Escherichia coli cells divide using a cytokinetic ring composed of polymers of the tubulin-like FtsZ. To function properly, these polymers must attach to the inner surface of the cytoplasmic membrane via two essential membrane-associated tethers, FtsA and ZipA. Both FtsA and ZipA contain peptide linkers that connect their membrane-binding domains with their FtsZ-binding domains. Although they are presumed to be crucial for cell division activity, the importance of these linkers has not yet been rigorously tested. Here, we show that large segments of these linkers can be removed with few consequences for cell division, although several subtle defects were uncovered. Our results suggest that ZipA, in particular, can function in cell division without an extended linker.
KEYWORDS: Escherichia coli, FtsA, FtsZ, ZipA, cell division
ABSTRACT
Bacteria such as Escherichia coli divide by organizing filaments of FtsZ, a tubulin homolog that assembles into dynamic treadmilling membrane-associated protein filaments at the cell midpoint. FtsA and ZipA proteins are required to tether these filaments to the inner face of the cytoplasmic membrane, and loss of either tether is lethal. ZipA from E. coli and other closely related species harbors a long linker region that connects the essential N-terminal transmembrane domain to the C-terminal globular FtsZ-binding domain, and part of this linker includes a P/Q-rich peptide that is predicted to be intrinsically disordered. We found unexpectedly that several large deletions of the ZipA linker region, including the entire P/Q rich peptide, had no effect on cell division under normal conditions. However, we found that the loss of the P/Q region made cells more resistant to excess levels of FtsA and more sensitive to conditions that displaced FtsA from FtsZ. FtsA also harbors a short ∼20-residue peptide linker that connects the main globular domain with the C-terminal amphipathic helix that is important for membrane binding. In analogy with ZipA, deletion of 11 of the central residues in the FtsA linker had little effect on FtsA function in cell division.
IMPORTANCE Escherichia coli cells divide using a cytokinetic ring composed of polymers of the tubulin-like FtsZ. To function properly, these polymers must attach to the inner surface of the cytoplasmic membrane via two essential membrane-associated tethers, FtsA and ZipA. Both FtsA and ZipA contain peptide linkers that connect their membrane-binding domains with their FtsZ-binding domains. Although they are presumed to be crucial for cell division activity, the importance of these linkers has not yet been rigorously tested. Here, we show that large segments of these linkers can be removed with few consequences for cell division, although several subtle defects were uncovered. Our results suggest that ZipA, in particular, can function in cell division without an extended linker.
INTRODUCTION
Escherichia coli cells divide using a large protein complex called the divisome (1). The divisome contains numerous proteins, 10 of which are essential (FtsZ, FtsA, ZipA, FtsK, FtsQ, FtsL, FtsB, FtsW, FtsI, and FtsN) (2). Assembly of the divisome begins with the accumulation of FtsZ at the future division site to form a Z ring. Cytoplasmic FtsZ polymers, which move dynamically by treadmilling (3, 4), are tethered to the inner membrane by two proteins, FtsA and ZipA (5, 6) (Fig. 1A). These proteins together form an early divisome structure specifically referred to as the protoring (7). While either membrane anchor is able to tether FtsZ polymers on its own (5), both FtsA and ZipA are required for formation of the protoring and the continued assembly of the divisome. The remainder of the Fts proteins and other nonessential divisome components (collectively known as downstream division proteins) are recruited to the protoring and septal peptidoglycan synthesis is activated, completing division (1, 2).
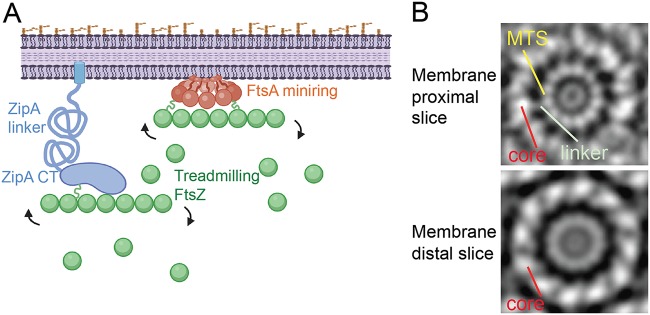
FIG 1.
