Abstract
Streptococcus spp. cause a wide range of diseases in animals and humans. A Streptococcus strain (FMD1) was isolated from forest musk deer lung. To identify the bacterium at the species level and investigate its pathogenicity, whole genome sequencing and experimental infections of mice were performed. The genome had 97.63% average nucleotide identity with the S. equinus strain. Through virulence gene analysis, a beta-hemolysin/cytolysin genome island was found in the FMD1 genome, which contained 12 beta-hemolysin/cytolysin-related genes. Hemolytic reaction and histopathological analysis established the strain’s pathogenicity in mice. This is the first report of a beta-hemolytic S. equinus strain in forest musk deer identified based on phenotypic and genotypic analyzes; this strategy could be useful for analyzing pathogens affecting rare animals.
Keywords: beta-hemolytic, forest musk deer, Streptococcus equinus, whole genome analysis
Forest musk deer (Moschus berezovskii) is a medium-sized mammal that inhabits alpine forests. This animal has a high economic value because of the musk secreted by adult males, which plays an important role in traditional Asian medicine and the international perfume industry [23]. Because of the wild origin of the forest musk deer, it is difficult for people (including breeders) to approach the animal, thus, noticing the onset of diseases is also difficult. In December 2018, a 10-year-old forest musk deer at the Sichuan Institute of Musk Deer Breeding (Chengdu, China) suddenly fell down, showed anorexia and purulent nasal secretion, and died before the veterinarian’s arrival. At autopsy, it was observed that the lungs were severely swollen, covered with petechial hemorrhages, and surrounded by a yellow peptone-like exudate (Supplementary Fig. 1). The lung tissue was collected and transported on ice to the laboratory for bacterial examinations.
First, the lung was streak-inoculated onto 5% sheep blood agar plates under aerobic and anaerobic conditions and incubated for 20 hr at 37°C. The single colony was selected and placed on 5% sheep blood agar plates based on the morphological characteristics. Then, the pure culture was subjected to Gram staining and biochemical identification. After this, the total DNA of the isolate was used as a template for a polymerase chain reaction (PCR) to amplify the 16S rRNA region with universal primers 27F and 1492R. The PCR procedure was the same as that described in a previous report [24]. The sequencing results were compared with the reference sequences from the NCBI database via BLAST. For the phylogenetic analysis, a dataset of 16S rRNA gene sequences was built, and all reference sequences were extracted from GenBank with high similarity based on the results of NCBI BLAST. The program used for the phylogenetic analysis was the same as that used in a previous article description [24]. In addition, the genome sequence of the isolate was sequenced at Novogene Bioinformatics Technology Co., Ltd., Beijing, 100000, China. The average nucleotide identity (ANI) and virulence genes of the isolate were identified using an ANI calculator (http://enve-omics.ce.gatech.edu/ani/index) [8] and virulence factor database (VFDB) (http://www.mgc.ac.cn/VFs/main.htm) [12], respectively. The names of the virulence genes were determined using NCBI BLASTn (e-value <1e-10, identity >80%, and coverage >90%) [21]. The complete genomes of S. equinus, S. lutetiensis, and S. infantarius were used as reference genomes for the ANI analysis.
Twelve BALB/c mice were divided into two groups: test and control. All experiments using mice were approved by the committee for the animal experimentation of the Sichuan Agricultural University. The bacterial suspension was diluted to 4.3 × 108 CFU/ml, and six mice were intraperitoneally injected with 0.2 ml of the bacterial suspension. At the same time, the control group was injected with physiological saline. The infected and healthy mice groups were sacrificed after seven days, and then the lungs were immersed in 4% neutral buffered formalin and stained with hematoxylin and eosin for pathological observation. Meanwhile, the lung tissue of dead forest musk deer was processed as outlined above.
