Abstract
The paradigm-changing opportunities of bio-logging sensors for ecological research, especially movement ecology, are vast, but the crucial questions of how best to match the most appropriate sensors and sensor combinations to specific biological questions, and how to analyse complex bio-logging data, are mostly ignored.
Here, we fill this gap by reviewing how to optimise the use of bio-logging techniques to answer questions in movement ecology and synthesise this into an Integrated Bio-logging Framework (IBF).
We highlight that multi-sensor approaches are a new frontier in bio-logging, whilst identifying current limitations and avenues for future development in sensor technology.
We focus on the importance of efficient data exploration, and more advanced multi-dimensional visualisation methods, combined with appropriate archiving and sharing approaches, to tackle the big data issues presented by bio-logging. We also discuss the challenges and opportunities in matching the peculiarities of specific sensor data to the statistical models used, highlighting at the same time the large advances which will be required in the latter to properly analyse bio-logging data.
Taking advantage of the bio-logging revolution will require a large improvement in the theoretical and mathematical foundations of movement ecology, to include the rich set of high-frequency multivariate data, which greatly expand the fundamentally limited and coarse data that could be collected using location-only technology such as GPS. Equally important will be the establishment of multi-disciplinary collaborations to catalyse the opportunities offered by current and future bio-logging technology. If this is achieved, clear potential exists for developing a vastly improved mechanistic understanding of animal movements and their roles in ecological processes, and for building realistic predictive models.
Keywords: Bio-logging, multi-disciplinary collaboration, movement ecology, multi-sensor approach, big data, data visualisation, Integrated Bio-logging Framework, accelerometer, GPS
Introduction
Movement is a fundamental aspect of life, intrinsically linked to almost every ecological and evolutionary process, from the acquisition of food, through reproduction and survival, to species distributions and community structure. Decades of technological developments have created vast possibilities in terms of data collection to study the movement of organisms, from VHF (Kenward, 2001), ARGOS and GPS technology (Kays, Crofoot, Jetz, & Wikelski, 2015; Tomkiewicz, Fuller, Kie, & Bates, 2010; Weimerskirch, 2009), to reverse GPS technology (Weiser et al., 2016) and dedicated satellite systems for tracking animals around the globe (ICARUS, see Wikelski et al., 2007), to sensor and acoustic receiver networks for animal tracking (Duda et al., 2018; Hoenner et al., 2018). In tandem, ecologists have driven a revolution in bio-logging sensor technology, motivated by the need to gather behavioural and ecological data that cannot be obtained through direct observation. This revolution has resulted in the development and use of a variety of sensors to observe the unobservable, including inter alia: accelerometers, magnetic field sensors, gyrometers, temperature and salinity sensors, further complemented by video cameras and proximity-loggers (Rutz & Hays, 2009) - see SI Table 1. The combined use of multiple sensors can provide indices of internal ‘state’ and behaviour, reveal intraspecific interactions, reconstruct fine-scale movements and even measure local environmental conditions (Rutz & Hays, 2009; Wilson et al., 2014). However, with increasing sensor possibilities comes a new challenge: pinpointing the appropriate information to collect, and finding efficient ways to do so.
It is hardly surprising, therefore, that there is an increasing number of high-profile reviews that showcase the paradigm-changing opportunities offered by animal-attached technology for ecological research (Hussey et al., 2015; Kays et al., 2015; Wilmers et al., 2015). Within these reviews, however, there exists scant treatment of how best to match the most appropriate sensors and sensor combinations to specific biological questions. As a result, ecologists have tended to use statistical methods post hoc to overcome the limitations of specific sensor data, including smoothing methods such as Kalman filtering and state-space models or machine-learning approaches applied to positional and accelerometer data. Similarly, although new analytical methods show great promise, such as the use of machine-learning to identify behaviours from tri-axial acceleration data (Nathan et al., 2012) or Hidden Markov Models (HMMs) to infer hidden behavioural states (Leos-Barajas et al., 2017), no clear guide exists to promote best practices. Such a guide would allow ecologists and statisticians to strike a balance between overly simplistic and complex models to deal with the vagaries of specific sensor data, for example the limitations of accelerometer data (see also Patterson et al., 2017). We aim to fill this gap by considering how to optimise the use of bio-logging techniques to answer key questions in movement ecology. In doing so, we identify four critical areas – questions, sensors, data, and analysis – and related opportunities for multi-disciplinary collaborations, and synthesise these into an Integrated Bio-logging Framework (IBF) to aid the decision-making process for ecologists. We then review the technologies and methodologies available to ecologists to make the links between nodes of the framework. We first consider how best to address biological questions using the most appropriate sensors while identifying current technological limitations. Second, we review the challenges and opportunities of linking new data types obtained from bio-logging sensors to the most adequate analytical techniques. We discuss issues relating to dealing with large, complex datasets, the fundamental properties of the new data types that can be collected, and the challenges of archiving and sharing bio-logging data. Finally, we discuss the value of multi-disciplinary collaborative links to optimise the opportunities offered by current and future bio-logging technology.
The Integrated Bio-logging Framework
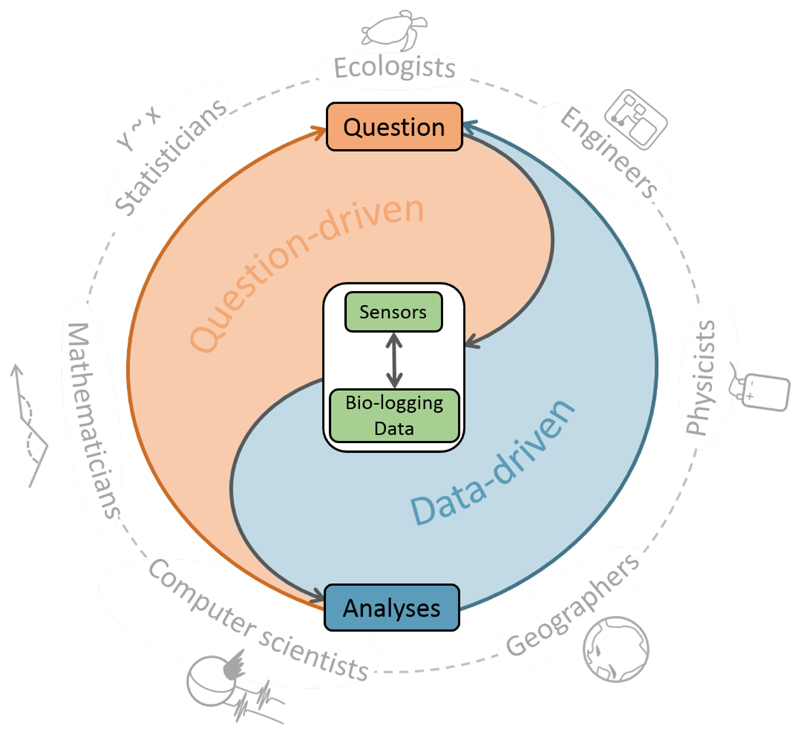
We connect the four areas critical for optimal study design via three-nodes in a cycle of feedback loops (Figure 1), linked by multi-disciplinary collaboration. Ecologists can work their way through the IBF to develop their study design – typically, this will start with the biological question, but the pathways will differ if, for example, using a question/hypothesis driven (blue) or data-driven (orange) approach. Figures 2 and 3 provide two such pathway examples.
Figure 1. The Integrated Bio-logging Framework (IBF) for optimal use of bio-logging in movement ecology.
Researchers may take a question-driven approach, beginning with a hypothesis, then selecting the appropriate sensor and analysis techniques. Alternatively, a data-driven approach can be taken, by allowing existing data to inform further hypotheses and data collection. The framework operates via collaboration between disciplines in a system of feedback loops, though these collaborative links are not exclusive to any particular node.
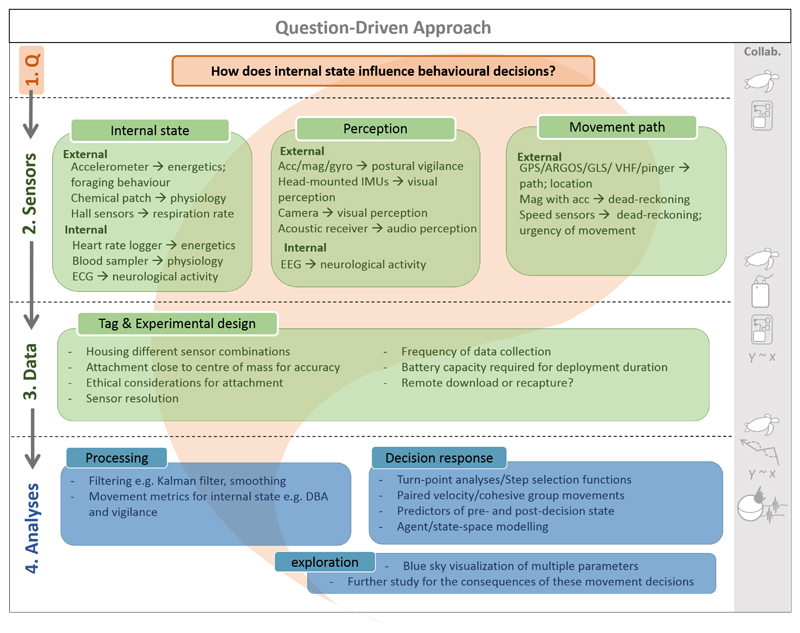
Figure 2. A question-driven approach to the IBF for optimal study design using bio-logging.
In this example, ecologists begin with their question of focus (top of Figure 1), in this case an investigation into the effect of internal state on movement decisions, and select the appropriate external and internal sensors for data collection. Here, sensors should be sensitive to different aspects of an animal’s movement that relate to their internal state, perceived information and the movement that may result from a particular decision. Selection of the sensors requires strong collaboration between ecologists and engineers (right-hand symbols). Simultaneously (bottom of Figure 1), ecologists should work with those analysing the data (e.g. physicists, mathematicians, statisticians, computer scientists) in the process of designing the data collection, to ensure the correct data are gathered that can answer the question using the analytic tools available.
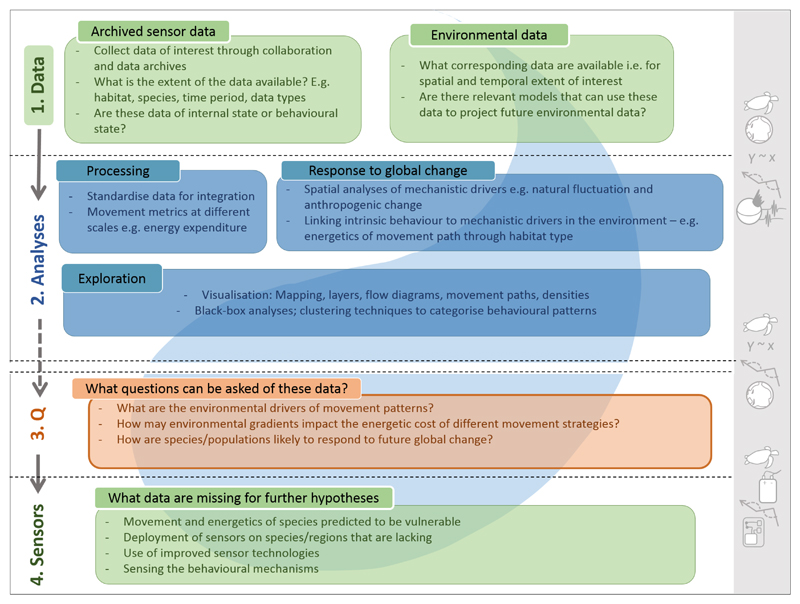
Figure 3. A data-driven approach to the IBF for optimal study design using archived bio-logging data.
In this example, ecologists begin by selecting appropriate data types for the study of movement patterns in relation to environmental measures at local and global scales. Understanding and predicting how animals respond to global change, including climate and land-use change, requires multiple data collected over a range of temporal and spatial scales. In this case, ecologists start at the central nodes of the IBF (Figure 1) to collate archived data and collaborate with mathematicians, statisticians and geographers (right-hand symbols) to implement the appropriate processing and analytical techniques to interrogate the data and identify patterns by which several questions may be approached. Following this, ecologists may work with other disciplines to deploy additional bio-logging sensors to collect data that complement the shared data.
Furthermore, bio-logging has become so multifaceted and complex that no-one can be a ‘master of all trades’, hence, establishing multi-disciplinary collaborations is key (as for other disciplines, Peters et al., 2018), and this idea is at the basis of the IBF. For example, at the study inception phase, dependent on the biological problem addressed, physicists and engineers can advise on sensor types, their limitations and power requirements, while mathematical ecologists and statisticians can aid in framing the study design and modelling requirements for specific questions (see Figure 2). Development of bio-logging tags is the result of collaborations between engineers, physicists and biologists, while visualisation and analytical methods for dealing with data are aided by interactions with computer scientists, geographers, statisticians and mathematicians (see Figure 3). On the other hand, ecologists can guide researchers from the other disciplines towards the key methodological hurdles and technological limitations which are hindering progress and need to be addressed.
We now review the literature regarding questions, sensors, data and analyses, and exemplify the links between the nodes of the IBF. We conclude by highlighting areas for future development.
1. From questions to sensors
Researchers can choose between an ever-increasing number of different bio-logging sensors (Table 1, SI Table 1). Following the adage that experimental design should be guided by the questions asked (e.g. Fieberg & Börger, 2012; Hebblewhite & Haydon, 2010), sensor choice is clearly critical. Here, we consider sensor selection within the general scheme of key movement ecology questions posed by Nathan et al. (2008) and provide an example for the use of the IBF in a question-driven approach to study design (Figure 2).
Table 1.
