Key Points
Question
Is there an association between genetically proxied inhibition of 3-hydroxy-3-methylglutaryl coenzyme A (HMG-CoA) reductase and epithelial ovarian cancer in the general population and among BRCA1/2 mutation carriers?
Findings
In this case-control study that included 63 347 participants, genetically proxied HMG-CoA reductase inhibition equivalent to a 1-mmol/L (38.7-mg/dL) reduction in low-density lipoprotein cholesterol was significantly associated with lower odds of epithelial ovarian cancer in the general population (odds ratio, 0.60) and among BRCA1/2 mutation carriers (hazard ratio, 0.69).
Meaning
There was a significant association between genetically proxied inhibition of HMG-CoA reductase and epithelial ovarian cancer, but further research is needed to understand whether there is a similar association with medications that inhibit HMG-CoA reductase.
Abstract
Importance
Preclinical and epidemiological studies indicate a potential chemopreventive role of statins in epithelial ovarian cancer risk.
Objective
To evaluate the association of genetically proxied inhibition of 3-hydroxy-3-methylglutaryl coenzyme A (HMG-CoA) reductase (ie, genetic variants related to lower function of HMG-CoA reductase, target of statins) with epithelial ovarian cancer among the general population and in BRCA1/2 mutation carriers.
Design, Setting, and Participants
Single-nucleotide polymorphisms (SNPs) in HMGCR, NPC1L1, and PCSK9 associated with low-density lipoprotein (LDL) cholesterol in a genome-wide association study (GWAS) meta-analysis (N ≤196 475) were used to proxy therapeutic inhibition of HMG-CoA reductase, Niemann-Pick C1-Like 1 (NPC1L1) and proprotein convertase subtilisin/kexin type 9 (PCSK9), respectively. Summary statistics were obtained for these SNPs from a GWAS meta-analysis of case-control analyses of invasive epithelial ovarian cancer in the Ovarian Cancer Association Consortium (OCAC; N = 63 347) and from a GWAS meta-analysis of retrospective cohort analyses of epithelial ovarian cancer among BRCA1/2 mutation carriers in the Consortium of Investigators of Modifiers of BRCA1/2 (CIMBA; N = 31 448). Across the 2 consortia, participants were enrolled between 1973 and 2014 and followed up through 2015. OCAC participants came from 14 countries and CIMBA participants came from 25 countries. SNPs were combined into multi-allelic models and mendelian randomization estimates representing lifelong inhibition of targets were generated using inverse-variance weighted random-effects models.
Exposures
Primary exposure was genetically proxied inhibition of HMG-CoA reductase and secondary exposures were genetically proxied inhibition of NPC1L1 and PCSK9 and genetically proxied circulating LDL cholesterol levels.
Main Outcomes and Measures
Overall and histotype-specific invasive epithelial ovarian cancer (general population) and epithelial ovarian cancer (BRCA1/2 mutation carriers), measured as ovarian cancer odds (general population) and hazard ratio (BRCA1/2 mutation carriers).
Results
The OCAC sample included 22 406 women with invasive epithelial ovarian cancer and 40 941 control individuals and the CIMBA sample included 3887 women with epithelial ovarian cancer and 27 561 control individuals. Median ages for the cohorts ranged from 41.5 to 59.0 years and all participants were of European ancestry. In the primary analysis, genetically proxied HMG-CoA reductase inhibition equivalent to a 1-mmol/L (38.7-mg/dL) reduction in LDL cholesterol was associated with lower odds of epithelial ovarian cancer (odds ratio [OR], 0.60 [95% CI, 0.43-0.83]; P = .002). In BRCA1/2 mutation carriers, genetically proxied HMG-CoA reductase inhibition was associated with lower ovarian cancer risk (hazard ratio, 0.69 [95% CI, 0.51-0.93]; P = .01). In secondary analyses, there were no significant associations of genetically proxied inhibition of NPC1L1 (OR, 0.97 [95% CI, 0.53-1.75]; P = .91), PCSK9 (OR, 0.98 [95% CI, 0.85-1.13]; P = .80), or circulating LDL cholesterol (OR, 0.98 [95% CI, 0.91-1.05]; P = .55) with epithelial ovarian cancer.
Conclusions and Relevance
Genetically proxied inhibition of HMG-CoA reductase was significantly associated with lower odds of epithelial ovarian cancer. However, these findings do not indicate risk reduction from medications that inhibit HMG-CoA reductase; further research is needed to understand whether there is a similar association with such medications.
This study uses mendelian randomization to estimate the associations between genetic variants related to reduced HMG-CoA reductase activity and epithelial ovarian cancer in the general population and in BRCA1/2 mutation carriers.
Introduction
In 2018 there were approximately 22 240 new cases of ovarian cancer diagnosed and 14 070 ovarian cancer deaths in the United States.1 The prognosis for ovarian cancer is generally poor because women typically present with advanced disease and because there is a lack of early detection tests.2 Given the limited success of screening strategies, primary prevention of ovarian cancer may present an important approach for reducing disease burden.
3-Hydroxy-3-methylglutaryl coenzyme A (HMG-CoA) reductase inhibitors (statins) are low-density lipoprotein (LDL) cholesterol–lowering drugs commonly prescribed for the prevention and management of cardiovascular disease. Along with their cardioprotective effects, statins have been hypothesized to protect against development of several cancers, including ovarian cancer.3 Laboratory studies have shown that statins can induce apoptosis and inhibit tumor proliferation, invasion, and metastasis.4,5,6 Observational studies have reported lower rates of ovarian cancer among statin users compared with nonusers.7,8 However, the clinical relevance of these findings is unclear because of the unknown generalizability of preclinical studies to humans and the susceptibility of conventional observational analyses to residual confounding and other biases.
Naturally occurring variation in genes encoding pharmacological targets can be used to proxy modulation of these targets to examine the potential effects of their molecular inhibition on disease outcomes (referred to as mendelian randomization).9 Because germline genetic variants are inherited approximately randomly and fixed at the time of conception, analyses using variants as proxies for intervention targets should be largely independent of confounding and cannot be influenced by reverse causation.
A mendelian randomization approach was used to examine the association of the drug target of statins (HMG-CoA reductase) with ovarian cancer and, in secondary analyses, to examine the association of other lipid-lowering drug targets, Niemann-Pick C1-Like 1 (NPC1L1; target of ezetimibe) and proprotein convertase subtilisin/kexin type 9 (PCSK9; target of PCSK9 inhibitors), along with LDL cholesterol directly, with ovarian cancer.
