Abstract
Congenital hypogonadotropic hypogonadism (CHH) is a rare genetic disorder characterized by infertility and the absence of puberty. Defects in GnRH neuron migration or altered GnRH secretion and/or action lead to a severe gonadotropin-releasing hormone (GnRH) deficiency. Given the close developmental association of GnRH neurons with the olfactory primary axons, CHH is often associated with anosmia or hyposmia, in which case it is defined as Kallmann syndrome (KS). The genetics of CHH are heterogeneous, and >40 genes are involved either alone or in combination. Several CHH-related genes controlling GnRH ontogeny encode proteins containing fibronectin-3 (FN3) domains, which are important for brain and neural development. Therefore, we hypothesized that defects in other FN3-superfamily genes would underlie CHH. Next-generation sequencing was performed for 240 CHH unrelated probands and filtered for rare, protein-truncating variants (PTVs) in FN3-superfamily genes. Compared to gnomAD controls the CHH cohort was statistically enriched for PTVs in neuron-derived neurotrophic factor (NDNF) (p = 1.40 × 10−6). Three heterozygous PTVs (p.Lys62∗, p.Tyr128Thrfs∗55, and p.Trp469∗, all absent from the gnomAD database) and an additional heterozygous missense mutation (p.Thr201Ser) were found in four KS probands. Notably, NDNF is expressed along the GnRH neuron migratory route in both mouse embryos and human fetuses and enhances GnRH neuron migration. Further, knock down of the zebrafish ortholog of NDNF resulted in altered GnRH migration. Finally, mice lacking Ndnf showed delayed GnRH neuron migration and altered olfactory axonal projections to the olfactory bulb; both results are consistent with a role of NDNF in GnRH neuron development. Altogether, our results highlight NDNF as a gene involved in the GnRH neuron migration implicated in KS.
Introduction
Congenital hypogonadotropic hypogonadism (CHH [MIM: 146110, MIM: 147950, MIM: 228300, MIM: 229070, MIM: 244200, MIM: 308700, MIM: 610628, MIM: 612370, MIM: 612702, MIM: 614837, MIM: 614838, MIM: 614839, MIM: 614840, MIM: 614841, MIM: 614842, MIM: 614858, MIM: 614880, MIM: 614897, MIM: 615266, MIM: 615267, MIM: 615269, MIM: 615270, MIM: 615271, and MIM: 616030]) is characterized by infertility and the absence of puberty. It is a rare genetic disorder caused by absent secretion or action of gonadotropin-releasing hormone (GnRH) and is often associated with non-reproductive phenotypes. The most commonly associated phenotype is anosmia, which when combined with CHH is defined as Kallmann syndrome (KS [MIM: 308700, MIM: 614897, and MIM: 613301). However, synkinesia, unilateral renal agenesis, sensorineural deafness, cleft palate, and other phenotypes are also observed.1,2
The genetics of CHH are complex, and mutations are described in more than 401,2 genes. Variable expressivity both within and between families, as well as incomplete penetrance, is often present.1,2 Although first described as a Mendelian disorder, both monogenic and oligogenic inheritance (i.e., mutations in more than one gene) occur in CHH.1, 2, 3 However, in approximately 50% of affected individuals, no mutations could be identified in the known CHH genes.3
To date, the genes implicated in CHH impact GnRH neuron fate, GnRH neuron migration and/or axon projection, GnRH neuron homeostasis, and/or gonadotrope defects. Notably, a unique feature of GnRH neurons is that they originate in the olfactory placode during embryonic development and migrate along the terminal nerve to reach their final destination in the hypothalamus.4, 5, 6 This complex migratory process is orchestrated by an intricate network of genes controlling cell signaling, adhesion, motility, and neurite and axonal elongation.
ANOS1 (formerly known as KAL1 [MIM: 300836]) was the first gene implicated in KS.7,8 ANOS1 encodes anosmin-1, which belongs to the fibronectin-3 (FN3) superfamily. Members of this family also include extracellular-matrix molecules and cell-surface hormone and cytokine receptors,9 and the evolutionarily conserved FN3 domain is typically involved in protein-protein interactions related to cell adhesion, migration, and embryonic development.10 Notably, mutations in several additional genes within this family have been associated with CHH; such genes include AXL [MIM: 109135], FLRT3 [MIM: 604808], LEPR [MIM: 601007], and most recently, DCC [MIM: 120470].2 Thus, we hypothesized that other proteins containing FN3 domains would also be involved in GnRH neuron ontogeny and contribute to the pathogenesis of CHH. Through a gene-based burden analysis of FN3-superfamily genes, we identified NDNF—a neurotrophic factor involved in neuron survival, migration, and neurite outgrowth11—as a locus for CHH.
Subjects and Methods
Subjects
A total of 240 unrelated CHH probands (140 KS and 100 normosmic CHH [nCHH], 179 males and 61 females) were included in the study. 222 individuals were from the Centre Hospitalier Universitaire Vaudois, and 85% were of European descent. The remaining 18 were from the Helsinki University Hospital and were of Finnish origin (See Supplemental Material and Methods). Diagnoses of nCHH and KS were made as previously described,1,12,13 and family members were recruited when possible. All subjects provided written informed consent, and the study was approved by the institutional ethics committee at the University of Lausanne and the Helsinki University Central Hospital.
DNA Sequencing and Bioinformatic Analyses
Genomic DNA was extracted from peripheral blood (or saliva) through the use of commercially available QIAGEN kits according to the manufacturer’s protocol. Whole-exome sequencing (WES), alignment, and variant calling were performed as previously described.14,15 All variants were then annotated with minor-allele frequencies (MAFs) from gnomAD (see Supplemental Material and Methods for details). Protein-truncating variants (PTVs; defined as stop gain, frameshift, and acceptor-donor splice sites ± 2 bp from the exon) with MAF ≤ 0.1% were identified in the 204 FN3-superfamily genes obtained from the Interpro database (IPR003961, see Table S1). A gene-based burden analysis for the FN3-superfamily genes was performed with a two-tailed Fisher’s exact test in CHH probands versus controls (gnomAD). To correct for multiple testing, we set a significance threshold at p < 2.45 × 10−4. Candidate FN3 genes were then prioritized according to the following criteria: (1) a significant enrichment of mutations in the CHH cohort as assessed with the gene-based burden test16 and (2) mutations in FN3 candidate genes that are present in individuals for whom the CHH genes do not contain any mutations that would fully explain their phenotype (see Supplemental Material and Methods). All variants in NDNF were confirmed with bi-directional Sanger sequencing.
Cell-Based Functional Assays
NDNF Protein Expression and Cell-Surface Expression
A double FLAG tag was added to the N terminus of human NDNF (GenBank: NM_024574.3) in the pCDNA3.1 expression plasmid. We used site-directed mutagenesis to introduce the mutations into the construct by using a QuickChange II Site-Directed Mutagenesis Kit (Stratagene) according to the manufacturer’s protocol. To assess the impact of the mutants on the expression and secretion of NDNF, we performed western-blot and cell-surface expression assays as previously described.14,17
Reporter Gene Assay
We used a nuclear factor of activated T cells (NFAT)-luciferase-based reporter bioassay to assess the activity of the wild-type (WT) and mutant NDNF in COS7 cells treated with increasing doses of FGF8 (addgene plasmid 10959).18,19 NFAT is a reporter for the PLCγ/Ca+2 cascade downstream of FGF8/FGFR1 signaling (see Supplemental Material and Methods).
Recombinant NDNF Protein Production
Full-length untagged 568 amino acid recombinant human NDNF protein (Uniprot: Q8TB73) was produced with the QMCF stable episomal expression system after cDNA codon optimization was performed in CHO cells by Icosagen (see Supplemental Material and Methods).
Cell-Culture and Migration Assays
Transwell experiments were performed with the GN11 cell line as previously reported.20 In brief, GN11 cells were seeded on the upper side of 8-mm-pore filters in transwell chambers and incubated for 8 h in 10% fetal bovine serum (FBS) DMEM with or without recombinant NDNF (10, 100, 200, 500 ng/mL; Icosagen). rNDNF was added either to both chambers (n = 5 for each dose) or only in the upper chamber (n = 5 for each dose). GN11 cells that migrated to the lower side of the filter were then fixed in cold 100% methanol for 5 min, and the nuclei were stained with DAPI for subsequent quantitative analysis. Data were compared by one-way ANOVA for multiple comparisons.
We performed three-dimensional matrix migration assays by co-culturing aggregates of GN11 cells with aggregates of COS7 cells21 transfected with plasmids encoding either mutated NDNF or WT NDNF as a control. We created cell aggregates by resuspending GN11 and COS7 cells in 2 μL or 5 μL of growth-factor-free Matrigel (BD Biosciences; 106 cells/mL), respectively, placing them on a culture dish, and inverting them until the gel had set. The droplets were then covered with a growth-factor-reduced Matrigel coating and kept in culture for 72 h. For analysis, the aggregates were fixed with paraformaldehyde (PFA) 4% for 40 min and stained with Alexa Fluor 594 Phalloidin (Invitrogen) for 30 min at room temperature. Photographs were taken for quantitative analysis with ImageJ. For illustration, photomicrographs have been digitally inverted, and a pseudo-color (purple) has been superimposed so that the cell distribution is highlighted.