Overview of potential protoring protein interactions. (A) ZipA, with its N-terminal transmembrane domain, unstructured linker domain, and C-terminal (CT) FtsZ-binding domain, is shown attached to an FtsZ protofilament undergoing subunit treadmilling. FtsZ has its own unstructured linker, shown as a green squiggle. FtsA, tethered to the cytoplasmic membrane with its C-terminal amphipathic helix and a putative linker domain (shown as an orange squiggle), forms oligomeric minirings (shown here) that, like ZipA, bind to treadmilling FtsZ protofilaments. The figure was created with Biorender. (B) An FtsA miniring on a lipid monolayer, shown as two tomographic slices from averaged negative-stain electron micrograph images. (Republished from Nature Communications [12].)
Recent studies suggest a model in which the protoring proteins regulate the transition from early to late division by acting on the oligomerization states of each other (8, 9). Genetic experiments suggest that disruption of the FtsA oligomeric state is important for the recruitment of downstream proteins and activation of division (10, 11). In support of this model, experiments with purified FtsA on lipid monolayers demonstrated that it assembles into dodecameric minirings (12). It has been proposed that these closed FtsA minirings act as a divisome checkpoint (9), occluding one or more binding sites for the downstream divisome protein FtsN (13, 14). According to this model, the checkpoint is relieved when the closed FtsA miniring is converted to an open oligomer with free termini, which allow FtsA-FtsN interactions that in turn promote divisome activation (9, 11). ZipA may play a role in this conversion, as it was shown recently to interact directly with FtsA in vivo (15). In support of this potential activation role for ZipA, FtsA* gain-of-function mutants can bypass the requirement for ZipA (10, 16), and when assembled on lipids, purified FtsA* proteins form open oligomers with free termini (9, 12, 17). In addition to its roles in FtsZ membrane tethering and perhaps in FtsA miniring disruption, ZipA is also important for activating synthesis of preseptal peptidoglycan by one of the class A penicillin-binding proteins (PBPs) in E. coli (18, 19). A recently published study proposed overlapping roles for ZipA and FtsN in linking the periplasmic peptidoglycan synthases PBP1A and PBP1B to the cytoplasmic Z ring (19).
All three protoring proteins, despite being functionally and structurally distinct, share one similar structural feature: a peptide linker predicted to be intrinsically disordered (Fig. 1A). E. coli FtsZ contains an ∼50-amino-acid peptide that links the N-terminal core globular domain with the C-terminal tail (20). FtsA and ZipA, as well as several other proteins, compete for binding to the conserved C-terminal tail of FtsZ (21). Several studies examined the effects of alterations of the FtsZ linker in different bacterial species (20, 22, 23). Surprisingly, the FtsZ linker is amenable to significant changes in length and charge, but it must remain flexible for proper function in cell division (24). This led to a model in which the constriction force exerted by FtsZ polymers is transmitted to the inner membrane through its unstructured linker as a signal for the divisome to activate septal peptidoglycan synthesis (24). Of course, any constriction transmitted to the membrane from the Z ring must also pass through the membrane tethers FtsA and ZipA, which prompted us to investigate the in vivo role of their peptide linkers.
FtsA harbors an ∼20-residue amino acid linker that connects its core globular domain, which binds to FtsZ (25), to its C-terminal amphipathic helix, which acts as a membrane-targeting sequence (MTS) required for FtsA function (6). Though short, the FtsA linker is predicted to have unstructured characteristics (see Fig. S1 in the supplemental material). In a subvolume-averaged structure of an FtsA miniring from a tomographic slice close to the membrane, each linker is visible as one of 12 spokes connecting an inner ring comprising 12 MTS domains and an outer ring comprising the 12 core domains of FtsA (Fig. 1B) (12). In a membrane-distal tomographic slice, the MTS and linkers are less prominent than the core (Fig. 1B), suggesting that the linkers allow the core domains to extend inward from the cytoplasmic membrane (Fig. 1A). In contrast to the model for FtsZ linker function, the short length of the FtsA linker may serve a structural role to maintain the integrity of the miniring structure, but no studies to date have examined this hypothesis. It is notable that FtsA proteins from Streptococcus pneumoniae and Thermotoga maritima form filaments on lipids instead of minirings (26, 27), and at least the linker region of S. pneumoniae FtsA is much longer than that of E. coli FtsA.