Methods
Study Design and Data Sources
All studies contributing data to this analysis had the relevant institutional review board approval from each country, in accordance with the Declaration of Helsinki, and all participants provided informed consent. Summary genetic association data were obtained from case-control genome-wide association study (GWAS) analyses of invasive epithelial ovarian cancer from the Ovarian Cancer Association Consortium (OCAC).10 OCAC data were also obtained for the following invasive epithelial ovarian cancer histotypes: high-grade serous carcinoma, low-grade serous carcinoma, endometrioid carcinoma, mucinous ovarian cancer, and clear cell carcinoma. The earliest start date for participants across OCAC studies was in 1976 and the latest date of follow-up was in 2015.
Summary data were also obtained from GWAS analyses of epithelial ovarian cancer among BRCA1 (Entrez Gene: 672) or BRCA2 (Entrez Gene: 675) mutation carriers from the Consortium of Investigators of Modifiers of BRCA1/2 (CIMBA), a retrospective cohort study.10 Participants were enrolled in CIMBA between 1973 and 2014.
In OCAC and CIMBA, participants with 5% or more missing genotype calls were excluded; otherwise, missing genotype data were imputed. Additional details about the OCAC and CIMBA analyses are available in the Supplement.
To generate genetic instruments to proxy HMG-CoA reductase, NPC1L1, and PCSK9 and to proxy LDL cholesterol levels directly, summary data were obtained from a GWAS meta-analysis of LDL cholesterol levels in the Global Lipids Genetics Consortium.11 To proxy HMG-CoA reductase, 5 single-nucleotide polymorphisms (SNPs) associated with LDL cholesterol at genome-wide significance level (P < 5.0 × 10−8) and within ±100 kb windows from the gene region for HMGCR (Entrez Gene: 3156; encoding HMG-CoA reductase) were obtained. To proxy NPC1L1, 3 SNPs associated with LDL cholesterol at genome-wide significance and within ±100 kb from NPC1L1 (Entrez Gene: 29881; encoding NPC1L1) were obtained. To proxy PCSK9, 11 SNPs associated with LDL cholesterol at genome-wide significance and within ±100 kb from PCSK9 (Entrez Gene: 255738; encoding PCSK9) were obtained. For each of these drug targets, SNPs used as proxies were permitted to be in weak linkage disequilibrium (r2<0.20) with each other to increase the proportion of variance in each respective drug target explained by the instrument, maximizing instrument strength.
To proxy LDL cholesterol levels, 76 independent (r2 <0.001) SNPs associated with LDL cholesterol at genome-wide significance level were obtained, irrespective of genomic position of variants. SNP selection procedures across drug targets and LDL cholesterol are presented in the Supplement.
Outcomes
The outcomes were overall and histotype-specific invasive epithelial ovarian cancer (in the general population) and epithelial ovarian cancer (in BRCA1/2 mutation carriers).
Statistical Analysis
Mendelian randomization analysis assumes that the genetic instrument used to proxy a risk factor (1) is associated with the risk factor (“relevance”), (2) does not share a common cause with the outcome (“exchangeability”), and (3) affects the outcome only through the risk factor (“exclusion restriction”).
SNPs used to proxy each of the risk factors were matched to ovarian cancer data sets by assigning them the same effect allele. For all risk factors, mendelian randomization estimates were first generated per individual SNP using the Wald ratio and standard errors were approximated using the delta method approximation. A random-effects inverse-variance weighted meta-analysis was then used to combine individual SNPs in an instrument. In analyses of drug targets, inverse-variance weighted models were adjusted for weak linkage disequilibrium (r2<0.20) between SNPs with reference to the 1000 Genomes Phase 3 reference panel.12 All mendelian randomization estimates (odds ratios [ORs] for OCAC analyses and hazard ratios [HRs] for CIMBA analyses) were scaled up from individual SNP-level effects on LDL cholesterol levels to reflect the equivalent of a 1-mmol/L (38.7-mg/dL) reduction in LDL cholesterol levels. HRs for CIMBA analyses were obtained from survival analyses performed using time to ovarian cancer diagnosis as an end point.
Primary analyses were the association of genetically proxied HMG-CoA reductase inhibition with overall and histotype-specific invasive epithelial ovarian cancer among women in the general population (OCAC) and epithelial ovarian cancer in BRCA1/2 mutation carriers (CIMBA). Secondary analyses were the association of genetically proxied inhibition of NPC1L1 and PCSK9 and genetically proxied circulating LDL cholesterol with these ovarian cancer outcomes. Primary and secondary analyses were prespecified.
The relevance mendelian randomization assumption was tested by generating estimates of the proportion of variance in each risk factor explained by the instrument (R2) and F statistics. As a convention, an F statistic of at least 10 is indicative of evidence against weak instrument bias (a reduction in statistical power to reject the null hypothesis when an instrument explains only a small proportion of variance in an exposure).13
The exchangeability mendelian randomization assumption was tested by performing co-localization, which examines whether SNPs associated with 2 traits are likely to share causal variants with each other.14 This can be used to assess whether risk factors and disease outcomes are influenced by distinct causal variants that are in linkage disequilibrium with each other (genetic confounding). This method generates a co-localization posterior probability that the same variant is causal for both traits and a 95% credible set that represents the minimum number of variants having a cumulative posterior probability greater than 0.95.
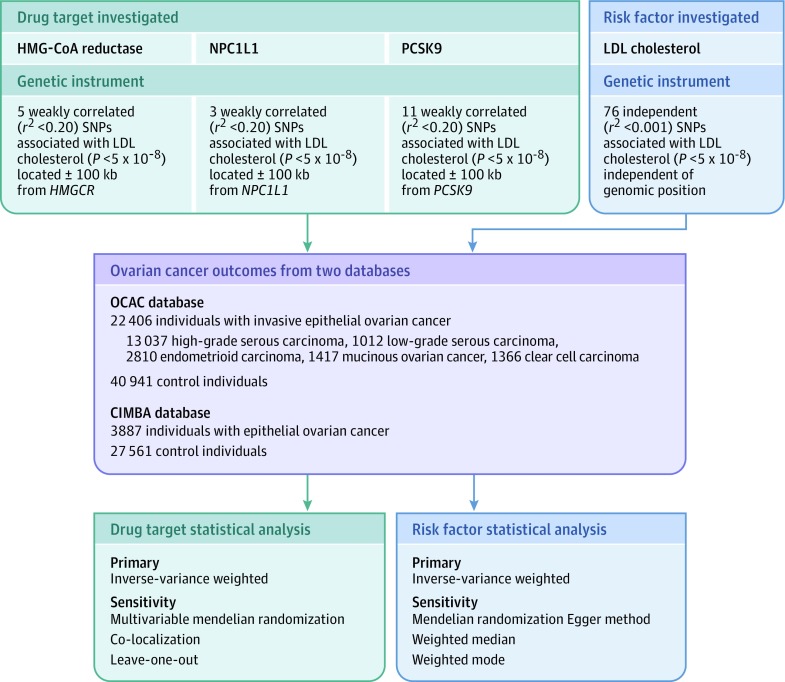
The exclusion restriction mendelian randomization assumption was tested using different methods for the analyses of genetically proxied drug targets and for the LDL cholesterol analyses. This use of different methods is because some of these methods require that variants used as genetic instruments are independent of each other, which was not the case for drug target analyses. In analyses of drug targets, violations of the exclusion restriction assumption were tested by examining associations of the genetic instruments with previously reported ovarian cancer risk factors (age at menarche, age at natural menopause, body mass index, genetic liability to endometriosis, ever use of oral contraceptives, and smoking initiation).15,16,17,18,19 The presence of an association between a drug target instrument and a previously reported ovarian cancer risk factor could provide evidence of horizontal pleiotropy (variants influencing 2 or more traits through independent biological pathways), a violation of the exclusion restriction criterion. If there was evidence of association of drug target instruments with a risk factor (P < .05), multivariable analyses were performed to examine associations between drug targets and ovarian cancer outcomes, adjusted for genetically proxied risk factors.20 Consistency of associations between analyses with and without adjustment for previously reported risk factors would suggest that analyses are unlikely to be substantially biased by horizontal pleiotropy. In analyses of LDL cholesterol levels, evidence of horizontal pleiotropy was examined via the following sensitivity analyses: mendelian randomization Egger method,21 weighted median analysis,22 and weighted mode analyses.23 In addition, iterative leave-one-out analyses were performed to examine whether results were driven by single influential SNPs in drug target proxies. Extended descriptions of these sensitivity analyses along with their assumptions are provided in the Supplement. A schematic overview of genetic instrument construction, data sources used, and primary and sensitivity analyses performed is presented in Figure 1.
