Abstract
Carriers of large recurrent copy number variants (CNVs) have a higher risk of developing neurodevelopmental disorders. The 16p11.2 distal CNV predisposes carriers to e.g., autism spectrum disorder and schizophrenia. We compared subcortical brain volumes of 12 16p11.2 distal deletion and 12 duplication carriers to 6882 non-carriers from the large-scale brain Magnetic Resonance Imaging collaboration, ENIGMA-CNV. After stringent CNV calling procedures, and standardized FreeSurfer image analysis, we found negative dose-response associations with copy number on intracranial volume and on regional caudate, pallidum and putamen volumes (β = −0.71 to −1.37; P < 0.0005). In an independent sample, consistent results were obtained, with significant effects in the pallidum (β = −0.95, P = 0.0042). The two data sets combined showed significant negative dose-response for the accumbens, caudate, pallidum, putamen and ICV (P = 0.0032, 8.9 × 10−6, 1.7 × 10−9, 3.5 × 10−12 and 1.0 × 10−4, respectively). Full scale IQ was lower in both deletion and duplication carriers compared to non-carriers. This is the first brain MRI study of the impact of the 16p11.2 distal CNV, and we demonstrate a specific effect on subcortical brain structures, suggesting a neuropathological pattern underlying the neurodevelopmental syndromes.
Subject terms: Psychiatric disorders, Neuroscience
Introduction
Carriers of large recurrent copy number variants (CNVs) are at increased risk for developing autism spectrum disorders (ASD), schizophrenia or intellectual disability [1]. While the same CNV may confer risk for each of these neurodevelopmental disorders, carriers show remarkable phenotypic variability [2–5]. Magnetic resonance imaging (MRI) can help unravel possible underlying brain consequences associated with carrying these CNVs and provide novel insight into neuropathological mechanisms [2, 6–10].
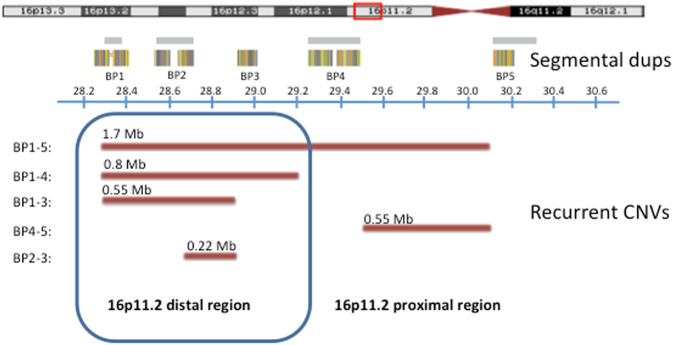
The neural correlates of a recurrent CNV at the distal 16p11.2 locus have remained unexplored, despite being potentially very informative. Low copy repeats at the 16p11.2 locus drive the formation of recurrent CNVs (Fig. 1) [5, 11–15] whose carriers experience increased risk for various neurodevelopmental disorders [3, 16–25] or somatic traits and diseases [17, 26–30]. Within the 16p11.2 region, the segment with breakpoints (BP) at 28.3 and 28.9 Mb (hg18, BP1–BP3) is referred to as the distal region. Within this region, there is a minimal core segment from 28.7 to 28.9 Mb (hg18, BP2–BP3) (Fig. 1) that contains nine genes. The deletion is associated with obesity [26, 27], intellectual disability [26] and schizophrenia [20, 22] and the duplication has been associated with lower body mass index (BMI) [17, 31]. Both 16p11.2 distal CNVs are associated with autism spectrum disorder [17] and have been found in individuals with epilepsy [23]. Several studies have been published about the microstructural effect on the brain [6, 8, 32] and cognition [30, 33, 34] of the 16p11.2 proximal CNV (29.5–30.1 MB) (Fig. 1). In contrast, the biological basis of the 16p11.2 distal phenotypes, including a possible effect on brain structure and cognition remains unknown.
Fig. 1.
Recurrent CNVs in the 16p11.2 region. CNVs are indicated with reddish lines. All coordinates (in MB) are from the human genome build hg18/NCBI 36. This study includes CNVs overlapping the core 16p11.2 distal region (BP2–BP3) of 220 kb (blue box). These CNVs include the 16p11.2 distal BP2–BP3 (~220 kb), the 16p11.2 distal BP1–BP3 (~550 kb), the 16p11.2 distal BP1–BP4 (~800 kb) and the 16p11.2 distal-proximal BP1–BP5 (~1.7 MB) CNVs
Large effect-size CNVs conferring risk for neurodevelopmental disorders including major psychiatric disorders [35] are rare (<0.25% in frequency). Assembling sufficiently powered MRI samples to detect effects of rare CNVs on brain morphometry is challenging. For instance, in the Icelandic population [2] and the UK Biobank [36], the frequencies of the 16p11.2 distal deletion are 0.019% and 0.012%, respectively. Likewise, the reciprocal duplication is found at a frequency of 0.038% and 0.030%, respectively. Hence, studying rare pathogenic CNVs like 16p11.2 distal calls for collaborative efforts. The ENIGMA-CNV consortium has collected a sample that currently includes 16,046 subjects with CNV and brain MRI data.
In recent studies, ‘mirrored phenotypes’ were described in 16p11.2 distal CNV allele carriers for both weight [17, 31] and head circumference [17]. On average, deletion carriers had increased BMI and head circumference, whereas duplication carriers had lower weight and smaller head circumference. Here we investigated gene dose response effects (i.e., effects dependent on the number of genomic copies at the 16p11.2 distal locus) on brain structural measures, including subcortical brain volumes, total surface area, mean cortical thickness and intracranial volume (ICV) in n = 6906 participants from primarily non-familial population samples, in addition to clinical cohorts, to resolve CNV effects relative to a general population average.
Material and methods
Discovery sample description
Supplementary Table 1 contains information on study design, participants, genotyping array, PFB-file and reference for previous description on individual inclusion and exclusion parameters for all 34 world-wide data sets in ENIGMA-CNV; altogether 16,046 individuals with CNV and MRI imaging data from the ENIGMA consortium. The 16p11.2 distal sample consisted of a subset of these individuals with twelve deletion carriers, twelve duplication carriers and 6882 non-carriers from eleven different cohorts and 14 scanner sites collected up until 1 July 2017. More demographic details are supplied in Supplementary Note 1 and Supplementary Table 2 (on CNV carriers).
CNV calls and validation
See Supplementary note 2 for details on CNV calling and quality control. In short, carriers in the 16p11.2 European consortium cohort were identified based on report from the cytogeneticist. All other cohorts had CNVs called in a unified manner using PennCNV [37]. Appropriate population frequency (PFB)-files (Human Genome Build NCBI36/hg18) and GC (content)-model files for each data set were selected from the PennCNV homepage (Supplementary Table 1).
Samples were filtered based on standardized quality control metrics and CNVs overlapping the region of interest (16p11.2 distal BP2–BP3 and BP1–BP3) were identified and visualized with the R package iPsychCNV. The minimally affected 16p11.2 distal region was covered well by all the arrays in the study (Supplementary Figure 1). No carriers carried other genomic imbalances (as defined by Supplementary Table 3) except six 16p11.2 distal-proximal CNV (BP1–BP5) carriers from the 16p11.2 European consortium sample (Supplementary Table 2).
Image acquisition and processing
Supplementary Table 4 outlines-specific technical details concerning scanners and acquisition parameters. The brain measures examined were obtained from structural MRI data collected at participating sites and processed locally following the ENIGMA protocol. The analysis was based on standardized image analysis, FreeSurfer, quality assurance and statistical methods as per the harmonized neuroimaging protocols developed within ENIGMA2 [38] and ENIGMA3 (http://enigma.ini.usc.edu/protocols/imaging-protocols). More details are supplied in Supplementary Note 3.
Statistical analysis
Imaging data processing and CNV calling were performed locally whereupon downstream analysis was performed centrally in a mega-analysis with de-identified data.
The primary analysis for this paper focused on the full set of subjects including family members and data sets with patients to maximize the power to detect effects. Only one of a pair of duplicates was kept. Individuals with a minimum overlap of 0.4 to regions (R package iPsychCNV) with known pathogenic CNVs (Supplementary Table 3) were excluded from the analysis regardless of copy number status. Only scanner sites with individuals carrying a 16p11.2 distal deletion or duplication were included. See Supplementary Note 4 for description of control analyses excluding either (a) individuals with an established neurodevelopmental diagnosis, (b) children below age 18, (c) first and second-degree relatives or (d) carriers of the 1.7 MB 16p11.2 distal-proximal (BP1–BP5) CNV or (e) matching each carrier with four controls or (f) testing the effect of ancestry.
Brain measures were normalized in R 3.2.3 by an inverse normal transformation of the residual of a linear regression on the phenotype correcting for covariates. The final covariance-corrected values (covariates = age, age squared, sex, scanner site and ICV) were used in downstream analysis and are reported for each measure. ICV was not included as a covariate in the analysis of ICV. For analytic purposes, total cortical surface area and total average thickness were normalized in the same way as subcortical volumes. We also performed analysis excluding ICV from the covariates.
For the dose response analysis (i.e., the effect on brain structure of 16p11.2 distal copy number variation), a linear regression on the copy number state of the individuals (deletion = 1, normal = 2, duplication = 3) was performed using the following model: covariance-corrected brain measure ~ copy number (deletion = 1, non-carrier = 2, duplication = 3).
For comparison between groups, a two sample two-sided t-test assuming equal variance in all carrier/non-carrier groups was employed (R 3.3.2) where deletion or duplication carriers were compared either to each other or to non-carriers. Results were considered statistically significant if they exceeded a Bonferroni-corrected P-value (P = 0.05/10 regions = 0.005). We report the uncorrected p-values throughout the manuscript.
Effect size is calculated as the absolute effect size (the difference in mean between the two copy number groups in the t-test—which, in this case, equals Cohen’s d as the standard deviation of the normalized brain measures is one) and the estimate of beta in the linear regression. Plots were generated using R library ggplot2 v2.2.1 [39].
deCODE replication sample
An independent sample of three 16p11.2 distal deletion and, six duplication carriers, as well as 832 non-carriers was obtained from deCODE Genetics, Iceland. CNVs were called with PennCNV as described previously and visually inspected. All 16p11.2 distal carriers were of the minimal 16p11.2 distal (BP2–BP3) CNV type. The individuals were scanned at one scanner site as previously described [7]. The statistical analysis was performed as for the primary discovery sample.
Meta-analysis
A fixed effects model was used to generate summary effect size estimates using a restricted maximum likelihood estimator in the R-package metafor-package [40] (version 1.9-9) using the effect size and calculated SD (for comparison between groups) or standard error (for dose response) from the discovery and replication sample as input. More details can be found in Supplementary Note 5.
IQ, BMI and gene expression analysis
Individuals aged 18–65 years were recruited for cognitive phenotyping based on a large genotyped sample from deCODE. The Icelandic version of the Wechsler Abbreviated Scale of Intelligence (WASIIS) [41, 42] was administered to 1693 non-carriers and all CNV carriers except one deletion carrier. Another 455 controls and one deletion carrier were tested with two subtests, Vocabulary and Matrix Reasoning, from the Wechsler Adult Intelligence Scale (WAIS-III) [43]. More details on tests are available in Supplementary Note 6. Carriers of known pathogenic CNVs (Supplementary Table 3) besides 16p11.2 distal as well as individuals with neurodevelopmental or psychiatric diagnoses were excluded from the analysis. IQ data were not normally distributed and, consequently, the non-parametric Kruskal–Wallis test (R, v3.2.3) was used to test differences in IQ between carrier groups. To test pairwise differences (deletion carriers versus non-carrier-controls, duplication carriers versus non-carrier-controls, deletion versus duplication carriers), we used Wilcoxon rank test in R. We applied a significance threshold of 0.05, without correction for multiple testing since this was secondary analyses. For description of BMI and gene expression analysis, see Supplementary Note 7.
Results
Study participants
In the ENIGMA-CNV discovery data set, we identified 12 16p11.2 distal deletion carriers and 12 duplication carriers scanned at 14 MRI scanners, and 6882 non-carriers investigated at the same MRI scanners. Demographic data are shown in Table 1. Most CNV carriers exhibited the minimal 16p11.2 distal CNV type (BP2–BP3) (Fig. 1), four CNVs were of the extended type (BP1–BP3) and six CNVs extended into the 16p11.2 proximal region (BP1–BP5) (Supplementary Table 2, Supplementary Figure 1). None of the participants carried additional known pathogenic CNVs (Supplementary Table 3).