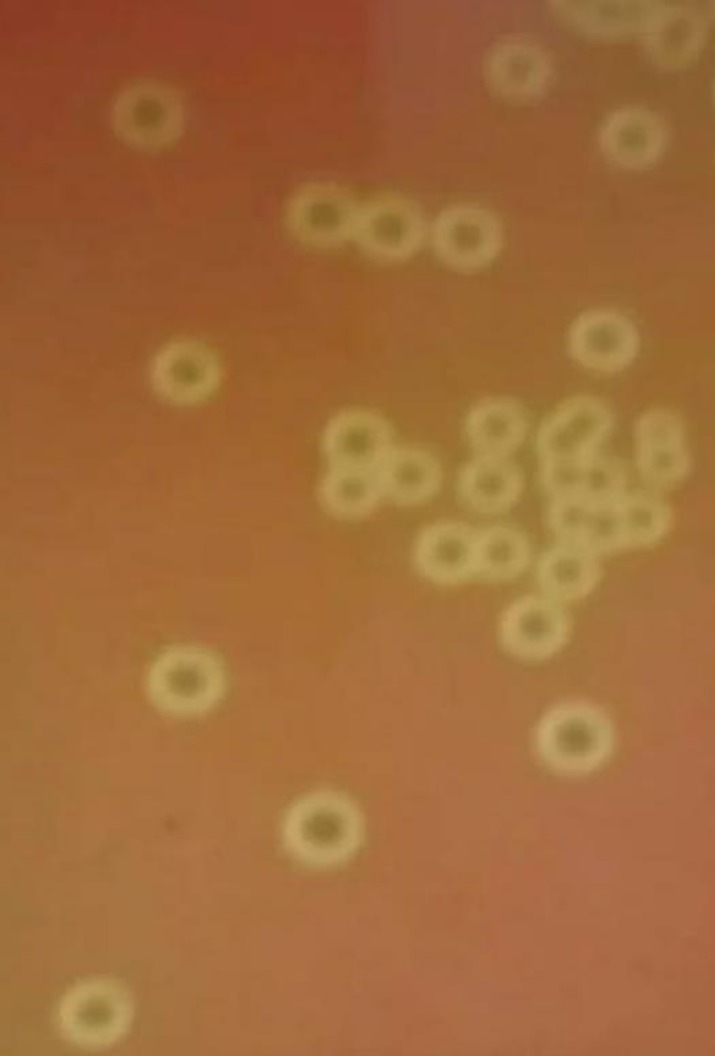
After 20 hr of incubation, smooth, grayish, and neat-edged colonies were found with a small zone of clear beta-hemolysis on the 5% sheep blood agar plates under both aerobic and anaerobic conditions (Fig. 1). Gram staining results showed that the isolate was a Gram-positive, round-shaped bacterium. Moreover, the isolate was negative in mannite, sorbitol, aesculin, inulin, catalase, and trehalose, positive in glucose, lactose, raffinose, and salicylic acid, and could not grow in Trypticase Soy Broth supplemented with 6.5% (w/v) NaCl. Electrophoresis indicated that the size of the PCR product of the 16S rRNA gene was 1,452 bp (Accession No. MK652875), which was highly similar (Ident >99.45%, Query cover=99%) to the 16S rRNA sequences of S. equinus, S. lutetiensis, and S. infantarius. In addition, the FMD1 strain was alone in a clade in the phylogenetic tree based on the 16S rRNA sequences of 60 Streptococci species (Fig. 2). Based on the above-mentioned characteristics, the FMD1 strain was difficult to identify at the species level based on conventional methods and 16S rRNA sequence analysis.
Fig. 1.
Hemolytic phenotype analyses of the Streptococcus equinus FMD1 strain on 5% sheep blood agar plates.
Fig. 2.
Phylogenetic comparisons of 60 Streptococci strains using 16S rRNA gene sequencing. This phylogenetic tree includes 7 Streptococcus bovis 16S rRNA gene sequences, 23 Streptococcus equinus 16S rRNA gene sequences, 3 Streptococcus infantarius 16S rRNA gene sequences, and 26 Streptococcus lutetiensis 16S rRNA gene sequences, along with the FMD1 16S rRNA gene sequence. Multiple sequence alignments were performed using Clustal X 2.1. The phylogenetic tree was constructed using the neighbor-joining method with the MEGA 6.0 program. Genetic distances were determined using Kimura’s 2-parameter model. The robustness of individual branches was estimated using bootstrapping with 1,000 replications. The scale bar corresponds to 0.001 estimated nucleotide substitutions per site.