Zebrafish Experiments
Animal Model
We used the OMP2k:gap-CFPrw034 and TRPC24.5k:gap-Venusrw037 transgenic zebrafish strains22, 23, 24 as reporters to visualize, respectively, the OMP+ and the Trpc2+ neurons and their axonal projections of the zebrafish olfactory system during embryonic development, as previously described.25 We used the GnRH3::GFP strain to visualize the zebrafish GnRH3+ neurons.25,26 A conventional antisense morpholino oligo (MO)-mediated strategy was used for down-modulation of z-ndnf.27,28 This involved two previously validated anti-z-ndnf MOs—one targeting the splice junction between intron 3 and exon 4 and leading to a premature stop codon upstream of the homeodomain and a second that anneals to the ATG start codon and prevents translation.29 As a control, a sequence previously shown to have no effect was used.30 Zebrafish embryos were injected at the one-cell stage with 4 ng of control or anti-z-ndnf MO, and then the number, morphology, and position relative to the olfactory placode of GFP+ cells in the frontonasal piece were examined at 72 hours post-fertilization (hpf), as previously described.25,30
Real-Time qPCR Analysis for Coding mRNAs
Relative mRNA levels of z-OMPa, z-OMPb, z-Trpc2, and z-HoxA10b were determined by real-time qPCR in total RNA samples extracted from the heads of z-ndnf- or control-MO injected zebrafish embryos, as previously described.25 The abundance of z-β actin mRNAs was used for normalization because doing so was previously shown to be the best method.31 The Roche UPS online tool was used for primer design (Supplemental Material and Methods), and the ΔΔCt formula (ABI software version 2.1, Applied Biosystems) was used for data analysis.25
Expression Studies in Mice
Mouse Model
GnRH-GFP32 mice were graciously provided by Dr. Daniel J. Spergel (Section of Endocrinology, Department of Medicine, University of Chicago). Mice were housed under specific pathogen-free conditions in a temperature-controlled room (21°C –22°C) with a 12 h light/dark cycle and ad libitum access to food and water. Animal studies were approved by the Institutional Ethics Committees of Care and Use of Experimental Animals of the University of Lille 2. All experiments were performed in accordance with the guidelines for animal use specified by the European Union Council Directive of September 22, 2010 (2010/63/EU).
Fluorescence-Activated Cell Sorting and Gene-Expression Analysis
GnRH-GFP neurons were isolated from embryos at embryonic days 14.5 and 18.5 (E14.5 and E18.5) through the use of a previously validated fluorescence-activated cell sorting (FACS) protocol.33 Reverse transcription and linear pre-amplification were performed, and Ndnf expression was measured by qRT PCR with Taqman expression assays. Data were compared by one-way ANOVA for multiple comparisons and a subsequent Tukey’s least-significant-difference post-hoc test. A detailed description is provided in the Supplemental Material and Methods.
Collection and Processing of Human Fetuses
Tissues were made available in accordance with French bylaws (Good Practice Concerning the Conservation, Transformation, and Transportation of Human Tissue to Be Used Therapeutically, published on December 29, 1998). The studies on human fetal tissue were approved by the French agency for biomedical research (Agence de la Biomédecine, Saint-Denis la Plaine, France, protocol n°: PFS16–002). Non-pathological human fetuses (7 and 11 gestational weeks (GW), n = 3) were obtained from voluntarily terminated pregnancies after written informed consent was obtained from the parents (Gynaecology Department, Jeanne de Flandre Hospital, Lille, France). Fetuses were fixed by immersion in 4% PFA at 4°C for 5 days. The tissues were then cryoprotected in PBS containing 30% sucrose at 4°C overnight, embedded in Tissue-Tek OCT compound (Sakura Finetek), frozen in dry ice, and stored at −80°C until sectioning. Frozen samples were cut serially at 20 μm with a Leica CM 3050S cryostat (Leica Biosystems Nussloch GmbH) and immunolabeled, as described above and as previously described,34,35 with the following primary antibodies: guinea pig anti-GnRH (dilution 1:10000), a kind gift from Erik Hrabovszky (Institute of Experimental Medicine, University of Budapest); monoclonal mouse TUJ1 (Sigma, 1:1000); and rabbit polyclonal NDNF (1:200; Cat: ARP68872_P050, Aviva Systems Biology).
Ndnf Knockout Mice
A Ndnf knockout (KO) mouse strain on the C57BL/6N genetic background was obtained from a NorCOMM B6N-derived C2 ES cell line generated by Geoffrey Hicks at the University of Manitoba (NorCOMM design ID N02206). Most of the coding part of Ndnf was deleted by homologous recombination. The targeting cassette replaced part of intron 3, the coding sequence of exon 4, and part of the 3′ UTR of the gene with an E. coli lacZ reporter construct, thus resulting in a non-functional gene product. Microinjections and production of KO mice were carried out at the Biocenter Oulu Transgenic Core Facility. Genotyping of the mice was performed as described in the Supplemental Material and Methods. The mice were housed with a 12 h light/dark cycle and ad libitum access to food and water. Both heterozygous and homozygous Ndnf KO mice appeared grossly normal and were able to produce viable pups.
All experiments were performed according to the guidelines for animal experiments at the University of Helsinki and under a license from the Finnish Review Board for Animal Experiments.
iDISCO Whole-Mount Staining
Experiments were performed as previously described36 and are detailed in Supplemental Material and Methods.
GnRH Cell Counting
Embryos were harvested at embryonic day 13.5 from Ndnf transgenic mice (wild-type, n = 5; heterozygous, n = 7; and homozygous, n = 4). Heads from the embryos were washed thoroughly in cold 0.01 M PBS, preserved in fixative solution (4% PFA, 0.01 M PBS [pH 7.4]] for 6–8 h at 4°C, and immunolabelled for GnRH and peripherin through the use of iDISCO as described above. Primary antibodies used for these experiments were guinea pig anti-GnRH (dilution 1:10000), which was a kind gift from Erik Hrabovszky (Institute of Experimental Medicine, University of Budapest) and rabbit polyclonal anti-peripherin (Millipore, 1:1000). Quantitative analysis of GnRH neuronal number and location was performed over four regions: nose, olfactory bulb, ventral forebrain, and cortex. Both coronal and sagittal views were used for confirming accurate GnRH neuron localization, and counting was performed every 20 planes on the 4× zoom tiff series with Fiji and the Cell Counter plug-in.
Statistical Analysis
Sample sizes for neuroanatomical studies, gene expression, and in vitro motility analyses were estimated on the basis of prior experience and similar published studies. For the mouse work, typically, embryos taken from at least two different litters for each group were used.
Quantitative RT-PCR gene-expression data were analyzed with SDS 2.4.1 and Data Assist 3.0.1 software (Applied Biosystems). All other analyses were performed with Prism 5 (GraphPad software). Datasets were assessed for normality (Shapiro-Wilk test) and variance. When appropriate, a one-way ANOVA followed by Bonferroni or Dunnets’s post-hoc correction was performed as indicated in the figure legends. Exact p values and adjusted p values are given in the figure legends where possible. α was set at 0.05 for all experiments except where otherwise specified.
Results
Gene-Based Burden Testing Identifies Neuron-Derived Neurotrophic Factor as a Putative CHH-Associated Gene
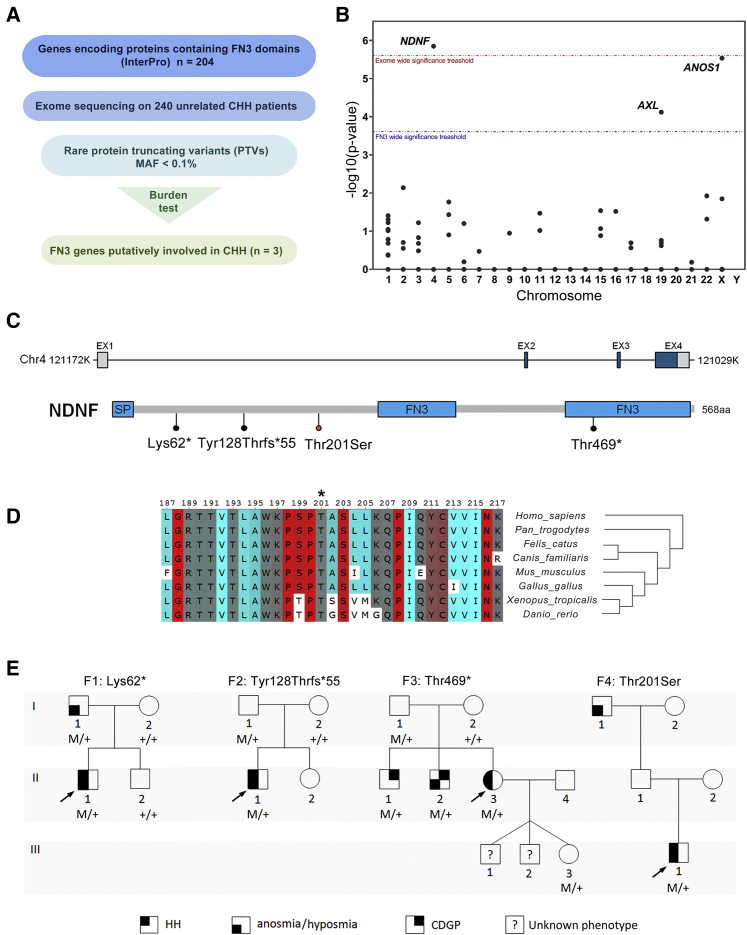
Given that several CHH-associated genes encode proteins containing FN3 domains, WES data from 240 unselected CHH-affected individuals (143 KS and 97 normosmic CHH) were filtered for rare (MAF < 0.1% in gnomAD) PTVs in the 204 FN3-superfamily genes from the Interpro database (Table S1). Next, a gene-based burden analysis comparing the frequency of PTVs in affected individuals to that of individuals in the gnomAD control database was performed. A statistical enrichment for PTVs in CHH-affected probands was demonstrated for three FN3 genes (Figures 1A and 1B), including two known CHH-associated genes (ANOS1, p = 2.88 × 10−6; AXL, p = 7.46 × 10−5) and one novel gene, NDNF [MIM: 616506] (p = 1.40 × 10−6). Notably, none of the probands carrying NDNF PTVs had mutations in the known CHH-affected genes.
