Abstract
Isolated complex III (CIII) deficiencies are among the least frequently diagnosed mitochondrial disorders. Clinical symptoms range from isolated myopathy to severe multi-systemic disorders with early death and disability. To date, we know of pathogenic variants in genes encoding five out of 10 subunits and five out of 13 assembly factors of CIII. Here we describe rare bi-allelic variants in the gene of a catalytic subunit of CIII, UQCRFS1, which encodes the Rieske iron-sulfur protein, in two unrelated individuals. Affected children presented with low CIII activity in fibroblasts, lactic acidosis, fetal bradycardia, hypertrophic cardiomyopathy, and alopecia totalis. Studies in proband-derived fibroblasts showed a deleterious effect of the variants on UQCRFS1 protein abundance, mitochondrial import, CIII assembly, and cellular respiration. Complementation studies via lentiviral transduction and overexpression of wild-type UQCRFS1 restored mitochondrial function and rescued the cellular phenotype, confirming UQCRFS1 variants as causative for CIII deficiency. We demonstrate that mutations in UQCRFS1 can cause mitochondrial disease, and our results thereby expand the clinical and mutational spectrum of CIII deficiencies.
Keywords: mitochondriopathy, mitochondrial complex III deficiency, Rieske iron-sulfur protein, cardiomyopathy, alopecia, mutation, microscale respiratory, mitochondrial import sequence, Q-cycle
Main Text
Complex III (CIII, also called ubiquinol:cytochrome c oxidoreductase or bc1 complex) is an electron transfer enzyme complex; it is the component of the mitochondrial respiratory chain that transports electrons from ubiquinol to cytochrome c. This redox reaction is coupled to proton translocation from the mitochondrial matrix to the intermembrane space via the “Q-cycle,” contributing to the proton gradient required for ATP synthesis.1 Mammalian CIII possesses a symmetrical dimeric structure (CIII2) in which each monomer is composed of 10 different subunits.2, 3, 4 Three of those subunits are catalytic and possess electron transfer properties: CYB, CYC1, and UQCRFS1, whose intermembrane-space-facing C terminus contains an iron-sulfur (2Fe-2S) cluster. UQCRFS1 is synthesized as a pre-protein in the cytosol, and its import into the mitochondrial matrix is directed by its cleavable N-terminal mitochondrial targeting sequence (MTS). Following import, UQCRFS1 is stabilized by the chaperone LYRM7.5,6 Direct binding of the co-chaperone HSC20 to LYRM7 is required for recruitment of the 2Fe-2S transfer complex to the LYRM7-UQCRFS1 intermediate, and ultimately for 2Fe-2S cluster incorporation into UQCRFS1.7 Once UQCRFS1 has acquired its 2Fe-2S cluster, BCS1L translocates and incorporates it into the pre-CIII2 complex, rendering it catalytically active.8,9 Interestingly, cleavage of the UQCRFS1 MTS is carried out only after its incorporation into CIII2.7 Although crystal structures from bovine heart CIII revealed the presence of UQCRFS1 N-terminal-derived peptides in between the core subunits UQCRC1 and UQCRC2,2,9,10 later research showed that their prominent accumulation in the absence of TTC19 would be detrimental to CIII function.11 Hence the current assumption is that during its incorporation, UQCRFS1 is processed in situ, and several peptides containing its MTS remain bound to CIII2. Later, these peptides have to be removed to preserve CIII2 structural integrity and function. This process is facilitated by TTC19.9,11 The functional importance of this turnover still needs to be elucidated, especially in the context of different metabolic demands.9 Within the mitochondrial respiratory chain, CIII2 is forming supercomplexes together with Complex I (CI) and Complex IV (CIV), and the most abundant of these supercomplexes (CI1III2IV1) is termed “respirasome.”12, 13, 14
Isolated CIII deficiencies are among the least frequently diagnosed mitochondrial disorders. They are associated with heterogeneous clinical presentations.15, 16, 17 To date, mutations in genes encoding five subunits, MT-CYB (MIM: 516020),18,19 CYC1 (MIM: 615453),20 UQCRC2 (MIM: 191329),21 UQCRB (MIM: 19330),22 and UQCRQ (MIM: 612080),23 and five assembly factors, UQCC2 (MIM: 614461),24,25 UQCC3 (MIM: 616097),26 LYRM7 (MIM: 615838),6 BCS1L (MIM: 124000, 603358, and 262000),27, 28, 29 and TTC19 (MIM: 615157),30 have been reported in more than 140 individuals.25
With more than 50 published cases, the most frequent causes of CIII deficiency are variants in MT-CYB, typically associated with myopathy and exercise intolerance. This is followed by more than 30 cases with variants in BCS1L that are associated with either Björnstad syndrome or GRACILE syndrome.17,25 Both of these syndromes are severe neurologic and multi-systemic diseases with neonatal onset. Similar phenotypes have been reported in individuals with pathogenic variants in UQCRB, UQCRC2, and CYC1, who presented with neonatal or early infantile onset, recurrent metabolic crises with elevated lactate levels and hypoglycemia, from which they completely and quickly recovered with intravenous glucose. All but one showed normal development and intellect.25 Based solely on clinical presentation, it is impossible to distinguish between subunit or assembly factor defects. All reported affected individuals are following an autosomal recessive mode of inheritance, apart from those with pathogenic variants in the mitochondrial DNA (mtDNA)-encoded MT-CYB that are maternally inherited or occur spontaneously. CIII defects, with the exception of TTC19 deficiency, often present as combined respiratory chain deficiencies in combination with CI and, in some cases, with CIV deficiencies.24,31 One reason could be the formation of supercomplexes, in which the presence of fully assembled CIII would be a prerequisite for the stability or assembly of CI and CIV.25,32
In this study, we report the clinical and molecular findings of two unrelated children with CIII deficiency, lactic acidosis, fetal bradycardia, lactic acidosis, hypertrophic cardiomyopathy, and alopecia totalis (Table 1). We recruited both individuals from the German network for mitochondrial disorders (MitoNET). Their parents provided written informed consent for all aspects of the study and for publication of facial photographs according to the Declaration of Helsinki. The ethical committees of both participating centers (Charité EA2/107/14 and TU Munich 5360/12S) have approved the study.
Table 1.
