Abstract
Purpose:
Emerging studies suggest that low-pass genome sequencing (GS) provides additional diagnostic yield of clinically significant copy-number variants (CNVs) compared with chromosomal microarray analysis (CMA). However, a prospective back-to-back comparison evaluating accuracy, efficacy, and incremental yield of low-pass GS compared with CMA is warranted.
Methods:
A total of 1,023 women undergoing prenatal diagnosis were enrolled. Each sample was subjected to low-pass GS and CMA for CNV analysis in parallel. CNVs were classified according to guidelines of the American College of Medical Genetics and Genomics.
Results:
Low-pass GS not only identified all 124 numerical disorders or pathogenic or likely pathogenic (P/LP) CNVs detected by CMA in 121 cases (11.8%, 121/1,023), but also defined 17 additional and clinically relevant P/LP CNVs in 17 cases (1.7%, 17/1,023). In addition, low-pass GS significantly reduced the technical repeat rate from 4.6% (47/1,023) for CMA to 0.5% (5/1,023) and required less DNA (50 ng) as input.
Conclusion:
In the context of prenatal diagnosis, low-pass GS identified additional and clinically significant information with enhanced resolution and increased sensitivity of detecting mosaicism as compared with the CMA platform used. This study provides strong evidence for applying low-pass GS as an alternative prenatal diagnostic test.
Keywords: molecular karyotyping, low-pass genome sequencing, copy-number variants, mosaicism
Introduction
DNA copy-number variants (CNVs) in the human genome represent a major genome diversity between two individuals1,2, and it is well known that some of these changes are etiologic factors in various human diseases3,4. The development and clinical implementation of array-based molecular cytogenetic techniques such as chromosomal microarray analysis (CMA) in the past decade have contributed to our understanding of disease-associated CNVs that may or may not be cryptic to conventional cytogenetics5. In addition, CMA provides options for clinical management in postnatal cases with precise prognostic information2 and also enables detection of fetuses affected with well-established genetic syndromes as early as in the first trimester6–8. Therefore, CMA has been recommended as the first-tier test for high-risk pregnancies for identifying fetal numerical disorders (such as trisomy 21) and microscopic/submicroscopic CNVs6–8.
However, CNV detection by CMA is based on probe density, which varies among different CMA platforms, versions and designs within the targeted regions9. In addition, discrepancies exist between the two different detection approaches10, i.e., array-based comparative genomic hybridization (aCGH)6,7 and single-nucleotide polymorphism arrays (SNP array)11 resulting in difficulties for cross-laboratory validation, clinical implementation and genetic counseling, given there are different sensitivities and specificities of detecting clinically relevant CNVs among different CMA platforms.
Recently, CNV analysis by utilizing data from next-generation sequencing platforms has demonstrated to be robust and is able to provide additional and cytogenetically relevant information for prenatal diagnosis12–14. Compared with capture-based assays such as exome sequencing (ES), genome sequencing (GS) provides better sensitivity and specificity of CNV detection due to the increased uniformly distributed/aligned reads15. In our previous study applying CNV analysis based on low-pass (or low-coverage) GS in different sample sources including abortuses, prenatal and postnatal samples, we showed the feasibility of its clinical implementation12. However, no prospective back-to-back comparison study evaluating accuracy, efficacy, and incremental yield of low-pass GS compared with CMA has been reported in routine prenatal diagnosis. Herein, we conducted a study of low-pass GS and CMA performed independently and simultaneously for 1,023 consecutive prenatal cases referred to two prenatal genetic diagnostic centers in the context of identifying numerical disorders and clinically relevant CNVs.
Materials and Methods
Subject Enrollment, Sample Recruitment and Preparation
This study was approved by the institutional review boards of the Chinese University of Hong Kong and Jinan University. Pregnant women requesting a prenatal diagnostic test referred to the two prenatal diagnosis centers were enrolled with written informed consent obtained from each participant. Prenatal samples including chorionic villi, amniotic fluid and cord blood were collected for DNA extraction and quality control (QC), while parental peripheral blood samples were collected whenever available either concurrently or after identification of a putative disease-associated variant for assessment of inheritance.
Genomic DNA was extracted with DNeasy Blood & Tissue Kit (cat. number/ID: 69506, Qiagen, Hilden, Germany) and treated with RNase (Qiagen, Hilden, Germany). DNA was subsequently quantified with the Qubit dsDNA HS Assay Kit (Invitrogen, Carlsbad, CA, USA) and DNA integrity was assessed by gel electrophoresis. Quantitative fluorescence polymerase chain reaction (QF-PCR) was conducted with 100 ng DNA and two panels of short tandem repeat (STR) markers (P1 and XY) located on chromosomes 18, X, and Y for exclusion of maternal cell admixture and polyploidy16. Subsequently, after exclusion, each DNA sample was subjected for routine CMA and low-pass GS in parallel.
CMA
For routine prenatal CMA testing, we employed two CMAs including a well-established customized 44K Fetal DNA Chip v1.0 (Agilent Technologies, Santa Clara, CA, USA) array-CGH based test and an updated version 8X60K Fetal DNA Chip v2.0 (Agilent Technologies) including SNP probes with equivalent performances in CNV detection given the same probe coverage for CNV analysis6,7. Each was performed according to the manufacturer’s protocols and CNVs were analyzed via CytoGenomics6,7 with a minimum requirement of three consecutive probes for a positive call17. Based on the manufacturer’s protocol, the CMA result was considered a technical failure requiring repeat testing when the derivative log ratio spread (DLRS) value was larger than 0.26,7.
Routine Low-pass GS
Low-pass GS was performed for each sample with a modified protocol12,18; in brief, 50 ng of genomic DNA was digested into small fragments (200–300 bp) by fragmentation-end-repair restriction enzyme (BGI-Wuhan, Wuhan, China). After end-repair, addition of an A overhang and adapter ligation, DNA fragments (without size selection) underwent seven cycles of PCR. PCR products from each library were subsequently purified with an Agencourt AMPure XP PCR Purification Kit (Beckman Coulter, Brea, CA, USA). Concentrations of each library were measured using the Qubit dsDNA HS Assay Kit (Invitrogen) and mixed with equal molality into a pool (20–24 samples per lane) and sequenced with a minimum of ~15 million reads per sample (single-end 50 bp) on a BGISeq-500 platform (BGI-Wuhan, Wuhan, China).
