Abstract
Methylotrophic yeasts such as Komagataella phaffii (syn. Pichia pastoris, Pp), Hansenula polymorpha (Hp), Candida boidinii (Cb) and Pichia methanolica (Pm) are widely used protein production platforms. Typically, strong, tightly regulated promoters of genes coding for their methanol utilization (MUT) pathways are used to drive heterologous gene expression. Despite highly similar open reading frames in the MUT pathways of the four yeasts, the regulation of the respective promoters varies strongly between species. While most endogenous Pp MUT promoters remain tightly repressed after depletion of a repressing carbon, Hp, Cb and Pm MUT promoters are derepressed to up to 70% of methanol induced levels, enabling methanol free production processes in their respective host background. Here, we have tested a series of orthologous promoters from Hp, Cb and Pm in Pp. Unexpectedly, when induced with methanol, the promoter of the HpMOX gene reached very similar expression levels as the strong methanol, inducible, and most frequently used promoter of the Pp alcohol oxidase 1 gene (PPpAOX1). The HpFMD promoter even surpassed PPpAOX1 up to three-fold, when induced with methanol, and reached under methanol-free/derepressed conditions similar expression as the methanol induced PPpAOX1. These results demonstrate that orthologous promoters from related yeast species can give access to otherwise unobtainable regulatory profiles and may even considerably surpass endogenous promoters in P. pastoris.
Keywords: Pichia pastoris, Komagataella phaffii, Orthologous promoters, Methanol-free, Derepression
Introduction
Recombinant proteins such as biopharmaceuticals or industrially relevant biocatalysts are commonly produced by heterologous gene expression in microorganisms. Escherichia coli, Saccharomyces cerevisiae, filamentous fungi, and cells of higher eukaryotes have been widely used as expression hosts since the advent of recombinant protein production. Over the past three decades, the methylotrophic yeasts Pichia pastoris (Pp), Hansenula polymorpha (Hp), Candida boidinii (Cb) and Pichia methanolica (Pm) have emerged as powerful alternatives, enabling high cell density fermentation and simple, pure secretion of heterologous proteins (Gellissen 2000; Hartner and Glieder 2006; Yurimoto et al. 2011; Vogl et al. 2013). The two Pichia species and H. polymorpha have phylogenetically been reassigned as Komagataella and Ogataea species, respectively resulting in the formal names Komagataella phaffii, Ogataea polymorpha, and Ogataea methanolica (Peña et al. 2018). Amongst these four methylotrophic yeasts, P. pastoris is most commonly applied for heterologous protein production, even surpassing S. cerevisiae according to a recent literature survey (Bill 2014).
All methylotrophic yeasts offer tightly regulated, strong promoters that are naturally regulating the expression of genes involved in the methanol utilization (MUT) pathway (Hartner and Glieder 2006). Typically, all promoters of MUT genes are tightly repressed on repressing carbon sources such as glucose and get strongly upregulated when shifted to methanol. However, derepression effects vary considerably between species (Hartner and Glieder 2006) and even within the same organism (Vogl et al. 2016). Derepression leads to activation of the promoter when the repressing carbon source is depleted or when a non-repressing carbon source is present. Under derepressed conditions, the promoter of the alcohol oxidase 1 gene in P. pastoris (PPpAOX1) is only activated at 2–4% compared to methanol induced levels (Vogl and Glieder 2013). Although some of the MUT promoters of P. pastoris showed substantial derepression effects, their efficiency was considerably lower than methanol induced AOX1, DAS1 or DAS2 promoters (Vogl et al. 2016). In contrast, the promoter of the orthologous gene (named differently: methanol oxidase, MOX) in H. polymorpha (PHpMOX) shows derepressed expression up to 70% of methanol induced levels, even in presence of glycerol whereas PPpAOX1 is fully repressed by glycerol. Also the promoters of the orthologous genes in C. boidinii (alcohol oxidase 1, abbreviated AOD1) and P. methanolica (methanol oxidase 1/2, abbreviated MOD1/2) were reported to be activated by derepression, reaching up to 70% of methanol induced levels (Hartner and Glieder 2006). However, the use of the orthologous AOX1 promoter of Pp in Hp indicated that the respective regulation is host specific rather than due to the specific promoter sequence since glycerol did not repress the PPpAOX1 in Hp (Rodriguez et al. 1996; Raschke et al. 1996). Note that the alcohol oxidase/methanol oxidase genes fulfilling the same function were assigned different three letter abbreviations in all four yeasts. We are keeping these identifiers in addition to the prefixes Pp, Hp, Cb and Pm to differentiate between the organisms.
Especially in large scale production processes and for biopharmaceutical production, induction with toxic and flammable methanol is unwanted due to safety issues, making strong derepressed promoters sought-after expression tools to enable methanol free processes. Derepressed promoters allow for regulated expression by simply varying the availability of the carbon source [i.e. repression is achieved with an excess of a repressing carbon source, subsequently reducing the feed rate to limiting amounts triggers activation e.g. (Hartner et al. 2008; Vogl et al. 2018c)]. PPpAOX1 variants (Hartner et al. 2008), alternative promoters (Prielhofer et al. 2013), novel MUT promoters (Vogl et al. 2016), synthetic bidirectional promoters (Vogl et al. 2018b) and altering the molecular regulation of PPpAOX1 (Shen et al. 2016a, b; Wang et al. 2017; Vogl et al. 2018c) showed derepression to varying extents in P. pastoris.
Recent studies in metazoans (Weirauch and Hughes 2010) and yeast (Zeevi et al. 2014) have shown that orthologous, highly divergent promoter sequences from different species can achieve similar expression. For example, the promoters of the genes coding for orthologous ribosomal proteins in various yeast species, showed high expression conservation in S. cerevisiae (Zeevi et al. 2014). We hypothesized that also MUT promoters of related methylotrophic yeasts may show some extent of conservation. Here we have tested a comprehensive series of commonly used MUT promoters from Hp, Cb and Pm in Pp and some of these promoters performed surprisingly well, even outperforming the most frequently used endogenous Pp promoters.