ZipA contains a much larger peptide linker, roughly 150 amino acids in length, consisting of a highly charged region followed by a P/Q-rich domain (28). This linker region is largely a disordered peptide and has been used as a tool for biophysical methods as verification for the behavior of unstructured peptides (29–31). Unlike the widely conserved FtsZ and FtsA, ZipA is confined to the gammaproteobacteria (1). Although the specific sequences of the ZipA linkers from different species are not well conserved, the various ZipA proteins share overall domain structure (see Fig. S2 in the supplemental material). The biological function of the ZipA linker in cell division has not yet been studied.
Here, we set out to examine the functional contributions of the FtsA and ZipA disordered peptide linkers in cell division. We were surprised to see that large portions of both the FtsA and ZipA linkers could be removed with no significant deleterious effects on cell viability. Focusing mainly on the ZipA linker, we stressed cells under several different conditions to determine the fitness cost of the linker deletions and found that altering FtsA activity revealed some effects of the linker deletions on cell viability.
RESULTS AND DISCUSSION
FtsA can tolerate deletions of up to 11 residues in its linker.
To investigate whether the postulated ∼20-residue linker is important for the function of FtsA, we constructed a library of deletions ranging from 1 to 8 amino acids. We chose to delete residues starting from the middle of the putative linker, reasoning that this would be less likely to perturb important residues within the N-terminal globular domain or the MTS (Fig. 2A). After verifying the linker deletion mutants by sequencing, we expressed them from the weakened isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible trc promoter in plasmid pDSW210F in a strain where the chromosomal copy of ftsA was replaced with the temperature-sensitive allele ftsA12. Promoter leakage from this plasmid normally allows a FLAG-tagged wild-type (WT) FtsA to complement ftsA12 without IPTG induction, whereas overproduction of FtsA by induction with 50 to 100 μM IPTG is toxic (Fig. 2B).
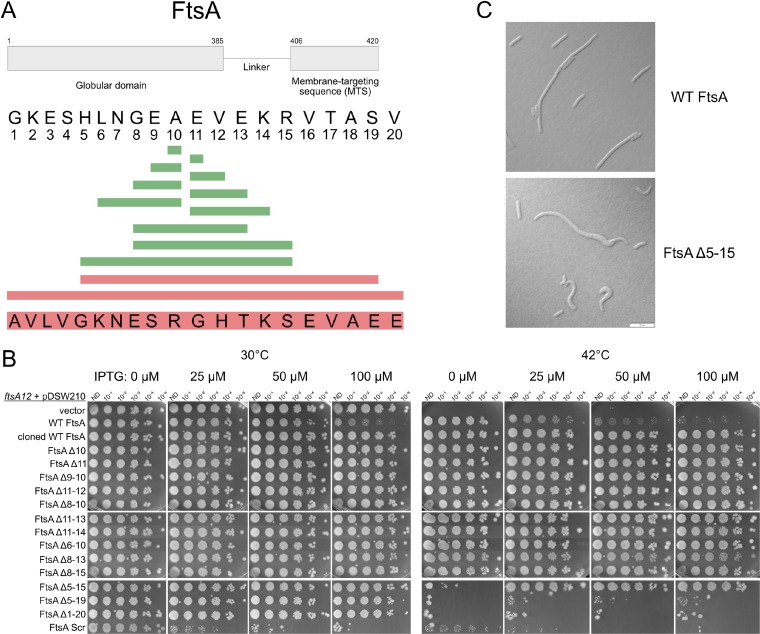
FIG 2.
Effects of deletions within the 20-residue region linking the core oligomeric and amphipathic helix domains of FtsA. (A) Schematic showing the amino acid sequence of the linker and its context within the entire FtsA protein (top), along with the residues deleted that had no effect on viability (green) or that abolished viability (red). The scrambled sequence that replaced the WT sequence is shown at the bottom. (B) After growth at 30°C, ftsA12(Ts) mutant strains containing the pDSW210F plasmid derivatives shown were serially diluted 10-fold and spotted on plates at either the permissive (30°C) or nonpermissive (42°C) temperature with the indicated amounts of IPTG and incubated overnight. The row labeled “cloned WT FtsA” represents a positive-control construct in which the WT linker sequence was cloned into a vector by the same method by which the smaller linker deletions were cloned. (C) Representative micrographs of ftsA null mutant cells harboring pDSW210F expressing either WT FtsA or FtsAΔ5–15 grown at 30°C and induced with 100 μM IPTG, highlighting aberrant cell morphologies. Scale bar, 10 μm.
