Abstract
Background
Toxoplasma gondii (T. gondii) is an obligate intracellular parasite, which can affect the pregnancy outcomes in infected females by damaging the uterus, and the intrauterine environment as well as and the hypothalamus resulting in hormonal imbalance. However, the molecular mechanisms underlying the parasite-induced poor pregnancy outcomes and the key genes regulating these mechanisms remain unclear. Therefore, this study aimed to analyze the gene expression in the mouse’s uterus following experimentally-induced acute infection with T. gondii RH strain. Three groups of female mice were intraperitoneally injected with tachyzoites as follow; 3 days before pregnancy (FBD6), after pregnancy (FAD6), and after implantation (FID8) as the experimental groups. Another corresponding three groups served as control, were injected with normal saline at the same time. Transcriptome analysis of the total RNA extracted from both infected and non-infected mouse uterus samples was performed using RNA sequencing (RNA-Seq).
Results
The three experimental groups (FBD6, FAD6, and FID8) had a total of 4,561, 2,345, and 2,997 differentially expressed genes (DEGs) compared to the controls. The significantly upregulated and downregulated DEGs were 2,571 and 1,990 genes in FBD6, 1,042 and 1,303 genes in FAD6 and 1,162 and 1,835 genes in FID8 group, respectively. The analysis of GO annotation, and KEGG pathway showed that DEGs were mainly involved in anatomical structure development, transport, cell differentiation, embryo development, hormone biosynthetic process, signal transduction, immune system process, phagosome, pathways in cancer, and cytokine-cytokine receptor interaction pathways.
Conclusion
T. gondii infection can induce global transcriptomic changes in the uterus that may cause pregnancy hypertension, destruct the intrauterine environment, and hinder the normal development of placenta and embryo. Our results may help to understand the molecular mechanisms of the acute T. gondii infection, which could promote the development of new therapeutics or prophylactics for toxoplasmosis in pregnancy.
Keywords: T. gondii, RNA-Seq, acute infection, abnormal pregnancy, DEGs
Introduction
Toxoplasmosis is a widespread zoonotic disease caused by the obligate intracellular protozoan parasite, Toxoplasma gondii (T. gondii). More than 30% of the human population has been exposed to T. gondii infections (Hill and Dubey, 2002). The main routes of T. gondii transmission are the oral and transplacental transmission However, T. gondii can be transmitted via blood transfusions and organ transplantation. T. gondii infections are usually asymptomatic but may cause serious complications, such as severe fever in infants, and fatal encephalitis in immunocompromised individuals such as AIDS patients (Luft et al., 1983). Moreover, immunosuppressed T. gondii-infected women during pregnancy are susceptible to abortion or stillbirth. Even if the fetus survives, there is a high probability of vertical transmission with subsequently expected serious sequelaein the congenitally infected fetuses such as hydrocephaly, retinochoroiditis, mental retardation, and seizures (Saadatnia and Golkar, 2012).
Females infected with T. gondii during pregnancy are three times more likely to experience a problematic pregnancy (Xiao et al., 2010). Stahl et al. (1985) reported a reproductive failure, caused by the hypothalamus dysfunction, in T. gondii-infected mice. As well as, these mice showed atrophy of the uterus and ovary, low mature oocytes number and oocyte chromosome aneuploidy, which increase the incidence of infertility (Stahl et al., 1985). At present, the hypothesis about the mechanism of infertility in T. gondii-infected women includes endometritis, and expulsion of the fetus. This hypothesis suggests that, during the placental formation, T. gondii cysts can be formed in the endometrium inducing non-suppurative inflammation (Remington et al., 1958). While this protozoan is being released, it irritates the placenta and causes miscarriages (Remington et al., 1961). Another hypothesis suggests that persistent infection of T. gondii in the hypothalamus results in infertility with follicle damage, and uterine atrophy (Aral et al., 2011). Barbosa et al. (2012) has shown that enoxacin, which is used for endometritis treatment in animals, has also a role in inhibiting the proliferation of T. gondii.
Previous research has shown that the incidence of uterine toxoplasmosis in the early stage of pregnancy is less than 6% (Dunn et al., 1999). However, an acute infection in early pregnancy can lead to serious clinical symptoms in 61% of the infected cases (Montoya and Remington, 2008). The offspring of mice infected with T. gondii in early pregnancy had a 40% survival rate and more stillbirths (Freeman et al., 2005; Rorman et al., 2006). Mice with acute infection (72 h after intraperitoneal injection) revealed tachyzoites in almost all nucleated cells with abnormal cell morphology in the uterus and placenta (Chen et al., 2003). In experimental acute vaginal infection set-up with T. gondii RH strain, tachyzoites and DNA of T. gondii were detected through biopsy and PCR assays, respectively in the uterine mucosa, and myometrium as well as the placental chorion of mice (Bayat et al., 2013). Several studies have investigated the effect of acute T. gondii infection during pregnancy, however, it focused principally on the immune responses associated with T. gondii infection (Nishimura et al., 2017; Xu et al., 2017). To best of our knowledge, research on the evidence of T. gondii-associated key genes, which are involved in the adverse pregnancy and disturbance of the pregnancy, is very limited and seldom.
Transcriptome analysis has been a valuable approach for investigating the global biological changes, gene expressions, and the transcription regulations in different body tissues of mice infected with T. gondii. For instance, Tanaka et al. used RNA-Seq approach to study the transcriptomic gene expressions in the brain of T. gondii-infected mice (Tanaka et al., 2013). Earlier to that, Knight et al. (2005) utilized the transcriptomics to reveal the role of inflammation in T. gondii-associated retinochoroiditis. More recently, He et al. (2016b) analyzed the transcriptomic changes in the spleen of infected mice to elucidate the immune responses against T. gondii infection. Later, they evaluated the transcriptomic gene expressions in the liver of infected mice during the early stages of T. gondii infection (He et al., 2016a). However, data on the global transcriptomic changes in uterine tissues of mouse with acute T. gondii infection are highly deficient, despite of its crucial importance.
To elucidate the underlying molecular mechanisms of the adverse pregnancy outcomes associated with the acute T. gondii infection, we utilized transcriptome sequencing technology (RNA-Seq) coupled with Bioinformatics approaches to analyze the transcriptomic gene expressions in the uterus of experimentally-infected mice with T. gondii RH strain during the acute stage of infection. Full understanding of these mechanisms will promote the development of effective therapeutics and/or vaccines to control and minimize the T. gondii infections.
Materials and Methods
Animals, Parasite Challenge and Sample Collection
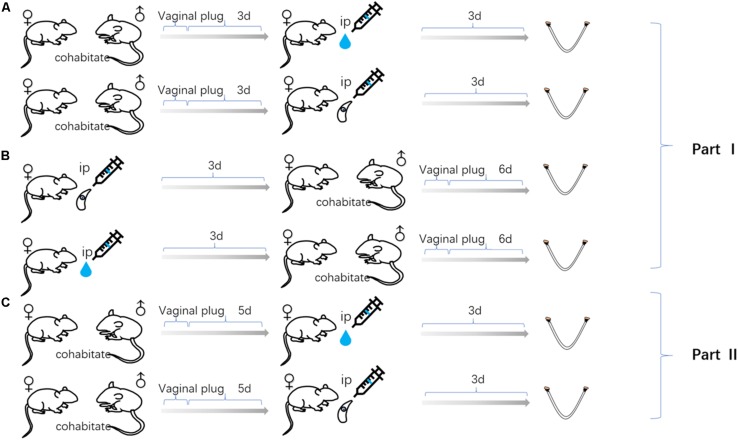
Specific-Pathogen Free (SPF) Kunming mice, purchased from Guangdong Medical Laboratory Animal Center (Guangdong, China), were subjected to carry out this study. Eight-week-old non-pregnant female Kunming mice were randomly divided into six equal groups (20 mice per group). Three groups of female mice were infected with tachyzoite, as follow; 3 days before pregnancy (FBD6; 3 days before cohabiting with male mice at a ratio of 2:1), 3 days after pregnancy (FAD6), and the last group, on the fifth day of pregnancy (FID8; 3 days after implantation). The vaginal plug, the indicator of pregnancy success, was carefully inspected to determine the start of pregnancy. Each mouse in the infected groups was intraperitoneally infected with 80 tachyzoites of T. gondii RH strain (Genotype I). Another three corresponding female mice groups, works as control groups, were injected with the normal saline at the same times as the former groups. All experiments were carried out in three replicates. All mice were humanely euthanized by CO2 asphyxiation on the sixth or eighth day of pregnancy for the groups of 3-days before and 3-days after pregnancy infected mice. Whereas, the third group, 3 days after implantation, were euthanized on the eighth day of pregnancy. Uterine tissues were quickly harvested. Each uterine sample was divided into three portions for the subsequent examinations including transcriptome sequencing, qPCR, and follow-up experiments, respectively. Afterward, the uterine samples were rapidly stored at −80°C until subsequent use. Animal experiments were applied in accordance to the standard feeding procedures and experimental procedures of South China Agricultural University. Figure 1 summarizes the time of infection and harvesting of uterine samples in all mice groups.
