Abstract
ESR1 mutations in breast cancer are known as one of the mechanisms of resistance to aromatase inhibitors. These mutations often occur in the hotspot regions in the ligand binding domain (LBD), but comprehensive mutational analysis has shown that mutations are observed throughout the whole LBD. We previously developed a molecular barcode sequencing (MB-NGS) technique to detect ESR1 hotspot mutations in plasma with high sensitivity. In this study, we have developed a multiplex MB-NGS assay that covers the whole LBD of ESR1. The assay demonstrated that the background errors in the plasma DNA of 10 healthy controls were below 0.1%; thus, the limit of detection was set at 0.1%. We analyzed the plasma DNA of 54 patients with estrogen receptor–positive metastatic breast cancer. Seventeen mutations were detected in 13 patients (24%), with variant allele frequencies ranging from 0.13% to 10.67%, including six rare mutations with a variant allele frequency <1.0% and a novel nonhotspot mutation (A312V). Three patients had double mutations located in the same amplicons, and it was revealed that the double mutations were located in different alleles. ESR1 hotspot mutations were associated with a longer duration of aromatase inhibitor treatment under metastatic conditions and to liver metastasis. The multiplex MB-NGS assay is useful for the sensitive and comprehensive detection of mutations throughout the whole LBD of ESR1. Our assay can be applied to any specific target region of interest using tailor-made primers and can result in minimized sequencing volume and cost.
Introduction
ESR1 mutations in breast cancer are known as one of the mechanisms of resistance to aromatase inhibitors (AIs). These mutations occur in approximately 30% of AI-treated metastatic breast cancers (MBCs) and accumulate in the hotspot regions at codons 536, 537, and 538 in the ligand binding domain (LBD) [[1], [2], [3]]. The majority of recent clinical studies have focused on these hotspot mutations, which are mostly assayed using cell-free DNA in plasma (liquid biopsy) by taking advantage of dPCR or BEAMing [[4], [5], [6], [7]]. However, comprehensive mutational analysis of the complete ESR1 gene has revealed that almost all ESR1 mutations are seen in its LBD and that 76% of them are located in the hotspots [2,3,[8], [9], [10], [11], [12], [13]], indicating that the analysis of only the hotspot mutations is not sufficient since 24% of ESR1 mutations might be overlooked. Thus, an assay for detecting the ESR1 mutations in the entire LBD with high sensitivity needs to be developed.
We have previously reported the use of conventional NGS for the detection of novel nonhotspot ESR1 mutations [14], but such a conventional NGS analysis is less sensitive than dPCR and thus is likely to miss a significant proportion of ESR1 mutations. Then, we introduced a molecular barcode technique [15,16] to achieve improved sensitivity and specificity (MB-NGS); with this technique, we could show that ESR1 mutations were detected with a high sensitivity (detection limit: 0.1%). However, only a single amplicon (114 bp) harboring the mutation hotspots at codons 536, 537, and 538 was analyzed in that study [17]. The analyzed region accounts for only 16% of the LBD, leaving the remaining 84% not screened. Therefore, in the present study, we attempted to develop multiplex MB-NGS for a comprehensive screening of ESR1 mutations in the LBD with high sensitivity.
Materials and Methods
Patients and Samples
Plasma samples were collected from 54 patients with MBC who were treated at Osaka University Hospital between 2000 and 2018. Twenty of these 54 patients were the same as those analyzed in our previous study [17]. Clinicopathological characteristics of the patients are shown in Table 1. Forty-nine patients had recurrent MBC, and five patients had primary MBC. Positive ER status (Allred score ≥ 3 [18]) was confirmed in primary or recurrent tumors for all cases. The median disease-free interval of recurrent MBCs was 42.8 months (range: 1.9-223.5). Forty-seven patients had been given AIs before sampling. Plasma samples were also collected from 10 healthy volunteers at Osaka Police Hospital. Informed consent was obtained before sampling, and this study was approved by the Ethical Review Board of Osaka University Hospital and Osaka Police Hospital.
Table 1.
