang Z, Zhai G. RETRACTED ARTICLE: Preparation and characterization of novel lipid nanocapsules of ropivacaine for transdermal delivery. Drug Deliv. 2016; 23:619–28. 10.3109/10717544.2014.930761 [DOI] [PubMed] [Google Scholar]
- 10.Fosså A, Siebert R, Aasheim HC, Maelandsmo GM, Berner A, Fosså SD, Paus E, Smeland EB, Gaudernack G. Identification of nucleolar protein No55 as a tumour-associated autoantigen in patients with prostate cancer. Br J Cancer. 2000; 83:743–9. 10.1054/bjoc.2000.1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Comtesse N, Zippel A, Walle S, Monz D, Backes C, Fischer U, Mayer J, Ludwig N, Hildebrandt A, Keller A, Steudel WI, Lenhof HP, Meese E. Complex humoral immune response against a benign tumor: frequent antibody response against specific antigens as diagnostic targets. Proc Natl Acad Sci U S A. 2005; 102:9601–6. 10.1073/pnas.0500404102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gruenwald K, Castagnola P, Besio R, Dimori M, Chen Y, Akel NS, Swain FL, Skinner RA, Eyre DR, Gaddy D, Suva LJ, Morello R. Sc65 is a novel endoplasmic reticulum protein that regulates bone mass homeostasis. J Bone Miner Res. 2014; 29:666–75. 10.1002/jbmr.2075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zimmerman SM, Besio R, Heard-Lipsmeyer ME, Dimori M, Castagnola P, Swain FL, Gaddy D, Diekman AB, Morello R. Expression characterization and functional implication of the collagen-modifying Leprecan proteins in mouse gonadal tissue and mature sperm. AIMS Genet. 2018; 5:24–40. 10.3934/genet.2018.1.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heard ME, Besio R, Weis M, Rai J, Hudson DM, Dimori M, Zimmerman SM, Kamykowski JA, Hogue WR, Swain FL, Burdine MS, Mackintosh SG, Tackett AJ, et al. Sc65-Null Mice Provide Evidence for a Novel Endoplasmic Reticulum Complex Regulating Collagen Lysyl Hydroxylation. PLoS Genet. 2016; 12:e1006002. 10.1371/journal.pgen.1006002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaboreanu AM, Hrstka R, Xu W, Shy M, Kamholz J, Lilien J, Balsamo J. Myelin protein zero/P0 phosphorylation and function require an adaptor protein linking it to RACK1 and PKC alpha. J Cell Biol. 2007; 177:707–16. 10.1083/jcb.200608060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Romcy-Pereira RN, Erraji-Benchekroun L, Smyrniotopoulos P, Ogawa S, Mello CV, Sibille E, Pavlides C. Sleep-dependent gene expression in the hippocampus and prefrontal cortex following long-term potentiation. Physiol Behav. 2009; 98:44–52. 10.1016/j.physbeh.2009.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crockett DK, Fillmore GC, Elenitoba-Johnson KS, Lim MS. Analysis of phosphatase and tensin homolog tumor suppressor interacting proteins by in vitro and in silico proteomics. Proteomics. 2005; 5:1250–62. 10.1002/pmic.200401046 [DOI] [PubMed] [Google Scholar]
- 18.Baldin V, Lukas J, Marcote MJ, Pagano M, Draetta G. Cyclin D1 is a nuclear protein required for cell cycle progression in G1. Genes Dev. 1993; 7:812–21. 10.1101/gad.7.5.812 [DOI] [PubMed] [Google Scholar]
- 19.Li C, Xue C, Yang Q, Low BC, Liou YC. NuSAP governs chromosome oscillation by facilitating the Kid-generated polar ejection force. Nat Commun. 2016; 7:10597. 10.1038/ncomms10597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raemaekers T, Ribbeck K, Beaudouin J, Annaert W, Van Camp M, Stockmans I, Smets N, Bouillon R, Ellenberg J, Carmeliet G. NuSAP, a novel microtubule-associated protein involved in mitotic spindle organization. J Cell Biol. 2003; 162:1017–29. 10.1083/jcb.200302129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sánchez-Tilló E, Liu Y, de Barrios O, Siles L, Fanlo L, Cuatrecasas M, Darling DS, Dean DC, Castells A, Postigo A. EMT-activating transcription factors in cancer: beyond EMT and tumor invasiveness. Cell Mol Life Sci. 2012; 69:3429–56. 10.1007/s00018-012-1122-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moreno-Bueno G, Portillo F, Cano A. Transcriptional regulation of cell polarity in EMT and cancer. Oncogene. 2008; 27:6958–69. 10.1038/onc.2008.346 [DOI] [PubMed] [Google Scholar]
- 23.Li W, Ye L, Chen Y, Chen P. P3H4 is correlated with clinicopathological features and prognosis in bladder cancer. World J Surg Oncol. 2018; 16:206. 10.1186/s12957-018-1507-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiao J, Yang S, Shen P, Wang Y, Sun H, Ji F, Zhou D. Phosphorylation of ETV4 at Ser73 by ERK kinase could block ETV4 ubiquitination degradation in colorectal cancer. Biochem Biophys Res Commun. 2017; 486:1062–68. 10.1016/j.bbrc.2017.03.163 [DOI] [PubMed] [Google Scholar]
- 25.Dumortier M, Ladam F, Damour I, Vacher S, Bièche I, Marchand N, de Launoit Y, Tulasne D, Chotteau-Lelièvre A. ETV4 transcription factor and MMP13 metalloprotease are interplaying actors of breast tumorigenesis. Breast Cancer Res. 2018; 20:73. 10.1186/s13058-018-0992-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tyagi N, Deshmukh SK, Srivastava SK, Azim S, Ahmad A, Al-Ghadhban A, Singh AP, Carter JE, Wang B, Singh S. ETV4 Facilitates Cell-Cycle Progression in Pancreatic Cells through Transcriptional Regulation of Cyclin D1. Mol Cancer Res. 2018; 16:187–96. 10.1158/1541-7786.MCR-17-0219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barros-Silva JD, Paulo P, Bakken AC, Cerveira N, Løvf M, Henrique R, Jerónimo C, Lothe RA, Skotheim RI, Teixeira MR. Novel 5′ fusion partners of ETV1 and ETV4 in prostate cancer. Neoplasia. 2013; 15:720–26. 10.1593/neo.13232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tomlins SA, Mehra R, Rhodes DR, Smith LR, Roulston D, Helgeson BE, Cao X, Wei JT, Rubin MA, Shah RB, Chinnaiyan AM. TMPRSS2:ETV4 gene fusions define a third molecular subtype of prostate cancer. Cancer Res. 2006; 66:3396–400. 10.1158/0008-5472.can-06-0168 [DOI] [PubMed] [Google Scholar]
- 29.Jiang J, Wei Y, Liu D, Zhou J, Shen J, Chen X, Zhang S, Kong X, Gu J. E1AF promotes breast cancer cell cycle progression via upregulation of Cyclin D3 transcription. Biochem Biophys Res Commun. 2007; 358:53–58. 10.1016/j.bbrc.2007.04.043 [DOI] [PubMed] [Google Scholar]
- 30.Qi M, Liu Z, Shen C, Wang L, Zeng J, Wang C, Li C, Fu W, Sun Y, Han B. Overexpression of ETV4 is associated with poor prognosis in prostate cancer: involvement of uPA/uPAR and MMPs. Tumour Biol. 2015; 36:3565–72. 10.1007/s13277-014-2993-7 [DOI] [PubMed] [Google Scholar]