Figure 1.
Exome Sequencing Identifies NDNF Mutations in KS Patients
(A) Filtering strategy for identifying novel candidate CHH-related genes.
(B) Manhattan plot of gene-based burden testing for FN3-superfamily genes. The red and blue dotted lines indicate the threshold for statistical significance after correction for multiple testing (red: n = 20,000, p < 2.5 × 10−6; blue: n = 204, p < 2.45 × 10−4).
(C) Schematic depiction of NDNF mRNA and protein; identified mutations are indicated. SP, signal peptide; FN3, fibronectin type 3 domain.
(D) Alignment of partial protein sequences of NDNF orthologs shows that the p.Thr201Ser mutation (asterisk) affects evolutionarily conserved amino acid residues across vertebrates (colored highlighting, turn propensity).
(E) Pedigrees of four families harboring NDNF mutations. Phenotypes are indicated by hatched symbols as shown in the legend (bottom), and arrows indicate the probands. HH, hypogonadotropic hypogonadism; CDGP, congenital delay of growth and puberty.
Individuals with KS Harbor NDNF Mutations
In addition to the three heterozygous PTVs identified in CHH probands in the burden analysis (GenBank: NM_024574.3; c.184A>T [p.Lys62∗], c.381del [p.Tyr128Thrfs∗55], c.1406G>A [p.Trp469∗]), we identified one additional heterozygous missense mutation (c.602C>G [p.Thr201Ser]) predicted to be deleterious in 4/6 programs (PolyPhen-2, MutationTaster, LRT, and CADD).
The genetic and clinical information of the probands harboring NDNF mutations is summarized in Table 1 and Figure 1E. Notably, all four probands exhibit severe GnRH deficiency and anosmia (related to KS). In addition to prepubertal testes, the male probands had cryptorchidism and micropenis, suggesting an abnormal GnRH activation during mini-puberty.2 The female proband presented with primary amenorrhea at age 17 (see the Supplemental Information for a description of affected individuals). Furthermore, three of these probands’ family members (F1:I,1; F3:II,1; and F3:II,2) also carry NDNF mutations and have partial phenotypes (i.e., anosmia or constitutional delay of puberty), suggesting an autosomal-dominant inheritance with variable expressivity (Figure 1E). The presence of asymptomatic carriers (F2:I,1; F3:I,1; and F3:III,3) is consistent with incomplete penetrance.
Table 1.
Phenotype and Genotype of Four Probands with Heterozygous NDNF Mutations
| Subject | Sex | NDNF Mutation | MAF | Diagnosis | Inheritance | Other Phenotypes |
|---|---|---|---|---|---|---|
| F1:II,1 | M | p.Lys62∗ | absent | KS | familial | micropenis crytorchidism |
| F2: II,1 | M | p.Tyr128Thrfs∗55 | absent | KS | sporadic | cryptorchidism |
| F2:II,3 | F | p.Trp469∗ | absent | KS | familial | |
| F3:III,1 | M | p.Thr201Ser | 0.10% | KS | familial | micropenis crytorchidism |
Abbreviations are as follows: MAF, minor-allele frequency in gnomAD; KS, congenital hypogonadotropic hypogonadism plus anosmia (Kallmann syndrome); N/A: no DNA available; M, male; F, female.
Overall, NDNF mutations are present in nearly 2% of the CHH cohort (4/240). When only KS probands are considered, the frequency increases to ~3% (4/140).
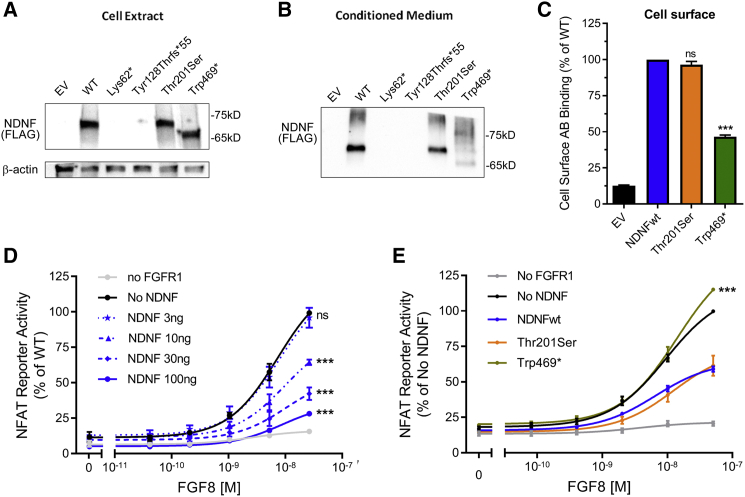
Truncating NDNF Mutations Result in Loss of Function In Vitro
To evaluate the functionality of NDNF mutants in vitro, we assessed COS7 cells that had been transiently transfected with WT and mutant tagged NDNF cDNA for ectopic NDNF protein synthesis and secretion. Western-blot analysis of WT-transfected COS7 cells showed that NDNF protein is detected in both the conditioned medium and in the cell extract (Figures 2A and 2B). A radiolabeled-antibody binding assay showed expression at the cell surface (Figure 2C), suggesting that NDNF has either a direct or an extracellular-matrix-mediated interaction with cell-surface proteins.
Figure 2.
Truncated NDNF Variants Result in a Loss of Function In Vitro
(A and B) Representative experiments showing ectopic expression and secretion of NDNF protein. COS7 cells were transiently transfected with 5 or 100 ng of WT or mutant NDNF and analyzed by western blotting. The Lys62∗ and the Tyr128Thrfs∗55 mutants were not detected. Thr201Ser and Trp469∗ were expressed at levels similar to those of the WT (means of five independent experiments).
(C) Cell-surface antibody binding of NDNF protein (means of four independent experiments). The expression of the truncated mutant Trp469∗ was decreased, whereas Thr201Ser was expressed at levels similar to those of the WT. ∗∗∗p < 0.001; ns, not significant.
(D) NDNF suppresses FGF8 signaling in a dose-dependent manner. COS7 cells were transiently co-transfected with NFAT-luciferase reporter, FGFR1, and increasing doses of WT NDNF and treated with FGF8. ∗∗∗p < 0.001; ns, not significant.
(E) Trp469∗ fails to inhibit FGF8 signaling. COS7 cells were transfected as described with 10 ng of WT or mutant NDNF. ∗∗∗p < 0.001; ns, not significant.
Among the PTVs, the Lys62∗ and Tyr128Thrfs∗55 mutants were not detected in whole cell extract or conditioned medium, suggesting disrupted protein synthesis or rapid degradation. In contrast, the Trp469∗ mutant is expressed in both the cell extract and conditioned medium but exhibits a reduced expression at the cell surface (50%). The Thr201Ser mutant displays no overall expression alterations (Figures 2A–2C).
We further examined the effect of WT NDNF on PLCgamma/Ca2+ pathway activation after FGF8 stimulation. As previously shown, FGF8 stimulation of FGFR1-transfected COS7 cells elicits a dose-dependent activation of the NFAT-luciferase reporter (Figure 2D).37 Notably, when WT NDNF is co-transfected, the FGF8-induced NFAT reporter activity is inhibited in a dose-dependent fashion (Figure 2D). This assay was then used for evaluating the functionality of the expressed NDNF mutants. Whereas WT NDNF suppressed FGF8 signaling by 30%, the Trp469∗ mutant had no effect on FGF8 signaling, consistent with loss of function (Figure 2E). The Thr201Ser mutant and the WT suppressed FGF8 signaling similarly. Taken together, these combined in vitro results validate the three truncated mutants as causing a loss of function.