Clinicopathological Features of Metastatic Breast Cancer Patients Analyzed in This Study According to ESR1 Mutation Status
| Total | Plasma ESR1 Mutation |
P | |||
|---|---|---|---|---|---|
| Positive | Negative | ||||
| Number | 54 | 13 | 41 | ||
| Age at blood sampling (years) | Mean (range) | 56.5 (30-74) | 56 (35-66) | 57 (30-74) | .348* |
| Estrogen receptor | Positive | 54 | 13 | 41 | |
| Negative | 0 | 0 | 0 | ||
| Progesterone receptor | Positive | 40 | 9 | 31 | .572† |
| Negative | 11 | 3 | 8 | ||
| Unknown | 3 | 1 | 2 | ||
| HER2 status | Positive | 5 | 0 | 5 | .321† |
| Negative | 49 | 13 | 36 | ||
| Primary or recurrent | Primary | 5 | 1 | 4 | 1.000† |
| Recurrent | 49 | 12 | 37 | ||
| DFI of recurrent breast cancer (month) | Median (range) | 42.8 (1.9-223.5) | 52.3 (12.7-177.1) | 35.6 (1.9-223.5) | .140* |
| AI treatment | No AI | 7 | 1 | 6 | .045† |
| Adjuvant AI only | 11 | 0 | 11 | ||
| AI for MBC | 36 | 12 | 24 | ||
| Duration (month) of AI for MBC | Median (range) | 5.6 (0.0-74.8) | 30.2 (0.0-74.8) | 2.9 (0.0-35.6) | <.001* |
HER2, human epidermal receptor 2; DFI, disease-free interval; AI, aromatase inhibitor; MBC, metastatic breast cancer.
Mann-Whitney U test.
Fisher's exact test.
DNA Extraction
Plasma was separated from whole blood by centrifugation for 10 minutes at 3000 (1840 G) rpm and stored at −80°C until further use. The samples were centrifuged again for 10 minutes at 13,300 (16,000 G) rpm prior to DNA extraction to remove debris. Cell-free DNA was isolated from 2 ml of plasma using the QIAamp Circulating Nucleic Acid Kit (Qiagen, Hilden, Germany) or the MagMAX Cell-Free DNA Isolation Kit (Thermo Fisher Scientific, Waltham, MA), according to the manufacturer's instructions, and eluted in 100 μl.
Library Preparation for MB-NGS
Assignment of MBs and adaptors (Rd1SP, Rd2SP, P5, and P7) was performed with PCR as previously reported [17], and the primers used for the library preparation are shown in Supplementary Table 1. The first PCR amplified the targeted region of ESR1-LBD and assigned a 15-base barcode (BDHVBDHVBDHVBDH). Eleven sets of primers covering ESR1-LBD (c.928-c.1641) were designed with a median amplicon size of 135 bp (range: 130-145) using Primer3 [http://bioinfo.ut.ee/primer3/] or DesignStudio [https://designstudio.illumina.com/]. Primers were divided into two sets including amplicons 01/03/04/07/08/11 and 02/05/06/09/10. The first PCR was performed in a 20-μl reaction containing 10 μl of template DNA, 5× Phusion HF buffer (NEB, Ipswich, MA), 0.9 U of Phusion polymerase (NEB), 250 μM dNTPs, and each set of primers. Each primer concentration was optimized to reduce primer dimer formation and to equalize the molecular-barcode family numbers of each amplicon (Supplementary Table 1). The cycling conditions were 1 cycle of 98°C for 30 seconds; 15 cycles of 98°C for 10 seconds, 60°C for 2 minutes, and 72°C for 30 seconds; and 1 cycle of 72°C for 10 minutes. The excess barcode primers were digested with 25 U of Exonuclease-I and 10× Exonuclease-I reaction buffer (NEB) in a 25-μl reaction at 37°C for 1 hour following heat inactivation at 98°C for 5 minutes. Adaptor primers with P5/P7 sequences (0.05 μM each) were added to the first PCR product, and the second PCR was performed in a 28-μl reaction volume. The second PCR was performed using forward and reverse primers including 2 and 24 different sample indices, respectively (Supplementary Table 2). The cycling conditions were 1 cycle of 98°C for 30 seconds; 10 cycles of 98°C for 10 seconds, 60°C for 30 seconds, and 72°C for 30 seconds; and 1 cycle of 72°C for 10 minutes. The second PCR products were purified using 0.7× AMPure XP (Beckman Coulter, Brea, CA), according to the manufacturer's instructions by Bravo (Agilent, Santa Clara, CA, USA), and eluted in 10 μl of nuclease-free water. The third PCR was performed using P5/P7 primers (0.5 μM each) in a 20-μl reaction containing 10 μl of the second PCR product, 5× Phusion HF buffer (NEB), 0.9 U of Phusion polymerase (NEB), and 250 μM dNTPs. The cycling conditions were 1 cycle of 98°C for 30 seconds; 30 cycles of 98°C for 10 seconds, 60°C for 30 seconds, and 72°C for 30 seconds; and 1 cycle of 72°C for 10 minutes. The third PCR products were purified with 0.7× AMPure XP and then 0.7× SPRIselect (Beckman) by Bravo to exclude nonspecific products of 200 bp or less and eluted in 10 μl of nuclease-free water. Libraries were quantified using Fragment Analyzer (Agilent). Four libraries were separately prepared for each sample and analyzed using HiSeq (Illumina, San Diego, CA).