Table 1.
Demographic data, discovery and replication data
| Discovery, ENIGMA-CNV | Replication, deCODE | |||||||
|---|---|---|---|---|---|---|---|---|
| del | con | dup | P | del | con | dup | P | |
| Individuals | 12 | 6882 | 12 | 3 | 832 | 6 | ||
| Age (mean (sd)) | 27.8 (20.4) | 43.5 (20.0) | 31.2 (19.6) | 0.003 | 48.7 (14) | 46.2 (12) | 47.0 (15) | NS |
| Sex = female (%) | 6 (50%) | 3724 (54%) | 3 (25%) | NS | 2 (67%) | 502 (60%) | 1 (17%) | NS |
| Established diagnosis, n (%) | 7 (58%) | 336 (4.9%) | 3 (25%) | <0.001 | 0 (0%) | 158 (19%) | 2 (33%) | NS |
| Diagnosis type (%)a | ||||||||
| ADHD | 1 (8.3%)b | 2 (<0.1%) | 149 (18%) | 2 (33%) | ||||
| Autism | 2 (17%) | 1 (0.1%) | ||||||
| Bipolar | 1 (8.3 %) | 100 (1.5 %) | 7 (0.8%) | |||||
| Dysthymia | 1 (8.3 %)c | 1 (<0.1%) | ||||||
| Epilepsy | 1 (8.3%) | 101 (1.5%) | ||||||
| Enuresis | 1 (8.3%) | |||||||
| Language disorder | 1 (8.3%)c | |||||||
| MDD | 13 (0.2%) | 1 (8.3%)d | ||||||
| Schizophrenia | 1 (8.3%) | 119 (1.7%) | 1 (0.1%) | |||||
| Scanner sites | 7 | 14 | 9 | 1 | 1 | 1 | ||
| Types of arrayse | 8 | 11 | 9 | Various | Various | Various | ||
| Children, n (%) | 5 (41.7%) | 788 (11.5%) | 5 (41.7%) | <0.001 | 0 (0) | 0 (0) | 0 (0) | |
| Close relative in data set, n (%) | 5 (41.7%) | 1994 (30.8%) | 2 (16.7%) | NS | 3 (0%) | 832 (0%) | 6 (0%) | |
Age, sex, established diagnosis, diagnosis type, number of scanner sites, types of arrays, children (number of individuals below 18 years) and individuals with close relatives (>3rd degree relatives) in the data set. P (p-value) is based on a χ2-test for categorical values and ANOVA for continuous values (R-package, TableOne)
ADHD attention deficit disorder, MDD major depressive disorder, SCZ schizophrenia, del deletion carrier, con non-carriers, dup duplication carrier, NS non-significant
aDiagnosis type specifies the total number within each carrier group with an established neurodevelopmental or psychiatric diagnosis
bIn addition to Specific Learning disorder
cIn addition to Moderate ID and Speech Sound disorder
dIn addition to Social Anxiety disorder
eThe arrays for ENIGMA-CNV are specified in Sup Table 1
Of 24 CNV carriers, 10 had an established neurodevelopmental diagnosis (Supplementary Table 2). The remaining carriers either did not have one or were recruited in studies from which diagnostic information was not available (Supplementary Table 2, Table 1).
There was a significant age difference between the groups (ANOVA, P = 0.003); the non-carriers were older (mean age 43.5 years) in comparison to the deletion (27.8 years) and duplication carriers (31.2 years). In addition, an established neurodevelopmental diagnosis was found in a significantly smaller proportion of non-carriers (4.9%) in comparison to deletion (58%) and duplication carriers (25%) (Table 1).
Brain imaging results in the discovery sample
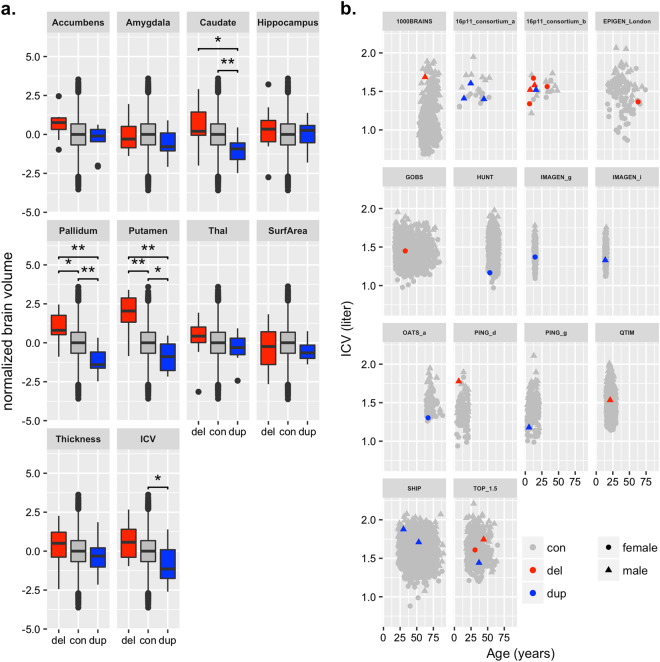
After correction for age, age squared, sex and scanner site, we found a significant negative correlation between the number of 16p11.2 distal copies (deletion = 1, non-carrier = 2, duplication = 3) and ICV (β = −0.71, P = 5.1 × 10−4) (Table 2, Fig. 2a) after correction for multiple testing (significance threshold P < 0.005 = 0.05/10 brain structures analysed), showing smaller ICV in duplication carriers compared to deletion carriers. The uncorrected ICV plotted against age stratified by scanner site are shown in Fig. 2b.
Table 2.
Dose response of 16p11.2 distal copy number on subcortical volumes
| ENIGMA-CNV, discovery | deCODE, replication | Combined (discovery + replication) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Brain measure | β | P | β | P | Q | p(Q) | I2 | β | CI, β, lower | CI, β, upper | P |
| Accumbens | −0.49 | 0.025 | −0.65 | 0.05 | 0.163 | 0.69 | 0 | −0.54 | −0.9 | −0.18 | 0.0032* |
| Amygdala | −0.21 | 0.31 | −0.13 | 0.7 | 0.043 | 0.84 | 0 | −0.19 | −0.52 | 0.15 | 0.27 |
| Caudate | −0.87 | 2.0E–05** | −0.46 | 0.17 | 1.13 | 0.29 | 11.4 | −0.76 | −1.1 | −0.42 | 8.9E–06** |
| Hippocampus | −0.14 | 0.48 | −0.03 | 0.94 | 0.0813 | 0.78 | 0 | −0.11 | −0.45 | 0.22 | 0.52 |
| Pallidum | −1.06 | 2.2E–07** | −0.95 | 0.0042* | 0.0813 | 0.78 | 0 | −1.03 | −1.37 | −0.7 | 1.7E–09** |
| Putamen | −1.37 | 1.8E–11** | −0.7 | 0.034 | 3.01 | 0.083 | 66.8 | −1.19 | −1.53 | −0.85 | 3.5E–12** |
| Thalamus | −0.33 | 0.11 | −0.37 | 0.27 | 0.0105 | 0.92 | 0 | −0.34 | −0.69 | 0.01 | 0.054 |
| Surface Area | −0.09 | 0.66 | 0.14 | 0.68 | 0.355 | 0.55 | 0 | −0.03 | −0.36 | 0.31 | 0.87 |
| Thickness | −0.29 | 0.16 | −0.02 | 0.94 | 0.49 | 0.48 | 0 | −0.22 | −0.55 | 0.12 | 0.2 |
| ICV | −0.71 | 5.1E–04* | −0.54 | 0.1 | 0.194 | 0.66 | 0 | −0.66 | −1 | −0.33 | 1.0E–04** |
The effect size (β of the linear regression) is presented. A linear regression based on the copy number state of the individuals (deletion = 1, non-carrier = 2, duplication = 3) was performed on normalized brain measures correcting for age [2], age, sex and scannersite (and ICV) in the ENIGMA-CNV (discovery) and deCODE (replication) cohorts. Results were considered statistically significant if they were below a Bonferroni-corrected P-value of 0.005 (0.05/10 regions). A final effect size estimate of the combined sample was obtained using a fixed effects meta-analysis framework
CI confidence interval, Q statistics for the test for heterogeneity, p(Q) p-value for the test for heterogeneity, I2 heterogeneity levels
*P < 0.005
**P < 0.0005
Fig. 2.
Measures of caudate, pallidum, putamen and ICV show a dose response to differences in copy number in the 16p11.2 distal region. All analyses were corrected for age, age squared, sex, scanner site and ICV (except for ICV). Deletion carriers (del) in red, non-carriers (con) in grey and duplication carriers (dup) in blue, respectively. a Boxplots of subcortical volumes, surface area and thickness and ICV. The normalized brain values are presented. Boxplots represent the mean. Significant differences after Bonferroni correction between groups are noted as *P < 0.005, **P < 0.0005. Centre line represents median, box limits are the upper and lower 25% quartiles, whiskers the 1.5 interquartile range and the points are the outliers. b Bivariate plot of age versus uncorrected ICV
We evaluated whether the 16p11.2 distal CNV affected seven subcortical (accumbens, caudate, putamen, pallidum, amygdala, hippocampus and thalamus) and two cortical (total surface area and mean cortical thickness) phenotypes. After adjusting subcortical and cortical volumes for age, age squared, sex, scanner site and ICV, the volumes of caudate, pallidum and putamen were negatively associated with the number of 16p11.2 distal copies with significance at the multiple testing threshold (β = −0.87, P = 2.0 × 10−5; β = −1.06, P = 2.2 × 10−7 and β = −1.37, P = 1.8 × 10−11, respectively) (Table 2, Fig. 2a). Plotting the unadjusted volumes of caudate, pallidum and putamen against the age of participants revealed a consistent pattern (deletion carriers with larger and duplication carriers with smaller subcortical volumes in comparison to non-carriers) for all scanner sites for putamen and pallidum volume and almost at all sites for caudate (Supplementary Figure 3a-c). This shows that our findings are robust and not dependent upon a few scanner sites.
To assess non-specific associations, we re-analyzed subcortical volumes without correcting for ICV. As expected, the absolute effect sizes of copy number on the volumes of caudate, pallidum and putamen increased (β = −1.18, P = 6.8 × 10−9; β = −1.27, P = 4.7 × 10−10 and β = −1.57, P = 1.4 × 10−14, respectively) and the association with the volumes of the rest of subcortical structures, except amygdala and hippocampus, became significant (Supplementary Table 5).
To test for the presence of nonlinear differences between deletion and duplication carriers, and deletion or duplication carriers and non-carriers, we conducted individual t-tests between these groups. We confirmed a negative dose response with increasing copy number for the volumes of the caudate, pallidum, putamen and for ICV but no additional structures revealed significant associations at significance threshold of <0.005 (Table 3, Supplementary Table 6, Supplementary Figure 3a-c).
Table 3.
