Abstract
Purpose
To develop an automated cone-beam computed tomography (CBCT) multi-organ segmentation method for potential CBCT-guided adaptive radiation therapy workflow.
Methods and materials
The proposed method combines the deep leaning-based image synthesis method, which generates magnetic resonance images (MRIs) with superior soft-tissue contrast from on-board setup CBCT images to aid CBCT segmentation, with a deep attention strategy, which focuses on learning discriminative features for differentiating organ margins. The whole segmentation method consists of 3 major steps. First, a cycle-consistent adversarial network (CycleGAN) was used to estimate a synthetic MRI (sMRI) from CBCT images. Second, a deep attention network was trained based on sMRI and its corresponding manual contours. Third, the segmented contours for a query patient was obtained by feeding the patient’s CBCT images into the trained sMRI estimation and segmentation model. In our retrospective study, we included 100 prostate cancer patients, each of whom has CBCT acquired with prostate, bladder and rectum contoured by physicians with MRI guidance as ground truth. We trained and tested our model with separate datasets among these patients. The resulting segmentations were compared with physicians’ manual contours.
Results
The Dice similarity coefficient and mean surface distance indices between our segmented and physicians’ manual contours (bladder, prostate, and rectum) were 0.95±0.02, 0.44±0.22 mm, 0.86±0.06, 0.73±0.37 mm, and 0.91±0.04, 0.72±0.65 mm, respectively.
Conclusion
We have proposed a novel CBCT-only pelvic multi-organ segmentation strategy using CBCT-based sMRI and validated its accuracy against manual contours. This technique could provide accurate organ volume for treatment planning without requiring MR images acquisition, greatly facilitating routine clinical workflow.
INTRODUCTION
Cone-beam computed tomography (CBCT) has been widely used in image-guided radiation therapy for prostate patients to improve treatment setup accuracy. In current clinical practice, it is acquired before treatment delivery and provides detailed anatomic information in the treatment position. The displacement of anatomic landmarks between CBCT images and the treatment planning CT images are then measured to quantitatively determine the error in patient setup (Barney et al., 2011).
In recent years, adaptive radiation therapy has been shown as a promising strategy to improve clinical outcomes by accommodating the inter-fraction variations (Yan et al., 1997; Kataria et al., 2016). In an adaptive radiation therapy workflow, CBCT plays an important role in providing the latest three-dimensional information of patient position and anatomy (Oldham et al., 2005). More demanding applications of CBCT have been proposed, such as daily estimation of target coverage and organs-at-risk (OARs) sparing for real-time CBCT-based treatment replanning (de la Zerda et al., 2007; Yoo and Yin, 2006).
These potential uses of CBCT require accurate and fast delineation of targets and OARs. Experienced physicians are able to manually contour multiple organs on CBCT images, but it is impractical in adaptive radiation therapy due to time constraints. Alternatively, it has been proposed that contours on planning CT images can be propagated to CBCT images by image registration (Thor et al., 2011; Xie et al., 2008). However, large local variations in patient anatomy and image content between CBCT and CT images is common, e.g. changes in bladder/rectum filling status in prostate cancer patients (Yang et al., 2014a). Such variations cannot be handled by rigid image registration, and also not readily by deformable image registration because of the lack of exact correspondence of image content between the two image sets (Yang et al., 2014b). The suboptimal registration result would lead to degraded accuracy of the propagated contours.
Automatic segmentation solely based on CBCT can avoid registration to the planning CT, but very few studies have been published. The contrast of some organs, such as prostate, is poor on CBCT images, which is further degraded by CBCT artifacts caused by scatter contamination (Lei et al., 2019b). In this study, we propose a novel method to automatically segment multiple organs on pelvic CBCT for prostate cancer patients. We synthesized sMRIs from CBCT images to provide superior soft-tissue contrast, and then used a deep attention network to automatically capture the significant features to differentiate the multi-organ margins in sMRI. With this sMRI-aided strategy, we aim to develop an automated and accurate segmentation method benefiting from the high soft-tissue contrast of MR images. Our method was evaluated in a retrospective study with 100 patients.
MATERIALS AND METHODS
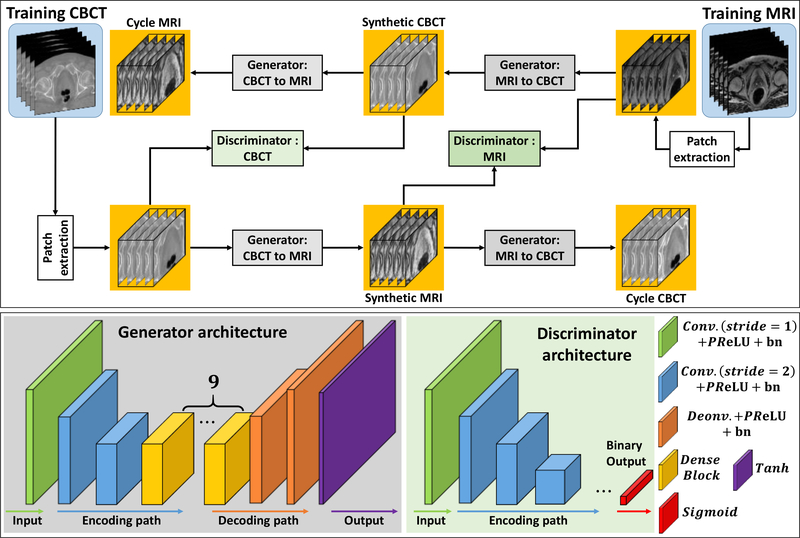
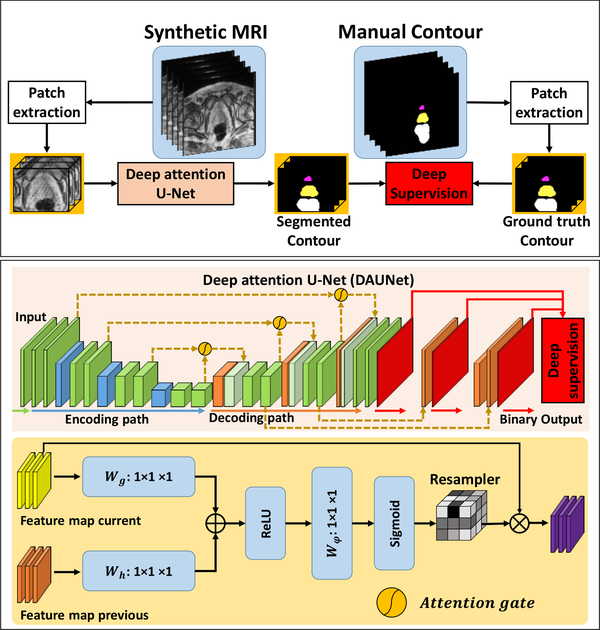
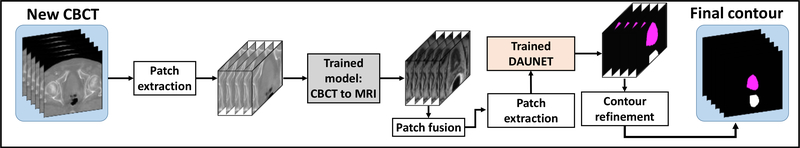