We assessed growth of the mutants on agar plates at 42°C with increasing levels of IPTG. We anticipated that the FtsA linker might be able to tolerate small deletions, and indeed, FtsA with as many as 8 residues in the middle of the linker deleted (FtsAΔ8–15) conferred full viability at 42°C, even with no IPTG induction (Fig. 2B). The plasmid carrying the recombinant FtsA likely has a promoter mutation preventing toxicity with full IPTG induction (Fig. 2B, row 3), because none of these smaller deletions derived from this plasmid (Fig. 2B, top two sets of spots) were toxic when induced with 100 μM IPTG. We were surprised to find that deletion of over half of the 20-amino-acid linker (FtsAΔ5–15) did not significantly affect cell viability (Fig. 2B), although it did require some IPTG induction for growth at 42°C.
At this point we wondered whether this linker was needed at all, but larger deletions, removing residues 5 to 19 or the entire linker, abolished viability (Fig. 2B, bottom set of spots). The ability to delete up to 11 residues from the heart of this domain strongly suggested that FtsA does not need its linker to be a specific length, but to bolster this idea, we replaced the 20-residue linker with a peptide carrying the same residues, but in random order, and found that including this scrambled linker abolished viability (Fig. 2B, bottom row), although instability of this chimeric protein at 42°C may be the reason for the inactivity (see Fig. S3 in the supplemental material). All of the other linker deletions were stably expressed from the vector at the permissive and nonpermissive temperatures, comparable to the case for WT FtsA (Fig. S3).
Although the cells expressing the viable linker deletions could clearly form colonies, we investigated whether there were any subtle effects on cell morphology that would not be detectable on spot plates. In log-phase culture under conditions of viability on plates, cells expressing all of the functional linker deletions had WT-like morphology (data not shown), with the exception of cells overexpressing the FtsAΔ5–15 mutant. These, particularly when the chromosomal ftsA gene was inactivated by transducing in an ftsA null allele linked to leuO::Tn10, showed morphological defects similar to those resulting from expression of FtsA mutants deficient in binding the cell membrane, including cell curving and bulging (Fig. 2C) (6, 32, 33). Several filamentous cells overexpressing WT FtsA are depicted in Fig. 2C to emphasize that even though these cells are much longer than normal, they still retain their rod shape and do not curve or bulge like the cells expressing the FtsAΔ5–15 mutant. This indicated that while still functional enough to support cell growth in rich medium, the FtsAΔ5–15 mutant may bind to the membrane less efficiently than WT FtsA. Nonetheless, our overall data show that the heart of the ∼20-amino-acid linker that connects the MTS with the core region of FtsA is largely dispensable for FtsA function in cell division.
The unstructured ZipA linker domain is dispensable.
