Abstract
Our understanding of the link between the human microbiome and disease, including obesity, inflammatory bowel disease, arthritis and autism, is rapidly expanding. Improvements in the throughput and accuracy of DNA sequencing of the genomes of microbial communities associated with human samples, complemented by analysis of transcriptomes, proteomes, metabolomes and immunomes, and mechanistic experiments in model systems, have vastly improved our ability to understand the structure and function of the microbiome in both diseased and healthy states. However, many challenges remain. In this Review, we focus on studies in humans to describe these challenges, and propose strategies that leverage existing knowledge to move rapidly from correlation to causation, and ultimately to translation.
Introduction
The microbial cells that colonize the human body, including in mucosal and skin environments, are at least as abundant as our somatic cells1, and certainly contain far more genes than our human genome (Box 1). An estimated 500–1000 species of bacteria exist in the human body at any one time2, although the number of unique genotypes (sub-species) could be orders of magnitude greater than this3. Each bacterial strain has a genome containing thousands of genes, offering substantially more genetic diversity and hence flexibility than the human genome. However, different people harbour radically different collections of microbes with substantially varying densities even among conserved taxa, and we understand little about what leads to and what regulates this variation. Importantly, we do not yet understand how the variation within a person over time, or between different people, influences wellness, the preservation of health, or the onset and progression of disease. However, we do know that changes in the microbiome, and microbial metabolome and their interaction with the immune, endocrine, and nervous system are correlated with a wide array of illnesses, ranging from inflammatory bowel disease 4–6 to cancer7 to major depressive disorder 8,9.
Box 1: How many microbial cells and genes colonize a human?
Although a frequently reported figure is that our microbes outnumber our own cells by 10:1, this number stems from a 1972 article which uses a ‘back of the envelope calculation’ to arrive at this ratio84. A more prosaic figure was provided by Rosner85 of between 5 and 724 × 1012 human cells, and between 30 and 400 × 1012 bacterial cells. More recently, a refined estimate based on experimental observation and extrapolation actually arrives at a ratio of 1.3 bacterial cells for every one human cell1. However, these estimates don’t take into consideration the viruses and phage present in various body environments, which could equal bacterial estimates or more likely outnumber them by at least an order of magnitude86. Although these estimates reduce the extent to which microbial cells outnumber human cells, they do not reduce the estimates associated with the diversity of microbial life associated with the human body. Bacteria and other microbes including archaea, fungi, and arguably, viruses, are extremely diverse. A similarly rough estimate of 1000 bacterial species in the gut with 2000 genes per species yields an estimate of 2,000,000 genes, 100 times the figure of approximately 20,000 human genes2. This agrees well with the actual size of microbial gene catalogues obtained by MetaHIT87 and the Human Microbiome Project13.
Human microbiome investigations have now reached a critical inflection point. We are transitioning from description and investigation to understanding mechanism, and developing novel clinical interventions based on this understanding10. These advances have also created a surge in translational research, resulting in substantial private investment not only in academic research, but also in the private sector, including so-called “Big Pharma”. This drive toward clinical microbiome studies is supported by a revolution in personalized medicine, in which, for example, the decline in cost of cancer genome sequencing is allowing the rapid identification of the precise treatment regimen that will lead to a positive outcome in an individual patient, for example, with colorectal cancer11. Our ability to rapidly and reproducibly characterize the microbiome, like cancer genomics, offers an opportunity to develop fundamentally new diagnostic biomarkers and therapeutics.
Here we present the current state of knowledge linking the microbiome to human disease. We have focused on human studies when possible, but also highlight select mouse studies when human studies are not available. This is to provide a platform from which to explore the future of applied clinical microbiome research.
We will strategize on how to progress from the correlative and biomarker studies towards studies that will reveal the underlying mechanisms and opportunities for new preventive and therapeutic modalities.
Factors influencing the human microbiome
To alter the microbiome deliberately for preventive or therapeutic purposes, or use it to understand a particular medical condition, we must first understand the factors that influence its composition. We have reviewed many of these factors in detail recently10,12, so we provide only a brief summary here.
Human genetics and immune interactions in early development
The composition of the human microbiome is unique in each individual, and the differences among individuals are large compared to the typical biochemical differences within a person over time13,14. Identical twins are barely more similar to one another in microbial composition and structure than are non-identical twins15, at least over the range of environmental factors captured in studies to date, suggesting that the effect of the human genome is limited, and that most of microbial community assembly may be determined by environmental factors. Early underpowered studies suggested that monozygotic twins were no more similar in terms of their overall gut microbiota than dizygotic twins16–18, although larger cohort sizes show a small but statistically significant effect of genetics on microbiome composition in twins with certain taxa being identified as highly heritable, such as Christensenella15. However, one way to rationalize this is that the number of bacteria that are able to successfully colonize humans is limited. Colonizing initially germ-free mice with diverse environmental samples demonstrates that very few bacteria present in the environment can survive in the mouse gut, and those that do are rapidly displaced by human- or mouse-derived bacteria on exposure19 Furthermore, human immune responses shape responses to changes in the microbiome and are involved in shaping the microbiome itself20.
Most human immunological studies regrettably still lack a microbiome component, that will be essential to untangling the relationship between the immune response and microbial colonisation and stability. The mammalian immune system has a complex and dynamic bidirectional relationship with the microbiome. Although, recent human cohort studies suggest that most of the variability in human immune response to stimulation derives from the genome, at least 10% of this immune response variability derives directly from interactions associated with the microbiome21.
The large majority of microbiome colonization occurs in the early years of life. This topic has been reviewed extensively22,23. During and shortly after birth, newborns are exposed to maternal and environmental microbes initiating gut microbiome establishment24. Within the first year of life, an estimated 1013 to 1014 microbes/ml comprising 500–1000 species colonize the gastrointestinal tract25. After weaning, the gut microbiota becomes firmly established, leading to a lifelong microbiome signature in healthy individuals26.