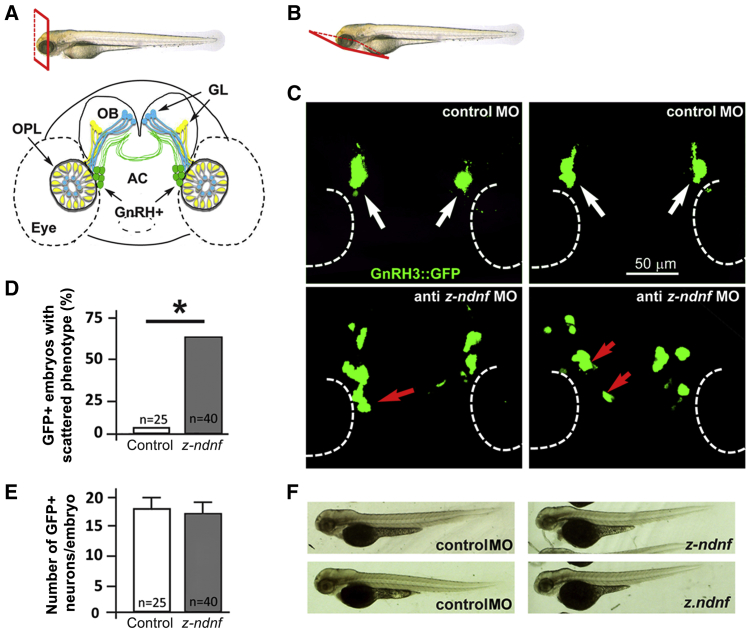
z-ndnf Contributes to GnRH Neuron Migration in Zebrafish
Using a previously validated MO-based strategy,29 we depleted z-ndnf in zebrafish embryos and observed no gross morphological defects. We therefore depleted z-ndnf in GnRH3::GFP transgenic embryos to specifically examine the number, position, and morphology of GnRH3-GFP+ neurons. Injection of anti-z-ndnf MO did not affect GnRH3-GFP+ cell number (average 16 ± 1 per embryo at 72 hpf) or GFP fluorescence intensity (used here as a reporter of GnRH3 promoter). However, it did affect GnRH3-GFP+ neuron positioning along the known migratory route of the terminal nerve (Figure 3). Indeed, GnRH3-GFP+ neurons in embryos treated with control MO form a discrete aggregate between the olfactory placodes and the olfactory bulbs. In contrast, the z-ndnf embryos frequently exhibit severely mis-positioned or scattered GnRH3-GFP+ neurons (i.e., > 2 mis-positioned neurons in one half-embryo; 22/40 embryos [54%] in z-NDNF-MO versus 1/25 embryos in controls [4%]), consistent with a specific role of NDNF in the migration of GnRH neurons.
Figure 3.
Depletion of z-ndnf in Zebrafish Resulted in Altered GnRH Neuron Position
(A) Schematic depiction showing the frontal view position of GnRH3::GFP+ neurons (green) relative to the olfactory placode (OPL), the olfactory bulb (OB), and the olfactory nerves (yellow and blue). The anterior commissure (AC) is shown at the base of the OB, GL, and glomeruli.
(B) Schematic depiction illustrating the ventral view used for fluorescence imaging.
(C) Micrographs of GnRH3:GFP zebrafish embryos at 72 hpf. The upper panels depict embryos injected with a control MO showing no significant alteration (white arrows), and the lower panels show embryos injected with anti-z-ndnf MO. The red arrows indicate mis-positioned GFP+ neurons.
(D) Percentage of embryos with “scattered GnRH neuron” phenotype in control versus anti-z-ndnf MOs, p < 0.05.
(E) Number of GFP+ neurons per embryo; there is no significant difference between the two groups.
(F) Whole-mount bright-field micrographs of control and injected embryos; The image shows normal embryonic morphology and size at 72 hpf.
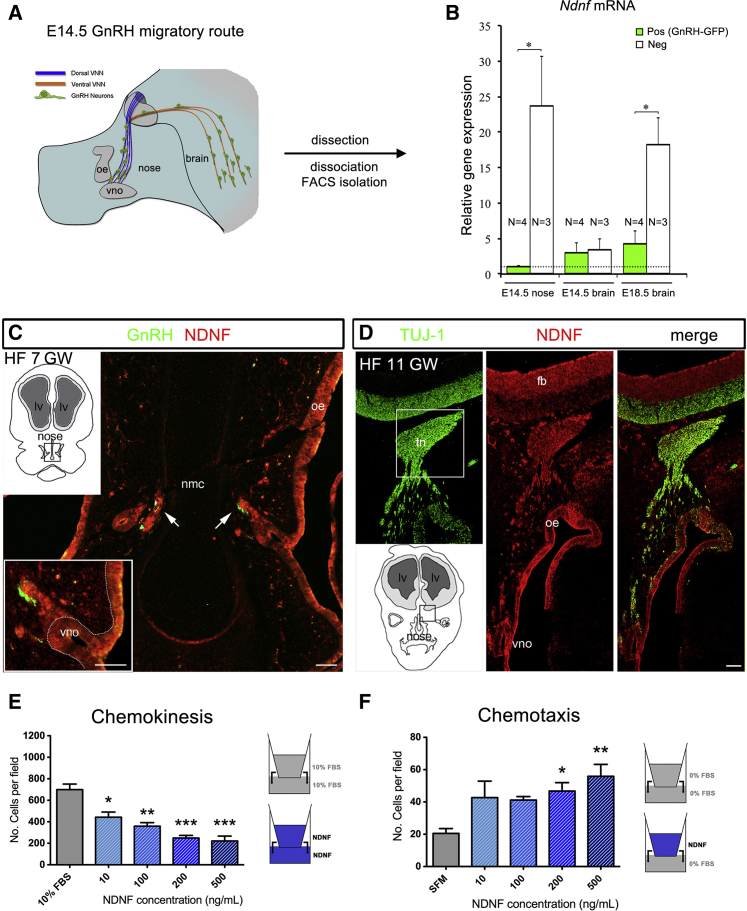
NDNF Is Expressed along the Migratory Route of Embryonic GnRH Cells in Mice and Humans
To determine whether Ndnf is expressed along the GnRH migratory pathway during mammalian embryonic development, we used a GnRH::GFP mouse model combined with FACS and qRT-PCR (Figures 4A and 4B). At E14.5, GnRH neurons are distributed throughout their migratory route, with approximately half in the nose and half in the brain (Figure 4A). Ndnf mRNA expression was very low in GnRH-GFP migratory cells in the nose, whereas Ndnf was strongly expressed by GFP-negative cells (e.g., olfactory and vomeronasal neurons, mesenchymal cells, and endothelial cells; Figure 4B). In the basal forebrain, Ndnf levels were low in both GnRH neurons and GFP-negative cells, suggesting that the major source of Ndnf at this stage is the nasal compartment. At E18.5, the GnRH migratory process is largely complete, and the majority of GnRH neurons have reached their target location in the hypothalamus. At this time point, Ndnf expression was significantly higher in GFP-negative cells than in GnRH neurons (Figure 4B).
Figure 4.
NDNF Expression Delineates the Migratory Route of Embryonic GnRH Cells in Mice and Humans and Repels Immortalized GnRH Cell Migration
(A) Schematic depiction of the GnRH migratory pathway in E14.5 mouse embryos (sagittal view).
(B) GFP-positive GnRH neurons and GFP-negative cells were isolated by FACS from the nasal region of E14.5 embryos (n = 3 or 4) and from the developing preoptic area of E14.5 (n = 3 or 4) and E18.5 (n = 3 or 4). Real-time PCR analysis helped to determine the expression levels of Nndf mRNA in GFP-positive and -negative cells. Values are expressed relative to values, set at 1, in GnRH neurons at E14.5 and are shown as means ± SEM. One-way ANOVA followed by Tukey’s Honest Significant Difference post-hoc analysis was used for comparing groups. ∗p < 0.05.
(C) Representative coronal section of a human fetus (HF) at 7 gestational weeks. The section is immunolabeled for GnRH and NDNF (n = 3). nmc, nasal midline cartilage; vno, vomeronasal organ.
(D) Representative coronal section of a human fetus at 11 gestational weeks. The section is immunolabeled for TUJ1 and NDNF (n = 3). tn, terminal nerve; fb, forebrain; oe, olfactory epithelium.
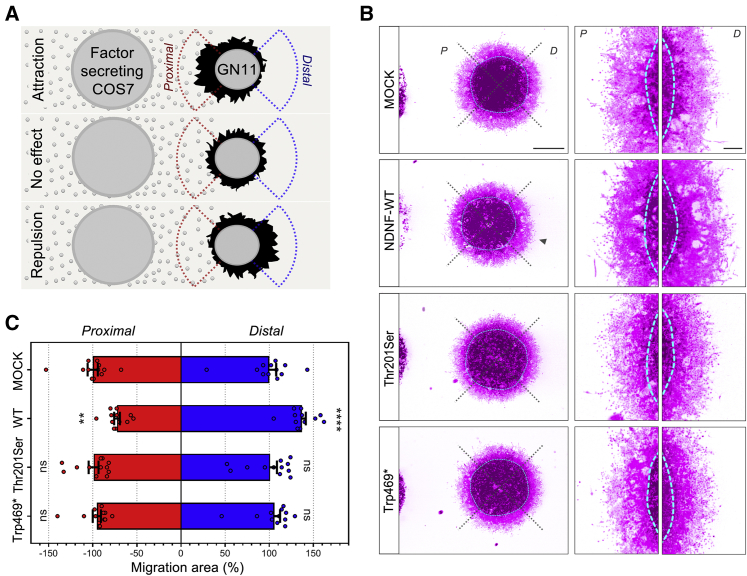
(E) For migration analysis, GN11 cells were used in a transwell assay in the presence or absence of recombinant NDNF (blue, both chambers).
(F) GN11 migration with NDNF in the upper chamber only (n = 5 for each treatment condition); ∗: p < 0.05, ∗∗: p < 0.01, ∗∗∗: p < 0.001, one-way ANOVA followed by Tukey’s post-hoc test. Scale bars in (C) represent 30 μm; scale bars in (D) represent 100 μm.
