Abstract
East Coast fever is caused by Theileria parva, and poses serious concerns for dairy farmers owing to massive economic losses. In the current study, we compared three methods (DNA extraction kits, FTA-NaOH and FTA-TENT) of DNA extraction to identify the most economical and reliable method. A survey for T. parva prevalence was conducted in dairy cattle in Mbarara, Uganda. Cytochrome C oxidase subunit I (COI) and T. parva-p104 genes were amplified to compare the methods. FTA-NaOH-based polymerase chain reaction (PCR) yielded the best detection rate for both COI gene and p104 gene. Prevalence of T. parva was 45.0% and 83.3% at animal and farm-level, respectively. FTA-NaOH based-PCR is simple, highly sensitive and cost-effective tool for T. parva diagnosis in resource constrained settings.
Keywords: dairy cow, East Coast fever, FTA card, sodium hydroxide, Theileria parva
East Coast fever (ECF) is one of the most severe bovine protozoan diseases, and it is caused by Theileria parva. It mainly occurs in East and Central Africa. In 16 African countries where it is endemic, the losses caused by ECF was estimated to be approximately US $300 million annually [5]. The parasite is transmitted by ticks, mainly Rhipicephalus appendiculatus, and causes fever, lymphadenopathy and high mortality in cattle, especially exotic dairy breeds [9].
Direct microscopy observation of parasites is a common routine diagnostic method, but reliably achieving accurate results using this method requires technical proficiency based on accumulated experience [9]. Other limitations of microscopy-based diagnosis include relatively low sensitivity and difficulty of detection in asymptomatic cases compared with molecular diagnostic methods [2]. In contrast, molecular diagnostic methods such as those based on the polymerase chain reaction (PCR) are highly sensitive and accurate, but require specialised equipment, and thus it is costly. Until after DNA extraction, the sample needs to be chilled or frozen, and several equipment such as water baths and centrifuges are needed to prepare the template DNA by using DNA extraction kit.
The FTA card manufactured by GE Healthcare (Chicago, IL, USA) utilises a unique type of filter paper that facilitates the isolation, purification and storage of nucleic acids on a membrane [3]. The main benefits of the FTA card are long-term stable storage of nucleic acid and easy preparation of DNA for the PCR template without a need for expensive specialised equipment.
In the present study, the efficiency of performing PCR using DNA templates obtained by using three different methods for the detection of T. parva infection in dairy cattle was compared in conjunction with an investigation of basic prevalence conducted in Mbarara District in southwestern Uganda, where dairy farming is popular. The results suggest that the FTA-NaOH based-PCR is useful for the detection of T. parva, and provide basic information about the ECF burden at the farm level in Mbarara District, Uganda.
Mbarara District is located in the southwestern region of Uganda. The district is bordered by Ibanda District to the north, Kiruhura District to the east, Isingiro District to the southeast, Ntungamo District to the southwest, Sheema District to the west and Buhweju District to the northwest. The coordinates of the district are 00°36´00˝S; 30°36´00˝E. Adult cattle and calf blood samples were collected from 30 selected dairy farms in Mbarara District from February 2017 to May 2017 inclusively (Fig. 1). The median number of cattle per farm was 59.5, and the range was 7–547. A total of 418 blood samples were collected from the farms. Approximately 125 µl of each blood sample collected was applied to an FTA Classic Card (GE Healthcare) in accordance with the manufacturer’s instructions, and the remainder of the sample was kept at −20°C prior to further use.
Fig. 1.
Map of the Republic of Uganda and the Mbarara district. (A) The map shows the location of the study site, Mbarara District, and capital city of the Republic of Uganda, Kampala. (B) Sub-countries with sampling farms in Mbarara District are shown in grey colour.