Body site
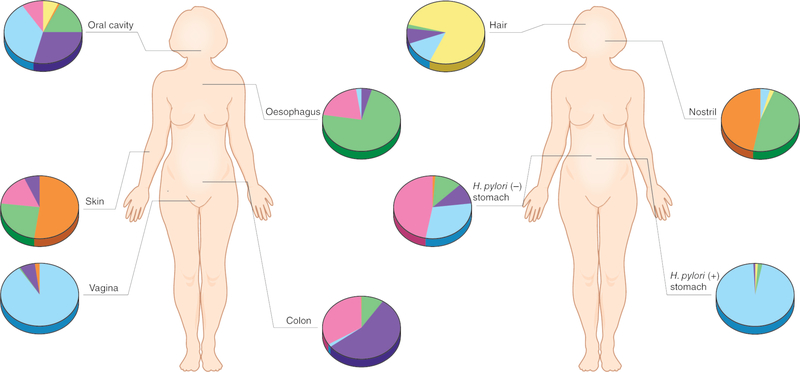
When the microbiomes of large cohorts of people at a given body site are compared, individuals fit on a continuum of microbial diversity within a human population, rather than clustering into discrete groups27,28. During human development, the human microbiome follows body site-specific trajectories, so that each body site develops a specific biogeography (Figure 1). The skin, for example, shows dramatic variation in microbiome composition and structure across different sites29. The physical and topographical characteristics of skin play a significant role in shaping the microbial community similarity between sites30. These factors also play a role in shaping the individuality of the microbiome, so that each person develops a unique microbial signature on their skin, irrespective of the differences between skin sites31. Similarly, although prolonged physical oral interaction between humans influences microbial community composition over time32, the oral microbiome still maintains a relatively unique composition in each person33. Longitudinal characterization of the human gut microbiome has shown that the adult microbiota remain relatively stable and unique to each person, compared to the drastic change over the first three years of life31,34.
Figure 1:
The human microbiome is highly personalized. Understanding the relevance of the differing microbiota between individuals is confounded by the uniqueness of an individuals’ microbiome. The different colours in the pie charts represent different species.
However, the microbiome is a living ecosystem, and consequently undergoes fluctuations in the growth rate and survival of each of its constituents. For example, changes in diet can profoundly impact the gut microbial community structure35,36, and vigorous cleaning can temporarily alter the skin microbiome. However, in both cases the original microbiota and structure re-emerge when the original conditions resume37. Transit time of food through the gut also influences the types of microbes which can proliferate within the gut, with rapid transit time selecting for functions associated with biofilm formation or rapid cell division38,39. Defining a microbiota based on the relative abundance of its members may therefore provide only a limited view of the microbial assemblage, and integrating more information about the function of each gene and genome in the context of the ecosystem and the host will provide increasingly important insights. Human microbiome variability makes blanket stratification difficult for particular disease states, although it is possible to identify biomarkers for some conditions [Box 2].
Box 2: The microbiome as novel biomarkers for disease.
There is extensive evidence for many diseases that the microbiome can be used to explain a substantially greater percentage of the variance in the relevant phenotypes for a given condition within a population than can human genetic factors. For example, in individuals with Clostridium difficile infection (CDI), the aberrant stool microbiome looks nothing like a healthy stool but rather more like the microbiome of a completely different body site. Fecal microbiota transplant is able to cure C. diff infection and after transplantation the restoration of the stool microbiome to a community that matches that of the healthy state is both rapid and visible88. The C. diff infection has a much larger impact on the stool microbiome composition than does any human genetic variation observed to date, which may explain the high efficacy of stool transplant relative to standard antibiotic treatments for C. diff89.
Obesity provides an example in which human genetics has failed to explain the obesity epidemic, in contrast, the gut microbiome can classify individuals as lean or obese with over 90% accuracy within the context of a given case-control study90, although this result depends on using the correct methods91,92. Conversely the abundance of Christensenella within the human gut are negatively correlated with BMI, and can induce weight loss when experimentally fed to mice15.
Autism Spectrum Disorder has a complex presentation of symptoms, and is difficult to attribute entirely to host genetics, mainly due to the number of confounding influences and variables93. Yet the environmental interaction, and potentially the microbiome, plays a substantial role in shaping the etiology of the disease94,95. Animal models have been used to indicate the ability of bacterial metabolites in mediating autism-like behaviours96, and fecal microbiota transplant in humans has been associated with improvement in behavioral and gastrointestinal symptoms of autism 97. In further work, the link between host genetics, behavior and the gut microbiome has been partially elucidated, identifying a strong association between Lactobacillus and memory formation98.
A host of allergic and immune diseases has increased in frequency in parallel to the above metabolic and cognitive diseases. These include childhood-onset asthma, and allergies, including food and cutaneous allergies. Similarly, inflammatory bowel disease and type 1 diabetes has been increasing globally, which cannot be explained by differences in assessment practices. A growing body of evidence is linking these with altered microbiota compositions, especially loss of diversity, seen in Inflammatory Bowel Disease patients99,100 and children at risk for T1D101. One hypothesis is that this might be linked to a microbiome perturbation, in general, rather than the acquisition or loss of specific microbes that modify phenotype 102. Perturbation of microbiomes during early life might be particularly important because that is when immunity, metabolism, and cognition are under development.
A large Canadian study of infant stool samples (n=319) collected over the first 90 days of life, compared the early-life (first 90 days) faecal microbiota of infants who went on to develop allergic sensitization and wheezing at age two, to subjects who did not. The depletion of certain bacterial taxa was characteristic of atopic children, and corresponded with reduced concentrations of faecal acetate and dysregulation of enterohepatic metabolites103, suggesting that the foundation for allergic disease development occurs in early life and is mediated at least in part by gut microbiome dysbiosis.
The vaginal microbiome has a similar degree of stability to the skin microbiome, and unlike the gut, classifying the vaginal microbiome into discrete states during disease has been possible. The vaginal microbiota of asymptomatic women tend to be dominated by individual species of Lactobacillus and diverse additional anaerobic taxa40. The Lactobacilli are believed to benefit the host by lowering vaginal pH through fermentation end-products, thereby reducing the likelihood of allochthonous microbial colonization or pathogen invasion. Microbial variation within an individual woman does occur over days to weeks41, although menstruation and pregnancy appear to result in a similar microbiome in different groups of women42. Diseases such as bacterial vaginosis result in disruption of the ‘normal’ vaginal ecosystem function, but also result in a highly similar microbial profile between women, thereby providing a generalized biomarker of disease43.