Figure 1. Genetic Instrument Construction, Data Sources, and Analysis Plan in a Study of the Association Between Genetically Proxied Inhibition of HMG-CoA Reductase and Epithelial Ovarian Cancer.
For each drug target or risk factor, genetic instruments were constructed by obtaining summary genetic association data on single-nucleotide polymorphisms (SNPs) associated with low-density lipoprotein (LDL) cholesterol located within or near the gene encoding the drug target (3-hydroxy-3-methylglutaryl coenzyme A [HMG-CoA] reductase, Niemann-Pick C1-Like 1 [NPC1L1], proprotein convertase subtilisin/kexin type 9 [PCSK9]) or independent of genomic position (LDL cholesterol) from Willer et al11 (N = 19 6465). Summary genetic association data for these SNPS were then extracted from genome-wide association studies of invasive epithelial ovarian cancer (Ovarian Cancer Association Consortium) and epithelial ovarian cancer in BRCA1/2 mutation carriers (Consortium of Investigators of Modifiers of BRCA1/2). After matching SNPs across data sets by assigning them the same effect allele, mendelian randomization analyses were performed using inverse-variance weighted random-effects models as primary analyses and various approaches as sensitivity analyses to test mendelian randomization assumptions (exchangeability and exclusion restriction). The bottom boxes represent the use of different sensitivity analyses to test mendelian randomization assumptions for drug target analyses and LDL cholesterol, because some of these sensitivity analyses require that variants used as genetic instruments are independent of each other, which was not the case for drug target analyses.
All statistical tests were 2-sided and a significance threshold was set at P < .05. For analyses of invasive epithelial ovarian cancer histotypes, evidence of heterogeneity of estimates across subtypes (P value for heterogeneity) was calculated from a Cochran Q test, where Q is distributed as a χ2 statistic with K (number of histotypes) – 1 degree of freedom and the percentage of variability across these estimates due to heterogeneity beyond chance was quantified using the I2 statistic.24 A significance threshold for evidence of heterogeneity across histotypes was set at P<.05.
As sensitivity analyses, Bonferroni corrections were applied to establish multiple testing–adjusted significance thresholds for the following analyses: overall invasive epithelial ovarian cancer analyses in OCAC (Bonferroni threshold of P < .01 [0.05/4 statistical tests {3 drug targets and LDL cholesterol tested against 1 ovarian cancer end point}]); epithelial ovarian cancer analyses in CIMBA (Bonferroni threshold of P < .01 [0.05/4 statistical tests {3 drug targets and LDL cholesterol tested against 1 ovarian cancer end point}]) and invasive epithelial ovarian cancer histotypes in OCAC (Bonferroni threshold of P < .003 [0.05/20 statistical tests {3 drug targets and LDL cholesterol tested against 5 ovarian cancer histotypes}]). All statistical analyses were performed using R version 3.3.1.
Results
After exclusion of 1936 participants because of missing genotype calls, the OCAC analytic sample included 22 406 women with invasive epithelial ovarian cancer and 40 941 control individuals of European ancestry along with individuals with the following invasive histotypes: high-grade serous carcinoma (n = 13 037), low-grade serous carcinoma (n = 1012), endometrioid carcinoma (n = 2810), mucinous ovarian cancer (n = 1417), and clear cell carcinoma (n = 1366). Individuals with invasive histotypes classified as “other” by OCAC (n = 2764) were included in analyses for invasive epithelial ovarian cancer but were not assessed separately. The median (interquartile range) ages of participants in overall invasive epithelial ovarian cancer analyses were 59.0 (51.0-67.0) years in the analytic sample and 57.0 (49.0-65.0) years in the control sample. Participants were enrolled from 14 countries (the United States, Australia, Belarus, Germany, Belgium, Denmark, Finland, Norway, Canada, Poland, the United Kingdom, Spain, the Netherlands, and Sweden). The CIMBA sample included 3887 women with epithelial ovarian cancer and 27 561 control individuals of European ancestry. The median (interquartile range) ages at censoring for CIMBA participants were as follows: 50.0 (45.0-57.0) years in individuals with BRCA1, 57.0 (50.0-63.2) years in individuals with BRCA2, 41.5 (34.1-50.0) years in BRCA1 control individuals, and 44.7 (36.5-54.0) years in BRCA2 control individuals. Participants in CIMBA came from 25 countries (including 20 European countries, the United States, Australia, Canada, Israel, and South Africa). The age range of participants across studies was 20 to 100 years. There was no overlap in participants across OCAC and CIMBA analyses. The Global Lipids Genetics Consortium sample consisted of less than or equal to 196 475 individuals who were primarily of European ancestry (96% of individuals).
Characteristics of genetic variants in HMGCR, NPC1L1, and PCSK9 used to proxy pharmacological targets are presented in Table 1. In brief, 5 SNPs in HMGCR (rs12916, rs10515198, rs12173076, rs3857388, rs7711235) were used to proxy HMG-CoA reductase inhibition, 3 SNPs in NPC1L1 (rs2073547, rs217386, rs7791240) proxied NPC1L1 inhibition, and 11 SNPs in PCSK9 (rs11591147, rs11206510, rs2479409, rs585131, rs11206514, rs2495477, rs572512, rs2479394, rs12067569, rs10493176, rs11583974) proxied PCSK9 inhibition. Effect allele frequencies for SNPs used to proxy drug targets were similar across GWASs for measured LDL cholesterol levels, invasive epithelial ovarian cancer in the general population (OCAC), and epithelial ovarian cancer among BRCA1/2 mutation carriers (CIMBA) (eTable 1 in the Supplement). Characteristics of 76 SNPs used to proxy circulating LDL cholesterol levels are presented in eTable 2 in the Supplement.