FTA-NaOH based-PCR was performed as described in previous reports [12, 13], with some modifications. Three 4-mm2 pieces of FTA card containing dry blood were cut with sterile surgical scalpel blades and transferred into a 0.2-ml PCR tube. To wash the pieces of FTA card, 200 µl of either modified Tris- ethylenediaminetetraacetic acid (EDTA)-NaCl-Triton X-100 (TENT) buffer composed of 10 mM Tris (hydroxymethyl) aminomethane (Tirs-HCl) (pH 8.0), 1 mM EDTA (pH 8.0), 12 mM sodium chloride and 1.25% Triton X-100 or 20 mM sodium hydroxide (NaOH) solution was added to the tube, followed by incubation for 5 min for TENT buffer or 20 min for NaOH solution at room temperature, with intermittent inverting. In the TENT buffer condition, two additional washes were performed. After discarding the solutions, the cards were washed with 200 µl TE0.1 buffer (10 mM Tris-HCl, 0.1 mM EDTA, pH 8.0) twice for TENT buffer condition or once for NaOH solution condition then dried in an incubator at 56°C for 10 min. The dried FTA cards were kept at room temperature and subjected to PCR testing within 1 week. Total DNA was extracted from 200 µl of ten randomly selected frozen blood samples using the QIAamp DNA Mini kit (Qiagen, Hilden, North Rhine-Westphalia, Germany), in accordance with the manufacturer’s instructions.
Bovine mitochondrial cytochrome C oxidase subunit I (COI) gene [1, 4] and T. parva-encoding p104 gene [8] were amplified via the PCR to evaluate the quality of template DNA and T. parva prevalence, respectively. The sequences of the primers used were as follows:
COI LCO1490, 5′-GGTCAACAAATCATAAAGATATTGG-3′
HCO2198, 5′-TAAACTTCAGGGTGACCAAAAAATCA-3′
p104 IL3231, 5′-ATTTAAGGAACCTGACGTGACTGC-3′
IL755, 5′-TAAGATGCCGACTATTAATGACACC-3′ (outer primer of nested PCR)
IL4234, 5′-GGCCAAGGTCTCCTTCAGAATACG-3′
IL3232, 5′-TGGGTGTGTTTCCTCGTCATCTGC-3′ (inner primer of nested PCR)
PCRs were performed using a 25 µl reaction mixture containing 2.5 µl of extracted DNA, 0.2 µM of each primer, 1.25 U of Blend Taq polymerase (Toyobo, Osaka, Japan) and 2.5 µl of 10x reaction buffer. In the PCR based on FTA card, DNA captured in three FTA card pieces treated with TENT or NaOH were put into the PCR tube instead of the 2.5 µl of extracted DNA. PCRs were conducted using the Simpli Amp™ thermal cycler (Applied Biosystems, Foster City, CA, USA) as previously described [4, 8]. For p104 gene amplification, 2.5 µl of primary PCR product was used as a template for nested PCR. Five microliters of each PCR product was run in a 1.5% agarose gel containing 0.01% ethidium bromide using x1 TBE buffer (50 mM Tris-HCl, 48.5 mM boric acid, 2 mM EDTA, pH 8.0) at 100 v for 30 min. The gels were visualized using an ultraviolet transilluminator, and the images were captured. The following three above mentioned DNA extraction methods were compared with regard to quality of DNA extracted and efficiency of PCR detection of the bovine mitochondrial COI gene and the p104 gene, using DNA obtained from ten samples randomly selected from a pool of 418 samples:
(i) DNA extraction kit
(ii) FTA card with TENT buffer (hereafter referred to as the FTA-TENT method), and
(iii) FTA card with NaOH (hereafter referred to as the FTA-NaOH method).
Among 3 methods, (iii) FTA-NaOH method showed highest efficacy of the PCRs and was used for the diagnoses of all 418 samples.
T. parva prevalence at the farm level and the 10th, 15th, 85th and 90th percentile within-herd prevalences were calculated. The 95% confidence interval (CI) for overall animal level prevalence based on beta distribution was calculated using a one-sample proportion test. All statistical analyses were performed using R software version 3.5.1 (R Core Team, 2018).