Our segmentation method consisted of 2 major steps: (1) sMRI synthesis from CBCT images; and (2) segmentation on sMRI. Fig. 1, 2 and 3 outline the schematic flow chart of the proposed method. First, a CycleGAN was trained to estimate sMRIs from CBCT images by introducing an inverse transformation, which is able to enforce the translation from CBCT to MRI to be one-to-one mapping, as is shown in Fig. 1. Second, a deep attention U-Net (DAUNet) was trained to segment multi organs on sMRIs, as is shown in Fig. 2. A deep attention network was introduced to retrieve the most relevant features to identify organ boundaries. Deep supervision was also incorporated into this DAUNet to enhance the features’ discriminative ability. Third, for a new patient, the contours were obtained by first feeding the CBCT image into the trained CycleGAN to generate the sMRI, and then feeding sMRI to the trained DAUNet to generate the segmentation, as is shown in Fig. 3.
Figure 1.
The first row shows the schematic flow of the training of synthetic MRI via CycleGAN. The second shows the generators’ and discriminators’ network architectures used in first row.
Figure 2.
The first row shows the schematic flow of the training of synthetic MRI pelvic segmentation via DAUNet. The second shows the DAUNet network architectures used in first row. The third row shows the attention gate architecture used in second row.
Figure 3.
The schematic workflow of segmenting of a new arrival patient’s CBCT image.
2.A. sMRI estimation
The first step of our method is to synthesize MRI from CBCT images using 3D CycleGAN. In this step, we first trained a CBCT-to-MRI transformation model using pairs of CBCT and MRI images from the training patient dataset. Note that the MRIs were deformably registered with CBCTs for each training patient using commercial software. The deformed MR images were used as the learning-based target of the CBCT images for the proposed sMRI-aided strategy. Because the two image modalities have fundamentally different properties, training a CBCT-to-MRI transformation model is difficult. To cope with this challenge, 3D CycleGAN architecture was used to learn this transformation model (Lei et al., 2019a), due to its ability to enforce the transformation to mimic target data distribution by incorporating an inverse transformation. Patient anatomy can vary significantly among individuals. In order to accurately predict each voxel in the anatomic region (bladder, prostate and rectum), we introduced several dense blocks to capture multi-scale information (including low-frequency structural information and high-frequency textural information) by extracting features from previous and following hidden layers (Lei et al., 2019a). Each 3D patch was extracted from paired CBCT and MRIs by sliding the window with overlap to its neighboring patches (Yang et al., 2017). This overlap ensures that a continuous whole-image output can be obtained and allows for increased training data for the network. The detailed 3D CycleGAN architecture is introduced in our previous study (Lei et al., 2019a).
2.B. Deep attention network
The second step of our method is to perform automatic segmentation using DAUNet on the sMRI generated from the first step. The DAUNet was trained on the sMRIs from the first step of training patient dataset, with their binary masks of corresponding manual prostate, bladder, and rectum contours of ground truth used as learning-based target. As shown in Fig. 2, the DAUNet architecture is implemented by introducing additional attention gates (AGs) (Mishra et al., 2018) and deep supervision (Wang et al., 2019a; Lei et al., 2019c; Wang et al., 2019c) on a basic U-Net architecture (Balagopal et al., 2018; Dong et al., 2019; Wang et al., 2019b). The U-Net architecture consists of an encoding path and a decoding path; the two paths were connected by several long skip connections. In our study, the long skip connection concatenated the feature maps from the current two decoding deconvolution operators and one previous encoding convolution operator by using AGs. Such concatenation with AGs encouraged the network to identify the most relevant semantic contextual information without a requirement to enlarge the receptive field, which is highly beneficial for organ localization (Mishra et al., 2018). We also use deep-supervision to force the intermediate feature maps to be semantically discriminative at each image scale (Wang et al., 2019a; Lei et al., 2019c). This helps to ensure that AGs, at different scales, have an ability to influence the responses to a large range of prostate content.
2.C. Database
In this retrospective study, we reviewed 100 patients with prostate malignancies treated with external beam radiation therapy in our clinic. All of the 100 patients underwent standard treatment planning workflow, i.e. CT simulation and at least one set of CBCT images acquired during treatment. The CBCT images were acquired using the Varian On-Board Imager CBCT system, with imaging spacing of 0.908 mm × 0.908 mm × 2.0 mm. We divided the 100 patients into two 50-patient groups. Among the first 50-patient group, the corresponding MR images used for fusion with planning CBCT images for prostate delineation were also retrieved for sMRI training. The MR images of all patients were acquired using a Siemens standard T2-weighted MRI scanner with 3D T2-SPACE sequence and 1.0×1.0×2.0 mm3 voxel size (TR/TE: 1000/123 ms, flip angle: 95°). The training MR and CBCT images were deformably registered using commercial software, Velocity AI 3.2.1 (Varian Medical Systems, Palo Alto, CA). Institutional review board approval was obtained; no informed consent was required for this HIPAA-compliant retrospective analysis.
