Abstract
Despite their small numbers, cancer stem cells play a central role in driving cancer cell growth, chemotherapeutic resistance, and distal metastasis. Previous studies mainly focused on how DNA or histone modification determines cell fate in cancer. However, it is still largely unknown how RNA modifications orchestrate cancer cell fate decisions. More than 170 distinct RNA modifications have been identified in the RNA world, while only a few RNA base modifications have been found in mRNA. Growing evidence indicates that three mRNA modifications, inosine, 5-methylcytosine, and N6-methyladenosine, are essential for the regulation of spatiotemporal gene expression during cancer stem cell fate transition. Furthermore, transcriptome-wide mapping has found that the aberrant deposition of mRNA modification, which can disrupt the gene regulatory network and lead to uncontrollable cancer cell growth, is widespread across different cancers. In this review, we try to summarize the recent advances of these three mRNA modifications in maintaining the stemness of cancer stem cells and discuss the underlying molecular mechanisms, which will shed light on the development of novel therapeutic approaches for eradicating cancer stem cells.
Keywords: RNA modification, Cancer stem cells, 5-methylcytosine, N6-methyladenosine, A-to-I editing
Introduction
With the rapid development of high-throughput sequencing technologies, more than 170 types of post-transcriptional RNA modifications have been detected so far [1]. RNA modifications were first identified in non-coding RNA elements like tRNA and rRNA [2] and have been historically regarded as irreversible decorations on RNA bases. However, subsequent investigations showed that some RNA modifications are actually reversible [3, 4]. Moreover, emerging evidence demonstrates that these dynamic and reversible RNA modifications are widely present in various RNA molecules, not only non-coding RNA but also mRNA. The multitude of RNA modifications led to the birth of “RNA epigenetics” in 2010 [5] and the “Epitranscriptome” in 2012 [6], which are analogous to the concept of epigenetic modulation mediated by DNA or histone modifications.
Emerging RNA immunoprecipitation-sequencing methods have provided a detailed understanding of the genome-wide landscape of RNA modifications in human cells [7–12]. However, the majority of these modifications are mapped to tRNA and rRNA [13]. So far, only a few forms of RNA modifications have been identified in mRNA, such as N6-methyladenosine (m6A), N1-methyladenosine(m1A), Inosine (I), Pseudouridine (Ψ), 5-methylcytosine (m5C), 5-hydroxymethylcytidine (hm5C), N6,2′-O-dimethyladenosine (m6Am), 7-methylguanosine (m7G), and N4-acetylcytidine (ac4C). Despite the low frequency in the human genome, they affect almost every step of mRNA biogenesis and degradation. For example, mRNA modifications extensively modulate a vast pool of biochemical events surrounding mRNA metabolisms, such as mRNA splicing [8, 14], RNA folding [15, 16], stability [17–21], mRNA translation [22–24], and RNA transport [25, 26].
Growing evidence indicates that mRNA modifications display dramatic and dynamic variations during lineage commitment and cell reprogramming [27–29], suggesting their biological significance in the maintenance of cell identity. As oncogenic transformation frequently accompanies activation of pluripotency genes like NANOG, MYC, and Oct4 [30–32], it is likely that mRNA modifications also actively participate in modulating cancer cells’ fate through controlling these oncogenic factors. Consistently, subsequent studies have found that mRNA modifications are also essential for maintaining the stemness and malignancy of cancer stem cells [33–36].
The concept of cancer stem cells (CSCs) was proposed in the 1970s [37]. Analogous to stem cells in healthy tissue, CSCs possess stem-like properties, including the capacity for self-renewal and the ability to enhanced tumor initiation upon experimental transplantation [38]. It is proposed that the existence of this small but aggressive cell population possesses a high risk of drug resistance and tumor relapse [39, 40]. The CSC hypothesis posits that tumors mirror the hierarchy as normal tissues and that the CSCs are located at the apex of this hierarchical organization [41, 42]. With the elevated capacity of persistent proliferation, CSCs undergo asymmetric division, leading to complicated tumor heterogeneity and resistance to chemotherapy [43].
A recent breakthrough of the high-throughput sequencing platform has illustrated the detailed epigenetic landscape in CSCs [44]. The epigenetic modifications of DNA or histones are fundamental to the maintenance of cancer stem cell identity [45–48]. For instance, transformed cells which escape the senescence checkpoint, possess elevated levels of DNA methylation, leading to enhanced self-renewal and pro-survival signals [45].
However, as a novel modification form in the field of epigenetics, the function of mRNA modification in controlling the stemness of CSCs is still poorly understood. Currently, the three widespread mRNA modification forms are inosine, 5-methylcytosine (m5C), and N6-methyladenosine (m6A). In this review, we will provide an update of how these three mRNA modifications orchestrate regulatory gene networks within CSCs (Table 1). In addition, we will discuss their underlying molecular mechanisms and potential novel therapeutic strategies based on mRNA modification profiles.
Table 1.
A summary of mRNA modification and cancer stem cells
| Cancer cell types | RNA modification | Expression profiles in CSC | Molecular mechanisms | References |
|---|---|---|---|---|
| Leukemia | A-to-I | Increased | A-to-I editing induced alternative splicing of GSK3β, resulting in enhanced β-catenin expression | [49, 50] |
| Multiple myeloma | A-to-I | Increased | A-to-I editing occurred in the exon of GLI1 mRNA, leading to a novel GLI1 protein with a point mutation | [51] |
| Leukemia | A-to-I | Increased | A-to-I editing occurred in the 3’UTR of MDM2 mRNA and miR-155 would no longer bind to the edited 3’UTR region | [52] |
| Leukemia | A-to-I | Increased | A-to-I editing in let-7 precursor impaired let-7 biogenesis | [36] |
| Skin cancer | m5C | Decreased | NSUN2-deletion impaired protein synthesis | [53] |
| Breast cancer | m6A | Decreased | ALKBH5 reduced m6A level of NANOG, which stabilized NANOG mRNA | [33] |
| Glioblastoma | m6A | Decreased | Knockdown of METTL3 or METTL14 in CSCs increased the expression of ADAM19 and EPHA3 | [34] |
| Glioblastoma | m6A | Decreased | ALKBH5 demethylated FOXM1 mRNA transcripts and stabilized FOXM1 | [35] |
| Glioblastoma | m6A | Increased | SOX2 was a target for METTL3 and methylated SOX2 mRNA displayed prolonged stability | [54] |
| Leukemia | m6A | Decreased | Treatment with FTO inhibitor R-2HG induced the degradation of MYC/CEBPA mRNAs | [55] |
| Leukemia | m6A | Increased | METTL14 catalyzed the m6A modification in oncogenic factors MYC and MYB, increasing their mRNA stability | [56] |
A-to-I modification and cancer stem cells
In eukaryotes, adenosine-to-inosine (A-to-I) editing is one of the most prevalent RNA modifications. This process involves hydrolytic deamination of adenosine, catalyzed by the ADAR family members (ADAR1, ADAR2, and ADAR3) [57]. The newly generated inosine base is interpreted by the ribosome as guanosine during mRNA translation, leading to altered protein products, if the modification occurs in the protein-coding region [58]. Among the ADAR family members, ADAR1 and ADAR2 are ubiquitously expressed in eukaryotic cells while ADAR3 is highly expressed in brain cells [59]. Genetic ablation of ADAR1 in mice led to embryonic lethality, at embryonic day E12.5, due to severe global interferon response and defects in hematopoiesis [60, 61]. ADAR2-deficient mice were born at the normal Mendelian ratio and appeared to develop normally, but these mice died within 3 weeks after birth, during or soon after weaning [62]. These results suggest that ADAR1 and ADAR2 are indispensable for embryonic development and normal growth. According to a large-scale study including 6236 patient samples from 17 cancer types, A-to-I modifications display distinct distribution patterns in tumors and normal tissues [63]. However, most modifications existed in the non-coding regions of the mRNA [63]. Despite their prevalence, the functional consequences of these aberrant patterns in tumorigenesis remain elusive.