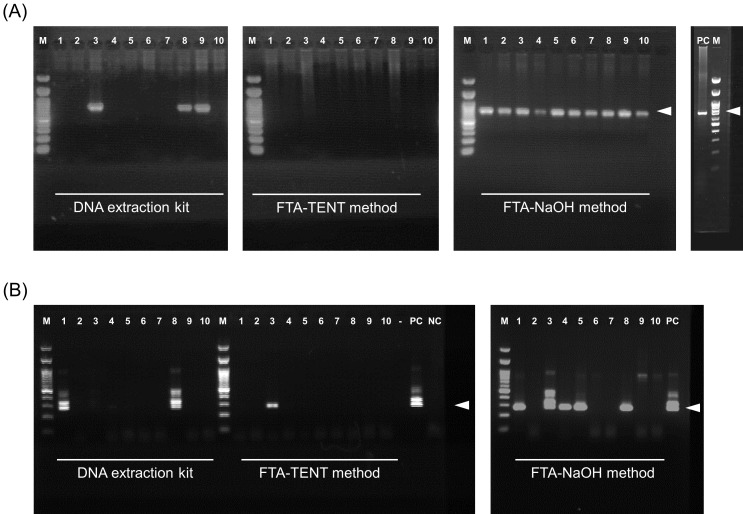
Figure 2A shows the results of PCRs of the bovine COI gene for ten selected samples (ID 1 to 10). The COI gene was amplified in three (3/10) and 10 (10/10) samples extracted by the DNA extraction kit and FTA-NaOH method, respectively. The COI gene was not amplified in any of the DNA samples extracted by the FTA-TENT method, whereas the p104 gene was amplified in two (2/10), one (1/10) and five (5/10) samples via the DNA extraction kit, FTA-TENT method and FTA-NaOH method, respectively (Fig. 2B). DNA preparation using the FTA-NaOH method yielded the best detection rate for both the COI gene and the p104 gene. Moreover, all p104-positive samples detected by the DNA extraction kit (ID 1 and 8) and the FTA-TENT method (ID 3) were successfully detected by the FTA-NaOH method, although there was some discrepancy between results derived from the DNA extraction kit method and the FTA-TENT method (Fig. 2B). The differences observed from the PCR results of the three methods could not have been due to the difference in template DNA concentration, because the volume equivalent to blood was approximately same in all methods (2.50 µl/reaction for DNA extraction kit and 3.06 µl/reaction for FTA-TENT method and FTA-NaOH method). Moreover the estimated amount of template DNA was sufficient to perform PCR using Blend Taq polymerase according to the manufacturer’s protocol (5 to 500 ng of genome DNA/reaction), although the concentration of DNA could not been confirmed. One of the possibilities for the observed difference in PCR results may be caused by the differences in the extent to which the various methods cause cell lysis to expose the nucleic acid. NaOH solution is a kind of strong protein denaturant and more effective in cell lysis, and our results suggested that NaOH solution has stronger effect on protein denaturation compared with the lysis buffer of DNA extraction kit and TENT buffer. However, there are many steps in the preparation of template DNA and it’s difficult to conclude on one factor causing the difference in this study.
Fig. 2.
Agarose gel electrophoresis of bovine Cytochrome C oxidase subunit I (COI) and Theileria parva p104 gene products derived from polymerase chain reactions conducted after three different DNA template preparation methods. Agarose gel electrophoresis images of the bovine COI gene (A) and the p104 gene (B). The polymerase chain reaction templates were prepared using three different methods. The white arrowheads indicate the position of the expected amplicon size. The numbers above the images indicate sample IDs (1 to 10). Positive control DNA of bovine COI gene were prepared from a commercially available bovine liver in Japan. M: 100 bp ladder marker; PC: positive control; NC: negative control; -: empty.
