Abstract
High-producing dairy cows are easily affected by left displacement of the abomasum (LDA) within 4 weeks postpartum. Although LDA is highly associated with metabolic disturbances, the related information on comprehensive metabolic changes, with the exception of some blood biochemical parameters, remains limited. In this study, the changes in plasma metabolites and in the metabolic profile of postpartum dairy cows with LDA were investigated through liquid chromatography coupled with quadrupole time of flight mass spectrometry (LC-Q/TOF-MS)-based metabolomics, and the metabolic networks related to LDA were constructed through metabolomics pathway analysis (MetPA). An obvious change in the metabolic profile was reflected by significant variations in 68 plasma metabolites in postpartum dairy cows with LDA, and these variations consequently altered 13 metabolic pathways (histidine metabolism, tyrosine metabolism, valine, leucine and isoleucine biosynthesis, phenylalanine, tyrosine and tryptophan biosynthesis, arginine and proline metabolism, tryptophan metabolism, synthesis and degradation of ketone bodies, linoleic acid metabolism, arachidonic acid metabolism, citrate cycle, butanoate metabolism, vitamin B6 metabolism and pyrimidine metabolism). This study shows that the more detailed information obtained by LC-Q/TOF-MS-based metabolomics and MetPA might contribute to a better understanding of the disordered metabolic networks in postpartum dairy cows with LDA.
Keywords: left displacement of the abomasum, liquid chromatography coupled with quadrupole time of flight mass spectrometry, metabolomics, pathway analysis, postpartum dairy cow
Left displacement of the abomasum (LDA) is often observed in high-producing dairy cows mainly within 4 weeks postpartum [2, 12, 14]. Specifically, LDA occurs when the abomasum migrates from its normal position to the left lateral abdominal wall, and this migration can cause anorexia and colic in dairy cows and even death in some cases [45]. Dairy cows with LDA exhibit a lower health status, decreased fertility, less milk production and hence a greater culling rate [11, 26]. Many studies have focused on factors related to LDA, and the findings suggest that the species, breed, gender, age, production level, nutrition, metabolism and concurrent diseases likely play an important role in the pathogenesis of LDA [13]. Marked variations in some blood biochemical parameters, such as amyloid A, haptoglobin, non-esterified fatty acids (NEFAs), beta-hydroxy-butyrate (BHBA), Ca, P, Mg, Cl, urea, glucose, aspartate aminotransferase and glutamate dehydrogenase, indicate that the metabolic status of lactating dairy cows with abomasum displacement is strikingly altered [18, 32, 34]. Nevertheless, the available information on comprehensive metabolic changes remains limited, and this topic should be further explored using omics technologies.
Metabolomics, as a branch of omics research, aims to identify and quantify small endogenous metabolites in biological samples using a high-throughput analytical platform, such as nuclear magnetic resonance (NMR), liquid chromatography/mass spectrometry (LC-MS) and gas chromatography-mass spectrometry (GC-MS) [49]. Metabolomics has been increasingly utilized in studies of diseases in postpartum dairy cows [27, 28, 47, 48, 50], and the relevant metabolites in dairy cows with a displaced abomasum have been determined through NMR-based metabolomics [3]. Considering the lower sensitivity of the NMR technique, further studies of dairy cows with a displaced abomasum should be performed using other high-throughput analysis technologies. LC-MS has the best sensitivity, selectivity and reproducibility among the analytical platforms used in metabolomics studies and is considered one of the most powerful tools for these studies [7, 53]. Furthermore, the LC-MS technique has been improved to separate and identify more lipid classes and is currently considered a prominent metabolomics technique for metabolite identification [52]. Faster and more reliable technology for liquid chromatography coupled with quadrupole time of flight mass spectrometry (LC-Q/TOF-MS) has been developed for the simultaneous analysis of a large number of blood metabolites in metabolomics studies [33].
To better understand the complex metabolic changes associated with LDA, this study designed an LC-Q/TOF-MS method for a comprehensive analysis of blood metabolites in postpartum dairy cows with LDA. Moreover, metabolomics pathway analysis (MetPA) was performed to clearly illuminate the system-level effects of the variation in these metabolites and construct the pathways related to LDA. To the best of our knowledge, this study constitutes the first application of LC-Q/TOF-MS-based metabolomics and MetPA for the analysis of the changes in blood metabolites and their related pathways in postpartum dairy cows with LDA, and the results might provide more detailed information for elucidating the metabolic responses and characterizing the metabolic pathways related to LDA.
MATERIALS AND METHODS
Selection of experimental dairy cows and plasma preparation
This research was performed in accordance to international, national and institutional guidelines for animal experiments and clinical studies and was authorized by the Animal Ethics Committee of Ningxia University (authorization number: 0126/2017). Over a period of 1 year, 15 Holstein dairy cows with LDA (Group D) and 10 clinically healthy dairy cows (Group H) were selected from the same modern dairy farm with approximately 5,000 cows in the Yinchuan suburbs of China. The LDA and healthy cows were fed the same total mixed rations (TMR) prior to delivery and another TMR after delivery (Supplementary Table 1), and blood samples collected at D 20~D 28 after delivery to avoid any possible effects of different physiological stages and feeding and management conditions. To identify LDA as early as possible, various clinical examinations, including food consumption, milk production, percussion and auscultation at the left rib cage, were conducted by an experienced veterinarian at the farm.
The blood samples from the LDA cows were collected immediately after suspicion of presence of the disease according to clinical signs, and then determined to be saved or not by later definitive diagnosis via surgery. Blood samples from the clinically healthy dairy cows were simultaneously collected at D 25 after delivery. All the blood samples were collected in vacuum tubes with 10 IU/ml heparin and immediately centrifuged at 1,500 × g and 4°C for 10 min. The plasma from all the blood samples was extracted into 2-ml Eppendorf tubes, snap-frozen in liquid nitrogen, and transferred to an ultralow-temperature freezer at −80°C until use for LC-Q/TOF-MS analysis.
Pretreatment of plasma samples
Prior to LC-Q/TOF-MS, the plasma samples were simultaneously thawed by immersing in a bath at 25°C and then completely mixed using a vortex oscillator. One hundred microliters were extracted with 300 µl of methanol to remove protein, and 10 µl of 2.8 mg/ml DL-o-chlorophenylalanine was added as an internal standard. Two hundred microliters of the supernatant obtained after centrifugation at 12,000 rpm and 4°C for 15 min was transferred to an autosampler injection vial for subsequent LC-Q/TOF-MS analysis. Every set of five consecutive samples was mixed together to obtain pooled quality control (QC) samples for validating the stability and repeatability of the LC-Q/TOF-MS system.
LC-Q/TOF-MS analysis
An Agilent 1290 Infinity LC System (Agilent Technologies, Santa Clara, CA, USA) with a C18 (Agilent, 2.1 mm × 100 mm, 1.8 µm) column was used for the LC-Q/TOF-MS analyses. The injection volume was set to 4 µl, and the column temperature was held at 40°C. The mobile phase consisted of water with 0.1% formic acid (A) and acetonitrile with 0.1% formic acid (B). The metabolites were subjected to gradient elution with a flow rate of 0.4 ml/min using the following program: 5% B from 0 to 1 min, 5% to 20% B from 1 to 6 min, 20% to 50% B from 6 to 9 min, 50% to 95% B from 9 to 13 min, and 95% B for 13 to 15 min [17]. The samples were maintained at 4°C throughout the analysis.