Table 1. Characteristics of LDL Cholesterol–Lowering Genetic Variants in HMGCR, NPC1L1, and PCSK9 .
| Single-Nucleotide Polymorphism | Effect Allele/Non-effect Allelea | Effect Allele Frequencyb | Effect (95% CI), mmol/L | P Value |
|---|---|---|---|---|
| HMGCR | ||||
| rs12916 | T/C | 0.57 | −0.073 (−0.081 to −0.065) | 7.8 × 10−78 |
| rs10515198 | G/A | 0.90 | −0.060 (−0.072 to −0.048) | 6.0 × 10−22 |
| rs12173076 | T/G | 0.88 | −0.065 (−0.077 to −0.053) | 2.3 × 10−27 |
| rs3857388 | T/C | 0.87 | −0.042 (−0.054 to −0.030) | 2.2 × 10−11 |
| rs7711235 | A/G | 0.73 | −0.038 (−0.050 to −0.026) | 5.0 × 10−10 |
| NPC1L1 | ||||
| rs2073547 | A/G | 0.81 | −0.049 (−0.059 to −0.039) | 1.9 × 10−21 |
| rs217386 | A/G | 0.41 | −0.036 (−0.044 to −0.028) | 1.2 × 10−19 |
| rs7791240 | T/C | 0.91 | −0.043 (−0.057 to −0.029) | 1.8 × 10−10 |
| PCSK9 | ||||
| rs11591147 | T/G | 0.02 | −0.497 (−0.532 to −0.462) | 8.6 × 10−143 |
| rs11206510 | C/T | 0.15 | −0.083 (−0.093 to −0.073) | 2.4 × 10−53 |
| rs2479409 | A/G | 0.67 | −0.064 (−0.072 to −0.056) | 2.5 × 10−50 |
| rs585131 | C/T | 0.18 | −0.064 (−0.074 to −0.054) | 2.7 × 10−35 |
| rs11206514 | C/A | 0.39 | −0.051 (−0.059 to −0.043) | 1.0 × 10−32 |
| rs2495477 | G/A | 0.40 | −0.064 (−0.074 to −0.054) | 7.3 × 10−30 |
| rs572512 | C/T | 0.65 | −0.048 (−0.058 to −0.038) | 5.3 × 10−26 |
| rs2479394 | A/G | 0.72 | −0.039 (−0.047 to −0.031) | 1.6 × 10−19 |
| rs12067569 | G/A | 0.97 | −0.089 (−0.109 to −0.069) | 2.0 × 10−17 |
| rs10493176 | G/T | 0.11 | −0.078 (−0.098 to −0.058) | 2.5 × 10−14 |
| rs11583974 | G/A | 0.97 | −0.065 (−0.089 to −0.041) | 4.0 × 10−09 |
SI conversion: To convert mmol/L to mg/dL, multiply by 38.7.
Low-density lipoprotein (LDL) cholesterol–lowering allele; effect represents the change in LDL cholesterol levels (mmol/L) per copy of the effect allele.
Estimates were obtained from the 1000 Genomes Phase 3 panel (European samples); single-nucleotide polymorphisms to act as genetic proxies for drug targets were obtained within a 100 kb window from HMGCR (chr5:74,632,154-74,657,929), NPC1L1 (chr7:44,552,134-44,580,914), and PCSK9 (chr1:55,505,221-55,530,525) using Genome Reference Consortium Human Build 37 coordinates.
Across the 4 risk factors examined, F statistics for their respective genetic instruments (used to examine the relevance mendelian randomization assumption) ranged from 71.7 to 196.4, suggesting that weak instrument bias was unlikely to contribute to the analyses. F statistics and power calculations are presented for each risk factor in eTable 3 in the Supplement.
Primary Analyses
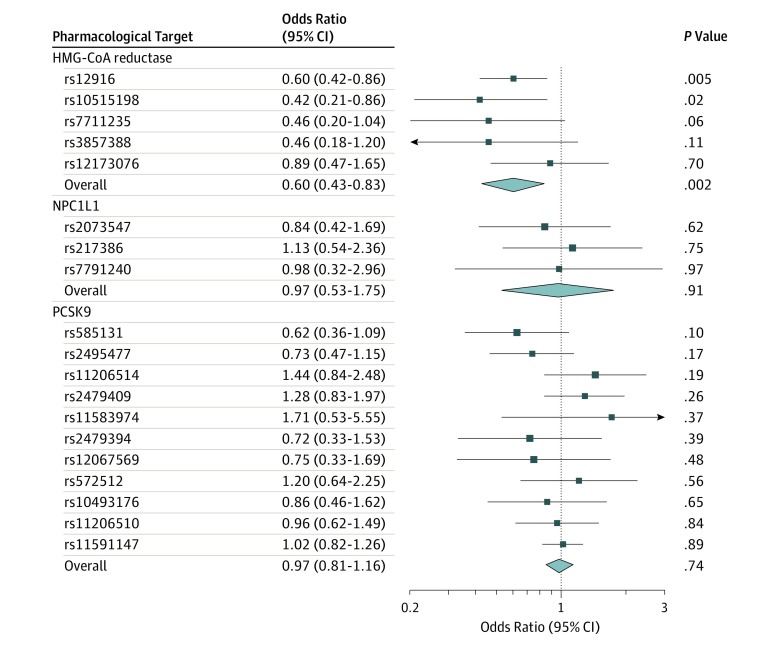
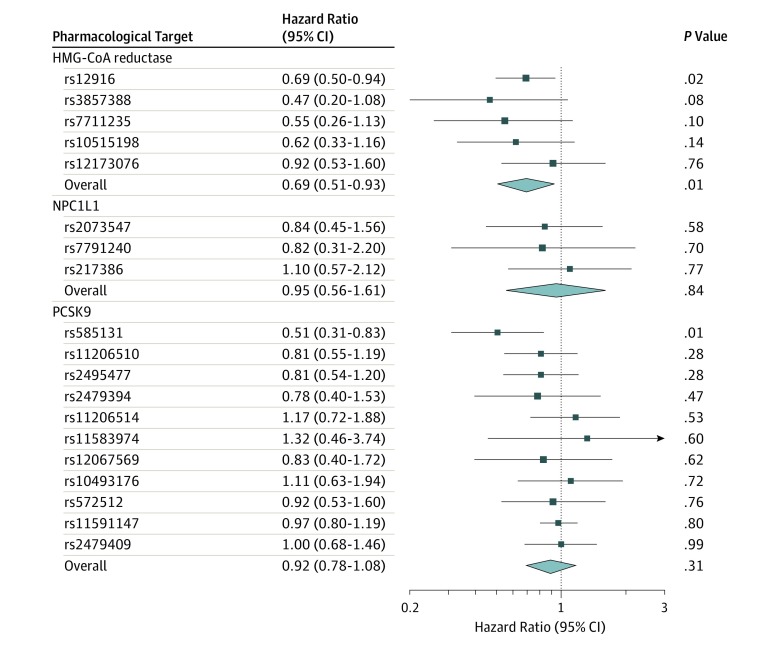
Genetically proxied HMG-CoA reductase inhibition equivalent to a 1-mmol/L (38.7-mg/dL) reduction in LDL cholesterol was significantly associated with invasive epithelial ovarian cancer in OCAC (OR, 0.60 [95% CI, 0.43-0.83]; P = .002; Table 2 and Figure 2). This association did not significantly differ across 5 invasive histotypes assessed (P value for heterogeneity = 0.84; I2 = 0.00%). In analyses among BRCA1 and BRCA2 mutation carriers combined in CIMBA, genetically proxied HMG-CoA reductase inhibition was significantly associated with epithelial ovarian cancer risk (HR, 0.69 [95% CI, 0.51-0.93]; P = .01; Figure 3).