Data Preparation and Quality Control
In this study, QC and CNV detection were performed following the methods reported in our previous study12. In brief, uniquely aligned reads (indicated by Burrows-Wheeler Aligner, [BWA]19) were classified into adjustable sliding windows [50-kb in length with 5-kb increments (windows determination described in Supplementary Methods)], in terms of their mapped locations (GRCh37/hg19). Coverage of each window was calculated by the read amount and underwent two-step bias correction (GC correction and population-scale normalization).
For the QC study, as described in our previous publication12, genome-wide standard deviation of the copy ratios from all windows (Supplementary Materials and Methods) excluding those located in the chromosomes of numerical disorders was used as a cutoff for quality control. In this study, to ensure the good quality of clinical application, a more rigorous QC cutoff value (genome-wide standard deviation) was set as 0.1 compared to 0.15 used in our previous publications12.
CNV Detection and Correction
Detection of homozygous/heterozygous deletion and duplication/triplication was performed by optimizing our reported method12. In brief, (1) aneuploidy detection was based on the average copy ratios for the particular chromosome with a mosaic level estimated by the differences of copy ratio compared with a normal copy ratio (expected as 1); (2) regions with putative CNVs were reported (with consecutive copy ratios >1.1 or <0.9) with precise boundaries identified by our reported algorithm Increment-Rate-of-Coverage based on the copy ratios from the adjustable non-overlapping windows; (3) segment recombination for reporting a CNV was performed.
Segment recombination was performed by combining two or more segments into one if the copy ratio difference was not larger than twofold of the standard deviation of this particular chromosome (Supplementary Figure S1). The start and end coordinates of this ‘new’ segment would be defined as the start coordinate of the first segment and the end coordinate of the last segment, respectively. In addition, the copy ratio of this ‘new’ segment would be calculated as the sum of copy ratio multplied by the number of windows in each segment and then divided by the total number of windows. Moreover, if more than 95% of windows with a copy ratio >1.1 or <0.9 were from the same chromosome, then the whole chromosome would be considered as one segment for further filtering (Supplementary Figure S1). The resolution for low-pass GS in reporting a CNV, except for homozygous or hemizygous deletions, was set as 50 kb in the present study.
As homozygous deletion of a certain region was revealed by the lack of aligned reads; detection of a homozygous deletion was based on the ratio values of the nonoverlapping windows throughout the genome. A homozygous deletion would be reported if there were more than one nonoverlapping window with an extremely low number of aligned reads (0.1 as copy ratio) or even absence of aligned read (copy ratio around 0, Supplementary Figure S2). On average, the minimal size of a reported homozygous deletion was approximately 10 kb.
For each CNV, a population-based U-test, whole-sample-based t-test and whole-chromosome-based t-test were performed to eliminate false positives and polymorphisms in the population.
Clinical Interpretation
CNV classification was performed based on the guidelines of the American College of Medical Genetics and Genomics (ACMG) with criteria, methods and in-house data sets described in our previous study12.
Validation of CNVs and Parental Confirmation
Quantitative PCR (qPCR) was performed for validation of additional pathogenic and likely pathogenic (P/LP) CNVs detected by low-pass GS. Primers specific for candidate regions were designed with Primer 3 Web, Primer-Blast (National Center for Biotechnology Information [NCBI]), and in silico PCR (University of California-Santa Cruz [UCSC]) based on the GRCh37/hg19 reference genome (Supplementary Table S1). Melt curve analysis was carried out for each pair of primers to ensure specificity of the PCR amplification, and the standard curve method was used to determine PCR efficiency (within a range from 95% to 105%). Each reaction was performed in triplicate in 10 μl of reaction mixture simultaneously in cases and control (in-house normal male and female controls) on a 7900HT Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) with SYBR Select Master Mix (Applied Biosystems) with the default setting for the reaction condition. The number of copies in each sample was determined by using the ΔΔ Ct method, which compares the difference in Ct (cycle threshold) of the region of interest with a reference primer pair targeting the universally conserved element between the case and the control. Two independent pairs of primers (Supplementary Table S1) were used in triplicate for validation of each CNV. Parental confirmation was performed whenever parental DNAs were available.
Code Availability
All relative programs are available at https://sourceforge.net/projects/increment-ratio-of-coverage-v2/files/Increment_Ratio_of_Coverage_V2.0.tar.gz/download.
Results
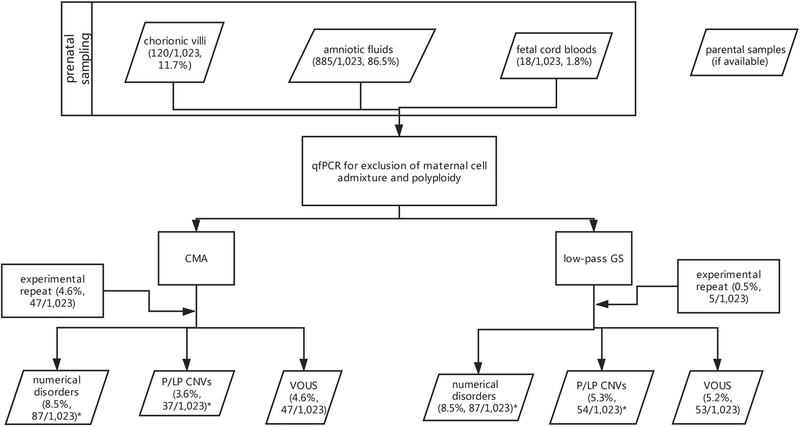
From late 2016 to early 2019, 1,023 women referred for prenatal genetic diagnosis in our clinical laboratories were enrolled in this study with informed consent obtained. There were 120 chorionic villi samples, 885 amniotic fluid samples and 18 fetal cord blood samples (Table 1, Figure 1). After exclusion of maternal cell admixture by QF-PCR16, all 1,023 cases were subjected to low-pass GS and an aCGH-based CMA platform6,7 for CNV analysis, independently and simultaneously.
Table 1.