T-test on subcortical volumes between different 16p11.2 distal copy number groups
| ENIGMA-CNV, discovery | deCODE, replication | Combined, discovery + replication | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Brain measure | Cohens D | P | Cohens D | P | n, CN1 | n, CN2 | Q | p(Q) | I2 | Cohens D | CI, lower | CI, upper | P |
| Deletion vs duplication | |||||||||||||
| Accumbens | −0.99 | 0.023 | −1.86 | 0.044 | 13 | 17 | 0.99 | 0.32 | 0 | −1.17 | −1.87 | −0.47 | 1.0E–03* |
| Amygdala | −0.42 | 0.32 | −0.27 | 0.73 | 15 | 18 | 0.031 | 0.86 | 0 | −0.38 | −1.09 | 0.32 | 0.28 |
| Caudate | −1.74 | 0.0013* | −1.25 | 0.25 | 15 | 18 | 0.251 | 0.62 | 0 | −1.63 | −2.44 | −0.82 | 8.0E–05** |
| Hippocampus | −0.29 | 0.57 | −0.16 | 0.79 | 15 | 18 | 0.034 | 0.85 | 0 | −0.22 | −0.91 | 0.47 | 0.53 |
| Pallidum | −2.11 | 1.8E–05** | −2.19 | 0.0038* | 15 | 18 | 0.0091 | 0.92 | 0 | −2.13 | −2.79 | −1.47 | 3.2E–10** |
| Putamen | −2.73 | 5.3E–06** | −1.67 | 0.089 | 15 | 18 | 0.77 | 0.38 | 0 | −2.58 | −3.41 | −1.74 | 1.4E–09** |
| Thalamus | −0.66 | 0.18 | −0.85 | 0.045 | 15 | 17 | 0.079 | 0.78 | 0 | −0.76 | −1.41 | −0.1 | 0.024 |
| Surface area | −0.18 | 0.69 | −0.04 | 0.96 | 15 | 18 | 0.012 | 0.91 | 0 | −0.16 | −0.98 | 0.66 | 0.7 |
| Thickness | −0.58 | 0.25 | 0.17 | 0.84 | 15 | 18 | 0.53 | 0.47 | 0 | −0.41 | −1.26 | 0.45 | 0.35 |
| ICV | −1.41 | 0.0084 | −0.70 | 0.4 | 15 | 18 | 0.65 | 0.42 | 0 | −1.19 | −1.99 | −0.4 | 0.0034* |
| Deletion vs non-carrier-controls | |||||||||||||
| Accumbens | −0.54 | 0.088 | −1.78 | 0.0019* | 13 | 2467 | 3.1 | 0.078 | 67.8 | −0.73 | −1.22 | −0.23 | 0.0039* |
| Amygdala | 0.08 | 0.79 | −0.15 | 0.8 | 15 | 3578 | 0.11 | 0.74 | 0 | 0.04 | −0.49 | 0.56 | 0.89 |
| Caudate | −0.63 | 0.027 | −1.12 | 0.05 | 15 | 3574 | 0.46 | 0.5 | 0 | −0.79 | −1.45 | −0.12 | 0.02 |
| Hippocampus | −0.26 | 0.37 | −0.23 | 0.69 | 15 | 3578 | 0.0031 | 0.96 | 0 | −0.24 | −0.75 | 0.27 | 0.35 |
| Pallidum | −0.82 | 0.0043* | −1.52 | 0.0081 | 15 | 3577 | 0.87 | 0.35 | 0 | −0.95 | −1.51 | −0.38 | 1.0E–03* |
| Putamen | −1.72 | 2.4E–09** | −1.24 | 0.031 | 15 | 3584 | 0.18 | 0.67 | 0 | −1.67 | −2.35 | −0.99 | 1.5E–06** |
| Thalamus | −0.3 | 0.3 | −0.59 | 0.3 | 15 | 3571 | 0.24 | 0.62 | 0 | −0.42 | −1 | 0.16 | 0.15 |
| Surface area | 0.24 | 0.4 | −0.50 | 0.38 | 15 | 4312 | 0.41 | 0.52 | 0 | 0.14 | −0.61 | 0.9 | 0.71 |
| Thickness | −0.28 | 0.32 | 0.43 | 0.45 | 15 | 4312 | 0.66 | 0.42 | 0 | −0.13 | −0.83 | 0.58 | 0.73 |
| ICV | −0.44 | 0.13 | 0.23 | 0.7 | 15 | 4314 | 0.97 | 0.33 | 0 | −0.26 | −0.85 | 0.32 | 0.38 |
| Non-carrier-controls vs duplication | |||||||||||||
| Accumbens | −0.34 | 0.26 | −0.08 | 0.84 | 4433 | 17 | 0.25 | 0.62 | 0 | −0.27 | −0.73 | 0.19 | 0.26 |
| Amygdala | −0.46 | 0.11 | −0.13 | 0.76 | 4533 | 18 | 0.41 | 0.52 | 0 | −0.36 | −0.83 | 0.11 | 0.13 |
| Caudate | −1.04 | 3.0E−04** | −0.12 | 0.77 | 4617 | 18 | 1.9 | 0.17 | 46.2 | −0.92 | −1.37 | −0.47 | 6.1E−05** |
| Hippocampus | 0.06 | 0.85 | 0.08 | 0.84 | 4557 | 18 | 0.0030 | 0.96 | 0 | 0.07 | −0.35 | 0.49 | 0.75 |
| Pallidum | −1.1 | 0.00013** | −0.66 | 0.1 | 4504 | 18 | 2.3 | 0.13 | 56.5 | −0.86 | −1.14 | −0.58 | 1.9E−09** |
| Putamen | −0.85 | 0.0031* | −0.44 | 0.28 | 4454 | 18 | 0.99 | 0.32 | 0 | −0.7 | −1.09 | −0.31 | 4.9E−04** |
| Thalamus | −0.33 | 0.27 | −0.25 | 0.54 | 4562 | 17 | 0.063 | 0.8 | 0 | −0.27 | −0.5 | −0.03 | 0.025 |
| Surface area | −0.33 | 0.25 | 0.46 | 0.26 | 4575 | 18 | 3.0 | 0.086 | 66.1 | −0.19 | −0.53 | 0.16 | 0.28 |
| Thickness | −0.28 | 0.33 | −0.24 | 0.55 | 4578 | 18 | 0.0054 | 0.94 | 0 | −0.27 | −0.75 | 0.21 | 0.28 |
| ICV | −0.88 | 0.0023* | −0.93 | 0.022 | 4690 | 18 | 0.0068 | 0.93 | 0 | −0.9 | −1.45 | −0.35 | 0.0014* |
Deletions versus duplication carriers, deletion carriers versus non-carriers and non-carriers versus duplication carriers in ENIGMA (discovery), deCODE (replication) and the combined sample. T-tests were performed on normalized values of brain measures correcting for age [2], age, sex and scanner site (and ICV). Results were considered significant if they were below a Bonferroni-corrected P-value of 0.005 (0.05/10 regions). A final Cohen’s d-effect size estimate of the combined sample was obtained using a fixed effects meta-analysis framework
CI confidence interval, Q statistics for the test for heterogeneity, p(Q) p-value for the test for heterogeneity, I2 heterogeneity levels
*P < 0.005
**P < 0.0005
To confirm the validity of the results, we carefully checked for the impact of removing subjects known to carry a neurodevelopmental diagnosis, children below age 18, first and second-degree relatives or CNV carriers whose CNVs extended into the 16p11.2 proximal region (Supplementary Tables 5 and 6). Likewise, we redid the analysis matching each carrier with four non-carriers for sex, age, diagnosis status (with/without neurodevelopmental diagnosis) and scanner site and finally in a separate analysis we controlled for population stratification in cohorts with available ancestry (Supplementary Table 7). None of these analyses changed the main results.
Replication in an independent cohort
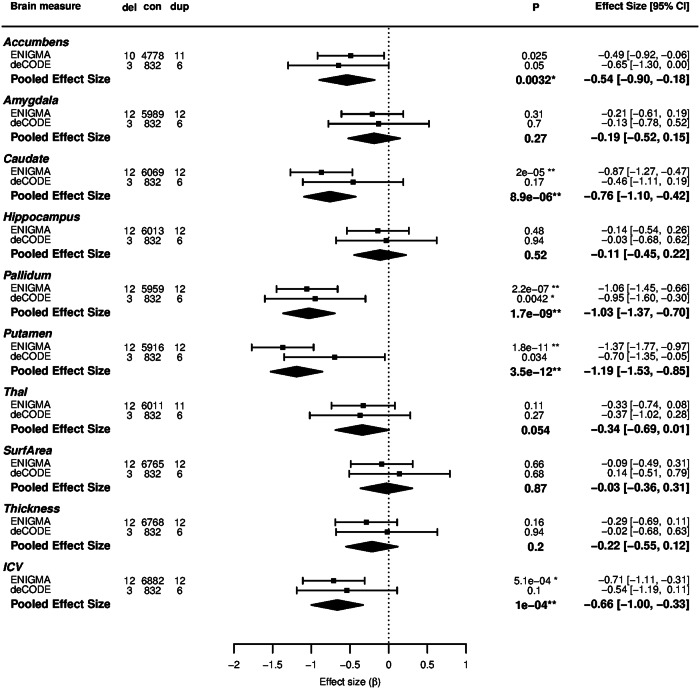
We performed replication of the subcortical findings in an Icelandic MRI sample (deCODE) comprising 841 individuals (3 deletion and 6 duplication carriers, 832 non-carriers) (Table 1). The negative correlation between the number of 16p11.2 distal copies and the volume of pallidum was confirmed (β = −0.95, P = 0.0042) (Fig. 3, Table 2) at a significance threshold of <0.005. For volumes of the caudate, putamen and for ICV, effects were in the same direction as in the discovery sample, albeit not significant (β = −0.46, P = 0.17; β = -0.70, P = 0.034; β = −0.54, P = 0.10, respectively) (Fig. 3, Table 2). Apart from cortical surface area, we observed the same direction of effect in the replication sample as in the discovery sample (Table 2, Fig. 3). For nonlinear differences, all directions of effect were the same for subcortical volumes in the discovery and replication data sets (Table 3, Supplementary Figure 4a-c).
Fig. 3.
Forest plots on the dose response of copy number on subcortical volumes, surface area, thickness and ICV. The effect size (β of the linear regression) at each site for each measure is shown by the position on the x-axis. Standard error is shown by the horizontal line. A summary polygon shows the results when fitting a random-effects model to the two groups: ENIGMA-CNV discovery and deCODE replication samples. del, con and dup denote the number of individuals in each analysis. *P < 0.005, **P < 0.0005. Effect size and confidence intervals are to the right
The combined analysis of the ENIGMA and the deCODE samples is shown in Table 2 and Fig. 3. Volumes of caudate, pallidum, putamen and ICV decreased with increasing number of 16p11.2 distal copies (β = −0.76, P = 8.9 × 10−6; β = −1,03, P = 1.7 × 10−9; β = −1.19, P = 3.5 × 10−12; β = −0.66, P = 1.0 × 10−4, respectively). In the combined analysis, the volume of the accumbens also revealed a significant association with the 16p11.2 distal copy number (β = −0.54, P = 0.0032) (Tables 2 and 3, Fig. 3, Supplementary Figure 4a-c).
Intelligence quotient (IQ)
Full scale IQ data were available for four 16p11.2 distal deletion and twelve duplication carriers and 2148 non-carriers from the Icelandic sample. None of these individuals had an established neurodevelopmental diagnosis, or other known pathogenic CNVs (as defined by Supplementary Table 3). Analysis showed a significant difference in IQ between groups (P = 0.0042). Both deletion (median IQ = 68.5) and duplication carriers (median IQ = 93) presented a significantly lower IQ (P = 0.011, P = 0.035) than non-carriers (median IQ = 101.5) (Supplementary Table 9, Supplementary Figure 5) at a significance threshold of P < 0.05.
Body mass index (BMI)
BMI data for mega-analysis were available for six cohorts from ENIGMA-CNV counting seven deletion and seven duplication carriers in addition to 1880 individuals without a 16p11.2 distal CNV (Supplementary Table 9). BMI z-scores were different between the carrier groups (Kruskal–Wallis, P = 0.009, Supplementary Figure 6, Supplementary Table 10). Duplication carriers had significantly lower BMI z-scores (median; SD = −0.65; 1.61) than the non-carriers (0.43; 1.19; P = 0.048). Also, the duplication carriers tended to have lower BMI z-scores than the deletions (−1.56; 1.1, P = 0.052; Supplementary Figure 6, Supplementary Table 10) and deletion carriers tended to have higher BMI z-scores than non-carriers (P = 0.18) (Supplementary Table 10). For detailed information on individual carriers, see Supplementary Table 2.
Discussion
This is the first study to determine the brain structure underpinnings of the 16p11.2 distal recurrent CNVs. We found a common denominator for 16p11.2 distal carriers across different clinical phenotypes in a dose response effect of copy number on the volumes of basal ganglia (caudate, putamen and pallidum) (Table 2, Figs. 2 and 3). The observed associations were independent of the presence of neurodevelopmental diagnosis and the ancestry of participants (Supplementary Tables 5 and 7). These effects were consistent in the independent replication sample (Fig. 3, Tables 2 and 3). Together with the result of lower IQ in carriers, these findings provide new insight into genetic mechanisms of brain structures and pathobiological processes involved in neurodevelopmental disorders.
There are nine genes in the core 16p11.2 distal (BP2–BP3) region. We tested the expression level of these in blood in available transcript data from two of our deletion carriers (BP1–BP3) and compared with 234 non-carriers (Supplementary Figure 7, Supplementary Table 11). Several transcripts of the nine genes were relatively decreased in blood. Due to the low numbers, only trends can be suggested from these data but we find the down-regulation of LAT gene expression to ~65% in 16p11.2 distal carriers (Supplementary Figure 7, Supplementary Table 11) particular interesting due to recent results in zebrafish: These showed that of the nine genes in 16p11.2 distal, only over-expression of LAT induced a decrease in cell proliferation in the brain with a concomitant microcephaly phenotype [44]. In parallel, LAT knockout mice showed brain anatomy changes [44]. According to the Allen Brain Atlas, LAT shows the highest expression in cerebellum and structures of the basal ganglia (data not shown). Thus, high expression of LAT overlaps with the position of brain structural changes identified in the present study. This further implicates LAT, an immune signaling adaptor, as a possible dosage-dependent driver of the CNV-associated brain phenotypes including the basal ganglia.