Streptococcus is a diverse genus, encompassing approximately 77 species of bacteria, which infect a host of different animals [9]. Many Streptococci species are well-known pathogens in humans and animals, including S. pneumoniae, S. suis, and S. equinus [9, 11], and cause a broad range of diseases [5]. In contrast, S. gallolyticus and S. infantarius play important roles in traditional fermented food products across the world [18]. Hence, there is a potential public health risk if Streptococci species are not accurately identified.
To better understand the species level and pathogenicity of the isolated Streptococcus strain, the genome sequence was used for ANI and virulence gene analysis. The whole genome sequence of FMD1 was submitted to GenBank under the accession number SPDR00000000. The FMD1 genome was aligned with the S. equinus NCTC8140 strain assemblies at 97.63% ANI, while the highest ANI with S. lutetiensis and S. infantarius was 87.15% and 86.95%, respectively. The ANI analysis results identified the FMD1 strain as S. equinus.
Conventional methods and PCR analyzes have been developed to improve the species identification of bacteria [2]. However, some studies have encountered challenges in identifying the bacterial species isolated from patients, such as Clostridium, Yersinia, Klebsiella and Streptococcus [10, 13, 15, 16]. The accurate identification of bacteria at the species level has become increasingly more important [6]. Thus, many reviews highly recommended whole genome sequencing and analysis to increase the confidence in the species identification accuracy [4]. ANI is a useful tool, used to improve the accuracy of bacterial identification, and has been proposed as the best option for determining species [4, 22].
Notably, in the FMD1 genome Scaffold 2, a beta-hemolysin/cytolysin genome island (SPDR01000002: 58,344–69,620) was found, which contains 12 beta-hemolysin/cytolysin related genes (Table 1), including cylX, cylD, cylG, acpC, cylZ, cylA, cylB, cylE, cylF, cylI, cylJ, and cylK. The gene structure agreed with the beta-hemolytic genome island, which was found in group B Streptococci and contributed to disease pathogenesis [3]. To the best of our knowledge, a beta-hemolysin S. equinus strain has been isolated from bovine milk by Babu [1]. Previous studies [14, 17] have reported that hemolytic reaction is a strong evidence of the pathogenic potential of microorganisms, and beta-hemolysin may direct tissue injury or the activation of the host inflammatory response. As an important intestinal bacterium, S. equinus was first isolated in 1906, was frequently detected in feces, and thought to be a nonpathogenic bacterium for a long time [19]. In recent years, some diseases caused by S. equinus have been reported [7]. This study is the first report of a beta-hemolytic S. equinus strain in the forest musk deer.
Table 1. Genetic constitution of the beta-hemolysin/cytolysin genome island in the Streptococcus. equinus FMD1 strain.
| Orf ID | Name | Identity (%) | CDS region in nucleotide | VFDB_ID | Product |
|---|---|---|---|---|---|
| Orf01544 | cylX | 96.44 | SPDR01000002:58344–58652 | VFG005761 | Acetyl coenzyme A CoA carboxylase CylX |
| Orf01545 | cylD | 98.22 | SPDR01000002:58649–59494 | VFG005764 | Malonyl-CoA-ACP transacylase CylD |
| Orf01546 | cylG | 97.65 | SPDR01000002:59491–60213 | VFG005766 | CylG protein |
| Orf01547 | acpC | 99.34 | SPDR01000002:60206–60508 | VFG005770 | AcpC acyl carrier protein AcpC |
| Orf01548 | cylZ | 97.69 | SPDR01000002:60495–60971 | VFG005773 | 3R-hydroxymyristoyl ACP dehydratase CylZ |
| Orf01549 | cylA | 98.28 | SPDR01000002:60961–61890 | VFG005776 | ABC ATP-binding cassette transporter CylA |
| Orf01550 | cylB | 98.63 | SPDR01000002:61883–62761 | VFG005779 | ABC ATP-binding cassette transporter CylB |
| Orf01551 | cylE | 97.96 | SPDR01000002:62758–64725 | VFG005780 | CylE protein |
| Orf01552 | cylF | 98.32 | SPDR01000002:64728–65678 | VFG005785 | Aminomethyltransferase CylF |
| Orf01553 | cylI | 97.67 | SPDR01000002:65678–67870 | VFG005788 | Putative 3-ketoacyl-ACP synthase CylI |
| Orf01554 | cylJ | 97.08 | SPDR01000002:67880–69106 | VFG005790 | CylJ protein |
| Orf01555 | cylK | 97.44 | SPDR01000002: 69114–69620 | VFG005793 | CylK protein |
Orf, open reading frame; CDS: coding sequence; VFDB: virulence factors database.