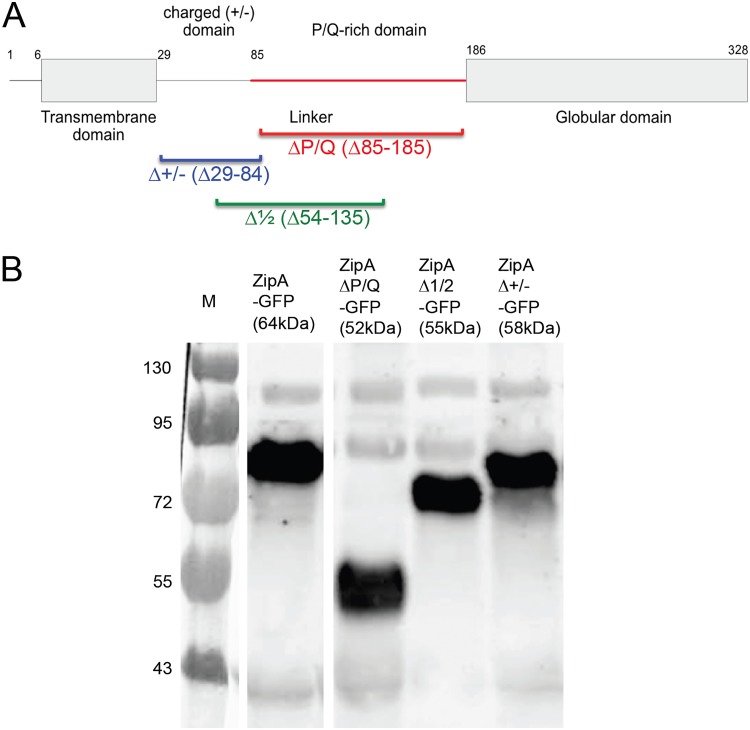
We next asked whether ZipA could similarly tolerate large deletions in its linker regions. We defined the ZipA linker and domain boundaries based on a previous study that found much of the linker to be unstructured (29) (Fig. S1). As a first step, large deletions in a functional ZipA-green fluorescent protein (GFP) fusion expressed from pDSW210 were made to remove the entire membrane-proximal charged domain (ZipAΔ+/−), the P/Q-rich domain (ZipAΔP/Q), or half of the linker (ZipAΔ1/2) (Fig. 3A). All linker deletions were stably expressed from the pDSW210 plasmid, with levels comparable to that for WT ZipA (see Fig. S4 in the supplemental material). It is known that full-length ZipA migrates aberrantly slowly in SDS-PAGE (28), running at ∼50 kDa instead of the predicted 37 kDa. We found that GFP fusions to ZipAΔ+/− and ZipAΔ1/2 also migrated more slowly than their predicted sizes of 58 and 55 kDa, respectively, but removal of the entire P/Q-rich domain restored migration to the predicted size of 52 kDa (Fig. 3B and S4). This result suggests that the P/Q domain is responsible for an unusual structure that persists even in the presence of SDS and that the loss of this domain might have significant physiological consequences.
FIG 3.
ZipA and the three deletions of its unstructured linker domain used in this study. (A) Schematic of ZipA domains, showing the extents of the linker deletions. (B) GFP fusions of ZipA, ZipAΔP/Q, ZipAΔ1/2, and ZipAΔ+/− expressed from pDSW210 were induced with 100 μM IPTG at 30°C, and proteins subjected to SDS-PAGE were probed on Western blots with anti-GFP. Molecular weight markers in kilodaltons are shown at the left. The expected size of each fusion protein is denoted in parentheses, with GFP at 27 kDa. All but ZipAΔP/Q-GFP run abnormally slowly, ∼16 kDa larger than predicted. All lanes were derived from the same blot but were spliced as shown.
To test for physiological effects, we transformed pDSW210 derivatives carrying ZipA and the linker deletion derivatives (fused to GFP at their C termini) into the thermosensitive zipA1 background to assess their ability to support cell growth at temperatures nonpermissive for zipA1 growth. As reported previously (34), cells with the zipA1 allele grow and divide at 30°C but are inviable at 34°C or higher (Fig. 4). However, the ZipA1 protein is likely partially functional at 34°C or 37°C (but not at 42°C), because other alleles, such as specific deletions of amino acid biosynthetic genes, can rescue zipA1 mutants at 34°C or 37°C (34).
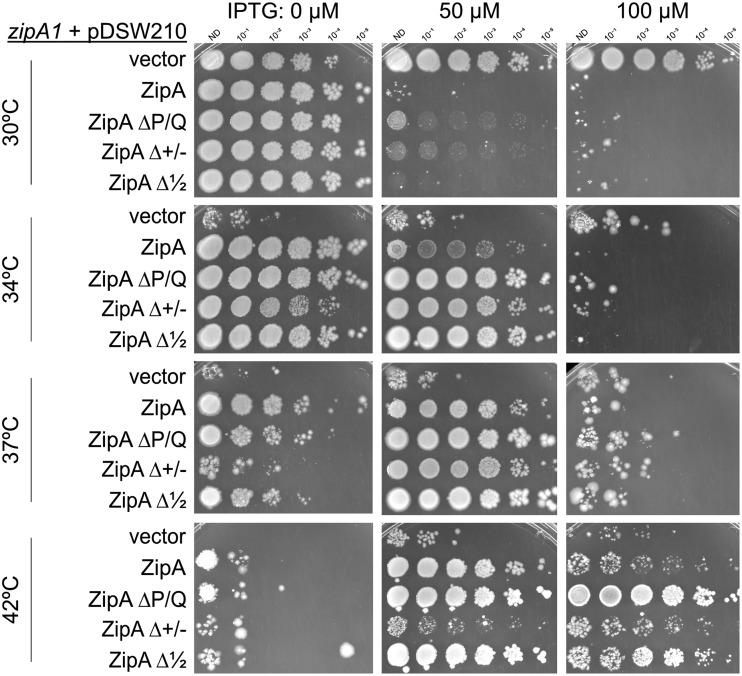
FIG 4.