Using double-immunofluorescence, we next evaluated the expression pattern of NDNF in the nasal compartment of human fetuses (7 and 11 GW) together with the expression of GnRH and TUJ1, a marker of the developing olfactory and vomeronasal systems and differentiated neurons (Figures 4C and 4D). NDNF was found in the developing olfactory epithelium, in the vomeronasal organ (VNO), and along the vomeronasal/terminal nerve (Figures 4C and 4D). At 7 GW, GnRH neurons were observed leaving the vomeronasal organ in close morphological association to NDNF-immunoreactive axons (Figure 4C). However, NDNF expression was absent from human migratory GnRH neurons.
NDNF Repels Migrating GnRH Cells in vitro
Since NDNF is highly expressed by olfactory and vomeronasal epithelia when GnRH neurons emerge from the VNO, we hypothesized that this soluble molecule could trigger GnRH directional migration by repelling these neurons from their site of origin and differentiation toward the forebrain (fb). We used GN11 cells, an in vitro model of migratory GnRH cells,38 in a transwell plate assay to evaluate the response to increasing doses of recombinant human NDNF (10, 100, 200, 500 ng/mL). We added NDNF to either the upper chamber alone or the upper and lower compartments of a transwell plate (Figures 4E and 4F) to specifically evaluate whether NDNF affects GN11 motility. This action can be through either chemokinesis or directional chemotaxis.
Exposure to increasing doses of NDNF plated in both chambers resulted in a significant reduction of GN11 cell migration to the bottom of the polycarbonate filter in comparison to what occurred in control conditions. This could be due to either an inhibition of cell motility or a repellent NDNF activity that neutralizes oriented migration when NDNF is present in both chambers. To investigate this further, we performed transwell migration assays by plating the same doses of NDNF in only the upper chamber in serum-free conditions (SFM). NDNF produced a significant increase in migration as compared to that in control conditions (Figure 4F). In combination with the expression data, these results suggest that NDNF acts as a chemorepellent molecule on GnRH cell migration.
To assess the functional consequences of NDNF mutants on GN11 directional migration, we performed three-dimensional matrix assays by co-culturing aggregates of GN11 cells for 72 h with aggregates of COS7 cells transiently transfected with a mock construct or with WT or mutant NDNF (Figure 5). Compared to the mock treatment, the WT NDNF GN11 cells displayed an asymmetrical distribution, with fewer cells on the proximal side and more cells on the distal side (Figure 5B). This asymmetrical migration confirmed the chemo-repulsive action of NDNF on immortalized GnRH neurons. However, this directional effect was prevented when GN11 cells were co-cultured with COS7 cells expressing any of the NDNF mutants. These results show that WT NDNF acts as a chemorepellent for GnRH cell migration and that this effect is lost in NDNF mutants, including the Thr201Ser mutant.
Figure 5.
NDNF Mutants Fail to Induce GnRH Cell Migration In Vitro
(A) Schematic illustration of the three-dimensional co-cultures of COS7 and GN11 cell aggregates. After 72 h in culture, the GN11 migration area is measured in the proximal (red) and distal (blue) quadrants as the surface covered by GN11 cells minus the aggregate inner mass area.
(B) Representative pictures of three-dimensional matrix co-cultures of GN11 cells with COS7 cells stained withDAPI and phalloidin red. For illustration, photographs have been digitally inverted, and a pseudo-color (purple) has been superimposed so that cell distribution is highlighted.
(C) Quantitative analysis of GN11 cell-migration aggregates (n = 12 per group) normalized and compared to the control condition (MOCK). Values are expressed as means ± SEM and analyzed by two-way ANOVA followed by Sidak’s multiple-comparisons test. ∗∗p < 0.01; ∗∗∗∗p < 0.0001. d, distal side; p, proximal side.
Loss of Ndnf in Murine Embryos Alters GnRH Neuron Migration and the Olfactory Axonal Scaffold
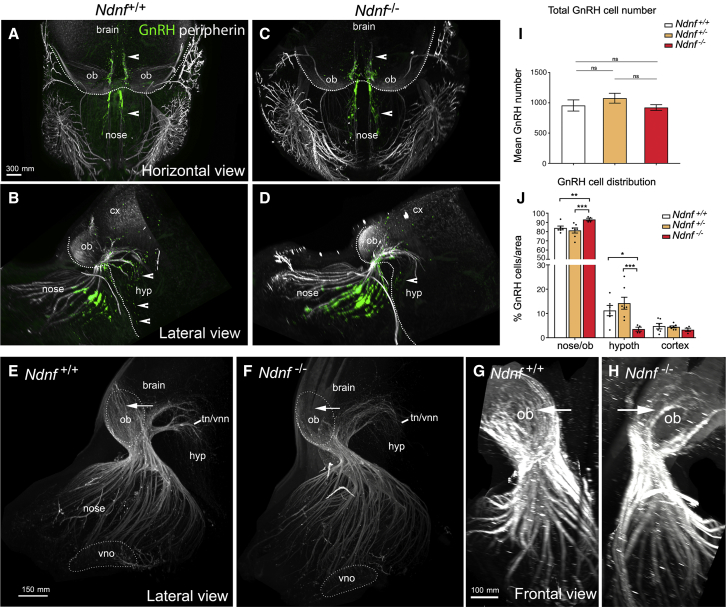
We then sought to determine whether genetic deletion of Ndnf in a murine model would lead to defects in GnRH neuronal migration. We performed a detailed analysis of E13.5 Ndnf WT, heterozygous, and homozygous mutants by using whole-mount immunostaining for GnRH and peripherin, a neuron-specific intermediate filament protein expressed by rodent sensory and autonomic axons,39 including the developing olfactory nerve (ON) and the vomeronasal nerve (VNN).35,40 This was followed by iDISCO tissue clearing36 and light-sheet microscopy (LSM) (Figures 6A–6D; Videos S1 and S2). In Ndnf+/+ embryos, GnRH neurons were more clustered in the nasal compartment, and fewer GnRH cells reached the developing hypothalamus at this embryonic stage (Figures 6A–6D, arrowheads; Videos S1 and S2). Moreover, in Ndnf+/+ embryos, peripherin-positive fibers were seen to almost completely innervate the olfactory bulb (Figures 6E and 6G, arrows), whereas in Ndnf−/− mice olfactory axons only partially innervate their target tissues (Figures 6F and 6H, arrows). The intracranial projections of the terminal and vomeronasal nerve (tn/vnn) did not show any abnormalities in Ndnf−/−embryos as compared to Ndnf+/+ embryos (Figures 6E and 6F).
Figure 6.
GnRH Migration and Olfactory Innervation Are Perturbed in Ndnf−/− Mice
(A–D) Representative whole-body iDISCO experiments in E13.5 Ndnf+/+and Ndnf−/−embryos immunolabelled for peripherin (white) and GnRH (green). (A and C) Horizontal projections of the embryo heads. (B and D) Lateral projections of the embryo heads. Arrowheads in A–D indicate GnRH cells located in the nasal and brain regions.
(E and F) High-magnification photomicrographs (lateral view) depicting olfactory and vomeronasal axon innervations (peripherin-positive) to the olfactory bulb and to the presumptive hypothalamus (terminal and vomeronasal nerve, tn/vnn) in E13.5 Ndnf+/+(E) and Ndnf−/−embryos (F).
(G and H) High-power view (frontal view) showing olfactory axon innervations of the OB in E13.5 Ndnf+/+(G) and Ndnf−/− (H) embryos. Arrows in (E)–(H) indicate noticeable differences in peripherin-positive fibers innervating the OB.
(I) Quantification of the total number of GnRH-immunoreactive neurons in Ndnf+/+(n = 6), Ndnf+/−(n = 7), and Ndnf−/− E13.5 embryos (n = 4). Data are represented as means ± SEM. Statistical analysis was performed by one-way ANOVA (F2,14 = 0.9, p = 0.4) followed by Fisher’s LSD post-hoc test.
(J) Quantitative analysis of GnRH neuronal distribution throughout the migratory pathway in the three experimental populations. Statistical analysis was performed by two-way ANOVA (F4,42 = 7.58, p = 0.0001) followed by Fisher’s LSD post-hoc test (∗p < 0.05, ∗∗p < 0.005, ∗∗∗p < 0.001). Scale bars (A–D) represent 200 μm.
We then counted the number of GnRH neurons in these embryos and found a comparable total number of GnRH neurons between all genotypes (Figure 6I), indicating that a lack of Ndnf has no effect on GnRH neuron survival. However, in Ndnf−/− mice the number of GnRH cells in the nasal and olfactory bulb region was significantly greater than in Ndnf+/+or Ndnf+/−mice (Figure 6J), respectively. As a result, fewer GnRH neurons reached the hypothalamic region in the Ndnf-null embryos (Figure 6J). The accumulation of GnRH neurons in the more rostral regions and decreased cell number within the ventral forebrain is suggestive of a delayed GnRH cell migration in Ndnf −/− mice.
Altogether, these experiments revealed that genetic invalidation of Ndnf leads to abnormal development of the olfactory system and defective GnRH migration to the hypothalamus.
Discussion
NDNF, the neuron-derived neurotrophic factor, was identified as a CHH gene via a strategy including WES and bioinformatics and focusing on genes in the FN3 superfamily. We first identified an enrichment of NDNF PTVs in our CHH cohort versus gnomAD. The three unrelated CHH probands (~2%) harboring heterozygous protein-truncating NDNF mutations had severe GnRH deficiency and anosmia consistent with Kallmann syndrome. Because Ndnf had been previously implicated in neuron structural plasticity, we then silenced z-ndnf in zebrafish, which resulted in a defect in GnRH neuron migration. In vitro studies established NDNF as a chemorepellent factor; combined with this protein’s high expression in the nasal compartment, this finding could indicate a role for NDNF in initiating GnRH neuron migration.