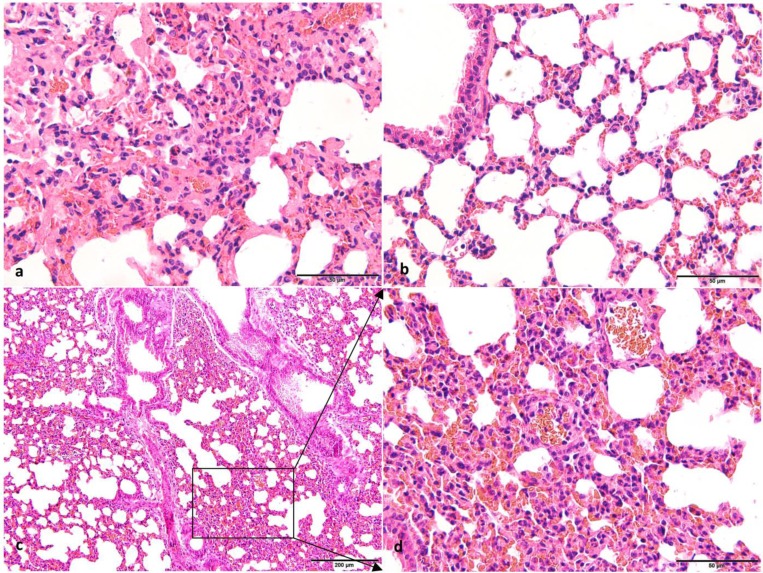
Forest musk deer is a first-class protected animal in China, and therefore animal experimentation using this species is forbidden. We have successfully established a mouse challenge model of the Escherichia coli O78 strain and established a foundation for future research on the pathogenesis of pathogens isolated from forest musk deer [20]. To better understand the pathogenesis of S. equinus FMD1, mice are an excellent experimental animal for replacing forest musk deer. The pathological features of the lungs of mice and forest musk deer showed different degrees of histopathological changes (Fig. 3). The histological lesions in the lung tissues of mice and forest musk deer showed infiltration of numerous erythrocytes and inflammatory cells in the alveolar lumen. The alveolar epithelial cells of mice proliferated, but there was no standard for the alveolar walls of healthy forest musk deer. Lung histological lesions and hemolytic reactions confirmed that the S. equinus FMD1 strain is a pathogenic bacterium affecting mice and forest musk deer.
Fig. 3.
Photomicrographs of lung tissues from mice and forest musk deer. (a) The histological structure of the lung of forest musk deer (H&E. 400×, Bar=50 μm). (b) The histological structure of the lung in the control group mice (H&E. 400×, Bar=50 μm). (c) The histological structure of the lung in the test group mice (H&E. 200×, Bar=200 μm). (d) The histological structure of the lung in the test group mice with high-expansion (H&E. 400×, Bar=50 μm). Histopathological observations showed infiltration of numerous erythrocytes, neutrophils, and monocyte in the alveolar lumen of forest musk deer and test group mice. In addition, an area of thickened alveolar wall was observed in the test group mice. There were no histopathological changes in the control group.