By comparing the effect of a range of CNVs on the brain, it is possible to identify patterns of effects related to the genes involved, and thus learn about biological mechanisms. All three CNVs previously shown to have an effect on ICV, 16p11.2 proximal [6, 8], 22q11 [9] and Williams Syndrome [45], have concomitantly identified an effect on either cortical surface area and/or cortical thickness. We observed no effect on cortical surface area or cortical thickness in 16p11.2 distal carriers (Table 2, Figs. 2 and 3). This does not rule out the presence of smaller effects on individual cortical areas. However, it may suggest a different impact on brain development mechanisms between these three CNVs [6, 8, 9, 45] and 16p11.2 distal.
CNVs in the two neighboring regions 16p11.2 distal and 16p11.2 proximal show overlapping phenotypes: they both dispose to various neurodevelopmental diseases and both show a positive dose response for head circumference and weight [17]. In addition, we found negative dose response effects for the 16p11.2 distal CNV on ICV, putamen and caudate volumes with effect size estimates comparable to those previously reported for the 16p11.2 proximal CNV [8]. Recently, a lymphoblastoid cell line study of chromosomal interaction in the 16p11.2 region suggested that the two adjacent 16p11.2 distal and 16p11.2 proximal regions (Fig. 1) interact [17]. The identified brain commonalities could further support a mechanism in which the similar phenotypic patterns are caused by disruption of the chromatin structure surrounding the entire 16p11.2 region [17, 32].
In this study, the CNVs (3 deletions and 3 duplications) of six carriers extend into the adjacent 16p11.2 proximal region. Redoing the analysis with exclusively 16p11.2 distal carriers did not result in a change in effect size (Supplementary Tables 5 + 6), suggesting that 1.7 MB distal-proximal (BP1–BP5) CNV carriers are not the main cause of the signal. A previous analysis from Loviglio et al. [44] suggests an additive effect of two 16p11.2 regions (distal + proximal) on human head circumference and weight. Together with our data, this indicates both separate and overlapping effects for the two CNVs and underlines the importance of studying specific CNVs independently despite overlapping phenotypes.
Interestingly, deletions and duplications of both 16p11.2 proximal and distal CNVs are associated with ASD [17, 19]. However, only the proximal duplication and the distal deletion are associated with schizophrenia [20, 22]. This difference in phenotype association between these bordering CNVs may indicate specific differences in the pathological mechanisms of ASD and schizophrenia.
We observed a decrease in absolute effect sizes for putamen and pallidum after removing individuals with a neurodevelopmental diagnosis (Supplementary Table 5). This is consistent with the enlargements of putamen and pallidum associated with duration of illness in schizophrenia [46], which may partly reflect the cumulative effect of antipsychotic medication on basal ganglia volumes [47]. The observed dose response effect on ICV is in agreement with previous findings on head circumference [17], as are the dose response effect on BMI (Supplementary Figure 6, Supplementary Table 10) [17, 31]. One of the strengths of this study is the inclusion of non-clinical samples allowing for estimates closer to the actual carrier population. Unfortunately, the small number of CNV cases does not provide enough power to investigate preferential alterations in deletion or duplication carriers.
To conclude, the present findings of negative dose-response effects of copy number on ICV and volumes of caudate, pallidum and putamen, with no effect on cortical measures, suggest a specific effect on basal ganglia structures of the 16p11.2 distal CNV. These results provide novel insight into genetic factors determining basal ganglia volumes and suggest specific pathobiological mechanisms involved in the development of neurodevelopmental disorders.
Electronic supplementary material
Acknowledgements
1000BRAINS: 1000BRAINS is a population-based cohort based on the Heinz-Nixdorf Recall Study and is supported in part by the German National Cohort. We thank the Heinz Nixdorf Foundation (Germany) for their generous support in terms of the Heinz Nixdorf Study. The HNR study is also supported by the German Ministry of Education and Science (FKZ 01EG940), and the German Research Council (DFG, ER 155/6-1). The authors are supported by the Initiative and Networking Fund of the Helmholtz Association (Svenja Caspers) and the European Union’s Horizon 2020 Research and Innovation Programme under Grant Agreement 7202070 (Human Brain Project SGA1; Katrin Amunts, Sven Cichon). This work was further supported by the German Federal Ministry of Education and Research (BMBF) through the Integrated Network IntegraMent (Integrated Understanding of Causes and Mechanisms in Mental Disorders) under the auspices of the e:Med Program (grant 01ZX1314A to M.M.N. and S.C.), and by the Swiss National Science Foundation (SNSF, grant 156791 to S.C.).
16p.11.2 European Consortium: B.D. is supported by the Swiss National Science Foundation (NCCR Synapsy, project grant Nr 32003B_159780) and Foundation Synapsis. LREN is very grateful to the Roger De Spoelberch and Partridge Foundations for their generous financial support. This work was supported by grants from the Simons Foundation (SFARI274424) and the Swiss National Science Foundation (31003A_160203) to A.R. and S.J.
Betula: The relevant Betula data collection and analyses were supported by a grant from the Knut & Alice Wallenberg (KAW) to L. Nyberg.
Brainscale: the Brainscale study was supported by the Netherlands Organization for Scientific Research MagW 480-04-004 (Dorret Boomsma), 51.02.060 (Hilleke Hulshoff Pol), 668.772 (Dorret Boomsma & Hilleke Hulshoff Pol); NWO/SPI 56-464-14192 (Dorret Boomsma), the European Research Council (ERC-230374) (Dorret Boomsma), High Potential Grant Utrecht University (Hilleke Hulshoff Pol), NWO Brain and Cognition 433-09-220 (Hilleke Hulshoff Pol).
Brain Imaging Genetics (BIG): This work makes use of the BIG database, first established in Nijmegen, The Netherlands, in 2007. This resource is now part of Cognomics (www.cognomics.nl), a joint initiative by researchers of the Donders Centre for Cognitive Neuroimaging, the Human Genetics and Cognitive Neuroscience departments of the Radboud university medical centre and the Max Planck Institute for Psycholinguistics in Nijmegen. The Cognomics Initiative has received supported from the participating departments and centres and from external grants, i.e., the Biobanking and Biomolecular Resources Research Infrastructure (the Netherlands) (BBMRI-NL), the Hersenstichting Nederland, and the Netherlands Organisation for Scientific Research (NWO). The research leading to these results also receives funding from the NWO Gravitation grant ‘Language in Interaction’, the European Community’s Seventh Framework Programme (FP7/2007–2013) under grant agreements n° 602450 (IMAGEMEND), n°278948 (TACTICS), and n°602805 (Aggressotype) as well as from the European Community’s Horizon 2020 programme under grant agreement n° 643051 (MiND) and from ERC-2010-AdG 268800-NEUROSCHEMA. In addition, the work was supported by a grant for the ENIGMA Consortium (grant number U54 EB020403) from the BD2K Initiative of a cross-NIH partnership.
COBRE: This work was supported by a NIH COBRE Phase I grant (1P20RR021938, Lauriello, PI and 2P20GM103472, Calhoun, PI) awarded to the Mind Research Network. We wish to express our gratitude to numerous investigators who were either external consultants to the Cores and projects, mentors on the projects, members of the external advisory committee and members of the internal advisory committee.
Decode: The research leading to these results has received financial contribution from the European Union’s Seventh Framework Programme (EU-FP7/2007–2013), EU-FP7 funded grant no. 602450 (IMAGEMEND) as well as support from the Innovative Medicines Initiative Joint Undertaking under grant agreement no.115300 (EUAIMS).
DemGene: Norwegian Health Association and Research Council of Norway.
Dublin: Work was supported by Science Foundation Ireland (SFI grant 12/IP/1359 to Gary Donohoe and SFI08/IN.1/B1916-Corvin to Aidan C Corvin) and the European Research Council (ERC-StG-2015-677467).
EPIGEN-UK (SMS, CL): The work was partly undertaken at UCLH/UCL, which received a proportion of funding from the UK Department of Health’s NIHR Biomedical Research Centres funding scheme. We are grateful to the Wolfson Trust and the Epilepsy Society for supporting the Epilepsy Society MRI scanner, and the Epilepsy Society for supporting CL.
Haavik: The work at the K.G.Jebsen center for neuropsychiatric disorders at the University of Bergen, Norway, was supported by Stiftelsen K.G. Jebsen, European Community’s Seventh Framework Program under grant agreement no 602805 and the H2020 Research and Innovation Program under grant agreement numbers 643051 and 667302.
HUNT: The HUNT Study is a collaboration between HUNT Research Centre (Faculty of Medicine, Norwegian University of Science and Technology), Nord-Trøndelag County Council, Central Norway Health Authority, and the Norwegian Institute of Public Health. HUNT-MRI was funded by the Liaison Committee between the Central Norway Regional Health Authority and the Norwegian University of Science and Technology, and the Norwegian National Advisory Unit for functional MRI.
IMAGEN: The work received support from the European Union-funded FP6Integrated Project IMAGEN (Reinforcement-related behaviour in normal brain function and psychopathology) (LSHM-CT- 2007-037286), the Horizon 2020 funded ERC Advanced Grant ‘STRATIFY’ (Brain network based stratification of reinforcement-related disorders) (695313), ERANID (Understanding the Interplay between Cultural, Biological and Subjective Factors in Drug Use Pathways) (PR-ST-0416-10004), BRIDGET (JPND: BRain Imaging, cognition Dementia and next generation GEnomics) (MR/N027558/1), the FP7 projects IMAGEMEND (602450; IMAging GEnetics for MENtal Disorders) and MATRICS (603016), the Innovative Medicine Initiative Project EU-AIMS (115300), the Medical Research Council Grant ‘c-VEDA’ (Consortium on Vulnerability to Externalizing Disorders and Addictions) (MR/N000390/1), the Swedish Research Council FORMAS, the Medical Research Council, the National Institute for Health Research (NIHR) Biomedical Research Centre at South London and Maudsley NHS Foundation Trust and King’s College London, the Bundesministeriumfür Bildung und Forschung (BMBF grants 01GS08152; 01EV0711; eMED SysAlc01ZX1311A; Forschungsnetz AERIAL), the Deutsche Forschungsgemeinschaft (DFG grants SM 80/7-1, SM 80/7-2, SFB 940/1). Further support was provided by grants from: ANR (project AF12-NEUR0008-01—WM2NA, and ANR-12-SAMA-0004), the Fondation de France, the Fondation pour la Recherche Médicale, the Mission Interministérielle de Lutte-contre-les-Drogues-et-les-Conduites-Addictives (MILDECA), the Assistance-Publique-Hôpitaux-de-Paris and INSERM (interface grant), Paris Sud University IDEX 2012; the National Institutes of Health, Science Foundation Ireland (16/ERCD/3797), USA (Axon, Testosterone and Mental Health during Adolescence; RO1 MH085772-01A1), and by NIH Consortium grant U54 EB020403, supported by a cross-NIH alliance that funds Big Data to Knowledge Centres of Excellence.
MCIC: This work was supported primarily by the Department of Energy DE-FG02-99ER62764 through its support of the Mind Research Network and the consortium as well as by the National Association for Research in Schizophrenia and Affective Disorders (NARSAD) Young Investigator Award (to SE) as well as through the Blowitz-Ridgeway and Essel Foundations, and through NWO ZonMw TOP 91211021, the DFG research fellowship (to SE), the Mind Research Network, National Institutes of Health through NCRR 5 month-RR001066 (MGH General Clinical Research Center), NIMH K08 MH068540, the Biomedical Informatics Research Network with NCRR Supplements to P41 RR14075 (MGH), M01 RR 01066 (MGH), NIBIB R01EB006841 (MRN), R01EB005846 (MRN), 2R01 EB000840 (MRN), 1RC1MH089257 (MRN), as well as grant U24 RR021992.
