Abstract
Orthoreoviruses have been indentified in several mammals, however, there is no information about orthoreoviruses in shrews. In this study, we screened wild animals in Zambia, including shrews, rodents, and bats for the detection of orthoreoviruses. Two orthoreovirus RNA genomes were detected from a shrew intestinal-contents (1/24) and a bat colon (1/96) sample by reverse-transcription (RT)-PCR targeting the RNA-dependent RNA polymerase gene of orthoreoviruses. Phylogenetic analyses revealed that each of the identified orthoreoviruses formed a distinct branch among members of the Orthoreovirus genus. This is the first report that shrews are susceptible to orthoreovirus infection. Our results suggest the existence of undiscovered orthoreoviruses in shrews and provide important information about the genetic diversity of orthoreoviruses.
Keywords: Crocidura hirta, Orthoreovirus, Rousettus aegyptiacus, Zambia
Orthoreoviruses, belonging to the genus Orthoreovirus in the family Reoviridae, are non-enveloped, icosahedral, segmented double-stranded RNA (dsRNA) viruses [8]. The orthoreovirus genome consists of 10 dsRNA segments, three large segments (L1–L3), three medium segments (M1–M3), and four small segments (S1–S4). Orthoreoviruses have been identified in vertebrates and invertebrates, describing the prevalence of orthoreoviruses in a wide range of hosts, and the Orthoreovirus genus is composed of seven species: Avian orthoreovirus (ARV), Baboon orthoreovirus (BRV), Mahlapitsi orthoreovirus (MAHLV), Mammalian orthoreovirus (MRV), Nelson Bay orthoreovirus (NBV), Piscine orthoreovirus (PRV), and Reptilian orthoreovirus (RRV) [3, 8]. The genus Orthoreovirus is divided into two distinct phylogenetic subgroups: the fusogenic orthoreoviruses such as NBV, ARV, BRV, MAHLV and RRV; and the nonfusogenic orthoreoviruses such as MRV and PRV [3, 6, 8]. MRV, a prototype of the genus Orthoreovirus, is nonfusogenic and has been identified in several mammals, including humans, bats, rodents, dogs, civet cats, cattle, pigs, minks and alpine chamois [1, 2, 4, 7, 17,18,19,20, 26]. In contrast, NBV, a bat-origin orthoreovirus, is a prototype of fusogenic orthoreovirus, inducing the formation of multinucleated syncytia in cell culture, and have also been isolated from humans with respiratory illness [5, 10, 27, 28, 35]. BRV isolated from baboons genetically differed from MRV and NBV, and assigned as the new species within the genus Orthoreovirus [9]. Broome virus, a recently discovered orthoreovirus from a bat, is divergent from any other species, and represents a new although it is not yet formally recognized as the species [29]. These reports have expanded our knowledge about the host range and genetic diversity of orthoreoviruses.
Shrews are small mammals with long, narrow, and pointed snouts, and insectivore, feeding on many invertebrates on the ground. The family Soricidae (shrews), belonging to order Soricomorpha, consists of three subfamilies: Crocidurinae (white-toothed shrews), Soricinae (red-toothed shrews) and Myosoricinae (African white-toothed shrews) [34]. Shrews are similar in size and appearance to rodents classified to order Rodentia, but there is a clear genetic and biological difference between these animals [11, 13]. Althougt tree shrews classified within order Scandentia are similar name to shrews, they are bigger than shrews and the animals inhabit forests on the tree [34]. Phylogenetic analyses based on mitochondrial cytochrome b gene sequences have shown a clear genetic difference between shrews, rodents and bats, and these animals are phylogenetically distinct from each other [11]. To date, various viruses identified in shrews, such as adenoviruses, arteriviruses, bufaviruses, hantaviruses, hepadnaviruses, herpesviruses and nairoviruses, are phylogenetically distinct from those in rodents and bats, indicating the unique shrew virome [11, 13, 21, 24, 31, 32, 36, 37]; however, there has been no report about the presence of orthoreoviruses in shrews.
Discovery of novel orthoreovirus in mammals, such as bats and baboons suggests that there may be a unrecognized orthoreoviruses circulating in a wide range of animal hosts [9, 29]. As part of the activities aimed at identifying potential pathogens in Zambian wildlife, we screened shrews, rodents and bats for the detection of orthoreoviruses and performed phylogenetic analysis of detected viruses.