Materials and methods
Cloning of promoters
The orthologous promoters were PCR amplified and cloned upstream of an eGFP reporter gene into a previously established reporter plasmid for P. pastoris [pPpT4mutZeoMlyI-intARG4-eGFP-BmrIstuffer, (Vogl et al. 2016)] based on the pPpT4 vector reported by Näätsaari et al. (2012). The promoters were cloned seamlessly, i.e. maintaining the natural sequence context to the start codon without additional restriction endonuclease sites or linker sequences. Primers were designed according to the literature (HpFMD/MOX promoters (Ledeboer et al. 1985; Song et al. 2003), CbAOD1 [Yurimoto et al. 2000) and CbFLD1 (Lee et al. 2002), Pm MOD1 and MOD2 (Raymond et al. 1998; Nakagawa et al. 2001, 2006)] and the primer sequences are provided in Additional file 1: S1. Genomic DNA of the strains Hp DSM 70277, Cb DSM 70026 and Pm DSM 2147 was isolated and used as template for the PCR reactions. The PCRs were cloned into the reporter vector by TA cloning as outlined previously (Vogl et al. 2015, 2016). The cloned promoters were verified by Sanger sequencing, showing in part minor differences to previously reported sequences (Additional file 1: S2). The control vectors of the P. pastoris endogenous AOX1, CAT1 and GAP promoters were available from previous studies (Vogl et al. 2016).
The alternative reporter vectors bearing HRP [isoenzyme A2A (Näätsaari et al. 2014)], CalB and MeHNL downstream of the respective promoters were in part available from previous studies (Vogl et al. 2016) or generated by cutting out the eGFP reporter gene from the above mentioned vectors (via NheI and NotI restriction sites) and seamlessly inserting PCR products of the GOIs by assembly cloning (Gibson et al. 2009). See Additional file 1: S1 for the primer sequences and Additional file 1: S4 for a list of the plasmids and strains used in this study. The HRP and CalB vectors previously reported (Vogl et al. 2016) were used as PCR templates, the MeHNL sequence was codon optimized for P. pastoris and ordered with overhangs to the AOX1 promoter and terminator for assembly cloning (Additional file 1: S1). This vector was sequenced and used as template for PCR amplification. Since the HRP and CalB genes were both fused to a mating factor alpha signal sequence, the same forward primer could be used for amplification (pHpFMD-MFalpha-Gib). The inserted genes were sequenced with primers binding to the AOX1 terminator and the respective promoters (Vogl et al. 2016), for PHpFMD primer seq-pHpHMD-149..126fwd was used to allow a new Sanger sequencing of the downstream gene.
Strains, materials, fluorescence measurements and enzyme assays
Materials and strains were used as previously reported in detail (Vogl et al. 2016). Deep well plate and shake flask cultivations were also performed as reported in the literature (Weis et al. 2004; Vogl et al. 2016). Fluorescence measurements, HRP and CalB activity assays were also performed as previously reported (Vogl et al. 2016). Culture supernatants for the HRP and CalB activity assays were obtained by centrifugation (3000g for 20 min) and carefully transferring the liquid without touching the pelleted cells. For MeHNL activity measurements, cell free extracts were generated in fourfold replicates from independently grown cultivations of the same strain (Vogl et al. 2018c) by centrifugation (3000g for 20 min), resuspending the pellet in 200 µL Y-PER (Thermo Scientific), shaking for 30 min followed by 30 min (3000g) centrifugation. The resulting supernatant was typically diluted at least tenfold for the MeHNL activity measurement [as described in (Hanefeld et al. 1999) using a mandelonitrile cyanogenesis assay (Wiedner et al. 2014) with a final mandelonitrile concentration of 15 mM]. For transformations of all basic promoter comparisons, the P. pastoris CBS7435 wildtype strain was used following the condensed protocol of Lin-Cereghino et al. (2005), see the following section for applied DNA amounts and the screening/rescreening procedure of transformants. Plasmids were linearized with SwaI prior to transformation (Vogl et al. 2018a). During transformation and selection of P. pastoris, we noticed for the CbAOD1 promoter transformation background (colonies showing no reporter protein expression when re-cultivated), as previously noticed for extended lengths of the P. pastoris CAT1 promoter (Vogl et al. 2016). As PCbAOD1 did not show any reporter protein fluorescence, we did not further investigate this phenomenon during this study. HRP and CalB were used for transformation of a mutS (methanol utilization slow, Δaox1) strain, as higher yields have been reported (Krainer et al. 2012) and the muts strain was also used for the control plasmids bearing these genes of interest under the control of P. pastoris endogenous promoters (Vogl et al. 2016).