Data Analysis
The variant detection analysis was performed in a similar manner to that described in the previous study [17], with some modifications. Briefly, the quality of sequence reads was confirmed using FastQC, and low-quality bases (Q < 30) and assembled sequences with an unideal length were trimmed using PEAR 0.9.6. The reads whose MB sequences, the first 15 bp of assembled sequences, did not match with “BDHVBDHVBDHVBDH” were removed. Extracted assembled sequences were clustered into each family containing sequences with the same MB sequence by CD-HIT-EST and the custom Ruby script. The threshold of choosing DNA families changed to 5 reads from 30 reads in our last study [17] to obtain sufficient MB families because the current assay included 11 amplicons with an expanded area which lead to a decreased and varied number of reads per amplicon. Barcode sequence was removed before alignment. For each family including over five reads, consensus sequences were mapped onto the ESR1 reference sequence using Bowtie2 ver.2.2.3, and the consensus sequence was constructed by the base accounting for over 80% at each position using SAMtools and the custom Ruby script. Insertion/deletion analysis was performed using lofreq2. Four amplified libraries of each sample were separately analyzed, and when at least one variant family was detected in all four libraries, they were considered as true mutations. Although two libraries per sample were analyzed in our last study [17], the number of libraries was increased from two to four to suppress the background errors to below 0.1% at all SNVs.
Statistics
R 3.5.1 was used for statistical processing. Fisher's exact test was used to compare 2×2 and 2×3 groups, Mann-Whitney U test was used to compare the duration of therapy, and log-rank test was used to analyze the prognosis. P < .05 was considered significant.
Results
Development of Multiplex MB-NGS for ESR1-LBD
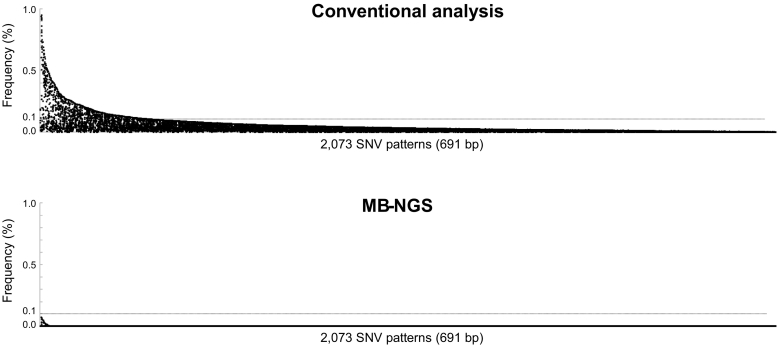
Figure 1 shows the scheme of 11 primers for multiplex MB-NGS that cover 96.8% (691 / 714 bp) of the whole LBD (c.928-1641 / a.310-547) of the ESR1 gene (accession; M12674.1/AAA52399.1). Two gaps (c.1236-1242 and c.1538-1553) included neither hotspots (c.1601-1613, a.V534-D538) nor other frequent mutations such as E380Q (G1138C) and S463P (T1387C). DNA samples from plasma (2 ml) of 10 healthy controls were analyzed by multiplex MB-NGS. The median read depth of each amplicon was 156,800 (range from 69,862 to 578,168), and the median number of MB families of each amplicon was 6043 (range from 2309 to 33,266), which was sufficient to obtain a detection sensitivity of 0.1%. In conventional analysis without MB, the background errors were observed in all of the possible 2073 SNVs, with a median variant allele frequency (VAF) of 0.014% (0.000-1.317%), and 16.1% (334 variations) of them were greater than 0.1%. After MB analyses, 98.8% (2049 SNVs) of these background SNVs were completely removed, and the frequencies of all the remaining 24 (1.2%) SNVs were below 0.1% (median: 0.017%; range: 0.003-0.074%) (Figure 2). Because the current assay targeted as many as 2073 SNVs, a sensitivity test by spike-in mutations could not be performed. Thus, the detection limit of the multiplex MB-NGS assay was set at 0.1% in the following analyses based on the result that the background errors were always <0.1% at any SNVs in controls.
Figure 1.
Scheme of primer design for multiplex MB-NGS.
Eleven sets of primers were designed to cover ESR1-LBD (the light gray box, c.928-1641), and their target regions were indicated by arrows. The two dark gray boxes showed the gaps not covered by these primers, and the coverage was 96.8% (691 / 714 bp). The hatching box represents the hotspot region (c.1606-1614) of ESR1 mutations.