With permission from Zambia Wildlife Authority, now the Department of National Parks and Wildlife (Act No. 12 of 1998), Ministry of Tourism and Arts [23,24,25], we captured 24 shrews and 48 rodents around houses and fields using sherman traps and cage traps in Mpulungu in 2012, and 96 bats captured using a harp trap or a shotgun in Lusaka, Ndola, Monze, Livingstone, and Kasanka National Park in Zambia during the period 2014–2015 (Supplementary Fig. 1). We collected intestinal contents from the shrews and the rodents, and colon samples from the bats . We used intestinal contents and colon samples for screening of the orthoreovirus genome, because some orthoreoviruses were isolated from intestine, feces, intestinal contents and rectal swab in various animals [1, 7, 12, 14, 16,17,18,19,20, 22, 26, 27]. These samples were kept at −80°C, and we isolated RNA from them to screen for the orthoreovirus genome (Table 1). In our previous studies, bat species were identified by morphology and sequencing of ribosomal RNA and the mitochondrial cytochrome b gene; sequencing of the latter was also used to identify rodent and shrew species [23, 25].
Table 1. Information of samples and reverse-transcription (RT)-PCR results.
| Animal species | No. of positive/ No. tested sample |
Sampling place | Sampling year | |
|---|---|---|---|---|
| Shrew | ||||
| Crocidura hirta | 1/23 | Mpulungu | 2012 | |
| Crocidura luna | 0/1 | |||
| Total | 1/24 | |||
| Bat | ||||
| Eidolon helvum | 0/9 | Kasanka NP | 2014 | |
| Eidolon helvum | 0/9 | Ndola | ||
| Epomophorus crypturus | 0/20 | Monze | ||
| Rousettus aegyptiacus | 1/9 | Lusaka | ||
| Hipposideros sp. | 0/10 | Lusaka | ||
| Miniopterus schreibersii | 0/9 | Lusaka | ||
| Nycteris sp. | 0/20 | Livingstone | ||
| Rousettus aegyptiacus | 0/10 | Lusaka | 2015 | |
| Total | 1/96 | |||
| Rodent | ||||
| Aethomys chrysophilus | 0/6 | Mpulungu | 2012 | |
| Cricetomys gambianus | 0/3 | |||
| Gerbilliscus leucogaster | 0/1 | |||
| Grammomys sp. | 0/1 | |||
| Mastomys natalensis | 0/28 | |||
| Paraxerus cepapi | 0/2 | |||
| Rattus rattus | 0/3 | |||
| Saccostomus sp. | 0/3 | |||
| Steatomys sp. | 0/1 | |||
| Total | 0/48 | |||
Intesitinal-contents suspensions in phosphate-buffered saline [10% (w/v)] and colon homogenates in Dulbecco’s Modified Eagle’s Medium containing 10% fetal bovine serum and 4% antibiotic–antimycotic solution (Life Technologies, Gibco, Waltham, MA, USA) [10% (w/v)] were prepared and briefly centrifuged. Total RNA was extracted from the supernatants of intesitinal-content suspensions and colon homogenates using the High Pure RNA Isolation Kit (Roche, Basel, Switzerland) and QIAamp Viral RNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacturers’ instructions, respectively. Nested reverse-transcription (RT)-PCR was performed using degenerate primers designed from a conserved sequence within the RNA-dependent RNA polymerase gene (RdRp) as previously described [33]. Briefly, viral gene fragments were amplified with PrimeScript One Step RT-PCR Kit Ver.2 (Takara Bio Inc., Kusatsu, Japan) using the forward primer 1607F (5′-CARMGNCGNSCHMGHTCHATHATGCC-3′) and the reverse primer 2608R (5′-TAVAYRAAVGWCCASMHNGGRTAYTG-3′) for first-round RT-PCR according to the manufacturers’ protocol. Nested PCR was performed with Takara Ex Taq hot start version (Takara Bio Inc.) using the forward primer 2090F (5′-GGBTCMACNGCYACYTCBACYGAGCA-3′) and the reverse primer 2334R (5′-CDATGTCRTAHWYCCANCCRAA- 3′) according to the manufacturers’ protocol. NBV RNA [35] was used as a positive control for the RT-PCR assay. PCR products of 226 bp were purified from an agarose gel and subjected to direct sequencing using the Big Dye Terminator v3.1 Cycle Sequencing kit (Applied Biosystems, Foster City, CA, USA).