Diet
Diet has been studied extensively in relation to the gut microbiome44, but less so with respect to other microbiomes at other sites across the body. Modulating diet is an ideal opportunity for low-risk, culturally and psychologically acceptable intervention to change the microbiome. Therefore, this avenue of research could yield novel therapeutic strategies through targeted dietary interventions should gut microbiota be shown to be causative for certain diseases. Evidence to date suggests that long-term diet has very large effects on gut microbiome composition45 although a sufficiently extreme short-term dietary change can cause people to resemble one another within days35. Fascinatingly, the effects of the same dietary ingredient on blood glucose measurements can vary in different people, an effect mediated by the microbiome46. Although we know that the microbiome can influence leptin concentrations in humans, and hence influence appetite47, an open question is whether the microbiome can influence dietary preferences, which could lead to positive feedback loops when these dietary changes in turn alter the microbiome.
Antibiotics
The effect of antibiotics on all microbiomes is expected to be large compared to other factors, and preliminary studies have been performed to determine the impact48. The gut microbiome in adults appears not to be resilient to repeated antibiotic administration49. The same antibiotic appears to affect particular microbes differently depending on the rest of the microbiome50, perhaps due to different growth phases, metabolic states, or contextual microbial network in which the microorganisms find themselves. An especially interesting area of research is the increasing evidence that antibiotics in early life have a profound effect of the gut microbiome that can result in the later development of obesity51, asthma, inflammatory bowel disease and other disorders.
Lifestyle
Lifestyle is also thought to have a strong influence on microbiome composition. Cohabitation with pets, such as dogs, has a statistically significant association with the microbiome. In one study, the skin microbiome of couples living together has a closer resemblance if the couple has a dog, but, intriguingly, a small child did not provide the same trend, so couples with a child but no dog were not significantly more similar to one another than couples without a child52. Pet ownership and exposure to livestock have been associated with decreased risk of asthma53. Interrupting this exposure in infants from human populations with a known ancestral history of interaction with animals has been shown to lead to a substantial increase in atopy, especially asthma54. If these results turn out to be caused by the microbiome, rather than simply correlative, they suggest potential new therapeutic strategies for disease intervention could come from microbial exposure focused on immune activation.
Other lifestyle traits have been shown to correlate with the composition of the microbiota. For example, exercise appears to influence the structure of the microbiome through reducing inflammation, with subtle changes in the microbial community composition correlated with changes in cytokine profiles55. Sleep deprivation correlates with changes in the gut microbiome, with a greater ratio of Firmicutes to Bacteroidetes and elevated abundance of Coriobacteriaceae and Erysipelotrichaceae associated with sleep loss56. Stress increases intestinal permeability, and is correlated with changes in Bacteroidetes and Actinobacteria, with corresponding shifts in metabolite concentrations and inflammatory markers57.
Occupation has primarily been assumed to influence the microbiome via exposure to different environments and place of residence. For example, farmers have a different microbiome than city workers58. However, very few microbiome studies have isolated occupation as a variable influencing composition. For example, the oral microbiota of sailors is significantly altered by their occupational activities, so that after 120 days at sea, they show a five-fold reduction in alpha diversity and an increase Streptococcus59. Similarly, sexual intercourse in heterosexual couples leads to an increased similarity of the penile and vaginal microbiota between sexual partners, which could potentially alter sexual disease ecology in the participants; there is emerging evidence that microbiome differences might affect transmission of STIs (Sexually Transmitted Infections)60. Finally, couples who physically interact have a more similar microbiota than people who share the same living quarters but do not physically interact14, indicating that physical interaction influence microbial sharing and hence microbiome similarity, highlighting the effects of social interaction on the microbiome.
Dynamics of the human microbiome
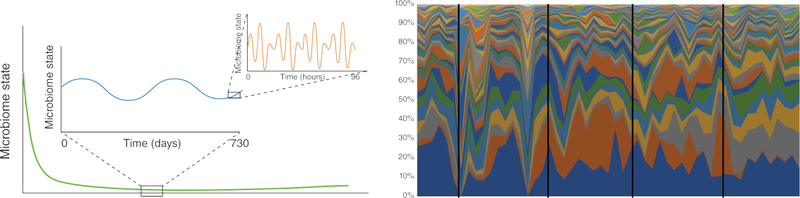
Human interaction with the environment, including with other people, creates the potential for specific microbial taxa to either act as an immune-stimulant that influences the microbiome through, for example, inflammation, or to act as a source for bacteria, fungi, and viruses that can colonize the human body. The identification of bacterial taxa in the gut that alter animal hormonal regulation, leading to obesity in mice61, suggests that such events may alter our physiology. The composition of the gut microbiome itself is influenced by circadian rhythm, and which also then affect host circadian cycles (Figure 2). Disruption of the microbial diurnal cycle can lead to disruption in host circadian rhythms, which can specifically alter hormone regulation in mice 62. The human microbiome demonstrates enormous plasticity, while also being extremely robust on longer timescales and to larger types of variation31,34,35, but experiments in mouse models have shown some of the ways in which it can be re-shaped.
Figure 2:
The dynamics of the human microbiome. The human microbiome has been shown to be highly dynamic. A) Taking a “representative” sample of a human microbiome at any given site is challenging because while the microbiome is known to settle after birth (green line), the composition can vary both over short term and long term timescales (orange line and blue line respectively). B) The effect of the rate of change of the varying species on the ability to take a representative sample as indicated by the black line is shown.