Figure 2.
Background errors of plasma samples from 10 healthy controls.
Plasma DNA from 10 healthy controls was analyzed by multiplex MB-NGS for ESR1-LBD, and the background errors per SNV were compared before (upper) and after (lower) barcode analysis. The frequency of background errors per 2073 different variances were plotted in descending order. Three hundred and thirty-four (16%) variances showed errors with frequency of >0.1% under conventional analysis, and these background errors were completely suppressed below 0.1% after molecular barcode analysis.
ESR1 Mutations in Plasma DNA from Patients with MBC
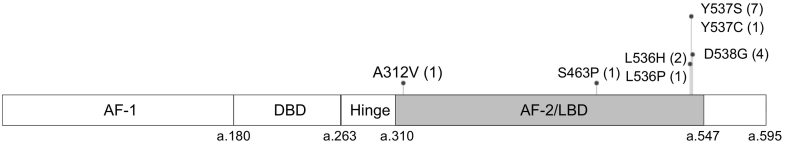
DNA from plasma (2 ml) of 54 patients with MBC was analyzed by multiplex MB-NGS for ESR1-LBD. Seventeen mutations were detected in 13 patients (24%), with VAFs ranging from 0.13 to 10.67%, including six mutations with VAF < 1% (Figure 3, Table 2). Although total sequence reads showed a great variation (range, 339,919-5,639,259) due to different sampling years, adequate MB families (from 9895 to 290,847) were obtained in all samples. Neither insertion nor deletion (In/Del) mutation was detected. The VAFs of 17 mutations obtained by multiplex MB-NGS and conventional analysis showed good correlation with each other (Spearman correlation: r = 0.973). Seventeen mutations included one A312V, one S463P, two L536Hs, one L536P, one Y537C, seven Y537Ss, and four D538Gs. A312V was an unreported mutation that located in a nonhotspot region (Figure 3).
Figure 3.
ESR1 mutations in plasma from 54 patients with MBC detected by MB-NGS.
Plasma DNA from 54 patients with MBC was analyzed by MB-NGS for ESR1-LBD (gray box). Seventeen mutations were detected in 13 patients and indicated by lollipops, including one A312V, one S463P, two L536H, one L536P, one Y537C, seven Y537S, and four D538G mutations. AF-1/2, activation function-1/2; DBD, DNA binding domain; LBD, ligand-binding domain.
Table 2.
Detailed NGS Results of 17 ESR1 Mutations
| Case | AA Change | Nt Change | COSMIC ID | MB-NGS |
Conventional Analysis |
||||
|---|---|---|---|---|---|---|---|---|---|
| Freq. (%) | Mutant Families | Total Families | Freq. (%) | Mutant Reads | Total Reads | ||||
| #026 | L536H | 1607 T > A | #6843697 | 4.47 | 13,003 | 290,847 | 5.46 | 308,101 | 5,639,259 |
| D538G | 1613A > G | #5413588 | 7.09 | 1921 | 27,083 | 7.43 | 99,014 | 1,332,645 | |
| #188 | Y537S | 1610A > C | #5413589 | 10.67 | 1056 | 9895 | 10.90 | 37,059 | 339,919 |
| #209 | S463P | 1387 T > C | #4771561 | 2.00 | 1428 | 71,553 | 2.48 | 100,830 | 4,059,243 |
| D538G | 1613A > G | #5413588 | 4.06 | 1318 | 32,498 | 4.27 | 65,895 | 1,542,075 | |
| #390 | D538G | 1613A > G | #5413588 | 6.82 | 15,173 | 222,588 | 9.11 | 292,095 | 3,206,519 |
| #424 | L536H | 1607 T > A | #6843697 | 3.43 | 1404 | 32,030 | 1.29 | 64,639 | 4,996,862 |
| #432 | Y537S | 1610A > C | #5413589 | 0.61 | 346 | 56,983 | (0.62) | 9847 | 1,596,141 |
| #482 | Y537S | 1610A > C | #5413589 | 0.31 | 313 | 100,469 | (0.31) | 7124 | 2,321,323 |
| #508 | Y537S | 1610A > C | #5413589 | 5.55 | 4256 | 76,715 | 6.06 | 106,362 | 1,755,541 |
| #F479 | A312V | 935C > T | N. A. | 0.26 | 82 | 31,764 | (0.83) | 8530 | 1,033,527 |
| #564 | Y537S | 1610A > C | #5413589 | 1.34 | 407 | 30,302 | 1.35 | 19,169 | 1,419,018 |
| #630 | Y537C | 1610A > G | #1074637 | 0.60 | 380 | 63,338 | (0.60) | 12,701 | 2,114,222 |
| Y537S | 1610A > C | #5413589 | 0.87 | 553 | 63,338 | 1.23 | 25,940 | 2,114,222 | |
| #654 | Y537S | 1610A > C | #5413589 | 5.76 | 8967 | 155,753 | 7.21 | 222,983 | 3,092,747 |
| #744 | L536P | 1607 T > C | #6906109 | 5.80 | 5723 | 98,681 | 6.29 | 303,965 | 4,833,854 |
| D538G | 1613A > G | #5413588 | 0.13 | 48 | 37,709 | (0.10) | 2463 | 2,446,431 | |
N.A., not assigned in breast cancer; Freq., frequency (Freq. < 1% of conventional analysis was described with parentheses since they could not be distinguished from background errors).
