Abstract
Currently, there are still no early biomarkers to detect infants with risk of autism spectrum disorder (ASD), which is mainly diagnosed based on behavioral observations at three or four years of age. Since intervention efforts may miss a critical developmental window after 2 years old, it is clinically significant to identify imaging-based biomarkers at an early stage for better intervention, before behavioral diagnostic signs of ASD typically arising. Previous studies on older children and young adults with ASD demonstrate altered developmental trajectories of the amygdala and hippocampus. However, our knowledge on their developmental trajectories in early postnatal stages remains very limited. In this paper, for the first time, we propose a volume-based analysis of the amygdala and hippocampal subfields of the infant subjects with risk of ASD at 6, 12, and 24 months of age. To address the challenge of low tissue contrast and small structural size of infant amygdala and hippocampal subfields, we propose a novel deep-learning approach, dilated-dense U-Net, to digitally segment the amygdala and hippocampal subfields in a longitudinal dataset, the National Database for Autism Research (NDAR). A volume-based analysis is then performed based on the segmentation results. Our study shows that the overgrowth of amygdala and cornu ammonis sectors (CA) 1–3 May start from 6 months of age, which may be related to the emergence of autistic spectrum disorder.
Keywords: Autism, Convolutional neural network, Trajectory, Amygdala, Hippocampus
1. Introduction
Autism, or autism spectrum disorder (ASD), refers to a range of conditions characterized by challenges with social skills, repetitive behaviors, speech, and nonverbal communication, as well as by unique strengths and differences. Globally, autism is estimated to affect 70 million people as of 20171. The diagnosis of ASD is mainly based on a thorough behavioral assessment. Studies demonstrate that behavioral signs can begin to emerge as early as 6 to 12 months [2]. However, most professionals who specialize in diagnosing the disorder won’t attempt to make a definite diagnosis until 2 or 3 years of age [3]. As a result, the time window of opportunity for effective intervention may have passed when the disorder is detected. Thus, it is of great importance to detect ASD earlier in life for better intervention.
Magnetic resonance (MR) examination allows researchers and clinicians to noninvasively examine brain anatomy. Structural MR examination is widely used to investigate brain morphology and plays an increasingly pivotal role in early diagnosis and intervention of ASD because of its high contrast sensitivity and spatial resolution [4]. Many neuroscience studies on older children and young adults with ASD demonstrate abnormalities in the amygdala [5, 6] and hippocampus [7, 8]. For example, some studies have reported increased amygdala and hippocampal volumes [9–11]. However, most of previous studies are based on cross-sectional subjects larger than 2 years of age. Hence, our knowledge on the volumetric growth of autistics in early postnatal stages remains very limited. Moreover, the studies on hippocampal subfields, i.e., the subiculum, the cornu ammonis sectors (CA) 1–3, and the dentate gyrus (DG) [8], are rare at early stages. In fact, each subfield has different functions. For example, CA3 and DG are involved in memory encoding and early retrieval, while CA1 is involved in late retrieval, consolidation, and recognition [12].
Therefore, longitudinal studies of the amygdala and hippocampal subfields development could identify critical periods of abnormal trajectory as a first step towards establishing neurobiological factors responsible for the autism. To characterize trajectories of the amygdala and hippocampal subfields at early stages, i.e., 6-, 12-, and 24-months of age, it is critically important to accurately segment them from MR images. Manual segmentation is often treated as a gold standard, but it is time-consuming and tedious, along with large inter- and intra-observer variability. In recent years, deep neural networks have been widely applied in medical image segmentation. Fully convolutional networks (FCNs) [13], as a natural extension of convolutional neural networks (CNNs), were developed for semantic segmentation of natural images and have been rapidly applied to biomedical images due to their powerful end-to-end training. 3D U-Net [14] extends the FCNs for volumetric segmentation by using skip connections to capture both the local and contextual information. To date, many network architectures further incorporate the residual connections [15] or dense connections [16] to get efficient improved flow of information and gradients throughout the network [17, 18]. A new convolutional network module, which is specifically designed for dense prediction, was proposed in [19]. The module uses dilated convolutions to systematically aggregate multiscale contextual information without losing resolution. Inspired by [20], in this paper, we propose a Dilated-Dense U-Net for accurate segmentation of amygdala and hippocampal subfields from around 6-, 12-, and 24-month-old infant brain MRI. Based on the segmentation results, for the first time, our study reveals that the amygdala and CA1–3 start to overgrow from 6 months of age, which may be related to the emergence of ASD.