This apparent dichotomy between dynamism and robustness of the microbiome at first glance seems difficult to resolve, until the ecological dynamics of the system are considered. All ecosystems undergo variation in species population density and assemblage diversity, but with differing magnitudes at different temporal scales. This variation includes competition among microbial taxa and shifting metabolic relationships, compounded and influenced by the state of the immune system, changing dietary patterns, and a constant exposure to bacteria from family and environment. Longitudinal characterization of the host microbiome and its sources is therefore essential to capture dynamic variance within an individual, and to determine the degree to which the system demonstrates predictable successional traits63.
The plasticity vs. stability dichotomy of the human microbiome is evident over a period of days, as was illustrated in the first dense time series analysis of the human microbiome 31 and confirmed in later ones 34. In that study, two subjects provided daily samples of their oral, skin and faecal microbiota, one for six months and the other for fifteen months. The results illustrated that at the sequencing depth studied only a tiny fraction of bacterial taxa were found to be consistently present across all (or even most) samples in an individual host. For the skin sites (the left and right palm) there were no species detected in all samples, while in the gut and the mouth, about 5% of the species were defined as belonging to a stable temporal core microbiome. Yet each person still maintained a personalized microbiome. The degree of personalization of the human microbiome vastly exceeds the host genome, which is over 99.5% identical between individuals, suggesting that only 0.5% of the genome is unique to an individual. However, based on current observations, two individuals can show no overlap in microbial species in their microbiome. This degree of personalization is so high that it may even have forensic applications64.
While we are now used to thinking about the composition of the human microbiome being personalized, it has also been shown that the rate of change of the human microbiome composition is personalized65. In that study, over an approximately three-month period, 85 college age adults donated weekly microbiome samples from gut, skin, and oral sites. Over this timeframe, the microbiome composition remained almost constant in some individuals, while that in other individuals abundances changed rapidly. These differing rates of temporal variability were identified at all of the body sites that were profiled (the palm of the dominant hand, the forehead, the tongue, and faeces), and the rate of change was not correlated across the different sites. On average, skin sites changed most rapidly, followed by the gut, and then oral (this pattern matches the relative sizes of the stable temporal core microbiome observed in the long-term survey mentioned above31). One potential reason for the dynamics in skin is that there are many species at low abundance. None of the information collected about the host correlated with the differing rates of change in the microbiome, so it was not possible to determine the underlying cause of these differences. However, one interesting observation was that individuals who self-reported taking antibiotics during (or in the week preceding) the sampling period did not change their microbiome composition more rapidly than subjects who did not report taking antibiotics. The absence of a difference may reflect that a one-week time frame does not fully capture the effects of recent or even lifetime antibiotic use. Nevertheless, on a per individual basis, in this study, reported antibiotic usage was typically associated with the largest change in an individual’s microbiome overall.
While most studies associate microbiome composition with host disease state, and likelihood of response to a treatment, at least one recent study suggests that the rate of change of the microbiome may itself be a clinical feature66, as also was observed in a mouse model of juvenile diabetes 67. The rate of change of the vaginal microbiome differed across women with bacterial vaginosis, and was predictive of the subtype of bacterial vaginosis affecting the women. That observation, paired with data indicating that individuals differ in the rate of change of their gut, skin, and oral microbiomes, suggests that characterizing temporal variability may be an important part of characterizing an individual’s microbiome.
Understanding traits such as variance in microbiome dynamics in individuals, and whether that relates to patterns of succession will simplify understanding of causal relationships between species and disease, and the interpretation of correlations among taxonomic groups68. By prospectively assessing the microbiomes of patients undergoing different procedures, we can determine the rate of change, and potentially the rate of recovery of the microbiome, if it is altered by the procedure or by the disease state that led to the procedure. Doing this in a human population will provide the statistical power to relate these measurements to remission of clinical symptoms. Examining the sources that shape the microbiome is key to determining this variance.
Bayesian statistics can also be used to map the relative contribution of a specific source to the human microbiome over time69, or to create artificial neural networks of conditional dependencies that can be used to capture predictive characteristics of a microbial network70,71. Using these methods, the dynamic nature of the human microbiome or metabolome both within an individual and within a population of individuals can be captured. Once gathered, the data can be harnessed to provide a predictive signature or characteristic biomarker for a given physiological, immunological or neurological condition. The application of machine learning algorithms have also proven to be valuable in identifying highly predictive characteristics of a microbial signature to map forensic relationships between humans and their built environments14.
Towards Mechanistic Studies of the Microbiome
Mechanistic studies of the microbiome are typically difficult to perform in humans, in part because of tremendous genetic and lifestyle heterogeneity, and there are ethical issues associated with colonizing human subjects with microbes that are hypothesized to cause disease. Therefore, most of what we know currently stems from experiments in animal models. However, recent studies that complement observations in humans with interventions in animal models have produced striking new insight into the microbial origins of disease that cannot be acquired from human studies alone.
The importance of strain-level resolution for microbiome studies
The field of host-pathogen interactions has long relied on culturing strains of pathogens including clinical isolates, and transferring these pathogens to isolated cells, tissues, or whole animals to perform intervention studies. Many components of the microbiome have been inaccessible to such techniques because the relevant organisms cannot be cultured, although recent advances greatly expand the repertoire of the organisms that can be grown from the human gut72 so this barrier may be temporary. However, the culturable component of the microbiome can still be extraordinarily useful, even if incomplete. For example, a recent study in which 53 strains of bacteria were isolated from the human gut and used to monocolonize previously germ-free mice revealed large differences in immunomodulatory properties of these bacteria, including closely related strains that affected production of cytokines such as IL10, IL17a, IL22, and interferon gamma with some promoting and others inhibiting production73. These results underscore the need to characterize microbial activity at the strain level, not just at the higher taxonomic levels that are typically provided by amplicon profiling, and will probably reveal important links between the microbiome and disease when extended to more complex communities.
Identifying disease - relevant strains from population studies
Population-based microbiome studies complemented with mechanistic experimental work in mice can use microbial associations with phenotype in humans to identify bacteria or compounds that then can be tested in intervention studies to reveal causal pathways. For example, a study of heritability of different taxa within the gut microbiome in twins in the UK revealed that one specific taxon, Christensenella, was highly heritable and associated with low BMI in this population15. Strains from this genus were cultured in the lab, and then transplanted into germ-free mice, resulting in decreased weight gain in these mice when compared to transplantation from an obese human, which would normally induce weight gain (as described above).