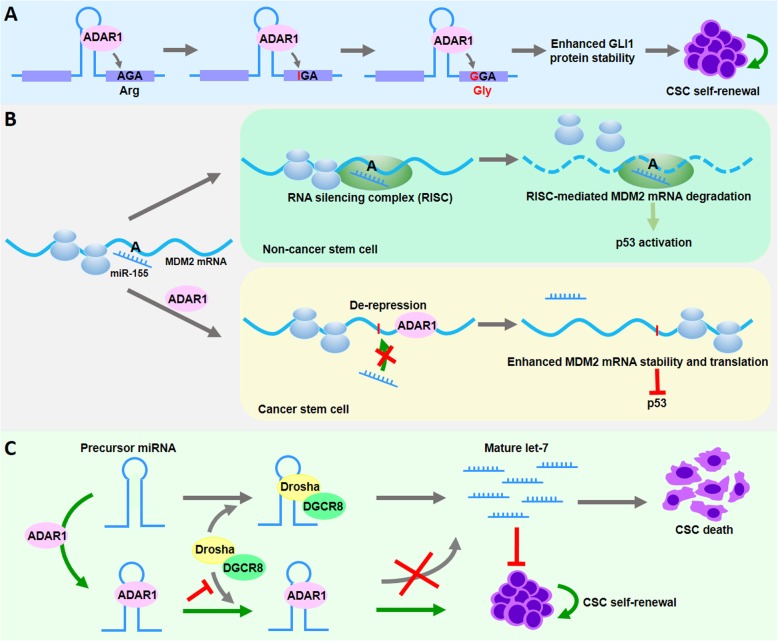
Previous studies demonstrated that A-to-I modifications could modulate the stemness of hematopoietic malignancies. For example, ectopic expression of ADAR1 potentiated malignant myeloid progenitor expansion through promoting alternative splicing of GSK3β, which enhanced the production of a misspliced form of GSK3β [49]. In vivo studies showed that leukemia progenitor cells harboring this misspliced GSK3β gene displayed enhanced β-catenin expression, which was required for the self-renewal of leukemia stem cells (LSCs) [50]. It was estimated that the genomic amplification of ADAR1 occurred in 30–50% of multiple myeloma patients and portended an unfavorable prognosis [51]. The silencing of ADAR1 attenuated in vivo engraftment of myeloma through suppression of the transcriptional activity of GLI1 [51], a self-renewal agonist and candidate marker for CSCs [64]. Further studies have shown that ADAR1 can edit exon 12 of the GLI1 transcript, leading to a novel GLI1 protein with a point mutation (Fig. 1a), which might stabilize the GLI1 protein by preventing the binding of a Hedgehog pathway negative regulator [51]. When comparing the A-to-I editing status, scientists revealed the elevated frequency of 3’UTR editing events in malignant progenitor cells. Interestingly, the majority of A-to-I events occurred in the 3’UTR of MDM2 RNA transcripts [52]. As an E3 ubiquitin ligase, MDM2 directly associates with and subsequently inactivates the transactivation domain of tumor suppressor p53. When the A-to-I editing occurred in the 3’UTR of MDM2 transcripts, miR-155 no longer bound to the edited 3’UTR (Fig. 1b), leading to the stabilization of MDM2 and inactivation of p53 [52].
Fig. 1.
Functional implication of ADAR1-mediated A-to-I editing in cancer stem cells. a A-to-I editing in the exon 12 of GLI1 transcript results in coding sequence change from Arg to Gly at position 701, which stabilizes GLI1 protein and enhances cancer stem cell renewal. b A-to-I editing at the 3′ UTR of MDM2 alters the interaction between MDM2 mRNA transcript and miR-155. c A-to-I RNA editing impairs the miRNA biogenesis of tumor suppressor let-7 through altering pre-miRNA secondary structures, leading to escape from let-7-mediated cancer cell death
In addition to mRNA, growing evidence demonstrates that ADAR1 also hinders the biogenesis of tumor-suppressive miRNAs, thereby driving leukemia stem cell self-renewal. Wild-type ADAR1, but not the editing-defective ADAR1E912 mutant, potentiates self-renewal gene expression and suppresses the biogenesis of stem cell inhibitory microRNA let-7 [36]. Subsequent studies found that A-to-G nucleotide changes altered RNA secondary structures at the Drosha/DGCR8 cleavage sites, leading to impaired let-7 miRNA biogenesis (Fig. 1c).
Recent studies have provided substantial new insights into how A-to-I modifications regulate RNA splicing. mRNA maturation involves serial processing steps which structurally alter the newly synthesized RNA transcripts, such as 5′ end capping, RNA splicing, RNA editing, and 3′ end polyadenylation. Among these molecular processes, RNA splicing is a well-documented molecular event that is tightly regulated by A-to-I modification [65, 66]. The creation or removal of splice sites by A-to-I editing plays a vital role in the RNA splicing process. In mammalian cells, A-to-I modifications preferentially occur at Alu elements in the introns of the transcribed gene and create novel splice sites, resulting in exonization of the noncoding sequence. According to high-throughput sequencing results, it was estimated that around 1.4% of total human mRNAs are subject to A-to-I editing and that the editing sites are closely associated with RNA splicing machinery [67]. Another mechanism by which A-to-I editing affects splicing is mediated through altering RNA secondary structures [68]. Because both ADAR proteins and splicing machinery act on double-stranded RNA, the substitution of adenosine by inosine may change the stability of the RNA duplex [69], and eventually alter the mutual interaction between the splicing machinery and the double-stranded RNAs.
In addition, emerging evidence suggests that A-to-I editing plays a role in regulating RNA stability. In human B cells, DNA and RNA sequencing data showed that the expression levels of thousands of genes were modulated by ADAR proteins [70]. Furthermore, ADAR1 strengthened target RNA stability through physically interacting with HuR, a potent RNA stabilizer. In mouse cells, a similar finding was also reported for ADAR2. The unedited Ctn RNA displayed a higher binding affinity with the RNA destabilizers HuR and PARN when compared to the ADAR2-edited Ctn RNA [71]. However, ADAR2-mediated A-to-I editing of the 3’UTR of Ctn RNA hampered the interaction between the RNA destabilizer and Ctn RNA transcript, thereby leading to a prolonged half-life of Ctn RNA. Although the current findings indicate that both ADAR1 and ADAR2 promote RNA stability through HuR, HuR can function both as an RNA destabilizer or an RNA stabilizer, the mechanism of which is still largely unknown.
In summary, ADAR1 plays a pivotal role in maintaining the stemness of hematopoietic malignancies. Through enhancing self-renewal gene expression and impairing the biogenesis of tumor-suppressive miRNAs, ADAR1 is indispensable for normal hematopoietic stem cell maintenance and leukemia stem cell self-renewal. This suggests that ADAR1 may play an important role in a wide spectrum of hematopoietic disorders which have acquired aberrant stem cell self-renewal features.
m5C modification regulates cancer stem cells
5-methylcytosine (m5C) was first identified in tRNA and rRNA [72, 73]. Recently, the transcriptome-wide landscape of the m5C profile has shown that m5C modifications are preferentially located in the vicinity of the translational start codon of mRNA [74]. In addition, the m5C modification is predominantly catalyzed by the RNA methyltransferase, NSUN2, and the m5C sites are recognized by the m5C reader protein, ALYREF (Fig. 2) [75]. Besides mRNA, NSUN2 also catalyzes tRNA methylation at the variable loop region (C47-C50) [76]. In NSUN2-null mice, the m5C modification was lost in tRNA at the following positions, tRNAGly, tRNALeu, tRNAAsp, and tRNAVal [77]. In addition to NSUN2, DNMT2 is another confirmed m5C RNA methyltransferase [78, 79], which mainly catalyzes tRNA methylation [77, 80].
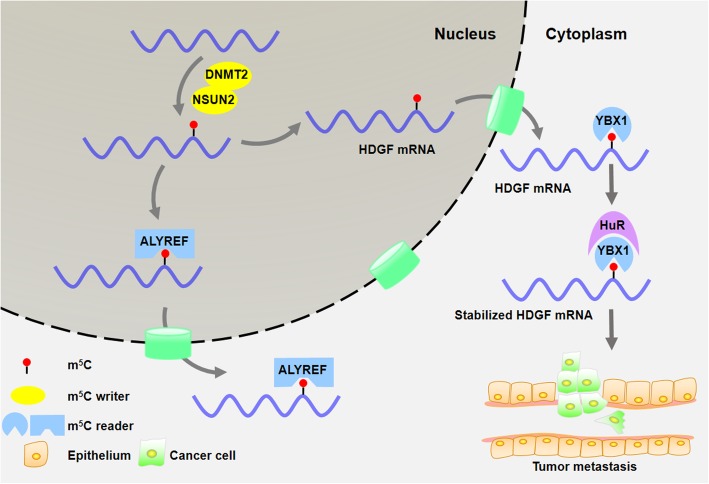
Fig. 2.
Roles of m5C RNA modifications in cancer. DNMT2 and NSUN2 are RNA methyltransferases responsible for m5C modification. The m5C reader protein ALYREF recognizes m5C- methylated mRNA and initiates transportation from nucleus towards cytoplasm. In bladder cancer, HDGF mRNA is methylated and captured by reader protein YBX1. By interacting with YBX1, HuR stabilizes HDGF mRNA and induces tumor metastasis
NSUN2 is highly expressed in various types of solid tumors [81–83] and is transcriptionally activated by the MYC oncogene [84]. In bladder cancer, many oncogenic RNAs harbor hyper-methylated m5C sites, which are catalyzed by NSUN2 [85]. YBX1 recognizes m5C-modified mRNA and then recruits mRNA stability maintainer, HuR (Fig. 2), which subsequently stabilizes the putative oncogene HDGF in an m5C-dependent manner [85].
However, paradoxically, in skin cancer, NSUN2 expression was downregulated and the depletion of NSUN2 increased the population of tumor-initiating cells [53]. By quantifying protein synthesis, it was found that tumor-initiating cells synthesized less protein when compared to their progeny [53]. Thus, a reduction in protein translation is beneficial for the generation of tumor-initiating cells as well as other stem cell types [53, 86].