Table 2. Association Between Genetically Proxied Inhibition of 3-Hydroxy-3-Methylglutaryl Coenzyme A (HMG-CoA) Reductase, Niemann-Pick C1-Like 1 (NPC1L1), and Proprotein Convertase Subtilisin/Kexin Type 9 (PCSK9) and Genetically Proxied LDL Cholesterol Levels and Overall and Histotype-Specific Invasive Epithelial Ovarian Cancer Among Women in the General Population.
| Outcome | Cases, No. | Odds Ratio (95% CI)a | P Value |
|---|---|---|---|
| HMG-CoA Reductase | |||
| Invasive epithelial ovarian cancer | 22 406 | 0.60 (0.43-0.83) | .002 |
| High-grade serous carcinoma | 13 037 | 0.70 (0.47-1.04) | .08 |
| Low-grade serous carcinoma | 1012 | 1.49 (0.22-10.05) | .68 |
| Mucinous carcinoma | 1417 | 0.53 (0.12-2.42) | .41 |
| Endometrioid carcinoma | 2810 | 0.40 (0.19-0.83) | .01 |
| Clear cell carcinoma | 1366 | 0.61 (0.19-1.92) | .40 |
| NPC1L1 | |||
| Invasive epithelial ovarian cancer | 22 406 | 0.97 (0.53-1.75) | .91 |
| High-grade serous carcinoma | 13 037 | 0.93 (0.46-1.88) | .83 |
| Low-grade serous carcinoma | 1012 | 0.34 (0.04-2.93) | .33 |
| Mucinous carcinoma | 1417 | 1.39 (0.23-8.24) | .72 |
| Endometrioid carcinoma | 2810 | 1.62 (0.44-5.92) | .47 |
| Clear cell carcinoma | 1366 | 0.43 (0.07-2.55) | .35 |
| PCSK9 | |||
| Invasive epithelial ovarian cancer | 22 406 | 0.97 (0.81-1.16) | .74 |
| High-grade serous carcinoma | 13 037 | 0.87 (0.79-1.20) | .82 |
| Low-grade serous carcinoma | 1012 | 1.21 (0.65-2.25) | .55 |
| Mucinous carcinoma | 1417 | 0.99 (0.58-1.66) | .96 |
| Endometrioid carcinoma | 2810 | 0.87 (0.55-1.39) | .57 |
| Clear cell carcinoma | 1366 | 1.05 (0.61-1.83) | .86 |
| LDL Cholesterol | |||
| Invasive epithelial ovarian cancer | 22 406 | 0.98 (0.91-1.05) | .55 |
| High-grade serous carcinoma | 13 037 | 1.00 (0.92-1.09) | .99 |
| Low-grade serous carcinoma | 1012 | 1.05 (0.85-1.29) | .68 |
| Mucinous carcinoma | 1417 | 0.80 (0.65-0.98) | .03 |
| Endometrioid carcinoma | 2810 | 0.92 (0.78-1.10) | .36 |
| Clear cell carcinoma | 1366 | 1.01 (0.85-1.21) | .90 |
The exponential change in odds of invasive epithelial ovarian cancer per genetically proxied inhibition of drug target equivalent to a 1-mmol/L (38.7-mg/dL) decrease in low-density lipoprotein (LDL) cholesterol or the exponential change in odds of invasive epithelial ovarian cancer per genetically proxied 1-mmol/L decrease in LDL cholesterol. Invasive histotypes classified as “other” by Ovarian Cancer Association Consortium (n = 2764) were included in analyses for invasive epithelial ovarian cancer but were not assessed separately. In histotype-stratified analyses, there was no statistically significant evidence of heterogeneity across the 5 histotypes assessed (P value for heterogeneity = .84; I2 = 0.00%). The P value for heterogeneity was calculated from a Cochran Q statistic where Q is distributed as a χ2 statistic with K (number of histotypes) – 1 degrees of freedom.
Figure 2. Mendelian Randomization Estimates of the Association Between 3-Hydroxy-3-Methylglutaryl Coenzyme A (HMG-CoA) Reductase, Niemann-Pick C1-Like 1 (NPC1L1), and Proprotein Convertase Subtilisin/Kexin Type 9 (PCSK9) With Invasive Epithelial Ovarian Cancer Among Women in the General Population.
Figure 3. Mendelian Randomization Estimates of the Association Between 3-Hydroxy-3-Methylglutaryl Coenzyme A (HMG-CoA) Reductase, Niemann-Pick C1-Like 1 (NPC1L1), and Proprotein Convertase Subtilisin/Kexin Type 9 (PCSK9) With Epithelial Ovarian Cancer Among BRCA1/2 Mutation Carriers.
