Abstract
The sediment marine samples were obtained from several places along the coastline of the Tuticorin shoreline, Tamil Nadu, India were separated for the presence of bioactive compound producing actinobacteria. The actinobacterial strain was subjected to 16Sr RNA sequence cluster analysis and identified as Nocardiopsis dassonvillei- DS013 NCBI accession number: KM098151. Bacterial mediated synthesis of nanoparticles gaining research attention owing its wide applications in nonmedical biotechnology. In the current study, a single step eco-friendly silver nanoparticles (AgNPs) were synthesized from novel actinobacteria Nocardiopsis dassonvillei- DS013 has been attempted. The actinobacterial mediated silver nanoparticles were characterized by TEM, UV–Visible, XRD, FT-IR spectroscopy. The initial detection of AgNPs was identified using UV–Vis spectrum and confirmed by the appearance of absorbance peak at 408 nm. A Fourier transform infrared spectroscopy (FT-IR) result reveals the presence of protein component in the culture supernatant may act as protecting agents. The XRD pattern indicated that the typical peaks reveal the presence of nanoparticles. The TEM morphology confirms the formation of circular and non uniform distributions of AgNPs with the size range from 30 to 80 nm. The antibacterial activity of both isolated actinobacterial (IA) and silver nanoparticles mediated actinobacterial (SNA) of Nocardiopsis dassonvillei- DS013 were done by well diffusion method against selected clinical isolates of bacteria, namely Escherichia coli, Enterococcus sp., Pseudomonas sp., Klebsiella sp., Proteus sp., Shigella sp., Bacillus subtilis, and Streptococcus sp. When compared to isolated actinobacteria, the SNA shows the better antibacterial activity against clinical isolates.
Keywords: Nocardiopsis dassonvillei, Marine actinobacteria, AgNPs, Marine sediments
1. Introduction
Existence of multidrug resistance pathogens in the environment is a serious problem around the globe. Which may cause different types of diseases and occasionally fatal to humans (Sondi and Salopek-Sondi, 2004, Pal et al., 2007, Ingle et al., 2008, Bonde et al., 2012, Begum et al., 2013, Gupta et al., 2013, Rai et al., 2015, Singh et al., 2015). To overcome this problem, there is a demand to find effective, affordable, and safe antimicrobials from the microorganisms present in the natural environment. Actinobacteria belong to the Gram-positive, free- living, saprophytic, filamentous bacteria that abound in the environment producing numerous well documented biologically active secondary metabolites having potential to be used as novel antibiotics that may have a significant role in therapeutic applications. They are predominantly associated with almost all soil, fresh water and marine environments (Gebreyohannes et al., 2013). The diverse group of marine actinobacteria has mostly been unexplored and thereby remains elusive. The highly complex marine environments prevalent in various regions were found to be like high pressures, varying salinity and diverse temperature zones in natural habitats that have resulted in the development of diversified group of microorganisms colonizing these ecological niches (Olano et al., 2009). The adaptation and survival of the marine actinobacteria in a complex ecosystem has been considerably linked to the inherent ability of the diversified group of actinobacteria that predominantly produce secondary metabolites with antibacterial activity. In this context, marine sediments have continuously been at the forefront as a prospective source for isolation of actinobacteria which may yield novel antimicrobial compounds (Goodfellow et al., 2013). Comparatively DNA of actinobacteria has higher percentage of GC bases. The highly conserved 16s rRNA encoding genes are regularly used as genetic markers to determine the taxonomy, phylogeny and species divergence rates among bacteria, and 16s rRNA sequence analysis should be used to construct phylogenetic trees for microorganisms (Manojkumar and Subbaiya, 2016). Screening and utilization of marine actinobacteria in bio nanotechnology has significant attention (Sadhasivam et al., 2010, Sadhasivam et al., 2012), which exhibit various effective and potential applications in drug delivery, cancer therapy, etc., (Willets and Van Duyne, 2007, Gittins et al., 2000, Jain et al., 2007). Recent focus has been on eco-friendly methods for the green synthesis of metal nanoparticles utilizing biomaterials (Kowshik et al., 2002, Senapati et al., 2005, Shahverdi et al., 2007). Many researchers have reported the effect of microbial mediated AgNPs against several multidrug resistant microbes (Chaudhari et al., 2012, Priyaragini et al., 2013, Saminathan, 2015). Previous investigator (Sastry et al., 2003) observed the antimicrobial and cytotoxic effects utilizing AgNPs from a newly isolated Nocardiopsis sp. MBRC. In this communication we report a single step eco synthesis of silver nanoparticles using novel actinobacteria Nocardiopsis dassonvillei-DS013 isolated and reported for the first time from marine sediments coastline of the Tuticorin shoreline, Tamil Nadu, India. The actinobacterial mediated AgNPs was utilized to study their antibacterial activities towards selected clinical bacterial isolates such as Escherichia coli, Enterococcus sp., Pseudomonas sp., Klebsiella sp., Proteus sp., Shigella sp., Bacillus subtilis, and Streptococcus sp.