Summary table of the current bio-logging sensors available, beyond classic location sensors. The detailed application and description of sensors is provided in SI Table 1.
| Sensor type | Examples | Description | Relevant questions | Optimisation |
|---|---|---|---|---|
| Location | Animal-borne radar, pressure, passive acoustic telemetry, proximity sensors | Location based on receiver location | Space use; interactions |
Use in combination with the behavioural sensors below; Create visualisations to facilitate interpretation of 3D space use and interactions |
| Intrinsic | Accelerometer, magnetometer, gyroscope, (gyrometer) | Patterns in body posture, dynamic movement, body rotation and orientation. | Behavioural identification; internal state; 3D movement reconstruction (dead-reckoning); energy expenditure; biomechanics; feeding activity; space use |
Use in combination with other intrinsic sensors to build up detail of behaviour and/or 3D path reconstruction; Increased sensitivity to detect micro-movements or stress-related activity; high resolution (temporal and spatial) environmental data may improve accuracy of path reconstruction (e.g. in relation to environmental flow, wind or current data) |
| Heart rate loggers, stomach temperature loggers, neurological sensors, flexible speed paddle, pitot tube, speed paddles |
Measures of activity. | |||
| Microphone, hall sensors, | Specific behaviour e.g. limb movement and vocalisations | |||
| Environment | Temperature | Ambient | Space use; energy expenditure; external factors; interactions |
In situ remote sensing; arrays to localise animals; visualisations to provide context and understanding of interactions |
| Microphone, proximity sensors, video loggers | Record external environment e.g. soundscape |
1.1. Where is the animal going?
ARGOS, GPS and related satellite and global navigation systems, as well as acoustic tracking arrays and geolocators, have revolutionised information on animal locations and movements (Kays et al., 2015). Bio-logging sensors, particularly in combination with such locational tracking-devices, can further help detect where animals move. For example, Hedenstrom et al. (2016) combined geolocator and accelerometer tags to record flight behaviour of migrating swifts, and Shipley, Kapoor, Dreelin, & Winkler (2018) used micro barometric pressure (altitude) sensors (<0.5 g) to uncover the aerial movements of migrating birds. A key limitation of telemetry devices is that transmission technology can fail, such as when canopy cover impedes GPS satellite fixes (Rempel, Rodgers, & Abraham, 1995). However, with the combined use of inertial measurement units (IMUs) and elevation/depth recording sensors it is possible to reconstruct animal movements in 2D and 3D using a dead-reckoning procedure, irrespective of transmission conditions (Bidder et al., 2015; Bramanti & Dall’Antonia, 1988). This uses the speed (including speed-dependent dynamic body acceleration (DBA) for terrestrial animals; Bidder, Qasem, & Wilson, 2012), combined with animal heading (from magnetometer data) and change in altitude/depth (pressure data) to calculate the successive movement vectors (oriented steps) from a known starting position.
The process gives extraordinarily finely resolved relative movement (it can, for example, determine how many times a dog has walked around a tree in scent-marking activity) but it can accumulate errors over time, especially in fluid media with current flow. Therefore, data used in dead-reckoning need correcting with frequent ground-truthing, such as by a GPS unit (Bidder et al., 2015). GPS-enabled dead-reckoning is an incredibly powerful combination of sensor systems which may become paradigm-shifting within animal movement studies. With this, researchers will have access to multiple scales of movement and seamless animal movement descriptors and will be able to identify true turn-points in the data (Potts et al., 2018; see Figure 2 and section 3.3 below). In turn, the improved track trajectory should allow us to connect behaviour to landscape ecology and population dynamics with increased confidence (Morales et al., 2010).
1.2. How is the animal moving?
At the smallest scale (locomotion), animals move according to their anatomy and the biomechanics that this engenders, with obvious differences between animals operating in fluid media (air or water) or on the ground (Biewener & Patek, 2018). In essence, locomotion is manifested by particular patterns of movement by the various body parts (most notably limbs) so that motion-sensitive transducers can provide critical information with respect to the pattern and intensity of movements and thereby derive critical whole-animal movement parameters such as speed and direction. The primary sensors used for this include accelerometers, magnetometers and gyrometers (often collectively grouped within inertial measurement units [IMUs]; e.g. Noda, Kawabata, Arai, Mitamura, & Watanabe, 2014). Accelerometers and magnetometers can be used to infer the 3D posture and orientation (i.e. azimuth, elevation angle and bank angles; see SI Table 1 for a glossary of terms) during locomotion, whereas gyrometers provide direct measures of yaw, pitch and roll (see Benhamou, 2018 for the mathematical relationships between these parameters). In addition, various iterations of speed-detecting systems, such as anteriorly mounted propellers (Ropert-Coudert et al., 2000; Watanabe et al., 2008), flexible paddles (Shepard, Wilson, Liebsch, et al., 2008), and Pitot tubes are also used (Taylor, Reynolds, & Thomas, 2016). Importantly, the speed at which an animal is moving provides information on the urgency with which the movement is being undertaken. When moving animals deviate from minimum cost of transport (cf. Schmidt-Nielsen, 1972), it indicates time-based selection pressures that incite animals to move non-optimally in energetic terms; the reasons for which may be critical for lifetime fitness and only become apparent post hoc (e.g. Shepard, Wilson, Quintana, Laich, & Forman, 2009). Sensors are thus required to record information from which we can quantify the energetics of animal locomotion, as well as the costs and benefits of behaviours. Several sensors provide proxies for oxygen consumption (VO2), including heart rate loggers (Green, 2011) and tri-axial accelerometers through the computation of dynamic body acceleration (DBA; reviewed in Wilson et al., 2019). Indeed, the continued refinement of these proxies of power use, one of the most fundamental currencies in the animal kingdom, will be pivotal in providing critical, missing information within previously established movement frameworks such as optimal foraging (McNamara & Houston, 1986; Pyke, 1984).
Sensors that detect body movements may also provide key information relating to biomechanical questions, such as how stroke frequency relates to stroke amplitude. For example, magnets used with Hall sensors (sensors detecting magnet-transducer paired magnetic field properties; Hall, 1879 - see S1 Table 1) can quantify the amplitude, angular velocity and frequency of limb movements of marine mammals (Wilson & Liebsch, 2003), providing insights into energy-saving mechanisms (Nassar, Jackson, & Carrier, 2001). Animal-borne video or audio may provide similar information, for example, being able to relate flipper beat frequencies in green turtles (Chelonia mydas; Hays, Marshall, & Seminoff, 2007) and emperor penguins (Aptenodytes forsteri; van Dam, Ponganis, Ponganis, Levenson, & Marshall, 2002), to dive strategies. Hall sensors can also measure respiration rates and extent of inhalation, heart rates, and even patterns of defaecation, providing information on the optimal breathing strategies and rates of digestion (Wilson et al., 2003, 2004), cases where mounted-accelerometers would be limited due to movement being mainly translocational. In addition, these behaviours can also be detected from on-board videos. Yet few studies use these techniques, perhaps because researchers find the magnetic field intensity drop off with distance intractable and because, at the time the studies were published, it was not possible to study angular changes between magnet and sensor, if distances were held constant. Inertial measurement units (IMUs) have changed this, so we think that the future of miniature IMUs holds promise for researchers to document minute changes in body movement and for quantifying motion capacity from limb movements.
1.3. What is the animal doing?
Allocating behaviours to space is key to understanding animal niche requirements and the link between behaviour and fitness consequences. Since the work by Yoda et al. (1999) using accelerometers to determine animal behaviour, there is a rich and varied literature that documents increasingly successful methodologies for determining animal behaviour from various sensor data, especially accelerometers (Nathan et al., 2012; Shepard, Wilson, Quintana, et al., 2008) and magnetometers (Williams et al., 2017). Thus, it is now possible to extract a remarkable amount of information regarding behaviour, beyond that of limb and body part movement as detected from tri-axial sensors.
In particular, quantifying the type and amount of food ingested by animals is essential to answering some of the “big questions” in movement ecology such as how animals manage their energy budgets in the wild (cf. Krebs & Davies, 1978). For example, combining GPS and DBA measures derived from tri-axial accelerometers, allows us to better understand the energetics underlying prey capture behaviour of large terrestrial predators (Wilmers, Isbell, Suraci, & Williams, 2017), while the drift and buoyancy inferred from time-depth recorders can provide information on the foraging success of marine predators (Abrahms et al., 2018). A further refinement is provided by indirect parameters such as those obtained by means of sensors that detect stomach, oesophageal or visceral temperature, which can provide invaluable insights into actual prey captures (Weimerskirch, Gault, & Cherel, 2005; Weimerskirch, Pinaud, Pawlowski, & Bost, 2007; Wilson, Cooper, & Plötz, 1992). An intriguing alternative is based on attaching a Hall sensor to one mandible opposite a magnet attached on the other mandible (but the ethical implications and feasibility must be well considered). The inter-mandibular angle can be determined by measuring changes in magnetic field strength (Wilson, Steinfurth, Ropert-Coudert, Kato, & Murita, 2002). This approach, which has been employed in several marine and terrestrial species, can provide information about both the number of food items and the type of food ingested (Ropert-Coudert et al., 2004). Indeed, such is the detail provided by these sensors that studies are now able to examine food acquisition within a probabilistic framework and thereby make predictions about how food abundance may affect populations (Wilson, Neate, et al., 2018).
Obtaining direct observations may sometimes be essential, either because robust calibration of bio-logging sensors is difficult, or because the study’s aim is to document particular behaviours in great detail (such as prey captures and social interactions; McInnes, McGeorge, Ginsberg, Pichegru, & Pistorius, 2017; Pagano et al., 2018; Watanabe & Takahashi, 2013) or to prospect for undiscovered behaviours (such as unusual foraging techniques; Rutz, Bluff, Weir, & Kacelnik, 2007). Under these circumstances, video loggers are the method of choice, or still-image loggers, if longer recording times are required and a lower frame rate is acceptable. Cameras may also offer the opportunity to assess what a wild animal sees in the field (Moll, Millspaugh, Beringer, Sartwell, & He, 2007) so that environmental information can be factored into foraging efficiency (Sutton, Hoskins, & Arnould, 2015) and movement patterns studied with respect to visual stimuli (Tremblay, Thibault, Mullers, & Pistorius, 2014). Video loggers can also be combined effectively with other sensors such as accelerometers (Watanabe & Takahashi, 2013), and are small enough to be fitted to a wide range of species (see below).
1.4. Why is the animal moving?
Animals make behavioural decisions based on their internal ‘state’ (physiological and psychological condition), and external biotic and abiotic factors (Nathan et al., 2008). Identifying and quantifying how internal state may drive behaviour is non-trivial, and can often only be indirectly inferred (Getz & Saltz, 2008). Some aspects of animals’ internal state have been investigated using accelerometers which are sensitive to micro-movements and postures indicative of chemical, disease, and affective states (Downey et al., 2017; Wilson et al., 2014), including vigilance behaviour, a stress-related response (Kröschel, Reineking, Werwie, Wildi, & Storch, 2017). Alongside accelerometers, other key sensors that can provide insights into internal state include heart rate, internal temperature, and neurological sensors (Rattenborg et al., 2016). For example, heart rate loggers to investigate the interplay between ecological pressures and energetic strategies were used by Bishop et al. (2015) in bar-headed geese (Anser indicus) and O’Mara et al. (2017) in fruit-eating bats (Uroderma bilobatum). As another example, Ditmer et al. (2018) used heart rate loggers to investigate how American black bears (Ursus americanus) perceive the risks of crossing roads. Research on humans has demonstrated that bio-loggers can measure a suite of physiological variables relating to internal state (Nikita, 2014; Yang, 2014) and the development of similar systems for wild animals is increasing; examples include animal-borne blood sample collection devices for stress hormones in seals (Takei et al., 2016), other hormonal sensors (Landry et al., 2014), and internal chemical detection nanosensors for freely moving animals (Lee et al., 2018).
The greatest insight into state-driven movement is likely to be gained from multi-sensor approaches (e.g. Wilson, Littman, Halpin, & Read, 2017), especially combining both physiological and/or neurological sensors with position-determining systems (Figure 2). For example, Vyssotski et al. (2006) simultaneously measured pigeon (Columba livia) movement and electrical brain activity using a miniaturised GPS combined with an electroencephalography logger, while Dunn et al. (2016) obtained a brain-wide mapping of neural activity of zebrafish (Danio rerio) during movement. The use of neurological sensors to monitor brain activity in freely moving animals is a relatively new advancement (e.g. Rattenborg et al., 2016; Skocek et al., 2018). Such multi-sensor developments are helping to meet the challenge of linking internal state, as a proximate cause of movement, to ultimate evolutionary causes (Nathan et al., 2008). However, there are important ethical considerations to be raised, especially for surgically implanted sensors (e.g. the example of frigatebirds, Rattenborg et al., 2016).
Alongside the internal state, what is happening in the environment is the other prime driver of animal movement. Global environmental data can be recorded through satellite remote sensing, and bio-loggers now routinely collect local environmental data both biotic and abiotic (Table 1; SI Table 1), thus a major aspiration is to link such data to movement. Though whilst ecologists can access an increasing amount of remote-sensed environmental data, linking them to location data is usually difficult, as environmental data are obtained at different, generally coarser, spatiotemporal scales than movement data (Dodge et al., 2013). Remelgado et al. (2019) recently developed a new pixel-based approach, combined with data mining and visualisation, to help ecologists efficiently deal with differences in the spatial, temporal and thematic resolutions between environmental data from remote sensing and GPS location data; yet the problem persists with high frequency bio-logging data.