The above contradictory findings raised two fundamental questions for NSUN2 and m5C methylation in cancer biology: (1) does NSUN2 exert opposite roles in different cancer types, and (2) does m5C methylation either promote or inhibit protein translation based on different microenvironments? It is not rare that a single gene can have dual roles as either a tumor suppressor or an oncogene. For instance, the stem cell marker gene, KLF4, has contrasting roles in various cancer types as reported in previous studies [87, 88]. Because different types of cancer possess distinct contexts and NSUN2 can target multiple RNAs simultaneously, it is likely that NSUN2 exerts its pleiotropic roles in a context-dependent pattern. In other words, if NSUN2 predominantly targets oncogenic RNA molecules in specific types of cancer, it would function as an oncogene. Otherwise, it would act as a tumor suppressor if it mainly affects tumor-suppressive RNA molecules.
Besides, whether the m5C modification promotes or suppresses mRNA translation is still under debate. It was reported that NSUN2-mediated m5C modification can either promote CDK1 and IL-17A translation or attenuate p27KIP1 translation [89–91]. In the DNMT2/NSUN2 double knockout cells, overall protein synthesis was dramatically reduced whereas protein translation in single knockout cells was not affected [77].
Taken together, these contrasting findings indicate that m5C has a sophisticated role in governing mRNA translation and further investigation is necessary to further clarify its mechanism of action.
m5C and the cellular fate of mRNA
Currently, it remains largely unknown how m5C modification alters mRNA expression. Recent findings suggest that m5C might enhance mRNA stability. It was found that YBX1 preferentially recognized mRNA with m5C modifications and subsequently stabilized target mRNAs. In zebrafish early embryos, m5C-modified maternal mRNAs displayed enhanced stability when compared to non-m5C-modified mRNAs [92]. Subsequent mechanistic studies showed that YBX1 enhanced the stability of m5C-modified mRNAs through cooperation with mRNA stabilizer Pabpc1a. This highlights an essential role of m5C modification in RNA metabolism and zebrafish embryo development. In human bladder cancer cells, it was reported that many oncogenic mRNAs were hypermethylated by NSUN2. As an m5C reader, YBX1 recognized m5C sites within HDGF mRNA transcripts and then recruited mRNA stabilizer HuR, leading to enhanced mRNA stability [85]. High expression of oncogenic HDGF mRNA subsequently promoted the pathogenesis of bladder cancer.
Besides, some recent papers indicate that m5C affects not only mRNA stability but also mRNA splicing. It was reported that the distribution of m5C sites partially overlapped with the binding sites of some RNA binding proteins. By analyzing the PAR-CLIP data from public databases, scientists uncovered that m5C sites were enriched in the binding regions of the mRNA splicing factors SRSF3 and SRSF4 [93], indicating a potent role of m5C modification in modulating mRNA alternative splicing. Interestingly, a recent publication revealed a previously unknown role of m5C modification in HIV infection through modulation of RNA splicing and translation [94]. It was found that HIV-1 RNA transcripts were highly methylated by m5C methyltransferase NSUN2. High-throughput sequencing data subsequently confirmed an m5C site located in the vicinity of the A2 splice site within the Vif gene. Knockout of NSUN2 reduced the use of the D1/A2 splice junction and altered the RNA splicing of HIV RNA transcript. Moreover, loss of NSUN2 reduced m5C occurrence on HIV RNA transcripts and hampered HIV RNA translation, suggesting an important role of m5C modification in the life cycle of HIV. Collectively, these data suggest that m5C modification may be involved in mRNA splicing, although there are many questions that must be addressed by scientists in the coming future. For instance, it is still unclear which reader proteins recognize m5C sites and thereby mediate mRNA splicing. Since only a small number of mRNA splicing factor binding sites overlap with the m5C region, is it a specific phenomenon that occurs in some particular RNA transcripts? Further studies are needed to elucidate the functional role of m5C in alternative splicing.
High-throughput sequencing has provided a detailed mapping of m5C sites in eukaryotic cells. Moreover, the identification of m5C writers and readers has aided our understanding of the functional roles of m5C modifications in the regulation of RNA stability, alternative splicing, and RNA translation. Although scientists have illustrated the genome-wide m5C distribution at single-nucleotide resolution, the role of m5C in mammalian cells remains unclear. For instance, the m5C eraser is still unknown and how m5C mediates other RNA processing steps is to be further explored. Therefore, it is necessary to identify novel m5C-interacting proteins or enzymes, which will further elucidate the functional roles of m5C in various biological events and human diseases.
The dual role of m6A in cancer stem cell
N6-methyladenosine (m6A), occurring at the N6 position of adenosine, is the most pervasive and abundant post-transcriptional modification in eukaryotic cells. By using the antibody-enrichment sequencing method, m6A sites were found in all areas of mRNA transcripts but displayed significant enrichment near the stop codon and 3’UTR region [95]. It was estimated that mRNA transcripts from 7676 mammalian genes have m6A modification [95]. Subsequent studies revealed that 77.29% of m6A sites are present in a consensus motif DRACH (D = A, G or U; R = A or G; H = A, C or U) [96]. Further bioinformatics analysis demonstrates that m6A RNA modification is evolutionarily conserved across different species [97].
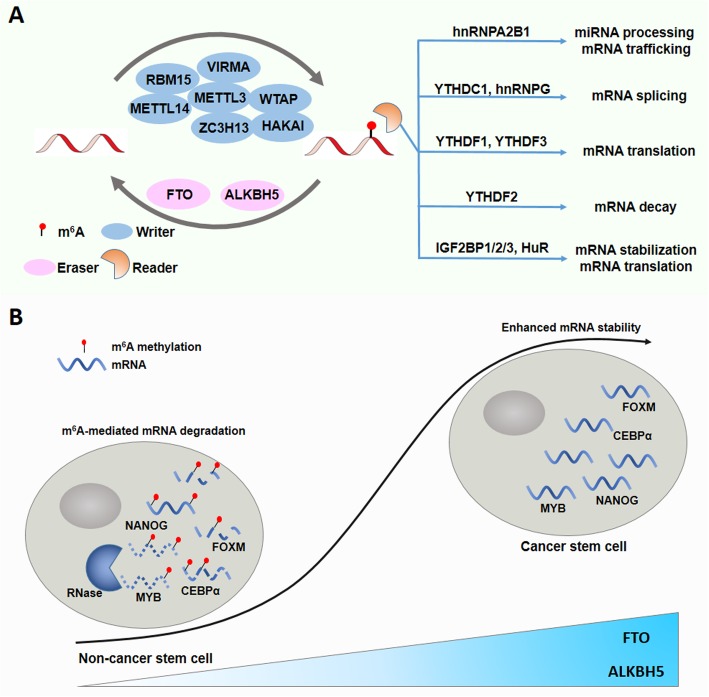
The deposition of m6A in mRNA is mediated by methyltransferase complexes, such as METTL3/14, VIRMA, RBM15/15B, WTAP, HAKAI, and ZC3H13, called ‘writers’. The removal of m6A from mRNA transcript is catalyzed by ‘eraser’ demethylases, FTO and ALKBH5 (Fig. 3a). Owing to the presence of writers and erasers, m6A modification is a dynamic and reversible process that can fine-tune the fate of mRNA transcripts within a short time. This important characteristic allows prompt adaptation to abrupt environmental changes, such as hypoxia and injury.
Fig. 3.
The functional role of m6A modification in cancer stem cells. a Summary of m6A modification machinery. The m6A effectors include the writer proteins (METTL3/METTL14/WTAP complex, probably also of VIRMA and RBM15, etc.), eraser proteins (m6A RNA demethylases: FTO and ALKBH5), and reader proteins (YTHDC1, YTHDF1/2/3, hnRNPA2B1, hnRNPG, IGF2BP1/2/3, HuR). b m6A affects mRNA stability and cancer stem cell differentiation. In cancer stem cells, FTO and ALKBH5 are highly expressed and remove m6A methylation on cancer stem cell marker genes like NANOG and MYB, leading to the stabilization of target mRNAs and enhanced self-renewal capacity
In the cytoplasm, m6A sites are recognized by m6A binding proteins, such as hnRNPA2B1, YTHDF1/2/3, YTHDC2, and IGF2BP1/2/3, called ‘readers’. Recent advances highlight m6A readers as fundamental players in the modulation of mRNA metabolism (Fig. 3a). It has been reported that the binding of hnRNPA2B1 to m6A sites promotes primary miRNA processing and mediates the nucleocytoplasmic trafficking of mRNAs [98, 99]. YTHDC1 selectively associates with m6A, marks and modulates mRNA alternative splicing, and recruitments of YTHDF1 and YTHDF3 to m6A sites enhance mRNA translation [100]. In addition, some readers also participate in the modulation of mRNA stability. YTHDF2 targets RNA transcripts that contain m6A modifications for degradation [101], while the binding of IGF2BP1/2/3 to m6A-modified mRNA promotes mRNA stability and translation [102]. In summary, m6A modification tightly modulates most aspects of mRNA processing, including mRNA stability, pre-mRNA splicing, mRNA transportation, and translation.
As the most prevalent post-transcriptional modification, m6A is essential for pluripotency and reprogramming [20]. Transcriptome-wide m6A profiling has shown that the majority of key pluripotent genes (e.g. NANOG, Oct3/4, SOX2, and KLF4) have abundant m6A modifications on their RNA transcripts, which eventually impairs mRNA stability and induces RNA degradation [103, 104]. METTL3 is the core component of the m6A methyltransferase complex. Complete depletion of m6A in METTL3-null mice led to early embryonic lethality owing to prolonged RNA half-life of core pluripotency genes, resulting in a delay in initiation of differentiation programs [103]. Therefore, the correct deposition of m6A in RNA transcripts is essential for the maintenance of self-renewal capacity during embryo development.