Clinical Phenotypes of Both Probands Encoded According to the Human Phenotype Ontology (HPO)
| Characteristics and Symptoms | HPO ID | Proband 1 | Proband 2 |
|---|---|---|---|
| Mutation in UQCRFS1 (NM_006003) | NA | homozygous c.215-1G>C | c.41T>A | c.610C>T |
| Effect on translation (NP_005994) | NA | p.Val72_Thr81del10 | p.Val14Asp | p.Arg204∗ |
| Origin | NA | Afghanistan | Germany |
| Gender | NA | male | male |
| Age at onset | NA | congenital | congenital |
| Age at last assessment | NA | 3.5 months | 9 years |
| Age at death | NA | 3.5 months | NA |
| Fetal Development | |||
| Intrauterine growth retardation (<P10) | HP:0001511 | - | + |
| Low birth weight | HP:0001518 | - (59th percentile) | + (3rd percentile) |
| Fetal bradycardia | HP:0001662 | + | + |
| Perinatal Development | |||
| Persistent fetal circulation | HP:0011726 | ND | + |
| Hypothermia | HP:0002045 | + | ND |
| Feeding difficulties | HP:0008872 | + | + |
| Hyperventilation | HP:0002883 | + | ND |
| Metabolism | |||
| Lactic acidosis [highest level] | HP:0003128 | 24 mmol/l | 15 mmol/l |
| Metabolic crises during febrile infections | HP:0004897 | ND | + |
| Cardiovascular System | |||
| Hypertrophic cardiomyopathy | HP:0001639 | + | + |
| Ventricular septal defect | HP:0001629 | - | + |
| Persistent left superior vena cava | HP:0005301 | - | + |
| Pericardial effusion | HP:0001698 | + | ND |
| Motor System | |||
| Muscular hypotonia | HP:0001252 | (+) | + |
| Muscular weakness | HP:0001324 | + | + |
| Delayed motor development | HP:0001270 | ND | + |
| Elevated creatine kinase levels [highest] | HP:0003236 | >5,000 U/l | ND |
| Hematologic System | |||
| Thrombocytopenia | HP:0001873 | (+) | + |
| Normochromic anemia | HP:0001895 | ND | + |
| Abnormality of blood coagulation | HP:0001928 | + | ND |
| Visual System | |||
| Bilateral papilledema | HP:0001085 | ND | + |
| Skin and Appendages | |||
| Alopecia totalis | HP:0007418 | + | + |
| Gastrointestinal System | |||
| Cholelithiasis | HP:0001081 | + | ND |
NA—not applicable, ND—not done or no information available
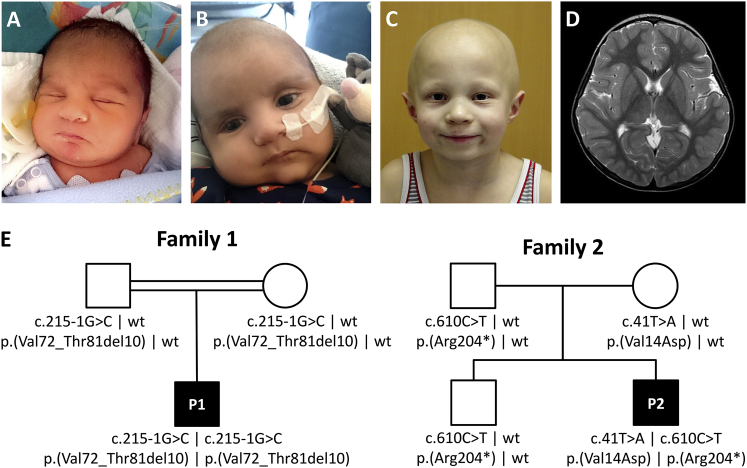
The male proband 1 (P1, Figure 1) was the first child of consanguineous Afghan parents, born at term by emergency Caesarean section due to fetal bradycardia. At the first day of life, physicians noted hypothermia, borderline thrombocytopenia (152/nL; N > 150), lactic acidosis (24 mmol/l; N < 2.0), and elevated creatine kinase levels (>5.000 IU/l; N < 190). Skin and skeletal muscle biopsies were performed on the first day of life. Measurements of respiratory chain enzyme activities showed normal values in the muscle and low-normal values in skin fibroblasts for combined CII+III activity (Tables S1 and S2). Metabolic urine tests revealed increased excretion of ketone bodies and of lactate. Plasma alanine was markedly elevated. Thiamine and coenzyme Q10 supplementation was initiated. Electroencephalogram (EEG) showed slightly pathologic baseline activity initially, but EEG results were normal at 7 weeks of age. Neonatal screening revealed hearing impairment. Echocardiography at the first day of life showed septum and right ventricle hypertrophy, increased right-ventricular pressure (≈65% of systemic pressure), and a patent ductus arteriosus with bi-directional shunting. At day 13, hypertrophic cardiomyopathy had progressed in severity and was treated with metoprolol. Echocardiography at age 2 months showed reduced biventricular function with severe ventricular hypertrophy. The size of the left ventricular posterior wall was 6–7 mm (Z score +5.3), and the left ventricular outflow tract was obstructed (Vmax of 2.9 m/s). Although P1 was born with scalp hair, 8 weeks after birth, his scalp hair had been lost entirely (Figure 1). P1 deceased at the age of 3.5 months from severe hypertrophic cardiomyopathy.
Figure 1.
Clinical Images of Both Probands and Family Pedigrees
(A and B) Proband 1 (P1) at the age of 1 week (A) and of 8 weeks (B) after loss of his scalp hair.
(C) Proband 2 (P2) at the age of 6 years with alopecia totalis of his scalp with only sparse hair growing around his eyebrows.
(D) Entirely normal cranial MRI scan of P2 without characteristic T2-signal-intense areas in the basal ganglia; these areas are often seen in Leigh syndrome and many other mitochondrial disorders.
(E) Pedigrees of both families with UQCRFS1 genotypes.
Proband 2 (P2, Figure 1) is a male and the youngest child of healthy unrelated German parents. An elder brother is healthy. During pregnancy, fetal growth retardation and a persistent left upper vena cava were diagnosed. He was born at 37 weeks of gestation by Caesarean section due to fetal bradycardia. Postnatal complications arose from hypertrophic cardiomyopathy, ventricular septum defect (VSD), persistent fetal circulation, and lactic acidosis, as well as thrombocytopenia and severe normochromic anemia. During early infancy, feeding difficulties, muscular hypotonia, and a moderately delayed psychomotor development became evident. Febrile infections triggered a series of more than 10 severe metabolic crises with high lactate levels of up to 15 mmol/l (N 0.5–2.2). Under normal conditions, serum lactate levels were within the reference range. Cranial MRI performed at the ages of 6 months and 5 years did not reveal any abnormality, especially no signs for Leigh syndrome (Figure 1). Subsequently, the boy‘s condition stabilized, and he was able to walk independently at 23 months of age, while language and cognitive development were adequate for his age. Both VSD and persistent fetal circulation resolved spontaneously in the first year of life while the hypertrophic cardiomyopathy remained stable without impairment of cardiac function. Normocytic anemia with blood hemoglobin of 6.25 mmol/l (N 7.4–9.1) and thrombocytopenia with 178/nL (N 285–510) are persistent, but do not require therapeutic intervention. Assessment of cultured skin fibroblasts revealed isolated CIII deficiency (Table S3). At birth, P2 had very fine and curly hair of the scalp, which he lost entirely during early infancy. Since then, he has total alopecia of the scalp with very fine and sparse hair intermittently growing only to be lost again. In contrast, his eyelashes and eyebrows are present (Figure 1). Microscopic hair analysis did not reveal any abnormalities. At 5 years of age, bilateral papilledema was diagnosed despite normal cerebrospinal fluid pressure (13 cm H2O) and without visual impairment. After initiation of Coenzyme Q10 supplementation with 8 mg/kg BW at the age of 6 years, the boy has remained in a good health for the last 3 years, without any further metabolic crises and with improved exercise tolerance. He displays slightly impaired gross and fine motor skills. His general muscle strength is reduced, but his walking ability in the plain is normal. The clinical phenotypes of both probands using the Human Phenotype Ontology33 terms are described on Table 1.