Depending on the question asked, it may be necessary to use modelling to derive high-accuracy dynamic maps of environmental conditions (e.g. vertical wind; see Scacco, Flack, Duriez, Wikelski, & Safi, 2019), or to use drones or LiDAR, to build ultra-high resolution, 2- and 3-dimensional maps of the study area (e.g. to investigate movement costs due to elevation or to quantify vegetation quality for optimal foraging questions). Importantly, bio-loggers allow the collection of high-frequency environmental data at the local scale experienced by the animals, such as temperature, light intensity, and wind or current velocity (Block, 2005; Dodge et al., 2013; Piersma & Lindström, 2004). This may be complemented by implanted sensors such as core body temperature sensors (e.g. when studying heat stress questions), combined with sophisticated use of meteorological data to estimate the so-called wet bulb globe temperature index (WBGT), a key measure of heat stress (Dimiceli, Piltz, & Amburn, 2011).
In terms of the biotic environment’, an animal’s movement decisions are likely to be influenced by interactions with conspecifics and heterospecifics and again, there are certain combinations of sensors that can record and help identify these interactions. There are two main approaches to remotely record the social contact between free-ranging animals: indirect and direct encounter mapping (see Bettaney, James, St Clair, & Rutz, 2015; Krause et al., 2013). Indirect encounter mapping can be achieved either with high-resolution tracking of subjects, or with the use of tags that transmit to, or that are detected by, fixed receiver stations at specific locations (e.g., coded VHF radio-tags or PIT/RFID tags). In both cases, the co-occurrence of animals is inferred at the data analysis stage. Direct encounter mapping, on the other hand, requires the use of proximity loggers (transceiver tags that both transmit and receive radio signals between animals) or camera tags (Hooker, Barychka, Jessopp, & Staniland, 2015), to create reciprocal records of social encounters (Bettaney et al., 2015; Krause et al., 2013). Proximity-loggers can be used for addressing a variety of biological questions, and have the advantage over cameras (e.g. Takahashi et al., 2004) that they survey in all directions (even though precise directional and distance information is often not collected), but their key strength lies in charting social associations of a large number of subjects of known identity, to reconstruct group, community, or even population-level social networks. Proximity sensors can also be used to record interspecific encounters, for example between predators and their prey, between different disease hosts, or in mixed groups of foraging or migrating animals. Some systems are set up as wireless sensor networks where animal-mounted sensors not only communicate with other sensors, but also with (a large number of) stationary receiver (base) stations (Rutz et al., 2012). This enables near real-time data transmission, which is key to evaluating system performance and to planning and monitoring experimental manipulations (St Clair et al., 2015).
A particular type of interspecific interaction occurs when animals interact with human activities, which can strongly affect animal movements (e.g. Tucker et al., 2018). An interesting development is animal-borne radar detectors, which detect signals from emitting radars in the surroundings and can be used in combination with a tracking device to log the occurrence of structures along an animal’s movement path (Table 1; SI Table 1). This has facilitated the study of seabird-fishing vessel interactions, quantifying attraction, attendance and foraging behaviour (Weimerskirch, Filippi, Collet, Waugh, & Patrick, 2018).
2. From sensors to data
Data collection and analysis issues must be addressed alongside sensor selection when approaching a specific ecological question. The first challenge concerns finding the most appropriate experimental/sampling design to answer a given ecological question. More broadly (see the internal data node of the IBF), this concerns the closely related issues of tag design and data management (which includes planning for data archiving and sharing) – all of which must be defined prior to tag deployment. The experimental design will strongly benefit from interdisciplinary collaborations to find the best solution, ensuring that the data-gathering is both feasible and will lead to sufficient data to answer the questions using available analytic techniques.
2.1. Experimental design
Consideration of an appropriate sampling regime prior to tag deployment, so as not to over-, or under-sample and maximise battery duration (and minimise tag weight), is a crucial aspect (note that battery power is required both to interrogate the sensors and write the data to memory, and possibly send the data). To do so, researchers should apply the Nyquist or sampling theorem, which states that the sampling frequency should be at least twice the fastest frequency of interest; e.g. consider wingbeat frequency vs. amplitude as focus of interest. This holds true in temporal and spatial domains (see discussion in Ropert-Coudert & Wilson, 2004). An obvious consequence of this trade-off is the use of smart sampling, whereby the sensors record at a frequency able to elucidate the relevant aspect properly, but no more. We do note, however, that highly prescribed, low frequency sampling may miss serendipitous observations of importance and may preclude the detection of new, never observed behaviours. Furthermore, derivation of body motion or measures of energy expenditure (DBA) requires smoothing of accelerometer data at an appropriate frequency (Shepard, Wilson, Halsey, et al., 2008), albeit the latter could indeed be processed on-board without storing the high frequency data (e.g. Cox et al., 2018). For example, a high frequency recording of raw data (> 20 Hz) may be necessary to compute animal posture and DBA (see also Brownscombe, Lennox, Danylchuk, & Cooke, 2018); however, higher frequencies draw more current (doubling the frequency from 20 Hz to 40 Hz might require to double or more the battery size/capacity, and hence weight, depending on the specifics of the sensors), thus a balance between behaviour resolution, information gain, and current draw is a key stage of experimental design. Equally important, when using IMUs featuring multiple sensors, might be to set different frequencies for different sensors, such as a higher frequency for accelerometers (40 Hz), a lower frequency for magnetometers (20 Hz), and an even lower frequency for temperature or pressure sensors (e.g. 4 Hz). Such settings can more than double the time a logger can record on a given battery size (note also that differences between battery types in the capacity to respond to peak current demands from the sensors can further affect the longevity of loggers), but preliminary studies for different study species, and interactions with engineers, might be required to find the best settings. Another area of current research (e.g. see Cox et al., 2018) is focussing on finding clever ways to store on-board only sub-sampled or summary data, rather than the raw high-frequency data, thereby reducing data storage requirements and, ideally, allow remote transmission of the data (often the latter is precluded for field studies due to the high power requirement). Closely related is the choice of sensor resolution (bit resolution, see discussion and examples in Ropert-Coudert & Wilson, 2004). The number of bits with which the data are stored directly determines the quality of the data obtained. For example, past loggers used an 8-bit resolution, meaning the sensor can obtain an absolute resolution given by the maximum resolution range divided by 256. In the case of a depth pressure transducer with a maximum range of 50 Bar this means a maximum resolution of circa 0.2 Bar, equal to resolving dive differences of 2 m (a 16-bit resolution allows instead to resolve steps of 0.008 m, see Ropert-Coudert & Wilson, 2004). Low resolution may preclude recording key information such as prey capture events. Equally important is the measurement range of the sensor. For example, an accelerometer which records up to 8 g will miss any data of animals moving more dynamically (e.g. head impacts) and unless the animals are known to be only relatively slow moving and good preliminary data exist, researchers should set the range to at least 16 g for initial studies (for terrestrial systems; a lower range may be sufficient for aquatic systems as, due to friction, movement speed may change less fast), and record this information in the metadata. Equally important are trade-offs between the quantity of data collected and the time a tag collects data on an individual, as well as trade-offs between the amount of data collected on single individuals against the number of different animals monitored across time and space (see also Hebblewhite & Haydon, 2010). Collaborations across disciplines are crucial to make such decisions.
2.2. Tag design
Reducing battery consumption not only extends the life of a bio-logging device, but has implications for tag size and attachment that should also be considered for both optimal study design and animal welfare. Reduction of tag size is paramount, yet even with recent advances in the reduction of sensor size, it is still battery size that limits that of the device. Note also that further evidence demonstrates that for some applications the total mass of the tag together with the animal is more important than the relative tag mass (Tomotani et al 2019). For cameras for example, current available loggers are small enough, at approximately 10 g, to be fitted to a wide range of species (Rutz et al., 2007). However, even state-of-the-art camera loggers remain severely battery limited, hence duty cycling is advisable for most applications, as this allows targeted data collection during periods of peak activity and/or repeated short-term recording over the course of several days (Rutz & Troscianko, 2013). An exciting recent development is the use of event-triggering technology that allows cameras to be switched on whenever particular behavioural states or environmental conditions are detected (see analysis section below). To provide an example, whilst miniature VHF tags weighing 0.5g may allow to track the movements of an animal for up to a month, a battery one or two orders of magnitude heavier would be required to record high-frequency accelerometer and magnetometer data for the same duration. On the other hand, a GPS running at 1 Hz may require between 30-50 mA of current, where as a modern IMU recording tri-axial acceleration and magnetometer data at 40 Hz requires only 5-10 mA of current (Bidder et al. 2014). More generally, close attention in required on how size, longevity, and attachment differ among different tags/sensors. Body et al (2017) provide a useful key for assessing device impacts prior to tag deployment, and a growing body of literature now highlights the importance of tag shape and attachment in terms of affecting an animals drag in aerial and marine environments (e.g. Kay et al 2019, Lear et al 2018, Vandenabeele et al. 2014).
2.3. Data management
A further consideration for optimal experimental design is that of data management and processing. The data provided by sensors often do not correspond directly to the information we look for, but to a proxy, which needs to be converted. For instance, a depth recorder is designed to provide a measure of pressure rather than a measure of depth, but underwater depth being linearly related to pressure, the conversion is straightforward. For other sensors, this is not so obvious, and raw data therefore require being pre-processed. For example, acceleration data do not provide a direct estimate of energy expenditure or oxygen consumed while moving. First, the dynamic component has to be extracted from the raw acceleration values, then converted to DBA, which finally has to be correlated with energy or oxygen through controlled lab experiments (reviewed in Wilson et al., 2019). Pre-processing is also required for integrating data provided by different sensors, possibly at different rates, and often based on separate clocks (exposing systems to clock drift); although inertial measurement units (IMUs) effectively deal with temporal synchronisation within any one logger. Notably, data recorded at high frequency are both noisy and highly serially auto-correlated. Noise can be reduced by filtering, e.g. by taking a running mean, or may involve more complex approaches such as Fourier transformations or Kalman filtering (e.g. Alam & Rohac, 2015). A simple and efficient solution consists of sub-sampling the processed data to a level (or deriving averages) to accord with the Nyquist frequency. Pre-processing should be performed before subsampling, although there is an element of feedback depending on the desired end-point, which may also need to be considered when selecting the sampling frequencies for the different sensors and their data types, which also has important implications for data archiving (see next section).
2.4. Data archiving and sharing
Bio-logging data also present considerable challenges for data sharing and replicability. One challenge lies in the lack of standardised protocols for data recording from animal-borne sensors (Campbell, Urbano, Davidson, Dettki, & Cagnacci, 2016). Logging data require very detailed metadata on the attachment type and position on the animal of the loggers, as otherwise, establishing a close relationship between the output from sensor data (such as triaxial accelerometer) and the orientation and posture of the animal, will be near impossible. Furthermore, whether or not to keep both the pre- and post-processed versions of the data (particularly before or after filtering and subsampling) is something to consider in terms of not only the current question and analyses, but also for the long-term goals of archiving data in the best format available to allow long-term use of those data. Thus, there is an urgent need to improve data protocols and database standards for bio-logging data. Indeed, the International Bio-Logging Society is actively working towards that goal. Efficient data sharing and archiving across many studies and authors will be key to answer the big questions in movement ecology, e.g. global responses to environmental change (Figure 3), and will reduce the need to collect new data (see also section 4).
3. From data to analysis
Data analysis issues must be addressed upfront alongside sensor selection and experimental design, to ensure the resulting data are sufficient for the proposed mathematical models and statistical tests used to infer biological information from the data. This requires strong inter-disciplinary collaborations between empiricists and theoreticians from the outset of the project. The first major challenge for the link between data collection and analyses in the IBF is the ‘big data’ problem. Rapid advances in bio-logging technology now provide information-rich, big data sets, even from single individuals, thus the challenges in data analyses are similar to those of ‘big data’ and ‘data science’ problems in ecology and other scientific disciplines (Hampton et al., 2013; Lewis, Vander Wal, & Fifield, 2018; Thums et al., 2018). There is an urgent need for the use and development of more sophisticated and computationally efficient data visualisation and exploration methods, as well as mathematical models that incorporate multidimensional bio-logging data.
3.1. Data Visualisation
A key part of exploratory data analysis consists in devising efficient ways to visualise and display quantitative information (Tukey, 1977), especially to aid exploration in this data-rich era. Data visualisation converts complex patterns in data into a visual display, capitalising on the extraordinary capacity of the human visual system to pick out patterns in complex landscapes, and thereby provide insights into data relations (Ware, 2012). While ecologists often develop their own visualisation tools, many methods come from other disciplines such as geographic information science (Demšar et al., 2015; Li, Wu, Song, & Zhou, 2016), medicine and neuroscience (with complex fMRI data e.g. de Ridder, Klein, & Kim, 2017).