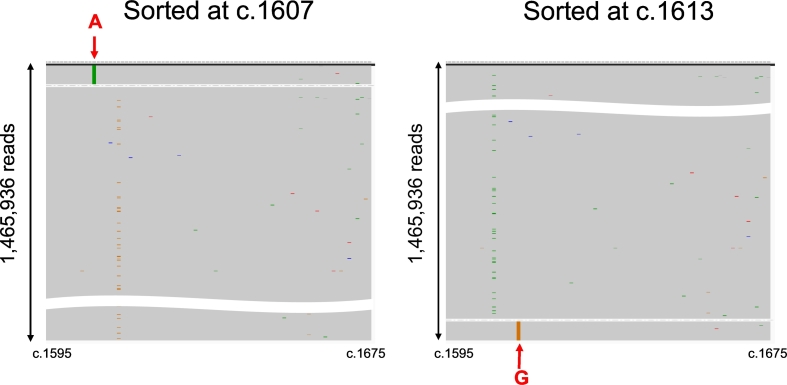
Four patients had double mutations (#026, L536H / D538G; #209, S463P / D538G; #630, Y537C / S; #744, L536P / D538G). Of these four patients, three had double mutations located in the same segment for PCR amplification (#026, #630, and #744), enabling us to analyze whether the double mutations located in the same read or not. We could demonstrate that the double mutations were exclusively located in different reads in all three patients, demonstrating that double mutations existed in different alleles. Representative results for case #26 (double mutations; L536H / D538G) are shown in Figure 4.
Figure 4.
Mutant read mapping of double mutant ctDNA (case #26; L536H / D538G).
Mutant read mappings of a representative case (#26) with double mutant ctDNA were shown. Sequence results were visualized by the Integrative Genomics Viewer and sorted at c.1607 (left) and c.1613 (right). The colored lines represented the mutations, i.e., L536H (c.1607 T > A) in green and D538G (c.1613A > G) in orange. These two mutations are exclusively located in different reads.
ESR1 Mutation and Clinical Course
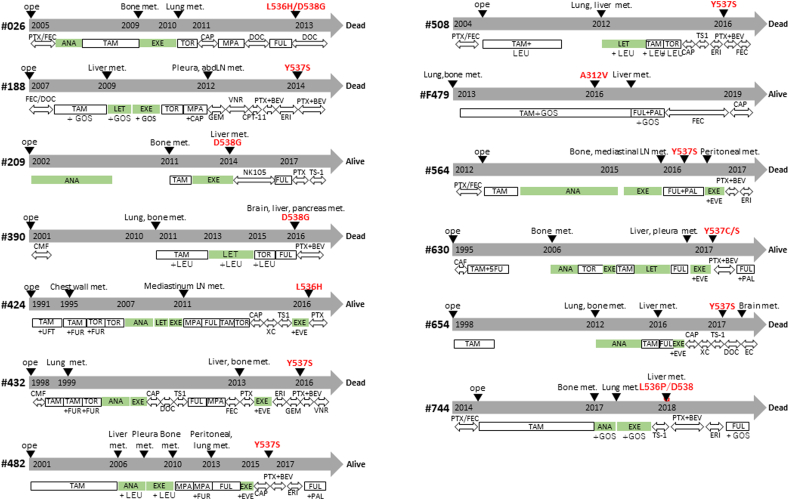
Clinicopathological features of the patients were assessed according to the status of ESR1 mutations (Table 1). The presence or absence of adjuvant AI treatment did not affect the frequency of ESR1 mutations, but they were significantly more frequent in patients with a longer duration of AI treatment under metastatic conditions (30.2 vs. 2.9 months, P < .001) (Table 1). ESR1 mutations tended to be more frequent in patients with liver metastasis, and this trend was significant for Y537S / D538G hotspot mutations (Supplementary Table 3). All ESR1 mutations except one (nonhotspot A312V mutation in patient #479) were detected after AI treatment under metastatic conditions (Supplementary Figure 1).