In our study, patients with MRI scans have manual contours delineated by physicians first. Such MRI was then registered with CBCT images using deformable registration, with the contours on MRI also propagated on CBCT images. The physicians then refined the propagated contours based on CBCT images to reduce error from registration. Such contours delineated on MRI and refined on CBCT were considered as ground truth contours and learning-based target in our study. For the deformable registration step, it should be noted that it is only performed on training dataset to match the CBCT and MR images. As long as the model is trained, deformable registration is no longer involved in segmenting a new patient. The new patient does not have MR images, and his sMRI predicted by our model shares same anatomy with his CBCT.
2.D. Reliability evaluation of the segmentation algorithm
The automatic segmentation results were compared with the gold standard of physicians’ manual contours. Dice similarity coefficient (DSC), sensitivity, specificity, Hausdorff distance (HD), mean surface distance (MSD), the residual mean square distance (RMSD), the center of mass distance (CMD), and volume difference (VD) were used to evaluate the accuracy of our segmentation method. The DSC, precision and recall scores are used to quantify volume similarity between two contours. The HD, MSD and RMSD metrics are used to quantify boundary similarity between two surfaces. The CMD metric is used to measure the distance between the center of segmented and manual contour, which is especially important for prostate contour because it determines the isocenter setup. The VD metric is used to measure the absolute volume difference between the segmented and manual contour, which dose volume histogram calculation depends on. More accurate segmentation results are associated with lower HD, MSD and RMSD scores and higher DSC, precision and recall scores.
Five-fold cross-validation method was used to evaluate the proposed segmentation algorithm among the first 50-patient group, because it gives the proposed model the opportunity to train on multiple train-test splits. This method split up data into five sub-groups. One of the sub-groups (10 patients’ data) was used as the test set and the rest (40 patients’ data) were used as the training set for both CBCT-to-sMRI transformation model (CycleGAN) and segmentation model. The models were trained on the training set and scored on the test set. Then the process was repeated until each unique sub-group had been used as the test set.
In addition, a hold-out method was used to evaluate the proposed segmentation algorithm how well it performs on unseen data. We used the first 50-patient group as the training set for both CBCT-to-sMRI transformation model (CycleGAN) and segmentation model, and used the second 50-patient group for testing. The second 50-patient group is not involved in any training steps.
By using both of the two validations, each of 100 patient data was exactly tested once. We used the numerical metrics of these patients to evaluate the proposed method’s performance. To illustrate the significant improvement of sMRI-aided strategy, a paired two-tailed t-test was used between the results of the other comparison methods.
RESULTS
3.A. Contribution of deep attention
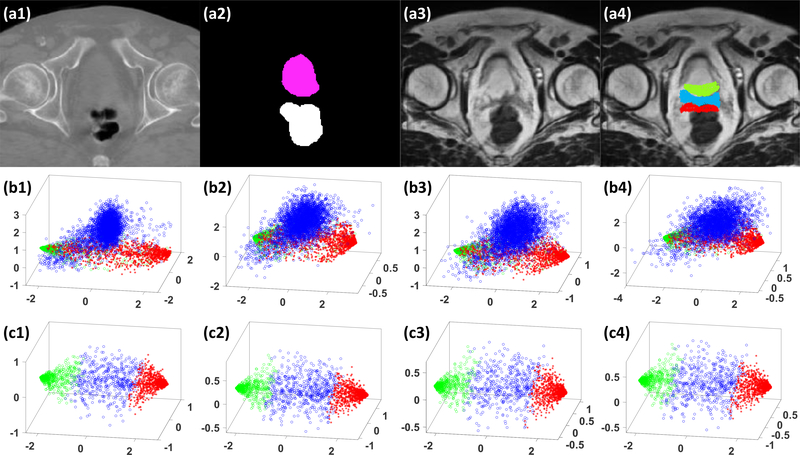
To demonstrate the efficacy of deep attention, we compared the results of our proposed DAUNet algorithm against the algorithm without using deep attention, i.e., deeply supervised U-Net (DSUNet). These two algorithms were both trained on sMRIs. Fig. 4 shows 3D scatter plots of the first three principal components of patch samples in the feature maps extracted from the last convolution layer of three deconvolution operators. To demonstrate the ability of AGs on differentiating tissues around organ margins, we randomly selected 1000 samples from the posterior margin of prostate, the anterior margin of rectum, and the region between prostate and rectum, as shown in the subfigure (a4). We selected the samples due to their location in the organ margin, proximity to an adjacent organ, and the resulting difficulty in accurate differentiation and identification. The scatter plots of a DSUNet in the subfigure (b1-b4) illustrates an overlap between the samples from the bladder, the region between bladder and rectum, and the rectum regions; thus these two regions cannot be easily separated. Whereas with DAUNet, as shown in the subfigure (c1-c4), the samples can be easily separated, demonstrating the significant enhanced discerning capability with the addition of a deep attention strategy. We also compared the numerical results of DAUNet with those of DSUNet on sMRI data in Table 1, 2 and 3. As is shown, DAUNet achieved better performance than DSUNet. Table 1 shows the numerical comparison for five-fold cross-validation experiments. Table 2 shows the numerical comparison for hold-out validation experiment. Table 3 shows the numerical results of all used 100 patients data.
Figure 4.
An illustrative example of the benefit of our DAUNet compared with DSUNet without using AGs, (a1) shows the original CBCT image in transverse plane. (a2) shows corresponding manual contours, (a3) shows the generated sMRI, (a4) shows the sample patches’ central positions drawn from sMRI, where the samples belonging to the bladder are highlighted by green circles, and the samples belonging to the rectum are highlighted by red asterisks, and the samples between bladder and rectum regions are highlighted by blue circles. (b1-b4) show the scatter plots of the first 3 principal components of corresponding patch samples in feature maps extracted by using a DSUNet, respectively. (c1-c4) show the scatter plots of first 3 principal components of corresponding patch samples in the feature maps extracted by DAUNet, respectively.