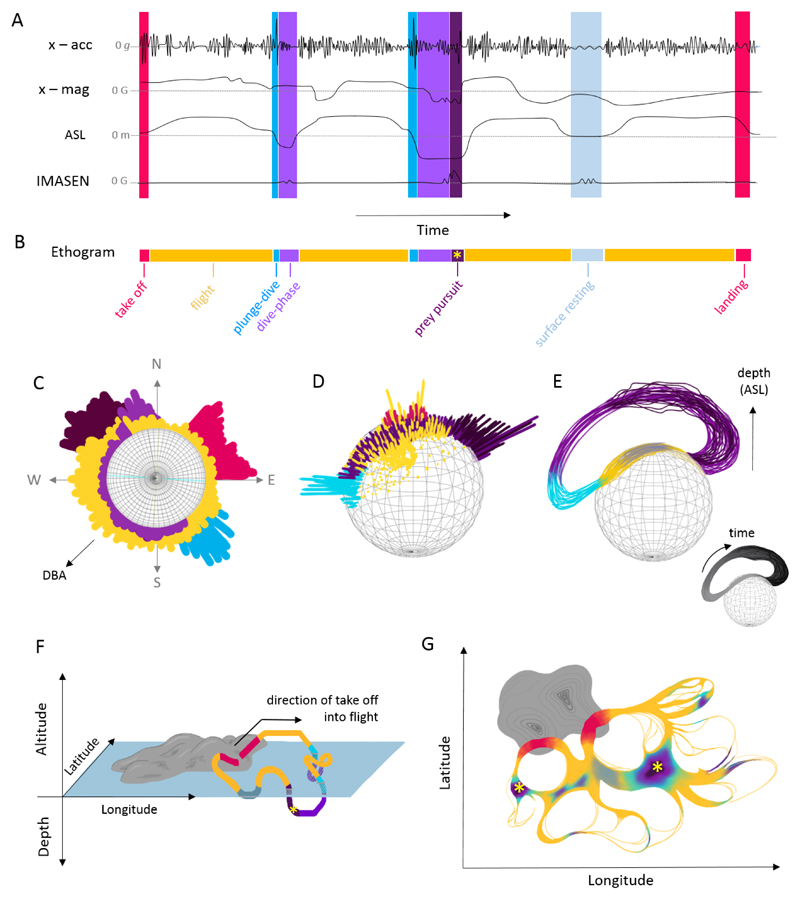
Conventionally, acceleration data tend to be visualised as time series plots (Figure 4A), with analyses based on summary statistics (derived from ethograms; Figure 4B) and the application of data transformations. While such approaches are useful for classification of time series data (Walker et al., 2015), integration of multi-sensor data are poorly covered by this approach (Lee & Jeong, 2017; Li et al., 2016; Walker, Borgo, & Jones, 2016), primarily due to time taking up one axis and constraining all other data to lie within its scaling and bounds. Other visualisations may bypass the time scaling factor by having spherical plots that present 3-dimensional scatterplots, histograms, clustering data by behavioural state (Grundy, Jones, Laramee, Wilson, & Shepard, 2009; Williams et al., 2017; Wilson et al., 2016; Figures 4C-E). The value in these spherical plots is that they are also multi-layer and present environmental data such as pressure and temperature as well as metrics of energetic expenditure (Roberts, Laramee, & Jones, 2015). In addition, time can also be represented, if necessary, by glyph or line colour (Figure 4E). Thus, such visual analytics systems can be linked interactively to allow different aspects of the same data to be explored, with and without temporal and spatial scales. In terms of sensor data this includes plots in tri-axial space with further dimensions related to movement and performance metrics (e.g. Roberts et al., 2015) and those that combine multi-dimensional trajectory visualisations on a map with environmental data (e.g. Buchin et al., 2015; Shamoun-Baranes et al., 2016; Figure 4F) and temporal visualisations (Demšar et al., 2015) such as DynamoVis (Dodge, Xavier, & Wong, 2018; Xavier & Dodge, 2014) and trackplot (Ware, Arsenault, Plumlee & Wiley, 2006) or flow visual analytics systems (Andrienko, Andrienko, Chen, Maciejewski, & Zhao, 2017; Graser, Schmidt, Roth, & Brändle, 2017; Figure 4G). Time is also commonly visualised through animation and there are two R packages that support this (albeit for traditional location-only data, not logger data): moveVis (Schwalb-Willmann, 2018) and anipaths (Scharf, 2018). See supplementary information for a detailed list of current visualisations (SI Table 3). Current developments indicate that it may be possible to bring these multi-dimensional plots into an interactive 3-dimensional lab space beyond a digital screen, which would dramatically help exploration of data and even advance behavioural studies through the manipulation of the virtual world (see Stowers et al., 2017). Equally important will be the development of improved ways to display results from machine-learning methods (see below); again, an area for which multi-disciplinary collaborations will be crucial.
Figure 4. Visualisation of sensor and location data.
A number of schematic plots of varying axes and information types to visualise data of a seabird in flight that plunge-dives in pursuit of prey. A) Logged sensor outputs (acceleration (g), magnetometry (µT), altitude above sea level (m) derived from pressure data (kPa) and the inter-mandibular angle sensor IMASEN output (µT)) in a time series plot. Peaks in dynamic acceleration are associated with wing beats during take-off (red) and in flight (yellow), as well on impact with the sea surface in plunge-dives (aqua blue). During the dive, as indicated by the negative altitude above sea level (ASL; purple) the bird may pursue prey (dark purple), as indicated by increased variation in acceleration and heading, from the magnetometer output. A successful prey capture attempt is evident in the peaks in the IMASEN signal output, as the bird opens its bill to capture the prey (yellow asterisk). B) The behaviours are classified and presented in an ethogram to show temporal variation in behaviour (this serves as a key for the schematic). Further to these time series plots, different sensor outputs can be combined, along with derived metrics, in various multi-axes visualisations to reveal patterns in behaviour. We present three examples (C-E) for data visualisation in multi-dimensional space and two for geographic space (F-G): C) a circular plot of heading on an m-sphere (magnetometry; Williams et al., 2017), where height of the bar is the magnitude of the extent of movement (DBA), the most active behaviours for this bird are foraging and diving, which occur at opposite headings; D) a g-sphere (static acceleration data) or Dubai plot, where a frequency histogram of static acceleration is resolved in tri-axial space (Wilson et al., 2016) and peaks show the most common postures for each behaviour; E) a g-sphere where distance from the surface of the sphere is relative to the depth below sea level, where colour indicates different behaviours in the dive, so that through the dive there is a shift in posture, and a greater variation in posture and depth during the prey pursuit (coloured by time in greyscale, bottom right); F) 3D movement path during for the foraging trip; G) 2D flow visualisation of foraging path, where thicker paths are more commonly used for the different behaviours (Verbeek, Buchin, & Speckmann, 2011).
3.2. Behavioural Classification
Behavioural classification involves identifying particular behaviour-linked signals within complex datasets, such as accelerometer and magnetometer data. This may involve searching for behaviour-linked thresholds, such as an increase in pressure to indicate diving (Kooyman, 1964) but more commonly will involve consideration of multiple data streams (Viviant, Trites, Rosen, Monestiez, & Guinet, 2010; Yoda et al., 2001), which makes the process more complex. For this reason, much emphasis has recently been placed on machine learning algorithms (including K-Nearest Neighbour [KNN], Support Vector Machines [SVMs], Classification and Regression Trees [CART], and Artificial Neural Networks [ANNs]) to classify behaviours automatically (Nathan et al., 2012). Supervised machine-learning models are trained with segments of data that have been manually labelled according to behaviour (Carroll, Slip, Jonsen, & Harcourt, 2014; Watanabe & Takahashi, 2013). The convenience of machine-learning systems is that they require little specialist knowledge and information about the data streams from the researcher. Against this, there is a tendency to put all primary data streams as well as derived elements (such as DBA metrics) into the process. Because the machine does not know which data streams are most relevant at the outset, processing times can be prohibitively long. An approach that attempts to deal with this uses a Boolean framework and requires that the researchers have enough specialist knowledge to be able to pick out a sequence of features in behaviours (systematic variation and direction in data streams over defined time periods) to be able to define the behaviour in a series of key elements. These are then defined in an algorithm and the computer made to search for exactly those conditions to define the behaviour (Wilson, Holton, et al., 2018). The obvious downside to this approach is the level of expertise of the user and familiarity with the meaning of the data streams, which highlights the crucial role ecologists and biologists have to play in making sure analyses remain biologically sound and relevant. On the other hand, bio-logging sensor data allow for the discovery of behaviours never seen before in animals (Wilson et al., 2014), thus both exploratory and confirmatory analyses, as well as supervised and non-supervised data analysis methods will be equally important for ecologists (see also Leos-Barajas et al., 2017). Behaviour classification using logger data can also inform the usage of limited, or previously collected, GPS and track data to identify different behaviours (e.g. Browning et al., 2018).
3.3. Movement analyses in the bio-logging era
There is a long history of theoretical investigation into the reasons and rules underpinning animal movement (Nathan et al., 2008) including, for example, optimal foraging theory (Houston, Clark, McNamara, & Mangel, 1988; Pyke, 1984). However, historically, there have been inadequate data on the energetics and the details of movements to embrace optimality properly. Consequently, theoretical movement ecology has tended to focus on statistical descriptions of movement that are agnostic to the underlying life-history needs that govern movement decisions. Step-selection analysis, for example, examines environmental features that are correlated to movements from one location to the next (Avgar, Potts, Lewis, & Boyce, 2016; Fortin et al., 2005; Thurfjell, Ciuti, & Boyce, 2014). As another example, there are a variety of techniques that use movement to infer changes in behaviour, by observing how features such as speed, or tortuosity change over time (Hooten, Johnson, McClintock, & Morales, 2017). These are categorised under various names such as state-space models (Jonsen et al., 2013; Morales, Haydon, Frair, Holsinger, & Fryxell, 2004; Patterson, Thomas, Wilcox, Ovaskainen, & Matthiopoulos, 2008), hidden Markov models (Langrock et al., 2012; McClintock & Michelot, 2018), continuous time models (reviewed in Patterson et al., 2017), and behavioural change-point analyses (Edelhoff, Signer, & Balkenhol, 2016; Gurarie, Andrews, & Laidre, 2009). Similarly, there has been significant interest in inferring broad-scale movement patterns, such as home range, migratory or dispersal patterns, from squared displacement statistics (Börger & Fryxell, 2012). There is also a long history of mathematical models for inferring space-use patterns from general features of movement, such as advective and diffusive components (Moorcroft & Lewis, 2006; Moorcroft, Lewis, & Crabtree, 1999; Potts & Lewis, 2014). All of these examples model movement in a descriptive fashion, where the biases and correlations (Benhamou, 2014; Codling, Plank, & Benhamou, 2008) represent hypothesised behavioural features of the movement path and the aspects of the movement that we either do not have direct knowledge of or are unable to test, as ‘random walks’, or ‘hidden states’. Incorporating high-resolution information from bio-logging studies can change this, as well as enable us to answer questions that link movement decisions to the life-history needs of animals.
Step selection analysis (SSA) is one of the most widely-used techniques for inferring the environmental drivers behind observed movement patterns. New bio-logging technologies enable us to build upon SSA in two important ways. First, the ultra-high frequency locations given by dead-reckoned IMU data enable us for the first time to find the precise points at which an animal changes direction (Potts et al., 2018), rather than assuming (implicitly) that changes in direction occur at the points where locations are acquired (which is typical in SSA studies based on GPS data, although there are exceptions; e.g. Merkle, Fortin, & Morales, 2014). Second, this approach can be extended to examine broader changes in the state of the animal, rather than simply its location, and without having to recur to statistical models trying to infer a ‘hidden state’. As such, we might parametrise a model containing not only the locations of the animal, but also any of the other aforementioned features that we can measure (or infer from metrics of movement) from bio-logging technology, such as head-position, heart-rate, movement “mode” (running/eating) or even interaction variables related to the movement of others in the environment (SI Box 1).
For example, by modifying step selection analysis and similar techniques to incorporate the energetic costs and benefits derived from detailed bio-logging data (acceleration and heart rate loggers), we may be able to uncover the bio-energetic reasons behind animal movement choices, rather than simply describing landscape aspects that co-vary with animal movement. This would help us re-visit old questions about the optimality of foraging decisions, and give important behavioural insights into animal decision-making at fine scales as they move through their energy landscape (Shepard et al., 2013). Quantifying the effects of the environment on movement costs in this way could help also derive a proxy of energy cost based on environmental conditions, to use with movement data without bio-logging information (e.g. Figure 3).
An interesting development in that direction is by Hooten, Scharf, & Morales (2018), who present a new approach to analyse movement data, including explicit mechanistic links to physiological dynamics, to better model decision making and movement in heterogeneous environments. Notably, this approach can be extended to accommodate additional data such as those provided by bio-loggers. Similarly, state-space models and behavioural change-point analysis would be enhanced greatly by careful incorporation of data on acceleration or energy expenditure. Indeed, the behavioural states in these models are often “hidden” (as in “hidden Markov model”) but the sort of bio-logging data described in this review may be able to shed light on these states more directly. This will be a major change in the field and allow markedly improved and biologically relevant understanding to be obtained; compared to any of the even most sophisticated modelling approaches currently used.
4. Future developments for optimising the use of bio-logging
So far, we have reviewed the current technologies and techniques available in the bio-logging toolbox, and how we may optimise their use to answer the big questions in ecology through collaborations within the IBF. Here we highlight potential key future developments, across all nodes of the IBF, which would markedly advance the fields of bio-logging and movement ecology.
New sensors: from speed measurement to skin-patches
As speed is a key parameter of movement, there is an urgent need for reliable speed sensors without the disadvantages (such as fouling) or limits of propellers, flexible paddles, and Pitot tubes (cf. Shepard, Wilson, Liebsch, et al., 2008). Speed of movement exposes animals differentially to conditions and equates to (the square root of) power. New sensors need not be limited to external sampling systems either. Animal skin-associated ‘patches’ are being increasingly used in lab scenarios to look at physiological variables such as stress hormones and other chemicals (Lee, Bakh, Bisker, Brown, & Strano, 2016), something that would find great resonance in wild animal studies. We see huge scope for cross-fostering between these fields, but there are substantial challenges as many of these applications, such as those developed for human studies (Nikita, 2014; Yang, 2014), require powerful readers that operate at close range, and tend to be severely battery limited. Finally, tags need to be able to drop off more routinely and be recovered reliably over large spatial scales, to obtain the large amount of recorded data. This may also save the animal the stress of being recaptured and having to carry the tags for longer than necessary, with all the tag detriment issues that this engenders.