Diagnostic yields in 1,023 prenatal samples by CMA and low-pass GS
| Sample type | Sample numbers | Low-Pass GSc | CMAc | ||||
|---|---|---|---|---|---|---|---|
| Aneuploidies | pCNVs | Overall | Aneuploidies | pCNVs | Overall | ||
| Amniotic Fluid | 885 | 62 (7.0%, 5.5 to 8.9) | 43 (4.9%, 3.6 to 6.5) | 103 (11.6%, 9.7 to 13.9)* | 62 (7.0%, 5.5 to 8.9) | 31 (3.5%, 2.5 to 4.9) | 91 (10.3%, 8.5 to 12.5)* |
| Chorionic Villi | 120 | 25 (20.8%, 14.5 to 28.9) | 9 (7.5%, 4.0 to 13.6) | 33 (27.5%, 20.3 to 36.1)# | 25 (20.8%, 14.5 to 28.9) | 6 (5%, 2.3 to 10.5) | 30 (25%, 18.1 to 33.4)# |
| Cord Blood | 18 | 0 (0%, 0 to 17.6) | 2 (11.1%, 3.1 to 32.8) | 2 (11.1%, 3.1 to 32.8) | 0 (0%, 0 to 17.6) | 0 (0%, 0 to 17.6) | 0 (0%, 0 to 17.6) |
| Total | 1,023 | 87 (8.5%, 6.9 to 10.4) | 54 (5.3%, 4.1 to 6.8) | 138 (13.5%, 11.5 to 15.7)*,# | 87 (8.5%, 6.9 to 10.4) | 37 (3.6%, 2.6 to 5.0) | 121 (11.8%, 10.0 to 14.0)*,# |
The data in each bracket refer to the detection rate and 95% confidence interval; pCNV, pathogenic or likely pathogenic copy number variants.
pCNVs includes pathogenic and likely pathogenic CNVs.
Percentage shows the frequency of samples with aneuploidy or pCNVs.
Diagnostic yields were calculated based on the number of cases detected with numerical disorders or pCNVs.
Multiple findings:
2 cases with aneuploidy and pCNV;
1 case with aneuploidy and pCNV
Figure 1. Flowchart of copy-number variant (CNV) analysis by low-pass genome sequencing (GS) versus chromosomal microarray (CMA) in 1023 prenatal cases.
All samples were referred to one of two prenatal diagnostic centers, and after exclusion of maternal cell admixture and polyploidy by QF-PCR with short tandem repeat (STR) makers, CNV analysis was performed by low-pass GS and CMA in parallel. Detailed methods and results are described in the main text. *Three cases with aneuploidy and P/LP CNV were detected simultaneously. VOUS variants of uncertain significance.
Comparison of Diagnostic Yields by Low-pass GS and CMA
Among all 1,023 cases, CMA identified 121 cases with 87 aneuploidies and 37 P/LP CNVs, providing a diagnostic yield of 11.8% (121/1,023, Supplementary Table S2). In comparison, low-pass GS not only detected all 124 events in these 121 cases defined by CMA, but also provided a 1.7% (17/1,023) increased yield (expressed as 17 events in 17 cases) compared with CMA (Figure 1, Supplementary Table S2). By using CMA results as reference, low-pass GS provided a 99.9% sensitivity (121/121) and an 87.7% specificity (121/138) of detecting numerical disorders and P/LP CNVs. Overall, low-pass GS provided a diagnostic yield of 13.5% (138/1,023).
Among those 17 cases with additionally diagnosis by low-pass GS (Table 2), the majority (82.4%, 14/17) were Southeast Asian (SEA) type alpha thalassemia due to a 19.3-kb homozygous deletion (Supplementary Figure S2). According to a literature review, affected fetuses would largely present with signs of fetal anemia, including increased middle cerebral artery peak systolic velocity, increased cardiothoracic ratio or an increase in placental thickness, eventually leading to hydrops fetalis. In this study, 6/14 cases were found with ultrasound anomalies with signs of fetal anemia, while the other 8 cases had a family history of thalassemia or chromosomal abnormalities found in previous pregnancies.
Table 2.
Additional pathogenic or likely pathogenic CNVs identified by low-pass GS
| Case ID | Gestational week | Primary indication# | Detailed clinical indication(s)*: | Low-Pass GS |
|---|---|---|---|---|
| 16BA0152 | 16 | DSS | High risk Down syndrome screen, and family history of thalassemia | seq[GRCh37] del(16)(p13.3) chr16:g.216271_235599del |
| 16BA0064 | 28 | AUS | Multiple congenital abnormalities, cardiomegaly, thickened myometrium, pericardial effusion, tricuspid regurgitation, abdominal and thoracic effusion | seq[GRCh37] del(16)(p13.3) chr16:g.216050_235599del |
| 18BA0454 | 17 | FHX | Family history of thalassemia | seq[GRCh37] del(16)(p13.3) chr16:g.216290_235599del |
| 18BA0351 | 17 | AMA | Previous pregnancy with chromosomal abnormality, advanced maternal age, and family history of thalassemia and cleft lip and palate in mother | seq[GRCh37] del(16)(p13.3) chr16:g.216050_229978del |
| 18BA0145 | 18 | FHX | Previous miscarriage and family history of thalassemia | seq[GRCh37] del(16)(p13.3) chr16:g.217556_231140del |
| 18BA0425 | 17 | FHX | Family history of thalassemia | seq[GRCh37] del(16)(p13.3) chr16:g.217215_231140del |
| 18BA0218 | 21+6 | FHX | Family history of thalassemia | seq[GRCh37] del(16)(p13.3) chr16:g.217215_231140del |
| 17BA0605 | 23+4 | AUS | Nuchal edema, echogenic bowel, pericardial effusion, oligohydramnios | seq[GRCh37] del(16)(p13.3) chr16:g.216530_235599del |
| 18BA0115 | 26 | AUS | Borderline risk for Down syndrome screen, structural abnormality | seq[GRCh37] del(16)(p13.3) chr16:g.216236_235599del |
| 18BA0376 | 23 | AUS | Cardiac structural abnormality | seq[GRCh37] del(16)(p13.3) chr16:g.217392_231140del |
| 18BA0453 | 17 | FHX | Family history of thalassemia | seq[GRCh37] del(16)(p13.3) chr16:g.216050_235599del |
| 18BA0442 | 16 | FHX | Family history of thalassemia | seq[GRCh37] del(16)(p13.3) chr16:g.217349_231140del |
| 19C0339-W | 21+4 | AUS | Cardiomegaly, pericardial effusion, thickened myometrium, enlarged liver | seq[GRCh37] del(16)(p13.3) chr16:g.216149_235599del |
| 19C0855-W | 11+5 | AUS | NT 4.75-mm, increased placental thickness, absent nasal bone | seq[GRCh37] del(16)(p13.3) chr16:g.216149_235599del |
| 17C1244-A | 13+5 | DSS | Fetal anomalies: NT 3.3-mm, positive Down syndrome screen: Risk: 1:6, T18 & T13 positive | seq[GRCh37] del(5)(q23.3)mat chr5:g.127783782_128082258del |
| 18BA0221 | 17+2 | AUS | NT 3.5-mm | seq[GRCh37] del(X)(p21.1)mat chrX:g.32311051_32342286del |
| 19C1149-W | 12 | AUS | NT 5.22-mm, high risk Down syndrome screen | seq[GRCh37] dup(8)(p21.2q24.3)dn chr8:g.23814437_146298884mos dup[0.3] |
NT, nuchal translucency, -SEA, α0-thalassemia of southeast Asian deletion.