In order to elucidate the genetic cause of the disease, we performed whole-exome sequencing (WES) on blood cell genomic DNA from both probands. This was done independently at the Institute of Human Genetics, Technical University Munich and at the NeuroCure Clinical Research Center, Charité–Universitätsmedizin Berlin, as described.34,35 Later, both groups connected via GeneMatcher.36
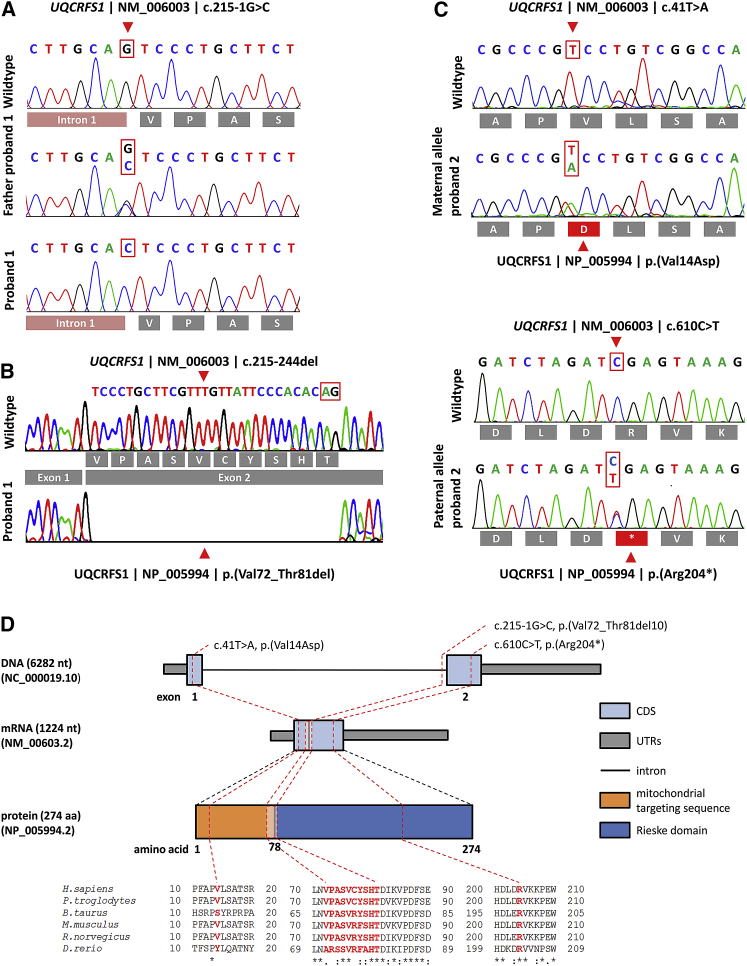
Clinical and biochemical data suggested a mitochondrial disorder with the autosomal recessive mode of inheritance. In P1, WES failed to identify likely pathogenic variants in genes associated with mitochondrial disease, but it did reveal a rare, potentially pathogenic homozygous splice-acceptor-site variant in UQCRFS1 (chr19:g.29,699,066C>G [GRCh37.p11] | c.215-1G>C [RefSeq accession number NM_006003.2]; Figure 2) as the only mitochondrial protein candidate gene.37 Segregation analysis via Sanger sequencing identified both parents as heterozygous carriers of this splice-site variant. To investigate the effect of this splice-site variant on the transcript level, we performed splicing analysis via RT-PCR and subsequent Sanger sequencing. This revealed a splicing defect resulting in the activation of a cryptic downstream splice-site and an in-frame deletion of 30 nucleotides and thus of 10 amino acids at a highly conserved region of the protein (Figure 2).
Figure 2.
Molecular Genetic Findings in Both Probands
(A) Electropherograms of the homozygous c.215-1G>C splice-site variant in P1, which was heterozygous in both parents.
(B) Loss of the splice acceptor site of intron 1 leads to activation of an alternative downstream cryptic splice site (highlighted by a red box) with subsequent loss of 30 bp from the cDNA.
(C) Electropherograms of the compound heterozygous variants, c.41T>A inherited from the mother and c.610C>T from the father. The effect of the mutation on the amino acid sequence is depicted below the electropherograms.
(D) Genomic organization of UQCRFS1 into two exons, the mRNA, and the protein structure depicting the location of the identified variants. The localization of the altered amino acids is highlighted on the protein domain structure. Phylogenetic conservation of the affected amino acid residues is presented in the alignment of homologs across different species. Positions of affected amino acids are highlighted in red. NB; the intronic region is not drawn to scale.
In P2, after assessing the pathogenic potential of variants with MutationTaster2,38 we identified two heterozygous UQCRFS1 variants (chr19:g.29,703,985A>T [GRCh37.p11] | c.41T>A [RefSeq NM_006003.2] | [p.Val14Asp] [RefSeq NP_005994.2] and chr19:g.29,698,670G>A | c.610C>T | [p.Arg204∗]; Figure 2). Segregation analysis via Sanger sequencing showed the c.41T>A variant to be inherited from the mother and the c.610C>T variant from the father. His elder sibling was also tested and discovered to be heterozygous for the c.610C>T variant only. RT-PCR verified that the c.610C>T mutant transcript evaded nonsense-mediated mRNA messenger (NMD) decay (Figure S1) because the premature termination codon (PTC) was located within 3′ of the last exon-exon boundary.39 No additional predicted pathogenic variants were detected for P2 in virtual gene panels for muscle diseases (n = 337 genes) and alopecia (n = 62 genes) (see Supplemental Information). Bioinformatic analysis of p.Val14Asp on the TargetP-2.0 Server40 showed that replacement of the non-polar valine by a negatively charged aspartic acid in the MTS reduces the probability that the MTS will fold correctly into an amphiphilic α-helix.41 All identified UQCRFS1 variants were submitted to ClinVar (see Accession Numbers). They affect regions that are highly conserved across vertebrates (Figure 2) and have neither been reported in public databases (gnomAD) nor in our in-house database, which contains >16,000 exome datasets of individuals with unrelated phenotypes.