2. Materials and Methods
Data Description.
Totally 276 subjects gathered from the National Database for Autism Research (NDAR) [21] were used in the study. More specifically, the dataset consists of 30 autistic subjects (25 males/5 females), 31 mild condition autism spectrum subjects (21 males/10 females), and 215 normal controls (133 males/82 females). In the experiment, we regard the first two types as one group. All images were acquired on a Siemens 3T scanner. T1-weighted MR images were acquired with 160 sagittal slices using parameters: TR/TE = 2400/3.16 ms and voxel resolution=1×1×1mm3. Then, in-house tools were used to perform skull stripping, intensity inhomogeneity correction, and histogram matching for MR images. There are 12, 13, and 15 MR images acquired at 6, 12, and 24 months, which were manually labeled by an experienced neuroscientist. These subjects were randomly selected, and, during manual annotations, diagnosis information is unknown to the neuroscientist. The longitudinal analyses were conducted on a subset of 29 ASD subjects and 113 normal control (NC) subjects, who have all the three longitudinal scans acquired at 6, 12 and 24 months of age. The age of participant did not differ significantly (p-value > 0.05) between ASD and NC group at each time point.
Dilated-Dense U-Net:
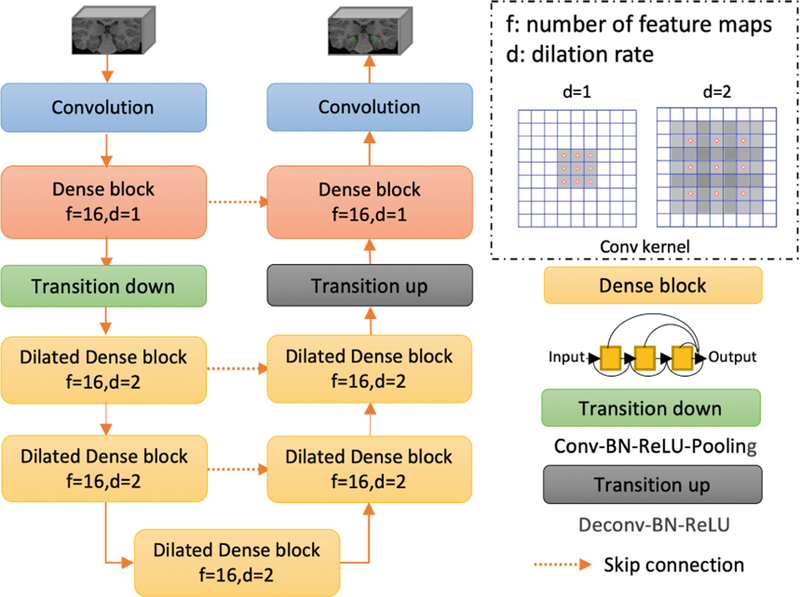
In this study, a novel network architecture, Dilated-Dense U-Net (DDUNET), is proposed to segment the amygdala and hippocampal subfields. Inspired by [20], the proposed network is a fully convolutional neural network taking advantages of the U-Net skip connections, dense blocks, and dilated convolutions. The U-Net skip connections facilitate the joint capturing of both the local and contextual information, while the dilated dense blocks bring a better flow of the gradient information without losing resolution. As the structure size of the amygdala and hippocampal subfields is very small, some details may be missed in max-pooling layer. Thus, dilated convolutions are used to support exponential expansion of the receptive field without loss of resolution or coverage.