AUS: Abnormal Ultrasound; DSS: 1st/2nd trimester aneuploidy screening high risk; AMA: Advanced Maternal Age; FHX: Family history.
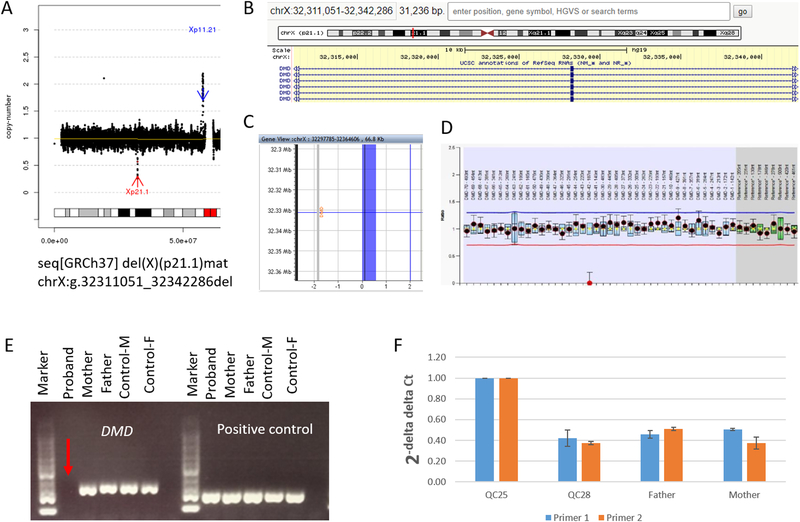
The other three cases involved two cryptic deletions and one mosaic partial aneuploidy. In case 18BA0221, with increased nuchal translucency (NT) (3.5 mm) detected in first trimester Down syndrome screening, low-pass GS detected a 31.2-kb cryptic hemizygous deletion in the male fetus involving the 42nd exon of DMD (Figure 2), resulting in an in-frame deletion (predicted by Leiden Muscular Dystrophy pages, http://www.dmd.nl). The deletion was further confirmed by multiplex ligation-dependent probe amplification (MLPA) and parental study revealed maternal inheritance. Because hemizygous deletion or disruption of DMD would potentially cause Duchenne muscular dystrophy (DMD) or Becker muscular dystrophy (BMD) (OMIM 300377), follow-up testing for creatine kinase (CK) and CK-MB (muscle/brain) in the infant at 8 months showed these two values were both increased. As DMD or BMD would be suspected at a later age, early diagnosis is imperative to provide precise prognostic information for genetic counseling and potential options for clinical management for the family.
Figure 2. Low-pass genome sequencing (GS) defines a cryptic deletion involving exon 42 of DMD in case 18BA0221.
(A) Low-pass GS identified a hemizygous deletion in Xp21.1 in a male fetus. The deletion is indicated by a red arrow and detailed coordinates are provided at the bottom in terms of the International System for Human Cytogenomic Nomenclature (ISCN) 2016. The X axis represents the genomic coordinates while the Y axis indicates the copy ratio of each window (shown as black dot). (B) The detailed gene component is shown in the UCSC genome browser. (C) Probe distribution of the CMA platform within this deleted region indicates absence of any probe. (D) MLPA validation. The deleted 42nd exon is indicated by a red dot at the bottom of the figure. (E) PCR validation with a pair of primers targeting the 42nd exon of DMD. Each column shows the band information after PCR amplification. In comparison with the mother, father, male control and female control, there is no amplified band (indicated by a red arrow) shown in case 18BA0221. Amplification of a pair of primers targeting an ultra-conserved region (https://ccg.epfl.ch/UCNEbase/view.php?data=ucne&entry=5530) was used as positive control. (F) Quantitative PCR with two independent pairs of primers targeting the 42nd exon in QC25 (female control), QC28 (male control), father and mother. Only one copy of exon 42 is observable in each DNA except the female control, confirming that the deletion in the male fetus was maternally inherited.
The second case, 17C1244, was referred for prenatal diagnosis for increased NT (3.3 mm) and high risk for Down syndrome (1:6). The mother was known to have a ventricular septal defect (VSD). Low-pass GS revealed a 298.7-kb maternally inherited heterozygous deletion involving exons 1–8 of FBN2 in the fetus (Supplementary Figure S3). Because pathogenic point variants and intragenic deletion in FBN2 are known to cause VSD or cardiovascular malformation in an autosomal dominant manner20–22, further follow-up study is being pursued.