FIGURE 1.
Uterine samples were collected from three mouse models of gestation: (A) The female mice infected with Toxoplasma gondii tachyzoites 3 days after pregnancy. (B) The female mice infected with tachyzoite 3 days before pregnancy. (C) The female mice infected with tachyzoite on the fifth day of pregnancy. Mice injected with normal saline (control group) at the same time as every experimental group.
mRNA Library Preparation and Sequencing
Experimental methods of the library preparation and RNA-Seq were conducted according to the standard procedures of Illumina. The total RNA was extracted from the preserved uterine samples using RNAiso Plus Total RNA extraction reagent following the manufacturer’s protocol (Takara, Dalian, China). The RNA quality was initially evaluated with 1.0% agarose gel electrophoresis and ultraviolet spectrophotometer (Biotek EPOCH, United States). Following that, the integrity of RNA samples was confirmed by Agilent Bioanalyzer 2100 and RNA 6000 Nano Lab Chip Kit (Agilent Technologies, Santa Clara, CA, United States). RNA integrity number (RIN) over 7.0 was considered as the purity standard. Approximately, 10 μg of total RNA was utilized to isolate Poly (A) mRNA using poly-T oligo-attached magnetic beads (Invitrogen, United States). The product was purified, and the mRNA was segmented into small fragments utilizing divalent cations at elevated temperature. After that, the sliced RNA fragments were reverse-transcribed to establish the final cDNA library according to the recommendations of the mRNA-Seq sample preparation kit (Illumina, San Diego, CA, United States). The average interposer dimension for the paired-end libraries was 300 bp (±50 bp). Finally, the paired-end sequencing was performed by the Illumina Hiseq2000/2500 (LC Sciences, United States) according to the manufacturer’s recommendations.
Processing of RNA-Seq Data
After the transcriptome sequencing using the Illumina paired-end RNA-Seq methodology, the quality and integrity of the raw reads data were analyzed. The adaptors in the sequenced reads, which have a ratio of base N > 5% and nucleotide with Q quality score < 20 were removed. Eventually, clean sequence reads were mapped to the mouse reference genome using TopHat2 aligner and gene transfer format (gtf) annotations data (Mus_musculus.GRCm38.69). A statistical analysis of the three aspects of TopHat2 including the reads statistics, the outline of regional distribution, and the chromosome distribution density was performed. For normalization of transcription profiles, genetic variance analysis was conducted based on the mapping results using the “cuffdiff” command in the cufflinks (Trapnell et al., 2010). The FPKM (Fragments Per Kilobase of exon model per Million mapped reads) value was used to calculate the abundance of gene expression.
Identification and Bioinformatic Analysis of Differentially Expressed Genes
Differentially expressed genes (DEGs) were identified by the EBSeq package and DESeq package of R software (Anders and Huber, 2010). The P-values were calculated using the Benjamini and Hochberg method (Ferreira, 2007). Significant DEGs were identified as those with | log 2 FC (fold-change) | ≥ 1 and an adjusted P-value of < 0.05. To understand the biological functions of these genes, firstly; UniProt database was used to expand the biological functions of DEGs. Afterward, we mapped all DEGs to the Gene Ontology (GO) database1 and classified these genes into three categories; biological process, molecular function, and cellular component. The Kyoto Encyclopedia of Genes and Genomes (KEGG)2 was used to predict molecular function, biological processes and significant DEGs pathways. Finally, the database for Annotation, Visualization, and Integrated Discovery (DAVID) online tool3 was used to systematically judge the affected pathways of DEGs.
Validation of RNA-Seq Results by Quantitative Real-Time PCR (qRT-PCR) Analysis
Gene expression data were verified by qRT-PCR. The total RNA was extracted from the remaining uterine specimen using the RNAiso Plus (Takara, Dalian, China) following the manufacturer’s protocols. The obtained RNA was resuspended in RNase-free water. The purity and concentration of total RNA were determined using ultramicrospectrophotometer (Biotek EPOCH, United States). Ten DEGs were selected to verify the gene expression data by qRT-PCR. Gene-specific qRT-PCR primers were designed, based on reference UniGene sequences, with Premier 5.0 software (Premier Biosoft International, Palo Alto, CA, United States). The genes and primers used in qRT-PCR are shown in Table 1. The first strand cDNA was synthesized by reverse transcription of RNA using RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, United States). Quantitative PCR was performed on a Rotor-Gene Q (Qiagen, Hilden, Germany) using AceQ qPCR SYBR® Green Master Mix (Vazyme, Nanjing, China) according to the manufacturer’s instructions. The thermal cycling program included 95°C for 5 min, 40 cycles of 95°C for 10 min, and 30 s of 60°C, followed 95°C for 15 s, 60°C for 1 min, and 95°C for 15 s as a final step. Each sample was examined in triplicate. Relative quantification of specific mRNA levels was performed using the cycle threshold (Ct) 2–ΔΔCt method (Livak and Schmittgen, 2001). For normalization of gene expression, β-actin was used as an internal standard of mRNA expression, and the blank control was used as a reference sample, which was set to a value 1.
TABLE 1.
Primer design table for qRT-PCR analysis.
| Gene | Description | Forward primer (5′–3′) | Reverse primer (5′–3′) | |
| 1 | Hsd11b2 | ENSMUSG00000031891 | ATAGCCCTGGTGCCCTAGAA | TCCAGAACACGGCTGATGTC |
| 2 | Slc39a4 | ENSMUSG00000063354 | CTCTGGATCGCCTGGAACTG | CTGTGGCTTGTCAGGTTTGC |
| 3 | Ifitm1 | ENSMUSG00000025491 | ATGGTGGGTGATACGACTGG | GGCACAGACAACGATGACGA |
| 4 | Krt8 | ENSMUSG00000049382 | TATGGGGGACTCACTAGCCC | CAGCTTCCCATCTCGGGTTT |
| 5 | Cd24a | ENSMUSG00000047139 | CCCACGCAGATTTACTGCAAC | CTGGTGGTAGCGTTACTTGG |
| 6 | Cndp2 | ENSMUSG00000024644 | GTGCCAGACATGATACCGGA | CCCGCCTGGTAATGAGGATG |
| 7 | Hsd17b7 | ENSMUSG00000026675 | GCGTGTAGGAACCTGAGCAA | GCTTGACTTCCTCTGCACCC |
| 8 | A2m | ENSMUSG00000030111 | AATATCGGTCTTCCGGCCAC | TGTGAGCAGTACGCTTTGGT |
| 9 | Stc1 | ENSMUSG00000014813 | ATAGGAGGCGCACAAATGAG | GGGAGGTGCGTTTGATGTGT |
| 10 | Hmgn5 | ENSMUSG00000031245 | CGACTGTCTGCTATGCCTGT | TGGCTTGGTTTCAGGAATTGG |
| 11 | Actb | ENSMUSG00000029580 | AAAGAGAAGCTGTGCTATGTTGCT | GCCTCAGGGCATCGGAAC |
Validations of RNA-Seq Results by Western Blotting
Western Blot was used to validate RNA-Seq results at the molecular level, by targeting the protein of two DEGs, Corticosteroid 11-beta-dehydrogenase isozyme 2 (Hsd11b2) and Interferon-induced transmembrane protein 1 (Ifitm1). Firstly, the pre-processed protein samples were run on a 5% stock gel at 80 V with 30 min, separated on a 12% SDS-PAGE in 60 min at 120 V, and then transferred into a nitrocellulose filter membrane (Millipore, Darmstadt, Germany) for 90 min at 150 mA. The membrane was blocked in 0.1% Tween 20-PBS containing 5% skimmed milk powder for 2 h and then incubated with β-actin mouse monoclonal antibodies (1:500 dilution, Neobioscience, Shenzhen, China), specific mouse anti-Hsd11b2 (1:500, Abcam, Cambridge, MA, United States), and anti-Ifitm1 (1:500, Abcam, Cambridge, MA, United States) antibodies overnight. The membranes were washed three times and probed with goat anti-mouse IgG conjugated with horseradish peroxidase (HRP) (Tiangen Biotech, Beijing, China) at 1:2500 dilution. Finally, the membranes were visualized with BeyoECL Plus (Beyotime, Shanghai, China) and the image was taken and analyzed by the Tanon5200 western blotting detection system (Tanon, Shanghai, China).