NCNG: this sample collection was supported by grants from the Bergen Research Foundation and the University of Bergen, the Dr Einar Martens Fund, the K.G. Jebsen Foundation, the Research Council of Norway, to SLH, VMS and TE. The Bergen group was supported by grants from the Western Norway Regional Health Authority (Grant 911593 to AL, Grant 911397 and 911687 to AJL).
NESDA: Funding for NESDA was obtained from the Netherlands Organization for Scientific Research (Geestkracht program grant 10-000-1002); the Center for Medical Systems Biology (CSMB, NWO Genomics), Biobanking and Biomolecular Resources Research Infrastructure (BBMRI-NL), VU University’s Institutes for Health and Care Research (EMGO+) and Neuroscience Campus Amsterdam, University Medical Center Groningen, Leiden University Medical Center, National Institutes of Health (NIH, R01D0042157-01A, MH081802, Grand Opportunity grants 1RC2 MH089951 and 1RC2 MH089995). Part of the genotyping and analyses were funded by the Genetic Association Information Network (GAIN) of the Foundation for the National Institutes of Health.Computing was supported by BiG Grid, the Dutch e-Science Grid, which is financially supported by NWO.
NTR: The NTR study was supported by the Netherlands Organization for Scientific Research (NWO), MW904-61-193 (Eco de Geus & Dorret Boomsma), MaGW-nr: 400-07- 080 (Dennis van ‘t Ent), MagW 480-04-004 (Dorret Boomsma), NWO/SPI 56-464-14192 (Dorret Boomsma), the European Research Council, ERC-230374 (Dorret Boomsma), and Amsterdam Neuroscience.
OATS: OATS (Older Australian Twins Study) was facilitated by access to Twins Research Australia, which is funded by a National Health and Medical Research Council (NHMRC) Enabling Grant 310667. OATS is also supported via a NHMRC/Australian Research Council Strategic Award (401162) and a NHMRC Project Grant (1045325). DNA extraction was performed by Genetic Repositories Australia, which was funded by a NHMRC Enabling Grant (401184). OATS genotyping was partly funded by a Commonwealth Scientific and Industrial Research Organisation Flagship Collaboration Fund Grant.
PAFIP: PAFIP data were collected at the Hospital Universitario Marqués de Valdecilla, University of Cantabria, Santander, Spain, under the following grant support: Carlos III Health Institute PIE14/00031 and SAF2013-46292-R and SAF2015-71526-REDT. We wish to acknowledge IDIVAL Neuroimaging Unit for imaging acquirement and analysis.We want to particularly acknowledge the patients and the BioBankValdecilla (PT13/0010/0024) integrated in the Spanish National Biobanks Network for its collaboration.
QTIM: The QTIM study was supported by grants from the US National Institute of Child Health and Human Development (R01 HD050735) and the Australian National Health and Medical Research Council (NHMRC) (486682, 1009064). Genotyping was supported by NHMRC (389875). Lachlan Strike is supported by an Australian Postgraduate Award (APA). AFM is supported by NHMRC CDF 1083656. We thank the twins and siblings for their participation, the many research assistants, as well as the radiographers, for their contribution to data collection and processing of the samples.
SHIP: SHIP is part of the Community Medicine Research net of the University of Greifswald, Germany, which is funded by the Federal Ministry of Education and Research (grants no. 01ZZ9603, 01ZZ0103, 01ZZ0403 and 01ZZ0701), the Ministry of Cultural Affairs as well as the Social Ministry of the Federal State of Mecklenburg-West Pomerania, and the network ‘Greifswald Approach to Individualized Medicine (GANI_MED)’ funded by the Federal Ministry of Education and Research (grant 03IS2061A). Genome-wide data have been supported by the Federal Ministry of Education and Research (grant no. 03ZIK012) and a joint grant from Siemens Healthineers, Erlangen, Germany and the Federal State of Mecklenburg- West Pomerania. Whole-body MR imaging was supported by a joint grant from Siemens Healthineers, Erlangen, Germany and the Federal State of Mecklenburg West Pomerania. The University of Greifswald is a member of the Caché Campus program of the InterSystems GmbH.
StrokeMRI: StrokeMRI has been supported by the Research Council of Norway (249795), the South-Eastern Norway Regional Health Authority (2014097, 2015044, 2015073) and the Norwegian ExtraFoundation for Health and Rehabilitation.
TOP: TOP is supported by the Research Council of Norway (223273, 213837, 249711), the South East Norway Health Authority (2017-112), the Kristian Gerhard Jebsen Stiftelsen (SKGJ‐MED‐008) and the European Community’s Seventh Framework Programme (FP7/2007–2013), grant agreement no. 602450 (IMAGEMEND). We acknowledge the technical support and service from the Genomics Core Facility at the Department of Clinical Science, the University of Bergen
for the 16p11.2 European Consortium; for the ENIGMA-CNV working group
Ida Elken Sønderby (NORMENT, K.G. Jebsen Centre for Psychosis Research, Institute of Clinical Medicine, University of Oslo and Division of Mental Health and Addiction, Oslo University Hospital, Oslo, Norway), Ómar Gústafsson (deCODE Genetics/Amgen, Reykjavik, Iceland), Nhat Trung Doan (NORMENT, K.G. Jebsen Centre for Psychosis Research, Institute of Clinical Medicine, University of Oslo and Division of Mental Health and Addiction, Oslo University Hospital, Oslo, Norway), Derrek Paul Hibar (Imaging Genetics Center, Mark and Mary Stevens Neuroimaging and Informatics Institute, Keck School of Medicine of the University of Southern California, Marina del Rey, USA), (Janssen Research and Development, La Jolla, CA USA, Imaging Genetics Center, Mark and Mary Stevens Neuroimaging and Informatics Institute, Keck School of Medicine of the University of Southern California, Marina del Rey, U.S.A), Sandra Martin-Brevet (Service of Medical Genetics, Centre Hospitalier Universitaire Vaudois and University of Lausanne, Rue du Bugnon 46, 1011 Lausanne, Switzerland), Abdel Abdellaoui (Biological Psychology, Vrije Universiteit Amsterdam, van Boechorststraat 1, 1081 BT Amsterdam, The Netherlands), (Department of Psychiatry, Academic Medical Center, Amsterdam, the Netherlands), David Ames (National Ageing Research Institute, Melbourne, Australia), (Academic Unit for Psychiatry of Old Age, University of Melbourne, Melbourne, Australia), Katrin Amunts (Institute of Neuroscience and Medicine (INM-1), Research Centre Juelich, Wilhelm-Johnen-Str., 52425 Juelich, Germany), (C. and O. Vogt Institute for Brain Research, Medical Faculty, University of Dusseldorf, Merowingerplatz 1A, 40225 Dusseldorf, Germany), (JARA-BRAIN, Juelich-Aachen Research Alliance, Wilhelm-Johnen-Str., 52425 Juelich, Germany), Michael Andersson (Umeå Center for Functional Brain Imaging (UFBI), Umeå University, 90187 Umeå, Sweden), Nicola J. Armstrong (Mathematics and Statistics, Murdoch University, Perth, Australia), Manon Bernard (The Hospital for Sick Children, University of Toronto, Toronto, M5G 1X8, Canada), Nicholas Blackburn (South Texas Diabetes and Obesity Institute, Department of Human Genetics, School of Medicine, University of Texas Rio Grande Valley, One West University Blvd., 78520 Brownsville, TX, USA), John Blangero (South Texas Diabetes and Obesity Institute, Department of Human Genetics, School of Medicine, University of Texas Rio Grande Valley, One West University Blvd., 78520 Brownsville, TX, USA), Dorret I Boomsma (Netherlands Twin Register, Vrije Universiteit, van der Boechorststraat 1, 1081BT Amsterdam, Netherlands), Janita Bralten (Department of Human Genetics, Radboud University Medical Center, Nijmegen, the Netherlands), Hans-Richard Brattbak (Department of Clinical Science, University of Bergen, Bergen, Norway), (Center for Medical Genetics and Molecular Medicine, Haukeland University Hospital, Bergen, Norway), Henry Brodaty (Centre for Healthy Brain Ageing and Dementia Collaborative Research Centre, UNSW, Sydney, Australia), Rachel M. Brouwer (Department of Psychiatry, Brain Center Rudolf Magnus, University Medical Center Utrecht, Utrecht, The Netherlands), Robin Bülow (Department of Diagnostic Radiology and Neuroradiology, University Medicine Greifswald, Greifswald, Germany), Vince Calhoun (The Mind Research Network, The University of New Mexico, Albuquerque, NM), Svenja Caspers (Institute of Neuroscience and Medicine (INM-1), Research Centre Juelich, Wilhelm-Johnen-Str., 52425 Juelich, Germany), (C. and O. Vogt Institute for Brain Research, Medical Faculty, University of Dusseldorf, Merowingerplatz 1A, 40225 Dusseldorf, Germany), (JARA-BRAIN, Juelich-Aachen Research Alliance, Wilhelm-Johnen-Str., 52425 Juelich, Germany), Gianpiero Cavalleri (The Royal College of Surgeons in Ireland, 123 St Stephen's Green, Dublin 2, Ireland), Chi-Hua Chen (Department of Radiology, University of California San Diego, La Jolla, USA), (Center for Multimodal Imaging and Genetics, University of California San Diego, La Jolla, USA), Sven Cichon (Institute of Neuroscience and Medicine (INM-1), Structural and Functional Organisation of the Brain, Genomic Imaging, Research Centre Juelich, Leo-Brandt-Strasse 5, 52425 Jülich, Germany), (Human Genomics Research Group, Department of Biomedicine, University of Basel, Hebelstrasse 20, 4031 Basel, Switzerland), (Institute of Medical Genetics and Pathology, University Hospital Basel, Schönbeinstrasse 40, 4031 Basel, Switzerland), Simone Ciufolini (Psychosis Studies, Insitute of Psychiatry, Psychology and Neuroscience, King's College London, 16 De Crespingy Park, SE5 8AF London, United Kingdom), Aiden Corvin (Neuropsychiatric Genetics Research Group, Discipline of Psychiatry, School of Medicine, Trinity College Dublin, Dublin 2, Ireland.), Benedicto Crespo-Facorro (Department of Medicine and Psychiatry, University Hospital Marque?s de Valdecilla, School of Medicine, University of Cantabria-IDIVAL, 39008 Santander, Spain), (CIBERSAM (Centro Investigacio?n Biome?dica en Red Salud Mental), Santander, 39011, Spain), Joanne E. Curran (South Texas Diabetes and Obesity Institute, Department of Human Genetics, School of Medicine, University of Texas Rio Grande Valley, One West University Blvd., 78520 Brownsville, TX, USA), Anders M Dale (Center for Multimodal Imaging and Genetics, University of California San Diego, La Jolla, USA), Shareefa Dalvie (Department of Psychiatry and Mental Health, Anzio Road, 7925 Cape Town, South Africa), Paola Dazzan (Department of Psychosis Studies, Institute of Psychiatry, Psychology and Neuroscience, King's College London, De Crespigny Park, SE5 8AF London, United Kingdom), (National Institute for Health Research (NIHR) Mental Health Biomedical Research Centre at South London and Maudsley NHS Foundation Trust and King's College London, United Kingdom), Eco JC de Geus (Department of Biological Psychology, Behavioral and Movement Sciences, Vrije Universiteit, van der Boechorststraat 1, 1081 BT Amsterdam, Netherlands), (Amsterdam Neuroscience, VU University medical center, van der Boechorststraat 1, 1081 BT Amsterdam, NH, Netherlands), Greig I. de Zubicaray (Faculty of Health and Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, Queensland, Australia), Sonja M.C. de Zwarte (Department of Psychiatry, Brain Center Rudolf Magnus, University Medical Center Utrecht, Utrecht, The Netherlands), Norman Delanty (The Royal College of Surgeons in Ireland, 123 St Stephen's