CHH is a complex genetic disorder for which >40 associated loci, each accounting for less than 10% of cases, have been identified to date. Importantly, about half of persons with CHH do not carry mutations in the known CHH-associated genes.3 Historically, cytogenetics,41, 42, 43, 44, 45 linkage studies,46,47 homozygosity mapping,48 candidate gene approaches,1,49 and other strategies have been used for the discovery of new genes related to CHH. Recently, Guo et al. applied collapsed gene-based burden testing to a population of 393 unrelated CHH probands and demonstrated significant associations to only three of the known CHH-related genes—FGFR1 [MIM: 136350], TACR3 [MIM: 162332], and GNRHR [MIM: 138850].16 Limitations of the study included a small population size and an inability to detect significant associations to most of the known CHH-related genes. Our current strategy combined collapsed gene-based burden testing with a targeted gene population (FN3 superfamily). This approach was based on the observation that several proteins that were involved in GnRH neuron migration and that were previously implicated in CHH (e.g., such proteins include ANOS1, AXL, DCC, and FLRT3) contain FN3 domains. Through the study of 240 CHH-unrelated probands, we demonstrated a statistical enrichment for PTVs in NDNF, which encodes a secreted glycosylated disulfide-bond protein containing FN3 domains. In addition, two known FN3-containing CHH-related genes (ANOS1 and AXL) also gave positive associations.16
The enrichment of PTVs in NDNF suggests that deletions in NDNF might explain some cases of CHH and will be further studied via whole-genome sequencing. Indeed, pathogenic deletions have been reported for other genes underlying CHH (e.g., such genes include ANOS1,50 SEMA3A,51 PROKR2,52 FGFR1,53 and, SOX1054).
The four probands carrying a heterozygous NDNF mutation exhibited both CHH and anosmia (i.e., KS), a condition related to defects in the embryonic development of GnRH neurons.1 Overall, ~3% of individuals with KS harbor NDNF mutations. Notably, several family members carrying NDNF mutations also exhibit a partial phenotype (i.e., delay of puberty and/or anosmia), consistent with an autosomal-dominant mode of inheritance with variable expressivity and incomplete penetrance—features seen in other CHH-related genes.1,2 None of the probands harbor mutations in other known CHH-related genes.1,2,55 As new research disentangles the NDNF signaling pathway (including its receptor), novel genetic interactions might be uncovered. Further investigations are needed to dissect the contribution of genetic (e.g., oligogenicity, structural variants, and eQTLs) and environmental factors (e.g., stress, nutrition, and endocrine disruptors) potentially modifying the phenotype of persons carrying NDNF loss-of-function mutations.
NDNF is a secreted neurotrophic factor that can promote neuron migration, growth, and survival, as well as neurite outgrowth.11 Ndnf is highly expressed in the olfactory bulb, Cajal-Retzius cells, hippocampus, and cerebellum during murine embryogenesis.11 Herein, we show that NDNF is expressed in the nasal region after formation of the olfactory placode in mice and humans, although not in GnRH neurons. Furthermore, the NDNF spatio-temporal expression pattern overlaps the GnRH neuron migratory route. In addition to the NDNF expression pattern, the positive effect of recombinant NDNF on GnRH neuron migration in vitro and the migratory defect of GnRH neurons in both z-ndnf MO-injected zebrafish and Ndnf-null mice provides strong evidence for a role of NDNF in GnRH neuron migration.
There are similarities between the two extracellular matrix proteins NDNF and anosmin-1, which is encoded by the first KS-related gene discovered, ANOS1. Indeed, both contain FN3 domains that are critical for binding to heparan sulfate proteoglycans.11,51 Anosmin-1 modulates FGFR1 signaling via a direct interaction of the anosmin-1-FN3 domain with heparan sulfate proteoglycan.56 Our in vitro studies demonstrate that NDNF also modulates FGFR1 signaling by inhibiting FGFR1-induced PLCγ/Ca2+ pathway activation after FGF8 stimulation. Thus, NDNF could interact in vivo with FGF8, a well-known morphogen acting during early developmental stages on the fate specification of GnRH neurons in the olfactory placode. Further studies investigating this process in vivo are needed.
Finally, NDNF promotes endothelial cell survival and vessel formation and, thus, is a powerful angiogenic factor.57,58 NDNF modulates endothelial plasticity through AKT/eNOS signaling,58 which is also known to be crucial for GnRH release in the median eminence.59, 60, 61 This suggests a possible role of NDNF in GnRH neuron biology beyond embryonic development.
In conclusion, we have identified NDNF as a novel factor involved in GnRH neuron ontogeny in humans and mice. Identifying the molecular signaling pathway used by NDNF in GnRH neuron ontogeny, along with the which particular receptors and upstream and downstream targets are involved, will be critical. Furthermore, it will also be crucial to study the role of NDNF in the adult GnRH system and homeostasis, specifically in regard to pubertal onset and fertility.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
We are grateful to the affected individuals and their families for their invaluable participation in this study, as well as to Erik Hrabovszky (Institute of Experimental Medicine, University of Budapest, Hungary) and Daniel J. Spergel (Section of Endocrinology, Department of Medicine, University of Chicago, IL) for providing anti-GnRH antibodies and GnRH-GFP mice, respectively. We also acknowledge Harri Niinikoski from the University of Turku for referring a CHH proband with an NDNF mutation, Kirsi Sainio from University of Helsinki for her guidance on mouse embryo harvesting, as well as M. Tardivel and A. Bongiovanni (microscopy core facility of Bioimaging Center Lille [BICeL], Lille University School of Medicine) for their technical assistance. We thank our colleagues Lucia Bartoloni, Alexia Spoerl, and Jenny Meylan of the Diagnostic Laboratory of Endocrine Genetic Diseases of the CHUV for their precious assistance in DNA sample preparation. This work was supported by Swiss National Foundation (N.P.) grant 310030_173260, the Institut National de la Santé et de la Recherche Médicale, Inserm, France (grant number U1172), the Agence Nationale de la Recherche, France (grant numbers ANR-14-CE12-0015-01 and ANR-18-CE14-0017-02 to P.G.), and Fondazione CRT (Torino, ITALY # 2014-0814 to G.R.M).
Published: December 26, 2019
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2019.12.003.
Web Resources
Interpro, https://www.ebi.ac.uk/interpro
OMIM, http://www.omim.org/.