The proposed network architecture is shown in Fig. 1. It consists of a contracting path and an expansive path going through 7 dense blocks. Each path consists of one standard dense block and two dilated dense blocks. After the first standard dense block in the contracting path, a max-pooling layer of size 2 × 2 × 2 is used to reduce the dimensionality and exploit the contextual information. Each dense block consists of three BN-ReLU-Conv-Dropout operations, in which each Conv includes 16 3 × 3 × 3 kernels and the dropout rate was 0.1during training. It should be noted that two dilated dense blocks with dilation rates d = 2 are used in the downsampling path to expand receptive fields. Then, a dense block with a dilation rate d = 2 is used to transfer the features from the contracting path to the expansive path. Skip connections are established between the contracting path and the expansive path to recover the spatial information lost during the downsampling.
Fig. 1.
DDUNET consists of two paths: a contracting path to capture contextual information and an expansive path to capture local information. There are seven dense blocks in total: two standard dense blocks (i.e., d = 1), and five dilated dense blocks (i.e., d = 2).
Network Implementation.
Due to large appearance differences in MRI between 6, 12, and 24 months, we trained three individual models for each age period. During training, we randomly extracted 3D patches from training images. The patch size was optimized as 16 × 64 × 16 based on cross-validation, by using cross-entropy as a loss function. We used SGD optimization strategy. The initial learning rate was 0.005, which was multiplied by 0.1 after each epoch. Training and test were performed on a NVIDIA Titan X GPU. Training a DDUNET takes around 72 h, and, in the application stage, segmenting a 3D image takes 60 s.
3. Experiments and Results
In this section, we present our segmentation results of the amygdala and hippocampal subfields using the proposed Dilated-Dense U-Net, by comparison with 3 state-of-the-art methods. Based on the segmentation results, we further measured the volumetric differences of the amygdala and hippocampal subfields between ASD group and NC group.
3.1. Segmentation Results and Performance
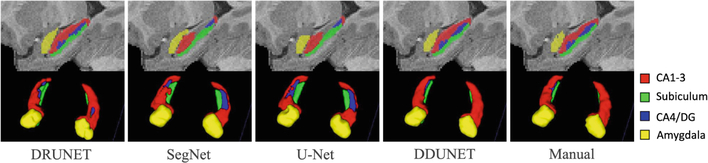
Leave-one-out cross-validation was used in the experiment, e.g., for 24 months, in each fold, we selected 12 subjects for training, 2 subjects for validation, and 1 subject for testing. Figure 2 shows the 2D and 3D views of segmented amygdala and hippocampal subfields for one randomly-selected subject acquired at 24 months old by different methods, including DRUNET [18], SegNet [22], U-Net [14], our proposed DDUNET, and manual segmentation. It can be seen that the comparison methods cannot accurately identify these ROIs, especially for CA1–3. By contrast, our method has achieved a consistent result with the manual result. Table 1 reports the Dice coefficients (mean ± std) of the segmentation results obtained by different networks. Our proposed method achieves the highest Dice coefficients for all age periods. We further calculate p-values between proposed results and any comparison results, and mark the significantly better performance in bold (p-value < 0.05). For most cases, our proposed method can achieve a significantly better result. We then apply our trained model to the remaining subjects and further perform ROI-based volumetric measurements.
Fig. 2.
Segmented amygdala and hippocampal subfields from one randomly-selected subject acquired at 24 months of age, by four different networks and manual segmentation.
Table 1.