Green, Dublin 2, Ireland), (Imaging of Dementia and Aging (IDeA) Laboratory, Department of Neurology and Center for Neuroscience, University of California at Davis, 4860 Y Street, Suite 3700, Sacramento, California 95817, USA.), Anouk den Braber (Department of Biological Psychology, Behavioral and Movement Sciences, Vrije Universiteit, van der Boechorststraat 1, 1081 BT Amsterdam, Netherlands), (Alzheimer Center and Department of Neurology, VU University Medical Center,, De Boelelaan 1105, 1081HV Amsterdam Amsterdam, Amsterdam), Sylvane Desrivières (Medical Research Council - Social, Genetic and Developmental Psychiatry Centre, Institute of Psychiatry, Psychology & Neuroscience, King's College London, United Kingdom), Gary Donohoe (Cognitive Genetics and Cognitive Therapy Group, Neuroimaging, Cognition & Genomics Centre (NICOG) & NCBES Galway Neuroscience Centre, School of Psychology and Discipline of Biochemistry, National University of Ireland Galway, H91 TK33, Galway, Ireland), (Neuropsychiatric Genetics Research Group, Department of Psychiatry and Trinity College Institute of Psychiatry, Trinity College Dublin, Dublin 8, Ireland), Bogdan Draganski (LREN - Département des neurosciences cliniques, Centre Hospitalier Universitaire Vaudois and University of Lausanne, Lausanne, Switzerland), (Max-Planck-Institute for Human Cognitive and Brain Sciences, Leipzig, Germany), Stefan Ehrlich (Division of Psychological and Social Medicine and Developmental Neurosciences, Faculty of Medicine, TU Dresden, 01307 Dresden, Germany), (Department of Psychiatry, Massachusetts General Hospital, Boston, Massachusetts 02114, USA), (Martinos Center for Biomedical Imaging, Massachusetts General Hospital, Charlestown, Massachusetts 02129, USA), Thomas Espeseth (NORMENT, K.G. Jebsen Centre for Psychosis Research, Institute of Clinical Medicine, University of Oslo and Division of Mental Health and Addiction, Oslo University Hospital, Oslo, Norway), (Department of Psychology, University of Oslo, Oslo, Norway), Simon E. Fisher (Language and Genetics Department, Max Planck Institute for Psycholinguistics, Wundtlaan 1, 6525 XD Nijmegen, Netherlands), (Donders Institute for Brain, Cognition and Behaviour, Radboud University, Nijmegen, Netherlands), Barbara Franke (Department of Human Genetics, Radboud University Medical Center, Nijmegen, the Netherlands), (Donders Institute for Brain, Cognition and Behaviour, Radboud University, Nijmegen, Netherlands), (Department of Psychiatry, Radboud University Medical Center, Nijmegen, The Netherlands), Vincent Frouin (NeuroSpin, CEA, Université Paris-Saclay, F-91191 Gif-sur-Yvette, France), Masaki Fukunaga (Division of Cerebral Integration, National Institute for Physiological Sciences, Aichi, Japan), Thomas Gareau (NeuroSpin, CEA, Université Paris-Saclay, F-91191 Gif-sur-Yvette, France), David C. Glahn (Yale University School of Medicine, 40 Temple Street, Suite 6E, 6511 New Haven, Vaud, USA), (Olin Neuropsychiatric Research Center, Institute of Living, Hartford Hospital, 300 George Street, 6106 Hartford, CT, USA), Hans Grabe (Department of Psychiatry und Psychotherapy, University Medicine Greifswald, Germany), Nynke A. Groenewold (Department of Psychiatry and Mental Health, Anzio Road, 7925 Cape Town, South Africa), Jan Haavik (K.G. Jebsen Centre for Neuropsychiatric Disorders, University of Bergen, Norway), Asta Håberg (Department of Neuroscience, Faculty of Medicine, Norwegian University of Science and Technology), Ryota Hashimoto (Molecular Research Center for Children's Mental Development, United Graduate School of Child Development, Osaka University, Suita, Osaka, Japan), Jayne Y. Hehir-Kwa (Princess Máxima Center for Pediatric Oncology, Lundlaan 6, 3584 EA Utrecht, The Netherlands), Andreas Heinz (Dept. of Psychiatry and Psychotherapie, Charite, Humboldt University, Chariteplatz 1, 10017 Berlin, Germany), Manon H.J. Hillegers (Department of Psychiatry, Brain Center Rudolf Magnus, University Medical Center Utrecht, Utrecht, The Netherlands), (Child and adolescent Psychiatry / Psychology, Erasmus medical center-Sophia's Childerens hospitaal, Rotterdam, Wytemaweg 8, 3000 CB Rotterdam, The Netherlands), Per Hoffmann (Institute of Neuroscience and Medicine (INM-1), Structural and Functional Organisation of the Brain, Genomic Imaging, Research Centre Juelich, Leo-Brandt-Strasse 5, 52425 Jülich, Germany), (Human Genomics Research Group, Department of Biomedicine, University of Basel, Hebelstrasse 20, 4031 Basel, Switzerland), (Institute of Human Genetics, University of Bonn, Sigmund-Freud-Str. 25, 53127 Bonn, Germany), Laurena Holleran (The Centre for Neuroimaging & Cognitive Genomics (NICOG) and NCBES Galway Neuroscience Centre, National University of Ireland Galway, Galway, Ireland), Jouke-Jan Hottenga (Biological Psychology, Vrije Universiteit Amsterdam, van Boechorststraat 1, 1081 BT Amsterdam, The Netherlands), Hilleke E. Hulshoff (Department of Psychiatry, Brain Center Rudolf Magnus, University Medical Center Utrecht, Utrecht, The Netherlands), Masashi Ikeda (Department of Psychiatry, Fujita Health University School of Medicine, Toyoake, Japan), Neda Jahanshad (Imaging Genetics Center, Mark and Mary Stevens Neuroimaging and Informatics Institute, Keck School of Medicine of the University of Southern California, Marina del Rey, USA), Terry Jernigan (Center for Human Development, University of California San Diego, CA, USA), Christiane Jockwitz (Institute of Neuroscience and Medicine (INM-1), Research Centre Juelich, Wilhelm-Johnen-Str., 52425 Juelich, Germany), (JARA-BRAIN, Juelich-Aachen Research Alliance, Wilhelm-Johnen-Str., 52425 Juelich, Germany), (Department of Psychiatry, Psychotherapy and Psychosomatics, RWTH Aachen University, Medical Faculty, Pauwelsstraße 30, 52074 Aachen, Germany), Stefan Johansson (Center for Medical Genetics and Molecular Medicine, Haukeland University Hospital, Bergen, Norway), (K.G. Jebsen Centre for Neuropsychiatric Disorders, University of Bergen, Norway), Gudrun A. Jonsdottir (deCODE Genetics/Amgen, Reykjavik, Iceland), Erik G. Jönsson (NORMENT, K.G. Jebsen Centre for Psychosis Research, Institute of Clinical Medicine, University of Oslo and Division of Mental Health and Addiction, Oslo University Hospital, Oslo, Norway), (Department of Clinical Neuroscience, Centre for Psychiatric Research, Karolinska Institutet, Karolinska University Hospital Solna, R5:00, SE-17176 Stockholm, Sweden), Rene Kahn (Department of Psychiatry, Brain Center Rudolf Magnus, University Medical Center Utrecht, Utrecht, The Netherlands), Tobias Kaufmann (NORMENT, K.G. Jebsen Centre for Psychosis Research, Institute of Clinical Medicine, University of Oslo and Division of Mental Health and Addiction, Oslo University Hospital, Oslo, Norway), Sinead Kelly (The Centre for Neuroimaging & Cognitive Genomics (NICOG) and NCBES Galway Neuroscience Centre, National University of Ireland Galway, Galway, Ireland), Masataka Kikuchi (Department of Genome Informatics, Graduate School of Medicine, Osaka University, 2-2, Yamadaoka, Suita, Osaka 565-0871, Japan), Emma EM. Knowles (Department of Psychiatry, Yale University, 40 Temple Street, 6515 New Haven, CT, USA), Knut K. Kolskår (NORMENT, K.G. Jebsen Centre for Psychosis Research, Institute of Clinical Medicine, University of Oslo and Division of Mental Health and Addiction, Oslo University Hospital, Oslo, Norway), (Department of Psychology, University of Oslo, Oslo, Norway), (Sunnaas Rehabilitation Hospital HT, Nesodden, Norway), John B. Kwok (Brain and Mind Centre, University of Sydney, Australia), Stephanie Le Hellard (NORMENT - KG Jebsen Centre, Department of Clinical Science, University of Bergen, Jonas Lies veg 87, 5021 Bergen, Norway), (Department of Clinical and Experimental Epilepsy, UCL Institute of Neurology, London, and Chalfont Centre for Epilepsy, UK), Costin Leu (Department of Clinical and Experimental Epilepsy, NIHR University College London Hospitals Biomedical Research Centre, UCL Institute of Neurology, 33 Queen Square, WC1N 3BG London, United Kingdom), Jingyu Liu (The Mind Research Network, 1101 Yale Blvd., 87106 Albuquerque, CT, USA), (Dept. of Electrical and Computer Engineering, University of New Mexico, 87131 Albuquerque, New Mexico, USA), Astri J. Lundervold (Yale University School of Medicine, 40 Temple Street, Suite 6E, 6511 New Haven, Vaud, USA), (Department of Biological and Medical Psychology, Jonas Lies vei 91, N-5009 Bergen, Norway), Arvid Lundervold (Department of Biomedicine, University of Bergen, 5009, Bergen, Norway), Nicholas G. Martin (QIMR Berghofer Medical Research Institute, Brisbane, Queensland, Australia), Karen Mather (Centre for Healthy Brain Ageing, School of Psychiatry, University of New South Wales, Sydney, New South Wales 2052, Australia), Samuel R. Mathias (Department of Psychiatry, Yale University, 40 Temple Street, 6515 New Haven, CT, USA), Mark McCormack (Molecular and Cellular Therapeutics, Royal College of Surgeons in Ireland, 123 St. Stephens Green, D02 YN77 Dublin, Ireland), (Centre for Molecular Medicine, University Medical Center Utrecht, Heidelberglaan 100, 3584 CX Utrecht, Netherlands), Katie L. McMahon (Centre for Advanced Imaging, University of Queensland, Brisbane, Queensland, Australia), Allan McRae (Program in Complex Trait Genomics, Institute for Molecular Bioscience, University of Queensland, St Lucia, Queensland, Australia), Yuri Milaneschi (Department of Psychiatry, Amsterdam Public Health and Amsterdam Neuroscience, VU University Medical Center/GGZ inGeest, Amsterdam, The Netherlands, Oldenaller 1, 1081 HJ Amsterdam, The Netherlands), Clara Moreau (CHU Sainte-Justine Research Center, Université de Montréal, Montréal, QC, Canada), Derek Morris (Cognitive Genetics and Cognitive Therapy Group, Neuroimaging, Cognition & Genomics Centre (NICOG) & NCBES Galway Neuroscience Centre, School of Psychology and Discipline of Biochemistry, National University of Ireland Galway, H91 TK33, Galway, Ireland), (Neuropsychiatric Genetics Research Group, Department of Psychiatry and Trinity College Institute of Psychiatry, Trinity College Dublin, Dublin 8, Ireland), David Mothersill (The Centre for Neuroimaging & Cognitive Genomics (NICOG) and NCBES Galway Neuroscience Centre, National University of Ireland Galway, Galway, Ireland), Thomas W. Mühleisen (Institute of Neuroscience and Medicine (INM-1), Structural and Functional Organisation of the Brain, Genomic Imaging, Research Centre Juelich, Leo-Brandt-Strasse 5, 52425 Jülich, Germany), (Human Genomics Research Group, Department of Biomedicine, University of Basel, Hebelstrasse 20, 4031 Basel, Switzerland), Robin Murray (Departments of Psychosis Studies, Institute of Psychiatry, Psychology & Neuroscience, King's College London, London, United Kingdom), Jan E. Nordvik (Sunnaas Rehabilitation Hospital HT, Nesodden, Norway), Lars Nyberg (Umeå Center for Functional Brain Imaging (UFBI), Umeå University, 90187 Umeå, Sweden), Loes M. Olde Loohuis (Center for Neurobehavioral Genetics, University of California, Los Angeles, California 90095, USA), Roel Ophoff (Department of Psychiatry, Brain Center Rudolf Magnus, University Medical Center Utrecht, Utrecht, The Netherlands), (Center for Neurobehavioral Genetics, University of California, Los Angeles, California 90095, USA), Tomas Paus (Rotman Research Institute, University