Secondary Analyses
Genetically proxied inhibition of NPC1L1 (OR, 0.97 [95% CI, 0.53-1.75]; P = .91) and PCSK9 (OR, 0.98 [95% CI, 0.85-1.13]; P = .80) were not significantly associated with overall or histotype-stratified ovarian cancer in the OCAC sample (Table 2 and Figure 2). Although genetically proxied circulating LDL cholesterol was not significantly associated with overall ovarian cancer (per 1-mmol/L reduction in LDL cholesterol: OR, 0.98 [95% CI, 0.91-1.05]; P = .55), a 1-mmol/L reduction in genetically proxied LDL cholesterol was significantly associated with invasive mucinous ovarian cancer (OR, 0.80 [95% CI, 0.65-0.98]; P = .03) in histotype-stratified analyses. Findings for associations of genetically proxied LDL cholesterol with overall and histotype-specific ovarian cancer were largely consistent in sensitivity analyses examining horizontal pleiotropy (eTable 4 in the Supplement). In analyses among BRCA1/2 mutation carriers in CIMBA, there were no significant associations of genetically proxied inhibition of NPC1L1 (HR, 0.95 [95% CI, 0.56-1.61]; P = .84), PCSK9 (HR, 0.92 [95% CI, 0.78-1.08]; P = .31), or genetically proxied circulating LDL cholesterol levels (HR, 0.96 [95% CI, 0.90-1.03]; P = .24) with epithelial ovarian cancer HR.
Sensitivity Analyses
In sensitivity analyses applying a Bonferroni correction to account for multiple testing across overall invasive epithelial ovarian cancer analyses in OCAC (P < .01), the association of genetically proxied HMG-CoA reductase inhibition with invasive epithelial ovarian cancer (P = .002) remained significant. When correcting for multiple tests performed across analyses examining epithelial ovarian cancer among BRCA1/2 mutation carriers in CIMBA (P < .01), the association of genetically proxied HMG-CoA reductase inhibition with epithelial ovarian cancer (P = .01) was no longer significant. Likewise, when applying a Bonferroni correction to multiple tests performed across invasive epithelial ovarian cancer histotypes in OCAC (P < .003), the associations of genetically proxied HMG-CoA reductase inhibition with endometrioid carcinoma (P = .01) and genetically proxied LDL cholesterol levels with invasive mucinous ovarian cancer (P = .03) were no longer significant.
Tests of Mendelian Randomization Assumptions
As a test for the exchangeability mendelian randomization assumption, co-localization analysis found evidence for co-localization of 1 variant (rs7703051) within HMGCR (co-localization posterior probability = 0.014) and generated a 95% credible set that incorporated 2 additional variants in perfect or near-perfect linkage disequilibrium with this variant (rs11749783 [r2 = 1.00] and rs3846663 [r2 = 0.98]) (eTable 5 in the Supplement). This top co-localized variant (rs7703051) was also in strong linkage disequilibrium with rs12916 (r2 = 0.92). Analyses examining associations of the top co-localized variant were consistent with primary analyses for ovarian cancer in OCAC (OR, 0.58 [95% CI, 41-0.84]) and in BRCA1/2 mutation carriers in CIMBA (HR, 0.69 [95% CI, 0.50-0.95]).
When testing associations of the genetic instrument for HMG-CoA reductase with ovarian cancer risk factors (examination of the exclusion restriction mendelian randomization assumption), there was evidence for an association of this instrument with age at menarche (change in age at onset per 1-mmol/L reduction in LDL cholesterol, −0.18 [95% CI, −0.33 to −0.03l]; P = .02) and body mass index (change in body mass index per 1-mmol/L reduction in LDL cholesterol, 0.20 [95% CI, 0.06-0.34]; P = .006) (eTable 6 in the Supplement). Multivariable mendelian randomization analyses adjusting for the association of these factors with epithelial ovarian cancer were consistent with results obtained in primary analyses among women in OCAC and in BRCA1/2 mutation carriers in CIMBA (eTable 7 in the Supplement). Additionally, results of analyses that iteratively removed 1 SNP at a time from the instrument and recalculated the overall mendelian randomization estimate were consistent, suggesting that associations were not being driven through individual influential SNPs (eTable 8 in the Supplement).
Discussion
In this mendelian randomization analysis of 22 406 women with invasive epithelial ovarian cancer and 40 941 control individuals, lifelong naturally randomized genetically proxied inhibition of HMG-CoA reductase was significantly associated with lower odds of ovarian cancer. Similar associations were observed when analyses were restricted to BRCA1/2 mutation carriers. There were no significant associations of genetically proxied NPC1L1 or PCSK9 inhibition or LDL cholesterol levels with epithelial ovarian cancer in the general population or among BRCA1/2 mutation carriers, supporting a possible mechanism-specific effect of HMG-CoA reductase inhibition.
Findings for HMG-CoA reductase inhibition and epithelial ovarian cancer are consistent with results from laboratory and most epidemiological studies. Statins have been shown to have antiproliferative and anti-invasive properties in cell lines and animal models of ovarian cancer.4,5,6 A meta-analysis of 5 observational studies with 624 ovarian cancer cases reported that statin use was significantly associated with a lower risk of ovarian cancer (relative risk, 0.79 [95% CI, 0.64-0.98]) with no significant evidence of heterogeneity across studies (P value for heterogeneity = 0.67; I2 = 0.0%).8 Two subsequent case-control studies yielded conflicting findings; a Danish registry-based analysis of 4103 individuals with epithelial ovarian cancer and 58 706 control individuals reported no significant evidence of an association between ever use of statins and cancer risk (OR, 0.98 [95% CI, 0.87-1.10]), but an analysis of the New England Case Control study (2040 individuals with epithelial ovarian cancer and 2100 control individuals) showed a significantly lower odds of ovarian cancer in women who self-reported statin use compared with nonusers (OR, 0.68 [95% CI, 0.54-0.85]).7,25
It has been hypothesized that the principal mechanism of a putative protective effect of statins on cancer is through prolonged lowering of circulating cholesterol levels which, in turn, modulates hyperplastic growth and neoplasia.26 The lack of association of genetically proxied inhibition of NPC1L1 and PCSK9 along with genetically proxied LDL cholesterol levels suggests that circulating cholesterol may not be driving the observed association of HMG-CoA reductase inhibition with ovarian cancer. The inhibition of mevalonate synthesis by HMG-CoA reductase inhibition in the cholesterol biosynthesis pathway has downstream consequences on various products in this pathway, including levels of steroid precursors, some of which have been implicated in ovarian cancer risk.27,28
The number of BRCA1 and BRCA2 mutation samples in the current study was too small to examine associations of pharmacological targets and LDL cholesterol with epithelial ovarian cancer stratified across common histotypes in these analyses; however, previous studies have demonstrated that the majority of cases in these populations are high-grade serous carcinoma.29 Women with mutations in BRCA1 have an estimated lifetime risk of ovarian cancer of 44% and women with mutations in BRCA2 have an estimated lifetime risk of 17%.30,31 Risk-reducing salpingo-oophorectomy is recommended for ovarian cancer prevention in these high-risk groups. However, adverse events associated with early-onset surgical menopause, including increased cardiovascular, osteoporotic, and overall mortality risks and potential deleterious effects on quality of life, discourage approximately 30% of BRCA1/2 mutation carriers from having this procedure.31,32,33
Strengths of this analysis include the use of genetic variants within genes that encode drug targets to proxy the potential effect of commonly prescribed LDL cholesterol–lowering therapies, which should minimize confounding and avoids reverse causation bias; the use of summary genetic association data, which permitted the exploitation of relatively precise estimates of SNP exposure and SNP outcome associations from several large GWAS meta-analyses of lipids and ovarian cancer, allowing statistical power and precision of analyses to be increased; and the combination of multiple variants into genetic proxies, allowing the proportion of variance in drug targets explained by instruments to be increased.