Using the combination of FTA-NaOH method followed by FTA-NaOH based-PCR, a total of 188/418 (45%, 95% CI 40.1–49.9%) of the samples tested were positive for the T. parva p104 gene. The farm-level prevalence was 83.3% (25/30), the median within-herd prevalence was 34.5% (10th, 15th, 85th and 90th percentiles were 0%, 2%, 97% and 100%, respectively), and the range was 0–100%, suggesting that even in the same district herd infection status can be diverse. Each farm-level prevalence is shown in Table 1. In a previous study investigating hemo-parasites conducted in Central and Western Uganda [7], a slightly higher prevalence of T. parva (56.2%) was detected in a region of western Uganda that included Mbarara than that detected in the current study. However, their data included results derived from seven districts and thus may not be directly comparable to the results of the present study. It is also notable that the results reported in that study were based on microscopy, which increases the possibility of false positives, and this may partly explain the higher prevalence reported in that study. Another study conducted in western regions of Uganda including Mbarara reported a T. parva prevalence of 36–39% [6], which is comparable to the results of the current study.
Table 1. Theileria parva prevalence at the farm level.
| Farm ID | No. of cattle | No. of tested | No. of positive | Prevalence (%) |
|---|---|---|---|---|
| 1 | 547 | 78 | 28 | 36 |
| 2 | 165 | 30 | 6 | 20 |
| 3 | 164 | 31 | 23 | 74 |
| 4 | 126 | 25 | 22 | 88 |
| 5 | 115 | 16 | 16 | 100 |
| 6 | 111 | 16 | 6 | 38 |
| 7 | 100 | 13 | 0 | 0 |
| 8 | 95 | 13 | 7 | 54 |
| 9 | 92 | 13 | 13 | 100 |
| 10 | 82 | 16 | 1 | 6 |
| 11 | 81 | 12 | 1 | 8 |
| 12 | 80 | 11 | 3 | 27 |
| 13 | 76 | 11 | 1 | 9 |
| 14 | 72 | 10 | 0 | 0 |
| 15 | 64 | 10 | 10 | 100 |
| 16 | 55 | 8 | 2 | 25 |
| 17 | 53 | 10 | 9 | 90 |
| 18 | 50 | 8 | 2 | 25 |
| 19 | 50 | 7 | 0 | 0 |
| 20 | 50 | 24 | 11 | 46 |
| 21 | 45 | 6 | 0 | 0 |
| 22 | 40 | 6 | 0 | 0 |
| 23 | 30 | 3 | 1 | 33 |
| 24 | 29 | 10 | 8 | 80 |
| 25 | 22 | 5 | 1 | 20 |
| 26 | 19 | 6 | 2 | 33 |
| 27 | 18 | 5 | 5 | 100 |
| 28 | 15 | 5 | 3 | 60 |
| 29 | 10 | 5 | 5 | 100 |
| 30 | 7 | 5 | 2 | 40 |
| Total | 2,463 | 418 | 188 | |
T. parva surveillance via PCR based on FTA card has been reported previously, but comparison of the efficiency of FTA-NaOH method and DNA extraction kit in this context had not been reported [10]. In the current study, the combination of FTA-NaOH method followed by PCR based on FTA card exhibited a substantial capacity to detect T. parva in bovine blood. Diagnosis based on PCR is reliable and objective, but it is often difficult to apply in the field due to the challenges relating to sample transportation and the resources available at the relevant diagnostic facilities. DNA preparation using FTA card does not require cold-chain DNA transportation, a water bath, or a centrifuge for the preparation of PCR template. Moreover, where resources are limited the cost of DNA extraction can be reduced by using a simple FTA-NaOH method, and this method can evidently yield sufficient DNA [13]. As a future prospect, a loop-mediated isothermal amplification (LAMP) system is currently available for T. parva diagnosis [11], and the combination of LAMP and FTA-NaOH method may be a practical and accurate point-of-care testing method for T. parva infection in the field.
Acknowledgments
We thank Mr. Fred Odua and all members of the RTC Laboratory, Department of Veterinary Pharmacy, Clinical and Comparative Medicine, COVAB, Makerere University, Uganda for their technical support. We thank Dr. William Mwebembezi of the Mbarara District Production Office, Mbarara District Local Government, Uganda for facilitating sample collection. This study was supported by the Rakuno Gakuen University Graduate School of Veterinary Medicine, and the ‘Safe Milk Promotion in Mbarara Project’ involving the Japan International Cooperation Agency Partnership Program.