A 6530 UHD with Accurate-Mass Q/TOF (Agilent Technologies, Santa Clara, CA, USA) was used for mass spectrometry in the positive and negative ion modes (ES+ and ES−, respectively). In both the ES+ and ES−, the source temperature was set to 100°C, the cone gas flow rate was 50 l/hr, and the extraction cone voltage was 4 V. The desolvation gas temperature was set to 350°C in the ES+ and 300°C in the ES−. The flow rates were 600 l/hr in the ES+ and 700 l/hr in the ES−. The capillary voltages in the ES+ and ES− were 4 and 3.5 kV, respectively, and the sampling cone voltages were 35 kV in the ES+ and 50 kV in the ES−. The scan range was 50~1,000 m/z with a scan time of 0.03 sec and an inter-scan time of 0.02 sec in both modes. Leucine-enkephalin was selected as the lock mass (556.2771 Da in the ES+ and 554.2615 Da in the ES−) to ensure the accuracy and reproducibility of all the analyses [51].
Data preprocessing and identification of the plasma metabolites
The raw data included in the LC-Q/TOF-MS chromatograms obtained in the ES+ and ES− were converted to mz format. The XCMS package (R program) was used for feature extraction and various processes, including nonlinear retention time (RT) alignment, matched filtration and detection/matching [40]. The preprocessed data were outputted into Excel 2007 software for the elimination of impurity peaks and duplicates. Finally, a 2D data matrix from the LC-Q/TOF-MS chromatograms containing the RT, MZ, samples and peak intensity was generated. According to the MZ value within a molecular weight tolerance of 30 ppm and the fragment information obtained from the LC-Q/TOF-MS analyses, the plasma metabolites were identified using the Metlin (http://metlin.scripps.edu/) and the Bovine Metabolome (http://www.cowmetdb.ca/) databases.
Metabolic profile analysis
The online software program MetaboAnalyst 4.0 (http://www.metaboanalyst.ca/) was used for the metabolomics analyses, and the metabolic profiles of the identified metabolites were analyzed through principal component analysis (PCA) and partial least-squares discriminant analysis (PLS-DA). PCA, which is an unsupervised multivariate statistical analysis method, was used to visualize the 2D data matrix from the LC-Q/TOF-MS chromatograms and display the similarities and differences among the plasma metabolic profiles. PLS-DA, a supervised multivariate statistical analysis method, was used to clarify the separation between different groups of samples and to identify the variables with class-discriminating ability [38].
The validity of PLS-DA was assessed through goodness-of-fit parameters (accuracy, R2 and Q2), which were calculated after 7-fold cross validation and varied from 0 to 1. The R2 value explains the variance in the ratio of the x to y variables, and Q2 describes the predictive class-discriminating ability [42]. The VIP (variable importance projection) value for each metabolite was also calculated using the PLS-DA model. Many metabolites with a VIP value greater than 1 are very likely to exert an important effect on the discrimination of metabolic profiles [5]. Hence, the metabolites that showed differences between Groups H and D were first selected based on their VIP values, and the degree of their variance was reflected by the corresponding fold changes.
The differences in the metabolites were further assessed by the Wilcoxon rank-sum test with MetaboAnalyst 4.0. The critical P-value was set to 0.05. The raw P-values were adjusted using the Benjamini and Hochberg procedure to control the false discovery rate (FDR). The adjusted P-value was considered significant if it was lower than the FDR [17].
Hierarchical cluster analysis of the differential plasma metabolites
A hierarchical cluster analysis (HCA) was performed using MetaboAnalyst 4.0 to assess whether the selected differential metabolites could accurately classify Groups H and D. Euclidean distance was selected as the similarity measure, and the ward linkage method was selected as the clustering algorithm. The clustering patterns among the samples and differential plasma metabolites were visualized using heat maps. The clustering results for the samples are provided at the top of the heat map, and those for the differential plasma metabolites are positioned on the left of the heat map.
Analysis of metabolic pathways related to LDA
The MetPA module of MetaboAnalyst 4.0 was applied to clarify the global changes in the metabolic pathways due to the variations in the differential plasma metabolites [23]. Pathway enrichment analysis needs to be integrated with pathway topology analysis for the analysis of metabolic pathways [8, 16].
In the functional enrichment analysis, a overrepresentation analysis (ORA) was performed using the hypergeometric test to evaluate whether a particular metabolite set was more representative than expected, and the P values were adjusted using the Benjamini and Hochberg procedure to control the FDR [24].
In the pathway topological analysis, the number of shortest paths traversing a matched metabolite node in the network was calculated, and the entire network structure, not just the immediate neighbor of the current node, was considered [1]. The relative between-ness and out-of-degree centrality measures were selected to assess the importance of the differential plasma metabolites and the cumulative percentage of the importance measures represented the impact of the matched metabolite in a given pathway.
A metabolome view was generated to illustrate the results from the metabolic pathway analysis, and in this graph, the vertical and horizontal coordinates represent the –log P value and the impact value, respectively. The significantly altered pathways were then selected according to their –log P or impact values. The threshold of the –log P value was set to 10, and that of the impact value was set to 0.10 [46].
RESULTS
Metabolic profiles of Groups H and D
The representative LC-Q/TOF-MS chromatograms obtained in the ES+ and ES− are shown in Supplementary Fig. 1a and 1b, and 1086 and 921 features were identified from the chromatograms obtained in the ES+ and ES−, respectively (Supplementary Tables 2 and 3). Further analysis using online biochemical databases successfully identified 101 and 84 metabolites from the ES+ and ES− chromatograms, respectively (Supplementary Tables 4 and 5).
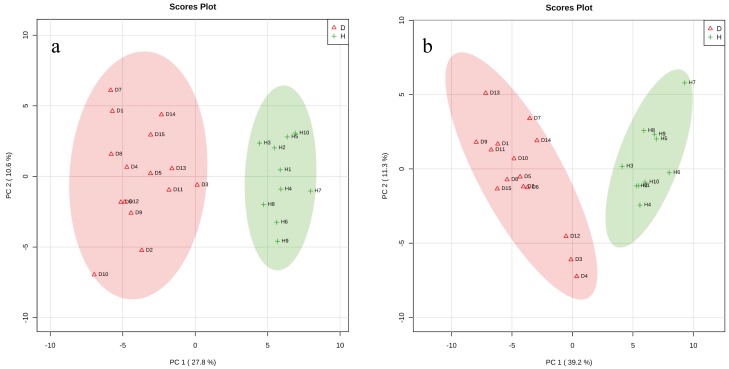
Two-dimensional score plots from the PCA revealed an obvious difference and separation in the blood metabolic profile between Groups H (green crosses) and D (red triangles) (Fig. 1a, 1b). The samples belonging to the same group were located within their respective 95% confidence region and shared marked similarities. PLS-DA models were then successfully developed from the ES+ and ES− spectra (Fig. 2a, 2b) and showed excellent abilities to discriminate Groups H (green crosses) and D (red triangles), as demonstrated by Q2 values higher than 0.9 (Fig. 3a, 3b). Thirty-nine metabolites from the ES+ spectra and 39 metabolites from the ES− spectra that differed between the two groups were selected according to their VIP values (>1), and 10 metabolites, namely, hippuric acid, tryptamine, tyrosine, oleic acid, isobutyrylglycine, citric acid, lysoPC (15:0), lysoPE (0:0/18:2 (9Z, 12Z)), stearic acid and tryptophan, were found in both spectra. The significance of the selected metabolites was further described using the Wilcoxon rank-sum test (P<0.05). Their VIP, fold change (FC) and statistical parameter values are shown in Tables 1 and 2.