Supplementary Figure 1.
The clinical course of patients with ESR1 mutations. The clinical course and the treatments are shown in the panel for the 13 patients with ESR1 mutations. Treatments with aromatase inhibitors are highlighted in green. LN, lymph node; met., metastasis; abd. abdominal; PTX, paclitaxel; FEC, fluorouracil, epirubicin and cyclophosphamide; ANA, anastrozole; TAM, tamoxifen; EXE, exemestane; TOR, toremifene; CAP, capecitabine; MPA, medroxyprogesterone acetate; DOC, docetaxel; FUL, fulvestrant; LET, letrozole; GEM, gemcitabine; VNR, vinorelbine; CPT-11, camptothecin 11; BEV, bevacizumab; ERI, eribulin; NK105, paclitaxel-incorporating micellar nanoparticle formulation; CMF, cyclophosphamide, methotrexate and fluorouracil; FUR, doxifluridine; XC, capecitabine and cyclophosphamide; EVE, everolimus; PAL, palbociclib.
Discussion
We have developed the multiplex MB-NGS assay to detect ESR1 mutations across the entire LBD, not limited to hotspots, with a detection sensitivity of 0.1%. In fact, of the 17 mutations detected in the present study, one mutation (A312V) was located outside the hotspots and would have been missed by the dPCR assay targeting the hotspots. Another advantage of the NGS-based assay is its ability to detect the In/Del mutations since In/Del of ESR1 is reported to account for approximately 2% of all ESR1 mutations [9,12,13]. However, in the present study, no In/Del mutations could be detected probably due to the limited number of patients. MB-NGS assay could not analyze unknown rearrangements because the both forward and reverse primers had to be designed on the ESR1 gene. However, the rearrangements of ESR1 gene is reportedly very rare (<1%) in breast cancer [19,20], so it was highly unlikely that it would be detected in our samples.
In line with the previous reports, the patients with hotspot mutations were all treated with AIs under metastatic conditions [4,21]. No ESR1 mutation was found in the patients treated with adjuvant AI only, including those who experienced relapse during or soon after adjuvant AI treatment, indicating that ESR1 mutations exclusively develop during the use of AI for clinically evident metastatic tumors. For the two mutations outside the hotpots, S463P coexisted with D538G, consistent with the previous report [2], and A312V was a noble mutation which was not reported previously and was unique in that it developed in a patient without a history of AI treatment. The biological function of this mutation must be investigated in the future, while in silico analysis (PolyPhen2, SHIT, PROVEAN, and PANTHER) suggests that this mutation is not functional.
It has been reported that ESR1 mutations are associated with liver metastasis [[22], [23], [24]]. Razavi's data [23] suggested that active hotspot mutations such as Y537S and D538G are more likely to occur in liver metastasis, and so we also examined which mutations were correlated with liver metastasis. Consistently with the previous report [23], ESR1 mutations, especially Y537S or D538G, were significantly correlated with liver metastasis, indicating the organotropism of these mutations.
It is reported that most of the double ESR1 mutations are located in different alleles, but they are occasionally located within the same allele [12]. NGS-based assay, but not dPCR, can differentiate whether double mutations are located in the same alleles or not — if they are located in the same segment for PCR amplification. Consistent with this report, in the present study, we could show that all three patients with double mutations had mutations in different alleles. Thus, it is strongly suggested that metastases are composed of a mixture of tumor cells with either mutation alone.
In conclusion, we have developed a multiplex MB-NGS assay for the sensitive and comprehensive detection of mutations across the whole ESR1-LBD and have been able to show the presence of ESR1 mutations outside the hotspots. It is thought that a future study on the detection of ESR1 mutations in cell-free DNA would be better conducted with the multiplex MB-NGS assay, which, unlike dPCR, can detect not only SNV but also In/Del mutations in the whole LBD of the ESR1 gene.
The following are the supplementary data related to this article.
Supplementary Tables
Acknowledgements
This work was supported by JSPS KAKENHI Grant Number JP19K09069. This study was also supported in part by research funding from Novartis. The authors would like to thank Katsuhide Yoshidome (Osaka Police Hospital, Osaka, Japan) for kindly providing the plasma samples of the healthy volunteers examined in this manuscript.