Improved ethical and animal welfare methodologies
Although sensor technology is advancing rapidly, the ethics of bio-logging is still a major concern both in terms of fitting the device, which often requires capture, and the effects of carrying a bio-logger for the study subject. Advancing methodology in capture and consideration of stress by the animal is something that ecologists can work on. Be it reducing handling times, protecting a nest from predators or competitors while the animal is unable to, or even advancing remote tagging methods where the animal does not need to be handled. An additional limitation, is that most devices store data on-board, necessitating recapture of animals and the recovery of the units. Improving the ability of these devices to remotely transmit data would improve their applicability and reduce invasiveness, though may require additional weight in terms of electronics and battery. Of greater concern are tags which require surgical implantation, including heartrate and internal temperature loggers. Though, recent advances have led to the development of surgically implanted sensors even measuring neurological activity, which may further our understanding of the mechanisms behind behaviour, but at what cost for the animal?
A related key limitation to current bio-logging devices is expressed by the ‘measurement effects performance’ paradigm (Wilson, Grant, & Duffy, 1986) via, for example, increased movement costs for the animal through additional mass loading or the ‘drag’ of the device (Barron, Brawn, & Weatherhead, 2010; Vandenabeele et al., 2015) producing non-representative data. There are also other important moral and ethical considerations to animal detriment (Cooke et al., 2017; Wilson et al., 2019), such as cumulative effects (as a result of re-tagging) and long-term effects (decreased survival and/or lifetime reproductive success, which may not be easily evident from short term changes in movement and activity patterns). Thus the current ‘rule of thumb’ based on 3-5% body weight (for aerial and terrestrial animals respectively) is naïve (Bodey et al., 2018), and will need to be improved using more comprehensive information on tag effects based on physical principles (e.g. via computational fluid dynamics to account for drag; Kay et al., 2019), considering also the often neglected effects of tag attachment itself (Vandenabeele et al., 2014). In the meantime, certainly researchers will have to better exploit the ongoing miniaturisation to reduce the relative mass of the devices attached to animals (Portugal & White, 2018). Equally important, researchers should consider if a new tagging study is necessary, or if the question can be answered using existing published data or through data sharing, which will require the development of markedly improved data standards for bio-logging data (see previous section; Figure 3).
Lifetime tracking, real-time processing and remote data transmission
As bio-logging technology continues to advance, the ability to study an individual or population throughout their entire life from conception to death becomes a more realistic possibility. Such large-scale tagging has major ethical implications, as not only a small subset is affected but an entire group, community or population. Especially for similar large-scale questions, researchers would benefit from enhanced bandwidth for transmitting data (cf. O’Donoghue & Rutz, 2016), an element that is already being trialled within the ICARUS system (Wikelski et al., 2007). In tandem with this comes smart on-board data-processing (e.g. Cox et al., 2018) which has the potential to markedly increase the temporal and taxonomic range of data which can be collected. The combination therein of real-time processing and transmission of data will not only enable scientists to dynamically adapt experiments, but has applications in conservation and management.
Improving the theoretical and mathematical foundations of movement ecology
Perhaps the most exciting aspect of bio-logging is that the data-rich approach driven by animals will not only help us to understand why animals do what they do, pinpointing drivers that range from internal state responses to pan-ocean basin atmospheric conditions, but thanks to an improved mechanistic understanding, we might actually be able to predict animal responses to future conditions. To do so will require a large improvement in the theoretical and mathematical foundations of movement ecology, to include the rich set of high-frequency multivariate data, which greatly expand the fundamentally limited and coarse data that could be collected using location-only technology such as GPS. In particular, there is a clear synergy between local (small-scale) information provided by sensors and large-scale information provided by, for example, remote sensing data. How to link and predict processes occurring across different scales is a central question in ecology (Levin, 1992) yet difficult to address, with the key issues being to identify the correct mesoscopic scale connecting microscopic processes to macroscopic patterns. This is the case even for ‘simple’ physical systems constituted of identical particles, whereas biological systems are instead fundamentally characterised by additional intra- and inter-specific heterogeneity. Movement ecologists therefore have to deal with processes which span multiple scales of spatio-temporal and biological complexity (Torney, Hopcraft, Morrison, Couzin, & Levin, 2018). Hence, demanding yet exciting challenges lie ahead for integrating novel bio-logging data with ecological questions. We may now have access to vastly improved information for wild animal biologists to predict processes.
Improved multi-disciplinary collaborations
Collaboration is key to the framework’s success as a tool for optimisation of bio-logging studies. At the same time, ecologists can feed new developments back to other disciplines, e.g. as inspiration for new theorems (Cohen, 2004; Sturmfels, 2005), or for biologically inspired engineering (Bionics), such as new models of navigation inspired by ants (Esterley, McCreery, & Nagpal, 2017) or improved collective decision making in robot swarms (Ebert, Gauci, & Nagpal, 2018). Indeed, actions to bring together multi-disciplinary groups of experts are gathering momentum in movement ecology; these include the EU COST actions from MOVE to develop improved methods for knowledge discovery from moving objects and big data (www.cost.eu/COST_Actions/ict/IC0903) with similar initiatives in the European Network for Radar Surveillance of Animal Movement (http://www.enram.eu/), the Special Interest Group in Movement Ecology of the British Ecological Society (www.britishecologicalsociety.org/membership-community/special-interest-groups/movement-ecology/) and the International Bio-Logging Society (www.bio-logging.net/).
Conclusion
We have i) reviewed how to optimise the use of bio-logging techniques for ecologists to be able to take full advantage of the paradigm-changing opportunities of bio-logging sensors for ecological research and ii) synthesised this into an Integrated Bio-logging Framework (IBF) for movement ecology research. We highlighted the many new and often unexplored opportunities to address biological questions using the most appropriate sensors and sensor-combinations, especially using multi-sensor approaches, a new frontier in bio-logging research. Given the technological complexities and rapid pace of advancement of the field, however, establishing multi-disciplinary collaborations will be paramount for ecologists – and at the same time, the latter can thereby more efficiently guide future technological and methodological advancements to address biological questions. Closely linked to the issue of matching ecological questions with sensors, is devising a good experimental design up front. This involves multiple closely connected challenges, from tag design and sampling regime, to the important related ethical and animal welfare considerations, and the challenges of data sharing. Linking new bio-logging data types to the most adequate analytical techniques presents many new and often unsolved issues in particular, and will require multi-disciplinary collaborations to tackle the ‘big data’ problem, and improve the theoretical and mathematical foundations of movement ecology. The tasks ahead are challenging, but a clear potential exists for a vastly improved mechanistic understanding of animal movements and their role in ecological processes, from which we can build unprecedented and realistic predictive models.
Supplementary Material
Acknowledgements & Funding
This review was written by the participants of a workshop on “linking bio-logging technology to biological questions and data analysis methods” at the British Ecological Society Movement Ecology Specialist Interest Group annual meeting (BES Move 2018). We acknowledge funding and support by the British Ecological Society, the Swansea Centre for Biomathematics, Wildbytes Ltd., and Swansea University. HJW was supported by the European Research Council under the European Union’s Horizon 2020 research and innovation program Grant 715874 (to E.L.C. Shepard); TAC is funded by a Human Frontier Research Program Young Investigator Grant (SeabirdSound - RGY0072/2017); SdG is funded by the European Regional Development Fund through the Ireland Wales Cooperation programme, BLUEFISH; WPK is supported by the Welsh Government’s European Social Fund (ESF), Natural Resources Wales (NRW) and SEACAMS2 – the latter supports also NF; JRP by the National Environmental Research Council (NERC) grant NE/R001669/1; and KFR is NERC funded PhD via EnvEast DTP. UD is supported by a Leverhulme Trust Research Project Grant (RPG-2018-258).
Footnotes
Author contributions
The manuscript was conceived by LB in discussion with all authors at BES Move 2018. All authors wrote the manuscript, led by HJW and LAT, aided by RPW, CR and LB.
The authors declare no conflicts of interest.
References
- Abrahms B, Scales KL, Hazen EL, Bograd SJ, Schick RS, Robinson PW, Costa DP. Mesoscale activity facilitates energy gain in a top predator. Proceedings of the Royal Society B: Biological Sciences. 2018;285 doi: 10.1098/rspb.2018.1101. 20181101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam M, Rohac J. Adaptive data filtering of inertial sensors with variable bandwidth. Sensors. 2015;15(2):3282–3298. doi: 10.3390/s150203282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrienko G, Andrienko N, Chen W, Maciejewski R, Zhao Y. Visual analytics of mobility and transportation: State of the art and further research directions. IEEE Transactions on Intelligent Transportation Systems. 2017;18(8):2232–2249. doi: 10.1109/TITS.2017.2683539. [DOI] [Google Scholar]
- Avgar T, Potts JR, Lewis MA, Boyce MS. Integrated step selection analysis: Bridging the gap between resource selection and animal movement. Methods in Ecology and Evolution. 2016;7(5):619–630. doi: 10.1111/2041-210X.12528. [DOI] [Google Scholar]
- Barron DG, Brawn JD, Weatherhead PJ. Meta-analysis of transmitter effects on avian behaviour and ecology. Methods in Ecology and Evolution. 2010;1(2):180–187. doi: 10.1111/j.2041-210X.2010.00013.x. [DOI] [Google Scholar]
- Benhamou S. Path integration and coordinate systems. Journal of Theoretical Biology. 2014;349:163–166. doi: 10.1016/j.jtbi.2014.02.012. [DOI] [PubMed] [Google Scholar]
- Benhamou S. Mean squared displacement and sinuosity of three-dimensional random search movements. ArXiv. 2018 1801.02435 Retrieved from http://arxiv.org/abs/1801.02435. [Google Scholar]
- Bettaney EM, James R, St Clair JJH, Rutz C. Processing and visualising association data from animal-borne proximity loggers. Animal Biotelemetry. 2015;3:27. doi: 10.1186/s40317-015-0065-4. [DOI] [Google Scholar]
- Bidder OR, Qasem LA, Wilson RP. On higher ground: how well can dynamic body acceleration determine speed in variable terrain? PLoS ONE. 2012;7(11):e50556. doi: 10.1371/journal.pone.0050556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidder OR, Walker JS, Jones MW, Holton MD, Urge P, Scantlebury DM, et al. Wilson RP. Step-by-step: reconstruction of terrestrial animal movement paths by dead-reckoning. Movement Ecology. 2015;3:23. doi: 10.1186/s40462-015-0055-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biewener AA, Patek SN. Animal locomotion. 2nd ed. Oxford, UK: Oxford University Press; 2018. [Google Scholar]
- Bishop CM, Spivey RJ, Hawkes LA, Batbayar N, Chua B, Frappell PB, et al. Butler PJ. The roller coaster flight strategy of bar-headed geese conserves energy during Himalayan migrations. Science. 2015;347(6219):250–254. doi: 10.1126/science.1258732. [DOI] [PubMed] [Google Scholar]
- Block BA. Physiological Ecology in the 21st Century: Advancements in Biologging Science. Integrative and Comparative Biology. 2005;45(2):305–320. doi: 10.1093/icb/45.2.305. [DOI] [PubMed] [Google Scholar]