Fig. 1.
Score plots of principal component analysis for Groups D (red triangles) and H (green crosses). (a) ES+ and (b) ES−.
Fig. 2.
Score plots of partial least-squares discriminant analysis for Groups D (red triangles) and H (green crosses). (a) ES+ and (b) ES−.
Fig. 3.
Goodness-of-fit parameters from 7-fold cross-validation of the partial least-squares discriminant analysis model. (a) ES+ and (b) ES−. R2 indicates the fraction of the variance in the x and y variables explained by the models, and Q2 represents the predictive performance of the models.
Table 1. Metabolites from ES+ spectra that differed between group H and group D.
| Metabolites | Variable importance projection valuea) | Fold changeb,c) | P-value | False discovery rate | Metabolites | Variable importance projection valuea) | Fold changeb,c) | P-value | False discovery rate |
|---|---|---|---|---|---|---|---|---|---|
| 1-Phenylethylamine | 1.81 | −2.07↓d) | 6.12E-07 | 1.17E-05 | Stearic acid | 1.35 | 1.07↑ | 4.03E-04 | 1.61E-03 |
| LysoPE (0:0/18:2 (9Z,12Z)) | 1.78 | −4.24↓ | 6.12E-07 | 1.17E-05 | Hippuric acid | 1.34 | −2.45↓ | 2.24E-04 | 9.77E-04 |
| L-Tryptophan | 1.74 | −2.24↓ | 1.22E-06 | 1.68E-05 | Glyceric acid | 1.34 | −2.06↓ | 5.94E-05 | 3.35E-04 |
| Cytosine | 1.71 | −2.00↓ | 6.12E-07 | 1.17E-05 | L-Histidine | 1.31 | 1.39↑ | 8.99E-04 | 3.32E-03 |
| Allantoin | 1.71 | −2.27↓ | 1.22E-06 | 1.68E-05 | Tryptamine | 1.29 | 1.44↑ | 1.15E-03 | 3.82E-03 |
| Cytidine | 1.69 | −2.29↓ | 6.12E-07 | 1.17E-05 | PGA2e) | 1.28 | 4.46↑ | 8.50E-05 | 4.30E-04 |
| Oleic Acid | 1.60 | 1.15↑ | 7.34E-06 | 7.05E-05 | Isobutyrylglycine | 1.24 | 2.95↑ | 5.32E-04 | 2.04E-03 |
| Sphingosine | 1.56 | −1.32↓ | 4.10E-05 | 2.62E-04 | Oleamide | 1.24 | 1.12↑ | 9.62E-03 | 2.10E-02 |
| Adenosine | 1.54 | −5.56↓ | 1.16E-05 | 9.30E-05 | LysoPC (15:0) | 1.21 | −1.60↓ | 1.15E-03 | 3.82E-03 |
| Homovanillic acid | 1.51 | −1.67↓ | 6.12E-07 | 1.17E-05 | L-Histidinol | 1.14 | 1.55↑ | 3.60E-03 | 9.89E-03 |
| 3-Hydroxyisobutyric acid | 1.48 | 3.54↑ | 1.19E-04 | 5.70E-04 | Citrulline | 1.14 | 1.10↑ | 4.44E-03 | 1.09E-02 |
| Citric acid | 1.47 | −3.49↓ | 4.10E-05 | 2.62E-04 | Ketoleucine | 1.11 | 1.53↑ | 2.91E-03 | 8.73E-03 |
| m-Coumaric acid | 1.45 | −1.77↓ | 1.16E-05 | 9.30E-05 | 3-Hydroxydodecanoic acid | 1.10 | −1.51↓ | 1.15E-03 | 3.82E-03 |
| L-Cystine | 1.45 | 3.04↑ | 1.65E-04 | 7.52E-04 | Alanine | 1.10 | 1.26↑ | 3.60E-03 | 9.89E-03 |
| Creatinine | 1.45 | 1.40↑ | 8.50E-05 | 4.30E-04 | L-Methionine | 1.09 | −1.35↓ | 1.47E-03 | 4.71E-03 |
| L-Tyrosine | 1.44 | −1.75↓ | 4.28E-06 | 5.14E-05 | Traumatic acid | 1.08 | 1.38↑ | 4.44E-03 | 1.09E-02 |
| Pyridoxine | 1.42 | −1.74↓ | 5.94E-05 | 3.35E-04 | Galactaric acid | 1.05 | −1.43↓ | 1.15E-02 | 2.46E-02 |
| Lactic acid | 1.42 | 1.80↑ | 1.84E-05 | 1.36E-04 | Docosanoic acid | 1.04 | −1.69↓ | 1.92E-02 | 3.92E-02 |
| L-Arginine | 1.41 | −1.66↓ | 7.34E-06 | 7.05E-05 | Uric acid | 1.03 | −2.45↓ | 6.60E-03 | 1.55E-02 |
| Kynurenine | 1.36 | −1.77↓ | 3.02E-04 | 1.26E-03 |
a) Variable importance projection value is calculated from the partial least-squares discriminant analysis model means. b) Fold change is the ratio of the relative peak intensity of the metabolite between the two groups. c) Positive value is the ration of Group D to Group H; negative value is the ration of Group H to Group D. d) ↑ or ↓ shows the change trend of the metabolites in Group D by comparing to Group H. e) PGA2 means prostaglandin A2.