Screening, rescreening procedures and culture conditions
To avoid clonal variation due to different copy numbers of integrated expression cassettes as well as different integration sites and genomic alterations that can bias expression strength comparisons in P. pastoris (Schwarzhans et al. 2016a, b; Vogl et al. 2018a), transformants from this study underwent the following screening and rescreening procedures [the section is adapted from the open access publication (Vogl et al. 2018b)]. P. pastoris cells were transformed with molar equivalents to 1 µg of the empty pPpT4_S vector SwaI linearized plasmids as 1 µg of the empty pPpT4_S vector was found to yield predominantly single copy integration (Vogl et al. 2014, 2018a). The screening and rescreening procedures to compare single P. pastoris strains have previously been reported (Vogl et al. 2014, 2016, 2018b) in detail. In brief, for each construct 42 transformants (approximately half a DWP) were screened to avoid clonal variation observed in P. pastoris (Schwarzhans et al. 2016a, 2016b; Vogl et al. 2018a). Four representative clones from the middle of the obtained expression landscape were streaked for single colonies and rescreened in biological 7-fold replicates (raw data provided as Additional file 1: S3) to avoid outliers of multi-copy integration or reduced expression because of deletions or undesired integration events (Schwarzhans et al. 2016a, b; Vogl et al. 2018a) were streaked for single colonies and rescreened in biological sevenfold replicates. Finally, one representative clone was selected and a final screening of all the promoters together was performed (data shown in the figures of the main manuscript). P. pastoris strains were grown for 60 h on 250 µL BMD1 media (buffered minimal dextrose with 1% glucose) and subsequently induced with methanol (250 µL BMM2 [1% methanol] at 60 h and 50 µL BMM10 [5% methanol] at 72 h followed by intervals of 24 h if applicable). Inoculation was performed with ~ 10 µL of frozen glycerol stocks (equaling to an approx. initial OD < 0.05). The BMD and BMM minimal media contain 0.2 M/L potassium phosphate buffer (pH 6), 13.4 g/L yeast nitrogen base and 0.4 mg/L biotin and only differ in the carbon source (as indicated above).
Accession numbers
Orthologous promoters (GenBank): MA887959, MA887960, MA887981, MA887982, MA887983, MA887984; Codon-optimized MeHNL gene: MA887980.
Results
Comparison of orthologous yeast promoters in P. pastoris
Based on their known high promoter activity and their frequent use in their native host (Hartner and Glieder 2006), we selected six orthologous promoters of the HpFMD, HpMOX, CbFLD1, CbAOD1, PmMOD1 and PmMOD2 genes for functional evaluation in P. pastoris (Table 1). These promoters have been reported to be amongst the strongest methanol inducible promoters and at the same time the most derepressed promoters in the respective organisms [reviewed by (Hartner and Glieder 2006)]. These promoters were compared to state of the art endogenous promoters which were so far most frequently used in P. pastoris, i.e. the methanol inducible PAOX1, constitutive PGAP, and derepressed/methanol inducible PCAT1 (Vogl et al. 2016) (Table 1). The orthologous promoters were PCR amplified from genomic DNA and cloned into a reporter vector previously established for promoter comparisons in P. pastoris (Vogl et al. 2016). The promoters were seamlessly fused (i.e. maintaining the natural transition of promoter to start codon without additional restriction sites or linker sequences in between) to an enhanced green fluorescent reporter gene (eGFP). DNA sequencing showed that the promoter sequences contained minor differences compared to previous reports (Additional file 1: S2). These differences are possibly arising from the use of genomic DNA from Hp, Cb and Pm strains from different strain collections than previously reported as PCR templates (see “Materials and methods” section).
Table 1.
Orthologous MUT promoters of related species and endogenous P. pastoris promoters used in this study
| Type | Abbreviation | Species | Gene name | Regulation in native species | Length (bp) | GC content (%) |
|---|---|---|---|---|---|---|
| Orthologous promoters | HpFMD | Hansenula polymorpha | Formate dehydrogenase | Strongly derepressed, methanol inducible | 623 | 53.3 |
| HpMOX | Hansenula polymorpha | Methanol oxidase | Strongly derepressed, methanol inducible | 1510 | 56.0 | |
| CbFLD1 | Candida boidinii | Formaldehyde dehydrogenase | Moderately derepressed, methanol inducible | 572 | 31.6 | |
| CbAOD1 | Candida boidinii | Alcohol oxidase 1 | Moderately derepressed, methanol inducible | 1652 | 28.6 | |
| PmMOD1 | Pichia methanolica | Methanol oxidase 1 | Strongly derepressed, methanol inducible | 1157 | 37.9 | |
| PmMOD2 | Pichia methanolica | Methanol oxidase 2 | Tightly repressed, methanol inducible | 1662 | 37.3 | |
| P. pastoris endogenous promoters | PpAOX1 | Pichia pastoris | Alcohol oxidase 1 | Tightly repressed, methanol inducible | 940 | 42.6 |
| PpCAT1 | Pichia pastoris | Catalase 1 | Moderately derepressed, methanol inducible | 500 | 40.8 | |
| PpGAP | Pichia pastoris | Glyceraldehyde 3-phosphate dehydrogenase | Constitutive | 486 | 46.7 |
Moderately derepressed: < 50% of methanol induced levels; strongly derepressed: > 50% of methanol induced levels [according to the data by (Hartner and Glieder 2006)]. Promoter lengths used in this study are listed and deviate in part slightly form values reported in the literature (see “Materials and methods” section and Additional file 1: S2)
The HpFMD promoter enables strong derepressed expression in P. pastoris
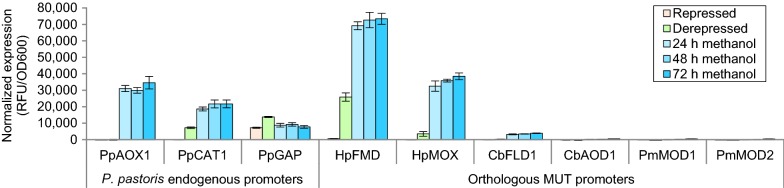
Pichia pastoris transformants of plasmids bearing CbAOD1, PmMOD1 and PmMOD2 promoters did not show any reporter protein fluorescence (Fig. 1). PCbFLD1 showed repression on glucose and weak methanol inducible expression of about 10% of PPpAOX1, in line with the initial expectation that host specific regulatory proteins and mechanisms are necessary for efficient transcription. However, both H. polymorpha promoters tested unexpectedly maintained their natural regulation and showed repression, derepression and methanol induction profiles. The HpMOX promoter showed weak derepressed reporter protein fluorescence and reached similar reporter protein fluorescence on methanol as PPpAOX1. The HpFMD promoter showed derepressed expression outperforming the constitutive PPpGAP and reaching approximately 75% of the methanol induced PPpAOX1 for the well expressible intracellular eGFP reporter. Derepressed expression from PHpFMD exceeded reporter protein fluorescence of the strongest derepressed endogenous MUT promoter from P. pastoris (PPpCAT1) considerably and upon methanol induction PHpFMD even outperformed PPpAOX1 ca. 2.1-fold.