Footnotes
Conflict of Interest
Shinzaburo Noguchi has received honoraria and research funding for other studies from AstraZeneca, Novartis, Pfizer, and Sysmex and has been an advisor for AstraZeneca and Novartis. Naofumi Kagara received honoraria from Novartis. Yasuto Naoi received honoraria from Sysmex and a research grant from AstraZeneca for another study. Masafumi Shimoda received honoraria from Novartis. Kenzo Shimazu received honoraria from AstraZeneca and Sysmex. Seung Jin Kim received honoraria from AstraZeneca, Novartis, and Pfizer. The other authors declare that they do not have a financial relationship with the organizations that sponsored the research.
References
- 1.Oesterreich S, Davidson NE. The search for ESR1 mutations in breast cancer. Nat Genet. 2013;45:1415–1416. doi: 10.1038/ng.2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Toy W, Shen Y, Won H, Green B, Sakr RA, Will M, Li Z, Gala K, Fanning S, King TA. ESR1 ligand-binding domain mutations in hormone-resistant breast cancer. Nat Genet. 2013;45:1439–1445. doi: 10.1038/ng.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Robinson DR, Wu YM, Vats P, Su F, Lonigro RJ, Cao X, Kalyana-Sundaram S, Wang R, Ning Y, Hodges L. Activating ESR1 mutations in hormone-resistant metastatic breast cancer. Nat Genet. 2013;45:1446–1451. doi: 10.1038/ng.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chandarlapaty S, Chen D, He W, Sung P, Samoila A, You D, Bhatt T, Patel P, Voi M, Gnant M. Prevalence of ESR1 Mutations in Cell-Free DNA and Outcomes in Metastatic Breast Cancer: A Secondary Analysis of the BOLERO-2 Clinical Trial. JAMA Oncol. 2016;2:1310–1315. doi: 10.1001/jamaoncol.2016.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fribbens C, O'Leary B, Kilburn L, Hrebien S, Garcia-Murillas I, Beaney M, Cristofanilli M, Andre F, Loi S, Loibl S. Plasma ESR1 Mutations and the Treatment of Estrogen Receptor-Positive Advanced Breast Cancer. J Clin Oncol. 2016;34:2961–2968. doi: 10.1200/JCO.2016.67.3061. [DOI] [PubMed] [Google Scholar]
- 6.O'Leary B, Hrebien S, Morden JP, Beaney M, Fribbens C, Huang X, Liu Y, Bartlett CH, Koehler M, Cristofanilli M. Early circulating tumor DNA dynamics and clonal selection with palbociclib and fulvestrant for breast cancer. Nat Commun. 2018;9:896. doi: 10.1038/s41467-018-03215-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spoerke JM, Gendreau S, Walter K, Qiu J, Wilson TR, Savage H, Aimi J, Derynck MK, Chen M, Chan IT. Heterogeneity and clinical significance of ESR1 mutations in ER-positive metastatic breast cancer patients receiving fulvestrant. Nat Commun. 2016;7:11579. doi: 10.1038/ncomms11579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Toy W, Weir H, Razavi P, Lawson M, Goeppert AU, Mazzola AM, Smith A, Wilson J, Morrow C, Wong WL. Activating ESR1 Mutations Differentially Affect the Efficacy of ER Antagonists. Cancer Discov. 2017;7:277–287. doi: 10.1158/2159-8290.CD-15-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jeselsohn R, Buchwalter G, De Angelis C, Brown M, Schiff R. ESR1 mutations-a mechanism for acquired endocrine resistance in breast cancer. Nat Rev Clin Oncol. 2015;12:573–583. doi: 10.1038/nrclinonc.2015.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Butler TM, Johnson-Camacho K, Peto M, Wang NJ, Macey TA, Korkola JE, Koppie TM, Corless CL, Gray JW, Spellman PT. Exome Sequencing of Cell-Free DNA from Metastatic Cancer Patients Identifies Clinically Actionable Mutations Distinct from Primary Disease. PLoS One. 2015;10 doi: 10.1371/journal.pone.0136407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chu D, Paoletti C, Gersch C, VanDenBerg DA, Zabransky DJ, Cochran RL, Wong HY, Toro PV, Cidado J, Croessmann S. ESR1 Mutations in Circulating Plasma Tumor DNA from Metastatic Breast Cancer Patients. Clin Cancer Res. 2016;22:993–999. doi: 10.1158/1078-0432.CCR-15-0943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chung JH, Pavlick D, Hartmaier R, Schrock AB, Young L, Forcier B, Ye P, Levin MK, Goldberg M, Burris H. Hybrid capture-based genomic profiling of circulating tumor DNA from patients with estrogen receptor-positive metastatic breast cancer. Ann Oncol. 