Table 2. Metabolites from ES− spectra that differed between group H and group D.
| Metabolites | Variable importance projection valuea) | Fold changeb,c) | P-value | False discovery rate | Metabolites | Variable importance projection valuea) | Fold changeb,c) | P-value | False discovery rate |
|---|---|---|---|---|---|---|---|---|---|
| Homogentisic acid | 1.55 | −7.97↓d) | 6.12E-07 | 3.91E-06 | LysoPE (0:0/20:0) | 1.26 | −1.60↓ | 5.94E-05 | 2.14E-04 |
| Uracil | 1.55 | 2.36↑ | 6.12E-07 | 3.91E-06 | Gentisic acid | 1.26 | −3.44↓ | 6.12E-07 | 3.91E-06 |
| R-3-Hydroxybutyric acid | 1.53 | 3.89↑ | 6.12E-07 | 3.91E-06 | L-Homocitrulline | 1.21 | −1.74↓ | 6.94E-04 | 1.60E-03 |
| L-Tryptophan | 1.51 | −2.12↓ | 6.12E-07 | 3.91E-06 | cis-9-palmitoleic acid | 1.20 | 4.13↑ | 2.75E-05 | 1.09E-04 |
| Tryptamine | 1.49 | 2.41↑ | 6.12E-07 | 3.91E-06 | LysoPC (15:0) | 1.19 | −1.48↓ | 3.02E-04 | 8.65E-04 |
| LysoPE (0:0/18:2 (9Z,12Z)) | 1.48 | −4.15↓ | 6.12E-07 | 3.91E-06 | Nonanedioic acid | 1.18 | −1.61↓ | 3.60E-03 | 6.50E-03 |
| Isobutyrylglycine | 1.45 | 5.69↑ | 6.12E-07 | 3.91E-06 | 2-Oxoarginine | 1.18 | 1.46↑ | 5.32E-04 | 1.34E-03 |
| LysoPE (0:0/16:0) | 1.45 | −1.64↓ | 6.12E-07 | 3.91E-06 | Alpha -Linolenic acid | 1.18 | 2.01↑ | 4.03E-04 | 1.08E-03 |
| L-Tyrosine | 1.44 | −2.58↓ | 6.12E-07 | 3.91E-06 | LysoPE (0:0/20:1 (11Z)) | 1.18 | −1.56↓ | 2.24E-04 | 6.64E-04 |
| D-Mannitol | 1.43 | −2.71↓ | 6.12E-07 | 3.91E-06 | Glycocholic acid | 1.13 | −1.56↓ | 8.50E-05 | 2.72E-04 |
| Pyridoxal | 1.40 | −13.24↓ | 6.12E-07 | 3.91E-06 | Aconitic acid | 1.11 | −1.91↓ | 1.84E-05 | 8.96E-05 |
| Oleic acid | 1.38 | 5.80↑ | 2.75E-05 | 1.09E-04 | Citric acid | 1.10 | −1.14↓ | 1.86E-03 | 3.51E-03 |
| LysoPE (0:0/20:2 (11Z,14Z)) | 1.37 | −3.53↓ | 6.12E-07 | 3.91E-06 | Stearic acid | 1.10 | 1.17↑ | 5.32E-04 | 1.34E-03 |
| Palmitic acid | 1.36 | 4.77↑ | 4.28E-06 | 2.54E-05 | Gluconic acid | 1.07 | −2.50↓ | 1.15E-03 | 2.39E-03 |
| L-Leucine | 1.33 | −1.95↓ | 7.34E-06 | 3.81E-05 | Hippuric acid | 1.07 | −2.56↓ | 8.99E-04 | 1.96E-03 |
| LysoPE (0:0/18:1 (11Z)) | 1.32 | −1.51↓ | 2.75E-05 | 1.09E-04 | PGE2e) | 1.06 | −2.73↓ | 6.94E-04 | 1.60E-03 |
| Acetoacetic acid | 1.30 | 1.64↑ | 5.94E-05 | 2.14E-04 | Arachidonic Acid | 1.06 | 2.75↑ | 6.12E-07 | 3.91E-06 |
| Linoleic acid | 1.29 | 2.28↑ | 1.65E-04 | 5.06E-04 | Cholic acid | 1.03 | −3.72↓ | 8.50E-05 | 2.72E-04 |
| Myristic acid | 1.28 | 3.07↑ | 8.50E-05 | 2.72E-04 | LTD4 | 1.01 | −0.30↓ | 1.15E-03 | 2.39E-03 |
| Ubiquinone Q2 | 1.28 | 2.62↑ | 2.75E-05 | 1.09E-04 |
a) Variable importance projection value is calculated from the partial least-squares discriminant analysis model. b) Fold change was calculated as a ratio of the relative peak intensities between different groups. c) Positive value, Group D vs. Group H; negative value, Group H vs. Group D. d) ↑ or ↓ means the trend of regulation for these metabolites in Group D vs. Group H. e) PGE2 means prostaglandin E2, LTD4 means Leukotriene D4.
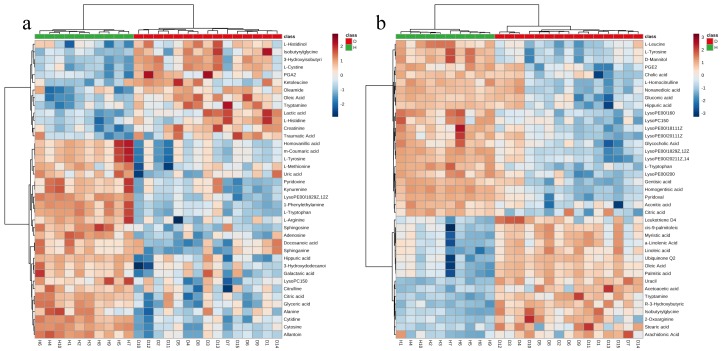
The HCA results are displayed in heat maps (Fig. 4a, 4b). The samples belonging to the same group were clustered together, which indicated that the selected differential metabolites could accurately classify Groups H and D. The trends obtained for the changes in the various metabolite between Groups H and D are also reflected by the variations in the color intensity (from light blue to dark red) on the heat maps.
Fig. 4.
Heatmaps showing the clustering of the differential metabolites. (a) ES+ and (b) ES−. Clustering was performed using ward linkage, and Euclidian distance was selected as the distance measure. A shorter Euclidean distance between tree clusters indicates higher similarity. The branch height represents the similarity between two samples and metabolites, and a subtree is more similar if a node is vertically lower in the graph. The dark red boxes indicate that the metabolite concentration is greater than the mean, and the light blue boxes show that the metabolite concentration was less than the mean.
Analysis of key metabolic pathways related to LDA
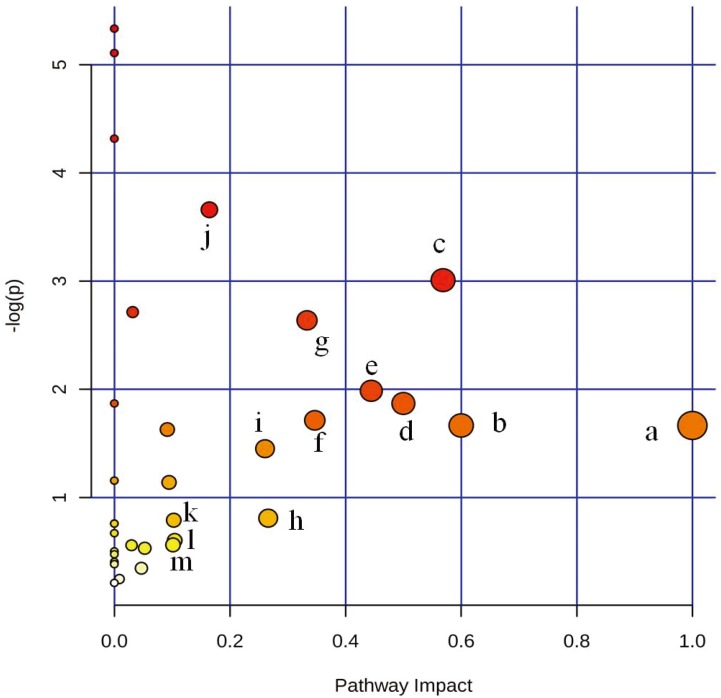
The detailed results from the metabolomics pathway analysis of the differential metabolites are provided in Supplementary Table 6. An interactive visualization framework was generated to display the altered metabolic pathways due to the variations in the blood concentrations of the differential metabolites, and 13 pathways were identified as the significant metabolic pathways related to LDA based on their corresponding impact value (Fig. 5). The importance of the metabolites on the 13 metabolic pathways is listed in Table 3.