For virus isolation, shrew intestinal-content suspensions and bat colon homogenates were prepared from orthoreovirus nested RT-PCR-positive samples and inoculated onto monolayers of African green monkey kidney cells (Vero E6) and baby hamster kidney (BHK) cells previously described for the isolation of orthoreoviruses [9, 17,18,19, 27, 29, 35].
Among 24 shrew intestinal-contents and 96 bat colon samples, one from a shrew (Crocidura hirta) and one specimen from a fruit bat (Rousettus aegyptiacus) were positive for orthoreovirus by nested RT-PCR assay (Table 1). We next attempted to amplify a different region of RdRp using semi-nested PCR with the first RT-PCR product as a template, and using degenerate primers and specific primers designed from the obtained sequence. A longer region of RdRp was amplified by semi-nested PCR from Zambian shrew orthoreovirus using the degenerate forward primer 1607F and the specific reverse primer 2140R (5′-GAAGAGCATTGTCTAGATGG-3′), and using the specific forward primer 2125F (5′-TACTATGATGCAATGCTTCC-3′) and the degenerate reverse primer 2608R with KOD FX Neo DNA polymerase (Toyobo, Osaka, Japan). Finally, a 1,000 bp length fragment of orthoreoviral RdRp was obtained from the shrew specimen. However, despite several attempts, we failed to amplify a longer RdRp fragment from the bat specimen. The two partial sequences of RdRp obtained from the bat colon and the shrew intesitinal-contents were deposited in the DNA Data Bank of Japan (DDBJ) under accession numbers (DDBJ/EMBL/GenBank LC486243 and LC486244, respectively).
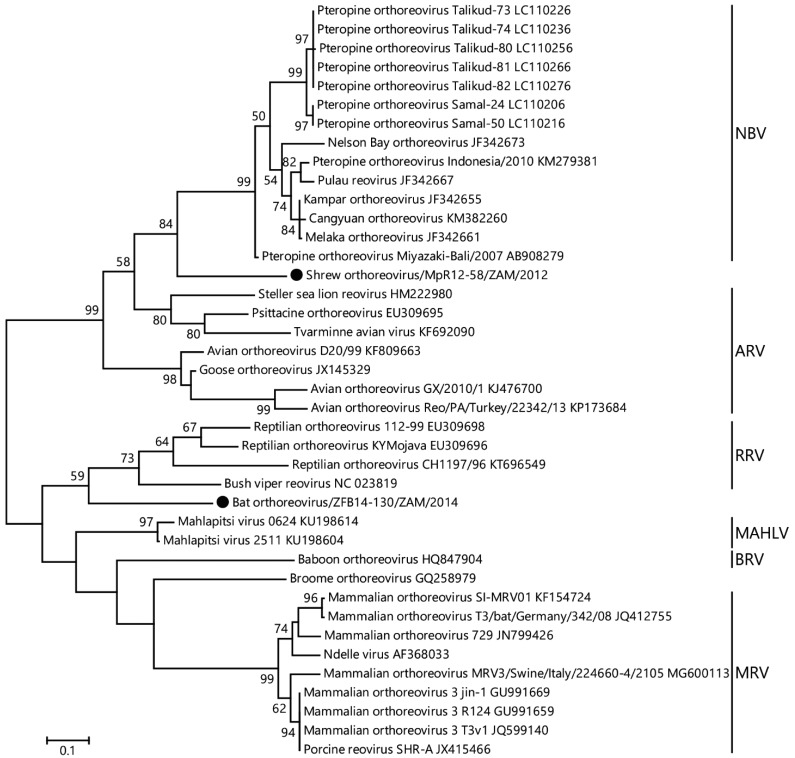
BLAST analyses indicated that the identified RdRp sequence of the Zambian shrew orthoreoviruses showed the highest similarity to RdRp of NBV. Pairwise comparison of the partial RdRp sequence from the shrew specimen showed 69.6% nucleotide sequence identity to that of NBV Miyazaki-Bali/2007 (GenBank accession no. AB908279), which was identified from humans returning to Japan from Bali, Indonesia [35]. The highest amino acid identity was 78.4% to ARV Pycno-1 (GenBank accession no. BAQ19494), which was identified from Hypsipetes amaurotis in Japan [22]. Phylogenetic analyses of the partial RdRp nucleotide sequences and amino acid sequences predicted based on obtained nucleotide sequences were conducted with representative orthoreovirus sequences available in GenBank (Supplementary Table 1). The sequences were aligned using the ClustalW protocol [30], and phylogenetic trees were generated using the maximum likelihood method based on the Tamura-Nei and JTT matrix-based model with 1,000 bootstrap replications by MEGA7 software [15]. In the phylogenetic tree based on 1,000 bp sequences from orthoreovirus RdRp, the Zambian shrew orthoreovirus segregated from other orthoreoviruses (Fig. 1). The Zambian shrew orthoreovirus shared a common origin with NBV and was located independently from any known orthoreoviruses. According to the phylogenetic analysis based on predicted 333 amino acid sequence of orthoreovirus RdRp, the Zambian shrew orthoreovirus formed an independent branch from the ancestor of NBV and ARV (Supplementary Fig. 2). To date, no information about orthoreoviruses in the shrew is available. Our results suggest that shrews are susceptible to orthoreovirus infection.