Fig. 1.
Orthologous MUT promoters outperform P. pastoris endogenous promoters. Reporter protein fluorescence of all orthologous and P. pastoris endogenous promoters tested. The orthologous MUT promoters of different methylotrophic yeasts were cloned upstream of an enhanced green fluorescent protein (eGFP) and transformed into P. pastoris. The strains were cultivated in deep well plates (DWPs) on BMD1 (glucose) media and subsequently induced with methanol (Weis et al. 2004; Vogl et al. 2016). Reporter protein fluorescence and OD600 were measured under glucose repressed (16 h) and derepressed (60 h) conditions and different time points of methanol induction. Fluorescence measurements were normalized per OD600. Mean values (MVs) and standard deviations (SDs) of biological quadruplicates are shown
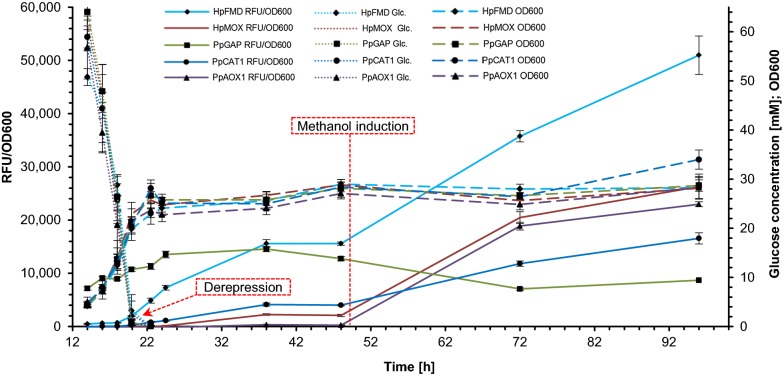
In deep well plate cultivations (Fig. 1) PHpFMD seemed to give also a very weak reporter fluorescence signal under glucose repressed conditions, hinting slight constitutive activity. Expression from the PHpMOX and P. pastoris PAOX1 and PCAT1 was undetectable. In experiments in shake flasks measuring also glucose levels (Fig. 2), PHpFMD showed very weak constitutive expression before full depletion of glucose. This result may suggest that the exceptional strength of PHpFMD, clearly outperforming even P. pastoris endogenous promoters, is at the expense of less tight repression at lower glucose concentrations. Constitutive activity of PHpFMD is less than 1% of fully induced levels, showing still induction over two logs.
Fig. 2.
PHpFMD enables strong derepression and exceeds the strength of methanol induced endogenous P. pastoris promoters. Strains bearing selected promoters from Fig. 1 (HpFMD, HpMOX, PpAOX1, PpCAT1, PpGAP) were cultivated in shake flasks and inoculated to a low starting OD600 of 0.05. Reporter protein fluorescence, OD600 and glucose levels were measured. Fluorescence/OD600 values at t = 0 are not shown, as the starting OD600 of 0.05 was outside the linear range of the spectrometer used. The initial glucose concentration of the media was 55.5 mM (10 g/L). MVs and SDs of biological triplicates are shown
Validation of PHpFMD promoter with additional reporter genes
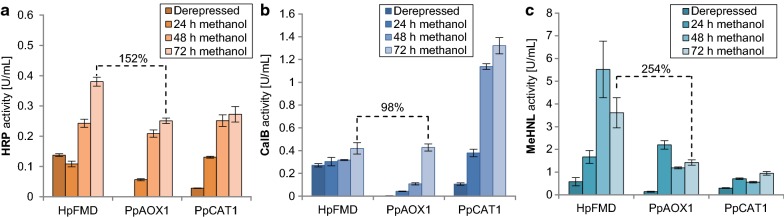
Since strong transcription not always favors expression of other proteins, especially when secreted, we were interested if the exceptionally strong expression of PHpFMD could also be reproduced with other proteins than eGFP. Therefore, the PHpFMD promoter was cloned upstream of the coding sequence of secretory proteins horseradish peroxidase (HRP) and Candida antarctica lipase B (CalB) and an intracellularly expressed hydroxynitrile lyase from Manihot esculenta (MeHNL) (Fig. 3). Yields obtained from PHpFMD were compared to the P. pastoris endogenous MUT promoters PPpCAT1 and PPpAOX1. Derepressed expression of HRP and CalB employing PHpFMD clearly outperformed derepressed expression from PPpCAT1. Methanol induced enzyme activities of PPpCAT1 and PPpAOX1 were similar, only for CalB expression PPpCAT1 outperformed all tested promoters, suggesting a specific beneficial effect. Methanol induced activities from PHpFMD outperformed methanol induced PPpAOX1 up to 2.5-fold. However, the effect was stronger for the intracellular expression of MeHNL (Fig. 3c) than the secretory expression of HRP and CalB (Fig. 3a, b). We assume that for the secretory proteins, not transcription but rather passage through the secretory pathway is the limiting factor. In line with this hypothesis, it has previously been shown that multicopy strains of CalB even show reduced activities compared to single copy if expressed without helper proteins (Abad et al. 2010). Similar effects were also noticed for HRP (Krainer et al. 2016), where maximum titers obtained so far are still in the several 100 mg/L range. Too strong overexpression of HRP and CalB by PHpFMD may overburden the secretion machinery [‘secretion saturation’ (Aw and Polizzi 2013)] or other factors such as cofactor synthesis might limit product titers, whereas intracellular expression of MeHNL appears more simple and well tolerated by the host.
Fig. 3.