Deletions of the P/Q domain (ZipAΔP/Q) or half of the ZipA linker (ZipAΔ1/2) fully suppress the thermosensitivity of the zipA1 temperature-sensitive mutant. WM 5337 (MG1655 zipA1) plus pDSW210F variants expressing WT ZipA, ZipA ΔP/Q, ZipAΔ+/−, or ZipAΔ1/2 on LB plus ampicillin were serially diluted and spotted onto plates containing various concentrations of IPTG at 30°C, 34°C, 37°C, or 42°C. Although ZipAΔ+/− could suppress zipA1 at the partially permissive temperatures of 34°C and 37°C, only ZipA, ZipAΔP/Q, and ZipAΔ1/2 were able to permit full viability at 42°C.
Surprisingly, ZipAΔP/Q or ZipAΔ1/2 allowed normal viability of zipA1 cells at any nonpermissive temperature, including 42°C (Fig. 4). Some IPTG induction was required for full viability at higher temperatures, although the reason for this is unclear, as the ZipA derivatives themselves were stable at 42°C (Fig. S4). ZipAΔ+/−, on the other hand, did not confer viability on zipA1 cells at 42°C, although it allowed viability at 34°C or 37°C, indicating that it retained some partial functionality (Fig. 4). Consistent with these results, cells producing ZipAΔP/Q and ZipAΔ1/2 from plasmids were able to survive transduction with the ΔzipA::kan allele, while ZipAΔ+/− was not able to confer viability (data not shown).
These results confirm that ZipA lacking most of its linker domain can function normally in cell division and indicate that deletion of the charged domain disrupts ZipA function. This disruption could be due to loss of a highly positively charged patch of amino acids in the charged domain immediately following the transmembrane domain, which could function to correctly orient ZipA in the membrane. In the ZipAΔ1/2 mutant, where only the second half of the charged domain is deleted (Fig. 2A), this positive patch is retained and may allow the ZipA transmembrane domain to orient itself properly.
The viability of cells with large deletions within the ZipA linker was unexpected. We surmised that perhaps under the rich growth conditions of the lab, the linker domains are dispensable for growth. The nature of the high-copy-number plasmid used to express the ZipA variants introduces heterogeneity in the cell population, where levels of the proteins in individual cells can vary widely with the number of plasmids per cell. We therefore decided that the best approach to detect potentially subtle changes in phenotype was to move the linker deletion mutants into their native locus on the chromosome.
We used the λRed system to recombineer the alleles into the chromosome (35), selecting either by conversion of the thermosensitive zipA1 allele to thermoresistance or by using ccd::kan-mediated selection-counterselection (see Materials and Methods). As insertion of the ccd::kan cassette disrupts the zipA reading frame in the second method, we used a host recombineering strain containing the ftsA* allele (WM2530), which permits growth and division even if ZipA is deleted completely. After confirming the recombinants by sequencing, the chromosomal linker mutant alleles were transferred to a WT (ftsA+) strain background by P1 transduction of a linked marker and confirmed again to be intact. Consistent with the data from plasmid-borne zipA alleles in zipA mutants, strains carrying zipAΔP/Q or zipAΔ1/2 in place of chromosomal WT zipA grew and divided normally at 30°C or 37°C (Fig. 5), with only a very slight increase in filamentous cells (see Fig. S5 in the supplemental material), confirming our conclusion that much of the ZipA linker is completely dispensable. To address the question of whether the unstructured linker of ZipA confers any fitness advantage, we used these engineered chromosomal zipA linker alleles to identify conditions in which the loss of the normal linker has a fitness cost.
FIG 5.