Fig. 1.
A phylogenetic tree based on 1,000 bp sequences from orthoreovirus RdRp corresponding to position 1601–2606 of the NBV L2 genome was constructed using the maximum likelihood method with 1,000 bootstrap replications. Bootstrap values greater than 50% based on 1,000 replications are shown on the interior nodes, and species names are indicated on the tree. Bar, 0.1 substitutions per site. A black circle represents the Zambian shrew orthoreovirus (GenBank accession no. LC486244) detected in this study. Species abbreviations of the genus orthoreovirus are as follows: ARV, Avian orthoreovirus; BRV, Baboon orthoreovirus; MAHLV, Mahlapitsi orthoreovirus; MRV, Mammalian orthoreovirus; NBV, Nelson Bay orthoreovirus; RRV, Reptilian orthoreovirus.
The identified RdRp sequences of the Zambian bat orthoreoviruses was related to that of RRV, and the highest identity is 64.0% to RRV 112-99 (GenBank accession no. EU309698), which was identified from a Boa constrictor in Germany [33]. The amino acid sequence of the Zambian bat orthoreovirus showed 60.0% to those of RRV CH1197/96 (GenBank accession no. ABY28252), which was identified from a Testudo graeca in Switzerland [33]. To infer phylogenetic relationship with the Zambian bat orthoreovirus, we constructed a phylogenetic tree based on 226 bp and predicted 75 amino acid sequences of orthoreovirus RdRp. The Zambian bat orthoreovirus also formed a distinct branch among members of the Orthoreovirus genus, and the Zambian shrew orthoreovirus showed the similar topology as shown in Fig. 1 (Fig. 2 and Supplementary Fig. 3). Although the Zambian bat orthoreovirus was phylogenetically located near the clade of RRV or Broome virus isolated from bats in Australia, the nucleotide and amino acid identities were less than 64.0% and 60.0% to any other known orthoreoviruses as described above, respectively. These results suggest that the Zambian bat orthoreovirus is genetically distict from other orthoreoviruses previously identified in bats. Throughout the experiment for virus isolation, no cytopathic effect was observed and no orthoreovirus genome was detected in the culture supernatants by nested RT-PCR.
Fig. 2.
A phylogenetic analysis based on 226 bp from the orthoreovirus RdRp corresponding to position 2084–2309 of NBV L2 genome was performed using the maximum likelihood method with 1,000 bootstrap replications. Bootstrap values greater than 50% based on 1,000 replications are shown on the interior nodes, and the species names are indicated on the tree. Bar, 0.1 substitutions per site. Black circles represent the Zambian shrew orthoreovirus (GenBank accession no. LC486244) and the Zambian bat orthoreovirus (GenBank accession no. LC486243) detected in this study. Species abbreviations of the genus orthoreovirus are as follows: ARV, Avian orthoreovirus; BRV, Baboon orthoreovirus; MAHLV, Mahlapitsi orthoreovirus; MRV, Mammalian orthoreovirus; NBV, Nelson Bay orthoreovirus; RRV, Reptilian orthoreovirus.