Our results indicate that whole genome analysis is a useful strategy for improving the accuracy of bacterial identification and mining genetic information. To the best of our knowledge, this report is the first to describe the beta-hemolytic S. equinus strain in forest musk deer. However, additional laboratory research investigating the beta-hemolytic genome island of the FMD1 strain is warranted.
Supplementary Material
Acknowledgments
This work was funded by the Transformation Program of Scientific and Technological Achievements of Sichuan Province in 2020.
REFERENCES
- 1.Babu V., Subathra Devi C.2015. Exploring the in vitro thrombolytic potential of streptokinase-producing β-hemolytic Streptococci isolated from bovine milk. J. Gen. Appl. Microbiol. 61: 139–146. doi: 10.2323/jgam.61.139 [DOI] [PubMed] [Google Scholar]
- 2.Birtles R. J., Rowbotham T. J., Raoult D., Harrison T. G.1996. Phylogenetic diversity of intra-amoebal legionellae as revealed by 16S rRNA gene sequence comparison. Microbiology 142: 3525–3530. doi: 10.1099/13500872-142-12-3525 [DOI] [PubMed] [Google Scholar]
- 3.Chou C. C., Lin M. C., Su F. J., Chen M. M.2019. Mutation in cyl operon alters hemolytic phenotypes of Streptococcus agalactiae. Infect. Genet. Evol. 67: 234–243. doi: 10.1016/j.meegid.2018.11.003 [DOI] [PubMed] [Google Scholar]
- 4.Ciufo S., Kannan S., Sharma S., Badretdin A., Clark K., Turner S., Brover S., Schoch C. L., Kimchi A., DiCuccio M.2018. Using average nucleotide identity to improve taxonomic assignments in prokaryotic genomes at the NCBI. Int. J. Syst. Evol. Microbiol. 68: 2386–2392. doi: 10.1099/ijsem.0.002809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clarke L. L., Fathke R. L., Sanchez S., Stanton J. B.2016. Streptococcus bovis/S. equinus complex septicemia in a group of calves following intramuscular vaccination. J. Vet. Diagn. Invest. 28: 423–428. doi: 10.1177/1040638716648364 [DOI] [PubMed] [Google Scholar]
- 6.Dekker J. P., Lau A. F.2016. An update on the Streptococcus bovis group: classification, identification, and disease associations. J. Clin. Microbiol. 54: 1694–1699. doi: 10.1128/JCM.02977-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dotis J., Petinaki E., Printza N., Stampouli S., Papachristou F.2017. Spontaneous peritonitis and bacteremia caused by Streptococcus equinus. Pediatric Dimensions 2: 1–2. doi: 10.15761/PD.1000139 [DOI] [Google Scholar]
- 8.Goris J., Konstantinidis K. T., Klappenbach J. A., Coenye T., Vandamme P., Tiedje J. M.2007. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 57: 81–91. doi: 10.1099/ijs.0.64483-0 [DOI] [PubMed] [Google Scholar]
- 9.Jiang Q., Zhou X., Cheng L., Li M.2019. The adhesion and invasion mechanisms of Streptococci. Curr. Issues Mol. Biol. 32: 521–560. doi: 10.21775/cimb.032.521 [DOI] [PubMed] [Google Scholar]
- 10.Kierzkowska M., Pędzisz P., Babiak I., Janowicz J., Kulig M., Majewska A., Sawicka-Grzelak A., Młynarczyk G.2018. Difficulties in identifying the bacterial species from the genus Clostridium in a case of injury-related osteitis. Folia Microbiol. (Praha) 63: 533–536. doi: 10.1007/s12223-018-0597-0 [DOI] [PubMed] [Google Scholar]
- 11.Li G., Wang G., Si X., Zhang X., Liu W., Li L., Wang J.2019. Inhibition of suilysin activity and inflammation by myricetin attenuates Streptococcus suis virulence. Life Sci. 223: 62–68. doi: 10.1016/j.lfs.2019.03.024 [DOI] [PubMed] [Google Scholar]