Results
Identification of Expressed Transcripts in the Mouse Uterus Transcriptome
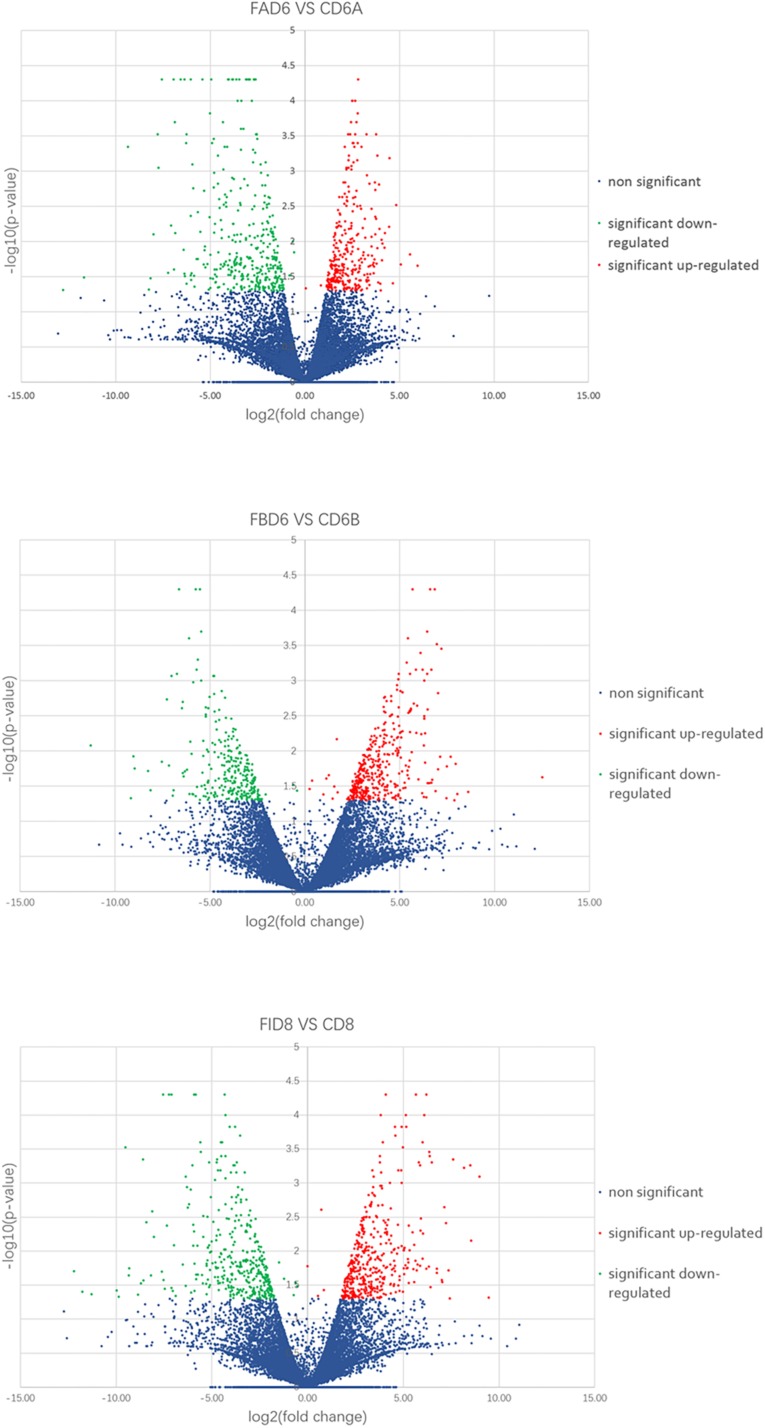
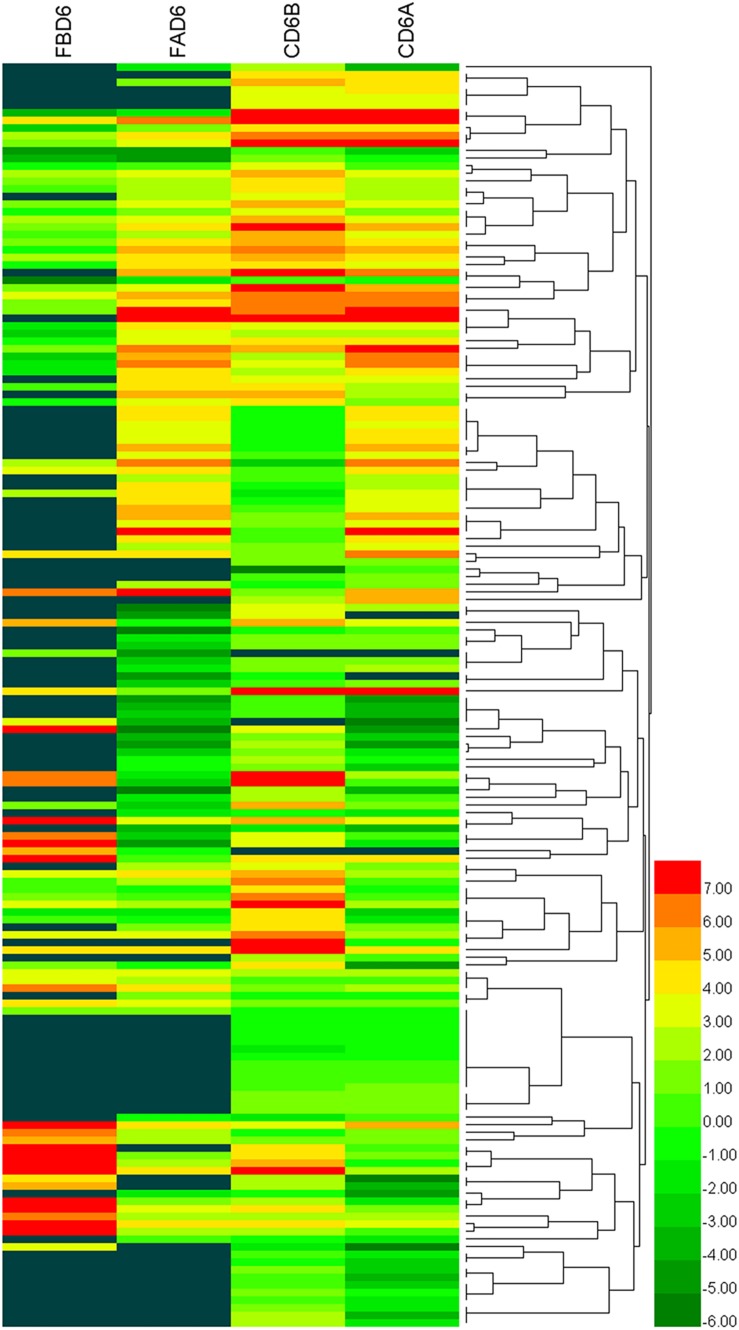
We established six different groups of mice, three experimental and three control groups, including mice infected with T. gondii 3 days after groups day of pregnancy (FAD6), mice infected with T. gondii 3 days before pregnancy (FBD6), mice infected with T. gondii on the fifth day of pregnancy (FID8), mice injected with normal saline (NS) 3 days after pregnancy (CD6A), mice injected with NS 3 days before pregnancy (CD6B), and lastly, mice injected with NS on the fifth day of pregnancy (CD8). Each group was tested in triplicates. The total RNA harvested from the uterus of all mice groups was subjected to high-throughput sequencing on Illumina Hiseq2000/2500. The number of the obtained raw reads was ranged from 56,804,874 (CD6B1 group) to 66,241,238 (FAD63 group). After filtration, the average number of clean reads of each mouse group was over 48,000,000, where 82.32–98.90% could be aligned to the reference mouse genome (Table 2). The sequence data of each group had more than 6G and Q20 > 99%. Therefore, the requirements of subsequent analysis were fulfilled. A filter criteria set at | log2 fold change| > 1 and P-value ≤ 0.05 to determine whether the difference was significant or not (shown in Supplementary Data Sheets 1, 2). Our findings showed that the FBD6 group produced a total of 4,561 DEGs compared to the control group B (CD6B). Out of them, 2,571 and 1,990 genes were identified as upregulated and downregulated, respectively. A total of 2,345 genes showed significant expression differences between the FAD6 group and its corresponding NS-treated group (CD6A). Out of this number, 1,042 and 1,303 genes were identified as upregulated and downregulated genes, respectively. As f for the third and last groupFID8 and its corresponding control group (CD8); 2,997 genes showed significantly different expression, where 1,162 and 1,835 of them were identified as upregulated and downregulated, respectively. The details of the top 10 DEGs in T. gondii-infected mouse uterine samples are shown in Table 3. Volcanic maps showed the distribution of gene expression differences between the experimental and control groups. The maps highlight the genes with an absolute fold change of ≥1 and an adjusted p-value of <0.05 after infection (Figure 2). Figure 3 shows a hierarchical clustering analysis of genes expressed in the mouse uterus infected with T. gondii in the various groups.
TABLE 2.