2. Materials and methods
2.1. Collection of marine sediments
Marine sediments were obtained in depth not more than 10 to 15 cm from 11 different places at intervals of 500 m from the coastline of Tuticorin, Tamil Nadu, India. The marine soil sediment samples from each of the locations were aseptically placed in sterile wide mouthed sample collection jars, appropriately labelled, stored in icebox and taken to the laboratory within 12 h. The materials were preserved by refrigeration, where required, until further processing. The sediment samples were air-dried for one week. The air-dried samples were sieved – using 1 mm mesh; and the fine grain samples were stored for pre-treatment in a hot air oven at 50 °C for 60 min. The dried marine soil sediment was then used for screening of actinobacteria.
2.2. Isolation of actinobacteria from marine sediments
Screening of actinobacteria from marine soil sediment sample was done by spread plate method (Manivasagan et al., 2013a, Manivasagan et al., 2013b). The soil suspension was subjected to standard serial decimal dilution to give dilutions of 10−1 to 10−10 using sterile 0.9% saline as diluents (Vimal et al., 2009). The actinobacteria from treated marine sediment samples was isolated by standard spread plating on the surface Starch Casein Agar (SCA; Hi-Media, India) (Selvakumar et al., 2012) supplemented with cycloheximide (25 mg/mL; SCAC) (Sivakumar et al., 2007) in level 1 bio safety cabinet. The whole procedure for each sample was done in triplicates to maximize the chance of isolating actinobacteria. The plates were dried for 25 min and incubated at 30 °C (4 weeks). Colonies showing typical actinobacteria growth characteristics were preserved by sub-culture on Yeast Extract Malt Extract Agar (Hi-Media) and stored under refrigerated conditions until further use.
2.3. Identification of actinobacteria by 16 s rRNA sequencing
2.3.1. DNA isolation
The isolated actinobacteria from marine sediments was aseptically transferred in Starch Casein Broth and incubated at 37 °C for 6 days. After obtained the significant growth (OD590 ≤ 0.9) the culture tube was centrifuged at 10,000 rpm for 15 min to separate the fungal mycelium. About 0.1g of mycelium was crushed with the help of liquid nitrogen. With addition of 50 µl TE buffer and 20 mg/ml of lysozyme the tube was incubated at 30 °C for 30 min. Later, the tube was added with each 20 µl of 10% SDS and proteinase K and kept for incubation at 50 °C for 30 min. The solution of Phenol - chloroform was added in 1:1 ratio and centrifuged at 10,000 rpm for 5.0 min to extract lysate. The upper layer solution was transferred into a new eppendorf tube and ethanol precipitation was carried out to separate the DNA using 70% of ethanol and then the tubes were kept for incubation at −20 °C for 30 min. To obtain the pellet the tubes were centrifuged at 10,000 rpm for 15 min and gently washed with 90% of ethanol before it suspended in 25 µl of TE buffer. The 20 µl RNase was added in the mixer tube and it was incubated at 37 °C for 1.0 h. The phenol chloroform extraction procedure was carried out to acquire pure product of nucleic acid (Saurav and Kannabiran, 2010).
2.3.2. PCR amplification and sequencing
Molecular identification procedure includes isolation and genomic DNA extraction, 16S rRNA amplification, PCR product purification, PCR amplicon gene product sequencing was performed in Acme Progen Biotech (India) Pvt. Ltd, Salem, Tamil Nadu, India. PCR amplification of the 16 s rRNA was achieved by adding final volume of the reaction mixture 25 µl includes PCR buffer (1X), MgCl2 (1.5 mM), Deoxyribonucleotide triphosphate dNTP (200 µM), primers 20 pmol, 2.5U Thermus aquaticus DNA polymerase and 100 ng of DNA template. The PCR amplification was carried out in Eppendorf Thermocycycler 96. The 30 cycles of PCR was carried out at the temperature of 94 °C for 1.0 min, 55 °C for 1.0 min. and 72 °C for 2.0 min. The PCR amplicon product was detected by electrophoresis - using 1.0% of agarose gel in Tris/Borate/EDTA buffer pH 8.5; at 150 V for 25 min, stained with ethidium bromide and visualized under ultraviolet transilluminator (Vijayakumar et al., 2010). The gel image was took using gel documentation and analysed. Later, the PCR amplicon products were eluted by the method given in Gel elution kit. In later PCR purification kit (Qiagen, Germany) were used to purify the elution mixture and finally the DNA sequences were obtained using an ABI xl 3730 DNA analyzer, Applied Biosystems. The obtained sequences were submitted to the NCBI and based upon the maximum score among the other organisms the query organism was confirmed.
2.3.3. Synthesis of silver nanoparticles using Nocardiopsis dassonvillei-DS013
About 3.0 ml culture of Nocardiopsis dassonvillei-DS013 was added into 47 ml of Silver nitrate (AgNO3) solution separately with continuous stirring the solution. The colour has transformed from pale yellow to dark brown color which illustrates the presence of SNPs (Heuer et al., 1997). The silver nanoparticles mediated actinobacterial (SNA) were studied by UV–Vis spectroscopy, XRD, FT-IR and TEM.