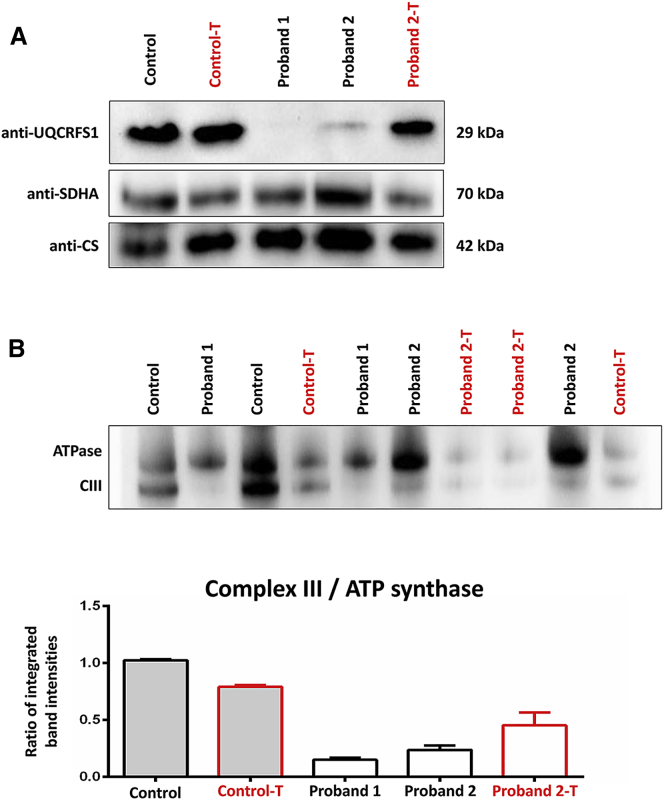
In order to characterize the consequence of the UQCRFS1 variants, we did functional investigations on primary fibroblast cell lines from both probands. Proband and control fibroblasts were cultivated in DMEM media supplemented with 10% FBS, 1% penicillin-streptomycin, and 200 μmol/l uridine at 37°C and 5% CO2. First, we assessed steady-state levels of UQCRFS1 protein by SDS-PAGE of fibroblast cell lysates. Immunoblotting using antibodies against UQCRFS1 (1:1,000, Abcam, ab14746), SDHA (1:3,000, Abcam, ab14715, a subunit of CII, the only respiratory chain complex not involved in the supercomplexes), and citrate synthase (CS) (1:3,000; THP, NBP2-43648) as controls for mitochondrial proteins showed strongly reduced UQCRFS1 band intensities in both probands, and hardly any signal was detected in P1 (Figure 3). Next, we wanted to gain insight into the consequences of UQCRFS1 depletion on CIII composition. To do this, we separated multi-protein complexes from lauryl-maltoside-solubilized fibroblast mitochondria in their native conformation through the use of Blue Native Polyacrylamide Gel Electrophoresis (BN-PAGE).25 In accordance with the crucial role of UQCRFS1 in CIII assembly and function, after we stained for CIII2 (UQCRC2; 1:1,500; Abcam, ab14745) and ATP synthase (ATP5F1A; 1:2,000; Abcam, ab14748) as control, assembled CIII2 was barely detectable in both probands' mitochondria (Figure 3). CIII2 depletion indicated impaired oxidative phosphorylation, which was later confirmed via microscale respirometry (Seahorse) in both fibroblast cell lines.42
Figure 3.
Protein Studies
(A) SDS-PAGE of fibroblast homogenates showing UQCRFS1 and SDHA. Citrate synthase (CS) was used as a mitochondrial loading control. UQCRFS1 protein levels were strongly reduced in P1 and P2 fibroblasts. Transduction rescued UQCRFS1 levels in P2 cells (Proband 2-T) and had no effect on control fibroblasts (Control-T).
(B) BN-PAGE stained for ATP synthase and complex III (CIII) (using anti-ATP5F1A and anti-UQCRC2 antibodies) with densitometric analysis of CIII/ATP synthase ratios in BN-PAGE. Relative CIII reduction is rescued by lentiviral transduction in P2 fibroblasts. P1 fibroblasts reached a high passage number after transduction and did not replicate sufficiently enough to be included in this experiment. Bars depict the mean and SD of two repeated measurements.
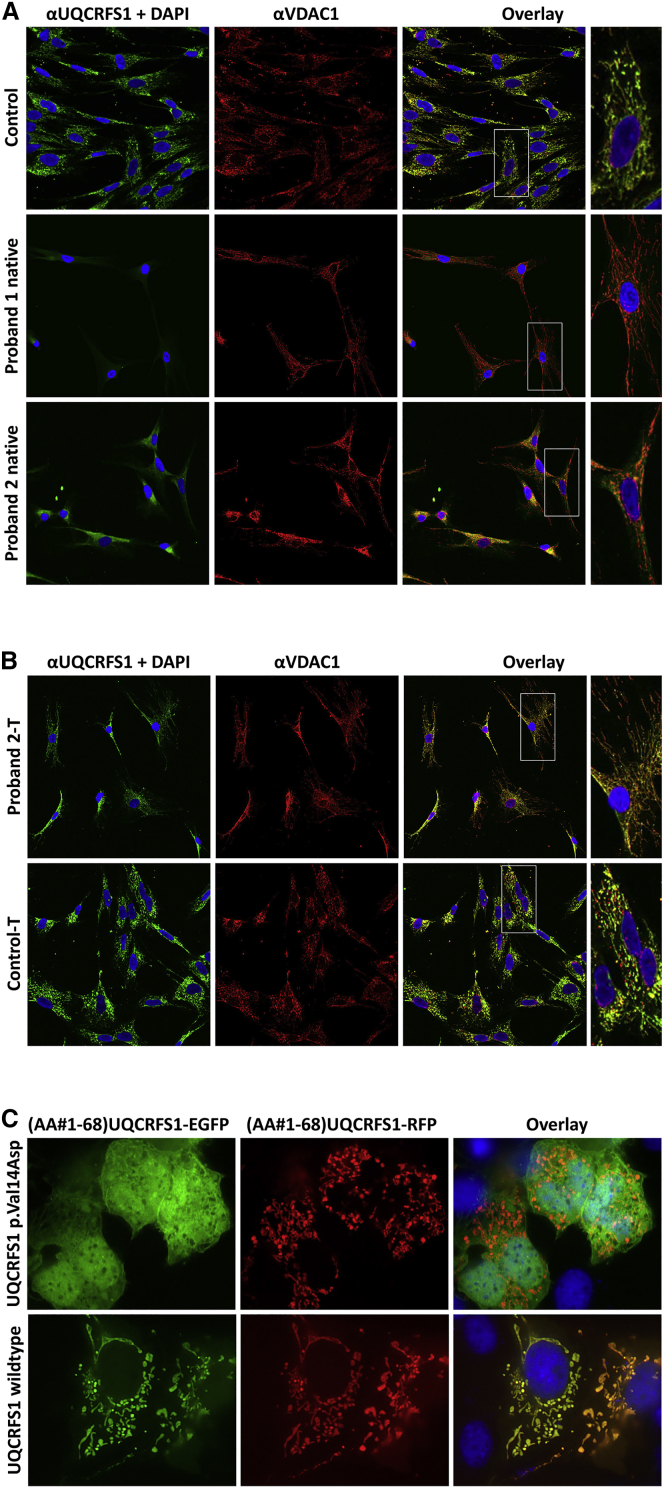
In order to investigate the subcellular distribution of wild-type (WT) and mutant UQCRFS1, we immunostained control and proband fibroblasts with anti-UQCRFS1 (1:100, Abcam, ab14746) and anti-VDAC1 (1:400, Abcam, ab15895) antibodies. The results confirmed a strong reduction of UQCRFS1 protein abundance in the mitochondria of both probands (Figure 4). For verification of the mitochondrial import defect caused by the UQCRFS1 c.41T>A variant, we cloned a set of protein expression plasmids by fusing the 5′ 204 bp of the WT and c.41T>A mutant UQCRFS2 cDNA (encoding the 68 N-terminal amino acids) to the coding sequence of EGFP (p.Val14-EGFP and p.Asp14-EGFP constructs). For controls, we fused the WT sequence to an RFP sequence (p.Val14-RFP construct). We transfected a mixture of p.Val14-EGFP + p.Val14-RFP and of p.Asp14-EGFP + p.Val14-RFP constructs into COS1 cells, and we followed the distributions of the fusion proteins through the use of fluorescence microscopy. The WT red and green fusion proteins had a normal mitochondrial distribution, while the mutant fusion protein failed to be imported and was found to be distributed over the entire cytosol and in the nucleus (Figure 4).