In this study, we detected two unique orthoreoviruses from a shrew and a bat in Zambia. Orthoreoviruses have been reported in various mammalian hosts, except for shrews [1, 2, 4, 5, 7, 9, 10, 17,18,19,20, 26,27,28,29, 35]. Although shrews are similar in size and appearance to rodents, they are biologically genetically different from rodents and classified to other order [34]. This is the first report for the detection of orthoreovirus genome in intesitinal contents of shrews captured in Zambia. In previous studies, clear phylogenetic divisions between various viruses from shrews, rodents, and bats were obsearved [11, 13, 21, 24, 31, 32, 36, 37]. The Zambian shrew orthoreovirus was also phylogenetically distinct from any other orthoreoviruses identified in rodents and bats (Fig. 1). Thus, there may be some differences in the genetic diversity of orthoreoviruses between shrews and other mammal. Bat-derived orthoreoviruses discovered in Australia, Asia and Europe belong to the clade of either NBV or MRV except Broome virus [5, 10, 20, 27,28,29, 35]. However, the Zambian bat orthoreovirus was located outside of the clades of NBVs and MRVs and formed a distinct branch among the members of Orthoreovirus. Our results suggest that orthoreoviruses in bats are genetically devided to at least four groups, showing the genetic diversity of bat orthoreoviruses. According to the species demarcation criteria in the genus Orthoreovirus of the International Committee on Taxonomy of Viruses [3], these two orthoreoviruses do not belong to any known orthoreovirus species, and we suppose that they may be novel orthoreovirus species. However, we need to obtained more information about their genomes. Additionally, phylogenetic analyses revealed that the Zambian shrew orthoreovirus shared a common origin with NBV, which was a causative agent of repiratory illness in humans (Fig. 1). Further epidemiological studies in humans and animals are required to clarify the distribution and risk of orthoreovirus infection in Zambia.
Supplementary Material
Acknowledgments
We would like to thank Dr. Masayuki Saijo and Dr. Satoshi Taniguchi from the National Institute of Infectious Diseases, Japan for for providing orthoreovirus RNA. This study was supported by grants for Grant-in-Aid Scientific Research (B) from the Ministry of Education, Culture, Sports, Science and Technology, Japan (MEXT)/Japan Society for the Promotion of Science (JSPS) KAKENHI (JP16H05805); and grants for Scientific Research on Innovative Areas and International Group from the MEXT/JSPS KAKENHI (JP16H06431, JP16H06429, JP16K21723), and the Japan Initiative for Global Research Network of Infectious Diseases (J-GRID) from Japan Agency for Medical Research and Development (AMED) (JP19fm0108008); and grants for the AMED and Japan International Cooperation Agency (JICA) within the framework of the Science and Technology Research Partnership for Sustainable Development (SATREPS) (JP19jm0110019).