Similarly, in a comparative study of different human populations in Finland, Russia and Estonia, which differ dramatically in the incidence of early-onset autoimmune diseases, Bacteroides sp. were especially common in the gut microbiomes of Finnish and Estonian children, in whom the incidence of the diseases were lowest, and were hypothesized to provide most of the LPS (lipopolysaccharide; a common marker of bacterial infection in the bloodstream) exposure in those populations. In contrast, the Russian children had high levels of E. coli. in their microbiomes. Tests of the effect injections of LPS from E. coli and B. dorei showed that the former, but not the latter, protected mice with a genetic defect from developing autoantibodies and diabetes symptoms, providing a potential explanation for the consequences of the different early-life microbiomes on development of autoimmune disease in humans74. A similar strategy was used to explain differences in asthma development between Amish and Hutterite children in the United States. Dust extracts from houses from each population, shown to differ in their microbiome content, were tested in a mouse model of asthma development that examines sensitivity to ovalbumin. The tests indicated that the dust from Amish but not Hutterite homes protected against asthma development54, which was attributed to differences in the bacterial content of the dust. These strategies are broadly applicable to many other situations in which differential exposure to environmental bacteria may play a role in disease etiology.
Identifying biomarkers in microbiome studies
Some studies are now performing these types of mechanistic experiments in humans directly. In one striking example, examining 500 European-ancestry individuals in the Netherlands, the authors tested the ability of the individual’s blood to produce cytokines after several antigen challenges, then paired these with data about their gut metagenome. The data suggest that the yeast Candida albicans had an especially large influence on the host’s TNF-alpha response21. This study also associated pathways active in bacteria such as palmitoleic acid metabolism with lower systemic inflammatory response; adding palmitoleic acid in challenge with C. albicans to an individual’s blood resulted in lower TNF-alpha, but unchanged IFN-gamma responses, as predicted from the association data. These types of studies are especially useful in conjunction with humans with naturally occurring genetic knockouts or variant alleles. These human genetic variants may enable microbially induced disease states that can be tested in mice with comparable null or variant genetic changes, as has been shown for Parkinson’s Disease.73
Characterizing microbial biomarkers has great potential for precision medicine, and is therefore a relatively simple way of translating microbiome research into clinical practice. For example, from groundbreaking animal studies, we know that bacterial probiotics (live bacteria deliberately introduced to the animal to produce a therapeutic effect) can be used to enhance immune checkpoint blockade therapy for melanoma patients75. Studying the microbiomes of melanoma patients prior to immune checkpoint blockade therapy has identified microorganisms in the gut to be biomarkers for diagnosis that can predict whether patients are at risk of developing checkpoint-blockade-induced colitis76.
These prospective studies are extremely important for linking microbial community structure, function, and metabolic products to health outcomes. Studies of the microbiome as infants develop are also key in this area, and many ongoing investigations, such as the NIH Common Core program Environmental Influences on Child Health Outcomes (ECHO: https://www.nih.gov/echo), now provide the infrastructure to sequence healthy, susceptible and diseased participants to examine how lifestyle and environmental experiences shape the development of immune, endocrine and neurological conditions. Although cross-sectional single time point studies of birth cohorts provide intriguing statistical associations77, longitudinal prospective studies complemented by mechanistic experiments in animal models are required to establish whether a certain microbiome causes disease.
Future studies: developing translational potential
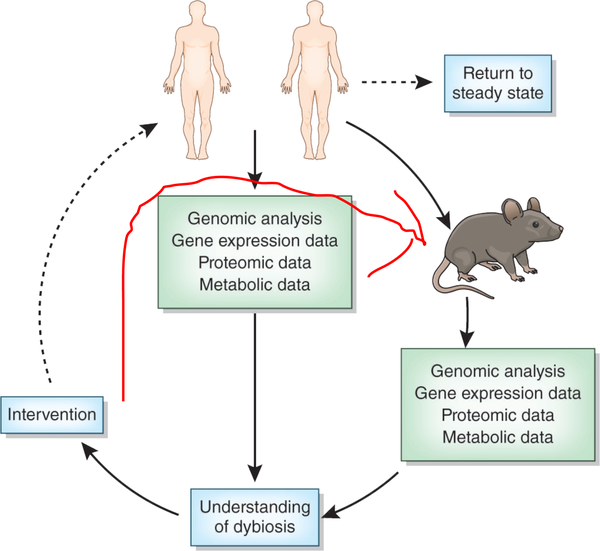
There remains much that we do not understand about the human microbiome. The sources of bacteria that colonize an infant include the mother and other caregivers (even one-day-old pre-term infants have unique microbiomes that differ from each other and from the mother but possibly derived from their mothers78), and human genetics shapes microbiome-immune interaction. Given these observations, why do monozygotic twins growing up in the same household develop microbiomes that are barely more similar than those of dizygotic twins? The role of exogenous immune stimulation in shaping the colonization efficiency of different strains must be investigated in more detail. Animal models have produced intriguing findings, but prospective longitudinal studies in human infants are required to better understand how human genetics influence the developing microbiome. These longitudinal investigations will also help us to understand the implication of ecological dynamics of the microbiome in health and disease. Microbiome stability (resistance to change) and resilience (return to the initial state following perturbation) are essential but poorly understood ecological characteristics that can be quantified through longitudinal studies by serial collection of DNA sequence data from the microbiome, perhaps complemented by metabolite and gene expression profiling. For example, performing weekly microbiome profiling of participants before, during and after surgery could help identify whether (and which) microbiome ecological dynamics are linked to response to surgery, complications, and recovery. Similarly, understanding the resistance and resilience of the microbiome to antibiotics requires larger-scale longitudinal studies of diverse cohorts (Figure 3). This is especially relevant in childhood, when the microbiome is in flux and may be less resistant, but more resilient to these stresses.
Figure 3.
Towards further understanding and developing therapies from microbiome data. The iterative cycle of analysis, interpretation and translational intervention that facilitate moving microbiome research out of correlative observation and into therapeutic treatments is shown.