Co-localization was used as a sensitivity analysis to test whether the association of genetically proxied HMG-CoA reductase with ovarian cancer reflected both of these traits being influenced by 1 or more shared causal variants or whether each trait is influenced by distinct causal variants that are in linkage disequilibrium with each other (ie, genetic confounding). Findings from co-localization analysis suggested that both HMG-CoA reductase and ovarian cancer shared a causal variant within the HMGCR locus, providing some evidence against genetic confounding accounting for the association between these traits. Multivariable mendelian randomization was used to test whether the association of HMG-CoA reductase inhibition with ovarian cancer was biased through horizontal pleiotropy by adjusting models for genetically proxied risk factors that have previously been linked to ovarian cancer. The consistency of associations of HMG-CoA reductase inhibition with ovarian cancer in models with and without adjustment for previously reported ovarian cancer risk factors suggested that findings were unlikely to be biased by horizontal pleiotropy through these risk factors. Therefore, results from sensitivity analyses that tested for potential confounding (co-localization) and pleiotropic effects (multivariable mendelian randomization) suggest that the findings of this study are compatible with a causal relationship between HMG-CoA reductase inhibition and ovarian cancer. However, the mendelian randomization approach used in this analysis cannot establish whether modulation of HMG-CoA reductase via drug treatment would reduce risk of ovarian cancer; establishing causality would require a randomized clinical trial.
Extension of the analyses presented in this study to a survival framework among women diagnosed with ovarian cancer could inform on the potential treatment efficacy of statins for ovarian cancer progression and survival. In addition, the design of randomized clinical trials within high-risk populations (BRCA1/2 mutation carriers and/or women at high polygenic risk) would be necessary to provide evidence for a role of statin use in ovarian cancer prevention in these groups.
Limitations
This study has several limitations. First, direct measures of drug targets to generate estimates of ovarian cancer risk reductions per unit change in function of these targets were not available. Consequently, estimates were scaled to represent reductions in LDL cholesterol levels under the assumption that drug target inhibition is proportional to LDL cholesterol lowering. Second, effect estimates for associations of SNPs with drug targets and LDL cholesterol along with estimates of genetic instrument strength for these risk factors within ovarian cancer data sets were derived from analyses performed in an independent GWAS of lipid levels in up to 196 475 individuals primarily of European ancestry. Effect estimates and measures of instrument strength can be transported across data sets under the condition that all data sets are representative of the same underlying population (the basis of the 2-sample mendelian randomization approach used in this analysis). Third, statistical power was limited to detect associations with less common invasive histotypes (low-grade serous, mucinous, and clear cell carcinomas), although there was no statistically significant evidence for heterogeneity of associations of drug targets across all subtypes assessed. Fourth, the analyses presented assume no interaction of the association of genetic proxies for drug targets and ovarian cancer (eg, gene-environment, gene-gene) and linear associations of drug target inhibition with cancer risk. Fifth, while these analyses did not account for previously reported associations of genetically proxied LDL cholesterol with lipid-lowering drug use in this analysis, such correction would be expected to strengthen, rather than attenuate, findings presented in this study. Sixth, the Bonferroni correction applied to multiple tests performed in sensitivity analyses suggests that some findings may represent false-positive findings. Seventh, if statin treatment were to lower the risk of ovarian cancer, the magnitude of risk lowering achieved through taking statins may not correspond to the effect size observed in this analysis. This is because mendelian randomization estimates represent the long-term modulation of drug targets on disease risk and therefore may suggest larger risk reductions per unit change in drug target compared with those obtained from drug administration over a relatively shorter duration. Eighth, findings among BRCA1/2 mutation carriers cannot be considered a replication of findings among women in the general population because of important clinical, histopathological, and molecular differences between hereditary and sporadic ovarian cancers.34 Ninth, valid causal inference in mendelian randomization analyses relies on several assumptions, some of which are not verifiable (ie, exchangeability and exclusion restriction assumptions). While the findings of this study were consistent in various sensitivity analyses testing these assumptions, the possibility that these findings were biased through confounding and/or horizontal pleiotropy cannot be definitively ruled out.
Conclusions
Genetically proxied inhibition of HMG-CoA reductase was significantly associated with lower odds of epithelial ovarian cancer. However, these findings do not indicate risk reduction from medications that inhibit HMG-CoA reductase; further research is needed to understand whether there is a similar association with such medications.
eMaterials
References
- 1.Torre LA, Trabert B, DeSantis CE, et al. Ovarian cancer statistics, 2018. CA Cancer J Clin. 2018;68(4):284-296. doi: 10.3322/caac.21456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Henderson JT, Webber EM, Sawaya GF. Screening for ovarian cancer: updated evidence report and systematic review for the US Preventive Services Task Force. JAMA. 2018;319(6):595-606. doi: 10.1001/jama.2017.21421 [DOI] [PubMed] [Google Scholar]
- 3.Boudreau DM, Yu O, Johnson J. Statin use and cancer risk: a comprehensive review. Expert Opin Drug Saf. 2010;9(4):603-621. doi: 10.1517/14740331003662620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Greenaway JB, Virtanen C, Osz K, et al. Ovarian tumour growth is characterized by mevalonate pathway gene signature in an orthotopic, syngeneic model of epithelial ovarian cancer. Oncotarget. 2016;7(30):47343-47365. doi: 10.18632/oncotarget.10121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kobayashi Y, Kashima H, Wu RC, et al. Mevalonate pathway antagonist suppresses formation of serous tubal intraepithelial carcinoma and ovarian carcinoma in mouse models. Clin Cancer Res. 2015;21(20):4652-4662. doi: 10.1158/1078-0432.CCR-14-3368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martirosyan A, Clendening JW, Goard CA, Penn LZ. Lovastatin induces apoptosis of ovarian cancer cells and synergizes with doxorubicin: potential therapeutic relevance. BMC Cancer. 2010;10:103. doi: 10.1186/1471-2407-10-103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Akinwunmi B, Vitonis AF, Titus L, Terry KL, Cramer DW. Statin therapy and association with ovarian cancer risk in the New England Case Control (NEC) study. Int J Cancer. 2019;144(5):991-1000. doi: 10.1002/ijc.31758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Y, Qin A, Li T, Qin X, Li S. Effect of statin on risk of gynecologic cancers: a meta-analysis of observational studies and randomized controlled trials. Gynecol Oncol. 2014;133(3):647-655. doi: 10.1016/j.ygyno.2014.04.007 [DOI] [PubMed] [Google Scholar]