Emerging evidence indicates that aberrant m6A profiles frequently occur in a variety of cancer types [105]. Unexpectedly, both elevated and depressed levels of m6A methylation have been reported in different types of cancer, such as liver cancer and acute myeloid leukemia (AML) [106]. In AML, elevated expression of METTL3 has been observed, which led to increased m6A methylation levels of BCL2 and c-MYC transcripts and thus enhanced their translation [107]. METTL3 also induced m6A modification within the mRNA transcript of SP1, an oncogene in AML which modulates c-MYC expression [108]. On the contrary, another group found that m6A demethylase FTO played an oncogenic role in AML through reducing the m6A levels of ABS2 and RARA, which led to decreased mRNA levels of these two targets and eventually contributed to leukemogenesis [109].
For CSCs, m6A demethylation actively helps to maintain the self-renewal capacity of cancer cells (Fig. 3b). In breast cancer cells, the m6A demethylase, ALKBH5, reduced the level of m6A modification in NANOG mRNA, which subsequently stabilized NANOG mRNA and thus promoted breast cancer stem cell phenotypes [33]. ALKBH5 was also highly expressed in glioblastoma stem-like cells (GSCs) and the knockdown of ALKBH5 attenuated the growth of patient-derived GSCs [35]. The mechanistic study revealed that ALKBH5 demethylated FOXM1 nascent RNA transcripts and enhanced FOXM1 expression, which ultimately maintained the self-renewal capacity of GSCs [35]. Similar to ALKBH5, another m6A demethylase FTO was reported to promote self-renewal and tumorigenesis in GSCs and suppression of FTO by its inhibitor MA2 attenuated GSC growth and self-renewal [34]. Consistently, treatment with another FTO inhibitor R-2HG significantly elevated global m6A modification in leukemia cells, which in turn induced the degradation of MYC/CEBPA RNA transcripts and inhibited the relevant pathways [55].
However, the opposite expression patterns of m6A exist in acute myeloid leukemia and glioblastoma. METTL14, a core component of the m6A methyltransferase complex, was dramatically elevated in normal hematopoietic stem/progenitor cells and acute myeloid leukemia cells [56]. METTL14 catalyzed the m6A modification in oncogenic factors MYC and MYB, increasing their mRNA stability and thus maintaining the stemness of leukemia stem cells [56]. In glioblastoma, m6A methyltransferase METTL3 was elevated in GSCs and its expression decreased during differentiation [54]. Subsequent studies found that SOX2 mRNA was methylated by METTL3 and that methylated SOX2 mRNA displayed prolonged stability, suggesting that HuR is essential for METTL3-mediated stabilization of SOX2 mRNA [54].
In summary, aberrant m6A modification frequently occurs in a variety of cancer types and m6A’s deregulation plays a vital role in modulating the stemness of CSCs. However, both elevated and depressed levels of m6A have been reported in CSCs, and the mechanisms by which m6A modification contributes to cell fate decisions remain elusive. Therefore, further studies are needed to explore the underlying molecular mechanisms.
The underlying mechanisms for m6A in RNA expression and splicing
Currently, the mechanisms by which m6A methylation modulates mRNA decay are still under debate. The majority of the current findings indicate that m6A methylation predominantly hampers mRNA stability [110]. It was reported that m6A-modified mRNA has shorter half-live in mammalian cells [111]. Complete depletion of METTL3, the core component of the m6A methyltransferase complex, led to prolonged mRNA half-live when compared to that of wild type cells [111]. On the other hand, knockdown of m6A demethylase, ALKBH5, impaired the stability of NANOG and FOXM1 mRNA transcripts in CSCs [33, 35], indicating that m6A methylation might destabilize mRNA transcripts. Interestingly, recent studies suggest that the destabilizing effect of m6A is attributed to the cytosolic m6A reader protein YTHDF2. The carboxy-terminal of YTHDF2 preferentially binds to m6A-modified mRNAs, and its amino-terminal is responsible for the translocation of the m6A-modified mRNAs towards the P-body, where the unwanted mRNAs are degraded [17]. Furthermore, YTHDF2 silencing results in a prolonged lifetime of its mRNA targets, suggesting that YTHDF2 may play a vital role in mRNA decay [17].
In contrast to the mRNA-decay-promoting role of m6A methylation, a few emerging studies indicate that m6A methylation also stabilizes mRNA by recruiting IGF2BP1/2/3 and HuR proteins. For example, SOX2 is an m6A target for METTL3 and methylated SOX2 mRNA displays prolonged stability. In addition, RNA stabilizer protein HuR is essential for METTL3-mediated SOX2 mRNA stabilization [54]. On the contrary, IGF2BP1/2/3 proteins can recognize m6A-modified mRNAs and enhance the RNA stability of their target mRNAs in an m6A-dependent manner, thereby modulating cancer cell proliferation [102].
Therefore, it seems that the cellular fate of m6A-modified mRNA depends on their binding proteins. YTHDF2 recognizes the m6A-modified mRNA transcripts and initiates RNA degradation [101]. However, the binding of IGF2BP1/2/3 or HuR to m6A-modified mRNA enhances mRNA stability and translation [54, 102].
In addition to mRNA decay, the presence of m6A may also participate in mRNA alternative splicing. In 2016, two independent groups reported that m6A sites within the intron affected the splicing of Sxl gene [112, 113], a master regulator of Drosophila sex determination. The m6A mapping results revealed Sxl as a major intronic m6A target and that disruption of the m6A pathway compromised the female-specific Sxl splicing [112, 113]. Further studies demonstrated that m6A reader YT521-B was a dominant m6A effector for female-specific Sxl alternative splicing [112–114]. In mammalian cells, a few studies have found that m6A affected RNA splicing by recruiting m6A reader YTHDC1 to m6A-modified mRNA. Mechanistic studies showed that YTHDC1 modulated RNA alternative splicing through interacting with splicing factors [14, 115]. During mouse oocyte development, YTHDC1 regulates m6A-dependent processing of pre-mRNA transcripts through the recruitment of splicing factors CPSF6, SRSF3, and SRSF7. YTHDC1-deficient oocytes displayed extensive alternative polyadenylation, leading to altered 3′-UTR length [14]. In mammalian cells, genome-wide m6A mapping and PAR-CLIP showed that the binding sites of YTHDC1 and SRSF3 co-localized with m6A sites [115]. Subsequent studies found that YTHDC1 promoted exon inclusion by interacting with pre-mRNA splicing regulator SRSF3. To further investigate whether m6A modulates RNA splicing, various high-resolution m6A mapping methods have been used to determine whether m6A sites are located in the vicinity of splice junctions. Some groups have found enrichment of m6A in the proximity of exonic and intronic splice sites [116–118], while another independent group found that the majority of m6A sites were not located close to splice sites [111]. These contradicting results raise concerns over the accuracy of current approaches to m6A mapping. More studies will be needed to provide a precise mapping of m6A distribution within the nascent RNAs, which will eventually elucidate the role of m6A in RNA splicing.
As the most prevalent RNA modification form in eukaryotic mRNAs, the m6A-interacting proteins (writers, erasers, and readers) have been identified by serial biochemical approaches. Subsequent studies have highlighted the biological and pathological importance of these proteins. However, the underlying molecular mechanisms of m6A modifications need to be further explored. In conclusion, the central questions remain about how m6A is added on or removed from target mRNAs, and how m6A modulates RNA metabolism.
Conclusion and perspectives
Previous studies highlighted mRNA modifications as key modulators in determining cell fate transition during embryonic development [103]. Recently, emerging evidence demonstrates that several mRNA modification forms are fundamental for maintaining the stemness of CSCs. A unique feature of CSCs is the efficient maintenance of their self-renewal capacity in response to external stimuli such as chemotherapy and radiotherapy. Therefore, depending on the distinct RNA modification profiles between CSCs and other tumor cells, we can exploit this unique feature to develop novel biomarkers to distinguish drug-resistant tumor cells from drug-responsive tumor cells. Furthermore, the dependency on RNA modifications to shift cancer cell fate may be able to be exploited as a powerful therapeutic strategy to specifically eliminate CSCs in cancer patients.
Recent breakthroughs in epitranscriptome sequencing technologies have enabled scientists to decode mRNA modifications in mammalian cells, which strengthen our current understanding of the distribution and function of various mRNA modifications. However, although more than 170 RNA modifications have been identified [105], only a few sequencing technologies have been established to decode RNA modifications. Moreover, many sequencing platforms fail to provide a precise transcriptome-wide RNA modification landscape at single-base resolution in eukaryotic cells. Thus, more robust and sensitive methods are urgently needed to decipher the epitranscriptome in mammalian cells. Recently, Nanopore technology, a novel single-molecule method, has displayed precise and single base-resolution detection of m6A in synthetic RNA molecules [13, 119]. This single-molecule approach might serve as a novel paradigm to detect different RNA modifications simultaneously.