Comparison between different networks in term of Dice coefficients (mean ± std) for each segmented structure. The best accuracy is shown in bold with p-value < 0.05.
| Age | Amygdala | CA1–3 | Subiculum | CA4/DG | |
|---|---|---|---|---|---|
| DRUNET | 6 months | 0.754 ± 0.013 | 0.728 ± 0.007 | 0.652 ± 0.020 | 0.685 ± 0.019 |
| 12 months | 0.804 ± 0.016 | 0.810 ± 0.009 | 0.825 ± 0.017 | 0.800 ± 0.015 | |
| 24 months | 0.846 ± 0.008 | 0.827 ± 0.008 | 0.842 ± 0.012 | 0.801 ± 0.002 | |
| SegNet | 6 months | 0.695 ± 0.015 | 0.688 ± 0.014 | 0.646 ± 0.018 | 0.638 ± 0.007 |
| 12 months | 0.792 ± 0.012 | 0.762 ± 0.012 | 0.692 ± 0.016 | 0.690 ± 0.004 | |
| 24 months | 0.834 ± 0.017 | 0.764 ± 0.021 | 0.705 ± 0.016 | 0.695 ± 0.006 | |
| U-Net | 6 months | 0.687 ± 0.006 | 0.624 ± 0.003 | 0.621 ± 0.006 | 0.600 ± 0.010 |
| 12 months | 0.754 ± 0.008 | 0.707 ± 0.004 | 0.679 ± 0.009 | 0.637 ± 0.012 | |
| 24 months | 0.795 ± 0.006 | 0.720 ± 0.010 | 0.695 ± 0.018 | 0.674 ± 0.008 | |
| DDUNET | 6 months | 0.882 ± 0.007 | 0.863 ± 0.009 | 0.832 ± 0.005 | 0.809 ± 0.010 |
| 12 months | 0.898 ± 0.010 | 0.878 ± 0.009 | 0.846 ± 0.009 | 0.820 ± 0.006 | |
| 24 months | 0.909 ± 0.014 | 0.880 ± 0.016 | 0.854 ± 0.006 | 0.815 ± 0.010 |
3.2. Volumetric Measures and Discussion
Cross-Sectional Studies:
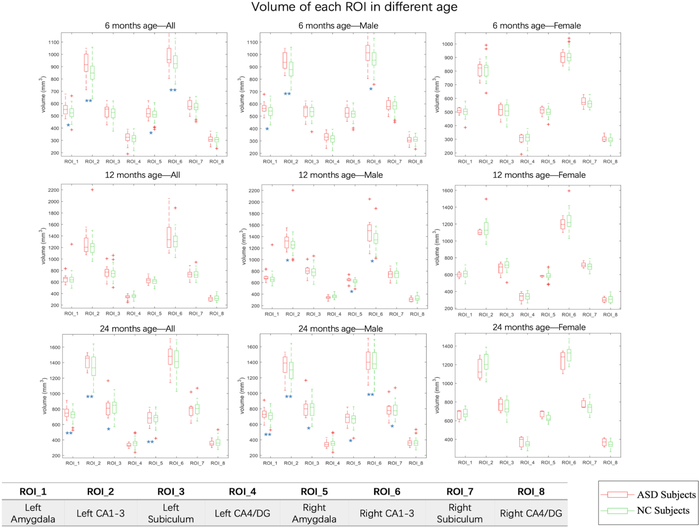
The volumetric measurements for all 276 subjects are provided in Fig. 3. At 6 months of age, compared with NC group, ASD group has significant enlargements (p < 0.05) in amygdala in both left and right hemispheres (with 4.7% and 3.4% enlargement, respectively), and in CA1–3 in left and right hemispheres (with 5.8% and 3.3% enlargement, respectively). At 12 months of age, for male subjects, the CA1–3 in both left and right hemisphere show 4.4% and 3.3% enlargement, respectively. For female subjects, there is no significant difference between ASD group and NC group, which may be caused by limited female subjects in the ASD group. At 24 months of age, compared with the NC group, the ASD group shows significant enlargement in amygdala (both hemispheres, p-value < 0.01), CA1–3 (left hemisphere, p-value < 0.01), and subiculum (left hemisphere, p-value < 0.05).
Fig. 3.
The volumetric data for all participants at different age stages. Significance of the p-value of each ROI is shown at the corner of each plot figure, with * indicating p-value < 0.05 and ** indicating p-value < 0.01.