Summary of clean reads mapped to the reference genome of mouse.
| Sample | Raw date read | Base | Valid date read | Base | Valid% read | Q20% | Q30% | GC% |
| CD6A-1 | 58748442 | 8.81G | 48364460 | 7.25G | 82.32 | 99.20 | 96.44 | 49.00 |
| CD6A-2 | 59452796 | 8.92G | 48959376 | 7.34G | 82.35 | 99.52 | 97.21 | 51.00 |
| CD6A-3 | 58563520 | 8.79G | 48696972 | 7.30G | 83.15 | 99.13 | 95.98 | 49.00 |
| FBD6-1 | 59970362 | 9.00G | 57652548 | 8.64G | 96.14 | 99.39 | 97.09 | 50.00 |
| FBD6-2 | 57907066 | 8.69G | 56827140 | 8.52G | 98.14 | 99.57 | 95.75 | 50.00 |
| FBD6-3 | 56905122 | 8.54G | 54152732 | 8.12G | 95.16 | 99.22 | 96.17 | 49.00 |
| FAD6-1 | 65608270 | 9.84G | 57597380 | 8.64G | 87.79 | 99.55 | 96.49 | 50.00 |
| FAD6-2 | 64061528 | 9.65G | 55865012 | 8.38G | 87.21 | 99.15 | 95.86 | 51.00 |
| FAD6-3 | 66241238 | 9.94G | 58153066 | 8.72G | 87.79 | 99.63 | 97.94 | 50.00 |
| CD6B-1 | 56804874 | 8.52G | 49590640 | 7.44G | 87.30 | 99.66 | 97.93 | 49.00 |
| CD6B-2 | 57290458 | 8.59G | 50295274 | 7.54G | 87.79 | 99.38 | 96.92 | 49.00 |
| CD6B-3 | 57516602 | 9.63G | 50243946 | 7.54G | 87.36 | 99.62 | 97.31 | 50.00 |
| CD8-1 | 51722480 | 7.76G | 51153928 | 7.67G | 98.90 | 99.68 | 96.19 | 50.00 |
| CD8-2 | 54269660 | 8.14G | 53547754 | 8.03G | 98.67 | 99.69 | 97.05 | 49.50 |
| CD8-3 | 48768472 | 7.32G | 48078448 | 7.21G | 98.59 | 99.69 | 96.90 | 49.00 |
| FID8-1 | 44641116 | 6.70G | 43937778 | 6.59G | 98.42 | 99.69 | 96.97 | 50.00 |
| FID8-2 | 53513324 | 8.03G | 52647430 | 7.90G | 98.38 | 99.65 | 96.94 | 50.00 |
| FID8-3 | 49853240 | 7.48G | 48944858 | 7.34G | 98.18 | 99.75 | 97.42 | 49.50 |
TABLE 3.
Partial list of differentially expressed genes in T. gondii-infected mice uterus.
| Gene symbol | Description |
Log2 (fold-change) |
Differential Expression | |||
| FAD6/CD6A | FBD6/CD6B | FID8/CD8 | ||||
| 1 | Hsd11b2 | Hydroxysteroid 11-Beta Dehydrogenase 2 | 2.40061 | 3.47496 | 2.71847 | Up |
| 2 | Slc39a4 | Solute Carrier Family 39 (Zinc Transporter), Member 4 | 2.72085 | 4.63169 | 1.28504 | Up |
| 3 | Ifitm1 | Interferon-induced transmembrane protein 1 | 1.75738 | 3.59516 | -0.23735 | Up (Down in FID8/CD8) |
| 4 | Krt8 | Keratin, type II cytoskeletal 8 | 1.01272 | 3.09309 | 3.46933 | Up |
| 5 | Cd24a | Signal transducer CD24 | 1.29161 | 1.19154 | 3.07802 | Up |
| 6 | Cndp2 | Cytosolic non-specific dipeptidase | 1.33972 | 3.14075 | 2.66401 | Up |
| 7 | Hsd17b7 | 3-keto-steroid reductase | −5.40267 | −6.46531 | −4.46804 | Down |
| 8 | A2m | Alpha-2-macroglobulin-P | −3.80101 | −5.8787 | −5.92206 | Down |
| 9 | Stc1 | Stanniocalcin-1 | −2.66361 | −3.95738 | −2.45878 | Down |
| 10 | Hmgn5 | High mobility group nucleosome-binding domain-containing protein 5 | −1.26749 | −3.44105 | −2.30626 | Down |
FIGURE 2.
Volcano maps of differentially expressed genes in the uterus of three infected mice groups compared to control groups. Significantly upregulated and downregulated genes are shown as a red and green dot, respectively. The blue dot represents genes without a significant difference in the expression level.
FIGURE 3.
Heat maps representing hierarchical clustering analysis of differentially expressed genes in the uterus of mice infected with T. gondii tachyzoites 3 days before pregnancy (FBD6) and 3 days after pregnancy (FAD6). Each row refers to the expression level of each gene in the various groups, and each column refers to the expression level of all DEGs in each group.
GO Enrichment of Differentially Expressed Genes
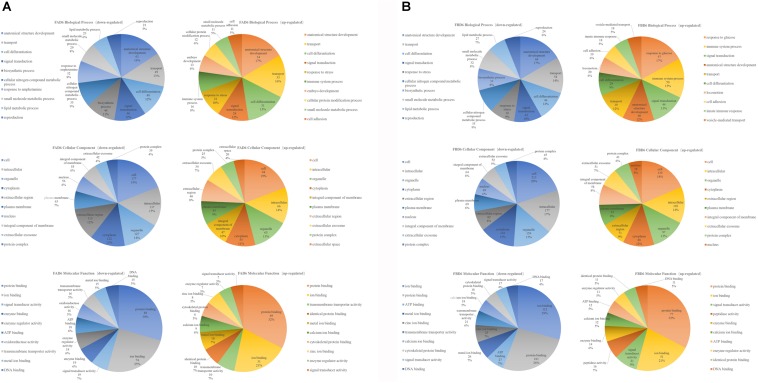
The top 10 enriched GO terms under each category in FAD6 and FBD6 groups are shown in Figure 4. In comparison to the control group, the upregulated genes in the FAD6 were annotated in 1,047 GO terms, which were mainly related to biological processes (such as anatomical structure development, transport, cell differentiation, response to stress), cellular components (such as cellular, intracellular, organelle and cytoplasmic component), and molecular functions (such as protein binding, ion binding, transmembrane transporter activity, and cytoskeletal protein binding). The number of GO terms of downregulated genes were 1,622. The most common identified biological process were lipid, small molecule, and cellular nitrogen compound metabolic processes, anatomical structure development, transport and response to amphetamine. The downregulated GO terms categorized under cellular component were similar to the functional classification of the upregulated genes. As for the molecular function, most of the downregulated genes were related to protein and enzyme activity besides binding to protein and ions. In general, there were 21 significant DEGs involved in embryonic development and one gene associated with embryo implantation in the FAD6 group (Figure 4A).
FIGURE 4.
Gene Ontology (GO) distribution of differentially expressed genes of the uterus transcriptome. The top 10 enriched GO terms under biological process, cellular component and molecular function in (A) mice infected with Toxoplasma gondii 3 days after pregnancy (FAD6) and (B) mice infected with T. gondii 3 days before pregnancy (FBD6).
Comparing FBD6 group to the control group, the upregulated genes involved 1,311 GO terms, whereas the downregulated genes were related to 1,744 GO terms. As for the biological process GO terms, the upregulated genes were mainly involved in glucose, immune system process, and signal transduction. Meanwhile, the downregulated genes were mostly related to anatomical structure development, transport, and cell differentiation. In the term of cellular component, the traits of FBD6 vs. CD6B groups were almost similar to those of FAD6 vs. CD6A groups. In respect to the molecular function, the DEGs showed mainly functions related to protein, ion and ATP binding (Figure 4B).
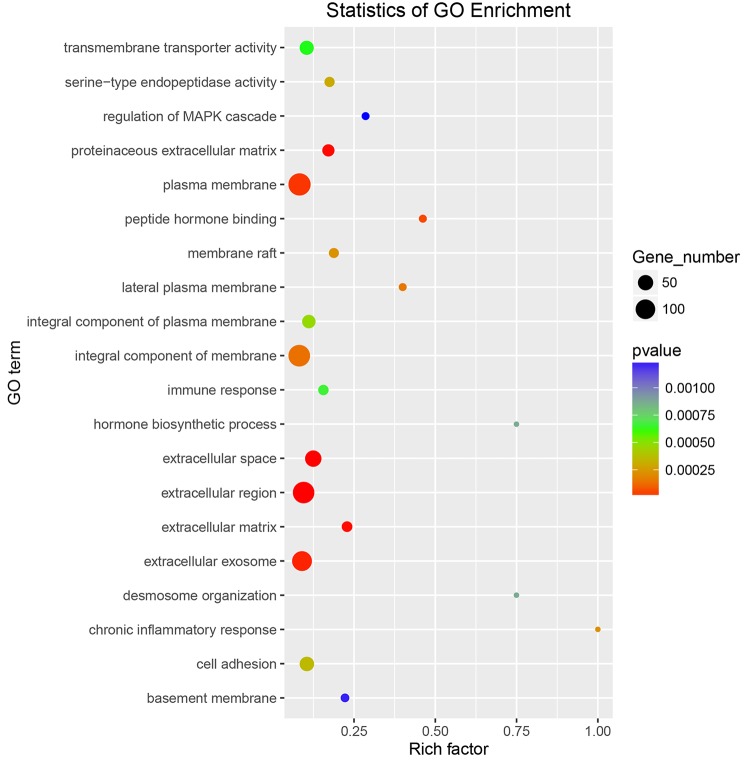
To explore whether the key genes related to embryo implantation and development are affected by inducing the experimental infection in the pregnant mice with T. gondii after implantation or not, we performed GO function classification of the DEGs in FID8 group using the scatter plot (Figure 5). As for the cellular component, DEGs were mostly associated with the plasma membrane, the integral component of the membrane, and extracellular region. Most DEGs were involved in peptide hormone binding, transmembrane transporter activity, and serine-type endopeptidase activity molecular functions. Moreover, DEGs were related to the central biological processes such as hormone biosynthetic process and immune response.