2.3.4. Antibacterial activity
The antibacterial activity of isolated actinobacterial (IA) and silver nanoparticles mediated actinobacterial (SNA) were evaluated by using well diffusion method described by (Naganathan and Thirunavukkarasu, 2017). In brief: sterile Mueller Hinton Agar (MHA; Hi-Media) plates were swabbed with overnight Mueller-Hinton Broth culture of any one of the selected test organism with approximately 106 CFU per ml - turbidity standard equal to 0.5 and 1.0 McFarland standard for Gram negative bacteria and Gram positive bacteria, respectively and labelled appropriately. The plates were dried for 25 min and 4.0 wells of 5.0 mm diameter were formed by utilizing sterile borer on MHA. The IA and SNA were cultivated in 50 ml Starch Casein Broth for 72 h at 30 °C and the culture filtrates were obtained by filtration through 45 µm membrane filter (Sartorius). 100 µl of culture filtrate from IA and SNA was respectively loaded into labelled well and incubated in upright position for 30 min and further incubated at 35 °C for 16–18 h. The results for preliminary screening of antibacterial activity by IA and SNA against test bacteria, if any, measured as zone of inhibition in mm and recorded.
3. Results and discussion
3.1. Isolation of actinobacteria from marine soil sediment
After incubation, a distinct actinobacteria were isolated and identified based on the appearance of powdery colonies from marine soil sediment samples collected from the Bay of Bengal along Tuticorin shoreline, Tamil Nadu, India. Based on characteristic appearance the strains of actinobacteria were purified by streak plate method using (SCA) Starch Casein Agar. Among the diverse groups of microbes in marine ecosystem, Actinobacteria have a significant niche as a prospective producer of distinctive metabolites and biological active compounds (Ganesan et al., 2017). Similarly, previous investigator Rathod et al. (2016) reported first time about the isolation of alkaliphilic actinobacterium N. valliformis OT1 strain from Central India, synthesis of AgNPs and its antibacterial activity against bacterial pathogens (Abirami and Kannabiran, 2016).
3.2. Molecular identification of Nocardiopsis dassonvillei-DS013 by 16s rRNA sequencing
The marine actinobacteria was identified as Nocardiopsis dassonvillei-DS013 based on the morphological, physiological, and various biochemical characteristics and also it was confirmed by 16s rRNA sequencing. The obtained DNA (genomic) was amplified and the PCR amplicon gene product (Fig. 1) was detected in 1.0% agarose gel with 1.0 kb ladder used as marker and determined that the amplicon was not degraded. The PCR amplified product yielded the molecular size of DNA 1.5 kb in length (Fig. 2). The automated sequencer was used to retrieve the sequence with help of appropriate primers. The obtained sequence was submitted to GenBank in NCBI with the accession number (KM098151).
Fig. 1.
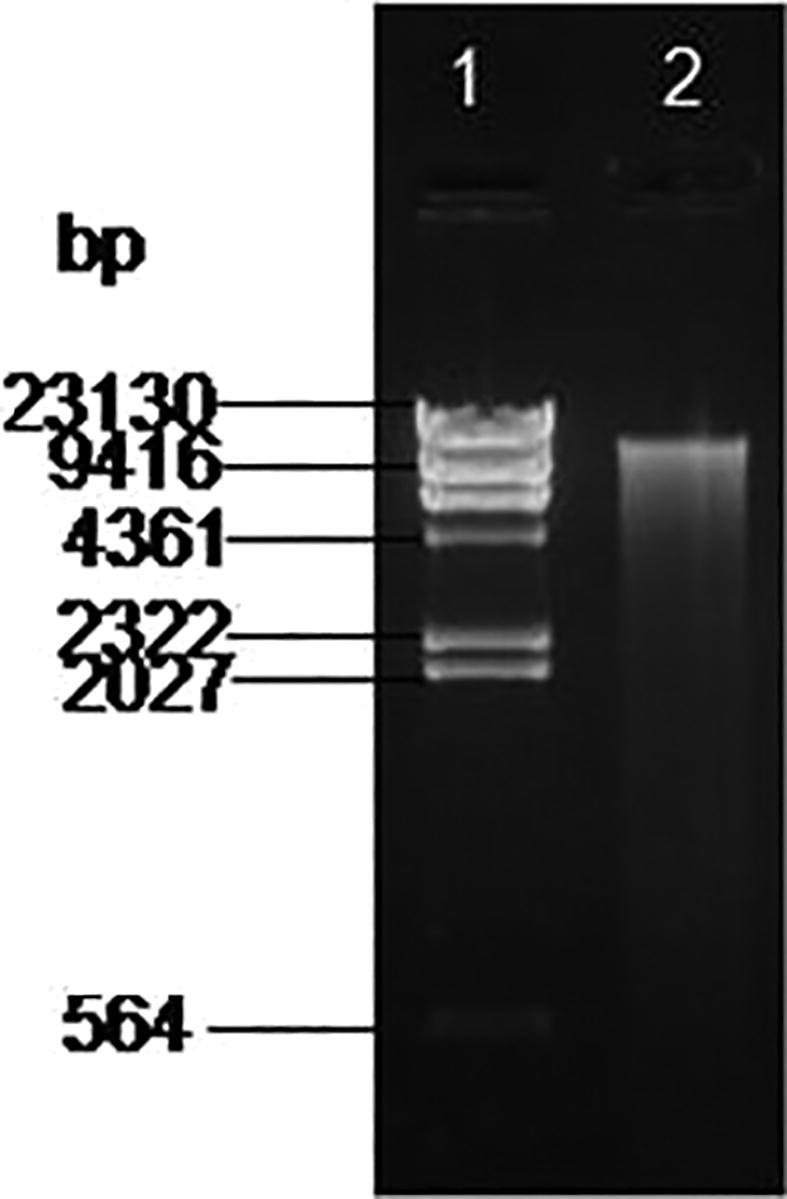
Agarose Gel (1.0%) showing (1) Lambda DNA/HindIII Digest Marker and (2) Genomic DNA product of Nocardiopsis dassonvillei-DS013.