In addition to novel sequencing strategies, the corresponding RNA modifying enzymes remain largely unknown. For instance, although the m5C methyltransferase NSUN2 has been characterized, we still do not know the parallel demethylases which are responsible for the removal of m5C [77]. Moreover, although the aberrant expression of RNA modifying enzyme has been identified in most aspects of cancer cells, it remains largely unknown how specific RNA modifications affect distinct cancer cell sub-populations. The functional consequences of RNA modification disruption remain unclear. Thus, a detailed understanding of how RNA modifications influence cancer cell fate is essential for harnessing these findings into novel cancer therapies.
In conclusion, the aberrant deposition of RNA modifications is tightly linked to the stemness of CSCs. The underlying molecular mechanisms show that RNA modifications orchestrate almost every step of mRNA metabolism, ranging from mRNA biogenesis to mRNA decay, which can eventually converge to determine the cancer stem cell’s fate and tumor progression.
Acknowledgements
Not applicable.
Abbreviations
- ALKBH5
Alkb homologue 5
- CRC
Colorectal cancer
- CSCs
Cancer stem cells
- D2-HG
D2-hydorxyglutarate
- DNMT 1
DNA methyltransferase 1
- DNMT2
DNA methyltransferase 2
- FOXM1
Forkhead box protein M1
- FOXM1-AS
Antisense to FOXM1
- FTO
Fat mass and obesity associated protein
- GSCs
Glioblastoma stem-like cells
- HCC
Hepatocellular carcinoma
- HuR
Human antigen R
- IGF2BP
IGF2 mRNA binding proteins
- lncRNAs
Long non-coding RNAs
- LSCs
Leukemia stem cells
- m5C
5-methylcytosine
- m6A
N 6-methyladenosine
- METTL3
Methyltransferase-like 3
- miRNAs
Micro RNAs
- mRNAs
Messenger RNAs
- ncRNAs
Noncoding RNAs
- PAR-CLIP
Photoactivatable-Ribonucleoside-Enhanced Crosslinking and Immunoprecipitation
- R-2HG
R-2-hydroxyglutarate
- Sxl
Sex lethal
- UTR
Untranslated terminal region
- WTAP
Wilms tumor 1-associated protein
- YBX1
Y-box binding protein 1
Authors’ contributions
W.L. and Q.Z. designed this study. W.L. drafted the manuscript. W.L. and C.D. prepared the figures. Z.L., D. Q, C.D., and Q.Z. revised this manuscript. The authors read and approved the final manuscript.
Funding
This work was supported by grants from the National Natural Science Foundation of China (81870449 and 81670601 to Q.Z., 81902886 to W.L.), Special Fund for Frontier and Key Technology Innovation of Guangdong Province (2015B020226004 to Q.Z.), Key Project Fund of Guangdong Natural Science Foundation (2017A030311034 to Q.Z.), Guangdong Province Universities and Colleges Pearl River Scholar Funded Scheme (Year 2017 to Q.Z.), National Key Point Research and Invention Program of the Thirteenth (2018ZX10723203 to Q.Z.).
Availability of data and materials
Not applicable.
Ethics approval and consent to participate
Not applicable.
Consent for publication
All of the authors approve the content of the paper and their co-authorship of the paper.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Boccaletto P, Machnicka MA, Purta E, Piatkowski P, Baginski B, Wirecki TK, de Crecy-Lagard V, Ross R, Limbach PA, Kotter A, et al. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018;46:D303–D307. doi: 10.1093/nar/gkx1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jonkhout N, Tran J, Smith MA, Schonrock N, Mattick JS, Novoa EM. The RNA modification landscape in human disease. RNA. 2017;23:1754–1769. doi: 10.1261/rna.063503.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li S, Mason CE. The pivotal regulatory landscape of RNA modifications. Annu Rev Genomics Hum Genet. 2014;15:127–150. doi: 10.1146/annurev-genom-090413-025405. [DOI] [PubMed] [Google Scholar]
- 4.Jia GF, Fu Y, Zhao X, Dai Q, Zheng GQ, Yang Y, Yi CQ, Lindahl T, Pan T, Yang YG, He C. N6-Methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7:885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He C. Grand challenge commentary: RNA epigenetics? Nat Chem Biol. 2010;6:863–865. doi: 10.1038/nchembio.482. [DOI] [PubMed] [Google Scholar]
- 6.Saletore Y, Meyer K, Korlach J, Vilfan ID, Jaffrey S, Mason CE. The birth of the Epitranscriptome: deciphering the function of RNA modifications. Genome Biol. 2012;13(10):175. doi: 10.1186/gb-2012-13-10-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khoddami V, Cairns BR. Identification of direct targets and modified bases of RNA cytosine methyltransferases. Nat Biotechnol. 2013;31:458–45+. doi: 10.1038/nbt.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, et al. Topology of the human and mouse m (6) A RNA methylomes revealed by m (6) A-seq. Nature. 2012;485:201–U284. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 9.Li XY, Xiong XS, Wang K, Wang LX, Shu XT, Ma SQ, Yi CQ. Transcriptome-wide mapping reveals reversible and dynamic N-1-methyladenosine methylome. Nat Chem Biol. 2016;12:311–31+. doi: 10.1038/nchembio.2040. [DOI] [PubMed] [Google Scholar]
- 10.Schwartz S, Bernstein DA, Mumbach MR, Jovanovic M, Herbst RH, Leon-Ricardo BX, Engreitz JM, Guttman M, Satija R, Lander ES, et al. Transcriptome-wide mapping reveals widespread dynamic-regulated Pseudouridylation of ncRNA and mRNA. Cell. 2014;159:148–162. doi: 10.1016/j.cell.2014.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delatte B, Wang F, Ngoc LV, Collignon E, Bonvin E, Deplus R, Calonne E, Hassabi B, Putmans P, Awe S, et al. Transcriptome-wide distribution and function of RNA hydroxymethylcytosine. Science. 2016;351:282–285. doi: 10.1126/science.aac5253. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Z, Chen LQ, Zhao YL, Yang CG, Roundtree IA, Zhang Z, Ren J, Xie W, He C, Luo GZ. Single-base mapping of m (6) A by an antibody-independent method. Sci Adv. 2019;5:eaax0250. doi: 10.1126/sciadv.aax0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li XY, Xiong XS, Yi CQ. Epitranscriptome sequencing technologies: decoding RNA modifications. Nat Methods. 2017;14:23–31. doi: 10.1038/nmeth.4110. [DOI] [PubMed] [Google Scholar]
- 14.Xiao W, Adhikari S, Dahal U, Chen YS, Hao YJ, Sun BF, Sun HY, Li A, Ping XL, Lai WY, et al. Nuclear m (6) A Reader YTHDC1 regulates mRNA splicing. Mol Cell. 2016;61:507–519. doi: 10.1016/j.molcel.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 15.Roost C, Lynch SR, Batista PJ, Qu K, Chang HY, Kool ET. Structure and thermodynamics of N-6-Methyladenosine in RNA: a spring-loaded base modification. J Am Chem Soc. 2015;137:2107–2115. doi: 10.1021/ja513080v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spitale RC, Flynn RA, Zhang QC, Crisalli P, Lee B, Jung JW, Kuchelmeister HY, Batista PJ, Torre EA, Kool ET, Chang HY. Structural imprints in vivo decode RNA regulatory mechanisms. Nature. 2015;519:486–48+. doi: 10.1038/nature14263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwartz S, Mumbach MR, Jovanovic M, Wang T, Maciag K, Bushkin GG, Mertins P, Ter-Ovanesyan D, Habib N, Cacchiarelli D, et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5′ sites. Cell Rep. 2014;8:284–296. doi: 10.1016/j.celrep.2014.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knuckles P, Carl SH, Musheev M, Niehrs C, Wenger A, Buhler M. RNA fate determination through cotranscriptional adenosine methylation and microprocessor binding. Nat Struct Mol Biol. 2017;24:561–56+. doi: 10.1038/nsmb.3419. [DOI] [PubMed] [Google Scholar]
- 20.Bertero A, Brown S, Madrigal P, Osnato A, Ortmann D, Yiangou L, Kadiwala J, Hubner NC, de los Mozos IR, Sadee C, et al. The SMAD2/3 interactome reveals that TGF beta controls m (6) A mRNA methylation in pluripotency. Nature. 2018;555:256–25+. doi: 10.1038/nature25784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mauer J, Luo X, Blanjoie A, Jiao X, Grozhik AV, Patil DP, Linder B, Pickering BF, Vasseur JJ, Chen Q, et al. Reversible methylation of m (6) Am in the 5′ cap controls mRNA stability. Nature. 2017;541:371–375. doi: 10.1038/nature21022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wickramasinghe VO, Laskey RA. Control of mammalian gene expression by selective mRNA export. Nat Rev Mol Cell Biol. 2015;16:431–442. doi: 10.1038/nrm4010. [DOI] [PubMed] [Google Scholar]