Figure 4.
Immunohistology and Mitochondrial Import
(A) Absent or reduced UQCRFS1 protein levels in both probands’ fibroblasts in comparison to wild-type (WT) control fibroblasts. The anti-VDAC1 antibody signal marks the mitochondria. In WT fibroblasts, anti-UQCRFS1 and anti-VDAC1 signals colocalized, thereby verifying the mitochondrial localization of UQCRFS1 (see magnified inset on the right). Nuclei stained with DAPI.
(B) Lentiviral transduction of UQCRFS1 in the fibroblasts of P2 (Proband 2-T) restores normal localization.
(C) The first 68 amino acids of WT and p.Val14Asp mutant mitochondrial import sequences of UQCRFS1 were fused to the enhanced green fluorescent protein (EGFP). For control, the WT sequence was fused to red fluorescent protein (RFP). After co-transfection of EGFP and RFP constructs into COS1 cells, only those EGFP constructs were imported into the mitochondria that carried the WT import sequence. The p.Val14Asp variant entirely prevented mitochondrial import.
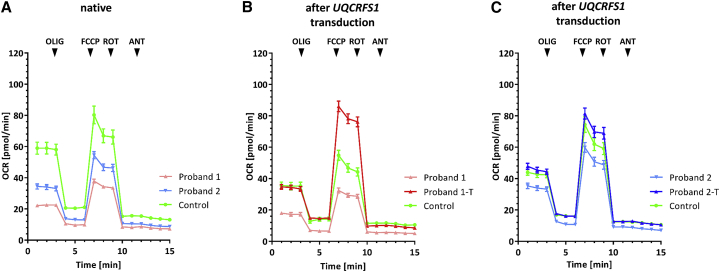
In order to determine whether the identified UQCRFS1 variants are indeed responsible for the CIII defect, we tested whether a WT copy of the UQCRFS1 cDNA could rescue the CIII deficiency in proband fibroblasts after lentiviral transduction.43 Only P2-transduced cells were growing sufficiently well to generate enough material to verify the rescue of all the various phenotypes; later, the growth potential these fibroblasts was also exhausted, which only allowed two repetitions of the experiment with comparative low cellular yields. Functional analysis confirmed that the transduction of cell lines with a WT UQCRFS1 cDNA transcript restored normal UQCRFS1 levels in P2 (Figure 3), restored the CIII assembly defect in P2 (Figure 3), and rescued the respiration defect in both probands’ cell lines (Figure 5 and Figures S2–S4). The Seahorse protocol allows calculation of various parameters pertinent to mitochondrial function (Figure S5). These show significant improvement of basal respiration, ATP production, maximum respiration, and spare respiratory capacity after lentiviral transduction. The maximum respiration could be augmented into the normal range (Figure S5).
Figure 5.
Microscale Respirometry Analysis in Fibroblasts
Combination of two replication experiments of normalized oxygen consumption rates (OCRs) of proband and control fibroblasts as measured with the Seahorse instrument (two biological replicates, 12–16 technical replicates for each condition). Each data point represents mean ± SEMs of measurements from 24–32 wells of technical replicates. The separate replication experiments are depicted in Figures S2–S4. OLIG, oligomycin; FCCP, carbonyl cyanide m-chlorophenyl hydrazone; ROT, rotenone; ANT; antimycine.
(A) Oxygen consumption rates of fibroblasts in the native state.
(B) Rescue of the OCR in fibroblasts of proband P1 after transduction with a lentivirus encoding wild-type (WT) UQCRFS1 (Proband 1-T).
(C) Rescue of the OCR of fibroblasts of P2 after transduction with a lentivirus encoding WT UQCRFS1 (Proband 2-T). NB, we normalized the OCRs of different wells in the Seahorse experiment separately for each plate. For that, we first determined cell numbers in each well through the use of the CyQUANT kit (ThermoFischer) and computed a correction factor for each well by dividing the number of cells in the individual well by the average number of cells in the wells of the whole plate. For normalization of OCR values, we then divided raw OCR readings from each well by its correction factor.
Because P2 had clinically benefitted from Coenzyme Q10 supplementation, we incubated his fibroblasts for 7 h in 0, 10, 30, and 100 μM Coenzyme Q10 prior to measurement of CIII activity and oxygen consumption rate, but we did not find any significant differences in the ensuing oxygen consumption rates (Figure S6).
Mitochondrial diseases associated with isolated CIII deficiency are rare, and due to general technical difficulties in measuring CIII activity in human tissues, they might be underdiagnosed.44 They may also be missed due to unremarkable or statistically insignificant reduction of CIII activity in the tissues under investigation. The increasing application of WES and whole-genome sequencing early in the diagnostic work-up will provide a more unbiased picture about the frequency of CIII deficiencies in the future.45
Considering the crucial role of UQCRFS1 for the catalytic function of CIII, it was no surprise that homozygous Uqcrfs1 loss-of-function variants are embryonically lethal in mice,46 and variants associated with impaired UQCRFS1 function are disease-causing. Here we describe two probands with pathogenic bi-allelic variants in UQCRFS1 leading to CIII deficiency. Their shared clinical features included fetal bradycardia, lactic acidosis, thrombocytopenia, hypertrophic cardiomyopathy, and alopecia totalis. It is estimated that cardiomyopathy occurs in 20%–40% of children with mitochondrial disorders, and hypertrophic cardiomyopathy is the most frequently observed of these cardiac manifestations.47,48 Hypertrophic cardiomyopathy was observed in two individuals with BCS1L variants.49 In addition, different forms of cardiomyopathy (hypertrophic, dilated, and histiocytoid) have been described in three individuals with MT-CYB variants.19,50,51 In one instance, the MT-CYB variant m.15243G>A (p.Gly166Glu) (RefSeq NC_012920.1) from an individual with cardiomyopathy50 disrupted the interaction between cytochrome b and UQCRFS1 in a yeast model of the disease.52
Early onset alopecia totalis is rarely observed in individuals with mitochondrial disease. In the past, 14 individuals were reported with specific hair and skin disorders in a group of 140 children with mitochondrial diseases, and three of those were CIII deficient.53 In addition, pili torti and secondary alopecia are common hair abnormalities of BSC1L-related Björnstad syndrome.29 Besides the above mentioned features, P2 suffers from chronic anemia and both probands suffered from thrombocytopenia, which is a known feature of mitochondrial disorders, especially of Pearson’s syndrome.54 Recent investigations in mice elucidated the importance of the Rieske iron-sulfur protein (RISP) and overall CIII function in hematopoiesis and immune response. Cell-specific loss of RISP in fetal hematopoietic stem cells impaired their differentiation, resulting in anemia and prenatal death.55 Cell-specific Uqcrfs1 knockout in regulatory T cells in mice led to an early-onset fatal inflammatory disease without any effect on the number of regulatory T cells. This suggests a role of CIII in the modulation of protein products relevant for regulation and suppression of immune responses.56
In summary, we have expanded the clinical and mutational spectrum of CIII deficiencies by reporting pathogenic variants in UQCRFS1 that lead to a mitochondrial disorder in two unrelated individuals. Persistent clinical improvement of individual P2 after high-dosage Coenzyme Q10 supplementation points toward a therapeutic option despite the fact that we did not see any increase in the oxygen consumption rate in his fibroblasts. Such supplementation might be beneficial if some residual CIII activity is present, but more research has to be done regarding the pharmacokinetics, subcellular distribution, and effect of Coenzyme Q10 supplementation.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
The authors thank the families for participation in the study. The project was funded by the German Research Foundation und Germany’s Excellence Strategy (EXC-2049—390688087) to M.S.; the E-Rare project GENOMIT (01GM1603 and 01GM1207 to H.P. and O1GML1207 to A.R.); the German network for mitochondrial disorders, MitoNET, (01GM1996B) to H.P.; and the Austrian Science Funds (FWF, I 2741-B26) to J.A.M. A Fellowship through the Graduate School of Quantitative Biosciences Munich (QBM) supports V.A.Y.