- Bodey TW, Cleasby IR, Bell F, Parr N, Schultz A, Votier SC, Bearhop S. A phylogenetically controlled meta-analysis of biologging device effects on birds: Deleterious effects and a call for more standardized reporting of study data. Methods in Ecology and Evolution. 2018;9(4):946–955. doi: 10.1111/2041-210X.12934. [DOI] [Google Scholar]
- Börger L, Fryxell J. Quantifying individual differences in dispersal using net squared displacement. In: Clobert J, Baguette M, Benton TG, Bullock JM, editors. Dispersal Ecology and Evolution. Oxford, UK: Oxford University Press; 2012. pp. 222–230. [Google Scholar]
- Bramanti M, Dall’Antonia L. A new technique to monitor the flight paths of birds. Journal of Experimental Biology. 1988;134:467–472. [Google Scholar]
- Browning E, Bolton M, Owen E, Shoji A, Guilford T, Freeman R. Predicting animal behaviour using deep learning: GPS data alone accurately predict diving in seabirds. Methods in Ecology and Evolution. 2018;9(3):681–692. doi: 10.1111/2041-210X.12926. [DOI] [Google Scholar]
- Brownscombe JW, Lennox RJ, Danylchuk AJ, Cooke SJ. Estimating fish swimming metrics and metabolic rates with accelerometers: the influence of sampling frequency. Journal of Fish Biology. 2018;93(2):207–214. doi: 10.1111/jfb.13652. [DOI] [PubMed] [Google Scholar]
- Buchin K, Sijben S, van Loon EE, Sapir N, Mercier S, Arseneau TJM, Willems EP. Deriving movement properties and the effect of the environment from the Brownian bridge movement model in monkeys and birds. Movement Ecology. 2015;3:18. doi: 10.1186/s40462-015-0043-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell HA, Urbano F, Davidson S, Dettki H, Cagnacci F. A plea for standards in reporting data collected by animal-borne electronic devices. Animal Biotelemetry. 2016;4:1. doi: 10.1186/s40317-015-0096-x. [DOI] [Google Scholar]
- Carroll G, Slip D, Jonsen I, Harcourt R. Supervised accelerometry analysis can identify prey capture by penguins at sea. Journal of Experimental Biology. 2014;217(24):4295–4302. doi: 10.1242/jeb.113076. [DOI] [PubMed] [Google Scholar]
- Codling EA, Plank MJ, Benhamou S. Random walk models in biology. Journal of the Royal Society Interface. 2008;5(25):813–834. doi: 10.1098/rsif.2008.0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen JE. Mathematics Is Biology’s Next Microscope, Only Better; Biology Is Mathematics’ Next Physics, Only Better. PLoS ONE. 2004;2(12):e439. doi: 10.1371/journal.pbio.0020439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke SJ, Nguyen VM, Kessel ST, Hussey NE, Young N, Ford AT. Troubling issues at the frontier of animal tracking for conservation and management. Conservation Biology. 2017;31(5):1205–1207. doi: 10.1111/cobi.12895. [DOI] [PubMed] [Google Scholar]
- Cox SL, Orgeret F, Gesta M, Rodde C, Heizer I, Weimerskirch H, Guinet C. Processing of acceleration and dive data on-board satellite relay tags to investigate diving and foraging behaviour in free-ranging marine predators. Methods in Ecology and Evolution. 2018;9(1):64–77. doi: 10.1111/2041-210X.12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Ridder M, Klein K, Kim J. Temporaltracks: visual analytics or exploration of 4D fMRI time-series coactivation. Proceedings of the Computer Graphics International Conference (CGI ’17); New York, USA: 2017. [DOI] [Google Scholar]
- Demšar U, Buchin K, Cagnacci F, Safi K, Speckmann B, Van de Weghe N, et al. Weibel R. Analysis and visualisation of movement: an interdisciplinary review. Movement Ecology. 2015;3:5. doi: 10.1186/s40462-015-0032-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimiceli VE, Piltz SF, Amburn SA. Estimation of Black Globe Temperature for Calculation of the Wet Bulb Globe Temperature Index. Proceedings of the World Congress on Engineering and Computer Science 2011; October 19-21, 2011; San Francisco, US: 2011. p. WCECS 2011. [Google Scholar]
- Ditmer MA, Rettler SJ, Fieberg JR, Iaizzo PA, Laske TG, Noyce KV, Garshelis DL. American black bears perceive the risks of crossing roads. Behavioral Ecology. 2018;29(3):667–675. doi: 10.1093/beheco/ary020. [DOI] [Google Scholar]
- Dodge S, Bohrer G, Weinzierl R, Davidson SC, Kays R, Douglas D, et al. Wikelski M. The environmental-data automated track annotation (Env - DATA) system: linking animal tracks with environmental data. Movement Ecology. 2013;1:3. doi: 10.1186/2051-3933-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodge S, Xavier G, Wong WY. DynamoVis - Dynamic Visualization of Animal Movement Data. Retrieved from the Data Repository for the University of Minnesota. 2018 doi: 10.13020/D6PH49. [DOI] [Google Scholar]
- Downey LA, Tysse B, Ford TC, Samuels AC, Wilson RP, Parrott AC. Psychomotor Tremor and Proprioceptive Control Problems in Current and Former Stimulant Drug Users: An Accelerometer Study of Heavy Users of Amphetamine, MDMA, and Other Recreational Stimulants. Journal of Clinical Pharmacology. 2017;57(10):1330–1337. doi: 10.1002/jcph.925. [DOI] [PubMed] [Google Scholar]
- Duda N, Nowak T, Hartmann M, Schadhauser M, Cassens B, Wägemann P, et al. Kölpin A. BATS: Adaptive Ultra Low Power Sensor Network for Animal Tracking. Sensors. 2018;18(10):3343. doi: 10.3390/s18103343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn TW, Mu Y, Narayan S, Radlett O, Naumann EA, Yang C-T, et al. Ahrens MB. Brain-wide mapping of neural activity controlling zebrafish exploratory locomotion. ELIFE. 2016;5:e12741. doi: 10.7554/eLife.12741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert J, Gauci M, Nagpal R. Multi-Feature Collective Decision Making in Robot Swarms. Proceedings of the 17th International Conference on Autonomous Agents and MultiAgent Systems; Stockholm, Sweden: 2018. pp. 1711–1719. [Google Scholar]
- Edelhoff H, Signer J, Balkenhol N. Path segmentation for beginners: an overview of current methods for detecting changes in animal movement patterns. Movement Ecology. 2016;4:21. doi: 10.1186/s40462-016-0086-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esterley EE, McCreery H, Nagpal R. Models of Adaptive Navigation, Inspired by Ant Cooperative Transport in the Presence of Obstacles. 2017 IEEE Symposium Series on Computational Intelligence (SSCI); Honolulu, USA: 2017. pp. 1–8. [DOI] [Google Scholar]
- Fieberg J, Börger L. Could you please phrase “home range” as a question? Journal of Mammalogy. 2012;93(4):890–902. doi: 10.1644/11-MAMM-S-172.1. [DOI] [Google Scholar]
- Fortin D, Beyer H, Boyce MS, Smith DW, Duchesne T, Mao J. Wolves influence elk movements: Behavior shapes a trophic cascade in Yellowstone national park. Ecology. 2005;86(5):1320–1330. doi: 10.1890/04-0953. [DOI] [Google Scholar]
- Getz WM, Saltz D. A framework for generating and analyzing movement paths on ecological landscapes. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(49):19066–19071. doi: 10.1073/pnas.0801732105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graser A, Schmidt J, Roth F, Brändle N. Untangling origin-destination flows in geographic information systems. Information Visualization. 2017;1(1):153–172. doi: 10.1177/1473871617738122. [DOI] [Google Scholar]
- Green JA. The heart rate method for estimating metabolic rate: review and recommendations. Comparative Biochemistry and Physiology. Part A, Molecular & Integrative Physiology. 2011;158(3):287–304. doi: 10.1016/j.cbpa.2010.09.011. [DOI] [PubMed] [Google Scholar]
- Grundy E, Jones MW, Laramee RS, Wilson RP, Shepard ELC. Visualisation of sensor data from animal movement. Computer Graphics Forum. 2009;28(3):815–822. doi: 10.1111/j.1467-8659.2009.01469.x. [DOI] [Google Scholar]
- Gurarie E, Andrews RD, Laidre KL. A novel method for identifying behavioural changes in animal movement data. Ecology Letters. 2009;12(5):395–408. doi: 10.1111/j.1461-0248.2009.01293.x. [DOI] [PubMed] [Google Scholar]
- Hall EH. On a New Action of the Magnet on Electric Currents. American Journal of Mathematics. 1879;2(3):287–292. [Google Scholar]
- Hampton SE, Strasser CA, Tewksbury JJ, Gram WK, Budden AE, Batcheller AL, et al. Porter JH. Big data and the future of ecology. Frontiers in Ecology and the Environment. 2013;11(3):156–162. doi: 10.1890/120103. [DOI] [Google Scholar]
- Hays GC, Marshall GJ, Seminoff JA. Flipper beat frequency and amplitude changes in diving green turtles, Chelonia mydas. Marine Biology. 2007;150(5):1003–1009. doi: 10.1007/s00227-006-0412-3. [DOI] [Google Scholar]
- Hebblewhite M, Haydon DT. Distinguishing technology from biology: a critical review of the use of GPS telemetry data in ecology. Philosophical Transactions of the Royal Society B: Biological Sciences. 2010;365(1550):2303–2312. doi: 10.1098/rstb.2010.0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedenström A, Norevik G, Warfvinge K, Andersson A, Bäckman J, Åkesson S. Annual 10-Month Aerial Life Phase in the Common Swift Apus apus. Current Biology. 2016;26(22):3066–3070. doi: 10.1016/j.cub.2016.09.014. [DOI] [PubMed] [Google Scholar]
- Hoenner X, Huveneers C, Steckenreuter A, Simpfendorfer C, Tattersall K, Jaine F, et al. Harcourt R. Data Descriptor: Australia’s continental-scale acoustic tracking database and its automated quality control process. Scientific Data. 2018;5 doi: 10.1038/sdata.2017.206. 170206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooker SK, Barychka T, Jessopp MJ, Staniland IJ. Images as proximity sensors: the incidence of conspecific foraging in Antarctic fur seals. Animal Biotelemetry. 2015;3:37. doi: 10.1186/s40317-015-0083-2. [DOI] [Google Scholar]
- Hooten MB, Johnson DS, McClintock BT, Morales JM. Animal movement: statistical models for telemetry data. Boca Raton, USA: CRC Press; 2017. [Google Scholar]
- Hooten M, Scharf H, Morales J. Running on empty: Recharge dynamics from animal movement data. Ecology Letters. 2018;22(2):377–389. doi: 10.1111/ele.13198. [DOI] [PubMed] [Google Scholar]
- Houston A, Clark C, McNamara J, Mangel M. Dynamic models in behavioural and evolutionary ecology. Nature. 1988;332(3):29–34. [Google Scholar]
- Hussey NE, Kessel ST, Aarestrup K, Cooke SJ, Cowley PD, Fisk AT, et al. Whoriskey FG. Aquatic animal telemetry: A panoramic window into the underwater world. Science. 2015;348(6240) doi: 10.1126/science.1255642. 1255642. [DOI] [PubMed] [Google Scholar]
- Jonsen ID, Basson M, Bestley S, Bravington MV, Patterson TA, Pedersen MW, et al. Wotherspoon SJ. Deep-Sea Research II State-space models for bio-loggers: A methodological road map. Deep-Sea Research Part II. 2013;88–89:34–46. doi: 10.1016/j.dsr2.2012.07.008. [DOI] [Google Scholar]
- Kay WP, Naumann DS, Bowen HJ, Withers S, Evans BJ, Wilson RP, et al. Börger L. Minimising the impact of biologging devices: Using Computational Fluid Dynamics for optimizing tag design and positioning. Methods in Ecology and Evolution. 2019 doi: 10.1111/2041-210X.13216. [DOI] [Google Scholar]
- Kays R, Crofoot MC, Jetz W, Wikelski M. Terrestrial animal tracking as an eye on life and planet. Science. 2015;348(6240) doi: 10.1126/science.aaa2478. aaa2478. [DOI] [PubMed] [Google Scholar]
- Kenward RE. A Manual for Wildlife Radiotracking. London, UK: Academic Press; 2001. [Google Scholar]
- Kooyman GL. Techniques used in measuring diving capacities of Weddel seals. The Polar Record. 1964;12(79):391–394. [Google Scholar]
- Krause J, Krause S, Arlinghaus R, Psorakis I, Roberts S, Rutz C. Reality mining of animal social systems. Trends in Ecology and Evolution. 2013;28(9):541–551. doi: 10.1016/j.tree.2013.06.002. [DOI] [PubMed] [Google Scholar]
- Krebs JR, Davies NB. Behavioural ecology: an evolutionary approach. Oxford, UK: Blackwell Scientific; 1978. [Google Scholar]
- Kröschel M, Reineking B, Werwie F, Wildi F, Storch I. Remote monitoring of vigilance behavior in large herbivores using acceleration data. Animal Biotelemetry. 2017;5(1):10. doi: 10.1186/s40317-017-0125-z. [DOI] [Google Scholar]
- Landry MP, Kruss S, Nelson JT, Bisker G, Iverson NM, Reuel NF, Strano MS. Experimental Tools to Study Molecular Recognition within the Nanoparticle Corona. Sensors. 2014;14(9):16193–16211. doi: 10.3390/s140916196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langrock R, King R, Matthiopoulos J, Thomas L, Fortin D, Morales JM. Flexible and practical modeling of animal telemetry data: hidden Markov models and extensions. Ecology. 2012;93(11):2336–2342. doi: 10.1890/11-2241.1. [DOI] [PubMed] [Google Scholar]
- Lear KO, Gleiss AC, Whitney MN. Metabolic rates and the energetic cost of external tag attachment in juvenile blacktip sharks Carcharhinus limbatus. Journal of Fish Biology. 2018;93(2):391–395. doi: 10.1111/jfb.13663. [DOI] [PubMed] [Google Scholar]
- Lee J, Jeong B-K. Understanding sensor data with uncertainty using a visual representation. IEEE 7th Annual Computing and Communication Workshop and Conference (CCWC); Las Vagas, USA: 2017. pp. 1–2. [DOI] [Google Scholar]
- Lee MA, Bakh N, Bisker G, Brown EN, Strano MS. A Pharmacokinetic Model of a Tissue Implantable Cortisol Sensor. Advanced Healthcare Materials. 2016;5(23):3004–3015. doi: 10.1002/adhm.201600650. [DOI] [PubMed] [Google Scholar]
- Lee MA, Nguyen FT, Scott K, Chan NYL, Bakh NA, Jones KK, et al. Strano MS. Implanted Nanosensors in Marine Organisms for Physiological Biologging: Design, Feasibility, and Species Variability. ACS Sensors. 2018;4(1):32–43. doi: 10.1021/acssensors.8b00538. research-article. [DOI] [PubMed] [Google Scholar]