- 9.Yarmolinsky J, Wade KH, Richmond RC, et al. Causal inference in cancer epidemiology: what is the role of mendelian randomization? Cancer Epidemiol Biomarkers Prev. 2018;27(9):995-1010. doi: 10.1158/1055-9965.EPI-17-1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Phelan CM, Kuchenbaecker KB, Tyrer JP, et al. ; AOCS study group; EMBRACE Study; GEMO Study Collaborators; HEBON Study; KConFab Investigators; OPAL study group . Identification of 12 new susceptibility loci for different histotypes of epithelial ovarian cancer. Nat Genet. 2017;49(5):680-691. doi: 10.1038/ng.3826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Willer CJ, Schmidt EM, Sengupta S, et al. ; Global Lipids Genetics Consortium . Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013;45(11):1274-1283. doi: 10.1038/ng.2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burgess S, Zuber V, Valdes-Marquez E, Sun BB, Hopewell JC. Mendelian randomization with fine-mapped genetic data: choosing from large numbers of correlated instrumental variables. Genet Epidemiol. 2017;41(8):714-725. doi: 10.1002/gepi.22077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burgess S, Thompson SG; CRP CHD Genetics Collaboration . Avoiding bias from weak instruments in mendelian randomization studies. Int J Epidemiol. 2011;40(3):755-764. doi: 10.1093/ije/dyr036 [DOI] [PubMed] [Google Scholar]
- 14.Hormozdiari F, van de Bunt M, Segrè AV, et al. Colocalization of GWAS and eQTL signals detects target genes. Am J Hum Genet. 2016;99(6):1245-1260. doi: 10.1016/j.ajhg.2016.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Day FR, Ruth KS, Thompson DJ, et al. ; PRACTICAL consortium; kConFab Investigators; AOCS Investigators; Generation Scotland; EPIC-InterAct Consortium; LifeLines Cohort Study . Large-scale genomic analyses link reproductive aging to hypothalamic signaling, breast cancer susceptibility and BRCA1-mediated DNA repair. Nat Genet. 2015;47(11):1294-1303. doi: 10.1038/ng.3412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Locke AE, Kahali B, Berndt SI, et al. ; LifeLines Cohort Study; ADIPOGen Consortium; AGEN-BMI Working Group; CARDIOGRAMplusC4D Consortium; CKDGen Consortium; GLGC; ICBP; MAGIC Investigators; MuTHER Consortium; MIGen Consortium; PAGE Consortium; ReproGen Consortium; GENIE Consortium; International Endogene Consortium . Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518(7538):197-206. doi: 10.1038/nature14177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perry JR, Day F, Elks CE, et al. ; Australian Ovarian Cancer Study; GENICA Network; kConFab; LifeLines Cohort Study; InterAct Consortium; Early Growth Genetics (EGG) Consortium . Parent-of-origin-specific allelic associations among 106 genomic loci for age at menarche. Nature. 2014;514(7520):92-97. doi: 10.1038/nature13545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tobacco GC; Tobacco and Genetics Consortium . Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat Genet. 2010;42(5):441-447. doi: 10.1038/ng.571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.GWAS results. UK Biobank website. http://www.nealelab.is/uk-biobank/. Updated August 1, 2018. Accessed 15 February, 2019.
- 20.Burgess S, Thompson SG. Multivariable mendelian randomization: the use of pleiotropic genetic variants to estimate causal effects. Am J Epidemiol. 2015;181(4):251-260. doi: 10.1093/aje/kwu283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44(2):512-525. doi: 10.1093/ije/dyv080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent estimation in mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. 2016;40(4):304-314. doi: 10.1002/gepi.21965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hartwig FP, Davey Smith G, Bowden J. Robust inference in summary data mendelian randomization via the zero modal pleiotropy assumption. Int J Epidemiol. 2017;46(6):1985-1998. doi: 10.1093/ije/dyx102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327(7414):557-560. doi: 10.1136/bmj.327.7414.557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baandrup L, Dehlendorff C, Friis S, Olsen JH, Kjær SK. Statin use and risk for ovarian cancer: a Danish nationwide case-control study. Br J Cancer. 2015;112(1):157-161. doi: 10.1038/bjc.2014.574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Solomon KR, Freeman MR. Do the cholesterol-lowering properties of statins affect cancer risk? Trends Endocrinol Metab. 2008;19(4):113-121. doi: 10.1016/j.tem.2007.12.004 [DOI] [PubMed] [Google Scholar]
- 27.Brown MS, Goldstein JL. Multivalent feedback regulation of HMG CoA reductase, a control mechanism coordinating isoprenoid synthesis and cell growth. J Lipid Res. 1980;21(5):505-517. [PubMed] [Google Scholar]
- 28.Ose J, Poole EM, Schock H, et al. ; Androgens Are Differentially Associated with Ovarian Cancer Subtypes in the Ovarian Cancer Cohort Consortium . Androgens are differentially associated with ovarian cancer subtypes in the ovarian cancer cohort consortium. Cancer Res. 2017;77(14):3951-3960. doi: 10.1158/0008-5472.CAN-16-3322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gilks CB, Prat J. Ovarian carcinoma pathology and genetics: recent advances. Hum Pathol. 2009;40(9):1213-1223. doi: 10.1016/j.humpath.2009.04.017 [DOI] [PubMed] [Google Scholar]
- 30.Kuchenbaecker KB, Hopper JL, Barnes DR, et al. ; BRCA1 and BRCA2 Cohort Consortium . Risks of breast, ovarian, and contralateral breast cancer for BRCA1 and BRCA2 mutation carriers. JAMA. 2017;317(23):2402-2416. doi: 10.1001/jama.2017.7112 [DOI] [PubMed] [Google Scholar]
- 31.Temkin SM, Bergstrom J, Samimi G, Minasian L. Ovarian cancer prevention in high-risk women. Clin Obstet Gynecol. 2017;60(4):738-757. doi: 10.1097/GRF.0000000000000318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parker WH, Broder MS, Chang E, et al. Ovarian conservation at the time of hysterectomy and long-term health outcomes in the nurses’ health study. Obstet Gynecol. 2009;113(5):1027-1037. doi: 10.1097/AOG.0b013e3181a11c64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parker WH, Feskanich D, Broder MS, et al. Long-term mortality associated with oophorectomy compared with ovarian conservation in the nurses’ health study. Obstet Gynecol. 2013;121(4):709-716. doi: 10.1097/AOG.0b013e3182864350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zweemer RP, Verheijen RH, Menko FH, et al. Differences between hereditary and sporadic ovarian cancer. Eur J Obstet Gynecol Reprod Biol. 1999;82(2):151-153. doi: 10.1016/S0301-2115(98)00218-8 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eMaterials