REFERENCES
- 1.Anbalagan S., Spaans T., Hause B. M.2014. Genome Sequence of the Novel Reassortant Mammalian Orthoreovirus Strain MRV00304/13, Isolated from a Calf with Diarrhea from the United States. Genome Announc. 2: e00451–e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Attoui H., Biagini P., Stirling J., Mertens P. P., Cantaloube J. F., Meyer A., de Micco P., de Lamballerie X.2001. Sequence characterization of Ndelle virus genome segments 1, 5, 7, 8, and 10: evidence for reassignment to the genus Orthoreovirus, family Reoviridae. Biochem. Biophys. Res. Commun. 287: 583–588. doi: 10.1006/bbrc.2001.5612 [DOI] [PubMed] [Google Scholar]
- 3.Attoui H., Mertens P., Becnel J., Belaganahalli S., Bergoin M., Brussaard C., Chappell J., Ciarlet M., del Vas M., Dermody T.2011. Orthoreovirus, Reoviridae pp. 546–554. In: Virus Taxonomy Classification and Nomenclature of Viruses: Ninth Report of the International Committee on the Taxonomy of Viruses, Elsevier Academic Press, London. [Google Scholar]
- 4.Besozzi M., Lauzi S., Lelli D., Lavazza A., Chiapponi C., Pisoni G., Viganò R., Lanfranchi P., Luzzago C.2019. Host range of mammalian orthoreovirus type 3 widening to alpine chamois. Vet. Microbiol. 230: 72–77. doi: 10.1016/j.vetmic.2019.01.012 [DOI] [PubMed] [Google Scholar]
- 5.Chua K. B., Crameri G., Hyatt A., Yu M., Tompang M. R., Rosli J., McEachern J., Crameri S., Kumarasamy V., Eaton B. T., Wang L. F.2007. A previously unknown reovirus of bat origin is associated with an acute respiratory disease in humans. Proc. Natl. Acad. Sci. USA 104: 11424–11429. doi: 10.1073/pnas.0701372104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Day J. M.2009. The diversity of the orthoreoviruses: molecular taxonomy and phylogentic divides. Infect. Genet. Evol. 9: 390–400. doi: 10.1016/j.meegid.2009.01.011 [DOI] [PubMed] [Google Scholar]
- 7.Decaro N., Campolo M., Desario C., Ricci D., Camero M., Lorusso E., Elia G., Lavazza A., Martella V., Buonavoglia C.2005. Virological and molecular characterization of a mammalian orthoreovirus type 3 strain isolated from a dog in Italy. Vet. Microbiol. 109: 19–27. doi: 10.1016/j.vetmic.2005.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dermody T. S., Parker J. S., Sherry B.2013. Orthoreoviruses. pp. 1304–1346. In: Fields Virology, vol. 2 6th edn. (Knipe, D. M., Howley, P. M., Cohen, J. I., Griffin, D. E., Lamb, R. A., Martin, M. A., Racaniello, V. R. and Roizman, B. eds.), Wolters Kluwer/Lippincott Williams, & Winlkins, Philadelphia. [Google Scholar]
- 9.Duncan R., Murphy F. A., Mirkovic R. R.1995. Characterization of a novel syncytium-inducing baboon reovirus. Virology 212: 752–756. doi: 10.1006/viro.1995.1536 [DOI] [PubMed] [Google Scholar]
- 10.Gard G., Compans R. W.1970. Structure and cytopathic effects of Nelson Bay virus. J. Virol. 6: 100–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo W. P., Lin X. D., Wang W., Tian J. H., Cong M. L., Zhang H. L., Wang M. R., Zhou R. H., Wang J. B., Li M. H., Xu J., Holmes E. C., Zhang Y. Z.2013. Phylogeny and origins of hantaviruses harbored by bats, insectivores, and rodents. PLoS Pathog. 9: e1003159. doi: 10.1371/journal.ppat.1003159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu T., Qiu W., He B., Zhang Y., Yu J., Liang X., Zhang W., Chen G., Zhang Y., Wang Y., Zheng Y., Feng Z., Hu Y., Zhou W., Tu C., Fan Q., Zhang F.2014. Characterization of a novel orthoreovirus isolated from fruit bat, China. BMC Microbiol. 14: 293. doi: 10.1186/s12866-014-0293-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kang H. J., Bennett S. N., Hope A. G., Cook J. A., Yanagihara R.2011. Shared ancestry between a newfound mole-borne hantavirus and hantaviruses harbored by cricetid rodents. J. Virol. 