Published: December 26, 2019
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2019.12.005.
Accession Numbers
The accession numbers for the data reported in this paper are:
• For proband 1, c.215-1G>C, ClinVar: VCV000619297, and
• For proband 2, c.41T>A, ClinVar: VCV000619501 and c.610C>T, ClinVar: VCV000619499
Web Resources
ExAC Browser, http://exac.broadinstitute.org/
GeneMatcher, https://www.genematcher.org
gnomAD Browser, https://gnomad.broadinstitute.org/
MutationTaster2, http://www.mutationtaster.org/
OMIM, https://www.omim.org/
TargetP-2.0 Server, http://www.cbs.dtu.dk/services/TargetP/
Supplemental Data
References
- 1.Crofts A.R., Holland J.T., Victoria D., Kolling D.R.J., Dikanov S.A., Gilbreth R., Lhee S., Kuras R., Kuras M.G. The Q-cycle reviewed: How well does a monomeric mechanism of the bc(1) complex account for the function of a dimeric complex? Biochim. Biophys. Acta. 2008;1777:1001–1019. doi: 10.1016/j.bbabio.2008.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Iwata S., Lee J.W., Okada K., Lee J.K., Iwata M., Rasmussen B., Link T.A., Ramaswamy S., Jap B.K. Complete structure of the 11-subunit bovine mitochondrial cytochrome bc1 complex. Science. 1998;281:64–71. doi: 10.1126/science.281.5373.64. [DOI] [PubMed] [Google Scholar]
- 3.Xia D., Esser L., Tang W.-K., Zhou F., Zhou Y., Yu L., Yu C.-A. Structural analysis of cytochrome bc1 complexes: implications to the mechanism of function. Biochim. Biophys. Acta. 2013;1827:1278–1294. doi: 10.1016/j.bbabio.2012.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith P.M., Fox J.L., Winge D.R. Reprint of: Biogenesis of the cytochrome bc(1) complex and role of assembly factors. Biochim. Biophys. Acta. 2012;1817:872–882. doi: 10.1016/j.bbabio.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 5.Sánchez E., Lobo T., Fox J.L., Zeviani M., Winge D.R., Fernández-Vizarra E. LYRM7/MZM1L is a UQCRFS1 chaperone involved in the last steps of mitochondrial Complex III assembly in human cells. Biochim. Biophys. Acta. 2013;1827:285–293. doi: 10.1016/j.bbabio.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Invernizzi F., Tigano M., Dallabona C., Donnini C., Ferrero I., Cremonte M., Ghezzi D., Lamperti C., Zeviani M. A homozygous mutation in LYRM7/MZM1L associated with early onset encephalopathy, lactic acidosis, and severe reduction of mitochondrial complex III activity. Hum. Mutat. 2013;34:1619–1622. doi: 10.1002/humu.22441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maio N., Kim K.S., Singh A., Rouault T.A. A Single Adaptable Cochaperone-Scaffold Complex Delivers Nascent Iron-Sulfur Clusters to Mammalian Respiratory Chain Complexes I-III. Cell Metab. 2017;25:945–953.e6. doi: 10.1016/j.cmet.2017.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nobrega F.G., Nobrega M.P., Tzagoloff A. BCS1, a novel gene required for the expression of functional Rieske iron-sulfur protein in Saccharomyces cerevisiae. EMBO J. 1992;11:3821–3829. doi: 10.1002/j.1460-2075.1992.tb05474.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fernandez-Vizarra E., Zeviani M. Mitochondrial complex III Rieske Fe-S protein processing and assembly. Cell Cycle. 2018;17:681–687. doi: 10.1080/15384101.2017.1417707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brandt U., Yu L., Yu C.A., Trumpower B.L. The mitochondrial targeting presequence of the Rieske iron-sulfur protein is processed in a single step after insertion into the cytochrome bc1 complex in mammals and retained as a subunit in the complex. J. Biol. Chem. 1993;268:8387–8390. [PubMed] [Google Scholar]
- 11.Bottani E., Cerutti R., Harbour M.E., Ravaglia S., Dogan S.A., Giordano C., Fearnley I.M., D’Amati G., Viscomi C., Fernandez-Vizarra E., Zeviani M. TTC19 Plays a Husbandry Role on UQCRFS1 Turnover in the Biogenesis of Mitochondrial Respiratory Complex III. Mol. Cell. 2017;67:96–105.e4. doi: 10.1016/j.molcel.2017.06.001. [DOI] [PubMed] [Google Scholar]
- 12.Gu J., Wu M., Guo R., Yan K., Lei J., Gao N., Yang M. The architecture of the mammalian respirasome. Nature. 2016;537:639–643. doi: 10.1038/nature19359. [DOI] [PubMed] [Google Scholar]
- 13.Letts J.A., Fiedorczuk K., Sazanov L.A. The architecture of respiratory supercomplexes. Nature. 2016;537:644–648. doi: 10.1038/nature19774. [DOI] [PubMed] [Google Scholar]
- 14.Wu M., Gu J., Guo R., Huang Y., Yang M. Structure of Mammalian Respiratory Supercomplex I1III2IV1. Cell. 2016;167:1598–1609.e10. doi: 10.1016/j.cell.2016.11.012. [DOI] [PubMed] [Google Scholar]
- 15.Bénit P., Lebon S., Rustin P. Respiratory-chain diseases related to complex III deficiency. Biochim. Biophys. Acta. 2009;1793:181–185. doi: 10.1016/j.bbamcr.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 16.Meunier B., Fisher N., Ransac S., Mazat J.-P., Brasseur G. Respiratory complex III dysfunction in humans and the use of yeast as a model organism to study mitochondrial myopathy and associated diseases. Biochim. Biophys. Acta. 2013;1827:1346–1361. doi: 10.1016/j.bbabio.2012.11.015. [DOI] [PubMed] [Google Scholar]
- 17.Fernández-Vizarra E., Zeviani M. Nuclear gene mutations as the cause of mitochondrial complex III deficiency. Front. Genet. 2015;6:134. doi: 10.3389/fgene.2015.00134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andreu A.L., Hanna M.G., Reichmann H., Bruno C., Penn A.S., Tanji K., Pallotti F., Iwata S., Bonilla E., Lach B. Exercise intolerance due to mutations in the cytochrome b gene of mitochondrial DNA. N. Engl. J. Med. 1999;341:1037–1044. doi: 10.1056/NEJM199909303411404. [DOI] [PubMed] [Google Scholar]