Table 1.
Numerical comparison of DAUNet trained and tested on sMRI (DSUNet sMRI), DAUNet trained and tested on CBCT (DSUNet CBCT), and DSUNet trained and tested on sMRI (DSUNet sMRI) for five-fold cross-validation.
| Method | DSC | Sensitivity | Specificity | HD (mm) | MSD (mm) | RMSD (mm) | CMD (mm) | VD (cc) | |
|---|---|---|---|---|---|---|---|---|---|
| Bladder | DSUNet sMRI | 0.93±0.04 | 0.95±0.032 | 0.91±0.07 | 5.91±4.90 | 0.52±0.29 | 0.85±0.62 | 1.25±1.00 | 9.17±6.30 |
| DAUNet CBCT | 0.91±0.06 | 0.96±0.048 | 0.87±0.08 | 12.79±14.82 | 0.67±0.33 | 1.07±0.64 | 1.4±1.49 | 16.57±18.61 | |
| DAUNet sMRI | 0.94±0.02 | 0.94±0.04 | 0.96±0.02 | 4.58±3.94 | 0.48±0.23 | 0.83±0.63 | 0.93±0.74 | 6.92±5.71 | |
| P-value DAUNet sMRI vs. DSUNet sMRI | 0.174 | 0.022 | 0.003 | 0.021 | 0.591 | 0.922 | 0.014 | 0.195 | |
| P-value DAUNet sMRI vs. DAUNet CBCT | 0.004 | 0.002 | <0.001 | <0.001 | 0.002 | 0.019 | 0.005 | 0.007 | |
| Prostate | DSUNet sMRI | 0.83±0.045 | 0.83±0.091 | 0.86±0.091 | 6.42±2.46 | 0.91±0.26 | 1.28±0.34 | 2.48±1.11 | 6.39±7.29 |
| DAUNet CBCT | 0.80±0.05 | 0.80±0.10 | 0.81±0.08 | 6.82±3.088 | 1.06±0.29 | 1.48±0.48 | 3.22±1.30 | 5.65±6.80 | |
| DAUNet sMRI | 0.86±0.04 | 0.86±0.06 | 0.87±0.09 | 5.36±2.46 | 0.76±0.26 | 1.14±0.36 | 1.98±1.14 | 3.22±2.55 | |
| P-value DAUNet sMRI vs. DSUNet sMRI | 0.002 | 0.541 | 0.074 | 0.065 | 0.017 | 0.091 | 0.147 | 0.049 | |
| P-value DAUNet sMRI vs. DAUNet CBCT | <0.001 | 0.010 | 0.043 | 0.034 | <0.001 | 0.006 | <0.001 | 0.103 | |
| Rectum | DSUNet sMRI | 0.86±0.042 | 0.85±0.07 | 0.89±0.07 | 9.96±5.52 | 1.32±2.27 | 2.63±5.17 | 4.83±4.99 | 7.14±9.95 |
| DAUNet CBCT | 0.83±0.055 | 0.81±0.098 | 0.87±0.07 | 15.51±22.13 | 1.40±1.06 | 3.07±3.21 | 5.28±5.88 | 9.60±8.96 | |
| DAUNet sMRI | 0.92±0.03 | 0.91±0.02 | 0.93±0.02 | 5.20±1.61 | 0.62±0.15 | 1.24±1.01 | 1.94±1.29 | 3.70±4.18 | |
| P-value DAUNet sMRI vs. DSUNet sMRI | <0.001 | 0.002 | 0.042 | <0.001 | 0.128 | 0.161 | 0.007 | 0.065 | |
| P-value DAUNet sMRI vs. DAUNet CBCT | <0.001 | <0.001 | 0.007 | 0.031 | <0.001 | 0.004 | 0.009 | 0.001 | |
Table 2.
Numerical comparison of DSUNet sMRI, DSUNet CBCT, and DSUNet sMRI for hold-out validation.
| Method | DSC | Sensitivity | Specificity | HD (mm) | MSD (mm) | RMSD (mm) | CMD (mm) | VD (cc) | |
|---|---|---|---|---|---|---|---|---|---|
| Bladder | DSUNet sMRI | 0.92±0.05 | 0.95±0.04 | 0.89±0.08 | 6.99±5.60 | 0.49±0.26 | 0.77±0.42 | 1.34±1.53 | 8.11±8.16 |
| DAUNet CBCT | 0.88±0.07 | 0.89±0.09 | 0.88±0.09 | 9.84±10.38 | 0.82±0.54 | 1.29±0.86 | 2.72±2.76 | 11.94±14.30 | |
| DAUNet sMRI | 0.95±0.03 | 0.96±0.02 | 0.93±0.04 | 4.79±5.83 | 0.40±0.20 | 0.78±0.76 | 0.86±0.63 | 4.41±4.45 | |
| P-value DAUNet sMRI vs. DSUNet sMRI | 0.004 | <0.001 | 0.005 | 0.138 | 0.105 | 0.974 | 0.008 | 0.005 | |
| P-value DAUNet sMRI vs. DAUNet CBCT | <0.001 | 0.001 | 0.015 | 0.048 | <0.001 | 0.023 | <0.001 | 0.005 | |
| Prostate | DSUNet sMRI | 0.84±0.09 | 0.87±0.11 | 0.82±0.10 | 4.55±2.36 | 0.68±0.39 | 0.96±0.50 | 2.04±2.03 | 1.87±1.67 |
| DAUNet CBCT | 0.84±0.11 | 0.82±0.10 | 0.85±0.14 | 4.84±3.26 | 0.78±0.57 | 1.12±0.78 | 1.95±2.23 | 2.31±1.84 | |
| DAUNet sMRI | 0.87±0.08 | 0.86±0.10 | 0.86±0.08 | 4.27±2.19 | 0.69±0.45 | 1.02±0.69 | 1.93±1.94 | 2.10±1.71 | |
| P-value DAUNet sMRI vs. DSUNet sMRI | 0.015 | 0.392 | 0.015 | 0.411 | 0.961 | 0.059 | 0.072 | 0.587 | |
| P-value DAUNet sMRI vs. DAUNet CBCT | <0.001 | <0.001 | 0.076 | 0.014 | <0.001 | 0.040 | 0.009 | 0.569 | |
| Rectum | DSUNet sMRI | 0.88±0.063 | 0.90±0.06 | 0.87±0.07 | 7.29±4.47 | 0.73±0.57 | 1.45±1.53 | 2.61±2.85 | 3.49±4.20 |
| DAUNet CBCT | 0.83±0.09 | 0.86±0.11 | 0.81±0.09 | 10.76±14.62 | 0.99±0.56 | 1.56±1.15 | 2.56±2.38 | 7.04±7.92 | |
| DAUNet sMRI | 0.90±0.05 | 0.92±0.06 | 0.88±0.07 | 5.72±4.25 | 0.81±0.90 | 2.00±3.18 | 2.02±2.32 | 4.16±3.58 | |
| P-value DAUNet sMRI vs. DSUNet sMRI | 0.004 | <0.001 | 0.018 | 0.003 | 0.194 | 0.194 | 0.003 | 0.171 | |
| P-value DAUNet sMRI vs. DAUNet CBCT | <0.001 | <0.001 | 0.002 | <0.001 | 0.003 | 0.486 | 0.003 | <0.001 | |
Table 3.