Applying the HpFMD promoter for expression of the enzymes HRP (a), CalB (b) and MeHNL (c) confirms the high expression observed with eGFP (Fig. 1). The strains were grown in DWPs on BMD1 media until glucose depletion for 60 h and were subsequently induced with methanol. HRP and CalB activities in the supernatants were measured and cells lysed to measure intracellular MeHNL activity. Mean values (MVs) and standard deviation (SDs) of biological quadruplicates are shown. The activities of the methanol induced HpFMD promoter compared to the state of the art AOX1 promoter are highlighted (after 72 h of methanol induction)
The strong expression from PHpFMD was consistently reproducible using four reporter genes (eGFP, HRP, CalB, MeHNL), demonstrating that orthologous promoters from related organisms can be valuable tools for protein production even exceeding endogenous promoters.
Discussion
Here we have shown that orthologous MUT promoters can be highly useful tools for single protein production, as demonstrated by up to 3.5-fold higher expression achieved form the PHpFMD compared to the strongest endogenous P. pastoris MUT promoters. Interestingly, although regulating genes of proteins with high sequence similarity and similar enzymatic function, the respective orthologous promoters show highly divergent sequences from P. pastoris. None of the orthologous promoters tested show clear identity to the P. pastoris genome when performing a BLAST search (using standard parameters) and also alignments to their P. pastoris orthologs did not exhibit clear identities (data not shown). A similar lack of sequence identity between similarly regulated promoters was reported for P. pastoris MUT promoters (Vogl et al. 2016), in metazoans (Weirauch and Hughes 2010) and S. cerevisiae (Zeevi et al. 2014). We assume that the expression from these MUT promoters is governed by short, partially degenerative transcription factor binding sites (TFBS), also conserved in some orthologous promoters. The P. pastoris methanol master regulator Mxr1p (Lin-Cereghino et al. 2006) binds for example a simple CYCCNY motif and this motif is dispersed over different positions in the P. pastoris AOX1, DAS2 and PEX8 promoter sequences (Kranthi et al. 2009, 2010).
Such high sequence diversity and lack of identity is especially advantageous if multiple genes should be co-expressed. The repeated use of identical sequences can results in ‘loop out’ recombination in yeast (Aw and Polizzi 2013), leading to loss of copies or parts of expression cassettes (Zhu et al. 2009; Geier et al. 2015; Schwarzhans et al. 2016a, b; Vogl et al. 2018a). To this end, orthologous promoters with similar regulation but dissimilar sequences may also become valuable tools for metabolic engineering and synthetic biology endeavors, requiring the expression of multiple genes from similarly regulated promoters (Vogl et al. 2016).
The strong derepressed activity of the HpFMD promoter is even more surprising in view of this large sequence diversity. It has previously been suggested, that regulation of derepression in methylotrophic yeasts is conferred primarily by the host regulatory machinery and not by the promoter sequences (Hartner and Glieder 2006). This assumption was taken, as the P. pastoris AOX1 promoter (tightly repressed in its natural host) did not maintain its tight repression if transferred to H. polymorpha. PPpAOX1 showed in Hp derepression similar to endogenous H. polymorpha promoters (Rodriguez et al. 1996; Raschke et al. 1996; Hartner and Glieder 2006). However, in our hands the HpFMD promoter exhibited strong derepressed expression in P. pastoris, unlike strong P. pastoris endogenous promoters. A possible explanation may be that P. pastoris contains unique repressors to maintain tight repression under derepressed conditions. It appears that this machinery does not exist in H. polymorpha [or at least does not act on the HpFMD and HpMOX promoters, as these promoters are naturally derepressed and also the PpAOX1 promoter is derepressed in presence of glycerol when applied in Hp (Raschke et al. 1996)]. So it is unlikely that the HpFMD and HpMOX promoters contain binding sites for the P. pastoris machinery to maintain tight repression, which would explain their derepressed regulation in P. pastoris. Alternatively, the effect may also be explained by an activating model: H. polymorpha may contain activators that start expression under derepressed conditions. P. pastoris may contain similar derepressed activators, as for example the PpCAT1 promoter is also moderately derepressed (Vogl et al. 2016). The HpFMD promoter may contain more TFBS for these activators than PPpCAT1, leading to stronger activation. However, these are just hypotheses and elucidating the exact mechanisms of the strong derepressed expression will require further studies.
Especially the use of the HpFMD promoter enables strong expression without employing methanol and provides several advantages over alternative strategies to achieve methanol-free, regulated expression in P. pastoris. Due to its sheer strength, PHpFMD surpasses under methanol-free conditions derepressed PPpAOX1 variants (Hartner et al. 2008) and naturally derepressed promoters such as PPpCAT1 (Vogl et al. 2016). In contrast to approaches of achieving derepressed expression from PPpAOX1 by overexpression of transcription factors or knockout of repressors (Shen et al. 2016a, b; Wang et al. 2017; Vogl et al. 2018c), the use of PHpFMD does not require genetic modifications of the production strains and can be readily applied in unmodified wildtype strains. However, transcription factor overexpression (Vogl et al. 2018c) does provide an advantage, as thereby existing high level production strains can be easily retrofitted for methanol free production. Despite the establishment of improved genome editing tools for P. pastoris (Weninger et al. 2015, 2016, 2018; Raschmanová et al. 2018), it is considerably more difficult to replace the promoters in existing strains. For the generation of novel expression strains the use of PHpFMD appears favorable and may even be boosted by molecular regulation alterations demonstrated for PPpAOX1 (Shen et al. 2016a, b; Wang et al. 2017; Vogl et al. 2018c). Also, the different strength of expression of PHpFMD under derepressed and methanol induced conditions also allow consecutive induction which might a reasonable explanation why final yields and titers are seemingly higher with such 2-step induction procedures for non-trivial secreted proteins such as CalB, where strong promoters or multicopy integration of expression cassettes are usually counteracting high titers of folded and active secreted product.