85: 7496–7503. doi: 10.1128/JVI.02450-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kosoltanapiwat N., Reamtong O., Okabayashi T., Ampawong S., Rungruengkitkun A., Thiangtrongjit T., Thippornchai N., Leaungwutiwong P., Mahittikorn A., Mori H., Yoohanngoa T., Yamwong P.2018. Mass spectrometry-based identification and whole-genome characterisation of the first pteropine orthoreovirus isolated from monkey faeces in Thailand. BMC Microbiol. 18: 135. doi: 10.1186/s12866-018-1302-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar S., Stecher G., Tamura K.2016. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 33: 1870–1874. doi: 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lelli D., Moreno A., Steyer A., Nagliˇc T., Chiapponi C., Prosperi A., Faccin F., Sozzi E., Lavazza A.2015. Detection and characterization of a novel reassortant mammalian orthoreovirus in bats in Europe. Viruses 7: 5844–5854. doi: 10.3390/v7112908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lelli D., Beato M. S., Cavicchio L., Lavazza A., Chiapponi C., Leopardi S., Baioni L., De Benedictis P., Moreno A.2016. First identification of mammalian orthoreovirus type 3 in diarrheic pigs in Europe. Virol. J. 13: 139. doi: 10.1186/s12985-016-0593-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Z., Shao Y., Liu C., Liu D., Guo D., Qiu Z., Tian J., Zhang X., Liu S., Qu L.2015. Isolation and pathogenicity of the mammalian orthoreovirus MPC/04 from masked civet cats. Infect. Genet. Evol. 36: 55–61. doi: 10.1016/j.meegid.2015.08.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lian H., Liu Y., Zhang S., Zhang F., Hu R.2013. Novel orthoreovirus from mink, China, 2011. Emerg. Infect. Dis. 19: 1985–1988. doi: 10.3201/eid1912.130043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Naglič T., Rihtarič D., Hostnik P., Toplak N., Koren S., Kuhar U., Jamnikar-Ciglenečki U., Kutnjak D., Steyer A.2018. Identification of novel reassortant mammalian orthoreoviruses from bats in Slovenia. BMC Vet. Res. 14: 264. doi: 10.1186/s12917-018-1585-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nie F. Y., Tian J. H., Lin X. D., Yu B., Xing J. G., Cao J. H., Holmes E. C., Ma R. Z., Zhang Y. Z.2019. Discovery of a highly divergent hepadnavirus in shrews from China. Virology 531: 162–170. doi: 10.1016/j.virol.2019.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ogasawara Y., Ueda H., Kikuchi N., Kirisawa R.2015. Isolation and genomic characterization of a novel orthoreovirus from a brown-eared bulbul (Hypsipetes amaurotis) in Japan. J. Gen. Virol. 96: 1777–1786. doi: 10.1099/vir.0.000110 [DOI] [PubMed] [Google Scholar]
- 23.Sasaki M., Muleya W., Ishii A., Orba Y., Hang’ombe B. M., Mweene A. S., Moonga L., Thomas Y., Kimura T., Sawa H.2014. Molecular epidemiology of paramyxoviruses in Zambian wild rodents and shrews. J. Gen. Virol. 95: 325–330. doi: 10.1099/vir.0.058404-0 [DOI] [PubMed] [Google Scholar]
- 24.Sasaki M., Orba Y., Anindita P. D., Ishii A., Ueno K., Hang’ombe B. M., Mweene A. S., Ito K., Sawa H.2015. Distinct lineages of bufavirus in wild shrews and nonhuman primates. Emerg. Infect. Dis. 21: 1230–1233. doi: 10.3201/eid2107.141969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sasaki M., Kajihara M., Changula K., Mori-Kajihara A., Ogawa H., Hang’ombe B. M., Mweene A. S., Simuunza M., Yoshida R., Carr M., Orba Y., Takada A., Sawa H.2018. Identification of group A rotaviruses from Zambian fruit bats provides evidence for long-distance dispersal events in Africa. Infect. Genet. Evol. 63: 104–109. doi: 10.1016/j.meegid.2018.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steyer A., Gutiérrez-Aguire I., Kolenc M., Koren S., Kutnjak D., Pokorn M., Poljšak-Prijatelj M., Racki N., Ravnikar M., Sagadin M., Fratnik Steyer A., Toplak N.2013. High similarity of novel orthoreovirus detected in a child hospitalized with acute gastroenteritis to mammalian orthoreoviruses found in bats in Europe. J. Clin. Microbiol. 51: 3818–3825. doi: 10.1128/JCM.01531-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takemae H., Basri C., Mayasari N. L. P. I., Tarigan R., Shimoda H., Omatsu T., Supratikno, Pramono D., Cahyadi D. D., Kobayashi R., Iida K., Mizutani T., Maeda K., Agungpriyono S., Hondo E.2018. Isolation of pteropine orthoreovirus from pteropus vampyrus in garut, Indonesia. Virus Genes 54: 823–827. doi: 10.1007/s11262-018-1603-y [DOI] [PubMed] [Google Scholar]