FIGURE 5.
Gene Ontology enrichment scatter plot for differentially expressed genes in the uterus of mice infected with T. gondii on the fifth day of pregnancy (FID8).
KEGG Enrichment Analysis
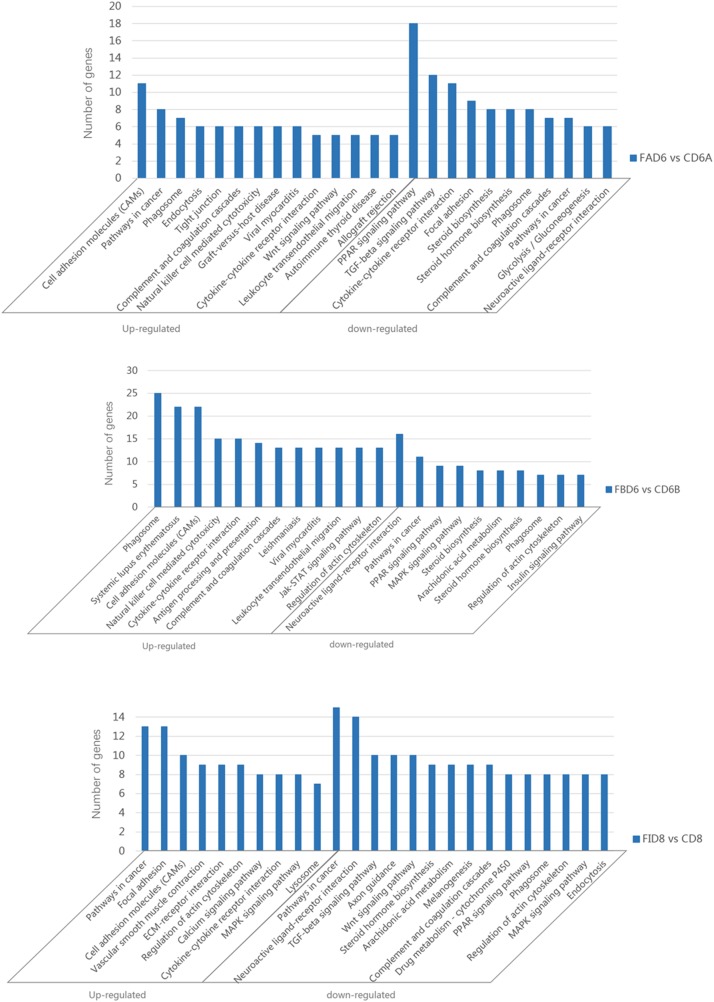
Enrichment analysis of the DEGs in the uteri of the infected versus the control groups’ pairs was performed by using the KEGG database, where the pathways that play an important role in the infection process can be furtherly explored. Of the 166 identified enriched pathways in the group FAD6 vs. CD6A, upregulated DEGs enriched in 107 pathways, and 136 pathways were enriched by downregulated DEGs. The top five enriched upregulated pathways were cell adhesion molecules (CAMs), pathways in cancer, phagosome, endocytosis, and tight junction. The top five identified enriched downregulated pathways were PPAR signaling pathway, TGF-beta signaling pathway, cytokine-cytokine receptor interaction, focal adhesion, and steroid biosynthesis (Figure 6A).
FIGURE 6.
Significantly enriched pathways in the uterus of T. gondii-infected mice. [A (Upper), B (Medium), and C (Lower)] represent KEGG pathway analysis of DEGs for FAD6 vs. CD6A, FBD6 vs. CD6B and FID8 vs. CD8 group, respectively.
Of the 175 identified enriched pathways in the FBD6 vs. CD6B group, 128 and 149 pathways were enriched with upregulated and downregulated DEGs, respectively. The major five enriched upregulated pathways were phagosome, systemic lupus erythematosus, cell adhesion molecules, cytokine-cytokine receptor interaction. The major five enriched downregulated pathways involved neuroactive ligand-receptor interaction, pathways in cancer, PPAR signaling pathway, MAPK signaling pathway, and steroid biosynthesis (Figure 6B).
In the FID8 vs. CD8 group, 169 pathways were enriched, including 134 upregulated DEGs enrichment pathways and 145 downregulated DEGs enrichment pathways. The top five enriched upregulated pathways were involving pathways related to cancer, focal adhesion, cell adhesion molecules (CAMs), vascular smooth muscle contraction, and ECM-receptor interactions. However, the top five enriched downregulated pathways were pathways related to cancer, neuroactive ligand-receptor interaction, TGF-beta signaling pathway, axon guidance, and Wnt signaling pathway (Figure 6C).
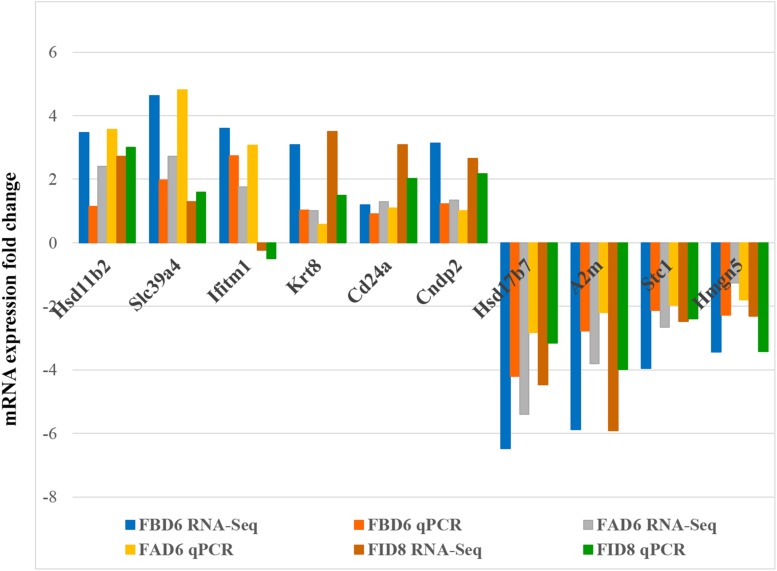
Validation of Ten Differentially Expressed Genes by qRT-PCR
In order to verify the reliability of RNA-Seq results, we selected 10 DEGs with different expression trends for qRT-PCR. Six genes, Hsd11b2, Slc39a4, Ifitm1, Krt8, Cd24a, and Cndp2 were upregulated, whereas Hsd17b7, A2m, Stc1, and Hmgn5 were downregulated. These results were consistent with the results of RNA-Seq (Figure 7).
FIGURE 7.
Validation of the relative abundance of 10 DEGs by q-PCR. The x-axis represents different DEGs in the uterus from different infected mouse groups and the y-axis represents the fold change in mRNA expression.
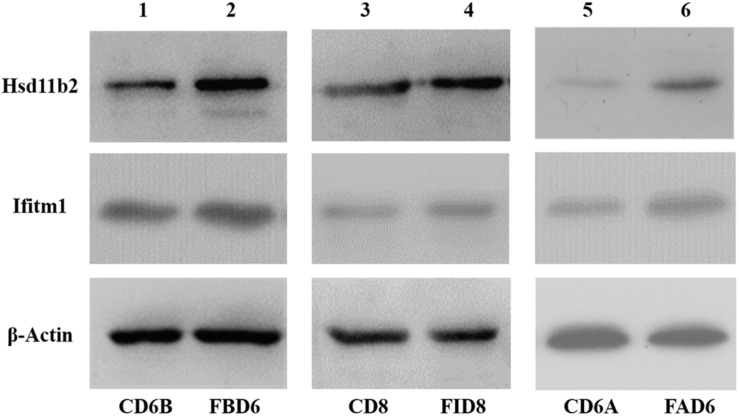
Verification of Hsd11b2 and Ifitm1 Expression Levels by Western Blotting
After analyzing the preliminary data and searching relevant literature, Hsd11b2 (UniprotKB: P51661) and Ifitm1 (UniprotKB: Q9D103) were selected as the research targets to explore their role in T. gondii infection. Therefore, we verified the expression changes of its protein level by western blot (Figure 8). The results were consistent with the sequencing results of the transcriptome, where these two proteins were overexpressed.
FIGURE 8.
Validation of expression level of Hsd11b2 and Ifitm1 in all three experimental groups by Western blot analysis.
Discussion
In this study, all the three experimental groups showed pregnancy failure confirming the adverse effect of T. gondii infection on pregnancy. Thus, we investigated the causes of T. gondii-induced pregnancy failure by analyzing the transcriptomic changes in the uterus of mice with acute T. gondii infection. A total of 4,561, 2,345, and 2,997 DEGs were identified in FBD6, FAD6, and FID8 groups compared to the corresponding control groups. Validation of our RNA-Seq results by qRT-PCR and Western blotting confirmed the differential expression, indicating that the RNA-seq results were reliable and subsequent further experiments and analyses can be conducted.