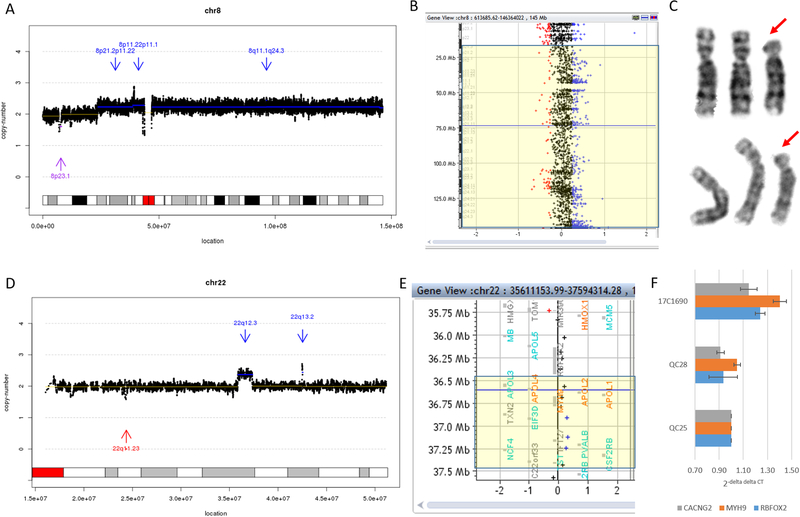
Among the 16 deletions not detected by CMA, the reason was attributed by insufficient probe coverage in the target regions on the CMA platform (Figure 2C and Supplementary Figure S3C). In contrast, in case 19C1149, cystic hygroma (NT=5.22 mm) was detected at gestational week 12 with positive Down syndrome screening, and high risk for both trisomy 13 and 18. CVS was collected and the CMA report was normal, while low-pass GS reported a mosaic partial aneuploidy of chromosome 8, dup(8)(p21.2q24.3), with mosaicism estimated at approximately 23% (Figure 3A)23. By reviewing the CMA result we found a copy ratio increase of 18% within the targeted region (Figure 3B). Karyotyping confirmed the finding of 46,XX[28]/47,XX,+del(8)(p21)[2] (Figure 3C) with a much higher percentage of cells with abnormal karyotypes (93.3%, 28/30). Further collection of amniotic fluid at 17 gestational weeks was conducted and tested with low-pass GS and karyotyping performed. Low-pass GS in the amniotic fluid (AF) sample revealed the same pathogenic duplication at an approximately 34% mosaic level, while karyotyping reported a similar percentage of abnormal cells (53.3%, 16/30). The discrepancy on the mosaic percentages in CVS was suspected to be due to different cell origins between the direct (uncultured) analysis (predominantly cytotrophoblasts) and cultured cells (mesenchymal core of villi)24. Overall, it indicated that low-pass GS would have higher sensitivity in detecting low-level mosaicism compared with CMA. The pregnancy is continuing and follow-up study is ongoing.
Figure 3. Low-level mosaicism defined by low-pass genome sequencing (GS).
(A) Low-pass GS identified a partial trisomy 8 (indicated by three blue arrows) at an approximately 23% mosaic level in 19C1149 involving the centromere. (B) Probe distribution in CMA with the candidate region reported by low-pass GS highlighted in yellow. (C) Two partial karyotypes of G-banded chromosomes showing two normal chromosome 8s and one copy of chromosome 8 with a deletion in the short (p) arm (red arrow). (D) Low-pass GS revealed a 1.4-Mb mosaic gain located in 22q12.3 (indicated by blue arrow and labeled with chromosome band). (E) Probe distribution in CMA. The candidate region is highlighted in yellow with seven probes (shown as ‘+’) mapped within this region. (F) Bar figure shows the result of qPCR validation. Three independent pairs of primers residing in the candidate region were used for validation and all showed approximately 40% increases compared with QC25 (normal control sample). In addition, results in QC28 (another normal control sample) are shown for comparison. In figures (A) and (D), the X-axis represents the genomic coordinates, and the Y-axis indicates the copy ratio of each window (shown as black dots). In figures (B) and (E), copy-number loss, neutral and gain are shown in red, black and blue, respectively.
Identification of Variants of Unknown Significance
Apart from reporting P/LP CNVs, CMA also identified 47 variants of uncertain significance (VOUS) in 47 cases (Supplementary Table S3). In comparison, low-pass GS not only detected all VOUS identified by CMA, but also revealed an additional six VOUS in six cases. Among these six cases, five were without a numerical disorder or P/LP CNV identified and the other case was reported as Turner syndrome (Supplementary Table S4).
Among these six VOUS not discovered by CMA, four were also due to the lack of sufficient probes within the corresponding regions, while the other two were due to low-level mosaicism (Figure 3A). Case 17C1690 was diagnosed with right aortic arch and the CMA result was normal. However, low-pass GS found a 1.4-Mb de novo mosaic gain seq[GRCh37] dup(22)(q12.3)dn chr22:g.35880201_37359631dup[0.4] (Figure 3D). The germline phenotype for this gain has been reported to be associated with multiple congenital anomalies, developmental delay and intellectual disability in ClinVar and DECIPHER databases. This gain was classified as a VOUS because it presented as a low-level mosaic and the reported associated phenotypes would not be diagnosed prenatally. The CMA possessed seven probes within this region and most of them showed an increased copy ratio although not indicated (Figure 3E). Three independent pairs of primers located in the targeted region were employed for qPCR validation experiments and confirmed an approximately 40% increase compared with controls (Figure 3F). We further applied a SNP CMA for the same sample source, and although allele differences were observed in the targeted region (Supplementary Figure S4), the SNP CMA still failed to detect this event.
In addition, low-pass GS provided better delineation of a VOUS in the context of a numerical disorder detected in 18BA0141 (Supplementary Figure S5). Noninvasive prenatal screening (NIPS) of this case indicated a loss of the X chromosome, and the CMA was interpreted as Turner syndrome (45,X). However, low-pass GS revealed a 15% mosaic dup(X)(p11.3q25), suspected to result from a ring chromosome X (r(X)) (Supplementary Figure 5A). Although in CMA, probes mapping in Xp11.3q25 showed a slightly increased copy ratio (Supplementary Figure 5B), CMA still failed to detect this event. Further confirmation with conventional G-banded chromosome analysis showed 17% (17/100) of cells with a r(X). Taken together, these data also support a higher sensitivity for low-pass GS in identifying low-level mosaicisms of both numerical disorders and CNVs compared with routine CMA.
Subgroup analysis
We further performed subgroup analyses of the diagnostic yields by CMA or low-pass GS based on different primary indications (Supplementary Table S5). Based on the interpretation by low-pass GS, the group with primary indications as high risk by NIPS had the highest diagnostic yield (37.0%, 54/146) of numerical disorders or P/LP CNVs, while the group with advanced maternal age had the lowest yield (6/99, 6.06%). In contrast, the group with a family history of fetal genetic anomaly in a previous pregnancy had the highest yield (7.0%, 6/86) of additional findings by low-pass GS. This is consistent with the finding of most additional P/LP CNVs defined by low-pass GS to be SEA type homozygous deletion, known to result in signs of fetal anemia and hydrops.