Fig. 2.
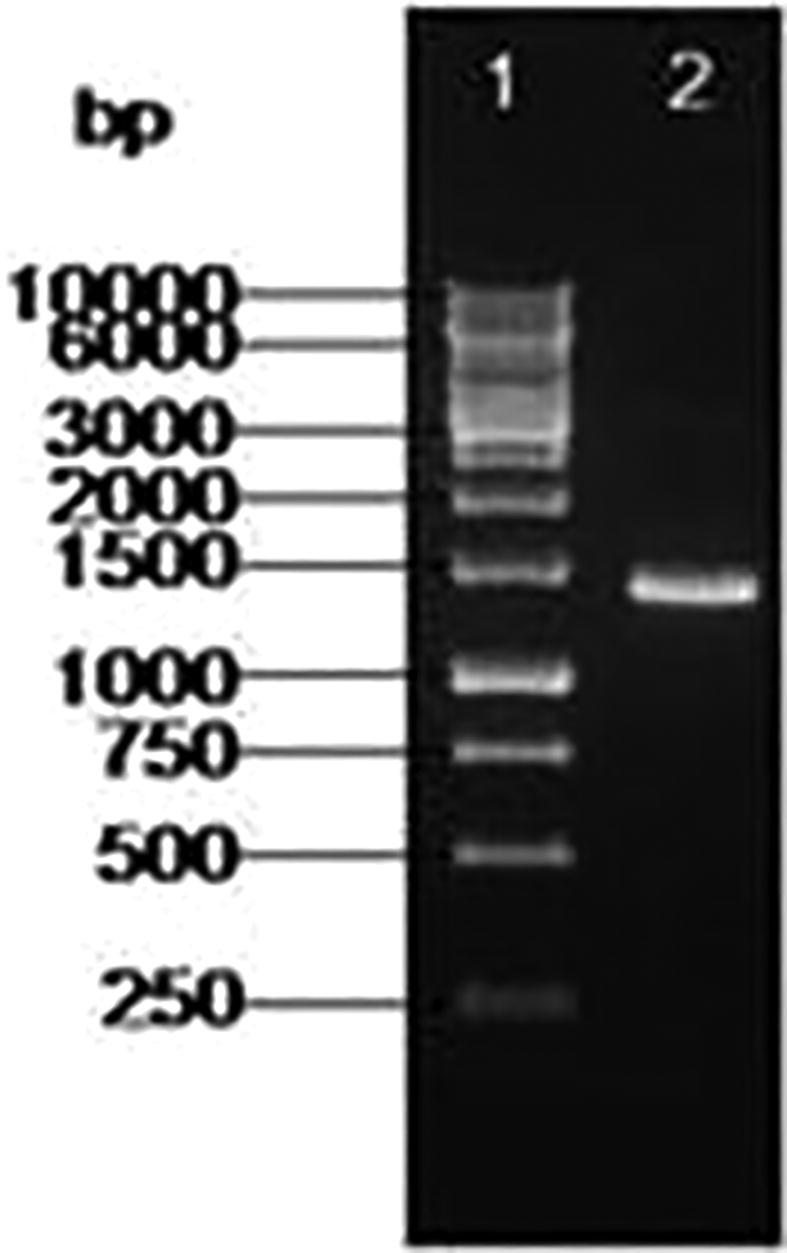
Agarose Gel (1.0%) showing (1) Lambda 1 Kb DNA ladder and (2) PCR product of Nocardiopsis dassonvillei-DS013.
3.3. Characterization of silver nanoparticle obtained from Nocardiopsis dassonvillei-DS013
3.3.1. UV–Vis spectrum of SNA
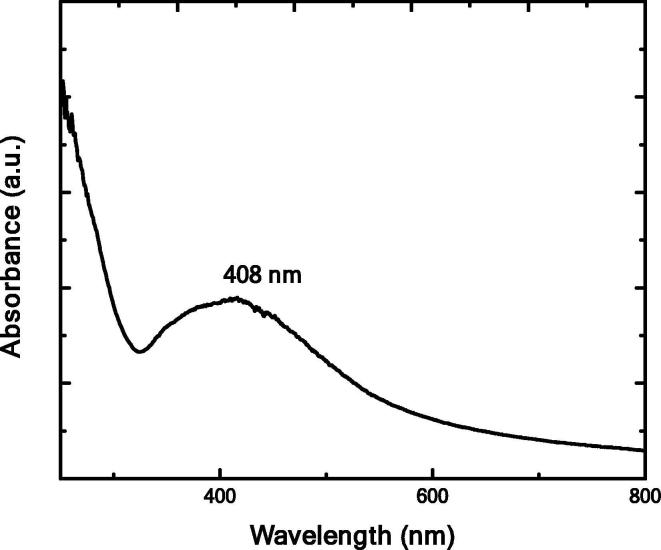
Fig. 3 illustrates the UV–Visible spectrum of AgNPs mediated Nocardiopsis dassonvillei-DS013, which shows absorbance peak at 408 nm. This is because of the effect of surface plasmon resonance intense peak. Nonappearance of peaks between 300 and 560 nm which clearly indicates the formed nanoparticles are without aggregation (Rathod et al., 2016, Banerjee and Rai, 2018, Balashanmugam et al., 2013).