As we move forward with transforming microbiome research from a descriptive to causal, and finally translational science, the ability to define biomarkers that can stratify patient populations within a disease state represents ‘low-hanging fruit’ (Box 2). Of course, the effort required to take advantage of these biomarkers is considerable. Clinical studies that recruit large and representative patient populations to examine the response to a new drug or therapeutic intervention should definitely consider the opportunity to collect data on both immune function and the microbiome. These additional variables may lead to new non-invasive diagnostic platforms. In the future, it may be possible to request a stool or vaginal sample, or even an saliva sample (which has been shown to yield effective microbial biomarkers for diseases not centered on the mouth such as rheumatoid arthritis79 (Box 2)) from a patient prior to a surgical intervention. Then, along with their genome and medical history, scientists could make a more accurate prediction about the likelihood of successful outcome and/or of complications for each proposed intervention. This additional information, if presented in a sufficiently clear format, would substantially aid clinicians by providing new data layers that enrich the decision-making process. To realize this vision, we must better understand the factors that influence the microbiome of a healthy individual, and how the microbiome is reshaped during different health and disease states.
Concluding Remarks
Microbiome analysis, and so-called Microbiome Wide Association Studies (MWAS)10, are revolutionizing clinical investigations by providing greater patient stratification and new biomarkers of disease. We are poised to make great advances in patient care over the next decade as we improve our ability to characterize and manipulate the microbiome and its metabolism. The omics tools available to perform this characterization have been developed independently, but now there is an ongoing concerted effort80,81 to better standardize and integrate methods and data resources to improve our ability to understand microbial dynamics in human systems. Systems microbiome medicine approaches are rapidly finding their way into clinical investigations, and this is producing a need to integrate traditional clinical statistics and epidemiology with microbial ecological statistics and theory. While these two concepts are not mutually exclusive, they are often treated as such; a new breed of data scientist is required as early-career clinician-scientists develop their new skills in this rapidly expanding field. This in turn increases the likelihood that patient cohort studies will be integrated with animal investigations that enable more accurate interpretation of observed host-microbiome traits.
It is a brave new world, where ecologists and data scientists are being integrated into clinical departments, but this paradigm shift is a necessary precondition to realize the potential of microbiome-informed and microbiome-based medicine. The societal need for improved medical interventions and preventive strategies is driving a sea-change in both the clinical and commercial world. The onus is on the basic and clinical translational research community to ensure that our experimental designs are robust and can deliver on the promises of this field. Just as important are the technical advances that must occur to ensure that we have the tools to derive the data to test our hypotheses. The microbial ecology community came together in 2015–2016 to support the proposal for a National Microbiome Initiative, which was in turn supported by the United States President’s Office of Science and Technology Policy82; one of the key outcomes of this effort was the identification of gaps in our technologies that would need to be filled to realize the full potential of microbiome science83. We have a long way to go, but with each new investigation we are moving closer to the realization of more effective diagnosis, treatment, and preventive modalities to improve human wellness and fight disease.
Acknowledgements
Many of the studies described here in our laboratories were supported by the NIH, NSF, DOE, and the Alfred P. Sloan Foundation. We thank numerous members of our laboratories for constructive criticism on drafts of this article.
References
- 1.Sender R, Fuchs S. & Milo R. Are We Really Vastly Outnumbered? Revisiting the Ratio of Bacterial to Host Cells in Humans. Cell 164, 337–340 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Turnbaugh PJ et al. The human microbiome project. Nature 449, 804–810 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Locey KJ & Lennon JT Scaling laws predict global microbial diversity. Proc. Natl. Acad. Sci. U. S. A 113, 5970–5975 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frank DN et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. U. S. A 104, 13780–13785 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gevers D. et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe 15, 382–392 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ni J. et al. A role for bacterial urease in gut dysbiosis and Crohn’s disease. Sci. Transl. Med 9, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kostic AD et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 14, 207–215 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang H. et al. Altered fecal microbiota composition in patients with major depressive disorder. Brain. Behav. Immun 48, 186–194 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Zheng P. et al. Gut microbiome remodeling induces depressive-like behaviors through a pathway mediated by the host’s metabolism. Mol. Psychiatry 21, 786–796 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Gilbert JA et al. Microbiome-wide association studies link dynamic microbial consortia to disease. Nature 535, 94–103 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Punt CJA, Koopman M. & Vermeulen L. From tumour heterogeneity to advances in precision treatment of colorectal cancer. Nat. Rev. Clin. Oncol 14, 235–246 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Debelius J. et al. Tiny microbes, enormous impacts: what matters in gut microbiome studies? Genome Biol. 17, 217 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Integrative HMP (iHMP) Research Network Consortium. The Integrative Human Microbiome Project: dynamic analysis of microbiome-host omics profiles during periods of human health and disease. Cell Host Microbe 16, 276–289 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lax S. et al. Longitudinal analysis of microbial interaction between humans and the indoor environment. Science 345, 1048–1052 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goodrich JK et al. Human Genetics Shape the Gut Microbiome. Cell 159, 789–799 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ridaura VK et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 341, 1241214 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Turnbaugh PJ et al. Organismal, genetic, and transcriptional variation in the deeply sequenced gut microbiomes of identical twins. Proc. Natl. Acad. Sci. U. S. A 107, 7503–7508 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turnbaugh PJ et al. A core gut microbiome in obese and lean twins. Nature 457, 480–484 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seedorf H. et al. Bacteria from diverse habitats colonize and compete in the mouse gut. Cell 159, 253–266 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karczewski J, Poniedziałek B, Adamski Z. & Rzymski P. The effects of the microbiota on the host immune system. Autoimmunity 47, 494–504 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Schirmer M. et al. Linking the Human Gut Microbiome to Inflammatory Cytokine Production Capacity. Cell 167, 1897 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK & Knight R. Diversity, stability and resilience of the human gut microbiota. Nature 489, 220–230 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O’Toole PW Changes in the intestinal microbiota from adulthood through to old age. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis 18 Suppl 4, 44–46 (2012). [DOI] [PubMed] [Google Scholar]