Fig. 5.
Metabolome view of the altered metabolic pathways in postpartum dairy cows with left displacement of the abomasum. Colors varying from yellow to red indicate the significance of the differential metabolites used in the enrichment analysis. The impact value from the pathway topology analysis indicates the degree of variability in a pathway, and the original P value from the enrichment analysis indicates whether a particular metabolite set is more representative than expected. (a) Linoleic acid metabolism, (b) synthesis and degradation of ketone bodies, (c) vitamin B6 metabolism, (d) phenylalanine, tyrosine and tryptophan biosynthesis, (e) citrate cycle, (f) arachidonic acid metabolism, (g) valine, leucine and isoleucine biosynthesis, (h) histidine metabolism, (i) tryptophan metabolism, (j) tyrosine metabolism, (k) pyrimidine metabolism, (l) arginine and proline metabolism, and (m) butanoate metabolism.
Table 3. The metabolic pathways related left displacement of the abomasum and the importance of the relevant metabolites.
| Metabolic pathway | −log (P) | Impact | Relevant metabolites | |
|---|---|---|---|---|
| Name | Importance | |||
| Synthesis and degradation of ketone bodies | 1.68 | 0.6 | Acetoacetic acid | 0.6 |
| Linoleic acid metabolism | 1.68 | 1 | Linoleic acid | 1 |
| Arachidonic acid metabolism | 1.75 | 0.35 | Arachidonic acid | 0.33 |
| Prostaglandin E2 | 0.02 | |||
| Leukotriene D4 | 0 | |||
| Histidine metabolism | 0.82 | 0.27 | L-Histidine | 0.27 |
| Vitamin B6 metabolism | 3.04 | 0.57 | Pyridoxine | 0.08 |
| Pyridoxal | 0.49 | |||
| Tyrosine metabolism | 3.73 | 0.16 | L-Tyrosine | 0.145 |
| Homogentisic acid | 0.017 | |||
| Gentisic acid | 0 | |||
| Homovanillic acid | 0.002 | |||
| Acetoacetic acid | 0 | |||
| Tryptophan metabolism | 1.49 | 0.26 | L-Tryptophan | 0.17 |
| Tryptamine | 0.06 | |||
| L-Kynurenine | 0.03 | |||
| Valine, leucine and isoleucine biosynthesis | 2.67 | 0.33 | L-Leucine | 033 |
| 4-Methyl-2-oxopentanoate | 0 | |||
| Citrate cycle | 2.02 | 0.44 | Citric acid | 0.294 |
| cis-Aconitic acid | 0.148 | |||
| Phenylalanine, tyrosine and tryptophan biosynthesis | 1.89 | 0.50 | L-Tyrosine | 0.5 |
| Pyrimidine metabolism | 0.81 | 0.10 | Cytidine | 0.01 |
| Uracil | 0.09 | |||
| Arginine and proline metabolism | 0.62 | 0.10 | Citrulline | 0.03 |
| L-Arginine | 0.07 | |||
| Butanoate metabolism | 0.57 | 0.10 | Acetoacetic acid | 0.10 |
–log (P) is calculated by the enrichment analysis, and Impact is generated by the pathway topology analysis.
Six, three and two of the 13 metabolic pathways are involved in amino acid metabolism (histidine metabolism, tyrosine metabolism, valine, leucine and isoleucine biosynthesis, phenylalanine, tyrosine and tryptophan biosynthesis, arginine and proline metabolism, and tryptophan metabolism), lipid metabolism (synthesis and degradation of ketone bodies, linoleic acid metabolism, and arachidonic acid metabolism), and carbohydrate metabolism (citrate cycle and butanoate metabolism), respectively. In addition, vitamin B6 metabolism and pyrimidine metabolism serve as cofactors in vitamin metabolism and nucleotide metabolism, respectively.
DISCUSSION
LDA is often observed in high-producing dairy cows during the first 4 weeks of lactation. Because the energy output (milk) exceeds the energy input, every postpartum high-producing dairy cow experiences NEB, but not every cow exhibits LDA [6]. Several previous studies have shown that LDA is accompanied by NEB and lower blood levels of succinate, which is an important biochemical intermediate of the TCA cycle [3, 9, 39]. In the present study, two important intermediate metabolites of the TCA cycle, citric acid and cis-aconitic acid, were found to be sharply decreased in postpartum dairy cows with LDA, which indicated that postpartum dairy cows with LDA suffer a more severe NEB than healthy postpartum dairy cows.
In this study, a significant increase in lactic acid blood concentration further confirmed that the postpartum dairy cows with LDA are suffering excessive NEB. In ruminant animals, gluconeogenesis supplies 90% of glucose, and 50 to 60% of this glucose is derived from propionate [36]. Thus, the supply of glucose is mainly dependent on the amount of propionate in gluconeogenesis. Lactate is mainly metabolized as volatile fatty acids (VFAs), including propionate, acetate and butyrate [22]. In dairy cows, more than 70% of lactic acid is fermented to propionic acid, and as a result of NEB, body fat is mobilized, which can potentially lead to ketosis or fatty liver. Dairy cows with ketosis have a higher lactate concentration and lower VFA concentrations than healthy dairy cows [21].
Compared with healthy postpartum dairy cows, a significantly elevated circulating level of acetoacetic acid was found in postpartum LDA dairy cows, and its involvement in the synthesis and degradation pathways of ketone bodies suggested a higher level of circulating ketone bodies. In addition, increases in some intermediate metabolites of lipid metabolism, including stearic acid, oleic acid, palmitic acid, linoleic acid, α-linolenic acid, myristic acid, arachidonic acid, and cis-9-palmitoleic acid, indicated elevated NEFAs in the blood of postpartum LDA dairy cows. The results further showed the LDA is associated with a metabolic status of excessive NEB, which is consistent with the published results [45].