The mechanism of poor pregnancy outcome, associated with T. gondii infection, can be discussed from two aspects, the DEGs, and the enrichment pathways. In respect to DEGs, Hsd11b2 gene was overexpressed in infected groups compared to controls. The Hsd11b2 gene is a marker for pregnancy hypertension (Bicho et al., 2012; Majchrzak-Celinska et al., 2017). When the Hsd11b2 gene is overexpressed, it can cause placental dysfunction, especially in early-onset preeclampsia. On top of that, it might affect the embryonic development, and cause even placental abruption (Majchrzak-Celinska et al., 2017). Therefore, pregnant women who are infected with T. gondii might have pregnancy hypertension and experience pregnancy failure. A gene named Ifitm1 was also upregulated in the uterus of infected mice. This gene is associated with endometrial tumor when it is overexpressed, thus it is a marker for endometrial tumor. Rojas et al. (2012) demonstrated that endometrial tumor can cause pregnancy failure. Kuang et al. (2009) concluded that after embryo implantation, the chemokine (C-X-C motif) ligand 14 (CXCL14) gene can promote the growth and development of trophoblast. However, CXCL14 gene was significantly downregulated in our transcriptome analysis, which may explain the adverse pregnancy outcome associated with T. gondii infection, but perhaps require further experimental confirmation on protein level.
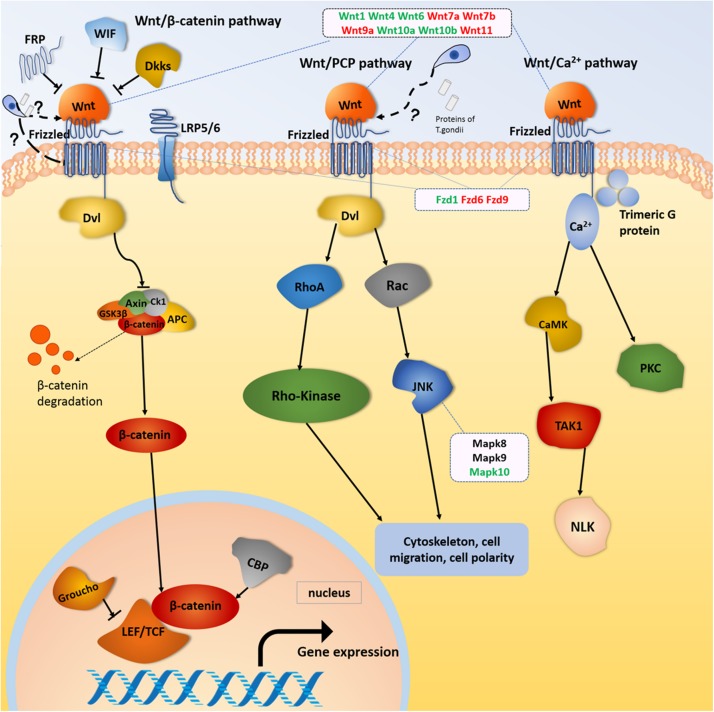
The mostly enriched DEGs and pathways in the uterus of the three experimental groups were identified and the signal pathways of pre-implantation and post-implantation were separately analyzed. Our results revealed that the number of DEGs in the cancer pathways was ranked in the top position of all the enrichment pathways. The cancer pathways included five DEGs (Wnt, Frizzled, CBL, JNK, and PPFP), which were shared by the three experimental groups and involved in the three related pathways, which are the wingless-type MMTV integration site family (Wnt) signaling pathway, the phosphatidylinositol 3-kinase-protein Kinase B (PI3K-Akt) signaling pathway, and the peroxisome proliferator-activated receptor (PPAR) signaling pathway. Among them, the DEGs related to the Wnt signaling pathway were the most abundant. The proteins expressed by the Wnt family genes are secreting morphogens, which play an important role in the growth and development of embryos. Such proteins and its products participate in the tissues and organs morphogenesis and the embryonic development and cell division (Moon et al., 2004). The Wnt signaling pathway includes three major transduction pathways (Figure 9), which regulate the growth and development of embryos. These three major transduction pathways are the Wnt/β-catenin classical pathway, the c-Jun NH2-terminal kinases (JNK) gene-mediated and participating planar cell polarity (PCP) pathway, and the Ca-mediated Wnt/Ca2+ pathways (Xing et al., 2003). In mice, the Wnt/β-catenin signaling pathway is essential for hormone-mediated uterine growth and implantation (Hou et al., 2004; Mohamed et al., 2005). This is agreement with previous research (Chen et al., 2009), which confirmed the important role of Wnt/β-catenin pathway in embryonic development, pre/post-implantation, and decidualization of the uterus.
FIGURE 9.
Wnt signaling pathway. Red and green color for genes indicate the upregulation and downregulation, respectively.
Our results showed that Wnt gene expression was altered during embryonic development in female mice infected with T. gondii. Wnt gene is not only related to the estrous cycle and steroid hormone levels, but also closely related to the morphological changes regulated by intrauterine epithelium (Park et al., 2011). The Wnt1 gene plays an important role in the development of the embryonic brain and central nervous system (Liu et al., 2005). In our study, the expression of Wnt1 genes in FBD6 and FID8 were inhibited. We believe that inhibition of the Wnt1 gene expression in early pregnancy by T. gondii infection had an adverse effect on embryonic development resulting in pregnancy failure. Wnt4 is important for the development of the female reproductive system, and it can be expressed in human trophoblast cells and the uterus of pregnant mice to regulate blastocyst implantation (Tranguch et al., 2005). Knocking out Wnt4 leads to embryo implantation and decidual abnormalities (Franco et al., 2011). Wnt4 expression in endometrial cancer is lower than that in normal tissues (Bui et al., 1997). In our study, Wnt4 was downregulated in the experimental groups with acute T. gondii infection. We hypothesize that acute T. gondii infection can affect the decidualization reaction and destroy the endometrium environment. Thus, it will hinder the implantation of embryos through reducing the expression of Wnt4. Wnt7a plays many roles, for instance, it stimulates embryo-implantation-associated responses, upregulates β-catenin in the nucleus of uterine epithelial cells, and activates the Wnt/β-catenin signaling pathway as well as improves embryo implantation success rates (Zhang et al., 2009). Wnt7a is also a tumor suppressor gene that inhibits cell proliferation and promotes apoptosis (Li et al., 2013; Bikkavilli et al., 2015). The mutation of Wnt7b can cause placental dysplasia and fetal death in mice (Biechele et al., 2011). In this study, the Wnt7a and Wnt7b gene were upregulated in all experimental groups compared to uninfected pregnant mice. Therefore, we hypothesize that the proliferation effect induced by T. gondii looks like the effect of cancer cells. In other words, acute T. gondii infection increases the expression of Wnt7a, which in turn, hinders the embryonic cell multiplication, and stimulates the apoptosis process. We think this fact beside other factors could explain the underlying mechanism of adverse pregnancy outcomes associated with acute T. gondii infection.
Frizzleds (seven-transmembrane proteins) act as receptors for the Wnt proteins and are involved in many functions such as regulation of embryonic development, cell differentiation, proliferation, apoptosis, and inflammation. The Frizzled gene family “Fzd9, Fzd6, and Fzd1genes,” were differentially expressed in the uterus of our experimental groups (FBD6, FAD6, and FID8). The former studies have shown that Fzd1 and Wnt7a combine to activate the Wnt classical pathway and play a key role in embryonic lung development. Fzd6 is expressed in endometrial epithelium and stromal cells, but it cannot be regulated directly by ovarian steroid hormones. Thus, the expression of Fzd6 is constant during the secretory and proliferative phases (Tulac et al., 2003). Therefore, we suggest that the differential expression of Frizzled genes in our study is caused by the acute infection of T. gondii. This finding was also consistent with the claim that the Wnt/Frizzled signaling pathway is involved in the inflammatory process (Neumann et al., 2010). We hypothesize that the Wnt genes expression is strongly influenced by T. gondii acute infection, which result in activation of the classical and non-canonical pathways of Wnt signaling pathway, and eventually altering the early embryonic development, the endometrial formation, and decidualization.
On the other hand, the analysis of the signaling pathways in mice infected with T. gondii after embryo implantation (FID8 group) showed that the infection had a marked influence on the vascular endothelial growth factor (VEGF) signaling and Wnt signaling pathways. In normal pregnancy, David (2017) demonstrated that placenta angiogenesis, and vascular expansion are enhanced by placental growth factor (PIGF), VEGF and insulin growth factor (IGF). However, VEGF was downregulated in our T. gondii-infected groups, indicating that it is responsible for the blockage of angiogenesis in placenta and even, miscarriage. As for the Wnt signaling pathway, we noticed that infection with T. gondii decreased the expression of glycogen synthase kinase beta (GSK-β), and β-catenin, which in turn, affect the neural development of the embryo. Wang et al. (2017) treated a group of mice with celecoxib and they found that the expression of GSK-β was changed, and β-catenin was decreased, which resulted in dysplasia of the fetus’ nerve cells. We propose that the reasons for pregnancy failure can be argued by many factors, which take place after the embryo implantation. In this regard, we think T. gondii infection can damage the placental tissues and impair the normal development of the embryo.