Numerical comparison of DSUNet sMRI, DSUNet CBCT, and DSUNet sMRI on 100 patients.
| Method | DSC | Sensitivity | Specificity | HD (mm) | MSD (mm) | RMSD (mm) | CMD (mm) | VD (cc) | |
|---|---|---|---|---|---|---|---|---|---|
| Bladder | DSUNet sMRI | 0.93±0.05 | 0.96±0.03 | 0.91±0.07 | 6.45±5.24 | 0.51±0.27 | 0.81±0.52 | 1.29±1.28 | 8.642±7.23 |
| DAUNet CBCT | 0.90±0.06 | 0.93±0.08 | 0.88±0.08 | 11.31±12.75 | 0.74±0.44 | 1.18±0.76 | 2.08±2.29 | 14.26±16.59 | |
| DAUNet sMRI | 0.95±0.02 | 0.95±0.04 | 0.95±0.03 | 4.69±4.92 | 0.44±0.22 | 0.80±0.69 | 0.90±0.68 | 6.55±6.68 | |
| P-value DAUNet sMRI vs. DSUNet sMRI | 0.003 | 0.180 | <0.001 | 0.049 | 0.154 | 0.960 | 0.023 | 0.096 | |
| P-value DAUNet sMRI vs. DAUNet CBCT | <0.001 | 0.169 | <0.001 | 0.002 | <0.001 | 0.009 | <0.001 | <0.001 | |
| Prostate | DSUNet sMRI | 0.84±0.07 | 0.85±0.10 | 0.84±0.096 | 5.48±2.57 | 0.80±0.35 | 1.12±0.47 | 2.26±1.63 | 4.13±5.71 |
| DAUNet CBCT | 0.82±0.09 | 0.81±0.10 | 0.84±0.11 | 5.83±3.30 | 0.9±0.47 | 1.30±0.66 | 2.58±1.92 | 3.98±5.21 | |
| DAUNet sMRI | 0.86±0.06 | 0.86±0.08 | 0.87±0.085 | 4.82±2.37 | 0.73±0.37 | 1.08±0.55 | 1.954±1.58 | 3.32±5.54 | |
| P-value DAUNet sMRI vs. DSUNet sMRI | <0.001 | 0.268 | 0.120 | 0.043 | 0.126 | 0.584 | 0.179 | 0.317 | |
| P-value DAUNet sMRI vs. DAUNet CBCT | <0.001 | <0.001 | 0.084 | 0.010 | <0.001 | 0.009 | 0.031 | 0.403 | |
| Rectum | DSUNet sMRI | 0.88±0.05 | 0.88±0.07 | 0.88±0.08 | 8.62±5.15 | 1.03±1.66 | 2.04±3.82 | 3.72±4.18 | 5.31±7.78 |
| DAUNet CBCT | 0.83±0.07 | 0.84±0.11 | 0.84±0.08 | 13.14±18.71 | 1.19±0.87 | 2.31±2.51 | 3.92±4.65 | 8.32±8.47 | |
| DAUNet sMRI | 0.91±0.04 | 0.92±0.04 | 0.90±0.05 | 5.46±3.19 | 0.71±0.65 | 1.62±2.36 | 1.98±1.86 | 3.93±3.86 | |
| P-value DAUNet sMRI vs. DSUNet sMRI | <0.001 | <0.001 | 0.014 | <0.001 | 0.192 | 0.434 | 0.002 | 0.155 | |
| P-value DAUNet sMRI vs. DAUNet CBCT | <0.001 | <0.001 | <0.001 | 0.006 | 0.001 | 0.130 | 0.005 | <0.001 | |
3.B. Efficacy of sMRI-aided strategy
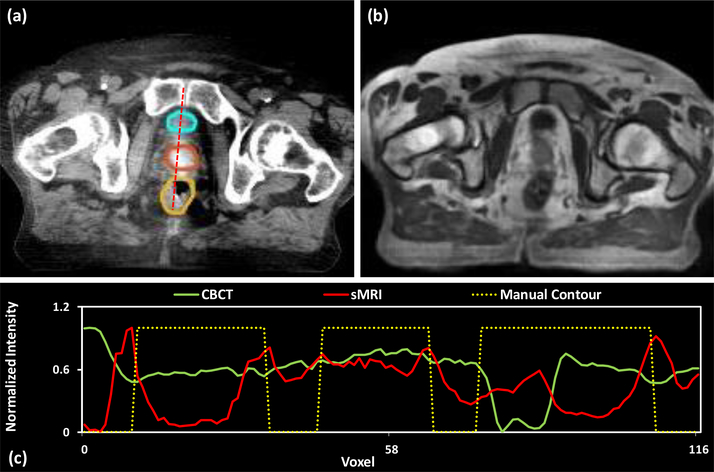
Fig. 5 shows axial views of the CBCT image, sMRI and deformed prostate manual contours at axial level for a patient. To better illustrate the contrast enhancement of sMRI-aided strategy, Fig. 5 (c) compare the profiles of the dashed red line for CBCT image (a), sMRIs (b) and manual contours (displayed overlaid on the CBCT image). The dash line passed through the bladder (cyan), prostate (orange), and rectum (yellow) manual contours. To make the boundary clear, we set 0 as the voxels without organs, and 1.0 as the voxels within the organs. Thus, the boundary of the prostate is the jump discontinuity on the plot profile. To provide a meaningful comparison, we use a normalization to scale voxel intensities on the dash line to [0, 1], where x denotes a voxel’s intensity on dash line, X denotes the all voxels’ intensity appeared on dash line. As is shown in subfigure (c), the middle jump of yellow dashed line depicts the region of prostate manual contour. It is shown that the sMRI’s plot profile (red line) in that region has three significant peaks, and two of them are correlated to the manual prostate contour’s boundary. However, from CBCT’s plot profile (green line), the peaks of prostate boundary are not very significant from other peaks within the prostate region. In addition, this situation occurs in first and third jumps of yellow dashed line, i.e., the region of bladder and rectum. As is shown in this subfigure, sMRI provides superior bladder and prostate contrast to CBCT image.