- 12.Liu B., Zheng D., Jin Q., Chen L., Yang J.2019. VFDB 2019: a comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 47D1: D687–D692. doi: 10.1093/nar/gky1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martínez-Romero E., Rodríguez-Medina N., Beltrán-Rojel M., Silva-Sánchez J., Barrios-Camacho H., Pérez-Rueda E., Garza-Ramos U.2018. Genome misclassification of Klebsiella variicola and Klebsiella quasipneumoniae isolated from plants, animals and humans. Salud Publica Mex. 60: 56–62. doi: 10.21149/8149 [DOI] [PubMed] [Google Scholar]
- 14.Nizet V., Rubens C. E.2000.. Pathogenic mechanisms and virulence factors of group B Streptococci. pp. 125–136. In: The Gram Positive Pathogens, American Society for Microbiology Press, Washington, DC. [Google Scholar]
- 15.Nishiura H., Yamazaki A., Wakakuri K., Sasaki J., Terajima J., Ochiai K.2019. Yersinia infection in two captive guereza colobus monkeys (Colobus guereza). J. Vet. Med. Sci. 81: 1201–1204. doi: 10.1292/jvms.19-0039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pompilio A., Di Bonaventura G., Gherardi G.2019. An overview on Streptococcus bovis/Streptococcus equinus complex isolates: identification to the species/subspecies level and antibiotic resistance. Int. J. Mol. Sci. 20: 480. doi: 10.3390/ijms20030480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Santos J. A., González C. J., Otero A., García-López M. L.1999. Hemolytic activity and siderophore production in different Aeromonas species isolated from fish. Appl. Environ. Microbiol. 65: 5612–5614. doi: 10.1128/AEM.65.12.5612-5614.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schoustra S. E., Kasase C., Toarta C., Kassen R., Poulain A. J.2013. Microbial community structure of three traditional zambian fermented products: mabisi, chibwantu and munkoyo. PLoS One 8: e63948. doi: 10.1371/journal.pone.0063948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schlegel L., Grimont F., Ageron E., Grimont P. A., Bouvet A.2003. Reappraisal of the taxonomy of the Streptococcus bovis/Streptococcus equinus complex and related species: description of Streptococcus gallolyticus subsp. gallolyticus subsp. nov., S. gallolyticus subsp. macedonicus subsp. nov. and S. gallolyticus subsp. pasteurianus subsp. nov. Int. J. Syst. Evol. Microbiol. 53: 631–645. doi: 10.1099/ijs.0.02361-0 [DOI] [PubMed] [Google Scholar]
- 20.Wang W. Y., Tian Q., Cheng J. G., Zhao W., Deng L., Luo Y.2018. Establishment and evaluation of BALB/c mice challenge model with lung pathogenic Escherichia coli O78 of forest musk deer origin. Microbiology China 45: 1333–1341. [Google Scholar]
- 21.Yan X., Fratamico P. M., Bono J. L., Baranzoni G. M., Chen C. Y.2015. Genome sequencing and comparative genomics provides insights on the evolutionary dynamics and pathogenic potential of different H-serotypes of Shiga toxin-producing Escherichia coli O104. BMC Microbiol. 15: 83. doi: 10.1186/s12866-015-0413-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoon S. H., Ha S. M., Lim J., Kwon S., Chun J.2017. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie van Leeuwenhoek 110: 1281–1286. doi: 10.1007/s10482-017-0844-4 [DOI] [PubMed] [Google Scholar]
- 23.Zhao K. L., Liu Y., Zhang X. Y., Palahati P., Wang H. N., Yue B. S.2011. Detection and characterization of antibiotic-resistance genes in Arcanobacterium pyogenes strains from abscesses of forest musk deer. J. Med. Microbiol. 60: 1820–1826. doi: 10.1099/jmm.0.033332-0 [DOI] [PubMed] [Google Scholar]
- 24.Zhao W., Tian Q., Luo Y., Wang Y., Yang Z. X., Yao X. P., Cheng J. G., Zhou X., Wang W. Y.2017. Isolation, identification, and genome analysis of lung pathogenic Klebsiella pneumoniae (LPKP) in forest musk deer. J. Zoo Wildl. Med. 48: 1039–1048. doi: 10.1638/2016-0241.1 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.