Supplemental Data
References
- 1.Boehm U., Bouloux P.M., Dattani M.T., de Roux N., Dodé C., Dunkel L., Dwyer A.A., Giacobini P., Hardelin J.P., Juul A. Expert consensus document: European Consensus Statement on congenital hypogonadotropic hypogonadism--pathogenesis, diagnosis and treatment. Nat. Rev. Endocrinol. 2015;11:547–564. doi: 10.1038/nrendo.2015.112. [DOI] [PubMed] [Google Scholar]
- 2.Young J., Xu C., Papadakis G.E., Acierno J.S., Maione L., Hietamäki J., Raivio T., Pitteloud N. Clinical management of congenital hypogonadotropic hypogonadism. Endocr. Rev. 2019;40:669–710. doi: 10.1210/er.2018-00116. [DOI] [PubMed] [Google Scholar]
- 3.Cassatella D., Howard S.R., Acierno J.S., Xu C., Papadakis G.E., Santoni F.A., Dwyer A.A., Santini S., Sykiotis G.P., Chambion C. Congenital hypogonadotropic hypogonadism and constitutional delay of growth and puberty have distinct genetic architectures. European J. Endocrinol. 2018;178:377–388. doi: 10.1530/EJE-17-0568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schwanzel-Fukuda M., Pfaff D.W. Origin of luteinizing hormone-releasing hormone neurons. Nature. 1989;338:161–164. doi: 10.1038/338161a0. [DOI] [PubMed] [Google Scholar]
- 5.Wray S. From nose to brain: development of gonadotrophin-releasing hormone-1 neurones. J. Neuroendocrinol. 2010;22:743–753. doi: 10.1111/j.1365-2826.2010.02034.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wierman M.E., Kiseljak-Vassiliades K., Tobet S. Gonadotropin-releasing hormone (GnRH) neuron migration: initiation, maintenance and cessation as critical steps to ensure normal reproductive function. Front. Neuroendocrinol. 2011;32:43–52. doi: 10.1016/j.yfrne.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwanzel-Fukuda M., Bick D., Pfaff D.W. Luteinizing hormone-releasing hormone (LHRH)-expressing cells do not migrate normally in an inherited hypogonadal (Kallmann) syndrome. Brain Res. Mol. Brain Res. 1989;6:311–326. doi: 10.1016/0169-328x(89)90076-4. [DOI] [PubMed] [Google Scholar]
- 8.del Castillo I., Cohen-Salmon M., Blanchard S., Lutfalla G., Petit C. Structure of the X-linked Kallmann syndrome gene and its homologous pseudogene on the Y chromosome. Nat. Genet. 1992;2:305–310. doi: 10.1038/ng1292-305. [DOI] [PubMed] [Google Scholar]
- 9.Bork P., Doolittle R.F. Proposed acquisition of an animal protein domain by bacteria. Proc. Natl. Acad. Sci. USA. 1992;89:8990–8994. doi: 10.1073/pnas.89.19.8990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bencharit S., Cui C.B., Siddiqui A., Howard-Williams E.L., Sondek J., Zuobi-Hasona K., Aukhil I. Structural insights into fibronectin type III domain-mediated signaling. J. Mol. Biol. 2007;367:303–309. doi: 10.1016/j.jmb.2006.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuang X.L., Zhao X.M., Xu H.F., Shi Y.Y., Deng J.B., Sun G.T. Spatio-temporal expression of a novel neuron-derived neurotrophic factor (NDNF) in mouse brains during development. BMC Neurosci. 2010;11:137. doi: 10.1186/1471-2202-11-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oliveira L.M., Seminara S.B., Beranova M., Hayes F.J., Valkenburgh S.B., Schipani E., Costa E.M., Latronico A.C., Crowley W.F., Jr., Vallejo M. The importance of autosomal genes in Kallmann syndrome: genotype-phenotype correlations and neuroendocrine characteristics. J. Clin. Endocrinol. Metab. 2001;86:1532–1538. doi: 10.1210/jcem.86.4.7420. [DOI] [PubMed] [Google Scholar]
- 13.Seminara S.B., Oliveira L.M., Beranova M., Hayes F.J., Crowley W.F., Jr. Genetics of hypogonadotropic hypogonadism. J. Endocrinol. Invest. 2000;23:560–565. doi: 10.1007/BF03343776. [DOI] [PubMed] [Google Scholar]
- 14.Xu C., Messina A., Somm E., Miraoui H., Kinnunen T., Acierno J., Jr., Niederländer N.J., Bouilly J., Dwyer A.A., Sidis Y. KLB, encoding β-Klotho, is mutated in patients with congenital hypogonadotropic hypogonadism. EMBO Mol. Med. 2017;9:1379–1397. doi: 10.15252/emmm.201607376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laitinen E.M., Vaaralahti K., Tommiska J., Eklund E., Tervaniemi M., Valanne L., Raivio T. Incidence, phenotypic features and molecular genetics of Kallmann syndrome in Finland. Orphanet J. Rare Dis. 2011;6:41. doi: 10.1186/1750-1172-6-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo M.H., Plummer L., Chan Y.M., Hirschhorn J.N., Lippincott M.F. Burden testing of rare variants identified through exome sequencing via publicly available control data. Am. J. Hum. Genet. 2018;103:522–534. doi: 10.1016/j.ajhg.2018.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raivio T., Sidis Y., Plummer L., Chen H., Ma J., Mukherjee A., Jacobson-Dickman E., Quinton R., Van Vliet G., Lavoie H. Impaired fibroblast growth factor receptor 1 signaling as a cause of normosmic idiopathic hypogonadotropic hypogonadism. J. Clin. Endocrinol. Metab. 2009;94:4380–4390. doi: 10.1210/jc.2009-0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ichida M., Finkel T. Ras regulates NFAT3 activity in cardiac myocytes. J. Biol. Chem. 2001;276:3524–3530. doi: 10.1074/jbc.M004275200. [DOI] [PubMed] [Google Scholar]
- 19.Crabtree G.R., Olson E.N. NFAT signaling: choreographing the social lives of cells. Cell. 2002;109:S67–S79. doi: 10.1016/s0092-8674(02)00699-2. [DOI] [PubMed] [Google Scholar]
- 20.Messina A., Ferraris N., Wray S., Cagnoni G., Donohue D.E., Casoni F., Kramer P.R., Derijck A.A., Adolfs Y., Fasolo A. Dysregulation of Semaphorin7A/β1-integrin signaling leads to defective GnRH-1 cell migration, abnormal gonadal development and altered fertility. Hum. Mol. Genet. 2011;20:4759–4774. doi: 10.1093/hmg/ddr403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giacobini P., Messina A., Morello F., Ferraris N., Corso S., Penachioni J., Giordano S., Tamagnone L., Fasolo A. Semaphorin 4D regulates gonadotropin hormone-releasing hormone-1 neuronal migration through PlexinB1-Met complex. J. Cell Biol. 2008;183:555–566. doi: 10.1083/jcb.200806160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoshida T., Ito A., Matsuda N., Mishina M. Regulation by protein kinase A switching of axonal pathfinding of zebrafish olfactory sensory neurons through the olfactory placode-olfactory bulb boundary. J. Neurosci. 2002;22:4964–4972. doi: 10.1523/JNEUROSCI.22-12-04964.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miyasaka N., Sato Y., Yeo S.Y., Hutson L.D., Chien C.B., Okamoto H., Yoshihara Y. Robo2 is required for establishment of a precise glomerular map in the zebrafish olfactory system. Development. 2005;132:1283–1293. doi: 10.1242/dev.01698. [DOI] [PubMed] [Google Scholar]
- 24.Sato Y., Miyasaka N., Yoshihara Y. Mutually exclusive glomerular innervation by two distinct types of olfactory sensory neurons revealed in transgenic zebrafish. J. Neurosci. 2005;25:4889–4897. doi: 10.1523/JNEUROSCI.0679-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garaffo G., Conte D., Provero P., Tomaiuolo D., Luo Z., Pinciroli P., Peano C., D’Atri I., Gitton Y., Etzion T. The Dlx5 and Foxg1 transcription factors, linked via miRNA-9 and -200, are required for the development of the olfactory and GnRH system. Mol. Cell. Neurosci. 2015;68:103–119. doi: 10.1016/j.mcn.2015.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abraham E., Palevitch O., Gothilf Y., Zohar Y. Targeted gonadotropin-releasing hormone-3 neuron ablation in zebrafish: effects on neurogenesis, neuronal migration, and reproduction. Endocrinology. 2010;151:332–340. doi: 10.1210/en.2009-0548. [DOI] [PubMed] [Google Scholar]
- 27.Flynt A.S., Li N., Thatcher E.J., Solnica-Krezel L., Patton J.G. Zebrafish miR-214 modulates Hedgehog signaling to specify muscle cell fate. Nat. Genet. 2007;39:259–263. doi: 10.1038/ng1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kloosterman W.P., Plasterk R.H. The diverse functions of microRNAs in animal development and disease. Dev. Cell. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 29.Melvin V.S., Feng W., Hernandez-Lagunas L., Artinger K.B., Williams T. A morpholino-based screen to identify novel genes involved in craniofacial morphogenesis. Dev. Dyn. 2013;242:817–831. doi: 10.1002/dvdy.23969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garaffo G., Provero P., Molineris I., Pinciroli P., Peano C., Battaglia C., Tomaiuolo D., Etzion T., Gothilf Y., Santoro M., Merlo G.R. Profiling, bioinformatic, and functional data on the developing olfactory/GnRH system reveal cellular and molecular pathways essential for this process and potentially relevant for the Kallmann Syndrome. Front. Endocrinol. (Lausanne) 2013;4:203. doi: 10.3389/fendo.2013.00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang R., Dodd A., Lai D., McNabb W.C., Love D.R. Validation of zebrafish (Danio rerio) reference genes for quantitative real-time RT-PCR normalization. Acta Biochim. Biophys. Sin. (Shanghai) 2007;39:384–390. doi: 10.1111/j.1745-7270.2007.00283.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spergel D.J., Krüth U., Hanley D.F., Sprengel R., Seeburg P.H. GABA- and glutamate-activated channels in green fluorescent protein-tagged gonadotropin-releasing hormone neurons in transgenic mice. J. Neurosci. 1999;19:2037–2050. doi: 10.1523/JNEUROSCI.19-06-02037.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parkash J., Cimino I., Ferraris N., Casoni F., Wray S., Cappy H., Prevot V., Giacobini P. Suppression of β1-integrin in gonadotropin-releasing hormone cells disrupts migration and axonal extension resulting in severe reproductive alterations. J. Neurosci. 2012;32:16992–17002. doi: 10.1523/JNEUROSCI.3057-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Giacobini P., Parkash J., Campagne C., Messina A., Casoni F., Vanacker C., Langlet F., Hobo B., Cagnoni G., Gallet S. Brain endothelial cells control fertility through ovarian-steroid-dependent release of semaphorin 3A. PLoS Biol. 2014;12:e1001808. doi: 10.1371/journal.pbio.1001808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Casoni F., Malone S.A., Belle M., Luzzati F., Collier F., Allet C., Hrabovszky E., Rasika S., Prevot V., Chédotal A., Giacobini P. Development of the neurons controlling fertility in humans: new insights from 3D imaging and transparent fetal brains. Development. 2016;143:3969–3981. doi: 10.1242/dev.139444. [DOI] [PubMed] [Google Scholar]