Prostaglandin E2 (PGE2) significantly decreases in the blood of LDA dairy cows [1], but leukotriene D4 (LTD4) has not been investigated. PGE2 and LTD4 are derived from arachidonic acid via the cyclooxygenase/lipoxygenase pathway [29]. PGE2 can regulate the gastrointestinal integrity, fertility and the inflammatory response [20], and LTD4 can act as a regulator of leukocyte chemotaxis, the inflammatory response and the contraction of gastrointestinal smooth muscle [31]. Previous studies have indicated that NEB and inflammation exhibit a strong cause-and-effect relationship [13]. In this study, however, the cyclooxygenase/lipoxygenase pathway was likely inhibited, and thus, the PGE2 and LTD4 levels were decreased in postpartum dairy cows with LDA, which suggests that the decrease in PGE2 and LTD4 might be related to the dilatation of abomasum smooth muscle, injury to the abomasum mucosa and decreased fertility in dairy cows with LDA but not to the inflammatory response. However, this conclusion needs to be verified through subsequent experiments.
Under energy deficiency, proteolysis processes occur in parallel to lipolysis, and muscle tissue can be used as a source of amino acid [30]. Thus, the elevated creatinine level measured in dairy cows with LDA in this study likely indicated an augmentation of proteolysis in response to excessive NEB. Previous studies have revealed that some amino acids are upregulated and others are downregulated in dairy cows with LDA [3, 19]. Similar trends have been uncovered in dairy cows with ketosis [30, 50]. The results of these studies indicate that increases in certain amino acids might be attributed to the production of glucose in response to ketosis, whereas decreases in some amino acids are due to their intensive use in both ketogenesis and gluconeogenesis processes. In the present study, some amino acids, including cystine, histidine, citrulline and alanine, were found to be upregulated in dairy cows with LDA, which indicated that proteolysis was strengthened to remedy the status of excessive NEB. Moreover, the downregulation of tryptophan, tyrosine, arginine, methionine and leucine in dairy cows with LDA mainly resulted from two factors: their intensive use to generate glucose and ketone bodies and the reduced dry matter intake under LDA (the latter was particularly relevant for the essential amino acids tryptophan, methionine and leucine). In addition, the decrease in arginine, which serves as a precursor of nitric oxide, is also possibly associated with increased nitric oxide synthesis by abomasal neurons, which is related to disordered abomasal muscle function in the displaced abomasum [15].
During gluconeogenesis and ketogenesis, NH3 obtained from the deamination of amino acids is used to generate carbamoyl phosphate, and this product can be decomposed into citrulline and pyrimidine. Certain amino acids involved in the urea cycle, such as proline, ornithine arginine and citrulline, have been confirmed to participate in the TCA cycle by affecting the formation of fumarate [29, 41]. Pyrimidine is used to synthesize uridine, which can be phosphorylated to produce uridine triphosphate. Uridine triphosphate acts with glucose-1-phosphate to generate uridine diphosphate glucose, which is involved in glycogen synthesis [10, 37]. Uridine is involved in insulin resistance and promotes gluconeogenesis by increasing protein glycosylation and reducing protein phosphorylation [35, 44]. In this study, citrulline and uracil were significantly increased in dairy cows with LDA and might thus contribute to the reinforcement of gluconeogenesis in response to excessive NEB.
Vitamin B6, as a co-factor for more than 150 enzymes, has multiple biological activities in the regulation of the metabolism and synthesis of carbohydrates, lipids and proteins [25]. In addition to its coenzyme role, this vitamin was recognized as an antioxidant and anti-inflammatory agent in recent years [4]. Low circulating levels of vitamin B6 are associated with increased risks of inflammation and inflammation-related chronic illnesses [43]. Concomitant diseases such as mastitis, endometritis and laminitis are considered to play an important role in the pathogenesis of LDA during the early postpartum period [12]. This study provided the first demonstration that the plasma pyridoxine and pyridoxal levels are significantly decreased in dairy cows with LDA, which indicates that the incidence of LDA is potentially related to postpartum oxidative stress and the inflammatory response.
In conclusion, a LC-Q/TOF-MS metabolomics technique combined with pathway analysis was applied to uncover a more comprehensive view of the metabolic changes initiated in response to LDA. Sixty-nine blood metabolites and 13 related metabolic pathways were found to be markedly altered in postpartum dairy cows with LDA. Although the present study uncovered useful information for understanding the detailed metabolic status in dairy cows with LDA, potential metabolic disturbances prior to LDA should also be investigated in further studies to better elucidate the pathogenesis of the disease.
Supplementary Material
Acknowledgments
This research was supported in part by funding from the National Natural Science Foundation of China (NSFC, Grant No. 31860719) and the Key Research & Development Plan of Ningxia (Grant No. 2017BY078). The authors particularly thank the veterinarians at the dairy cow farms who graciously assisted with this project.
REFERENCES
- 1.Abbasi A., Hossain L., Leydesdorff L.2012. Betweenness centrality as a driver of preferential attachment in the evolution of research collaboration networks. J. Informetrics 6: 403–412. doi: 10.1016/j.joi.2012.01.002 [DOI] [Google Scholar]
- 2.Al-Rawashdeh O., Ismail Z. B., Talafha A., Al-Momani A.2017. Changes of hematological and biochemical parameters and levels of pepsinogen, histamine and prostaglandins in dairy cows affected with left displacement of the abomasum. Pol. J. Vet. Sci. 20: 13–18. doi: 10.1515/pjvs-2017-0002 [DOI] [PubMed] [Google Scholar]
- 3.Başoğlu A., Başpinar N., Coşkun A.2014. NMR-based metabolomic evaluation in dairy cows with displaced abomasum. Turk. J. Vet. Anim. Sci. 38: 325–330. doi: 10.3906/vet-1310-52 [DOI] [Google Scholar]
- 4.Bird R. P.2018. The emerging role of vitamin B6 in inflammation and carcinogenesis. Adv. Food Nutr. Res. 83: 151–194. doi: 10.1016/bs.afnr.2017.11.004 [DOI] [PubMed] [Google Scholar]
- 5.Cajka T., Smilowitz J. T., Fiehn O.2017. Validating Quantitative Untargeted Lipidomics Across Nine Liquid Chromatography-High-Resolution Mass Spectrometry Platforms. Anal. Chem. 89: 12360–12368. doi: 10.1021/acs.analchem.7b03404 [DOI] [PubMed] [Google Scholar]