Fig.3.
UV–Visible Spectrum of SNA.
3.3.2. XRD analysis of SNA
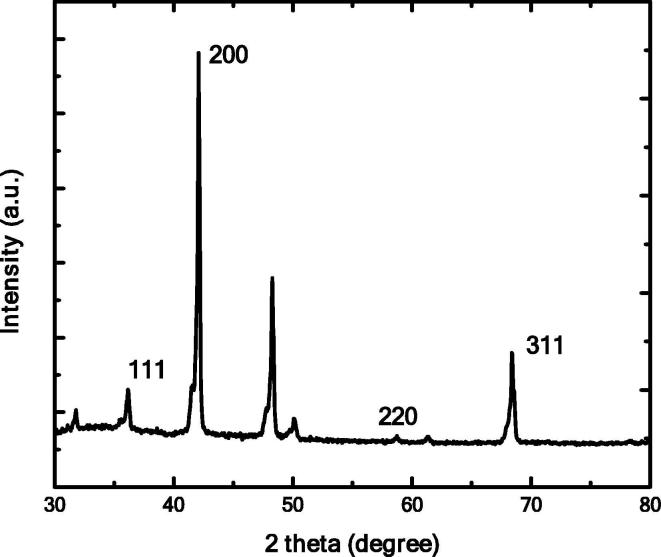
XRD pattern of SNA show intensive peaks at 2θ = 37.10°, 42.10°, 59.42° and 69.21° these indexed peaks are (1 1 1), (2 0 0), (2 2 0) and (3 1 1) lattice, respectively (Fig. 4). AgNPs The indexed plane indicates the FCC structure of Ag.
Fig.4.
XRD pattern of SNA.
3.3.3. FTIR analysis of SNA
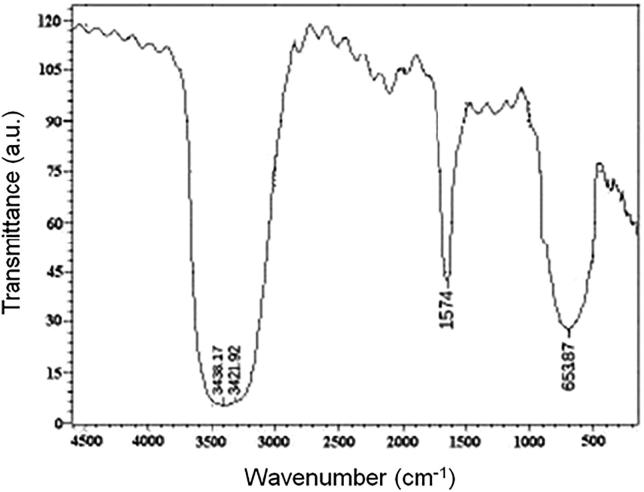
To understand the contribution of the bio molecules which reduces the Ag + ions into AgNPs, the FT-IR spectroscopy was performed. Fig. 5 represents the FT-IR spectrum of SNA, the absorbance peaks at 3336 cm−1 and 3370 cm−l leads to the extending OH groups of vibrations. A strong peaks of 1574 cm−1Check table 1 and relates the extending vibration of C O which affirms the formation of AgNPs of Nocardiopsis dassonvillei-DS013. A peaks at 653 cm−1 and 504 cm−1 relates to extending vibration of C—Cl and C—Br bond, respectively. The strong broad absorbance at 653 cm−1 is the characteristics of alkynes group (Verma et al., 2014, Sivalingam et al., 2012, Kumar and Mamidyala, 2011).
Fig.5.
FTIR analysis of SNA.
3.3.4. TEM image of SNA
Fig. 6 shows the TEM image of AgNPs mediated Nocardiopsis dassonvillei-DS013 with circular in morphology with non uniform distributions in the dimension range of 30–80 nm.
Fig.6.
TEM analysis of SNA.
The obtained results were correlated among previous investigations (Krishnaraj et al., 2010, Shahverdi et al., 2007).
3.4. Antibacterial potential of IA and SNA of Nocardiopsis dassonvillei-DS013 against tested clinical isolates of bacteria
In this work, the antibacterial activity of silver nanoparticles mediated actinobacterial (SNA) were assessed against the selected clinical isolates of bacteria such as Escherichia coli, Enterococcus sp., Pseudomonas sp., Klebsiella sp., Proteus sp., Shigella sp., Bacillus subtilis, and Streptococcus sp. In this assessment the most interesting results were observed that SNA had the highest activity showing remarkable activity against all test organisms compare to IA. The IA from marine sediment demonstrated slight (1.0–8.0 mm) antibacterial activity and SNA showed the moderate to maximum activity (more than 13 mm) against all clinical isolates as determined by zone diameter in mm (Table 1) this is because of the reduction of size of Ag+ to Ag0 utilizing the marine actinobacteria (Kalishwaralal et al., 2008, Manivasagan et al., 2013a, Manivasagan et al., 2013b).