- 19.Schuelke M., Krude H., Finckh B., Mayatepek E., Janssen A., Schmelz M., Trefz F., Trijbels F., Smeitink J. Septo-optic dysplasia associated with a new mitochondrial cytochrome b mutation. Ann. Neurol. 2002;51:388–392. doi: 10.1002/ana.10151. [DOI] [PubMed] [Google Scholar]
- 20.Gaignard P., Menezes M., Schiff M., Bayot A., Rak M., Ogier de Baulny H., Su C.-H., Gilleron M., Lombes A., Abida H. Mutations in CYC1, encoding cytochrome c1 subunit of respiratory chain complex III, cause insulin-responsive hyperglycemia. Am. J. Hum. Genet. 2013;93:384–389. doi: 10.1016/j.ajhg.2013.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miyake N., Yano S., Sakai C., Hatakeyama H., Matsushima Y., Shiina M., Watanabe Y., Bartley J., Abdenur J.E., Wang R.Y. Mitochondrial complex III deficiency caused by a homozygous UQCRC2 mutation presenting with neonatal-onset recurrent metabolic decompensation. Hum. Mutat. 2013;34:446–452. doi: 10.1002/humu.22257. [DOI] [PubMed] [Google Scholar]
- 22.Haut S., Brivet M., Touati G., Rustin P., Lebon S., Garcia-Cazorla A., Saudubray J.M., Boutron A., Legrand A., Slama A. A deletion in the human QP-C gene causes a complex III deficiency resulting in hypoglycaemia and lactic acidosis. Hum. Genet. 2003;113:118–122. doi: 10.1007/s00439-003-0946-0. [DOI] [PubMed] [Google Scholar]
- 23.Barel O., Shorer Z., Flusser H., Ofir R., Narkis G., Finer G., Shalev H., Nasasra A., Saada A., Birk O.S. Mitochondrial complex III deficiency associated with a homozygous mutation in UQCRQ. Am. J. Hum. Genet. 2008;82:1211–1216. doi: 10.1016/j.ajhg.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tucker E.J., Wanschers B.F.J., Szklarczyk R., Mountford H.S., Wijeyeratne X.W., van den Brand M.A.M., Leenders A.M., Rodenburg R.J., Reljić B., Compton A.G. Mutations in the UQCC1-interacting protein, UQCC2, cause human complex III deficiency associated with perturbed cytochrome b protein expression. PLoS Genet. 2013;9:e1004034. doi: 10.1371/journal.pgen.1004034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feichtinger R.G., Brunner-Krainz M., Alhaddad B., Wortmann S.B., Kovacs-Nagy R., Stojakovic T., Erwa W., Resch B., Windischhofer W., Verheyen S. Combined Respiratory Chain Deficiency and UQCC2 Mutations in Neonatal Encephalomyopathy: Defective Supercomplex Assembly in Complex III Deficiencies. Oxid. Med. Cell. Longev. 2017;2017:7202589. doi: 10.1155/2017/7202589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wanschers B.F.J., Szklarczyk R., van den Brand M.A.M., Jonckheere A., Suijskens J., Smeets R., Rodenburg R.J., Stephan K., Helland I.B., Elkamil A. A mutation in the human CBP4 ortholog UQCC3 impairs complex III assembly, activity and cytochrome b stability. Hum. Mol. Genet. 2014;23:6356–6365. doi: 10.1093/hmg/ddu357. [DOI] [PubMed] [Google Scholar]
- 27.de Lonlay P., Valnot I., Barrientos A., Gorbatyuk M., Tzagoloff A., Taanman J.W., Benayoun E., Chrétien D., Kadhom N., Lombès A. A mutant mitochondrial respiratory chain assembly protein causes complex III deficiency in patients with tubulopathy, encephalopathy and liver failure. Nat. Genet. 2001;29:57–60. doi: 10.1038/ng706. [DOI] [PubMed] [Google Scholar]
- 28.Visapää I., Fellman V., Vesa J., Dasvarma A., Hutton J.L., Kumar V., Payne G.S., Makarow M., Van Coster R., Taylor R.W. GRACILE syndrome, a lethal metabolic disorder with iron overload, is caused by a point mutation in BCS1L. Am. J. Hum. Genet. 2002;71:863–876. doi: 10.1086/342773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hinson J.T., Fantin V.R., Schönberger J., Breivik N., Siem G., McDonough B., Sharma P., Keogh I., Godinho R., Santos F. Missense mutations in the BCS1L gene as a cause of the Björnstad syndrome. N. Engl. J. Med. 2007;356:809–819. doi: 10.1056/NEJMoa055262. [DOI] [PubMed] [Google Scholar]
- 30.Ghezzi D., Arzuffi P., Zordan M., Da Re C., Lamperti C., Benna C., D’Adamo P., Diodato D., Costa R., Mariotti C. Mutations in TTC19 cause mitochondrial complex III deficiency and neurological impairment in humans and flies. Nat. Genet. 2011;43:259–263. doi: 10.1038/ng.761. [DOI] [PubMed] [Google Scholar]
- 31.Morán M., Marín-Buera L., Gil-Borlado M.C., Rivera H., Blázquez A., Seneca S., Vázquez-López M., Arenas J., Martín M.A., Ugalde C. Cellular pathophysiological consequences of BCS1L mutations in mitochondrial complex III enzyme deficiency. Hum. Mutat. 2010;31:930–941. doi: 10.1002/humu.21294. [DOI] [PubMed] [Google Scholar]
- 32.Ghezzi D., Zeviani M. Human diseases associated with defects in assembly of OXPHOS complexes. Essays Biochem. 2018;62:271–286. doi: 10.1042/EBC20170099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Groza T., Köhler S., Moldenhauer D., Vasilevsky N., Baynam G., Zemojtel T., Schriml L.M., Kibbe W.A., Schofield P.N., Beck T. The Human Phenotype Ontology: Semantic Unification of Common and Rare Disease. Am. J. Hum. Genet. 2015;97:111–124. doi: 10.1016/j.ajhg.2015.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schottmann G., Seelow D., Seifert F., Morales-Gonzalez S., Gill E., von Au K., von Moers A., Stenzel W., Schuelke M. Recessive REEP1 mutation is associated with congenital axonal neuropathy and diaphragmatic palsy. Neurol. Genet. 2015;1:e32. doi: 10.1212/NXG.0000000000000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kremer L.S., Bader D.M., Mertes C., Kopajtich R., Pichler G., Iuso A., Haack T.B., Graf E., Schwarzmayr T., Terrile C. Genetic diagnosis of Mendelian disorders via RNA sequencing. Nat. Commun. 2017;8:15824. doi: 10.1038/ncomms15824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Elstner M., Andreoli C., Klopstock T., Meitinger T., Prokisch H. Methods in Enzymology. Academic Press; 2009. Chapter 1: The Mitochondrial Proteome Database: MitoP2; pp. 3–20. [DOI] [PubMed] [Google Scholar]