- 23.Fustin JM, Doi M, Yamaguchi Y, Hida H, Nishimura S, Yoshida M, Isagawa T, Morioka MS, Kakeya H, Manabe I, Okamura H. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell. 2013;155:793–806. doi: 10.1016/j.cell.2013.10.026. [DOI] [PubMed] [Google Scholar]
- 24.Zheng GQ, Dahl JA, Niu YM, Fedorcsak P, Huang CM, Li CJ, Vagbo CB, Shi Y, Wang WL, Song SH, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X, Zhao BS, Roundtree IA, Lu ZK, Han DL, Ma HH, Weng XC, Chen K, Shi HL, He C. N-6-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161:1388–1399. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shi HL, Wang X, Lu ZK, Zhao BXS, Ma HH, Hsu PJ, Liu C, He C. YTHDF3 facilitates translation and decay of N-6-methyladenosine-modified RNA. Cell Res. 2017;27:315–328. doi: 10.1038/cr.2017.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aguilo F, Zhang F, Sancho A, Fidalgo M, Di Cecilia S, Vashisht A, Lee DF, Chen CH, Rengasamy M, Andino B, et al. Coordination of m (6) A mRNA methylation and gene transcription by ZFP217 regulates pluripotency and reprogramming. Cell Stem Cell. 2015;17:689–704. doi: 10.1016/j.stem.2015.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Batista PJ, Molinie B, Wang J, Qu K, Zhang J, Li L, Bouley DM, Lujan E, Haddad B, Daneshvar K, et al. m (6) A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell. 2014;15:707–719. doi: 10.1016/j.stem.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wen J, Lv RT, Ma HH, Shen HJ, He CX, Wang JH, Jiao FF, Liu H, Yang PY, Tan L, et al. Zc3h13 regulates nuclear RNA m (6) A methylation and mouse embryonic stem cell self-renewal. Mol Cell. 2018;69:1028–102+. doi: 10.1016/j.molcel.2018.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jeter CR, Liu B, Liu X, Chen X, Liu C, Calhoun-Davis T, Repass J, Zaehres H, Shen JJ, Tang DG. NANOG promotes cancer stem cell characteristics and prostate cancer resistance to androgen deprivation. Oncogene. 2011;30:3833–3845. doi: 10.1038/onc.2011.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumar SM, Liu S, Lu H, Zhang H, Zhang PJ, Gimotty PA, Guerra M, Guo W, Xu X. Acquired cancer stem cell phenotypes through Oct4-mediated dedifferentiation. Oncogene. 2012;31:4898–4911. doi: 10.1038/onc.2011.656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sancho P, Burgos-Ramos E, Tavera A, Kheir TB, Jagust P, Schoenhals M, Barneda D, Sellers K, Campos-Olivas R, Grana O, et al. MYC/PGC-1 alpha balance determines the metabolic phenotype and plasticity of pancreatic cancer stem cells. Cell Metab. 2015;22:590–605. doi: 10.1016/j.cmet.2015.08.015. [DOI] [PubMed] [Google Scholar]
- 33.Zhang C, Samanta D, Lu H, Bullen JW, Zhang H, Chen I, He X, Semenza GL. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m (6) A-demethylation of NANOG mRNA. Proc Natl Acad Sci U S A. 2016;113:E2047–E2056. doi: 10.1073/pnas.1602883113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cui Q, Shi H, Ye P, Li L, Qu Q, Sun G, Sun G, Lu Z, Huang Y, Yang CG, et al. m (6) A RNA methylation regulates the self-renewal and tumorigenesis of glioblastoma stem cells. Cell Rep. 2017;18:2622–2634. doi: 10.1016/j.celrep.2017.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang S, Zhao BS, Zhou A, Lin K, Zheng S, Lu Z, Chen Y, Sulman EP, Xie K, Bogler O, et al. m (6) A demethylase ALKBH5 maintains Tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell. 2017;31:591–606.e596. doi: 10.1016/j.ccell.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zipeto MA, Court AC, Sadarangani A, Delos Santos NP, Balaian L, Chun HJ, Pineda G, Morris SR, Mason CN, Geron I, et al. ADAR1 activation drives leukemia stem cell self-renewal by impairing Let-7 biogenesis. Cell Stem Cell. 2016;19:177–191. doi: 10.1016/j.stem.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Visvader JE, Lindeman GJ. Cancer stem cells in solid tumors: accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8:755–768. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- 38.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 39.Lathia JD, Mack SC, Mulkearns-Hubert EE, Valentim CLL, Rich JN. Cancer stem cells in glioblastoma. Genes Dev. 2015;29:1203–1217. doi: 10.1101/gad.261982.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saygin C, Matei D, Majeti R, Reizes O, Lathia JD. Targeting cancer stemness in the clinic: from hype to hope. Cell Stem Cell. 2019;24:25–40. doi: 10.1016/j.stem.2018.11.017. [DOI] [PubMed] [Google Scholar]
- 41.Qi W, Liang W, Jiang H, Miuyee Waye M. The function of miRNA in hepatic cancer stem cell. Biomed Res Int. 2013;2013:358902. doi: 10.1155/2013/358902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu J, Mirshahidi S, Simental A, Lee SC, De Andrade Filho PA, Peterson NR, Duerksen-Hughes P, Yuan X. Cancer stem cell self-renewal as a therapeutic target in human oral cancer. Oncogene. 2019;38:5440–5456. doi: 10.1038/s41388-019-0800-z. [DOI] [PubMed] [Google Scholar]
- 43.Lytle NK, Barber AG, Reya T. Stem cell fate in cancer growth, progression and therapy resistance. Nat Rev Cancer. 2018;18:669–680. doi: 10.1038/s41568-018-0056-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Latil M, Nassar D, Beck B, Boumahdi S, Wang L, Brisebarre A, Dubois C, Nkusi E, Lenglez S, Checinska A, et al. Cell-type-specific chromatin states differentially prime squamous cell carcinoma tumor-initiating cells for epithelial to mesenchymal transition. Cell Stem Cell. 2017;20:191–19+. doi: 10.1016/j.stem.2016.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xie WB, Kagiampakis I, Pan LX, Zhang YW, Murphy L, Tao Y, Kong XQ, Kang B, Xia LM, Carvalho FLF, et al. DNA methylation patterns separate senescence from transformation potential and indicate cancer risk. Cancer Cell. 2018;33:309–30+. doi: 10.1016/j.ccell.2018.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kagara N, Huynh KT, Kuo C, Okano H, Sim MS, Elashoff D, Chong K, Giuliano AE, Hoon DSB. Epigenetic regulation of cancer stem cell genes in triple-negative breast cancer. Am J Pathol. 2012;181:257–267. doi: 10.1016/j.ajpath.2012.03.019. [DOI] [PubMed] [Google Scholar]
- 47.Gopisetty G, Xu J, Sampath D, Colman H, Puduvalli VK. Epigenetic regulation of CD133/PROM1 expression in glioma stem cells by Sp1/myc and promoter methylation. Oncogene. 2013;32:3119–3129. doi: 10.1038/onc.2012.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gonzalez ME, Moore HM, Li X, Toy KA, Huang W, Sabel MS, Kidwell KM, Kleer CG. EZH2 expands breast stem cells through activation of NOTCH1 signaling. Proc Natl Acad Sci U S A. 2014;111:3098–3103. doi: 10.1073/pnas.1308953111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jiang QF, Crews LA, Barrett CL, Chun HJ, Court AC, Isquith JM, Zipeto MA, Goff DJ, Minden M, Sadarangani A, et al. ADAR1 promotes malignant progenitor reprogramming in chronic myeloid leukemia. Proc Natl Acad Sci U S A. 2013;110:1041–1046. doi: 10.1073/pnas.1213021110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Abrahamsson AE, Geron I, Gotlib J, Dao KHT, Barroga CF, Newton IG, Giles FJ, Durocher J, Creusot RS, Karimi M, et al. Glycogen synthase kinase 3 beta missplicing contributes to leukemia stem cell generation. Proc Natl Acad Sci U S A. 2009;106:3925–3929. doi: 10.1073/pnas.0900189106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lazzari E, Mondala PK, Delos Santos N, Miller AC, Pineda G, Jiang QF, Leu H, Ali SA, Ganesan AP, Wu CN, et al. Alu-dependent RNA editing of GLI1 promotes malignant regeneration in multiple myeloma. Nat Commun. 2017;8(1):1922. doi: 10.1038/s41467-017-01890-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jiang QF, Isquith J, Zipeto MA, Diep RH, Pham J, Delos Santos N, Reynoso E, Chau J, Leu H, Lazzari E, et al. Hyper-editing of cell-cycle regulatory and tumor suppressor RNA promotes malignant progenitor propagation. Cancer Cell. 2019;35:81–8+. doi: 10.1016/j.ccell.2018.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Blanco S, Bandiera R, Popis M, Hussain S, Lombard P, Aleksic J, Sajini A, Tanna H, Cortes-Garrido R, Gkatza N, et al. Stem cell function and stress response are controlled by protein synthesis. Nature. 2016;534:335–33+. doi: 10.1038/nature18282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Visvanathan A, Patil V, Arora A, Hegde AS, Arivazhagan A, Santosh V, Somasundaram K. Essential role of METTL3-mediated m (6) A modification in glioma stem-like cells maintenance and radioresistance. Oncogene. 2018;37:522–533. doi: 10.1038/onc.2017.351. [DOI] [PubMed] [Google Scholar]