Table.1.
Showing antibacterial activity of IA and SNA against clinical bacterial isolates.
| Clinical bacterial isolates | Antibacterial activity of IA and SNA strain | |
|---|---|---|
| Zone diameter (mm) |
||
| IA | SNA | |
| E. coli | 4 | 14 |
| Enterococcus sp., | 0 | 18 |
| Pseudomonas sp., | 0 | 15 |
| Klebsiella sp., | 3 | 24 |
| Proteus sp., | 2 | 16 |
| Bacillus subtilis sp., | 2 | 18 |
| Shigella sp., | 2 | 13 |
| Streptococcus sp., | 9 | 17 |
IA – Isolated Actinobacteria; SNA – Silver Nanoparticles Mediated Actinobacteria.
4. Conclusions
All among the microorganisms in the marine ecosystem; marine actinomycetes have a highest metabolic and genetic diversity. An exceptional perceives and efforts are need to explore on marine actinobacteria as source for the novel metabolites. Actinomycetes not only exist in the ocean, but also widely present in the different marine ecosystem. Precious secondary metabolites from marine actinomycetes and its potential should not be overlooked. In the present study we isolated and identified a novel marine actinobacteria Nocardiopsis dassonvillei-DS013 present in the soil sediments from different places along the coastline of the Bay of Bengal along the Tuticorin shoreline, Tamil Nadu, India. The marine actinobacteria Nocardiopsis dassonvillei-DS013 mediated AgNPs was synthesised. The XRD indexed plane indicates the FCC structure of AgNPs and further the structure and size of the synthesized SNA were confirmed by TEM which clearly indicates the dimension range of 30–80 nm with circular in morphology with non uniform distributions. Compared to isolated actinobacteria (IA), the AgNPs mediated marine actinobacteria (SNA) showed excellent antibacterial activity against the various selected clinical isolates of bacteria such as Escherichia coli, Enterococcus sp., Pseudomonas sp., Klebsiella sp., Proteus sp., Shigella sp., Bacillus subtilis, and Streptococcus sp. From this work we have recognized that the single step eco – synthesis of AgNPs from marine actinobacteria Nocardiopsis dassonvillei-DS013 which plays a crucial role in bionanomedicine applications.
Acknowledgements
This project was supported by Researchers Supporting Project number (RSP-2019/5) King Saud University, Riyadh, Saudi Arabia.
Footnotes
Peer review under responsibility of King Saud University.
Contributor Information
Arunachalam Chinnathambi, Email: carunachalam@ksu.edu.sa.
Sulaiman Ali Alharbi, Email: sharbi@ksu.edu.sa.
References
- Abirami M., Kannabiran K. Streptomyces ghanaensis VITHM1 mediated green synthesis of silver nanoparticles: Mechanism and biological applications. Front. Chem. Sci. Eng. 2016;10(4):542–551. [Google Scholar]
- Balashanmugam P., Santhosh S., Giyaullah H., Balakumaran M.D., Kalaichelvan P.T. Mycosynthesis, characterization and antibacterial activity of silver nanoparticles from Microporus xanthopus: a macro Mushroom. Int. J. Innov. Res. Sci. Eng. Technol. 2013;2:1–9. [Google Scholar]
- Banerjee K., Rai V.R. A review on mycosynthesis, mechanism, and characterization of silver and gold nanoparticles. Bionanosci. 2018;8(1):17–31. [Google Scholar]
- Begum S., Hasan F., Hussain S., Shah A.A. Prevalence of multi drug resistant Acinetobacter baumannii in the clinical samples from Tertiary Care Hospital in Islamabad. Pakistan. Pak. J. Med. Sci. 2013;29(5):1253. doi: 10.12669/pjms.295.3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonde S.R., Rathod D.P., Ingle A.P., Ade R.B., Gade A.K., Rai M.K. Murraya koenigii-mediated synthesis of silver nanoparticles and its activity against three human pathogenic bacteria. Nanosci. Method. 2012;1(1):25–36. [Google Scholar]
- Chaudhari P.R., Masurkar S.A., Shidore V.B., Kamble S.P. Effect of biosynthesized silver nanoparticles on Staphylococcus aureus biofilm quenching and prevention of biofilm formation. Nano. Micro, Lett. 2012;4(1):34–39. doi: 10.1049/iet-nbt.2011.0061. [DOI] [PubMed] [Google Scholar]
- Ganesan P., Reegan A.D., David R.H.A., Gandhi M.R., Paulraj M.G., Al-Dhabi N.A., Ignacimuthu S. Antimicrobial activity of some actinomycetes from Western Ghats of Tamil Nadu, India. Alexandria. Med. J. 2017;53(2):101–110. [Google Scholar]