- 38.Schwarz J.M., Cooper D.N., Schuelke M., Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat. Methods. 2014;11:361–362. doi: 10.1038/nmeth.2890. [DOI] [PubMed] [Google Scholar]
- 39.Thermann R., Neu-Yilik G., Deters A., Frede U., Wehr K., Hagemeier C., Hentze M.W., Kulozik A.E. Binary specification of nonsense codons by splicing and cytoplasmic translation. EMBO J. 1998;17:3484–3494. doi: 10.1093/emboj/17.12.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Emanuelsson O., Brunak S., von Heijne G., Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- 41.von Heijne G., Steppuhn J., Herrmann R.G. Domain structure of mitochondrial and chloroplast targeting peptides. Eur. J. Biochem. 1989;180:535–545. doi: 10.1111/j.1432-1033.1989.tb14679.x. [DOI] [PubMed] [Google Scholar]
- 42.Yépez V.A., Kremer L.S., Iuso A., Gusic M., Kopajtich R., Koňaříková E., Nadel A., Wachutka L., Prokisch H., Gagneur J. OCR-Stats: Robust estimation and statistical testing of mitochondrial respiration activities using Seahorse XF Analyzer. PLoS ONE. 2018;13:e0199938. doi: 10.1371/journal.pone.0199938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kremer L.S., Prokisch H. Identification of disease-causing mutations by functional complementation of patient-derived fibroblast cell lines. In: Mokranjac D., Perocchi F., editors. Mitochondria: Practical Protocols. Springer New York; New York, NY: 2017. pp. 391–406. [DOI] [PubMed] [Google Scholar]
- 44.Chretien D., Slama A., Brière J.-J., Munnich A., Rötig A., Rustin P. Revisiting pitfalls, problems and tentative solutions for assaying mitochondrial respiratory chain complex III in human samples. Curr. Med. Chem. 2004;11:233–239. doi: 10.2174/0929867043456151. [DOI] [PubMed] [Google Scholar]
- 45.Stenton S.L., Prokisch H. Advancing genomic approaches to the molecular diagnosis of mitochondrial disease. Essays Biochem. 2018;62:399–408. doi: 10.1042/EBC20170110. [DOI] [PubMed] [Google Scholar]
- 46.Hughes B.G., Hekimi S. A mild impairment of mitochondrial electron transport has sex-specific effects on lifespan and aging in mice. PLoS ONE. 2011;6:e26116. doi: 10.1371/journal.pone.0026116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scaglia F., Towbin J.A., Craigen W.J., Belmont J.W., Smith E.O., Neish S.R., Ware S.M., Hunter J.V., Fernbach S.D., Vladutiu G.D. Clinical spectrum, morbidity, and mortality in 113 pediatric patients with mitochondrial disease. Pediatrics. 2004;114:925–931. doi: 10.1542/peds.2004-0718. [DOI] [PubMed] [Google Scholar]
- 48.Yaplito-Lee J., Weintraub R., Jamsen K., Chow C.W., Thorburn D.R., Boneh A. Cardiac manifestations in oxidative phosphorylation disorders of childhood. J. Pediatr. 2007;150:407–411. doi: 10.1016/j.jpeds.2006.12.047. [DOI] [PubMed] [Google Scholar]
- 49.Al-Owain M., Colak D., Albakheet A., Al-Younes B., Al-Humaidi Z., Al-Sayed M., Al-Hindi H., Al-Sugair A., Al-Muhaideb A., Rahbeeni Z. Clinical and biochemical features associated with BCS1L mutation. J. Inherit. Metab. Dis. 2013;36:813–820. doi: 10.1007/s10545-012-9536-4. [DOI] [PubMed] [Google Scholar]
- 50.Valnot I., Kassis J., Chretien D., de Lonlay P., Parfait B., Munnich A., Kachaner J., Rustin P., Rötig A. A mitochondrial cytochrome b mutation but no mutations of nuclearly encoded subunits in ubiquinol cytochrome c reductase (complex III) deficiency. Hum. Genet. 1999;104:460–466. doi: 10.1007/s004390050988. [DOI] [PubMed] [Google Scholar]
- 51.Hagen C.M., Aidt F.H., Havndrup O., Hedley P.L., Jespersgaard C., Jensen M., Kanters J.K., Moolman-Smook J.C., Møller D.V., Bundgaard H., Christiansen M. MT-CYB mutations in hypertrophic cardiomyopathy. Mol. Genet. Genomic Med. 2013;1:54–65. doi: 10.1002/mgg3.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fisher N., Bourges I., Hill P., Brasseur G., Meunier B. Disruption of the interaction between the Rieske iron-sulfur protein and cytochrome b in the yeast bc1 complex owing to a human disease-associated mutation within cytochrome b. Eur. J. Biochem. 2004;271:1292–1298. doi: 10.1111/j.1432-1033.2004.04036.x. [DOI] [PubMed] [Google Scholar]
- 53.Bodemer C., Rötig A., Rustin P., Cormier V., Niaudet P., Saudubray J.-M., Rabier D., Munnich A., de Prost Y. Hair and skin disorders as signs of mitochondrial disease. Pediatrics. 1999;103:428–433. doi: 10.1542/peds.103.2.428. [DOI] [PubMed] [Google Scholar]
- 54.Finsterer J. Hematological manifestations of primary mitochondrial disorders. Acta Haematol. 2007;118:88–98. doi: 10.1159/000105676. [DOI] [PubMed] [Google Scholar]
- 55.Ansó E., Weinberg S.E., Diebold L.P., Thompson B.J., Malinge S., Schumacker P.T., Liu X., Zhang Y., Shao Z., Steadman M. The mitochondrial respiratory chain is essential for haematopoietic stem cell function. Nat. Cell Biol. 2017;19:614–625. doi: 10.1038/ncb3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weinberg S.E., Singer B.D., Steinert E.M., Martinez C.A., Mehta M.M., Martínez-Reyes I., Gao P., Helmin K.A., Abdala-Valencia H., Sena L.A. Mitochondrial complex III is essential for suppressive function of regulatory T cells. Nature. 2019;565:495–499. doi: 10.1038/s41586-018-0846-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.