- 55.Su R, Dong L, Li C, Nachtergaele S, Wunderlich M, Qing Y, Deng X, Wang Y, Weng X, Hu C, et al. R-2HG exhibits anti-tumor activity by targeting FTO/m (6) A/MYC/CEBPA signaling. Cell. 2018;172:90–105.e123. doi: 10.1016/j.cell.2017.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weng H, Huang H, Wu H, Qin X, Zhao BS, Dong L, Shi H, Skibbe J, Shen C, Hu C, et al. METTL14 inhibits hematopoietic stem/progenitor differentiation and promotes Leukemogenesis via mRNA m (6) A modification. Cell Stem Cell. 2018;22:191–205.e199. doi: 10.1016/j.stem.2017.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zipeto MA, Jiang QF, Melese E, Jamieson CHM. RNA rewriting, recoding, and rewiring in human disease. Trends Mol Med. 2015;21:549–559. doi: 10.1016/j.molmed.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 58.Xu Li-Di, Öhman Marie. ADAR1 Editing and its Role in Cancer. Genes. 2018;10(1):12. doi: 10.3390/genes10010012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jiang QF, Crews LA, Holm F, Jamieson CHM. RNA editing-dependent epitranscriptome diversity in cancer stem cells. Nat Rev Cancer. 2017;17:381–392. doi: 10.1038/nrc.2017.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mannion NM, Greenwood SM, Young R, Cox S, Brindle J, Read D, Nellaker C, Vesely C, Ponting CP, McLaughlin PJ, et al. The RNA-editing enzyme ADAR1 controls innate immune responses to RNA. Cell Rep. 2014;9:1482–1494. doi: 10.1016/j.celrep.2014.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hartner JC, Walkley CR, Lu J, Orkin SH. ADAR1 is essential for the maintenance of hematopoiesis and suppression of interferon signaling. Nat Immunol. 2009;10:109–115. doi: 10.1038/ni.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Higuchi M, Stefan M, Single FN, Hartner J, Rozov A, Burnashev N, Feldmeyer D, Sprengel R, Seeburg PH. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature. 2000;406:78–81. doi: 10.1038/35017558. [DOI] [PubMed] [Google Scholar]
- 63.Han L, Diao L, Yu S, Xu X, Li J, Zhang R, Yang Y, Werner HMJ, Eterovic AK, Yuan Y, et al. The genomic landscape and clinical relevance of A-to-I RNA editing in human cancers. Cancer Cell. 2015;28:515–528. doi: 10.1016/j.ccell.2015.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ni W, Yang Z, Qi W, Cui C, Cui Y, Xuan Y. Gli1 is a potential cancer stem cell marker and predicts poor prognosis in ductal breast carcinoma. Hum Pathol. 2017;69:38–45. doi: 10.1016/j.humpath.2017.08.038. [DOI] [PubMed] [Google Scholar]
- 65.Hsiao YE, Bahn JH, Yang Y, Lin X, Tran S, Yang EW, Quinones-Valdez G, Xiao X. RNA editing in nascent RNA affects pre-mRNA splicing. Genome Res. 2018;28:812–823. doi: 10.1101/gr.231209.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bazak L, Haviv A, Barak M, Jacob-Hirsch J, Deng P, Zhang R, Isaacs FJ, Rechavi G, Li JB, Eisenberg E, Levanon EY. A-to-I RNA editing occurs at over a hundred million genomic sites, located in a majority of human genes. Genome Res. 2014;24:365–376. doi: 10.1101/gr.164749.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lev-Maor G, Sorek R, Levanon EY, Paz N, Eisenberg E, Ast G. RNA-editing-mediated exon evolution. Genome Biol. 2007;8:R29. doi: 10.1186/gb-2007-8-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Savva YA, Rieder LE, Reenan RA. The ADAR protein family. Genome Biol. 2012;13:252. doi: 10.1186/gb-2012-13-12-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rieder LE, Reenan RA. The intricate relationship between RNA structure, editing, and splicing. Semin Cell Dev Biol. 2012;23:281–288. doi: 10.1016/j.semcdb.2011.11.004. [DOI] [PubMed] [Google Scholar]
- 70.Wang IX, So E, Devlin JL, Zhao Y, Wu M, Cheung VG. ADAR regulates RNA editing, transcript stability, and gene expression. Cell Rep. 2013;5:849–860. doi: 10.1016/j.celrep.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Anantharaman A, Tripathi V, Khan A, Yoon JH, Singh DK, Gholamalamdari O, Guang S, Ohlson J, Wahlstedt H, Ohman M, et al. ADAR2 regulates RNA stability by modifying access of decay-promoting RNA-binding proteins. Nucleic Acids Res. 2017;45:4189–4201. doi: 10.1093/nar/gkw1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Helm M. Post-transcriptional nucleotide modification and alternative folding of RNA. Nucleic Acids Res. 2006;34:721–733. doi: 10.1093/nar/gkj471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Agris PF. Bringing order to translation: the contributions of transfer RNA anticodon-domain modifications. EMBO Rep. 2008;9:629–635. doi: 10.1038/embor.2008.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Amort T, Rieder D, Wille A, Khokhlova-Cubberley D, Riml C, Trixl L, Jia XY, Micura R, Lusser A. Distinct 5-methylcytosine profiles in poly(A) RNA from mouse embryonic stem cells and brain. Genome Biol. 2017;18:1. doi: 10.1186/s13059-016-1139-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yang X, Yang Y, Sun BF, Chen YS, Xu JW, Lai WY, Li A, Wang X, Bhattarai DP, Xiao W, et al. 5-methylcytosine promotes mRNA export-NSUN2 as the methyltransferase and ALYREF as an m (5) C reader. Cell Res. 2017;27:606–625. doi: 10.1038/cr.2017.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Blanco S, Dietmann S, Flores JV, Hussain S, Kutter C, Humphreys P, Lukk M, Lombard P, Treps L, Popis M, et al. Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. EMBO J. 2014;33:2020–2039. doi: 10.15252/embj.201489282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tuorto F, Liebers R, Musch T, Schaefer M, Hofmann S, Kellner S, Frye M, Helm M, Stoecklin G, Lyko F. RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat Struct Mol Biol. 2012;19:900–905. doi: 10.1038/nsmb.2357. [DOI] [PubMed] [Google Scholar]
- 78.Brzezicha B, Schmidt M, Makalowska I, Jarmolowski A, Pienkowska J, Szweykowska-Kulinska Z. Identification of human tRNA:m5C methyltransferase catalysing intron-dependent m5C formation in the first position of the anticodon of the pre-tRNA Leu (CAA) Nucleic Acids Res. 2006;34:6034–6043. doi: 10.1093/nar/gkl765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Goll MG, Kirpekar F, Maggert KA, Yoder JA, Hsieh CL, Zhang X, Golic KG, Jacobsen SE, Bestor TH. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science. 2006;311:395–398. doi: 10.1126/science.1120976. [DOI] [PubMed] [Google Scholar]
- 80.Schaefer M, Pollex T, Hanna K, Tuorto F, Meusburger M, Helm M, Lyko F. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev. 2010;24:1590–1595. doi: 10.1101/gad.586710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Okamoto M, Hirata S, Sato S, Koga S, Fujii M, Qi G, Ogawa I, Takata T, Shimamoto F, Tatsuka M. Frequent increased gene copy number and high protein expression of tRNA (cytosine-5-)-methyltransferase (NSUN2) in human cancers. DNA Cell Biol. 2012;31:660–671. doi: 10.1089/dna.2011.1446. [DOI] [PubMed] [Google Scholar]
- 82.Lu L, Zhu G, Zeng H, Xu Q, Holzmann K. High tRNA Transferase NSUN2 gene expression is associated with poor prognosis in head and neck squamous carcinoma. Cancer Investig. 2018;36:246–253. doi: 10.1080/07357907.2018.1466896. [DOI] [PubMed] [Google Scholar]
- 83.Frye M, Dragoni I, Chin SF, Spiteri I, Kurowski A, Provenzano E, Green A, Ellis IO, Grimmer D, Teschendorff A, et al. Genomic gain of 5p15 leads to over-expression of Misu (NSUN2) in breast cancer. Cancer Lett. 2010;289:71–80. doi: 10.1016/j.canlet.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 84.Frye M, Watt FM. The RNA methyltransferase Misu (NSun2) mediates Myc-induced proliferation and is upregulated in tumors. Curr Biol. 2006;16:971–981. doi: 10.1016/j.cub.2006.04.027. [DOI] [PubMed] [Google Scholar]
- 85.Chen X, Li A, Sun BF, Yang Y, Han YN, Yuan X, Chen RX, Wei WS, Liu YC, Gao CC, et al. 5-methylcytosine promotes pathogenesis of bladder cancer through stabilizing mRNAs. Nat Cell Biol. 2019;21:978–97+. doi: 10.1038/s41556-019-0361-y. [DOI] [PubMed] [Google Scholar]
- 86.Signer RAJ, Magee JA, Salic A, Morrison SJ. Haematopoietic stem cells require a highly regulated protein synthesis rate. Nature. 2014;509:49–4+. doi: 10.1038/nature13035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rowland BD, Peeper DS. KLF4, p21 and context-dependent opposing forces in cancer. Nat Rev Cancer. 2006;6:11–23. doi: 10.1038/nrc1780. [DOI] [PubMed] [Google Scholar]