Figure 5.
Visual results of generated sMRI. (a) shows the original CBCT image at axial level, the display window of CBCT image was set to [−160, 240] HU. The manual contours of bladder (cyan), prostate (orange) and rectum (yellow) were displayed overlaid on the CBCT image. (b) shows the generated sMRI. (c) shows the normalized plot profile of CBCT, sMRI, and manual contour of the red dashed line in (a), respectively.
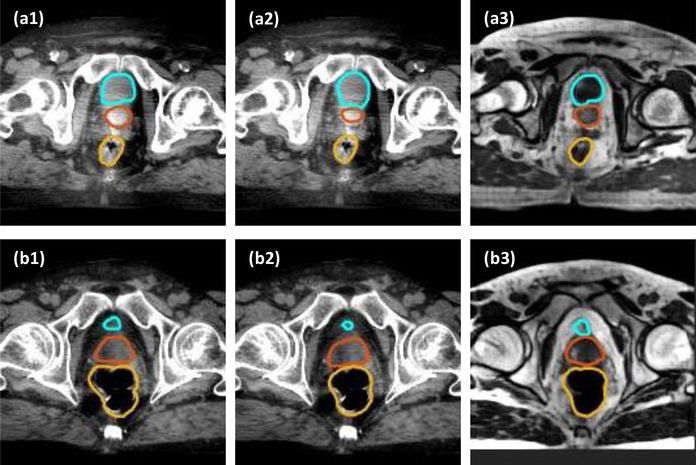
Fig. 6 compares the segmentation results with and without using the sMRI-aided strategy. In the first row, the segmented contour on sMRI (a3) is quite close to the manual contours, while the segmentation results on CBCT show different size and shape for bladder and prostate from manual contours. In the second row, the segmented prostate contour of both CBCT (b2) and sMRI (b3) images are different from the manual prostate contour. The segmentation error of sMRI (b3) may be caused by the synthesis error caused by CycleGAN during synthesis process. Even so, the segmented prostate contour of sMRI (b3) is closer to manual contour as compared to the segmented prostate contour of CBCT (b2).
Figure 6.
Comparison of the proposed method on sMRI data and CBCT data. (a1) and (b1) show the CBCT images of two patients, with manual contour of bladder (cyan), prostate (orange) and rectum (yellow) overlaid on the CBCT images. (a2) and (b2) show the CBCT images of two patients, with the segmented contour of DAUNet trained on CBCT data overlaid on CBCT images. (a3) and (b3) show the sMRI images with segmented contours of DAUNet trained on sMRI data. The display window of all CBCT images was set to [−160, 240] HU.
To evaluate the influence of sMRI-aided strategy on the segmentation, we compared the results obtained with the proposed DAUNet tested separately on CBCT and sMRIs. The numerical comparison between DAUNet using CBCT (DAUNet CBCT) and DAUNet using sMRI (DAUNet sMRI) for five-fold cross-validation is shown in Table 1. The numerical comparison for hold-out validation is shown in Table 2. In addition, the performance of proposed method on two validations has no significant difference in large amount of metrics. We show the numerical results of all 100 patients in Table 3. As shown in these tables, compared CBCT segmentation, our sMRI-aided segmentation demonstrates superior performances on all metrics.
DISCUSSION
We proposed a new pelvic multi-organ segmentation method which incorporates both a sMRI-aided strategy and a deep attention strategy into a U-Net architecture for automatic multi-organ segmentation of CBCT images. The proposed method makes use of the superior soft-tissue contrast of sMRI, bypasses MR acquisition and has the potential to generate accurate and consistent pelvic multi-organ segments using CBCT images alone.
This paper is the first study of automatic multi-organ segmentation for pelvic CBCT images using deep learning-based method. Compared to deformable registration-based methods, our method achieves higher accuracy. Thor et al. reported the average DSC (range) was 0.80 (0.65–0.87) for prostate, 0.77 (0.63–0.87) for rectum and 0.73 (0.34–0.91) for bladder among 36 patient datasets when propagating the corresponding contours from planning CT to CBCT with Demons deformable registration algorithm (Thor et al., 2011). Similar results can also be found in other deformable registration-based studies (Woerner et al., 2017; Rubeaux et al., 2013; Thörnqvist et al., 2010; Gardner et al., 2015), and our method demonstrates 6%, 14%, and 22% higher DSC for prostate, rectum and bladder, respectively.