Experimental Repeat Rate and DNA Usage
In this study, after QF-PCR (100 ng) for each sample, amounts of DNA for low-pass GS and CMA were set as 50 ng and 300 ng, respectively. Among these samples, the initial DNA extraction yielded sufficient amounts for QF-PCR, CMA and low-pass GS. However, there were four cases (all of which were amniotic fluid samples) that yielded insufficient amounts of DNA, and only QF-PCR and low-pass GS were performed. CMA was pursued after extracting additional DNA from cell cultures (0.4%, 4/1023).
According to the clinical QC setting for low-pass GS and CMA, 5 and 47 cases failed in the initial experiments, respectively. Thus, the experimental repeat rate of 0.5% (5/1,023) for low-pass GS was significantly lower than that in CMA (4.6%, 47/1,023, Chi-Square Test: P<0.0001, Figure 1). For all cases requiring repeat testing, all were subsequently successful.
Discussion
Genome sequencing techniques allow identification of additional and clinically relevant chromosomal abnormalities compared with CMA. We performed a prospective back-to-back study evaluating the accuracy, experimental repeat rate and additional yield of low-pass GS by using CMA as a reference. Overall, our study demonstrated that low-pass GS provided a diagnostic yield of 13.5% (138/1,023), including all aneuploidies and P/LP CNVs defined by CMA (11.8%) and 17 cases with additional diagnostic findings (Figure 1, Supplementary Table S1).
Low-pass GS not only showed the advantage of identifying cryptic CNVs located in regions with insufficient probe coverage on CMA platforms, but also demonstrated its increased sensitivity in detecting low-level mosaicism. The resolution for CNV analysis in low-pass GS was optimized to be 10 kb for homozygous or hemizygous deletions and 50 kb for all other types of CNVs, respectively12, both of which were higher than the reported prenatal study with the resolution set as 100 kb14. The value of GS in prenatal studies is demonstrated by identification of the SEA type alpha thalassemia 19.3-kb homozygous deletion in 1.4% (14/1,023) of cases, given the high prevalence of carriers in the South Chinese population25. In addition, apart from identifying cryptic CNVs <100 kb, low-pass GS also defined a pathogenic 298.7-kb deletion (affecting FBN2) that was missed by CMA. This indicates that cryptic CNVs missed by CMA are not only due to size but also due to nonuniformly distributed probes on the CMA platform9. With genome-wide resolution, low-pass GS shows the potential advantage in identifying genetic etiologies in human diseases attributed by Mendelian disorders or monogenic disorders such as DMD. Moreover, the increased sensitivity in identifying low-level mosaicism ranges from cryptic CNVs (Figure 3D) to partial aneuploidy (Figure 3A).
The CMA platform used in this study was designed by removing the probes located in regions known to have copy-number polymorphisms (defined by the Database of Genomic Variants [DGV] and DECIPHER), and adding probes within regions known to have reported P/LP CNVs. However, given there are various CMA platforms applied in the clinical setting in different laboratories, we assessed the feasibility and reliability of applying the CMA platform used in this study as a standard for prenatal diagnosis. To that aim, we further calculated probe numbers within the 428 reported P/LP CNVs from a study summarizing the CMA detection results among over 23,865 prenatal cases26. The result shows that there are 422 regions (422/428, 98.6%, Supplementary Figure S6) with a minimum of three probes, which is the minimal cutoff of contiguous probes for identification of a CNV in this particular CMA platform17. It indicated that this CMA is able to detect >98% of the reported P/LP CNVs in prenatal diagnosis. Moreover, among those six regions with insufficient probes, although the sizes ranged from 81.6 kb to 298.1 kb, five were larger than 100 kb and all of these five were located in regions without an OMIM disease-causing gene or with an OMIM autosomal recessive disease-causing gene. This indicates that these five “P/LP CNVs” defined previously might be reclassified as VOUS or even benign. Therefore, it demonstrates the reliability of applying this CMA platform for routine prenatal diagnosis compared with those CMA platforms with higher genome-wide probe density. In addition, we further compared probe distributions within those P/LP CNVs only identified by low-pass GS and the results showed a variable probe density in the targeted region among different CMA platforms (Supplementary Figure S7 and Table S6). Furthermore, insufficient probes were also found in some of these CMA platforms with these regions indicating the importance of applying low-pass GS for CNV analysis, a technology relying on genome-wide uniformly distributed reads/windows.
Emerging studies raise concerns regarding the increased rate of detecting VOUS by GS, subsequently increasing the challenge of genetic counseling and maternal anxiety, compared with CMA, a target-based assay27. In this study, low-pass GS yielded a total number of 53 VOUS, resulting in only a 0.6% (six additional VOUS) increased rate compared with CMA. The first line of clinical interpretation in this study was to review the prevalence of each rare CNV not only in public databases (such as ClinVar, DECIPHER and DGV), but also present in the in-house data sets with most data derived from the same ethnic populations12. Data for in-house data sets consisted of the former testing data from CMA and/or low-pass GS with well-characterized phenotypes in each tested sample. By comparison of the spectrum of the deletions/duplications detected among these samples with ones reported by DGV (Supplementary Figure S8AB), we were able to identify events confined to our cohort (Supplementary Figure S9). For instance, seq[GRCh37] del(1)(p36.22) chr1:g.10003418_10118326del involving GeneReviews and OMIM disease causing gene NMNAT1 were detected in two cases, 18C1031 with cardiac defects (Ebstein) and fetal hydrops reported and 18C0156 with intrauterine death at 32+2 gestational weeks in this study (Supplementary Figure S9A). However, there were no similar deletions reported in DGV (Supplementary Figure S9B). When comparing with our in-house data sets, there were three samples with similar deletions detected but none of them had similar phenotypes compared with the indications from these two cases. In addition, as NMNAT1 was reported to cause autosomal recessive Leber congenital amaurosis 9 (OMIM 608553), this deletion was further classified as benign. The reason for the absence of this deletion in DGV might be due to the ethnic differences. In addition, DGV is composed from curation of “control” data sets with specific phenotypes/conditions confined to particular studies. Therefore, this indicates the importance of building up in-house data sets with well-characterized phenotypic data derived from the same population, which would further inform CNV classification. Particularly, parental confirmation and detailed clinical assessment of the parental phenotypes/features are important for confirmation of the inheritance. This emphasizes that the clinical utility of applying low-pass GS for CNV analysis would not only rely on the accuracy of variant detection but also build on the accumulated data sets for interpretation18.