of Toronto, Toronto, M6A 2E1, Canada), (Department of Psychiatry, University of Toronto, Toronto, M5S 1A1, Canada), (Center for Developing Brain, Child Mind Institute, New York, NY, 10022, USA), (Department of Psychology, University of Toronto, Toronto, M5S 1A1, Canada), Zdenka Pausova (The Hospital for Sick Children, University of Toronto, Toronto, M5G 1X8, Canada), Brenda Penninx (Department of Psychiatry, Amsterdam Public Health and Amsterdam Neuroscience, VU University Medical), Juan M. Peralta (South Texas Diabetes and Obesity Institute, Department of Human Genetics, School of Medicine, University of Texas Rio Grande Valley, One West University Blvd., 78520 Brownsville, TX, USA), Bruce Pike (Departments of Radiology & Clinical Neuroscience, University of Calgary, Calgary, T2N 1N4, Canada), Carlos Prieto (Bioinformatics Service, Nucleus, University of Salamanca (USAL), 37007 Salamanca, Spain), Sara Pudas (Umeå Center for Functional Brain Imaging (UFBI), Umeå University, 90187 Umeå, Sweden), (Department of Integrative Medical Biology, Linnéus väg 9, 901 87 Umeå, Sweden), Erin Quinlan (Centre for Population Neuroscience and Stratified Medicine, Social, Genetic and Development Psychiatry Centre, Institute of Psychiatry, Psychology and Neuroscience, King's College London, 16 De Crespigny Park, SE5 8AF London, UK), Daniel S. Quintana (NORMENT, K.G. Jebsen Centre for Psychosis Research, Institute of Clinical Medicine, University of Oslo and Division of Mental Health and Addiction, Oslo University Hospital, Oslo, Norway), (Department of Psychology, University of Oslo, Oslo, Norway), Céline S. Reinbold (Human Genomics Research Group, Department of Biomedicine, University of Basel, Hebelstrasse 20, 4031 Basel, Switzerland), (Institute of Medical Genetics and Pathology, University Hospital Basel, Schönbeinstrasse 40, 4031 Basel, Switzerland), Tiago Reis Marques (Department of Psychosis Studies, Institute of Psychiatry, Psychology and Neuroscience, King's College London, De Crespigny Park, SE5 8AF London, United Kingdom), (Psychiatry Imaging Group, MRC London Institute of Medical Sciences, Faculty of Medicine, Imperial College London, Hammersmith Hospital, Du Cane Road, W12 0NN London, UK), Alexandre Reymond (Center for Integrative Genomics, University of Lausanne, Genopode building, CH-1015 Lausanne, Switzerland), Genevieve Richard (NORMENT, K.G. Jebsen Centre for Psychosis Research, Institute of Clinical Medicine, University of Oslo and Division of Mental Health and Addiction, Oslo University Hospital, Oslo, Norway), (Department of Psychology, University of Oslo, Oslo, Norway), (Sunnaas Rehabilitation Hospital HT, Nesodden, Norway), Borja Rodriguez-Herreros (Service of Medical Genetics, Centre Hospitalier Universitaire Vaudois and University of Lausanne, Rue du Bugnon 46, 1011 Lausanne, Switzerland), (CHU Sainte-Justine Research Center, Université de Montréal, Montréal, QC, Canada), Roberto Roiz-Santiañez (Department of Medicine and Psychiatry, University Hospital Marque?s de Valdecilla, School of Medicine, University of Cantabria-IDIVAL, 39008 Santander, Spain), (CIBERSAM (Centro Investigacio?n Biome?dica en Red Salud Mental), Santander, 39011, Spain), James Rucker (National Institute for Health Research (NIHR) Mental Health Biomedical Research Centre at South London and Maudsley NHS Foundation Trust and King's College London, United Kingdom), (Medical Research Council - Social, Genetic and Developmental Psychiatry Centre, Institute of Psychiatry, Psychology & Neuroscience, King's College London, United Kingdom), Perminder Sachdev (Centre for Healthy Brain Ageing, School of Psychiatry, University of New South Wales, Sydney, New South Wales 2052, Australia), Anne-Marthe Sanders (NORMENT, K.G. Jebsen Centre for Psychosis Research, Institute of Clinical Medicine, University of Oslo and Division of Mental Health and Addiction, Oslo University Hospital, Oslo, Norway), (Department of Psychology, University of Oslo, Oslo, Norway), (Sunnaas Rehabilitation Hospital HT, Nesodden, Norway), Sigrid B. Sando (Department of Neuroscience, Faculty of Medicine, Norwegian University of Science and Technology), (Department of Neurology, University Hospital of Trondheim, Edvard Griegs gate 8, N-7006 Trondheim, Norway), Lianne Schmaal (Orygen, The National Centre of Excellence in Youth Mental Health, 35 Poplar Road, 3502 Parkville, New Mexico, Australia), (Centre for Youth Mental Health, The University of Melbourne, 35 Poplar Road, 3502 Parkville, Victoria, Australia), (Department of Psychiatry, VU University Medical Center, 1007 MB Amsterdam, The Netherlands), Peter R. Schofield (Neuroscience Research Australia, Randwick, Australia), (School of Medical Sciences, University of New South Wales, Sydney, Australia), Andrew J. Schork (Center for Multimodal Imaging and Genetics, University of California San Diego, La Jolla, USA), Gunter Schumann (Medical Research Council - Social, Genetic and Developmental Psychiatry Centre, Institute of Psychiatry, Psychology & Neuroscience, King's College London, United Kingdom), Jean Shin (The Hospital for Sick Children, University of Toronto, Toronto, M5G 1X8, Canada), (Rotman Research Institute, University of Toronto, Toronto, M6A 2E1, Canada), Elena Shumskaya (Department of Human Genetics, Radboud University Medical Center, Nijmegen, the Netherlands), (Donders Institute for Brain, Cognition and Behaviour, Radboud University, Nijmegen, Netherlands), Sanjay Sisodiya (Department of Clinical and Experimental Epilepsy, UCL Institute of Neurology, London, and Chalfont Centre for Epilepsy, UK), Vidar M. Steen (NORMENT - KG Jebsen Centre, Department of Clinical Science, University of Bergen, Jonas Lies veg 87, 5021 Bergen, Norway), (Dr. Einar Martens Research Group for Biological Psychiatry, Center for Medical Genetics and Molecular Medicine, Haukeland University Hospital, Jonas Lies veg 87, 5021 Bergen, Norway), Dan J. Stein (Dept of Psychiatry, University of Cape Town, Groote Schuur Hospital, Anzio Rd, 7925 Cape Town, South Africa), Stacy Steinberg (deCODE Genetics/Amgen, Reykjavik, Iceland), Lachlan Strike (Queensland Brain Institute, University of Queensland, St Lucia, Queensland, Australia), Alexander Teumer (Institute for Community Medicine, University Medicine Greifswald, Greifswald, Germany), Anbu Thalamuthu (Centre for Healthy Brain Ageing, School of Psychiatry, University of New South Wales, Sydney, New South Wales 2052, Australia), Diana Tordesillas-Gutierrez (CIBERSAM (Centro Investigacio?n Biome?dica en Red Salud Mental), Santander, 39011, Spain), (Neuroimaging Unit, Technological Facilities. Valdecilla Biomedical Research Institute IDIVAL, Santander, Cantabria, 39011, Spain), Jessica Turner (Department of Psychology, Georgia State University, Atlanta, GA), Torill Ueland (NORMENT, K.G. Jebsen Centre for Psychosis Research, Institute of Clinical Medicine, University of Oslo and Division of Mental Health and Addiction, Oslo University Hospital, Oslo, Norway), Anne Uhlmann (Department of Psychiatry and Mental Health, Anzio Road, 7925 Cape Town, South Africa), (Department of Psychiatry, Stellenbosch University, TBH Francie van Zijl Avenue, 7500 Cape Town, South Africa), (Department of Psychiatry, 1 South Prospect Street, 5401 Burlington, Vermont, USA), Magnus O. Ulfarsson (deCODE Genetics/Amgen, Reykjavik, Iceland), (Faculty of Electrical and Computer Engineering, University of Iceland, Reykjavik, Iceland), Dennis van 't Ent (Biological Psychology, Vrije Universiteit Amsterdam, van Boechorststraat 1, 1081 BT Amsterdam, The Netherlands), Dennis van der Meer (NORMENT, K.G. Jebsen Centre for Psychosis Research, Institute of Clinical Medicine, University of Oslo and Division of Mental Health and Addiction, Oslo University Hospital, Oslo, Norway), Neeltje E.M. van Haren (Department of Psychiatry, Brain Center Rudolf Magnus, University Medical Center Utrecht, Utrecht, The Netherlands), Anja Vaskinn (NORMENT, K.G. Jebsen Centre for Psychosis Research, Institute of Clinical Medicine, University of Oslo and Division of Mental Health and Addiction, Oslo University Hospital, Oslo, Norway), Evangelos Vassos (MRC Social, Genetic and Developmental Psychiatry Centre, Institute of Psychiatry, Psychology and Neuroscience, King's College London, 16 De Crespigny Park, SE5 8AF London, UK), G. Bragi Walters (deCODE Genetics/Amgen, Reykjavik, Iceland), Yunpeng Wang (NORMENT, K.G. Jebsen Centre for Psychosis Research, Institute of Clinical Medicine, University of Oslo and Division of Mental Health and Addiction, Oslo University Hospital, Oslo, Norway), Wei Wen (Centre for Healthy Brain Ageing, School of Psychiatry, University of New South Wales, Sydney, New South Wales 2052, Australia), Christopher D. Whelan (Molecular and Cellular Therapeutics, Royal College of Surgeons in Ireland, 123 St. Stephens Green, D02 YN77 Dublin, Ireland), Katharina Wittfeld (German Center for Neurodegenerative Diseases (DZNE), Rostock, Greifswald, Greifswald, Germany), Margie Wright (QIMR Berghofer Medical Research Institute, Brisbane, Queensland, Australia), (Centre for Advanced Imaging, University of Queensland, St Lucia, Queensland, Australia), Hidenaga Yamamori (Department of Psychiatry, Osaka University Graduate School of Medicine, Suita, Osaka, Japan), Tetyana Zayats (K.G. Jebsen Centre for Neuropsychiatric Disorders, University of Bergen, Norway), (Department of Biomedicine, University of Bergen, 5009, Bergen, Norway), Ingrid Agartz (NORMENT, K.G. Jebsen Centre for Psychosis Research, Institute of Clinical Medicine, University of Oslo and Division of Mental Health and Addiction, Oslo University Hospital, Oslo, Norway), Lars T. Westlye (NORMENT, K.G. Jebsen Centre for Psychosis Research, Institute of Clinical Medicine, University of Oslo and Division of Mental Health and Addiction, Oslo University Hospital, Oslo, Norway), (Department of Psychology, University of Oslo, Oslo, Norway), Sébastien Jacquemont (CHU Sainte-Justine Research Center, Université de Montréal, Montréal, QC, Canada), (Department of Pediatrics, University of Montreal, Montreal H3C 3J7, Canada), Srdjan Djurovic (NORMENT - KG Jebsen Centre, Department of Clinical Science, University of Bergen, Jonas Lies veg 87, 5021 Bergen, Norway), (Department of Medical Genetics, Oslo University Hospital, Kirkeveien 166, 424 Oslo, Norway), Hreinn Stefánsson (deCODE Genetics/Amgen, Reykjavik, Iceland), Kári Stefánsson (deCODE Genetics/Amgen, Reykjavik, Iceland), (Faculty of Medicine, University of Iceland, Reykjavik, Iceland), Paul Thompson (Imaging Genetics Center, Mark and Mary Stevens Neuroimaging and Informatics Institute, Keck School of Medicine of the University of Southern California, Marina del Rey, USA), Ole A. Andreassen (NORMENT, K.G. Jebsen Centre for Psychosis Research, Institute of Clinical Medicine, University of Oslo and Division of Mental Health and Addiction, Oslo University Hospital, Oslo, Norway)
Compliance with ethical standards
Conflict of interest
D.P.H. is a full-time employee of Janssen R&D. Decode: O.G., GB Walters, Gudrun A Jonsdottir, Stacy Steinberg, Magnus O Ulfarsson, Hreinn Stefansson and Kari Stefansson are employees of deCODE genetics/Amgen.
Electronic supplementary material
The online version of this article (10.1038/s41380-018-0118-1) contains supplementary material, which is available to authorized users.