The results of the proposed method can be further examined by comparing it to inter-observer variation of manual contouring on CBCT. Gardner et al. reported an average DSC of 0.872 and HD of 5.22 mm of prostate contours among five radiation oncologists on ten patients’ dataset when compared to the consensus contour (Gardner et al., 2015; Gardner et al., 2019). Choi et al. showed that the mean center of mass distances from averaged prostate contour was 1.73 mm among three observers contouring on ten patients (Choi et al., 2011). White et al. reported the mean standard deviation of prostate volume among five observers on five patients was 8.93 cc with a large range (3.98 – 19.00 cc) (White et al., 2009). Our results are similar or better when compared to the above findings, indicating manual contouring is prone to errors from significant inter-observer variation. The method proposed here can provide an observer-independent segmentation method to improve reproducibility and efficiency with comparable accuracy.
There are several limitations in the implementation of this study. As we stated in 2.C, the prostate and organ contours in this study were delineated on MR first, and then propagated to CBCT with proper refinement. The refinement aims to correct obvious error from registration when keeping the majority of contours. Thus, such contour for prostate would mostly avoid including periprostatic region. However, we noticed that during this process, it may introduce registration errors which cannot be fully corrected by the refinement, and the error in refinement which may inevitably include some of the periprostatic region since the poor contrast on CBCT. The registration error may affect the performance in sMRI stage by poorly matched MRI-CBCT training dataset. Thus, in order to reduce the errors, we review each deformable registration very carefully to make sure that all MRI-CBCT registration in our training database matched very well. Secondly, the refinement was done by one physician, which may have been affected by the physician’s contouring style. A potential solution is to include consensus contours by multiple observers on the refinement of contours to reduce the observer bias on segmentation training dataset.
In addition, the sMRI quality may affect the segmentation accuracy. On the one hand, the CBCT image quality is often affected by the physical imaging characteristics, namely a large scatter-to-primary ratio leading to image artifacts such as streaking, shading, cupping, and reduced image contrast, which will inherently influence sMRI quality and thus accuracy of the segmentation. Applying deep learning-based CBCT image quality enhancement to first improve the CBCT image quality will be our future work. On the other hand, the sMRI synthesis error may also introduce the segmentation error, to improve the accuracy of the sMRI synthesis model and define a more specific loss function for CBCT-to-MRI supervision will be our future work.
The computation complexity of proposed method is higher than U-Net algorithms due to the use of another CycleGAN-based sMRI generation model. The learning rate for Adam optimizer was set to 2e-4, and the model was trained and tested on 2 NVIDIA Tesla V100 with 32 GB of memory for each GPU. A batch size of 20. 12 GB CPU memory and 58 GB GPU memory was used for each batch optimization. The training was stopped after 100000 iterations. Model training takes approximately 30 hours. For testing, sMRI patches were generated by feeding CBCT patches into the trained CycleGAN model. Pixel values were averaged when overlapping exists. The sMRI estimation for one patient takes about 1–2 minutes. Tensorflow was used to implement both CycleGAN and DAUNet network architecture. During segmentation, binary cross entropy was used as loss function, and the learning rate for Adam optimizer was set to 1e-3. The training was stopped after 180 epochs. For each epoch, the batch size was set to 20. The training of DAUNet model takes about 1.7 hours.
In this study, we demonstrated the feasibility of our method with 100 clinical patient cases. A comprehensive evaluation with a larger cohort of patients with diverse disease characteristic and image quality would be involved in the future. This study validated the proposed method by quantifying the shape similarity of contours, while small differences from ground truth were observed. Its potential clinical impact due to such differences in dose estimation and treatment replanning is not yet fully understood. Thus, a further investigation in clinical outcomes of the proposed method in adaptive radiation therapy would be of great interest and is needed for eventual adoption in general clinical use.
CONCLUSIONS
In summary, we developed an accurate pelvic multi-organ segmentation strategy on CBCT images with CBCT-based sMRIs. This technique could provide real-time accurate target and organ contours for adaptive radiation therapy, which would greatly facilitate clinical workflow.
ACKNOWLEDGEMENTS
This research is supported in part by the National Cancer Institute of the National Institutes of Health under Award Number R01CA215718 (XY), the Department of Defense (DoD) Prostate Cancer Research Program (PCRP) Award W81XWH-17-1-0438 (TL) and W81XWH-17-1-0439 (AJ) and Dunwoody Golf Club Prostate Cancer Research Award (XY), a philanthropic award provided by the Winship Cancer Institute of Emory University.
Footnotes
DISCLOSURES
The authors declare no conflicts of interest.