In this study, QF-PCR with STR markers was employed as the first-tier method for exclusion of maternal cell admixture and determination of polyploidy. However, none of these 1,023 cases were reported to have polyploidy. Although fetuses with triploidy are less likely to survive to term28, a combination of low-pass GS and QF-PCR would be essential in the clinical setting. In this study, the read-depth used for low-pass GS was only 0.25-fold, which is unable to detect absence of heterozygosity (AOH) attributed to the absence of genotypic information. However, AOH of multiple chromosomes or for a single chromosome was observed in 2 cases among 455 cases with the 8X60K CMA platform that included SNP probes. The multiple AOH detected in case 17C0705 resulted from parental consanguinity (Supplementary Figure S10A), while trisomy rescue was likely in case 18C1493 (Supplementary Figure S10B). If parental consanguinity were known prior to testing, it would be appropriate to pursue testing on a platform with SNPs. In addition, AOH such as that due to uniparental disomy (UPD) in 18C1493 is an aberration known to be disease associated29. Because UPD testing is only recommended if a clinically relevant chromosome (chromosomes 6, 7, 11, 14, 15, or 20) is involved in a prenatally detected chromosomal abnormality such as trisomy mosaicism30, this case could be pursued by additional testing31 if such an event were identified by low-pass GS (Supplementary Table S2). Nonetheless, future studies to increase the sequencing read-depth to obtain genotypic information is warranted. In addition, because single-end sequencing with 50 bp was employed in this study, low-pass GS was unable to detect balanced translocations and inversions. Given increasing numbers of studies demonstrating the pathogenicity of structural rearrangements32,33 particularly for those in the “blind spot” of current conventional technologies such as G-banded chromosome analysis, further study with increased read depth and paired-end sequencing approach or even high read-depth GS is also warranted34–36.
A variety of aspects favor utilizing low-pass GS in prenatal diagnosis. Because CMA requires a larger quantity of DNA (300 ng) compared to low-pass GS (50 ng), CMA assay was pending for cell culture in four cases, all of which were amniotic fluids. The significantly decreased amount of DNA required would make possible migration of testing on amniotic fluids to earlier gestational weeks, thus, reducing maternal anxiety. In addition, low-pass GS significantly reduced the technical repeat rates from 4.6% in CMA (47/1,023) to 0.5% (5/1,023). The majority of failed cases in the initial experiment on the CMA platform were due to limited amounts of DNA extracted from amniotic fluid cells (95.7%, 45/47). Another cause for failure may be attributed to the presence of abundant cell-free messenger RNA (mRNA) in amniotic fluids37. Lower repeat rate in low-pass GS would decrease the experimental and labor costs compared with CMA, thus, reducing the burden for clinical diagnostic laboratories. Moreover, low-pass GS has much higher throughput than CMA (40–48 samples per run).
In conclusion, our study demonstrates that the performance of CNV analysis in low-pass GS is equivalent and surpasses routine CMA in the context of prenatal diagnostic testing. Given a comparable turnaround-time, higher throughput, and significant reduction in the technical repeat rate and amount of DNA required for the assay, our study provides compelling evidence for applying low-pass GS as an alternative prenatal diagnostic test.
Supplementary Material
Acknowledgments:
This project is supported by the National Natural Science Foundation of China (81741004,81671470 and 31801042), the Health and Medical Research Fund (04152666), National Key R&D Program of China (2018YFC1002702), Sanming Project of Medicine in Shenzhen (Project No.: SZSM201406007; SZSM201606088). C.C.M is supported by the National Institute for Health Research (NIHR) Manchester Biomedical Research Centre.
Footnotes
Conflicting of interest: The authors declare no conflicting of interest.
Availability of data
Genome sequencing data used in this study has been made available in the China National GeneBank (CNGB) Nucleotide Sequence Archive (CNSA: https://db.cngb.org/cnsa) under the accession number CNP0000507.
References:
- 1.1000 Genomes Project Consortium. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sudmant PH, Rausch T, Gardner EJ, et al. An integrated map of structural variation in 2,504 human genomes. Nature. 2015;526(7571):75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cooper GM, Coe BP, Girirajan S, et al. A copy number variation morbidity map of developmental delay. Nat Genet. 2011;43(9):838–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chong WW, Lo IF, Lam ST, et al. Performance of chromosomal microarray for patients with intellectual disabilities/developmental delay, autism, and multiple congenital anomalies in a Chinese cohort. Mol Cytogenet. 2014;7:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miller DT, Adam MP, Aradhya S, et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet. 2010;86(5):749–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leung TY, Vogel I, Lau TK, et al. Identification of submicroscopic chromosomal aberrations in fetuses with increased nuchal translucency and apparently normal karyotype. Ultrasound Obstet Gynecol. 2011;38(3):314–319. [DOI] [PubMed] [Google Scholar]
- 7.Huang J, Poon LC, Akolekar R, Choy KW, Leung TY, Nicolaides KH. Is high fetal nuchal translucency associated with submicroscopic chromosomal abnormalities on array CGH? Ultrasound Obstet Gynecol. 2014;43(6):620–624. [DOI] [PubMed] [Google Scholar]
- 8.Yang X, Li R, Fu F, Zhang Y, Li D, Liao C. Submicroscopic chromosomal abnormalities in fetuses with increased nuchal translucency and normal karyotype. J Matern Fetal Neonatal Med. 2017;30(2):194–198. [DOI] [PubMed] [Google Scholar]