- 24.Koenig JE et al. Succession of microbial consortia in the developing infant gut microbiome. Proc. Natl. Acad. Sci. U. S. A 108 Suppl 1, 4578–4585 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weng M. & Walker WA The role of gut microbiota in programming the immune phenotype. J. Dev. Orig. Health Dis 4, 203–214 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maynard CL, Elson CO, Hatton RD & Weaver CT Reciprocal interactions of the intestinal microbiota and immune system. Nature 489, 231–241 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knights D. et al. Rethinking ‘enterotypes’. Cell Host Microbe 16, 433–437 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jeffery IB, Claesson MJ, O’Toole PW & Shanahan F. Categorization of the gut microbiota: enterotypes or gradients? Nat. Rev. Microbiol 10, 591–592 (2012). [DOI] [PubMed] [Google Scholar]
- 29.Grice EA & Segre JA The skin microbiome. Nat. Rev. Microbiol 9, 244–253 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grice EA et al. Topographical and temporal diversity of the human skin microbiome. Science 324, 1190–1192 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Caporaso JG et al. Moving pictures of the human microbiome. Genome Biol 12, R50 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kort R. et al. Shaping the oral microbiota through intimate kissing. Microbiome 2, 41 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lazarevic V, Whiteson K, Hernandez D, François P. & Schrenzel J. Study of inter- and intra-individual variations in the salivary microbiota. BMC Genomics 11, 523 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.David LA et al. Host lifestyle affects human microbiota on daily timescales. Genome Biol. 15, R89 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.David LA et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maier TV et al. Impact of Dietary Resistant Starch on the Human Gut Microbiome, Metaproteome, and Metabolome. mBio 8, e01343–17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hannigan GD et al. The human skin double-stranded DNA virome: topographical and temporal diversity, genetic enrichment, and dynamic associations with the host microbiome. mBio 6, e01578–01515 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vandeputte D. et al. Quantitative microbiome profiling links gut community variation to microbial load. Nature 551, 507–511 (2017). [DOI] [PubMed] [Google Scholar]
- 39.Vandeputte D. et al. Stool consistency is strongly associated with gut microbiota richness and composition, enterotypes and bacterial growth rates. Gut 65, 57–62 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma B, Forney LJ & Ravel J. Vaginal microbiome: rethinking health and disease. Annu. Rev. Microbiol 66, 371–389 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ravel J. et al. Daily temporal dynamics of vaginal microbiota before, during and after episodes of bacterial vaginosis. Microbiome 1, 29 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Romero R. et al. The composition and stability of the vaginal microbiota of normal pregnant women is different from that of non-pregnant women. Microbiome 2, 4 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xiao B. et al. Predictive value of the composition of the vaginal microbiota in bacterial vaginosis, a dynamic study to identify recurrence-related flora. Sci. Rep 6, 26674 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Albenberg LG & Wu GD Diet and the intestinal microbiome: associations, functions, and implications for health and disease. Gastroenterology 146, 1564–1572 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu GD et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 334, 105–108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zeevi D. et al. Personalized Nutrition by Prediction of Glycemic Responses. Cell 163, 1079–1094 (2015). [DOI] [PubMed] [Google Scholar]
- 47.Zhang C. et al. Dietary Modulation of Gut Microbiota Contributes to Alleviation of Both Genetic and Simple Obesity in Children. EBioMedicine 2, 968–984 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Modi SR, Collins JJ & Relman DA Antibiotics and the gut microbiota. J. Clin. Invest 124, 4212–4218 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dethlefsen L. & Relman DA Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc. Natl. Acad. Sci. U. S. A 108 Suppl 1, 4554–4561 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maurice CF, Haiser HJ & Turnbaugh PJ Xenobiotics shape the physiology and gene expression of the active human gut microbiome. Cell 152, 39–50 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trasande L. et al. Infant antibiotic exposures and early-life body mass. Int. J. Obes 2005 37, 16–23 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Song SJ et al. Cohabiting family members share microbiota with one another and with their dogs. eLife 2, e00458 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.von Mutius E. The microbial environment and its influence on asthma prevention in early life. J. Allergy Clin. Immunol 137, 680–689 (2016). [DOI] [PubMed] [Google Scholar]
- 54.Stein MM et al. Innate Immunity and Asthma Risk in Amish and Hutterite Farm Children. N. Engl. J. Med 375, 411–421 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cook MD et al. Exercise and gut immune function: evidence of alterations in colon immune cell homeostasis and microbiome characteristics with exercise training. Immunol. Cell Biol 94, 158–163 (2016). [DOI] [PubMed] [Google Scholar]
- 56.Benedict C. et al. Gut microbiota and glucometabolic alterations in response to recurrent partial sleep deprivation in normal-weight young individuals. Mol. Metab 5, 1175–1186 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Karl JP et al. Changes in intestinal microbiota composition and metabolism coincide with increased intestinal permeability in young adults under prolonged physiological stress. Am. J. Physiol. Gastrointest. Liver Physiol 312, G559–G571 (2017). [DOI] [PubMed] [Google Scholar]