Eventually, PHpFMD can also be induced with methanol, if the derepressed yields should be exceeded and methanol induction is feasible. In this setup, it represents to the best of our knowledge the strongest promoter reported in P. pastoris so far, exceeding the state of the art AOX1 promoter up to three-fold. In a broader view, our work demonstrates that orthologous promoters from related yeast species can give access to otherwise unobtainable regulatory profiles and may even considerably surpass endogenous promoters, suggesting that this strategy may also be generalized for the discovery of potent, orthologous promoters in other eukaryotic hosts.
Supplementary information
Additional file 1. Supporting information including S1 (primer sequences), S2 (promoter alignments), S3 (screening data), and S4 (list of plasmids and strains).
Acknowledgements
Not applicable.
Authors’ contributions
TV and AG conceived of the study. TV designed all plasmids and experiments. LS performed the molecular cloning of the orthologous promoters. PH and LS generated the P. pastoris strains and performed the eGFP reporter measurements. RW performed the molecular cloning of the HpFMD promoter with CalB, HRP and MeHNL genes. JEF performed the CalB, HRP and MeHNL measurements and the time series experiment. TV and AG wrote the manuscript and supervised the research. All authors read and approved the final manuscript.
Funding
T.V.: Austrian Science Fund (FWF) project number W901 (DK ‘Molecular Enzymology’ Graz).
Availability of data and materials
References for all DNA sequences used are provided, for the primary promoter sequences changes to previously reported sequences are provided in Additional file.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
TV. and A.G. are inventors on a related patent application entitled “Yeast cell” (WO2017109082). The other authors declare no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jasmin Elgin Fischer, Patrick Hyden and Richard Wasmayer contributed equally to this work
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13568-020-00972-1.
References
- Abad S, Kitz K, Hörmann A, Schreiner U, Hartner FS, Glieder A. Real-time PCR-based determination of gene copy numbers in Pichia pastoris. Biotechnol J. 2010;5:413–420. doi: 10.1002/biot.200900233. [DOI] [PubMed] [Google Scholar]
- Aw R, Polizzi KM. Can too many copies spoil the broth? Microb Cell Fact. 2013;12:128. doi: 10.1186/1475-2859-12-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bill RM. Playing catch-up with Escherichia coli: using yeast to increase success rates in recombinant protein production experiments. Front Microbiol. 2014;5:85. doi: 10.3389/fmicb.2014.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geier M, Fauland P, Vogl T, Glieder A. Compact multi-enzyme pathways in P. pastoris. Chem Commun. 2015;51:1643–1646. doi: 10.1039/C4CC08502G. [DOI] [PubMed] [Google Scholar]
- Gellissen G. Heterologous protein production in methylotrophic yeasts. Appl Microbiol Biotechnol. 2000;54:741–750. doi: 10.1007/s002530000464. [DOI] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang R, Venter JC, Hutchison CA, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- Hanefeld U, Straathof AJJ, Heijnen JJ. Study of the (S)-hydroxynitrile lyase from Hevea brasiliensis: mechanistic implications. Biochim Biophys Acta. 1999;1432:185–193. doi: 10.1016/S0167-4838(99)00108-9. [DOI] [PubMed] [Google Scholar]
- Hartner FS, Glieder A. Regulation of methanol utilisation pathway genes in yeasts. Microb Cell Fact. 2006;5:39–59. doi: 10.1186/1475-2859-5-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartner FS, Ruth C, Langenegger D, Johnson SN, Hyka P, Lin-Cereghino GP, Lin-Cereghino J, Kovar K, Cregg JM, Glieder A. Promoter library designed for fine-tuned gene expression in Pichia pastoris. Nucleic Acids Res. 2008;36:e76. doi: 10.1093/nar/gkn369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krainer FW, Dietzsch C, Hajek T, Herwig C, Spadiut O, Glieder A. Recombinant protein expression in Pichia pastoris strains with an engineered methanol utilization pathway. Microb Cell Fact. 2012;11:22. doi: 10.1186/1475-2859-11-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krainer FW, Gerstmann MA, Darnhofer B, Birner-Gruenberger R, Glieder A. Biotechnological advances towards an enhanced peroxidase production in Pichia pastoris. J Biotechnol. 2016;233:181–189. doi: 10.1016/j.jbiotec.2016.07.012. [DOI] [PubMed] [Google Scholar]
- Kranthi BV, Kumar R, Kumar NV, Rao DN, Rangarajan PN. Identification of key DNA elements involved in promoter recognition by Mxr1p, a master regulator of methanol utilization pathway in Pichia pastoris. Biochim Biophys Acta. 2009;1789:460–468. doi: 10.1016/j.bbagrm.2009.05.004. [DOI] [PubMed] [Google Scholar]
- Kranthi BV, Kumar HRV, Rangarajan PN. Identification of Mxr1p-binding sites in the promoters of genes encoding dihydroxyacetone synthase and peroxin 8 of the methylotrophic yeast Pichia pastoris. Yeast. 2010;27:705–711. doi: 10.1002/yea.1766. [DOI] [PubMed] [Google Scholar]
- Ledeboer AM, Edens L, Maat J, Visser C, Bos JW, Verrips CT, Janowicz Z, Eckart M, Roggenkamp R, Hollenberg CP. Molecular cloning and characterization of a gene coding for methanol oxidase in Hansenula polymorpha. Nucleic Acids Res. 1985;13:3063–3082. doi: 10.1093/nar/13.9.3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B, Yurimoto H, Sakai Y, Kato N. Physiological role of the glutathione-dependent formaldehyde dehydrogenase in the methylotrophic yeast Candida boidinii. Microbiology. 2002;148:2697–2704. doi: 10.1099/00221287-148-9-2697. [DOI] [PubMed] [Google Scholar]