References
- 1.Kirov G, Rees E, Walters J. What a psychiatrist needs to know about copy number variants. Br J Psych Adv. 2015;21:157–63. [Google Scholar]
- 2.Stefansson H, Meyer-Lindenberg A, Steinberg S, Magnusdottir B, Morgen K, Arnarsdottir S, et al. CNVs conferring risk of autism or schizophrenia affect cognition in controls. Nature. 2014;505:361–6. doi: 10.1038/nature12818. [DOI] [PubMed] [Google Scholar]
- 3.Bijlsma EK, Gijsbers AC, Schuurs-Hoeijmakers JH, van Haeringen A, Fransen van de Putte DE, Anderlid BM, et al. Extending the phenotype of recurrent rearrangements of 16p11.2: deletions in mentally retarded patients without autism and in normal individuals. Eur J Med Genet. 2009;52:77–87. doi: 10.1016/j.ejmg.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 4.Shen Y, Chen X, Wang L, Guo J, Shen J, An Y, et al. Intra-family phenotypic heterogeneity of 16p11.2 deletion carriers in a three-generation Chinese family. Am J Med Genet B Neuropsychiatr Genet. 2011;156:225–32. doi: 10.1002/ajmg.b.31147. [DOI] [PubMed] [Google Scholar]
- 5.Tabet AC, Pilorge M, Delorme R, Amsellem F, Pinard JM, Leboyer M, et al. Autism multiplex family with 16p11.2p12.2 microduplication syndrome in monozygotic twins and distal 16p11.2 deletion in their brother. Eur J Human Genet. 2012;20:540–6. doi: 10.1038/ejhg.2011.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qureshi AY, Mueller S, Snyder AZ, Mukherjee P, Berman JI, Roberts TP, et al. Opposing brain differences in 16p11.2 deletion and duplication carriers. J Neurosci. 2014;34:11199–211. doi: 10.1523/JNEUROSCI.1366-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ulfarsson MO, Walters GB, Gustafsson O, Steinberg S, Silva A, Doyle OM, et al. 15q11.2 CNV affects cognitive, structural and functional correlates of dyslexia and dyscalculia. Transl Psychiatry. 2017;7:e1109. doi: 10.1038/tp.2017.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maillard AM, Ruef A, Pizzagalli F, Migliavacca E, Hippolyte L, Adaszewski S, et al. The 16p11.2 locus modulates brain structures common to autism, schizophrenia and obesity. Mol Psychiatry. 2015;20:140–7. doi: 10.1038/mp.2014.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin A, Ching CRK, Vajdi A, Sun D, Jonas RK, Jalbrzikowski M, et al. Mapping 22q11.2 gene dosage effects on brain morphometry. J Neurosci. 2017;37:6183–99. doi: 10.1523/JNEUROSCI.3759-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fan CC, Brown TT, Bartsch H, Kuperman JM, Hagler DJ, Jr., Schork A, et al. Williams syndrome-specific neuroanatomical profile and its associations with behavioral features. NeuroImage Clin. 2017;15:343–7. doi: 10.1016/j.nicl.2017.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ballif BC, Hornor SA, Jenkins E, Madan-Khetarpal S, Surti U, Jackson KE, et al. Discovery of a previously unrecognized microdeletion syndrome of 16p11.2-p12.2. Nat Genet. 2007;39:1071–3. doi: 10.1038/ng2107. [DOI] [PubMed] [Google Scholar]
- 12.Battaglia A, Novelli A, Bernardini L, Igliozzi R, Parrini B. Further characterization of the new microdeletion syndrome of 16p11.2-p12.2. Am J Med Genet A. 2009;149a:1200–4. doi: 10.1002/ajmg.a.32847. [DOI] [PubMed] [Google Scholar]
- 13.Hempel M, Rivera Brugues N, Wagenstaller J, Lederer G, Weitensteiner A, Seidel H, et al. Microdeletion syndrome 16p11.2-p12.2: clinical and molecular characterization. Am J Med Genet A. 2009;149a:2106–12. doi: 10.1002/ajmg.a.33042. [DOI] [PubMed] [Google Scholar]
- 14.Barber JC, Hall V, Maloney VK, Huang S, Roberts AM, Brady AF, et al. 16p11.2-p12.2 duplication syndrome; a genomic condition differentiated from euchromatic variation of 16p11.2. Eur J Human Genet. 2013;21:182–9. doi: 10.1038/ejhg.2012.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pebrel-Richard C, Debost-Legrand A, Eymard-Pierre E, Greze V, Kemeny S, Gay-Bellile M, et al. An unusual clinical severity of 16p11.2 deletion syndrome caused by unmasked recessive mutation of CLN3. Eur J Human Genet. 2014;22:369–73. doi: 10.1038/ejhg.2013.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumar RA, KaraMohamed S, Sudi J, Conrad DF, Brune C, Badner JA, et al. Recurrent 16p11.2 microdeletions in autism. Hum Mol Genet. 2008;17:628–38. doi: 10.1093/hmg/ddm376. [DOI] [PubMed] [Google Scholar]
- 17.Loviglio MN, Leleu M, Mannik K, Passeggeri M, Giannuzzi G, van der Werf I, et al. Chromosomal contacts connect loci associated with autism, BMI and head circumference phenotypes. Mol Psychiatry. 2016;22:836–49. doi: 10.1038/mp.2016.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanders SJ, Ercan-Sencicek AG, Hus V, Luo R, Murtha MT, Moreno-De-Luca D, et al. Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron. 2011;70:863–85. doi: 10.1016/j.neuron.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weiss LA, Shen Y, Korn JM, Arking DE, Miller DT, Fossdal R, et al. Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med. 2008;358:667–75. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 20.Guha S, Rees E, Darvasi A, Ivanov D, Ikeda M, Bergen SE, et al. Implication of a rare deletion at distal 16p11.2 in schizophrenia. JAMA Psychiatry. 2013;70:253–60. doi: 10.1001/2013.jamapsychiatry.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCarthy SE, Makarov V, Kirov G, Addington AM, McClellan J, Yoon S, et al. Microduplications of 16p11.2 are associated with schizophrenia. Nat Genet. 2009;41:1223–7. doi: 10.1038/ng.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marshall CR, Howrigan DP, Merico D, Thiruvahindrapuram B, Wu W, Greer DS, et al. Contribution of copy number variants to schizophrenia from a genome-wide study of 41,321 subjects. Nat Genet. 2017;49:27–35. doi: 10.1038/ng.3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mefford HC, Muhle H, Ostertag P, von Spiczak S, Buysse K, Baker C, et al. Genome-wide copy number variation in epilepsy: novel susceptibility loci in idiopathic generalized and focal epilepsies. PLoS Genet. 2010;6:e1000962. doi: 10.1371/journal.pgen.1000962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reinthaler EM, Lal D, Lebon S, Hildebrand MS, Dahl HH, Regan BM, et al. 16p11.2 600 kb duplications confer risk for typical and atypical Rolandic epilepsy. Hum Mol Genet. 2014;23:6069–80. doi: 10.1093/hmg/ddu306. [DOI] [PubMed] [Google Scholar]
- 25.Zufferey F, Sherr EH, Beckmann ND, Hanson E, Maillard AM, Hippolyte L, et al. A 600 kb deletion syndrome at 16p11.2 leads to energy imbalance and neuropsychiatric disorders. J Med Genet. 2012;49:660–8. doi: 10.1136/jmedgenet-2012-101203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bachmann-Gagescu R, Mefford HC, Cowan C, Glew GM, Hing AV, Wallace S, et al. Recurrent 200-kb deletions of 16p11.2 that include the SH2B1 gene are associated with developmental delay and obesity. Genet Med. 2010;12:641–7. doi: 10.1097/GIM.0b013e3181ef4286. [DOI] [PubMed] [Google Scholar]
- 27.Bochukova EG, Huang N, Keogh J, Henning E, Purmann C, Blaszczyk K, et al. Large, rare chromosomal deletions associated with severe early-onset obesity. Nature. 2010;463:666–70. doi: 10.1038/nature08689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jacquemont S, Reymond A, Zufferey F, Harewood L, Walters RG, Kutalik Z, et al. Mirror extreme BMI phenotypes associated with gene dosage at the chromosome 16p11.2 locus. Nature. 2011;478:97–102. doi: 10.1038/nature10406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sampson MG, Coughlin CR, 2nd, Kaplan P, Conlin LK, Meyers KE, Zackai EH, et al. Evidence for a recurrent microdeletion at chromosome 16p11.2 associated with congenital anomalies of the kidney and urinary tract (CAKUT) and Hirschsprung disease. Am J Med Genet Part A. 2010;152a:2618–22. doi: 10.1002/ajmg.a.33628. [DOI] [PubMed] [Google Scholar]
- 30.D’ Angelo D, Lebon S, Chen Q, Martin-Brevet S, Snyder LG, Hippolyte L, et al. Defining the effect of the 16p11.2 duplication on cognition, behavior, and medical comorbidities. JAMA Psychiatry. 2016;73:20–30. doi: 10.1001/jamapsychiatry.2015.2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mace A, Tuke MA, Deelen P, Kristiansson K, Mattsson H, Noukas M, et al. CNV-association meta-analysis in 191,161 European adults reveals new loci associated with anthropometric traits. Nat Commun. 2017;8:744. doi: 10.1038/s41467-017-00556-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martin-Brevet S, Rodriguez-Herreros B, Nielsen JA, Moreau C, Modenato C, Maillard AM, et al. Quantifying the effects of 16p11.2 copy number variants on brain structure: a multi-site ‘genetic-first’ study. Biol Psychiatry. 2018; 10.1016/j.biopsych.2018.02.1176. [DOI] [PubMed]
- 33.Hippolyte L, Maillard AM, Rodriguez-Herreros B, Pain A, Martin-Brevet S, Ferrari C, et al. The number of genomic copies at the 16p11.2 locus modulates language, verbal memory, and inhibition. Biol Psychiatry. 2016;80:129–39. doi: 10.1016/j.biopsych.2015.10.021. [DOI] [PubMed] [Google Scholar]
- 34.Hanson E, Bernier R, Porche K, Jackson FI, Goin-Kochel RP, Snyder LG, et al. The cognitive and behavioral phenotype of the 16p11.2 deletion in a clinically ascertained population. Biol Psychiatry. 2015;77:785–93. doi: 10.1016/j.biopsych.2014.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Insel TR. Rethinking schizophrenia. Nature. 2010;468:187–93. doi: 10.1038/nature09552. [DOI] [PubMed] [Google Scholar]
- 36.Kendall KM, Rees E, Escott-Price V, Einon M, Thomas R, Hewitt J, et al. Cognitive performance among carriers of pathogenic copy number variants: analysis of 152,000 UK Biobank Subjects. Biol Psychiatry. 2016;1–12. [DOI] [PubMed]
- 37.Wang K, Li M, Hadley D, Liu R, Glessner J, Grant SF, et al. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17:1665–74. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hibar DP, Stein JL, Renteria ME, Arias-Vasquez A, Desrivieres S, Jahanshad N, et al. Common genetic variants influence human subcortical brain structures. Nature. 2015;520:224–9. doi: 10.1038/nature14101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wickham H. ggplot2: Elegant Graphics for Data Analysis. New York: Springer-Verlag; 2009. [Google Scholar]
- 40.Viechtbauer W. Conducting meta-analyses in R with the metafor package. J Stat Softw. 2010;36:1–48. [Google Scholar]
- 41.Wechsler D. Wechsler Abbreviated Scale of Intelligence. Harcourt Brace and Company, New York, NY, 1999.
- 42.Gudmundsson E. Mat á greind fullorðinna: WASIIS [The Assessment of Intelligence in Adults: WASIIS]. Menntamalastofnun, Reykjavik, Iceland, 2015.
- 43.Wechsler D. Wechsler Adult Intelligence Scale. The Psychological Corporation, San Antonio, TX, 1997.
- 44.Loviglio MN, Arbogast T, Jonch AE, Collins SC, Popadin K, Bonnet CS, et al. The immune signaling adaptor LAT contributes to the neuroanatomical phenotype of 16p11.2 BP2-BP3 CNVs. Am J Hum Genet. 2017;101:564–77. doi: 10.1016/j.ajhg.2017.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meda SA, Pryweller JR, Thornton-Wells TA. Regional brain differences in cortical thickness, surface area and subcortical volume in individuals with Williams syndrome. PLoS ONE. 2012;7:e31913. doi: 10.1371/journal.pone.0031913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Erp TG, Hibar DP, Rasmussen JM, Glahn DC, Pearlson GD, Andreassen OA, et al. Subcortical brain volume abnormalities in 2028 individuals with schizophrenia and 2540 healthy controls via the ENIGMA consortium. Mol Psychiatry. 2016;21:547–53. doi: 10.1038/mp.2015.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jorgensen KN, Nesvag R, Gunleiksrud S, Raballo A, Jonsson EG, Agartz I. First- and second-generation antipsychotic drug treatment and subcortical brain morphology in schizophrenia. Eur Arch Psychiatry Clin Neurosci. 2016;266:451–60. doi: 10.1007/s00406-015-0650-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.