Longitudinal Study:
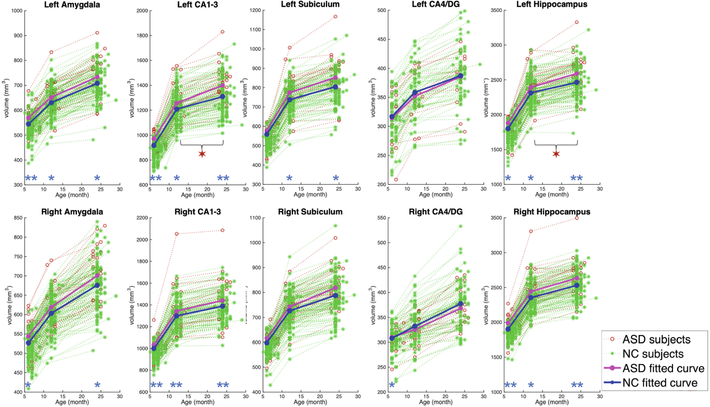
Figure 4 shows the longitudinal trajectory of each ROI volume from a subset of 29 ASD subjects and 113 NC subjects with all three longitudinal MR images acquired at around 6, 12, and 24 months of age. At 6 months of age, compared with the NC group, the ASD group shows significant enlargement in amygdala (both left and right hemispheres, p-value < 0.05), and hippocampus in CA1–3 (both left and right hemispheres, p-value < 0.01). At 12 months of age, compared with the NC group, the ASD group shows significant enlargement in amygdala (left hemisphere, p-value < 0.05), and hippocampus in CA1–3 (both left and right hemispheres, p-value < 0.05). At 24 months of age, the difference between ASD and NC groups becomes larger, and it is significantly different in the amygdala (both left and right hemispheres, p-value < 0.05), and CA1–3 (both left and right hemispheres, p-value < 0.01).
Fig. 4.
The growth trajectory of each ROI. Significance of the p-value of each ROI at different ages is indicated by * with p-value < 0.05, and by ** with p-value < 0.01. While, significance of the p-value of growth rate is indicated by ✶ with p-value < 0.05.
We further investigate the growth rates from 6 to 12 months of age and find that the growth rates of the two groups are not significantly different. However, in the second year, the average ROI volumetric growth rates of ASD group become larger than the NC group in CA1–3 (left hemisphere, p-value = 0.0101), and hippocampus (left hemisphere, p-value = 0.0365). We further normalized the ROI volumes with total brain volumes (ROI volume/total brain volume) and still find that the ASD group shows significant increase of growth rate in CA1–3 (left hemisphere, p-value = 0.0053), and hippocampus (left hemisphere, p-value = 0.0146).
The degree of amygdala enlargement at the early age is associated with the severity of social and communication and emotional perception [5]. The enlargement of CA1–3 may represent up-regulation, strengthen emotion of fear to communicate with surrounding or others. These findings suggest that there are developmental abnormalities in amygdala and hippocampus (especially for CA1–3) in the early age of ASD, which is also confirmed by previous reports on older children and young adults [5, 6].
In conclusion, for the first time, our study finds that the overgrowth of amygdala and CA1–3 in hippocampal subfields starts from 6 months of age, which may be linked to early emergence of ASD.
Acknowledgements.
Part of this work was done when Guannan Li was in UNC (supported in part by NIH grants MH109773). Gang Li and Dinggang Shen were supported in part by NIH grants (MH117943). Li Wang was supported by NIH grants MH109773 and MH117943.