- 6.Cameron R. E., Dyk P. B., Herdt T. H., Kaneene J. B., Miller R., Bucholtz H. F., Liesman J. S., Vandehaar M. J., Emery R. S.1998. Dry cow diet, management, and energy balance as risk factors for displaced abomasum in high producing dairy herds. J. Dairy Sci. 81: 132–139. doi: 10.3168/jds.S0022-0302(98)75560-2 [DOI] [PubMed] [Google Scholar]
- 7.Chen H., Cao G., Chen D. Q., Wang M., Vaziri N. D., Zhang Z. H., Mao J. R., Bai X., Zhao Y. Y.2016. Metabolomics insights into activated redox signaling and lipid metabolism dysfunction in chronic kidney disease progression. Redox Biol. 10: 168–178. doi: 10.1016/j.redox.2016.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chong J., Soufan O., Li C., Caraus I., Li S., Bourque G., Wishart D. S., Xia J.2018. MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 46W1: W486–W494. doi: 10.1093/nar/gky310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Civelek T., Sevinc M., Boydak M., Basoglu A.2006. Serum apolipoprotein B100 concentrations in dairy cows with left displaced abomasum. Rev. Med. Vet. 157: 361–365. [Google Scholar]
- 10.Connolly G. P., Duley J. A.1999. Uridine and its nucleotides: biological actions, therapeutic potentials. Trends Pharmacol. Sci. 20: 218–225. doi: 10.1016/S0165-6147(99)01298-5 [DOI] [PubMed] [Google Scholar]
- 11.Coppock C. E.1974. Displaced abomasum in dairy cattle: etiological factors. J. Dairy Sci. 57: 926–933. doi: 10.3168/jds.S0022-0302(74)84988-X [DOI] [PubMed] [Google Scholar]
- 12.Doll K., Sickinger M., Seeger T.2009. New aspects in the pathogenesis of abomasal displacement. Vet. J. 181: 90–96. doi: 10.1016/j.tvjl.2008.01.013 [DOI] [PubMed] [Google Scholar]
- 13.Esposito G., Irons P. C., Webb E. C., Chapwanya A.2014. Interactions between negative energy balance, metabolic diseases, uterine health and immune response in transition dairy cows. Anim. Reprod. Sci. 144: 60–71. doi: 10.1016/j.anireprosci.2013.11.007 [DOI] [PubMed] [Google Scholar]
- 14.Fiore F., Musina D., Cocco R., Di Cerbo A., Spissu N.2018. Association between left-displaced abomasum corrected with 2-step laparoscopic abomasopexy and milk production in a commercial dairy farm in Italy. Ir. Vet. J. 71: 20–20. doi: 10.1186/s13620-018-0132-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geishauser T., Gronostay S.1998. Arginine in liquid contents of displaced abomasal in dairy cows. Berl. Munch. Tierarztl. Wochenschr. 111: 146–149. [PubMed] [Google Scholar]
- 16.Guo Y. S., Tao J. Z.2018. 1H nuclear magnetic resonance-based plasma metabolomics provides another perspective of response mechanisms of newborn calves upon the first colostrum feeding. J. Anim. Sci. 96: 1769–1777. doi: 10.1093/jas/sky078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo Y. S., Tao J. Z.2018. Metabolomics and pathway analyses to characterize metabolic alterations in pregnant dairy cows on D 17 and D 45 after AI. Sci. Rep. 8: 5973. doi: 10.1038/s41598-018-23983-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guzelbektes H., Sen I., Ok M., Constable P. D., Boydak M., Coskun A.2010. Serum amyloid A and haptoglobin concentrations and liver fat percentage in lactating dairy cows with abomasal displacement. J. Vet. Intern. Med. 24: 213–219. doi: 10.1111/j.1939-1676.2009.0444.x [DOI] [PubMed] [Google Scholar]
- 19.Hamana M., Ohtsuka H., Oikawa M., Kawamura S.2010. Blood free amino acids in the postpartum dairy cattle with left displaced abomasum. J. Vet. Med. Sci. 72: 1355–1358. doi: 10.1292/jvms.09-0239 [DOI] [PubMed] [Google Scholar]
- 20.Harizi H., Gualde N.2006. Pivotal role of PGE2 and IL-10 in the cross-regulation of dendritic cell-derived inflammatory mediators. Cell. Mol. Immunol. 3: 271–277. [PubMed] [Google Scholar]
- 21.Henning P.H., Horn C.H., Leeuw K.J., Meissner H.H., Hagg F.M.J.A.F.S. and Technology. 2010. Effect of ruminal administration of the lactate-utilizing strain Megasphaera elsdenii (Me) NCIMB 41125 on abrupt or gradual transition from forage to concentrate diets. Anim. Feed Sci. Technol. 157: 20–29. doi: 10.1016/j.anifeedsci.2010.02.002 [DOI] [Google Scholar]
- 22.Hino T., Shimada K., Maruyama T.1994. Substrate Preference in a Strain of Megasphaera elsdenii, a Ruminal Bacterium, and Its Implications in Propionate Production and Growth Competition. Appl. Environ. Microbiol. 60: 1827–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson C. H., Ivanisevic J., Siuzdak G.2016. Metabolomics: beyond biomarkers and towards mechanisms. Nat. Rev. Mol. Cell Biol. 17: 451–459. doi: 10.1038/nrm.2016.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaever A., Landesfeind M., Feussner K., Morgenstern B., Feussner I., Meinicke P.2014. Meta-analysis of pathway enrichment: combining independent and dependent omics data sets. PLoS One 9: e89297. doi: 10.1371/journal.pone.0089297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klein M. S., Almstetter M. F., Nürnberger N., Sigl G., Gronwald W., Wiedemann S., Dettmer K., Oefner P. J.2013. Correlations between milk and plasma levels of amino and carboxylic acids in dairy cows. J. Proteome Res. 12: 5223–5232. doi: 10.1021/pr4006537 [DOI] [PubMed] [Google Scholar]
- 26.Klevenhusen F., Humer E., Metzler-Zebeli B., Podstatzky-Lichtenstein L., Wittek T., Zebeli Q.2015. Metabolic profile and inflammatory responses in dairy cows with left displaced abomasum kept under small-scaled farm conditions. Animals (Basel) 5: 1021–1033. doi: 10.3390/ani5040396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Y., Xu C., Xia C., Zhang H., Sun L., Gao Y.2014. Plasma metabolic profiling of dairy cows affected with clinical ketosis using LC/MS technology. Vet. Q. 34: 152–158. doi: 10.1080/01652176.2014.962116 [DOI] [PubMed] [Google Scholar]
- 28.Lu J., Antunes Fernandes E., Páez Cano A. E., Vinitwatanakhun J., Boeren S., van Hooijdonk T., van Knegsel A., Vervoort J., Hettinga K. A.2013. Changes in milk proteome and metabolome associated with dry period length, energy balance, and lactation stage in postparturient dairy cows. J. Proteome Res. 12: 3288–3296. doi: 10.1021/pr4001306 [DOI] [PubMed] [Google Scholar]
- 29.Luo Z. Z., Shen L. H., Jiang J., Huang Y. X., Bai L. P., Yu S. M., Yao X. P., Ren Z. H., Yang Y. X., Cao S. Z.2019. Plasma metabolite changes in dairy cows during parturition identified using untargeted metabolomics. J. Dairy Sci. 102: 4639–4650. doi: 10.3168/jds.2018-15601 [DOI] [PubMed] [Google Scholar]