Concerning the time before the embryo implantation, we found that DEGs were mostly related to cytokines pathway. Transforming growth factor beta (TGF-β) is highly expressed in the normal endometrium epithelium and trophoblastic layer. It plays an important role several process such as endometrial inflammation, maternal support for embryonic development, immune-regulating maternal-fetal interface, and maintaining normal pregnancy. In this study, TGF gene family and the cytokines pathway were downregulated in the infected groups compared to the control. This is consistent with previous studies, which reported that downregulation of TGF could cause abortion or fetal death (Pazmany and Tomasi, 2006; Norris et al., 2011). Therefore, we conclude that infection with T. gondii affects the development and implantation of the embryo.
KEGG pathway analysis of DEGs showed upregulation of pathways related to diseases in infected mice such as systemic lupus erythematosus, leishmaniasis, viral myocarditis, and autoimmune thyroid disease. This finding indicates that the abortion caused by T. gondii infection during pregnancy is complicated, and may not be exclusively caused by Toxoplasma infection, where other diseases may be involved as a secondary sequelae of T. gondii infections. Previous research showed a close relationship between T. gondii infection and the occurrence of systemic lupus erythematosus in 1990 (Wilcox et al., 1990) and rheumatoid arthritis (Fischer et al., 2013).
Conclusion
The adverse pregnancy caused by acute toxoplasmosis can be attributed to three factors: (i) direct injury to tissues and cells due to T. gondii proliferation, (ii) disruption of the early maternal uterine environment, which impair the normal embryo implantation, and (iii) poor development of embryos before and after the implantation. Our finding indicates that acute T. gondii infection altered the Wnt genes expression and signaling pathway, leading to the failure of early embryonic development, endometrial formation, and decidualization. Acute T. gondii infection is disturbing the normal implantation of the embryo. Acute infection of T. gondii is associated with gestational hypertension, systemic lupus erythematosus, and possibly endometrial tumor resulting in adverse effects on the pregnancy outcome. Our findings uncover the role of the acute T. gondii infection and associated mechanisms in the pregnancy failure which could promote the development of a new treatment for toxoplasmosis in pregnancy.
Data Availability Statement
The sequencing data generated in this study has been deposited into BioProject (accession no. PRJNA593831, https://www.ncbi.nlm.nih.gov/bioproject/PRJNA593831).
Ethics Statement
Experimental animals were handled according to the Animal Ethics Procedures and Guidelines of the People’s Republic of China. All animal experimental procedures were reviewed and approved by the Experimental Animal Welfare Ethics Committee, the Laboratory Animal Center of Guangdong Province (Reference No. 2016-10).
Author Contributions
X-ML and Z-GY conceived and designed the experiments. XZ performed the experiments, analyzed the data, and wrote the manuscript. X-XZ, JH, and YM revised the manuscript. XZ, G-FL, W-YH, Y-PW, and Y-XZ contributed to data collection and laboratory work. All authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Thank for Zhanhong Zheng’s experimental technical support. We also thanks for the student Junjie Wang to submit the data to SRA database.
Abbreviations
- CD6A
mice injected with normal saline (NS) 3 days after pregnancy
- CD6B
mice injected with NS 3 days before pregnancy
- CD8
mice injected with NS in the fifth day of pregnancy
- DEGs
differentially expressed genes
- FAD6
mice infected with T. gondii 3 days after pregnancy
- FBD6
mice infected with T. gondii 3 days before pregnancy
- FID8
mice infected with T. gondii on the fifth day of pregnancy.
Funding. This work was supported, in part, by grants from the National Key Research and Development Programme of China (2018YFD0501200), National Natural Science Foundation of China (31972707), the Key Research and Development Programme of Guangdong Province (2019B020218004), the Natural Science Foundation of Guangdong Province (2019A1515011534), and Major Scientific Research Projects (Characteristic Innovation) of Guangdong Province (2017KTSCX018).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2020.00115/full#supplementary-material
References
- Anders S., Huber W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11:R106. 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aral A. G., Elhan H. A., Akarsu C. (2011). Retrospective evaluation of Toxoplasma gondii seropositivity in fertile and infertile women. Mikrobiyol. Bull. 45 174–180. [PubMed] [Google Scholar]
- Barbosa B. F., Gomes A. O., Ferro E. A., Napolitano D. R., Mineo J. R., Silva N. M. (2012). Enrofloxacin is able to control Toxoplasma gondii infection in both in vitro and in vivo experimental models. Vet. Parasitol. 187 44–52. 10.1016/j.vetpar.2011.12.039 [DOI] [PubMed] [Google Scholar]
- Bayat P. D., Eslamirad Z., Shojaee S. (2013). Toxoplasmosis: experimental vaginal infection in NMRI mice and its effect on uterine, placenta and fetus tissues. Iran Red Crescent. Med. J. 15 595–599. 10.5812/ircmj.11427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bicho M. D., Matos A., Silva A. P., Macedo C., Afonso C., Areias M. J., et al. (2012). PP159. In women with previous pregnancy hypertension, cardiovascular risk biomarkers may be modulated by haptoglobin genotype. Preg. Hypertens. 2:325. 10.1016/j.preghy.2012.04.270 [DOI] [PubMed] [Google Scholar]
- Biechele S., Cox B. J., Rossant J. (2011). Porcupine homolog is required for canonical Wnt signaling and gastrulation in mouse embryos. Dev. Biol. 355 275–285. 10.1016/j.ydbio.2011.04.029 [DOI] [PubMed] [Google Scholar]
- Bikkavilli R. K., Avasarala S., Van Scoyk M., Arcaroli J., Brzezinski C., Zhang W., et al. (2015). Wnt7a is a novel inducer of beta-catenin-independent tumor-suppressive cellular senescence in lung cancer. Oncogene 34:5406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui T. D., Zhang L., Rees M. C., Bicknell R., Harris A. L. (1997). Expression and hormone regulation of Wnt2, 3, 4, 5a, 7a, 7b and 10b in normal human endometrium and endometrial carcinoma. Br. J. Cancer. 75 1131–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B., Wang H., Wang L., Lu Q., Zhou L., Cheng B., et al. (2003). Studies on the Effect of IFN-γ on Toxoplasma gondii-infected pregnancy mice in ultramicrostructure of uterus and placenta. Jiang. Pharm. Clin. Res. 11 3–6. [Google Scholar]
- Chen Q., Zhang Y., Lu J., Wang Q., Wang S., Cao Y., et al. (2009). Embryo-uterine cross-talk during implantation: the role of Wnt signaling. Mol. Hum. Reprod. 15 215–221. 10.1093/molehr/gap009 [DOI] [PubMed] [Google Scholar]
- David A. L. (2017). Maternal uterine artery VEGF gene therapy for treatment of intrauterine growth restriction. Placenta 59(Suppl. 1), S44–S50. 10.1016/j.placenta.2017.09.011 [DOI] [PubMed] [Google Scholar]
- Dunn D., Wallon M., Peyron F., Petersen E., Peckham C., Gilbert R. (1999). Mother-to-child transmission of toxoplasmosis: risk estimates for clinical counselling. Lancet 353 1829–1833. [DOI] [PubMed] [Google Scholar]
- Ferreira J. A. (2007). The benjamini-hochberg method in the case of discrete test statistics. Int. J. Biostat. 3:11. 10.2202/1557-4679.1065 [DOI] [PubMed] [Google Scholar]
- Fischer S., Agmon-Levin N., Shapira Y., Bat-Sheva P. K., Eduard G., Ricard C., et al. (2013). Toxoplasma gondii: bystander or cofactor in rheumatoid arthritis. Immunol. Res. 56 287–292. 10.1007/s12026-013-8402-2 [DOI] [PubMed] [Google Scholar]
- Franco H. L., Dai D., Lee K. Y., Cory A. R., Dennis R., Derek B., et al. (2011). WNT4 is a key regulator of normal postnatal uterine development and progesterone signaling during embryo implantation and decidualization in the mouse. FASEB J. 25 1176–1187. 10.1096/fj.10-175349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman K., Oakley L., Pollak A., Semprini A. E., Salt A., Gilbert R. (2005). Association between congenital toxoplasmosis and preterm birth, low birthweight and small for gestational age birth. BJOG 112 31–37. [DOI] [PubMed] [Google Scholar]