- 36.Renier N., Wu Z., Simon D.J., Yang J., Ariel P., Tessier-Lavigne M. iDISCO: a simple, rapid method to immunolabel large tissue samples for volume imaging. Cell. 2014;159:896–910. doi: 10.1016/j.cell.2014.10.010. [DOI] [PubMed] [Google Scholar]
- 37.Goetz R., Mohammadi M. Exploring mechanisms of FGF signalling through the lens of structural biology. Nat. Rev. Mol. Cell Biol. 2013;14:166–180. doi: 10.1038/nrm3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Radovick S., Wray S., Lee E., Nicols D.K., Nakayama Y., Weintraub B.D., Westphal H., Cutler G.B., Jr., Wondisford F.E. Migratory arrest of gonadotropin-releasing hormone neurons in transgenic mice. Proc. Natl. Acad. Sci. USA. 1991;88:3402–3406. doi: 10.1073/pnas.88.8.3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parysek L.M., Goldman R.D. Distribution of a novel 57 kDa intermediate filament (IF) protein in the nervous system. J. Neurosci. 1988;8:555–563. doi: 10.1523/JNEUROSCI.08-02-00555.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fueshko S., Wray S. LHRH cells migrate on peripherin fibers in embryonic olfactory explant cultures: an in vitro model for neurophilic neuronal migration. Dev. Biol. 1994;166:331–348. doi: 10.1006/dbio.1994.1319. [DOI] [PubMed] [Google Scholar]
- 41.Meitinger T., Heye B., Petit C., Levilliers J., Golla A., Moraine C., Dalla Piccola B., Sippell W.G., Murken J., Ballabio A. Definitive localization of X-linked Kallman syndrome (hypogonadotropic hypogonadism and anosmia) to Xp22.3: close linkage to the hypervariable repeat sequence CRI-S232. Am. J. Hum. Genet. 1990;47:664–669. [PMC free article] [PubMed] [Google Scholar]
- 42.Legouis R., Hardelin J.P., Levilliers J., Claverie J.M., Compain S., Wunderle V., Millasseau P., Le Paslier D., Cohen D., Caterina D. The candidate gene for the X-linked Kallmann syndrome encodes a protein related to adhesion molecules. Cell. 1991;67:423–435. doi: 10.1016/0092-8674(91)90193-3. [DOI] [PubMed] [Google Scholar]
- 43.Dodé C., Levilliers J., Dupont J.M., De Paepe A., Le Dû N., Soussi-Yanicostas N., Coimbra R.S., Delmaghani S., Compain-Nouaille S., Baverel F. Loss-of-function mutations in FGFR1 cause autosomal dominant Kallmann syndrome. Nat. Genet. 2003;33:463–465. doi: 10.1038/ng1122. [DOI] [PubMed] [Google Scholar]
- 44.de Roux N., Young J., Misrahi M., Genet R., Chanson P., Schaison G., Milgrom E. A family with hypogonadotropic hypogonadism and mutations in the gonadotropin-releasing hormone receptor. N. Engl. J. Med. 1997;337:1597–1602. doi: 10.1056/NEJM199711273372205. [DOI] [PubMed] [Google Scholar]
- 45.Franco B., Guioli S., Pragliola A., Incerti B., Bardoni B., Tonlorenzi R., Carrozzo R., Maestrini E., Pieretti M., Taillon-Miller P. A gene deleted in Kallmann’s syndrome shares homology with neural cell adhesion and axonal path-finding molecules. Nature. 1991;353:529–536. doi: 10.1038/353529a0. [DOI] [PubMed] [Google Scholar]
- 46.de Roux N., Genin E., Carel J.C., Matsuda F., Chaussain J.L., Milgrom E. Hypogonadotropic hypogonadism due to loss of function of the KiSS1-derived peptide receptor GPR54. Proc. Natl. Acad. Sci. USA. 2003;100:10972–10976. doi: 10.1073/pnas.1834399100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seminara S.B., Messager S., Chatzidaki E.E., Thresher R.R., Acierno J.S., Jr., Shagoury J.K., Bo-Abbas Y., Kuohung W., Schwinof K.M., Hendrick A.G. The GPR54 gene as a regulator of puberty. N. Engl. J. Med. 2003;349:1614–1627. doi: 10.1056/NEJMoa035322. [DOI] [PubMed] [Google Scholar]
- 48.Topaloglu A.K., Reimann F., Guclu M., Yalin A.S., Kotan L.D., Porter K.M., Serin A., Mungan N.O., Cook J.R., Imamoglu S. TAC3 and TACR3 mutations in familial hypogonadotropic hypogonadism reveal a key role for Neurokinin B in the central control of reproduction. Nat. Genet. 2009;41:354–358. doi: 10.1038/ng.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stamou M.I., Cox K.H., Crowley W.F., Jr. Discovering genes essential to the hypothalamic regulation of human reproduction using a human disease model: Adjusting to life in the "-omics" era. Endocr. Rev. 2016;2016:4–22. doi: 10.1210/er.2015-1045.2016.1. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 50.Ballabio A., Bardoni B., Carrozzo R., Andria G., Bick D., Campbell L., Hamel B., Ferguson-Smith M.A., Gimelli G., Fraccaro M. Contiguous gene syndromes due to deletions in the distal short arm of the human X chromosome. Proc. Natl. Acad. Sci. USA. 1989;86:10001–10005. doi: 10.1073/pnas.86.24.10001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Young J., Metay C., Bouligand J., Tou B., Francou B., Maione L., Tosca L., Sarfati J., Brioude F., Esteva B. SEMA3A deletion in a family with Kallmann syndrome validates the role of semaphorin 3A in human puberty and olfactory system development. Hum. Reprod. 2012;27:1460–1465. doi: 10.1093/humrep/des022. [DOI] [PubMed] [Google Scholar]
- 52.Pitteloud N., Zhang C., Pignatelli D., Li J.D., Raivio T., Cole L.W., Plummer L., Jacobson-Dickman E.E., Mellon P.L., Zhou Q.Y., Crowley W.F., Jr. Loss-of-function mutation in the prokineticin 2 gene causes Kallmann syndrome and normosmic idiopathic hypogonadotropic hypogonadism. Proc. Natl. Acad. Sci. USA. 2007;104:17447–17452. doi: 10.1073/pnas.0707173104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Trarbach E.B., Teles M.G., Costa E.M., Abreu A.P., Garmes H.M., Guerra G., Jr., Baptista M.T., de Castro M., Mendonca B.B., Latronico A.C. Screening of autosomal gene deletions in patients with hypogonadotropic hypogonadism using multiplex ligation-dependent probe amplification: detection of a hemizygosis for the fibroblast growth factor receptor 1. Clin. Endocrinol. (Oxf.) 2010;72:371–376. doi: 10.1111/j.1365-2265.2009.03642.x. [DOI] [PubMed] [Google Scholar]
- 54.Amato L.G.L., Montenegro L.R., Lerario A.M., Jorge A.A.L., Guerra Junior G., Schnoll C., Renck A.C., Trarbach E.B., Costa E.M.F., Mendonca B.B. New genetic findings in a large cohort of congenital hypogonadotropic hypogonadism. Eur. J. Endocrinol. 2019;181:103–119. doi: 10.1530/EJE-18-0764. [DOI] [PubMed] [Google Scholar]
- 55.Pitteloud N., Quinton R., Pearce S., Raivio T., Acierno J., Dwyer A., Plummer L., Hughes V., Seminara S., Cheng Y.Z. Digenic mutations account for variable phenotypes in idiopathic hypogonadotropic hypogonadism. J. Clin. Invest. 2007;117:457–463. doi: 10.1172/JCI29884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.González-Martínez D., Kim S.H., Hu Y., Guimond S., Schofield J., Winyard P., Vannelli G.B., Turnbull J., Bouloux P.M. Anosmin-1 modulates fibroblast growth factor receptor 1 signaling in human gonadotropin-releasing hormone olfactory neuroblasts through a heparan sulfate-dependent mechanism. J. Neurosci. 2004;24:10384–10392. doi: 10.1523/JNEUROSCI.3400-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ohashi K., Enomoto T., Joki Y., Shibata R., Ogura Y., Kataoka Y., Shimizu Y., Kambara T., Uemura Y., Yuasa D. Neuron-derived neurotrophic factor functions as a novel modulator that enhances endothelial cell function and revascularization processes. J. Biol. Chem. 2014;289:14132–14144. doi: 10.1074/jbc.M114.555789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Joki Y., Ohashi K., Yuasa D., Shibata R., Kataoka Y., Kambara T., Uemura Y., Matsuo K., Hayakawa S., Hiramatsu-Ito M. Neuron-derived neurotrophic factor ameliorates adverse cardiac remodeling after experimental myocardial infarction. Circ Heart Fail. 2015;8:342–351. doi: 10.1161/CIRCHEARTFAILURE.114.001647. [DOI] [PubMed] [Google Scholar]
- 59.Aguan K., Mahesh V.B., Ping L., Bhat G., Brann D.W. Evidence for a physiological role for nitric oxide in the regulation of the LH surge: effect of central administration of antisense oligonucleotides to nitric oxide synthase. Neuroendocrinology. 1996;64:449–455. doi: 10.1159/000127151. [DOI] [PubMed] [Google Scholar]
- 60.De Seranno S., Estrella C., Loyens A., Cornea A., Ojeda S.R., Beauvillain J.C., Prevot V. Vascular endothelial cells promote acute plasticity in ependymoglial cells of the neuroendocrine brain. J. Neurosci. 2004;24:10353–10363. doi: 10.1523/JNEUROSCI.3228-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.de Seranno S., d’Anglemont de Tassigny X., Estrella C., Loyens A., Kasparov S., Leroy D., Ojeda S.R., Beauvillain J.C., Prevot V. Role of estradiol in the dynamic control of tanycyte plasticity mediated by vascular endothelial cells in the median eminence. Endocrinology. 2010;151:1760–1772. doi: 10.1210/en.2009-0870. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.