- 30.Marczuk J., Brodzki P., Brodzki A., Kurek Ł.2018. The concentration of free amino acids in blood serum of dairy cows with primary ketosis. Pol. J. Vet. Sci. 21: 149–156. [DOI] [PubMed] [Google Scholar]
- 31.Mezhybovska M., Wikström K., Ohd J. F., Sjölander A.2005. Pro-inflammatory mediator leukotriene D4 induces transcriptional activity of potentially oncogenic genes. Biochem. Soc. Trans. 33: 698–700. doi: 10.1042/BST0330698 [DOI] [PubMed] [Google Scholar]
- 32.Mokhber Dezfouli M., Eftekhari Z., Sadeghian S., Bahounar A., Jeloudari M.2013. Evaluation of hematological and biochemical profiles in dairy cows with left displacement of the abomasum. Comp. Clin. Pathol. 22: 175–179. doi: 10.1007/s00580-011-1382-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nybo S. E., Lamberts J. T.2019. Integrated use of LC/MS/MS and LC/Q-TOF/MS targeted metabolomics with automated label-free microscopy for quantification of purine metabolites in cultured mammalian cells. Purinergic Signal. 15: 17–25. doi: 10.1007/s11302-018-9643-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ospina P. A., Nydam D. V., Stokol T., Overton T. R.2010. Association between the proportion of sampled transition cows with increased nonesterified fatty acids and β-hydroxybutyrate and disease incidence, pregnancy rate, and milk production at the herd level. J. Dairy Sci. 93: 3595–3601. doi: 10.3168/jds.2010-3074 [DOI] [PubMed] [Google Scholar]
- 35.Park S. Y., Ryu J., Lee W.2005. O-GlcNAc modification on IRS-1 and Akt2 by PUGNAc inhibits their phosphorylation and induces insulin resistance in rat primary adipocytes. Exp. Mol. Med. 37: 220–229. doi: 10.1038/emm.2005.30 [DOI] [PubMed] [Google Scholar]
- 36.Reynolds C. K., Huntington G. B., Tyrrell H. F., Reynolds P. J.1988. Net metabolism of volatile fatty acids, D-β-hydroxybutyrate, nonesterifield fatty acids, and blood gasses by portal-drained viscera and liver of lactating Holstein cows. J. Dairy Sci. 71: 2395–2405. doi: 10.3168/jds.S0022-0302(88)79824-0 [DOI] [PubMed] [Google Scholar]
- 37.Roach P. J., Depaoli-Roach A. A., Hurley T. D., Tagliabracci V. S.2012. Glycogen and its metabolism: some new developments and old themes. Biochem. J. 441: 763–787. doi: 10.1042/BJ20111416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saccenti E., Timmerman M. E.2016. Approaches to Sample Size Determination for Multivariate Data: Applications to PCA and PLS-DA of Omics Data. J. Proteome Res. 15: 2379–2393. doi: 10.1021/acs.jproteome.5b01029 [DOI] [PubMed] [Google Scholar]
- 39.Sevinc M., Ok M., Basoglu A.2002. Liver function in dairy cows with abomasal displacement. Rev. Med. Vet. 153: 447–480. [Google Scholar]
- 40.Smith C. A., Want E. J., O’Maille G., Abagyan R., Siuzdak G.2006. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 78: 779–787. doi: 10.1021/ac051437y [DOI] [PubMed] [Google Scholar]
- 41.Sugiyama K., Ebinuma H., Nakamoto N., Sakasegawa N., Murakami Y., Chu P. S., Usui S., Ishibashi Y., Wakayama Y., Taniki N., Murata H., Saito Y., Fukasawa M., Saito K., Yamagishi Y., Wakita T., Takaku H., Hibi T., Saito H., Kanai T.2014. Prominent steatosis with hypermetabolism of the cell line permissive for years of infection with hepatitis C virus. PLoS One 9: e94460. doi: 10.1371/journal.pone.0094460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Triba M. N., Le Moyec L., Amathieu R., Goossens C., Bouchemal N., Nahon P., Rutledge D. N., Savarin P.2015. PLS/OPLS models in metabolomics: the impact of permutation of dataset rows on the K-fold cross-validation quality parameters. Mol. Biosyst. 11: 13–19. doi: 10.1039/C4MB00414K [DOI] [PubMed] [Google Scholar]
- 43.Ueland P. M., McCann A., Midttun Ø., Ulvik A.2017. Inflammation, vitamin B6 and related pathways. Mol. Aspects Med. 53: 10–27. doi: 10.1016/j.mam.2016.08.001 [DOI] [PubMed] [Google Scholar]
- 44.Urasaki Y., Pizzorno G., Le T. T.2014. Uridine affects liver protein glycosylation, insulin signaling, and heme biosynthesis. PLoS One 9: e99728. doi: 10.1371/journal.pone.0099728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Van Winden S. C., Kuiper R.2003. Left displacement of the abomasum in dairy cattle: recent developments in epidemiological and etiological aspects. Vet. Res. 34: 47–56. doi: 10.1051/vetres:2002060 [DOI] [PubMed] [Google Scholar]
- 46.Wang X., Yang B., Sun H., Zhang A.2012. Pattern recognition approaches and computational systems tools for ultra performance liquid chromatography-mass spectrometry-based comprehensive metabolomic profiling and pathways analysis of biological data sets. Anal. Chem. 84: 428–439. doi: 10.1021/ac202828r [DOI] [PubMed] [Google Scholar]
- 47.Xi X., Kwok L. Y., Wang Y., Ma C., Mi Z., Zhang H.2017. Ultra-performance liquid chromatography-quadrupole-time of flight mass spectrometry MSE-based untargeted milk metabolomics in dairy cows with subclinical or clinical mastitis. J. Dairy Sci. 100: 4884–4896. doi: 10.3168/jds.2016-11939 [DOI] [PubMed] [Google Scholar]
- 48.Zhang G., Deng Q., Mandal R., Wishart D. S., Ametaj B. N.2017. DI/LC-MS/MS-Based Metabolic Profiling for Identification of Early Predictive Serum Biomarkers of Metritis in Transition Dairy Cows. J. Agric. Food Chem. 65: 8510–8521. doi: 10.1021/acs.jafc.7b02000 [DOI] [PubMed] [Google Scholar]
- 49.Zhang H., Tong J., Zhang Y., Xiong B., Jiang L.2019. Metabolomics reveals potential biomarkers in the rumen fluid response to different milk production of dairy cows. Asian-Australas. J. Anim. Sci.doi: 10.5713/ajas.19.0214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang H., Wu L., Xu C., Xia C., Sun L., Shu S.2013. Plasma metabolomic profiling of dairy cows affected with ketosis using gas chromatography/mass spectrometry. BMC Vet. Res. 9: 186. doi: 10.1186/1746-6148-9-186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang L., Li M., Zhan L., Lu X., Liang L., Su B., Sui H., Gao Z., Li Y., Liu Y., Wu B., Liu Q.2015. Plasma metabolomic profiling of patients with diabetes-associated cognitive decline. PLoS One 10: e0126952. doi: 10.1371/journal.pone.0126952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao Y. Y., Liu J., Cheng X. L., Bai X., Lin R. C.2012. Urinary metabonomics study on biochemical changes in an experimental model of chronic renal failure by adenine based on UPLC Q-TOF/MS. Clin. Chim. Acta 413: 642–649. doi: 10.1016/j.cca.2011.12.014 [DOI] [PubMed] [Google Scholar]
- 53.Zhao Y. Y., Wu S. P., Liu S., Zhang Y., Lin R. C.2014. Ultra-performance liquid chromatography-mass spectrometry as a sensitive and powerful technology in lipidomic applications. Chem. Biol. Interact. 220: 181–192. doi: 10.1016/j.cbi.2014.06.029 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.