REFERENCES
- Balagopal A, Kazemifar S, Nguyen D, Lin MH, Hannan R, Owrangi A and Jiang S 2018. Fully automated organ segmentation in male pelvic CT images Physics in Medicine and Biology 63 [DOI] [PubMed] [Google Scholar]
- Barney BM, Lee RJ, Handrahan D, Welsh KT, Cook JT and Sause WT 2011. Image-Guided Radiotherapy (IGRT) for Prostate Cancer Comparing kV Imaging of Fiducial Markers with Cone Beam Computed Tomography (CBCT) Int J Radiat Oncol 80 301–5 [DOI] [PubMed] [Google Scholar]
- Choi HJ, Kim YS, Lee SH, Lee YS, Park G, Jung JH, Cho BC, Park SH, Ahn H, Kim C-S, Yi SY and Ahn SD 2011. Inter- and intra-observer variability in contouring of the prostate gland on planning computed tomography and cone beam computed tomography Acta Oncologica 50 539–46 [DOI] [PubMed] [Google Scholar]
- de la Zerda A, Armbruster B and Xing L 2007. Formulating adaptive radiation therapy (ART) treatment planning into a closed-loop control framework Physics in Medicine and Biology 52 4137–53 [DOI] [PubMed] [Google Scholar]
- Dong X, Lei Y, Wang T, Thomas M, Tang L, Curran WJ, Liu T and Yang X 2019. Automatic multiorgan segmentation in thorax CT images using U-net-GAN Medical physics 46 2157–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner S J, Mao W, Liu C, Aref I, Elshaikh M, Lee JK, Pradhan D, Movsas B, Chetty IJ and Siddiqui F 2019. Improvements in CBCT Image Quality Using a Novel Iterative Reconstruction Algorithm: A Clinical Evaluation Advances in Radiation Oncology 4 390–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner S J, Wen N, Kim J, Liu C, Pradhan D, Aref I, Cattaneo R, Vance S, Movsas B, Chetty IJ and Elshaikh MA 2015. Contouring variability of human- and deformable-generated contours in radiotherapy for prostate cancer Physics in Medicine and Biology 60 4429–47 [DOI] [PubMed] [Google Scholar]
- Kataria T, Gupta D, Goyal S, Bisht S S, Basu T, Abhishek A, Narang K, Banerjee S, Nasreen S, Sambasivam S and Dhyani A 2016. Clinical outcomes of adaptive radiotherapy in head and neck cancers British Journal of Radiology 89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Y, Harms J, Wang T, Liu Y, Shu HK, Jani AB, Curran WJ, Mao H, Liu T and Yang X 2019a. MRI-only based synthetic CT generation using dense cycle consistent generative adversarial networks Medical physics 46 3565–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Y, Tang X, Higgins K, Lin J, Jeong J, Liu T, Dhabaan A, Wang T, Dong X, Press R, Curran W J and Yang X 2019b. Learning-based CBCT correction using alternating random forest based on auto-context model Medical physics 46 601–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Y, Tian S, He X, Wang T, Wang B, Patel P, Jani A B, Mao H, Curran W J, Liu T and Yang X 2019c. Ultrasound Prostate Segmentation Based on Multi-Directional Deeply Supervised V-Net Medical physics In press [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra D, Chaudhury S, Sarkar M and Soin A S 2018. Ultrasound Image Segmentation: A Deeply Supervised Network with Attention to Boundaries IEEE Trans Biomed Eng 66 1637–48. [DOI] [PubMed] [Google Scholar]
- Oldham M, Letourneau D, Watt L, Hugo G, Yan D, Lockman D, Kim LH, Chen PY, Martinez A and Wong JW 2005. Cone-beam-CT guided radiation therapy: A model for on-line application Radiotherapy and Oncology 75 271–8 [DOI] [PubMed] [Google Scholar]
- Rubeaux M, Simon A, Gnep K, Colliaux J, Acosta O, Crevoisier R d and Haigron P 2013. Evaluation of non-rigid constrained CT/CBCT registration algorithms for delineation propagation in the context of prostate cancer radiotherapy SPIE Medical Imaging 8671 867106 [Google Scholar]
- Thor M, Petersen JB, Bentzen L, Hoyer M and Muren LP 2011 Deformable image registration for contour propagation from CT to cone-beam CT scans in radiotherapy of prostate cancer Acta Oncol 50 918–25 [DOI] [PubMed] [Google Scholar]
- Thörnqvist S, Petersen JBB, Høyer M, Bentzen LN and Muren LP 2010. Propagation of target and organ at risk contours in radiotherapy of prostate cancer using deformable image registration Acta Oncologica 49 1023–32 [DOI] [PubMed] [Google Scholar]
- Wang B, Lei Y, Tian S, Wang T, Liu Y, Patel P, Jani AB, Mao H, Curran WJ, Liu T and Yang X 2019a. Deeply supervised 3D fully convolutional networks with group dilated convolution for automatic MRI prostate segmentation Medical physics 46 1707–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Lei Y, Tang H, He Z, Castillo R, Wang C, Li D, Higgins K, Liu T, Curran W J, Zhou W and Yang X 2019b. A learning-based automatic segmentation and quantification method on left ventricle in gated myocardial perfusion SPECT imaging: A feasibility study JNucl Cardiol In press, doi: 10.1007/s12350-019-01594-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Lei Y, Tian S, Jiang X, Zhou J, Liu T, Dresser S, Curran WJ, Shu HK and Yang X 2019c. Learning-based Automatic Segmentation of Arteriovenous Malformations on Contrast CT Images in Brain Stereotactic Radiosurgery Medical physics 46 3133–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White EA, Brock KK, Jaffray DA and Catton CN 2009. Inter-observer Variability of Prostate Delineation on Cone Beam Computerised Tomography Images Clinical Oncology 21 32–8 [DOI] [PubMed] [Google Scholar]
- Woerner A J, Choi M, Harkenrider M M, Roeske J C and Surucu M 2017. Evaluation of Deformable Image Registration-Based Contour Propagation From Planning CT to Cone-Beam CT Technology in Cancer Research & Treatment 16 801–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Chao M, Lee P and Xing L 2008. Feature-based rectal contour propagation from planning CT to cone beam CT Medical physics 35 4450–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D, Vicini F, Wong J and Martinez A 1997. Adaptive radiation therapy Phys Med Biol 42 123–32 [DOI] [PubMed] [Google Scholar]
- Yang X, Rossi P, Ogunleye T, Marcus DM, Jani AB, Mao H, Curran WJ and Liu T 2014a. Prostate CT segmentation method based on nonrigid registration in ultrasound-guided CT-based HDR prostate brachytherapy Medical physics 41 111915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XF, Lei Y, Shu HK, Rossi P, Mao H, Shim H, Curran WJ and Liu T 2017. Pseudo CT Estimation from MRI Using Patch-based Random Forest Proc SPIE Medical Imaging 10133 101332Q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XF, Wu N, Cheng GH, Zhou ZY, Yu DS, Beitler JJ, Curran WJ and Liu T 2014b. Automated Segmentation of the Parotid Gland Based on Atlas Registration and Machine Learning: A Longitudinal MRI Study in Head-and-Neck Radiation Therapy Int J Radiat Oncol 90 1225–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S and Yin FF 2006. Dosimetric feasibility of cone-beam CT-based treatment planning compared to CT-based treatment planning Int J Radiat Oncol 66 1553–61 [DOI] [PubMed] [Google Scholar]