- He J. J., Ma J., Elsheikha H. M., Song H. Q., Huang S. Y., Zhu X. Q. (2016a). Transcriptomic analysis of mouse liver reveals a potential hepato-enteric pathogenic mechanism in acute Toxoplasma gondii infection. Parasit. Vectors 9:427. 10.1186/s13071-016-1716-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J. J., Ma J., Song H. Q., Zhou D. H., Wang J. L., Huang S. Y., et al. (2016b). Transcriptomic analysis of global changes in cytokine expression in mouse spleens following acute Toxoplasma gondii infection. Parasitol. Res. 115 703–712. 10.1007/s00436-015-4792-5 [DOI] [PubMed] [Google Scholar]
- Hill D., Dubey J. P. (2002). Toxoplasma gondii: transmission, diagnosis and prevention. Clin. Microbiol. Infect. 8 634–640. [DOI] [PubMed] [Google Scholar]
- Hou X., Tan Y., Li M., Dey S. K., Das S. K. (2004). Canonical Wnt signaling is critical to estrogen-mediated uterine growth. Mol. Endocrinol. 18 035–3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight B., Brunton C., Modi N., Wallace G., Stanford M. (2005). The effect of Toxoplasma gondii infection on expression of chemokines by rat retinal vascular endothelial cells. J. Neuroimmunol. 160 41–47. [DOI] [PubMed] [Google Scholar]
- Kuang H., Chen Q., Fan X., Zhang Y., Zhang L., Peng H., et al. (2009). CXCL14 inhibits trophoblast outgrowth via a paracrine/autocrine manner during early pregnancy in mice. J. Cell Physiol. 221 448–457. 10.1002/jcp.21877 [DOI] [PubMed] [Google Scholar]
- Li J., Xu X., Kook-Joo N., Chan C. (2013). “The role of Wnt7a gene in the proliferation, invasion and metastasis of non-small cell lung cancer,” in Proceedings of the Thirteenth National Lung Cancer Conference, Changchun. [Google Scholar]
- Liu Y., Shi J., Lu C. C., Zheng-Bei W., Anna I. L., Xue-Jun S., et al. (2005). Ryk-mediated Wnt repulsion regulates posterior-directed growth of corticospinal tract. Nat. Neurosci. 8 1151–1159. [DOI] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25 402–408. [DOI] [PubMed] [Google Scholar]
- Luft B. J., Conley F., Remington J. S., Laverdiere M., Wagner K. F., Levine J. F., et al. (1983). Outbreak of central-nervous-system toxoplasmosis in western Europe and North America. Lancet 1 781–784. [DOI] [PubMed] [Google Scholar]
- Majchrzak-Celinska A., Kosicka K., Paczkowska J., Główka F. K., Brêborowicz G. H., Krzyścin M., et al. (2017). HSD11B2, RUNX3, and LINE-1 methylation in placental DNA of hypertensive disorders of pregnancy patients. Reprod. Sci. 24 1520–1531. 10.1177/1933719117692043 [DOI] [PubMed] [Google Scholar]
- Mohamed O. A., Jonnaert M., Labelle-Dumais C., Kazuki K., Hugh J. C., Daniel D. (2005). Uterine Wnt/beta-catenin signaling is required for implantation. Proc. Natl. Acad. Sci. U.S.A. 102 8579–8584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montoya J. G., Remington J. S. (2008). Management of Toxoplasma gondii infection during pregnancy. Clin. Infect. Dis. 47 554–566. [DOI] [PubMed] [Google Scholar]
- Moon R. T., Kohn A. D., De Ferrari G. V., Kaykas A. (2004). WNT and beta-catenin signalling: diseases and therapies. Nat. Rev. Genet. 5 691–701. [DOI] [PubMed] [Google Scholar]
- Neumann J., Schaale K., Farhat K., Tobias E., Artur J. U., Stefan E., et al. (2010). Frizzled1 is a marker of inflammatory macrophages, and its ligand Wnt3a is involved in reprogramming Mycobacterium tuberculosis-infected macrophages. FASEB J. 24 4599–4612. 10.1096/fj.10-160994 [DOI] [PubMed] [Google Scholar]
- Nishimura M., Umeda K., Suwa M., Furuoka H., Nishikawa Y. (2017). CCR5 is involved in interruption of pregnancy in mice infected with toxoplasma gondii during early pregnancy. Infect. Immun. 85:e00257-17. 10.1128/IAI.00257-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris W., Nevers T., Sharma S., Kalkunte S. (2011). Review: hCG, preeclampsia and regulatory T cells. Placenta 32(Suppl. 2), S182–S185. 10.1016/j.placenta.2011.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H. J., Kuk I. S., Kim J. H., Kim J. H., Song S. J., Choi B. C., et al. (2011). Characterisation of mouse interferon-induced transmembrane protein-1 gene expression in the mouse uterus during the oestrous cycle and pregnancy. Reprod. Fertil. Dev. 23 798–808. 10.1071/RD10086 [DOI] [PubMed] [Google Scholar]
- Pazmany T., Tomasi T. B. (2006). The major histocompatibility complex class II transactivator is differentially regulated by interferon-gamma and transforming growth factor-beta in microglial cells. J. Neuroimmunol. 172 18–26. [DOI] [PubMed] [Google Scholar]
- Remington J. S., Jacobs L., Kaufman H. E. (1958). Studies on chronic toxoplasmosis; the relation of infective dose to residual infection and to the possibility of congenital transmission. Am. J. Ophthalmol. 46 261–268. [PubMed] [Google Scholar]
- Remington J. S., Jacobs L., Melton M. L. (1961). Congenital transmission of toxoplasmosis from mother animals with acute and chronic infections. J. Infect. Dis. 108 163–173. [DOI] [PubMed] [Google Scholar]
- Rojas D. B., de Andrade R. B., Gemelli T., Oliveira L. S., Campos A. G., Dutra-Filho C. S., et al. (2012). Effect of histidine administration to female rats during pregnancy and lactation on enzymes activity of phosphoryltransfer network in cerebral cortex and hippocampus of the offspring. Metab. Brain Dis. 27 595–603. 10.1007/s11011-012-9319-5 [DOI] [PubMed] [Google Scholar]
- Rorman E., Zamir C. S., Rilkis I., Ben-David H. (2006). Congenital toxoplasmosis–prenatal aspects of Toxoplasma gondii infection. Reprod. Toxicol. 21 458–472. [DOI] [PubMed] [Google Scholar]
- Saadatnia G., Golkar M. (2012). A review on human toxoplasmosis. Scand. J. Infect. Dis. 44 805–814. 10.3109/00365548.2012.693197 [DOI] [PubMed] [Google Scholar]
- Stahl W., Dias J. A., Turek G. (1985). Hypothalamic-adenohypophyseal origin of reproductive failure in mice following chronic infection with Toxoplasma gondii. Proc. Soc. Exp. Biol. Med. 178 246–249. [DOI] [PubMed] [Google Scholar]
- Tanaka S., Nishimura M., Ihara F., Yamagishi J., Suzuki Y., Nishikawa Y. (2013). Transcriptome analysis of mouse brain infected with Toxoplasma gondii. Infect. Immun. 81 3609–3619. 10.1128/IAI.00439-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tranguch S., Daikoku T., Guo Y., Wang H., Dey S. K. (2005). Molecular complexity in establishing uterine receptivity and implantation. Cell Mol. Life Sci. 62 1964–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Williams B. A., Pertea G., Mortazavi A., Kwan G., van Baren M. J., et al. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28 511–515. 10.1038/nbt.1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulac S., Nayak N. R., Kao L. C., Van Waes M., Huang J., Lobo S., et al. (2003). Identification, characterization, and regulation of the canonical Wnt signaling pathway in human endometrium. J. Clin. Endocrinol. Metab. 88 3860–3866. [DOI] [PubMed] [Google Scholar]
- Wang R., Tian S., Yang X., Liu J., Wang Y., Sun K. (2017). Celecoxib-induced inhibition of neurogenesis in fetal frontal cortex is attenuated by curcumin via Wnt/beta-catenin pathway. Life Sci. 185 95–102. 10.1016/j.lfs.2017.07.028 [DOI] [PubMed] [Google Scholar]
- Wilcox M. H., Powell R. J., Pugh S. F., Balfour A. H. (1990). Toxoplasmosis and systemic lupus erythematosus. Ann. Rheum. Dis. 49 254–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y., Yin J., Jiang N., Mei X., Lili H., Huijun L., et al. (2010). Seroepidemiology of human Toxoplasma gondii infection in China. BMC Infect. Dis. 10:4. 10.1186/1471-2334-10-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y., Clements W. K., Kimelman D., Xu W. (2003). Crystal structure of a beta-catenin/axin complex suggests a mechanism for the beta-catenin destruction complex. Genes Dev. 17 2753–2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Zhang J., Zhan S., Li Z., Liu X., Zhang H., et al. (2017). TGF-β1 improving abnormal pregnancy outcomes induced by Toxoplasma gondii infection: regulating NKG2D/DAP10 and killer subset of decidual NK cells. Cell Immunol. 317 9–17. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Peng S., Kuang H., Chen Q., Liu S., Zhang L., et al. (2009). Expression and regulation of Dickkopf2 during periimplantation in mice. J. Reprod. Dev. 55 17–22. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data generated in this study has been deposited into BioProject (accession no. PRJNA593831, https://www.ncbi.nlm.nih.gov/bioproject/PRJNA593831).