- Leos-Barajas V, Photopoulou T, Langrock R, Patterson TA, Watanabe YY, Murgatroyd M, Papastamatiou YP. Analysis of animal accelerometer data using hidden Markov models. Methods in Ecology and Evolution. 2017;8(2):161–173. doi: 10.1111/2041-210X.12657. [DOI] [Google Scholar]
- Levin SA. The problem of pattern and scale in ecology. Ecology. 1992;73(6):1943–1967. doi: 10.2307/1941447. [DOI] [Google Scholar]
- Lewis KP, Vander Wal E, Fifield DA. Wildlife biology, big data, and reproducible research. Wildlife Society Bulletin. 2018;42:172–179. doi: 10.1002/wsb.847. [DOI] [Google Scholar]
- Li W, Wu S, Song M, Zhou X. A scalable cyberinfrastructure solution to support big data management and multivariate visualization of time-series sensor observation data. Earth Science Informatics. 2016;9(4):449–464. doi: 10.1007/s12145-016-0267-1. [DOI] [Google Scholar]
- McClintock BTBT, Michelot T. momentuHMM: R package for generalized hidden Markov models of animal movement. Methods in Ecology and Evolution. 2018;9(6):1518–1530. doi: 10.1111/2041-210X.12995. [DOI] [Google Scholar]
- McInnes AM, McGeorge C, Ginsberg S, Pichegru L, Pistorius PA. Group foraging increases foraging efficiency in a piscivorous diver, the African penguin. Royal Society Open Science. 2017;4 doi: 10.1098/rsos.170918. 170918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara JM, Houston AI. The Common Currency for Behavioral Decisions. The American Naturalist. 1986;127(3):358–378. [Google Scholar]
- Merkle JA, Fortin D, Morales JM. A memory-based foraging tactic reveals an adaptive mechanism for restricted space use. Ecology Letters. 2014;17(8):924–931. doi: 10.1111/ele.12294. [DOI] [PubMed] [Google Scholar]
- Moll RJ, Millspaugh JJ, Beringer J, Sartwell J, He Z. A new ‘view’ of ecology and conservation through animal-borne video systems. Trends in Ecology and Evolution. 2007;22(12):660–668. doi: 10.1016/j.tree.2007.09.007. [DOI] [PubMed] [Google Scholar]
- Moorcroft PR, Lewis MA. Mechanistic home range analysis. Princeton, USA: Princeton University Press; 2006. [Google Scholar]
- Moorcroft PR, Lewis MA, Crabtree RL. Home range analysis using a mechanistic home range model. Ecology. 1999;80(5):1656–1665. doi: 10.1890/0012-9658(1999)080[1656:HRAUAM]2.0.CO;2. [DOI] [Google Scholar]
- Morales JM, Haydon DT, Frair J, Holsinger KE, Fryxell JM. Extracting more out of relocation data: building movement models as mixtures of random walks. Ecology. 2004;85(9):2436–2445. doi: 10.1890/03-0269. [DOI] [Google Scholar]
- Morales JM, Moorcroft PR, Matthiopoulos J, Frair JL, Kie JG, Powell RA, et al. Haydon DT. Building the bridge between animal movement and population dynamics. Philosophical Transactions of the Royal Society B: Biological Sciences. 2010;365(1550):2289–2301. doi: 10.1098/rstb.2010.0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassar PN, Jackson AC, Carrier DR. Entraining the natural frequencies of running and breathing in guinea fowl (Numida meleagris) The Journal of Experimental Biology. 2001;204(9):1641–1651. doi: 10.1242/jeb.204.9.1641. [DOI] [PubMed] [Google Scholar]
- Nathan R, Getz WM, Revilla E, Holyoak M, Kadmon R, Saltz D, Smouse PE. A movement ecology paradigm for unifying organismal movement research. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(49):19052–19059. doi: 10.1073/pnas.0800375105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan R, Spiegel O, Fortmann-Roe S, Harel R, Wikelski M, Getz WM. Using tri-axial acceleration data to identify behavioral modes of free-ranging animals: general concepts and tools illustrated for griffon vultures. The Journal of Experimental Biology. 2012;215(6):986–996. doi: 10.1242/jeb.058602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikita KS. Handbook of Biomedical Telemetry. Hoboken, USA: John Wiley & Sons, Inc; 2014. [Google Scholar]
- Noda T, Kawabata Y, Arai N, Mitamura H, Watanabe S. Animal-mounted gyroscope/accelerometer/magnetometer: In situ measurement of the movement performance of fast-start behaviour in fish. Journal of Experimental Marine Biology and Ecology. 2014;451:55–68. doi: 10.1016/j.jembe.2013.10.031. [DOI] [Google Scholar]
- O’Donoghue P, Rutz C. Real-time anti-poaching tags could help prevent imminent species extinctions. Journal of Applied Ecology. 2016;53(1):5–10. doi: 10.1111/1365-2664.12452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Mara MT, Wikelski M, Voigt CC, Ter Maat A, Pollock HS, Burness G, et al. Dechmann DKN. Cyclic bouts of extreme bradycardia counteract the high metabolism of frugivorous bats. ELIFE. 2017;6:e26686. doi: 10.7554/eLife.26686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagano AM, Durner GM, Rode KD, Atwood TC, Atkinson SN, Peacock E, et al. Williams TM. High-energy, high-fat lifestyle challenges an Arctic apex predator, the polar bear. Science. 2018;359(6375):568–572. doi: 10.1126/science.aan8677. [DOI] [PubMed] [Google Scholar]
- Patterson TA, Parton A, Langrock R, Blackwell PG, Thomas L, King R. Statistical modelling of individual animal movement: an overview of key methods and a discussion of practical challenges. AStA Advances in Statistical Analysis. 2017;101(4):399–438. doi: 10.1007/s10182-017-0302-7. [DOI] [Google Scholar]
- Patterson TA, Thomas L, Wilcox C, Ovaskainen O, Matthiopoulos J. State-space models of individual animal movement. Trends in Ecology and Evolution. 2008;23(2):87–94. doi: 10.1016/j.tree.2007.10.009. [DOI] [PubMed] [Google Scholar]
- Peters DPC, Burruss ND, Rodriguez LL, McVey DS, Elias EH, Pelzel-McCluskey AM, et al. Vivoni ER. An Integrated View of Complex Landscapes: A Big Data-Model Integration Approach to Transdisciplinary Science. BioScience. 2018;68(9):653–669. doi: 10.1093/biosci/biy069. [DOI] [Google Scholar]
- Piersma T, Lindström Å. Migrating shorebirds as integrative sentinels of global environmental change. Ibis. 2004;146(Suppl. 1):61–69. doi: 10.1111/j.1474-919X.2004.00329.x. [DOI] [Google Scholar]
- Portugal SJ, White CR. Miniaturization of biologgers is not alleviating the 5% rule. Methods in Ecology and Evolution. 2018;9(7):1662–1666. doi: 10.1111/2041-210X.13013. [DOI] [Google Scholar]
- Potts JR, Börger L, Scantlebury DM, Bennett NC, Alagaili A, Wilson RP. Finding turning-points in ultra-high-resolution animal movement data. Methods in Ecology and Evolution. 2018;9(10):2091–2101. doi: 10.1111/2041-210X.13056. [DOI] [Google Scholar]
- Potts JR, Lewis MA. How do animal territories form and change? Lessons from 20 years of mechanistic modelling. Proceedings of the Royal Society B: Biological Sciences. 2014;281(1784) doi: 10.1098/rspb.2014.0231. 20140231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyke GH. Optimal Foraging Theory: A Critical Review. Annual Review of Ecology and Systematics. 1984;15:523–575. [Google Scholar]
- Qasem L, Cardew A, Wilson A, Griffiths I, Halsey LG, Shepard ELC, et al. Wilson R. Tri-axial dynamic acceleration as a proxy for animal energy expenditure; should we be summing values or calculating the vector? PLoS ONE. 2012;7(2):e31187. doi: 10.1371/journal.pone.0031187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattenborg NC, Voirin B, Cruz SM, Tisdale R, Dell’Omo G, Lipp H, et al. Vyssotski AL. Evidence that birds sleep in mid-flight. Nature Communications. 2016;7 doi: 10.1038/ncomms12468. 12468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remelgado R, Wegmann M, Safi K. rsMove – An R package to bridge Remote Sensing and Movement Ecology. Methods in Ecology and Evolution. 2019 doi: 10.1111/2041-210X.13199. [DOI] [Google Scholar]
- Rempel RS, Rodgers AR, Abraham KF. Performance of a GPS Animal Location System under Boreal Forest Canopy. The Journal of Wildlife Management. 1995;59(3):543–551. [Google Scholar]
- Roberts RC, Laramee RS, Jones MW. Multivariate hybrid visualisation of ornithological sensor data. In: Borgo R, Turkay C, editors. Proceedings of Computer Graphics and Visual Computing (CGVC) 2015. Eurographics Association; 2015. pp. 1–6. [DOI] [Google Scholar]
- Ropert-Coudert Y, Kato A, Liebsch N, Wilson RP, Müller G, Baubet E. Monitoring jaw movements: A cue to feeding activity. Game and Wildlife Science. 2004;20(4):1–19. [Google Scholar]
- Ropert-Coudert Y, Sato K, Kato A, Charrassin J-B, Bost C-A, Le Maho Y, Naito Y. Preliminary investigations of prey pursuit and capture by king penguins at sea. Polar Bioscience. 2000;13:101–112. [Google Scholar]
- Ropert-Coudert Y, Wilson RP. Subjectivity in bio-logging science: do logged data mislead? Memoirs of the National Institute of Polar Research: Special Issue. 2004;58:23–33. [Google Scholar]
- Rutz C, Bluff LA, Weir AAS, Kacelnik A. Video Cameras on Wild Birds. Science. 2007;318(5851):765. doi: 10.1126/science.1146788. [DOI] [PubMed] [Google Scholar]
- Rutz C, Burns ZT, James R, Ismar SMH, Burt J, Otis B, et al. St Clair JJH. Automated mapping of social networks in wild birds. Current Biology. 2012;22(17):R669–R671. doi: 10.1016/j.cub.2012.06.037. [DOI] [PubMed] [Google Scholar]
- Rutz C, Hays GC. New frontiers in biologging science. Biology Letters. 2009;5(3):289–292. doi: 10.1098/rsbl.2009.0089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutz C, Troscianko J. Programmable, miniature video-loggers for deployment on wild birds and other wildlife. Methods in Ecology and Evolution. 2013;4:114–122. doi: 10.1111/2041-210x.12003. [DOI] [Google Scholar]
- Scacco M, Flack A, Duriez O, Wikelski M, Safi K. Static landscape features predict uplift locations for soaring birds across Europe. Royal Society Open Science. 2019;5 doi: 10.1098/rsos.181440. 181440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf H. anipaths: Animation of Observed Trajectories Using Spline-Based Interpolation. R package version 0.9.6. 2018 https://CRAN.R-project.org/package=anipaths. [Google Scholar]
- Schmidt-Nielsen K. Locomotion: Energy Cost of Swimming, Flying, and Running. Science. 1972;177(4045):222–228. doi: 10.1126/science.177.4045.222. [DOI] [PubMed] [Google Scholar]
- Schwalb-Willmann J. moveVis: Movement Data Visualization. R package version 0.9.8. 2018 https://CRAN.R-project.org/package=moveVis. [Google Scholar]
- Shamoun-Baranes J, Farnsworth A, Aelterman B, Alves JA, Azijn K, Bernstein G, et al. van Gasteren H. Innovative visualizations shed light on avian nocturnal migration. PLoS ONE. 2016;11(8):e0160106. doi: 10.1371/journal.pone.0160106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard ELC, Wilson RP, Halsey LG, Quintana F, Laich AG, Gleiss AC, et al. Norman B. Derivation of body motion via appropriate smoothing of acceleration data. Aquatic Biology. 2008;4(3):235–241. doi: 10.3354/ab00104. [DOI] [Google Scholar]
- Shepard ELC, Wilson RP, Liebsch N, Quintana F, Laich AG, Lucke K. Flexible paddle sheds new light on speed: a novel method for the remote measurement of swim speed in aquatic animals. Endangered Species Research. 2008;4:157–164. doi: 10.3354/esr00052. [DOI] [Google Scholar]
- Shepard ELC, Wilson RP, Quintana F, Laich AG, Forman DW. Pushed for time or saving on fuel: fine-scale energy budgets shed light on currencies in a diving bird. Proceedings of the Royal Society B: Biological Sciences. 2009;276(1670):3149–3155. doi: 10.1098/rspb.2009.0683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard ELC, Wilson RP, Quintana F, Laich AG, Liebsch N, Albareda DA, et al. Macdonald DW. Identification of animal movement patterns using tri-axial accelerometry. Endangered Species Research. 2008;10(1):47–60. doi: 10.3354/esr00084. [DOI] [Google Scholar]
- Shepard ELC, Wilson RP, Rees WG, Grundy E, Lambertucci SA, Vosper SB. Energy Landscapes Shape Animal Movement Ecology. The American Naturalist. 2013;182(3):298–312. doi: 10.1086/671257. [DOI] [PubMed] [Google Scholar]
- Shipley JR, Kapoor J, Dreelin RA, Winkler DW. An open-source sensor-logger for recording vertical movement in free-living organisms. Methods in Ecology and Evolution. 2018;9(3):465–471. doi: 10.1111/2041-210X.12893. [DOI] [Google Scholar]
- Skocek O, Nöbauer T, Weilguny L, Traub FM, Xia CN, Molodtsov MI, et al. Vaziri A. High-speed volumetric imaging of neuronal activity in freely moving rodents. Nature Methods. 2018;15:429–432. doi: 10.1038/s41592-018-0008-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Clair JJH, Burns ZT, Bettaney EM, Morrissey MB, Otis B, Ryder TB, et al. Rutz C. Experimental resource pulses influence social-network dynamics and the potential for information flow in tool-using crows. Nature Communications. 2015;6:7197. doi: 10.1038/ncomms8197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowers JR, Hofbauer M, Bastien R, Griessner J, Higgins P, Farooqui S, et al. Straw AD. Virtual reality for freely moving animals. Nature Methods. 2017;14(10):995–1002. doi: 10.1038/nmeth.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturmfels B. Can biology lead to new theorems? Annual Report of the Clay Mathematics Institute. 2005;13:26. [Google Scholar]