REFERENCES
- 1.Agrizzi J., Loss A. C., Farro A. P. C., Duda R., Costa L. P., Leite Y. L. R.2012. Molecular diagnosis of atlantic forest mammals using mitochondrial DNA sequences: didelphid marsupials. Open Zool. J. 5: 2–9. doi: 10.2174/1874336601205010002 [DOI] [Google Scholar]
- 2.Criado-Fornelio A.2007. A review of nucleic-acid-based diagnostic tests for Babesia and Theileria, with emphasis on bovine piroplasms. Parassitologia 49Suppl 1: 39–44. [PubMed] [Google Scholar]
- 3.Dobbs L. J., Madigan M. N., Carter A. B., Earls L.2002. Use of FTA gene guard filter paper for the storage and transportation of tumor cells for molecular testing. Arch. Pathol. Lab. Med. 126: 56–63. [DOI] [PubMed] [Google Scholar]
- 4.Folmer O., Black M., Hoeh W., Lutz R., Vrijenhoek R.1994. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 3: 294–299. [PubMed] [Google Scholar]
- 5.ILRI. The project goal is to design subunit vaccines for the control of East Coast fever. https://www.ilri.org/research/projects/improved-vaccines-control-east-coast-fever-cattle-africa[accessed on December 16 2019].
- 6.Kabi F., Masembe C., Muwanika V., Kirunda H., Negrini R.2014. Geographic distribution of non-clinical Theileria parva infection among indigenous cattle populations in contrasting agro-ecological zones of Uganda: implications for control strategies. Parasit. Vectors 7: 414. doi: 10.1186/1756-3305-7-414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kasozi K. I., Matovu E., Tayebwa D. S., Natuhwera J., Mugezi I., Mahero M.2014. Epidemiology of increasing hemo-parasite burden in Ugandan cattle. Open J. Vet. Med. 4: 220–231. doi: 10.4236/ojvm.2014.410026 [DOI] [Google Scholar]
- 8.Konnai S., Imamura S., Nakajima C., Witola W. H., Yamada S., Simuunza M., Nambota A., Yasuda J., Ohashi K., Onuma M.2006. Acquisition and transmission of Theileria parva by vector tick, Rhipicephalus appendiculatus. Acta Trop. 99: 34–41. doi: 10.1016/j.actatropica.2006.06.008 [DOI] [PubMed] [Google Scholar]
- 9.Mans B. J., Pienaar R., Latif A. A.2015. A review of Theileria diagnostics and epidemiology. Int. J. Parasitol. Parasites Wildl. 4: 104–118. doi: 10.1016/j.ijppaw.2014.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muhanguzi D., Picozzi K., Hatendorf J., Thrusfield M., Welburn S. C., Kabasa J. D., Waiswa C.2014. Prevalence and spatial distribution of Theileria parva in cattle under crop-livestock farming systems in Tororo District, Eastern Uganda. Parasit. Vectors 7: 91. doi: 10.1186/1756-3305-7-91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thekisoe O. M., Rambritch N. E., Nakao R., Bazie R. S., Mbati P., Namangala B., Malele I., Skilton R. A., Jongejan F., Sugimoto C., Kawazu S., Inoue N.2010. Loop-mediated isothermal amplification (LAMP) assays for detection of Theileria parva infections targeting the PIM and p150 genes. Int. J. Parasitol. 40: 55–61. doi: 10.1016/j.ijpara.2009.07.004 [DOI] [PubMed] [Google Scholar]
- 12.Thompson M. M., Hrabak E. M.2018. Capture and storage of plant genomic DNA on a readily available cellulose matrix. Biotechniques 65: 285–287. doi: 10.2144/btn-2018-0109 [DOI] [PubMed] [Google Scholar]
- 13.Zhou H., Hickford J. G., Fang Q.2006. A two-step procedure for extracting genomic DNA from dried blood spots on filter paper for polymerase chain reaction amplification. Anal. Biochem. 354: 159–161. doi: 10.1016/j.ab.2006.03.042 [DOI] [PubMed] [Google Scholar]