- 9.Wang JC, Radcliff J, Coe SJ, Mahon LW. Effects of platforms, size filter cutoffs, and targeted regions of cytogenomic microarray on detection of copy number variants and uniparental disomy in prenatal diagnosis: Results from 5026 pregnancies. Prenat Diagn. 2019;39(3):137–156. [DOI] [PubMed] [Google Scholar]
- 10.Shah MS, Cinnioglu C, Maisenbacher M, Comstock I, Kort J, Lathi RB. Comparison of cytogenetics and molecular karyotyping for chromosome testing of miscarriage specimens. Fertil Steril. 2017;107(4):1028–1033. [DOI] [PubMed] [Google Scholar]
- 11.Lin SB, Xie YJ, Chen Z, et al. Improved assay performance of single nucleotide polymorphism array over conventional karyotyping in analyzing products of conception. J Chin Med Assoc. 2015;78(7):408–413. [DOI] [PubMed] [Google Scholar]
- 12.Dong Z, Zhang J, Hu P, et al. Low-pass whole-genome sequencing in clinical cytogenetics: a validated approach. Genet Med. 2016;18(9):940–948. [DOI] [PubMed] [Google Scholar]
- 13.Liang D, Peng Y, Lv W, et al. Copy number variation sequencing for comprehensive diagnosis of chromosome disease syndromes. J Mol Diagn. 2014;16(5):519–526. [DOI] [PubMed] [Google Scholar]
- 14.Wang J, Chen L, Zhou C, et al. Prospective chromosome analysis of 3429 amniocentesis samples in China using copy number variation sequencing. Am J Obstet Gynecol. 2018;219(3):287 e281–287 e218. [DOI] [PubMed] [Google Scholar]
- 15.Belkadi A, Bolze A, Itan Y, et al. Whole-genome sequencing is more powerful than whole-exome sequencing for detecting exome variants. Proc Natl Acad Sci U S A. 2015;112(17):5473–5478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choy KW, Kwok YK, Cheng YK, et al. Diagnostic accuracy of the BACs-on-Beads assay versus karyotyping for prenatal detection of chromosomal abnormalities: a retrospective consecutive case series. BJOG. 2014;121(10):1245–1252. [DOI] [PubMed] [Google Scholar]
- 17.Brown KH, Dobrinski KP, Lee AS, et al. Extensive genetic diversity and substructuring among zebrafish strains revealed through copy number variant analysis. Proc Natl Acad Sci U S A. 2012;109(2):529–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dong Z, Xie W, Chen H, et al. Copy-Number Variants Detection by Low-Pass Whole-Genome Sequencing. Curr Protoc Hum Genet. 2017;94:8 17 11–18 17 16. [DOI] [PubMed] [Google Scholar]
- 19.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang M, Clericuzio CL, Godfrey M. Familial occurrence of typical and severe lethal congenital contractural arachnodactyly caused by missplicing of exon 34 of fibrillin-2. Am J Hum Genet. 1996;59(5):1027–1034. [PMC free article] [PubMed] [Google Scholar]
- 21.Kolble N, Wisser J, Babcock D, Maslen C, Huch R, Steinmann B. Prenatal ultrasound findings in a fetus with congenital contractural arachnodactyly. Ultrasound Obstet Gynecol. 2002;20(4):395–399. [DOI] [PubMed] [Google Scholar]
- 22.Lavillaureix A, Heide S, Chantot-Bastaraud S, et al. Mosaic intragenic deletion of FBN2 and severe congenital contractural arachnodactyly. Clin Genet. 2017;92(5):556–558. [DOI] [PubMed] [Google Scholar]
- 23.Abu-Amero KK, Kondkar AA, Salih MA, et al. Ophthalmologic observations in a patient with partial mosaic trisomy 8. Ophthalmic Genet. 2013;34(4):249–253. [DOI] [PubMed] [Google Scholar]
- 24.Wapner RJ, Martin CL, Levy B, et al. Chromosomal microarray versus karyotyping for prenatal diagnosis. N Engl J Med. 2012;367(23):2175–2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lai K, Huang G, Su L, He Y. The prevalence of thalassemia in mainland China: evidence from epidemiological surveys. Sci Rep. 2017;7(1):920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chau MHK, Cao Y, Yvonne Kwok KY, et al. Characteristics and mode of inheritance of pathogenic copy number variants in prenatal diagnosis. Am J Obstet Gynecol. 2019. [DOI] [PubMed] [Google Scholar]
- 27.Westerfield L, Darilek S, van den Veyver IB. Counseling Challenges with Variants of Uncertain Significance and Incidental Findings in Prenatal Genetic Screening and Diagnosis. J Clin Med. 2014;3(3):1018–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McFadden DE, Robinson WP. Phenotype of triploid embryos. J Med Genet. 2006;43(7):609–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu S, Zhang K, Song F, et al. Uniparental Disomy of Chromosome 15 in Two Cases by Chromosome Microarray: A Lesson Worth Thinking. Cytogenet Genome Res. 2017;152(1):1–8. [DOI] [PubMed] [Google Scholar]
- 30.Eggermann T, Soellner L, Buiting K, Kotzot D. Mosaicism and uniparental disomy in prenatal diagnosis. Trends Mol Med. 2015;21(2):77–87. [DOI] [PubMed] [Google Scholar]
- 31.EUCROMIC. Trisomy 15 CPM: probable origins, pregnancy outcome and risk of fetal UPD: European Collaborative Research on Mosaicism in CVS (EUCROMIC). Prenat Diagn. 1999;19(1):29–35. [PubMed] [Google Scholar]
- 32.Zepeda-Mendoza CJ, Ibn-Salem J, Kammin T, et al. Computational Prediction of Position Effects of Apparently Balanced Human Chromosomal Rearrangements. Am J Hum Genet. 2017;101(2):206–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lupiáñez DG, Kraft K, Heinrich V, et al. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell. 2015;161(5):1012–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dong Z, Wang H, Chen H, et al. Identification of balanced chromosomal rearrangements previously unknown among participants in the 1000 Genomes Project: implications for interpretation of structural variation in genomes and the future of clinical cytogenetics. Genet Med. 2018;20(7):697–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dong Z, Jiang L, Yang C, et al. A robust approach for blind detection of balanced chromosomal rearrangements with whole-genome low-coverage sequencing. Hum Mutat. 2014;35(5):625–636. [DOI] [PubMed] [Google Scholar]
- 36.Dong Z, Zhao X, Li Q, et al. Development of coupling controlled polymerizations by adapter-ligation in mate-pair sequencing for detection of various genomic variants in one single assay. DNA Res. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Edlow AG, Bianchi DW. Tracking fetal development through molecular analysis of maternal biofluids. Biochim Biophys Acta. 2012;1822(12):1970–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.