Footnotes
References
- 1.Newschaffer CJ, et al. : The epidemiology of autism spectrum disorders. Dev. Disabil. Res. Rev 8, 151–161 (2002) [Google Scholar]
- 2.Filipek PA, et al. : The screening and diagnosis of autistic spectrum disorders. J. Autism Dev. Disord 29, 439–484 (1999) [DOI] [PubMed] [Google Scholar]
- 3.Baird G, et al. : Diagnosis of autism. BMJ Br. Med. J 327, 488–493 (2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen R, et al. : Structural MRI in autism spectrum disorder. Pediatr. Res 69, 63R (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Avino TA, et al. : Neuron numbers increase in the human amygdala from birth to adulthood, but not in autism. Proc. Natl. Acad. Sci U. S. A 115, 3710–3715 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aylward E, et al. : MRI volumes of amygdala and hippocampus in non–mentally retarded autistic adolescents and adults. Neurology 53, 2145 (1999) [DOI] [PubMed] [Google Scholar]
- 7.Schumann CM, et al. : The amygdala is enlarged in children but not adolescents with autism; the hippocampus is enlarged at all ages. J. Neurosci 24, 6392 (2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duvernoy HM: The Human Hippocampus: Functional Anatomy, Vascularization and Serial Sections with MRI Springer, Heidelberg: (2005). 10.1007/b138576 [DOI] [Google Scholar]
- 9.Howard MA, et al. : Convergent neuroanatomical and behavioural evidence of an amygdala hypothesis of autism. NeuroReport 11, 2931–2935 (2000) [DOI] [PubMed] [Google Scholar]
- 10.Sparks B, et al. : Brain structural abnormalities in young children with autism spectrum disorder. Neurology 59, 184–192 (2002) [DOI] [PubMed] [Google Scholar]
- 11.Avino TA, et al. : Neuron numbers increase in the human amygdala from birth to adulthood, but not in autism. Proc. Natl. Acad. Sci 115, 3710–3715 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mueller SG, et al. : Evidence for functional specialization of hippocampal subfields detected by MR subfield volumetry on high resolution images at 4 T. Neuroimage 56, 851–857 (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Long J, et al. : Fully convolutional networks for semantic segmentation. In: CVPR, pp. 3431–3440 (2016) [DOI] [PubMed] [Google Scholar]
- 14.Ronneberger O, Fischer P, Brox T: U-Net: convolutional networks for biomedical image segmentation. In: Navab N, Hornegger J, Wells, William M, Frangi, Alejandro F (eds.) MICCAI 2015, Part III. LNCS, vol. 9351, pp. 234–241. Springer, Cham: (2015). 10.1007/978-3-319-24574-4_28 [DOI] [Google Scholar]
- 15.He K, et al. : Deep residual learning for image recognition. In: CVPR, pp. 770–778 (2018) [Google Scholar]
- 16.Huang G, et al. : Densely connected convolutional networks. In: CVPR, p. 3 (2017) [Google Scholar]
- 17.Jégou S, et al. : The one hundred layers tiramisu: fully convolutional DenseNets for semantic segmentation. In: CVPRW, pp. 1175–1183 (2017) [Google Scholar]
- 18.Devalla SK, et al. : DRUNET: a dilated-residual U-Net deep learning network to digitally stain optic nerve head tissues in optical coherence tomography images. arXiv preprint: arXiv: 1803.00232 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu F, Koltun V: Multi-scale context aggregation by dilated convolutions. arXiv preprint arXiv:1511.07122 (2015) [Google Scholar]
- 20.Wang L, et al. : Volume-Based Analysis of 6-Month-Old Infant Brain MRI for Autism Biomarker Identification and Early Diagnosis In: Frangi Alejandro F., Schnabel Julia A., Davatzikos C, Alberola-López C, Fichtinger G (eds.) MICCAI 2018, Part III. LNCS, vol. 11072, pp. 411–419. Springer, Cham (2018). 10.1007/978-3-030-00931-1_47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Payakachat N, et al. : National Database for Autism Research (NDAR): big data opportunities for health services research and health technology assessment. PharmacoEconomics 34, 127–138 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Badrinarayanan V, et al. : SegNet: a deep convolutional encoder-decoder architecture for image segmentation. arXiv preprint: arXiv:1511.00561 (2015) [DOI] [PubMed] [Google Scholar]