- 28.Taniguchi S., Maeda K., Horimoto T., Masangkay J. S., Puentespina R., Jr, Alvarez J., Eres E., Cosico E., Nagata N., Egawa K., Singh H., Fukuma A., Yoshikawa T., Tani H., Fukushi S., Tsuchiaka S., Omatsu T., Mizutani T., Une Y., Yoshikawa Y., Shimojima M., Saijo M., Kyuwa S.2017. First isolation and characterization of pteropine orthoreoviruses in fruit bats in the Philippines. Arch. Virol. 162: 1529–1539. doi: 10.1007/s00705-017-3251-2 [DOI] [PubMed] [Google Scholar]
- 29.Thalmann C. M., Cummins D. M., Yu M., Lunt R., Pritchard L. I., Hansson E., Crameri S., Hyatt A., Wang L. F.2010. Broome virus, a new fusogenic Orthoreovirus species isolated from an Australian fruit bat. Virology 402: 26–40. doi: 10.1016/j.virol.2009.11.048 [DOI] [PubMed] [Google Scholar]
- 30.Thompson J. D., Higgins D. G., Gibson T. J.1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680. doi: 10.1093/nar/22.22.4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vanmechelen B., Vergote V., Laenen L., Koundouno F. R., Bore J. A., Wada J., Kuhn J. H., Carroll M. W., Maes P.2018. Expanding the arterivirus host spectrum: olivier’s shrew virus 1, a novel arterivirus discovered in African giant shrews. Sci. Rep. 8: 11171. doi: 10.1038/s41598-018-29560-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walker P. J., Widen S. G., Firth C., Blasdell K. R., Wood T. G., Travassos da Rosa A. P., Guzman H., Tesh R. B., Vasilakis N.2015. Genomic characterization of yogue, kasokero, issyk-kul, keterah, gossas, and thiafora viruses: nairoviruses naturally infecting bats, shrews, and ticks. Am. J. Trop. Med. Hyg. 93: 1041–1051. doi: 10.4269/ajtmh.15-0344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wellehan J. F., Jr, Childress A. L., Marschang R. E., Johnson A. J., Lamirande E. W., Roberts J. F., Vickers M. L., Gaskin J. M., Jacobson E. R.2009. Consensus nested PCR amplification and sequencing of diverse reptilian, avian, and mammalian orthoreoviruses. Vet. Microbiol. 133: 34–42. doi: 10.1016/j.vetmic.2008.06.011 [DOI] [PubMed] [Google Scholar]
- 34.Wilson D. E., Reeder D. M.2005. Mammal Species of the World: A Taxonomic and Geographic Reference, 3rd edn. Johns Hopkins University Press, Baltimore. [Google Scholar]
- 35.Yamanaka A., Iwakiri A., Yoshikawa T., Sakai K., Singh H., Himeji D., Kikuchi I., Ueda A., Yamamoto S., Miura M., Shioyama Y., Kawano K., Nagaishi T., Saito M., Minomo M., Iwamoto N., Hidaka Y., Sohma H., Kobayashi T., Kanai Y., Kawagishi T., Nagata N., Fukushi S., Mizutani T., Tani H., Taniguchi S., Fukuma A., Shimojima M., Kurane I., Kageyama T., Odagiri T., Saijo M., Morikawa S.2014. Imported case of acute respiratory tract infection associated with a member of species nelson bay orthoreovirus. PLoS One 9: e92777. doi: 10.1371/journal.pone.0092777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zheng X. Y., Qiu M., Ke X. M., Guan W. J., Li J. M., Huo S. T., Chen S. W., Zhong X. S., Zhou W., Xiong Y. Q., Ge J., Chen Q.2016. Detection of novel adenoviruses in fecal specimens from rodents and shrews in southern China. Virus Genes 52: 417–421. doi: 10.1007/s11262-016-1315-0 [DOI] [PubMed] [Google Scholar]
- 37.Zheng X. Y., Qiu M., Ke X. M., Zhou W., Guan W. J., Chen S. W., Li J. M., Huo S. T., Chen H. F., Jiang L. N., Zhong X. S., Xiong Y. Q., Ma S. J., Ge J., Chen Q.2016. Molecular detection and phylogenetic characteristics of herpesviruses in rectal swab samples from rodents and shrews in southern China. Vector Borne Zoonotic Dis. 16: 476–484. doi: 10.1089/vbz.2015.1908 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.