- Gebreyohannes G., Moges F., Sahile S., Raja N. Isolation and characterization of potential antibiotic producing actinomycetes from water and sediments of Lake Tana. Ethiopia. Asian. Pac. J. Trop. Biomed. 2013;3(6):426–435. doi: 10.1016/S2221-1691(13)60092-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gittins D.I., Bethell D., Schiffrin D.J., Nichols R.J. A nanometre-scale electronic switch consisting of a metal cluster and redox-addressable groups. Nature. 2000;408(6808):67. doi: 10.1038/35040518. [DOI] [PubMed] [Google Scholar]
- Goodfellow M., Brown R., Ahmed L., Pathom-Aree W., Bull A.T., Jones A.L., Wang J. Verrucosispora fiedleri sp. nov., an actinomycete isolated from a fjord sediment which synthesizes proximicins. Anton. Leeuw. Int. J. 2013;103(3):493–502. doi: 10.1007/s10482-012-9831-y. [DOI] [PubMed] [Google Scholar]
- Gupta A., Bonde S.R., Gaikwad S., Ingle A., Gade A.K., Rai M. Lawsonia inermis-mediated synthesis of silver nanoparticles: activity against human pathogenic fungi and bacteria with special reference to formulation of an antimicrobial nanogel. IET Nano. Biotechnol. 2013;8(3):172–178. doi: 10.1049/iet-nbt.2013.0015. [DOI] [PubMed] [Google Scholar]
- Heuer H., Krsek M., Baker P., Smalla K., Wellington E.M. Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Appl. Environ. Microbiol. 1997;63(8):3233–3241. doi: 10.1128/aem.63.8.3233-3241.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingle A., Gade A., Pierrat S., Sonnichsen C., Rai M. Mycosynthesis of silver nanoparticles using the fungus Fusarium acuminatum and its activity against some human pathogenic bacteria. Curr. Nanosci. 2008;4(2):141–144. [Google Scholar]
- Jain P.K., El-Sayed I.H., El-Sayed M.A. Au nanoparticles target cancer. Nano Today. 2007;2(1):18–29. [Google Scholar]
- Kalishwaralal K., Deepak V., Ramkumarpandian S., Nellaiah H., Sangiliyandi G. Extracellular biosynthesis of silver nanoparticles by the culture supernatant of Bacillus licheniformis. Mater. Lett. 2008;62(29):4411–4413. [Google Scholar]
- Kowshik M., Ashtaputre S., Kharrazi S., Vogel W., Urban J., Kulkarni S.K., Paknikar K.M. Extracellular synthesis of silver nanoparticles by a silver-tolerant yeast strain MKY3. Nanotech. 2002;14(1):95. [Google Scholar]
- Krishnaraj C., Jagan E.G., Rajasekar S., Selvakumar P., Kalaichelvan P.T., Mohan N. Synthesis of silver nanoparticles using Acalypha indica leaf extracts and its antibacterial activity against water borne pathogens. Colloids. Surf. B Biointerfaces. 2010;76(1):50–56. doi: 10.1016/j.colsurfb.2009.10.008. [DOI] [PubMed] [Google Scholar]
- Kumar C.G., Mamidyala S.K. Extracellular synthesis of silver nanoparticles using culture supernatant of Pseudomonas aeruginosa. Colloids. Surf. B Biointerfaces. 2011;84(2):462–466. doi: 10.1016/j.colsurfb.2011.01.042. [DOI] [PubMed] [Google Scholar]
- Manivasagan P., Venkatesan J., Senthilkumar K., Sivakumar K., Kim S.K. Biosynthesis, antimicrobial and cytotoxic effect of silver nanoparticles using a novel Nocardiopsis sp. MBRC-1. Biomed. Res. 2013 doi: 10.1155/2013/287638. Article ID 287638 pp 1–9.A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manivasagan P., Venkatesan J., Senthilkumar K., Sivakumar K., Kim S.K. Biosynthesis, antimicrobial and cytotoxic effect of silver nanoparticles using a novel Nocardiopsis sp. MBRC-1. Biomed. Res. Article ID. 2013;287638:1–9. doi: 10.1155/2013/287638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manojkumar S., Subbaiya R. Isolation and Identification of Soil derived Actinomycetes Nocardiopsis alba. Res. J. Pharm. Biol. Chem. Sci. 2016;7(3):485–493. [Google Scholar]
- Naganathan K., Thirunavukkarasu S. Green way genesis of silver nanoparticles using multiple fruit peels waste and its antimicrobial, anti-oxidant and anti-tumor cell line studies. Mat. Sci. Eng. 2017;191(1) [Google Scholar]
- Olano C., Méndez C., Salas J. Antitumor compounds from marine actinomycetes. Mar. Drugs. 2009;7(2):210–248. doi: 10.3390/md7020210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal S., Tak Y.K., Song J.M. Does the antibacterial activity of silver nanoparticles depend on the shape of the nanoparticle? A study of the gram-negative bacterium Escherichia coli. Appl. Environ. Microbiol. 2007;73(6):1712–1720. doi: 10.1128/AEM.02218-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priyaragini S., Sathishkumar S.R., Bhaskararao K.V. Biosynthesis of silver nanoparticles using actinobacteria and evaluating its antimicrobial and cytotoxicity activity. Int. J. Pharm. Pharm. Sci. 2013;5(2):709–712. [Google Scholar]