- Sutton GJ, Hoskins AJ, Arnould JPY. Benefits of Group Foraging Depend on Prey Type in a Small Marine Predator, the Little Penguin. PLoS ONE. 2015;10(12):e0144297. doi: 10.1371/journal.pone.0144297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Nakai A, Shimoyama I. Waterproof airflow sensor for seabird bio-logging using a highly sensitive differential pressure sensor and nano-hole array. Sensors and Actuators A: Physical. 2018;281:243–249. [Google Scholar]
- Takahashi A, Sato K, Naito Y, Dunn MJ, Trathan PN, Croxall JP. Penguin-mounted cameras glimpse underwater group behaviour. Proceedings of the Royal Society B: Biological Sciences. 2004;271(Suppl_5):S281–S282. doi: 10.1098/rsbl.2004.0182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takei Y, Suzuki I, Wong MKS, Milne R, Moss S, Sato K, Hall A. Development of an animal-borne blood sample collection device and its deployment for the determination of cardiovascular and stress hormones in phocid seals. American Journal of Physiology-Regulatory, Integrative and Comparative Physiology. 2016;311(4):R788–R796. doi: 10.1152/ajpregu.00211.2016. [DOI] [PubMed] [Google Scholar]
- Taylor GK, Reynolds KV, Thomas ALR. Soaring energetics and glide performance in a moving atmosphere. Philosophical Transactions of the Royal Society of London B: Biological Sciences. 2016;371(1704) doi: 10.1098/rstb.2015.0398. 20150398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thums M, Fernández-Gracia J, Sequeira AMM, Eguíluz VM, Duarte CM, Meekan MG. How Big Data Fast Tracked Human Mobility Research and the Lessons for Animal Movement Ecology. Frontiers in Marine Science. 2018;5:21. doi: 10.3389/fmars.2018.00021. [DOI] [Google Scholar]
- Thurfjell H, Ciuti S, Boyce MS. Applications of step-selection functions in ecology and conservation. Movement Ecology. 2014;2:4. doi: 10.1186/2051-3933-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomkiewicz SM, Fuller MR, Kie JG, Bates KK. Global positioning system and associated technologies in animal behaviour and ecological research. Philosophical Transactions of the Royal Society B: Biological Sciences. 2010;365(1550):2163–2176. doi: 10.1098/rstb.2010.0090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomotani BM, Bil W, van der Jeugd HP, Pieters RPM, Muijres FT. Carrying a logger reduces escape flight speed in a passerine bird, but relative logger mass may be a misleading measure of this flight performance detriment. Methods in Ecology and Evolution. 2019;10(1):70–79. [Google Scholar]
- Torney CJ, Hopcraft JGC, Morrison TA, Couzin ID, Levin SA. From single steps to mass migration: The problem of scale in the movement ecology of the Serengeti wildebeest. Philosophical Transactions of the Royal Society B: Biological Sciences. 2018;373(1746) doi: 10.1098/rstb.2017.0012. 20170012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay Y, Thibault A, Mullers R, Pistorius P. Bird-Borne Video-Cameras Show That Seabird Movement Patterns Relate to Previously Unrevealed Proximate Environment, Not Prey. PLoS ONE. 2014;9(2):e88424. doi: 10.1371/journal.pone.0088424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker MA, Böhning-Gaese K, Fagan WF, Fryxell JM, Van Moorter B, Alberts SC, et al. Mueller T. Moving in the Anthropocene: Global reductions in terrestrial mammalian movements. Science. 2018;359(6374):466–469. doi: 10.1126/science.aam9712. [DOI] [PubMed] [Google Scholar]
- Tukey JW. Exploratory Data Analysis. New York: Addison-Wesley; 1977. [Google Scholar]
- van Dam RP, Ponganis PJ, Ponganis KV, Levenson DH, Marshall G. Stroke frequencies of emperor penguins diving under sea ice. The Journal of Experimental Biology. 2002;205:3769–3774. doi: 10.1242/jeb.205.24.3769. [DOI] [PubMed] [Google Scholar]
- Vandenabeele SP, Grundy E, Friswell MI, Grogan A, Votier SC, Wilson RP. Excess baggage for birds: Inappropriate placement of tags on gannets changes flight patterns. PLoS ONE. 2014;9(3):e92657. doi: 10.1371/journal.pone.0092657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenabeele SP, Shepard ELC, Grémillet D, Butler PJ, Martin GR, Wilson RP. Are bio-telemetric devices a drag? Effects of external tags on the diving behaviour of great cormorants. Marine Ecology Progress Series. 2015;519:239–249. doi: 10.3354/meps11058. [DOI] [Google Scholar]
- Verbeek K, Buchin K, Speckmann B. Flow Map Layout via Spiral Trees. IEEE Transactions on Visualization and Computer Graphics. 2011;17(12):2536–2544. doi: 10.1109/TVCG.2011.202. [DOI] [PubMed] [Google Scholar]
- Viviant M, Trites AW, Rosen DAS, Monestiez P, Guinet C. Prey capture attempts can be detected in Steller sea lions and other marine predators using accelerometers. Polar Biology. 2010;33(5):713–719. doi: 10.1007/s00300-009-0750-y. [DOI] [Google Scholar]
- Vyssotski AL, Serkov AN, Itskov PM, Dell’Omo G, Latanov AV, Wolfer DP, Lipp H-P. Miniature Neurologgers for Flying Pigeons: Multichannel EEG and Action and Field Potentials in Combination With GPS Recording. Journal of Neurophysiology. 2006;95(2):1263–1273. doi: 10.1152/jn.00879.2005. [DOI] [PubMed] [Google Scholar]
- Walker J, Borgo R, Jones M. TimeNotes: A study on effective chart visualization and interaction techniques for time-series data. IEEE Transactions on Visualization and Computer Graphics. 2016;22(1):549–558. doi: 10.1109/TVCG.2015.2467751. [DOI] [PubMed] [Google Scholar]
- Walker JS, Jones MW, Laramee RS, Bidder OR, Williams HJ, Scott R, et al. Wilson RP. TimeClassifier: a visual analytic system for the classification of multi-dimensional time series data. The Visual Computer. 2015;31(6):1067–1078. doi: 10.1007/s00371-015-1112-0. [DOI] [Google Scholar]
- Ware C. Information Visualization: Perception for Design. 3rd ed. San Francisco, USA: Morgan Kaufmann Publishers; 2012. [Google Scholar]
- Ware C, Arsenault R, Plumlee M, Wiley D. Visualizing the Underwater Behavior of Humpback Whales. IEEE Computer Graphics and Applications. 2006;July/August:14–18. doi: 10.1109/mcg.2006.93. [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Wei Q, Yang D, Chen X, Du H, Yang J, et al. Miyazaki N. Swimming behavior in relation to buoyancy in an open swimbladder fish, the Chinese sturgeon. Journal of Zoology. 2008;275:381–390. doi: 10.1111/j.1469-7998.2008.00451.x. [DOI] [Google Scholar]
- Watanabe YY, Takahashi A. Linking animal-borne video to accelerometers reveals prey capture variability. Proceedings of the National Academy of Sciences. 2013;110(6):2199–2204. doi: 10.1073/pnas.1216244110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weimerskirch H. In: 30 years of wildlife tracking with ARGOS. Ferro H, editor. Cape Town, South Africa: CLS; 2009. p. 3. [Google Scholar]
- Weimerskirch H, Filippi DP, Collet J, Waugh SM, Patrick SC. Use of radar detectors to track attendance of albatrosses at fishing vessels. Conservation Biology. 2018;32(1):240–245. doi: 10.1111/cobi.12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weimerskirch H, Gault A, Cherel Y. Prey distribution and patchiness: factors in foraging success and efficiency of wandering albatrosses. Ecology. 2005;86(10):2611–2622. doi: 10.1093/schbul/sbt124. [DOI] [Google Scholar]
- Weimerskirch H, Pinaud D, Pawlowski F, Bost C. Does Prey Capture Induce Area-Restricted Search? A Fine-Scale Study Using GPS in a Marine Predator, the Wandering Albatross. The American Naturalist. 2007;170(5):734–743. doi: 10.1086/522059. [DOI] [PubMed] [Google Scholar]
- Weiser AW, Orchan Y, Nathan R, Charter M, Weiss AJ, Toledo S. Characterizing the Accuracy of a Self-Synchronized Reverse-GPS Wildlife Localization System. 2016 15th ACM/IEEE International Conference on Information Processing in Sensor Networks (IPSN); Vienna, Austria: 2016. pp. 1–12. [DOI] [Google Scholar]
- Wikelski M, Kays RW, Kasdin NJ, Thorup K, Smith JA, Swenson GW. Going wild: what a global small-animal tracking system could do for experimental biologists. Journal of Experimental Biology. 2007;210:181–186. doi: 10.1242/jeb.02629. [DOI] [PubMed] [Google Scholar]
- Williams HJ, Holton MD, Shepard ELC, Largey N, Norman B, Ryan PG, et al. Wilson RP. Identification of animal movement patterns using tri-axial magnetometry. Movement Ecology. 2017;5:6. doi: 10.1186/s40462-017-0097-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmers CC, Isbell LA, Suraci JP, Williams TM. Energetics-informed behavioral states reveal the drive to kill in African leopards. Ecosphere. 2017;8(6):e01850. doi: 10.1002/ecs2.1850. [DOI] [Google Scholar]
- Wilmers CC, Nickel B, Bryce CM, Smith JA, Wheat RE, Yovovich V. The golden age of bio-logging: how animal-borne sensors are advancing the frontiers of ecology. Ecology. 2015;96(7):1741–1753. doi: 10.1890/14-1401.1. [DOI] [PubMed] [Google Scholar]
- Wilson K, Littman C, Halpin P, Read A. Integrating multiple technologies to understand the foraging behaviour of Hawaiian monk seals. Royal Society Open Science. 2017;4 doi: 10.1098/rsos.160703. 160703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RP, Börger L, Holton MD, Scantlebury DM, Gómez-Laich A, Quintana F, et al. Shepard ELC. Estimates for energy expenditure in free-living animals using acceleration proxies; a reappraisal. Journal of Animal Ecology. 2019 doi: 10.1111/1365-2656.13040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RP, Cooper J, Plötz J. Can we determine when marine endotherms feed? A case study with seabirds. The Journal of Experimental Biology. 1992;167:267–275. [Google Scholar]
- Wilson RP, Grant WS, Duffy DC. Recording Devices on Free-Ranging Marine Animals: Does Measurement Affect Foraging Performance? Ecology. 1986;67(4):1091–1093. [Google Scholar]
- Wilson RP, Grundy E, Massy R, Soltis J, Tysse B, Holton M, et al. Butt T. Wild state secrets: ultra-sensitive measurement of micro-movement can reveal internal processes in animals. Frontiers in Ecology and the Environment. 2014;12(10):582–587. doi: 10.1890/140068. [DOI] [Google Scholar]
- Wilson RP, Holton MD, di Virgilio A, Williams H, Shepard ELC, Lambertucci S, et al. Duarte CM. Give the machine a hand: A Boolean time-based decision-tree template for rapidly finding animal behaviours in multisensor data. Methods in Ecology and Evolution. 2018;9:2206–2215. doi: 10.1111/2041-210X.13069. [DOI] [Google Scholar]
- Wilson RP, Holton MD, Walker JS, Shepard ELC, Scantlebury DM, Wilson VL, et al. Jones MW. A spherical-plot solution to linking acceleration metrics with animal performance, state, behaviour and lifestyle. Movement Ecology. 2016;4:22. doi: 10.1186/s40462-016-0088-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RP, Holton M, Wilson VL, Gunner R, Tysse B, Wilson GI, et al. Scantlebury DM. Towards informed metrics for examining the role of human-induced animal responses in tag studies on wild animals. Integrative Zoology. 2019;14:17–29. doi: 10.1111/1749-4877.12328. [DOI] [PubMed] [Google Scholar]
- Wilson RP, Liebsch N. Up-beat motion in swinging limbs: New insights into assessing movement in free-living aquatic vertebrates. Marine Biology. 2003;142(3):537–547. doi: 10.1007/s00227-002-0964-9. [DOI] [Google Scholar]
- Wilson RP, Neate A, Holton MD, Shepard ELC, Scantlebury DM, Lambertucci SA, et al. Marks N. Luck in Food Finding Affects Individual Performance and Population Trajectories. Current Biology. 2018;28(23):3871–3877.e5. doi: 10.1016/j.cub.2018.10.034. [DOI] [PubMed] [Google Scholar]
- Wilson RP, Scolaro A, Quintana F, Siebert U, thor Straten M, Mills K, et al. Müller G. To the bottom of the heart: cloacal movement as an index of cardiac frequency, respiration and digestive evacuation in penguins. Marine Biology. 2004;144:813–827. doi: 10.1007/s00227-003-1247-9. [DOI] [Google Scholar]
- Wilson RP, Simeone A, Luna-Jorquera G, Steinfurth A, Jackson S, Fahlman A. Patterns of respiration in diving penguins: is the last gasp an inspired tactic? The Journal of Experimental Biology. 2003;206:1751–1763. doi: 10.1242/jeb.00341. [DOI] [PubMed] [Google Scholar]
- Wilson RP, Steinfurth A, Ropert-Coudert Y, Kato A, Murita M. Lip-reading in remote subjects: an attempt to quantify and separate ingestion, breathing and vocalisation in free-living animals using penguins as a model. Marine Biology. 2002;140:17–27. doi: 10.1007/s002270100659. [DOI] [Google Scholar]
- Xavier G, Dodge S. An exploratory visualization tool for mapping the relationships between animal movement and the environment. Proceedings of the 2nd ACM SIGSPATIAL International Workshop on Interacting with Maps - MapInteract ’14; 2014. pp. 36–42. [DOI] [Google Scholar]
- Yang G-Z. Body Sensor Networks. London, UK: Springer-Verlag; 2014. [Google Scholar]
- Yoda K, Naito Y, Sato K, Takahashi A, Nishikawa J, Ropert-Coudert Y, et al. Le Maho Y. A new technique for monitoring the behaviour of free-ranging Adélie penguins. The Journal of Experimental Biology. 2001;204:685–690. doi: 10.1016/s0304-3959(97)00161-9. [DOI] [PubMed] [Google Scholar]
- Yoda K, Sato K, Niizuma Y, Kurita M, Bost C-A, Le Maho Y, Naito Y. Precise monitoring of porpoising behaviour of Adélie penguins determined using acceleration data loggers. The Journal of Experimental Biology. 1999;202:3121–3126. doi: 10.1242/jeb.202.22.3121. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.