- Lin-Cereghino Joan, Wong William W., Xiong See, Giang William, Luong Linda T., Vu Jane, Johnson Sabrina D., Lin-Cereghino Geoff P. Condensed protocol for competent cell preparation and transformation of the methylotrophic yeast Pichia pastoris. BioTechniques. 2005;38(1):44–48. doi: 10.2144/05381BM04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin-Cereghino GP, Godfrey L, de la Cruz BJ, Johnson S, Khuongsathiene S, Tolstorukov I, Yan M, Lin-Cereghino J, Veenhuis M, Subramani S, Cregg JM. Mxr1p, a key regulator of the methanol utilization pathway and peroxisomal genes in Pichia pastoris. Mol Cell Biol. 2006;26:883–897. doi: 10.1128/MCB.26.3.883-897.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Näätsaari L, Mistlberger B, Ruth C, Hajek T, Hartner FS, Glieder A. Deletion of the Pichia pastoris KU70 homologue facilitates platform strain generation for gene expression and synthetic biology. PLoS ONE. 2012;7:e39720. doi: 10.1371/journal.pone.0039720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Näätsaari L, Krainer FW, Schubert M, Glieder A, Thallinger GG. Peroxidase gene discovery from the horseradish transcriptome. BMC Genom. 2014;15:227. doi: 10.1186/1471-2164-15-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Sakai Y, Mukaiyama H, Mizumura T, Miyaji T, Yurimoto H, Kato N, Tomizuka N. Analysis of alcohol oxidase isozymes in gene-disrupted strains of methylotrophic yeast Pichia methanolica. J Biosci Bioeng. 2001;91:225–227. doi: 10.1016/S1389-1723(01)80071-2. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Inagaki A, Ito T, Fujimura S, Miyaji T, Yurimoto H, Kato N, Sakai Y, Tomizuka N. Regulation of two distinct alcohol oxidase promoters in the methylotrophic yeast Pichia methanolica. Yeast. 2006;23:15–22. doi: 10.1002/yea.1334. [DOI] [PubMed] [Google Scholar]
- Peña DA, Gasser B, Zanghellini J, Steiger MG, Mattanovich D. Metabolic engineering of Pichia pastoris. Metab Eng. 2018 doi: 10.1016/j.ymben.2018.04.017. [DOI] [PubMed] [Google Scholar]
- Prielhofer R, Maurer M, Klein J, Wenger J, Kiziak C, Gasser B, Mattanovich D. Induction without methanol: novel regulated promoters enable high-level expression in Pichia pastoris. Microb Cell Fact. 2013;12:5. doi: 10.1186/1475-2859-12-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raschke WC, Neiditch BR, Hendricks M, Cregg JM. Inducible expression of a heterologous protein in Hansenula polymorpha using the alcohol oxidase 1 promoter of Pichia pastoris. Gene. 1996;177:163–167. doi: 10.1016/0378-1119(96)00293-4. [DOI] [PubMed] [Google Scholar]
- Raschmanová H, Weninger A, Glieder A, Kovar K, Vogl T. Implementing CRISPR-Cas technologies in conventional and non-conventional yeasts: current state and future prospects. Biotechnol Adv. 2018;36:641–665. doi: 10.1016/j.biotechadv.2018.01.006. [DOI] [PubMed] [Google Scholar]
- Raymond CK, Bukowski T, Holderman SD, Ching AF, Vanaja E, Stamm MR. Development of the methylotrophic yeast Pichia methanolica for the expression of the 65 kilodalton isoform of human glutamate decarboxylase. Yeast. 1998;14:11–23. doi: 10.1002/(SICI)1097-0061(19980115)14:1<11::AID-YEA196>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Rodriguez L, Narciandi RE, Roca H, Cremata J, Montesinos R, Rodriguez E, Grillo JM, Muzio V, Herrera LS, Delgado JM. Invertase secretion in Hansenula polymorpha under the AOX1 promoter from Pichia pastoris. Yeast. 1996;12:815–822. doi: 10.1002/(SICI)1097-0061(199607)12:9<815::AID-YEA916>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Schwarzhans J-P, Wibberg D, Winkler A, Luttermann T, Kalinowski J, Friehs K. Integration event induced changes in recombinant protein productivity in Pichia pastoris discovered by whole genome sequencing and derived vector optimization. Microb Cell Fact. 2016;15:84. doi: 10.1186/s12934-016-0486-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzhans J-P, Wibberg D, Winkler A, Luttermann T, Kalinowski J, Friehs K. Non-canonical integration events in Pichia pastoris encountered during standard transformation analysed with genome sequencing. Sci Rep. 2016;6:38952. doi: 10.1038/srep38952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W, Kong C, Xue Y, Liu Y, Cai M, Zhang Y, Jiang T, Zhou X, Zhou M. Kinase screening in Pichia pastoris identified promising targets involved in cell growth and alcohol oxidase 1 promoter (PAOX1) regulation. PLoS ONE. 2016;11:e0167766. doi: 10.1371/journal.pone.0167766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W, Xue Y, Liu Y, Kong C, Wang X, Huang M, Cai M, Zhou X, Zhang Y, Zhou M. A novel methanol-free Pichia pastoris system for recombinant protein expression. Microb Cell Fact. 2016;15:178. doi: 10.1186/s12934-016-0578-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H, Li Y, Fang W, Geng Y, Wang X, Wang M, Qiu B. Development of a set of expression vectors in Hansenula polymorpha. Biotechnol Lett. 2003;25:1999–2006. doi: 10.1023/B:BILE.0000004392.87179.29. [DOI] [PubMed] [Google Scholar]
- Vogl T, Glieder A. Regulation of Pichia pastoris promoters and its consequences for protein production. N Biotechnol. 2013;30:385–404. doi: 10.1016/j.nbt.2012.11.010. [DOI] [PubMed] [Google Scholar]