2017;28:2866–2873. doi: 10.1093/annonc/mdx490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O'Leary B, Cutts RJ, Liu Y, Hrebien S, Huang X, Fenwick K, Andre F, Loibl S, Loi S, Garcia-Murillas I. The Genetic Landscape and Clonal Evolution of Breast Cancer Resistance to Palbociclib plus Fulvestrant in the PALOMA-3 Trial. Cancer Discov. 2018;8:1390–1403. doi: 10.1158/2159-8290.CD-18-0264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yanagawa T, Kagara N, Miyake T, Tanei T, Naoi Y, Shimoda M, Shimazu K, Kim SJ, Noguchi S. Detection of ESR1 mutations in plasma and tumors from metastatic breast cancer patients using next-generation sequencing. Breast Cancer Res Treat. 2017;163:231–240. doi: 10.1007/s10549-017-4190-z. [DOI] [PubMed] [Google Scholar]
- 15.Kinde I, Wu J, Papadopoulos N, Kinzler KW, Vogelstein B. Detection and quantification of rare mutations with massively parallel sequencing. Proc Natl Acad Sci U S A. 2011;108:9530–9535. doi: 10.1073/pnas.1105422108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kukita Y, Matoba R, Uchida J, Hamakawa T, Doki Y, Imamura F, Kato K. High-fidelity target sequencing of individual molecules identified using barcode sequences: de novo detection and absolute quantitation of mutations in plasma cell-free DNA from cancer patients. DNA Res. 2015;22:269–277. doi: 10.1093/dnares/dsv010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Masunaga N, Kagara N, Motooka D, Nakamura S, Miyake T, Tanei T, Naoi Y, Shimoda M, Shimazu K, Kim SJ. Highly sensitive detection of ESR1 mutations in cell-free DNA from patients with metastatic breast cancer using molecular barcode sequencing. Breast Cancer Res Treat. 2018;167:49–58. doi: 10.1007/s10549-017-4487-y. [DOI] [PubMed] [Google Scholar]
- 18.Allred DC, Harvey JM, Berardo M, Clark GM. Prognostic and predictive factors in breast cancer by immunohistochemical analysis. Mod Pathol. 1998;11:155–168. [PubMed] [Google Scholar]
- 19.Hartmaier RJ, Trabucco SE, Priedigkeit N, Chung JH, Parachoniak CA, Vanden Borre P, Morley S, Rosenzweig M, Gay LM, Goldberg ME. Recurrent hyperactive ESR1 fusion proteins in endocrine therapy-resistant breast cancer. Ann Oncol. 2018;29:872–880. doi: 10.1093/annonc/mdy025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lei JT, Shao J, Zhang J, Iglesia M, Chan DW, Cao J, Anurag M, Singh P, He X, Kosaka Y. Functional Annotation of ESR1 Gene Fusions in Estrogen Receptor-Positive Breast Cancer. Cell Rep. 2018;24:1434–1444. doi: 10.1016/j.celrep.2018.07.009. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schiavon G, Hrebien S, Garcia-Murillas I, Cutts RJ, Pearson A, Tarazona N, Fenwick K, Kozarewa I, Lopez-Knowles E, Ribas R. Analysis of ESR1 mutation in circulating tumor DNA demonstrates evolution during therapy for metastatic breast cancer. Sci Transl Med. 2015;7 doi: 10.1126/scitranslmed.aac7551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jeselsohn R, Yelensky R, Buchwalter G, Frampton G, Meric-Bernstam F, Gonzalez-Angulo AM, Ferrer-Lozano J, Perez-Fidalgo JA, Cristofanilli M, Gomez H. Emergence of constitutively active estrogen receptor-alpha mutations in pretreated advanced estrogen receptor-positive breast cancer. Clin Cancer Res. 2014;20:1757–1767. doi: 10.1158/1078-0432.CCR-13-2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Razavi P, Chang MT, Xu G, Bandlamudi C, Ross DS, Vasan N, Cai Y, Bielski CM, Donoghue MTA, Jonsson P. The Genomic Landscape of Endocrine-Resistant Advanced Breast Cancers. Cancer Cell. 2018;34:427–438. doi: 10.1016/j.ccell.2018.08.008. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sokol ES, Feng YX, Jin DX, Basudan A, Lee AV, Atkinson JM, Chen J, Stephens PJ, Frampton GM, Gupta PB. Loss of function of NF1 is a mechanism of acquired resistance to endocrine therapy in lobular breast cancer. Ann Oncol. 2019;30:115–123. doi: 10.1093/annonc/mdy497. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Tables