- 58.Ying S. et al. The Influence of Age and Gender on Skin-Associated Microbial Communities in Urban and Rural Human Populations. PloS One 10, e0141842 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zheng W. et al. Metagenomic sequencing reveals altered metabolic pathways in the oral microbiota of sailors during a long sea voyage. Sci. Rep 5, 9131 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zozaya M. et al. Bacterial communities in penile skin, male urethra, and vaginas of heterosexual couples with and without bacterial vaginosis. Microbiome 4, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fei N. & Zhao L. An opportunistic pathogen isolated from the gut of an obese human causes obesity in germfree mice. ISME J. 7, 880–884 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Leone V. et al. Effects of Diurnal Variation of Gut Microbes and High-Fat Feeding on Host Circadian Clock Function and Metabolism. Cell Host Microbe 17, 681–689 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Knight R. et al. Unlocking the potential of metagenomics through replicated experimental design. Nat. Biotechnol 30, 513–520 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fierer N. et al. Forensic identification using skin bacterial communities. Proc. Natl. Acad. Sci. U. S. A 107, 6477–6481 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Flores GE et al. Temporal variability is a personalized feature of the human microbiome. Genome Biol. 15, 531 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gajer P. et al. Temporal dynamics of the human vaginal microbiota. Sci. Transl. Med 4, 132ra52 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Livanos AE et al. Antibiotic-mediated gut microbiome perturbation accelerates development of type 1 diabetes in mice. Nat. Microbiol 1, 16140 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sugihara G. et al. Detecting Causality in Complex Ecosystems. Science 338, 496–500 (2012). [DOI] [PubMed] [Google Scholar]
- 69.Knights D. et al. Bayesian community-wide culture-independent microbial source tracking. Nat. Methods 8, 761–763 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Larsen PE, Field D. & Gilbert JA Predicting bacterial community assemblages using an artificial neural network approach. Nat. Methods 9, 621–625 (2012). [DOI] [PubMed] [Google Scholar]
- 71.Larsen PE & Dai Y. Metabolome of human gut microbiome is predictive of host dysbiosis. GigaScience 4, 42 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Browne HP et al. Culturing of ‘unculturable’ human microbiota reveals novel taxa and extensive sporulation. Nature 533, 543–546 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Geva-Zatorsky N. et al. Mining the Human Gut Microbiota for Immunomodulatory Organisms. Cell 168, 928–943.e11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vatanen T. et al. Variation in Microbiome LPS Immunogenicity Contributes to Autoimmunity in Humans. Cell 165, 842–853 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sivan A. et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 350, 1084–1089 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dubin K. et al. Intestinal microbiome analyses identify melanoma patients at risk for checkpoint-blockade-induced colitis. Nat. Commun 7, 10391 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mueller NT et al. Does vaginal delivery mitigate or strengthen the intergenerational association of overweight and obesity? Findings from the Boston Birth Cohort . Int. J. Obes 2005 41, 497–501 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Raveh-Sadka T. et al. Gut bacteria are rarely shared by co-hospitalized premature infants, regardless of necrotizing enterocolitis development. eLife 4, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang X. et al. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat. Med 21, 895–905 (2015). [DOI] [PubMed] [Google Scholar]
- 80.Sinha R. et al. Assessment of variation in microbial community amplicon sequencing by the Microbiome Quality Control (MBQC) project consortium. Nat. Biotechnol 35, 1077–1086 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Costea PI et al. Towards standards for human fecal sample processing in metagenomic studies. Nat. Biotechnol 35, 1069–1076 (2017). [DOI] [PubMed] [Google Scholar]
- 82.Alivisatos AP et al. A unified initiative to harness Earth’s microbiomes. Science 350, 507–508 (2015). [DOI] [PubMed] [Google Scholar]
- 83.Biteen JS et al. Tools for the Microbiome: Nano and Beyond. ACS Nano 10, 6–37 (2016). [DOI] [PubMed] [Google Scholar]
- 84.Luckey TD Introduction to intestinal microecology. Am. J. Clin. Nutr 25, 1292–1294 (1972). [DOI] [PubMed] [Google Scholar]
- 85.Rosner JL Ten Times More Microbial Cells than Body Cells in Humans? Microbe Mag. 9, 47–47 (2014). [Google Scholar]
- 86.Reyes A. et al. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature 466, 334–338 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Qin J. et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Weingarden A. et al. Dynamic changes in short- and long-term bacterial composition following fecal microbiota transplantation for recurrent Clostridium difficile infection. Microbiome 3, 10 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kassam Z, Lee CH, Yuan Y. & Hunt RH Fecal microbiota transplantation for Clostridium difficile infection: systematic review and meta-analysis. Am. J. Gastroenterol 108, 500–508 (2013). [DOI] [PubMed] [Google Scholar]
- 90.Knights D, Parfrey LW, Zaneveld J, Lozupone C. & Knight R. Human-associated microbial signatures: examining their predictive value. Cell Host Microbe 10, 292–296 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Walters WA, Xu Z. & Knight R. Meta-analyses of human gut microbes associated with obesity and IBD. FEBS Lett. 588, 4223–4233 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sze MA & Schloss PD Looking for a Signal in the Noise: Revisiting Obesity and the Microbiome. mBio 7, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sahin M. & Sur M. Genes, circuits, and precision therapies for autism and related neurodevelopmental disorders. Science 350, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.McDonald D. et al. Towards large-cohort comparative studies to define the factors influencing the gut microbial community structure of ASD patients. Microb. Ecol. Health Dis 26, 26555 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kang D-W et al. Reduced Incidence of Prevotella and Other Fermenters in Intestinal Microflora of Autistic Children. PLoS ONE 8, e68322 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hsiao EY et al. Microbiota Modulate Behavioral and Physiological Abnormalities Associated with Neurodevelopmental Disorders. Cell 155, 1451–1463 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kang D-W et al. Microbiota Transfer Therapy alters gut ecosystem and improves gastrointestinal and autism symptoms: an open-label study. Microbiome 5, 10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Snijders AM et al. Influence of early life exposure, host genetics and diet on the mouse gut microbiome and metabolome. Nat. Microbiol 2, 16221 (2016). [DOI] [PubMed] [Google Scholar]
- 99.Knights D. et al. Complex host genetics influence the microbiome in inflammatory bowel disease. Genome Med. 6, 107 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Halfvarson J. et al. Dynamics of the human gut microbiome in inflammatory bowel disease. Nat. Microbiol 2, 17004 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Uusitalo U. et al. Association of Early Exposure of Probiotics and Islet Autoimmunity in the TEDDY Study. JAMA Pediatr. 170, 20–28 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Blaser MJ The theory of disappearing microbiota and the epidemics of chronic diseases. Nat. Rev. Immunol 17, 461–463 (2017). [DOI] [PubMed] [Google Scholar]
- 103.Arrieta M-C et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci. Transl. Med 7, 307ra152 (2015). [DOI] [PubMed] [Google Scholar]