- Vogl T, Hartner FS, Glieder A. New opportunities by synthetic biology for biopharmaceutical production in Pichia pastoris. Curr Opin Biotechnol. 2013;24:1094–1101. doi: 10.1016/j.copbio.2013.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogl T, Ruth C, Pitzer J, Kickenweiz T, Glieder A. Synthetic core promoters for Pichia pastoris. ACS Synth Biol. 2014;3:188–191. doi: 10.1021/sb400091p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogl T, Ahmad M, Krainer FW, Schwab H, Glieder A. Restriction site free cloning (RSFC) plasmid family for seamless, sequence independent cloning in Pichia pastoris. Microb Cell Fact. 2015;14:103. doi: 10.1186/s12934-015-0293-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogl T, Sturmberger L, Kickenweiz T, Wasmayer R, Schmid C, Hatzl A-M, Gerstmann MA, Pitzer J, Wagner M, Thallinger GG, Geier M, Glieder A. A toolbox of diverse promoters related to methanol utilization: functionally verified parts for heterologous pathway expression in Pichia pastoris. ACS Synth Biol. 2016;5:172–186. doi: 10.1021/acssynbio.5b00199. [DOI] [PubMed] [Google Scholar]
- Vogl T, Gebbie L, Palfreyman RW, Speight R. Effect of plasmid design and type of integration event on recombinant protein expression in Pichia pastoris. Appl Environ Microbiol. 2018 doi: 10.1128/aem.02712-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogl T, Kickenweiz T, Pitzer J, Sturmberger L, Weninger A, Biggs BW, Köhler E-M, Baumschlager A, Fischer JE, Hyden P, Wagner M, Baumann M, Borth N, Geier M, Ajikumar PK, Glieder A. Engineered bidirectional promoters enable rapid multi-gene co-expression optimization. Nat Commun. 2018;9:3589. doi: 10.1038/s41467-018-05915-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogl T, Sturmberger L, Fauland PC, Hyden P, Fischer JE, Schmid C, Thallinger GG, Geier M, Glieder A. Methanol independent induction in Pichia pastoris by simple derepressed overexpression of single transcription factors. Biotechnol Bioeng. 2018;115:1037–1050. doi: 10.1002/bit.26529. [DOI] [PubMed] [Google Scholar]
- Wang J, Wang X, Shi L, Qi F, Zhang P, Zhang Y, Zhou X, Song Z, Cai M. Methanol-independent protein expression by AOX1 promoter with trans-acting elements engineering and glucose-glycerol-shift induction in Pichia pastoris. Sci Rep. 2017;7:41850. doi: 10.1038/srep41850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weirauch MT, Hughes TR. Conserved expression without conserved regulatory sequence: the more things change, the more they stay the same. Trends Genet. 2010;26:66–74. doi: 10.1016/j.tig.2009.12.002. [DOI] [PubMed] [Google Scholar]
- Weis R, Luiten R, Skranc W, Schwab H, Wubbolts M, Glieder A. Reliable high-throughput screening with Pichia pastoris by limiting yeast cell death phenomena. FEMS Yeast Res. 2004;5:179–189. doi: 10.1016/j.femsyr.2004.06.016. [DOI] [PubMed] [Google Scholar]
- Weninger A, Glieder A, Vogl T. A toolbox of endogenous and heterologous nuclear localization sequences for the methylotrophic yeast Pichia pastoris. FEMS Yeast Res. 2015;15:fov082. doi: 10.1093/femsyr/fov082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weninger A, Hatzl A-M, Schmid C, Vogl T, Glieder A. Combinatorial optimization of CRISPR/Cas9 expression enables precision genome engineering in the methylotrophic yeast Pichia pastoris. J Biotechnol. 2016;235:139–149. doi: 10.1016/j.jbiotec.2016.03.027. [DOI] [PubMed] [Google Scholar]
- Weninger A, Fischer JE, Raschmanová H, Kniely C, Vogl T, Glieder A. Expanding the CRISPR/Cas9 toolkit for Pichia pastoris with efficient donor integration and alternative resistance markers. J Cell Biochem. 2018;119:3183–3198. doi: 10.1002/jcb.26474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedner R, Gruber-Khadjawi M, Schwab H, Steiner K. Discovery of a novel (R)-selective bacterial hydroxynitrile lyase from Acidobacterium capsulatum. Comput Struct Biotechnol J. 2014;10:58–62. doi: 10.1016/j.csbj.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurimoto H, Komeda T, Lim CR, Nakagawa T, Kondo K, Kato N, Sakai Y. Regulation and evaluation of five methanol-inducible promoters in the methylotrophic yeast Candida boidinii. Biochim Biophys Acta. 2000;1493:56–63. doi: 10.1016/S0167-4781(00)00157-3. [DOI] [PubMed] [Google Scholar]
- Yurimoto H, Oku M, Sakai Y. Yeast methylotrophy: metabolism, gene regulation and peroxisome homeostasis. Int J Microbiol. 2011;2011:101298. doi: 10.1155/2011/101298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeevi D, Lubliner S, Lotan-Pompan M, Hodis E, Vesterman R, Weinberger A, Segal E. Molecular dissection of the genetic mechanisms that underlie expression conservation in orthologous yeast ribosomal promoters. Genome Res. 2014;24:1991–1999. doi: 10.1101/gr.179259.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu T, Guo M, Sun C, Qian J, Zhuang Y, Chu J, Zhang S. A systematical investigation on the genetic stability of multi-copy Pichia pastoris strains. Biotechnol Lett. 2009;31:679–684. doi: 10.1007/s10529-009-9917-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Supporting information including S1 (primer sequences), S2 (promoter alignments), S3 (screening data), and S4 (list of plasmids and strains).
Data Availability Statement
References for all DNA sequences used are provided, for the primary promoter sequences changes to previously reported sequences are provided in Additional file.