- 88.Rowland BD, Bernards R, Peeper DS. The KLF4 tumor suppressor is a transcriptional repressor of p53 that acts as a context-dependent oncogene. Nat Cell Biol. 2005;7:1074–1082. doi: 10.1038/ncb1314. [DOI] [PubMed] [Google Scholar]
- 89.Wang N, Tang H, Wang X, Wang WG, Feng J. Homocysteine upregulates interleukin-17A expression via NSun2-mediated RNA methylation in T lymphocytes. Biochem Biophys Res Commun. 2017;493:94–99. doi: 10.1016/j.bbrc.2017.09.069. [DOI] [PubMed] [Google Scholar]
- 90.Tang H, Fan XQ, Xing JY, Liu ZY, Jiang B, Dou Y, Gorospe M, Wang WG. NSun2 delays replicative senescence by repressing p27 (KIP1) translation and elevating CDK1 translation. Aging-Us. 2015;7:1143–1158. doi: 10.18632/aging.100860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Xing JY, Yi J, Cai XY, Tang H, Liu ZY, Zhang XT, Martindale JL, Yang XL, Jiang B, Gorospe M, Wang WG. NSun2 promotes cell growth via elevating Cyclin-dependent kinase 1 translation. Mol Cell Biol. 2015;35:4043–4052. doi: 10.1128/MCB.00742-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yang Y, Wang L, Han X, Yang WL, Zhang M, Ma HL, Sun BF, Li A, Xia J, Chen J, et al. RNA 5-Methylcytosine facilitates the maternal-to-zygotic transition by preventing maternal mRNA decay. Mol Cell. 2019;75:1188–1202.e1111. doi: 10.1016/j.molcel.2019.06.033. [DOI] [PubMed] [Google Scholar]
- 93.Squires JE, Patel HR, Nousch M, Sibbritt T, Humphreys DT, Parker BJ, Suter CM, Preiss T. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 2012;40:5023–5033. doi: 10.1093/nar/gks144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Courtney DG, Tsai K, Bogerd HP, Kennedy EM, Law BA, Emery A, Swanstrom R, Holley CL, Cullen BR. Epitranscriptomic addition of m (5) C to HIV-1 transcripts regulates viral gene expression. Cell Host Microbe. 2019;26:217–227.e216. doi: 10.1016/j.chom.2019.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Linder B, Grozhik AV, Olarerin-George AO, Meydan C, Mason CE, Jaffrey SR. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat Methods. 2015;12:767–U114. doi: 10.1038/nmeth.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Schwartz S, Agarwala SD, Mumbach MR, Jovanovic M, Mertins P, Shishkin A, Tabach Y, Mikkelsen TS, Satija R, Ruvkun G, et al. High-resolution mapping reveals a conserved, widespread, dynamic mRNA methylation program in yeast meiosis. Cell. 2013;155:1409–1421. doi: 10.1016/j.cell.2013.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Alarcon CR, Goodarzi H, Lee H, Liu X, Tavazoie S, Tavazoie SF. HNRNPA2B1 is a mediator of m (6) A-dependent nuclear RNA processing events. Cell. 2015;162:1299–1308. doi: 10.1016/j.cell.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wang Lei, Wen Mingyue, Cao Xuetao. Nuclear hnRNPA2B1 initiates and amplifies the innate immune response to DNA viruses. Science. 2019;365(6454):eaav0758. doi: 10.1126/science.aav0758. [DOI] [PubMed] [Google Scholar]
- 100.Patil DP, Pickering BF, Jaffrey SR. Reading m (6) A in the Transcriptome: m (6) A-binding proteins. Trends Cell Biol. 2018;28:113–127. doi: 10.1016/j.tcb.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ivanova I, Much C, Di Giacomo M, Azzi C, Morgan M, Moreira PN, Monahan J, Carrieri C, Enright AJ, O'Carroll D. The RNA m (6) A Reader YTHDF2 is essential for the post-transcriptional regulation of the maternal transcriptome and oocyte competence. Mol Cell. 2017;67:1059–105+. doi: 10.1016/j.molcel.2017.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, Zhao BS, Mesquita A, Liu C, Yuan CL, et al. Recognition of RNA N (6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. 2018;20:285–295. doi: 10.1038/s41556-018-0045-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Geula S, Moshitch-Moshkovitz S, Dominissini D, Mansour AA, Kol N, Salmon-Divon M, Hershkovitz V, Peer E, Mor N, Manor YS, et al. Stem cells. m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science. 2015;347:1002–1006. doi: 10.1126/science.1261417. [DOI] [PubMed] [Google Scholar]
- 104.Wang Y, Li Y, Toth JI, Petroski MD, Zhang Z, Zhao JC. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat Cell Biol. 2014;16:191–198. doi: 10.1038/ncb2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Delaunay S, Frye M. RNA modifications regulating cell fate in cancer. Nat Cell Biol. 2019;21:552–559. doi: 10.1038/s41556-019-0319-0. [DOI] [PubMed] [Google Scholar]
- 106.Liu J, Harada BT, He C. Regulation of gene expression by N (6)-methyladenosine in cancer. Trends Cell Biol. 2019;29:487–499. doi: 10.1016/j.tcb.2019.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Vu LP, Pickering BF, Cheng Y, Zaccara S, Nguyen D, Minuesa G, Chou T, Chow A, Saletore Y, MacKay M, et al. The N (6)-methyladenosine (m (6) A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat Med. 2017;23:1369–1376. doi: 10.1038/nm.4416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Barbieri I, Tzelepis K, Pandolfini L, Shi J, Millan-Zambrano G, Robson SC, Aspris D, Migliori V, Bannister AJ, Han N, et al. Promoter-bound METTL3 maintains myeloid leukaemia by m (6) A-dependent translation control. Nature. 2017;552:126–131. doi: 10.1038/nature24678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Li Z, Weng H, Su R, Weng X, Zuo Z, Li C, Huang H, Nachtergaele S, Dong L, Hu C, et al. FTO plays an oncogenic role in acute myeloid leukemia as a N (6)-Methyladenosine RNA demethylase. Cancer Cell. 2017;31:127–141. doi: 10.1016/j.ccell.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zaccara S, Ries RJ, Jaffrey SR. Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell Biol. 2019;20:608–624. doi: 10.1038/s41580-019-0168-5. [DOI] [PubMed] [Google Scholar]
- 111.Ke S, Pandya-Jones A, Saito Y, Fak JJ, Vagbo CB, Geula S, Hanna JH, Black DL, Darnell JE., Jr Darnell RB: m (6) A mRNA modifications are deposited in nascent pre-mRNA and are not required for splicing but do specify cytoplasmic turnover. Genes Dev. 2017;31:990–1006. doi: 10.1101/gad.301036.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lence T, Akhtar J, Bayer M, Schmid K, Spindler L, Ho CH, Kreim N, Andrade-Navarro MA, Poeck B, Helm M, Roignant JY. m (6) A modulates neuronal functions and sex determination in Drosophila. Nature. 2016;540:242–247. doi: 10.1038/nature20568. [DOI] [PubMed] [Google Scholar]
- 113.Haussmann IU, Bodi Z, Sanchez-Moran E, Mongan NP, Archer N, Fray RG, Soller M. m (6) A potentiates Sxl alternative pre-mRNA splicing for robust Drosophila sex determination. Nature. 2016;540:301–304. doi: 10.1038/nature20577. [DOI] [PubMed] [Google Scholar]
- 114.Kan L, Grozhik AV, Vedanayagam J, Patil DP, Pang N, Lim KS, Huang YC, Joseph B, Lin CJ, Despic V, et al. The m (6) A pathway facilitates sex determination in Drosophila. Nat Commun. 2017;8:15737. doi: 10.1038/ncomms15737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kasowitz SD, Ma J, Anderson SJ, Leu NA, Xu Y, Gregory BD, Schultz RM, Wang PJ. Nuclear m6A reader YTHDC1 regulates alternative polyadenylation and splicing during mouse oocyte development. PLoS Genet. 2018;14:e1007412. doi: 10.1371/journal.pgen.1007412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T. N (6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015;518:560–564. doi: 10.1038/nature14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Louloupi A, Ntini E, Conrad T, Orom UAV. Transient N-6-Methyladenosine transcriptome sequencing reveals a regulatory role of m6A in splicing efficiency. Cell Rep. 2018;23:3429–3437. doi: 10.1016/j.celrep.2018.05.077. [DOI] [PubMed] [Google Scholar]
- 118.Zhou KI, Shi H, Lyu R, Wylder AC, Matuszek Z, Pan JN, He C, Parisien M, Pan T. Regulation of co-transcriptional pre-mRNA splicing by m (6) A through the low-complexity protein hnRNPG. Mol Cell. 2019;76:70–81.e79. doi: 10.1016/j.molcel.2019.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Garalde DR, Snell EA, Jachimowicz D, Sipos B, Lloyd JH, Bruce M, Pantic N, Admassu T, James P, Warland A, et al. Highly parallel direct RNA sequencing on an array of nanopores. Nat Methods. 2018;15:201–206. doi: 10.1038/nmeth.4577. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.