- Rai M., Ingle A.P., Gade A.K., Duarte M.C.T., Duran N. Three Phoma spp. synthesised novel silver nanoparticles that possess excellent antimicrobial efficacy. IET Nano Biotechnol. 2015;9(5):280–287. doi: 10.1049/iet-nbt.2014.0068. [DOI] [PubMed] [Google Scholar]
- Rathod D., Golinska P., Wypij M., Dahm H., Rai M. A new report of Nocardiopsis valliformis strain OT1 from alkaline Lonar crater of India and its use in synthesis of silver nanoparticles with special reference to evaluation of antibacterial activity and cytotoxicity. Med. Microbiol. Immunol. 2016;205(5):435–447. doi: 10.1007/s00430-016-0462-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadhasivam S., Shanmugam P., Yun K. Biosynthesis of silver nanoparticles by Streptomyces hygroscopicus and antimicrobial activity against medically important pathogenic microorganisms. J. Colloid. Interface. Sci. 2010;81(1):358–362. doi: 10.1016/j.colsurfb.2010.07.036. [DOI] [PubMed] [Google Scholar]
- Sadhasivam S., Shanmugam P., Veerapandian M., Subbiah R., Yun K. Biogenic synthesis of multidimensional gold nanoparticles assisted by Streptomyces hygroscopicus and its electrochemical and antibacterial properties. Biometals. 2012;25(2):351–360. doi: 10.1007/s10534-011-9506-6. [DOI] [PubMed] [Google Scholar]
- Saminathan K. Biosynthesis of silver nanoparticles using soil Actinomycetes Streptomyces sp. Int. J. Curr. Microbiol. App. Sci. 2015;4(3):1073–1083. [Google Scholar]
- Sastry M., Ahmad A., Khan M.I., Kumar R. Biosynthesis of metal nanoparticles using fungi and actinomycete. Curr. Sci. 2003;85(2):162–170. [Google Scholar]
- Saurav K., Kannabiran K. Diversity and optimization of process parameters for the growth of Streptomyces VITSVK9 spp isolated from Bay of Bengal. India. Int. J. Environ. Sci. 2010;1(2):56–65. [Google Scholar]
- Selvakumar P., Viveka S., Prakash S., Jasminebeaula S., Uloganathan R. Antimicrobial activity of extracellularly synthesized silver nanoparticles from marine derived Streptomyces rochei. Int. J. Pharm. Biol. Sci. 2012;3(3):188–197. [Google Scholar]
- Senapati S., Ahmad A., Khan M.I., Sastry M., Kumar R. Extracellular biosynthesis of bimetallic Au–Ag alloy nanoparticles. Small. 2005;1(5):517–520. doi: 10.1002/smll.200400053. [DOI] [PubMed] [Google Scholar]
- Shahverdi A.R., Minaeian S., Shahverdi H.R., Jamalifar H., Nohi A.A. Rapid synthesis of silver nanoparticles using culture supernatants of Enterobacteria: a novel biological approach. Process. Biochem. 2007;42(5):919–923. [Google Scholar]
- Singh A., Prasad K.N., Misra R., Rahman M., Singh S.K., Rai R.P., Srivastava J.K. Increasing trend of heterogeneous vancomycin intermediate Staphylococcus aureus in a Tertiary Care Center of Northern India. Microb. Drug. Resist. 2015;21(5):545–550. doi: 10.1089/mdr.2015.0004. [DOI] [PubMed] [Google Scholar]
- Sivakumar K., Sahu M.K., Thangaradjou T., Kannan L. Research on marine actinobacteria in India. Ind. J. Microbiol. 2007;47(3):186–196. doi: 10.1007/s12088-007-0039-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivalingam P., Antony J.J., Siva D., Achiraman S., Anbarasu K. Mangrove Streptomyces sp. BDUKAS10 as nano-factory for fabrication of bactericidal silver nanoparticles. Colloids. Surf. B Biointerfaces. 2012;98:12–17. doi: 10.1016/j.colsurfb.2012.03.032. [DOI] [PubMed] [Google Scholar]
- Sondi I., Salopek-Sondi B. Silver nanoparticles as antimicrobial agent: a case study on E. coli as a model for Gram-negative bacteria. J. Colloid. Interface. Sci. 2004;275(1):177–182. doi: 10.1016/j.jcis.2004.02.012. [DOI] [PubMed] [Google Scholar]
- Verma H.N., Singh P., Chavan R.M. Gold nanoparticle: synthesis and characterization. Vet. World. 2014;7(2):72. [Google Scholar]
- Vijayakumar R., Murugesan S., Panneerselvam A. Isolation, characterization and antimicrobial activity of actinobacteria from point calimere coastal region, east coast of India. Int. Res J. Pharam. 2010;1:358–365. [Google Scholar]
- Vimal V., Rajan B.M., Kannabiran K. Antimicrobial activity of marine actinomycete, Nocardiopsis sp. VITSVK 5 (FJ973467) Asian. J. Med. Sci. 2009;1(2):57–63. [Google Scholar]
- Willets K.A., Van Duyne R.P. Localized surface plasmon resonance spectroscopy and sensing. Annu. Rev. Phys. Chem. 2007;58:267–297. doi: 10.1146/annurev.physchem.58.032806.104607. [DOI] [PubMed] [Google Scholar]