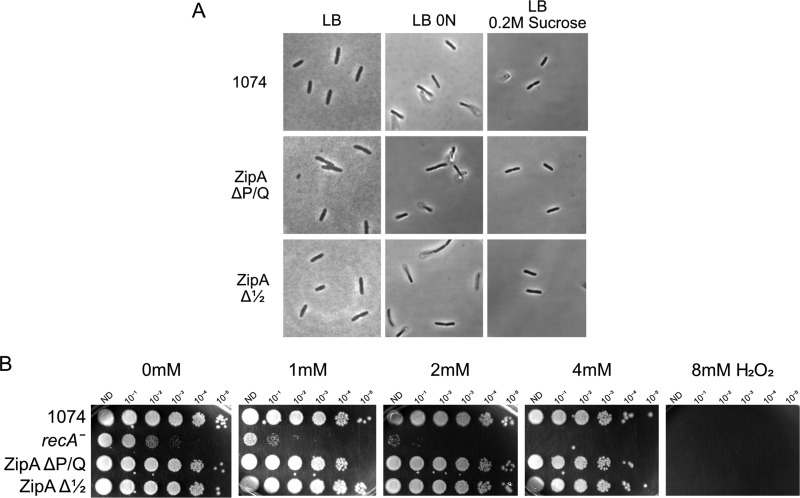
Effects of osmotic or oxidative stress on the ZipA linker mutants. (A) Cells carrying either WT zipA, zipAΔP/Q, or zipAΔ1/2 at the native zipA chromosomal locus were grown to mid-exponential phase in regular LB (0.5% NaCl), LB with no added NaCl (LB 0N), or LB with 0.2 M sucrose and imaged by DIC microscopy. (B) The same strains used for panel A, along with a recA mutant control, were grown in LB supplemented with the indicated concentrations of hydrogen peroxide for 2.5 h at 30°C, and then 10-fold dilutions were spotted onto LB plates and incubated overnight at 30°C.
Deletion of the ZipA P/Q domain has no effect under osmotic or oxidative stress conditions.
The sensitivity of several fts cell division mutants to low-salt conditions (36) prompted us to test whether cells with ZipA linker deletions would grow more poorly in medium with no added NaCl. However, the growth and division of strains with chromosomal zipAΔP/Q or zipAΔ1/2 were virtually indistinguishable from those of the zipA+ strain WM1074 (Fig. 5A), again with only a very slight increase in filamentous cells (Fig. S5). Growth in hypertonic medium containing 0.2 M sucrose yielded similar results (this time, WM1074 cells included a few filamentous cells compared with the mutants), suggesting that deletions of the ZipA linker do not sensitize cells to extremes in osmality.
The divisome is also known to be sensitive to oxidative stress (37). To test whether ZipA linker deletions make the divisome more sensitive to oxidative stress, we subjected the chromosomal linker mutants to various levels of H2O2 for 2 h in liquid culture and then assessed their viabilities on agar plates. As expected, cells lacking a functional recA gene were hypersensitive to peroxide stress (38), with ∼100-fold killing in 1 mM peroxide compared with no peroxide and zero viability in 4 mM peroxide (Fig. 5B). However, cells carrying either WT zipA, zipAΔP/Q, or zipAΔ1/2 were equally viable in 1 to 4 mM peroxide, and all were killed when subjected to 8 mM peroxide. Therefore, we conclude that the loss of most of the ZipA linker domains has no significant effect on the sensitivity or resistance to oxidative stress.
Loss of the ZipA P/Q domain confers resistance to excess FtsA.
As ZipA and FtsA both bind to the same short conserved C-terminal peptide of FtsZ and are required to tether FtsZ to the membrane, we asked whether deletions of the ZipA linker might be affected when FtsA levels are perturbed. It is well known that overproduction of FtsA strongly inhibits cell division, but the inhibition can be suppressed by increasing levels of FtsZ (39, 40) or by the hypermorphic allele FtsZ* (L169R), which promotes lateral interactions between FtsZ protofilaments (41) and seems to disassemble FtsA minirings on membranes in vitro. As ZipA interacts directly with FtsA in cells (15) but can be bypassed by oligomerization-deficient versions of FtsA (FtsA*) (10, 16), it has been inferred that ZipA itself may help to break up FtsA minirings and activate the divisome (12). FtsA also likely competes with ZipA for binding to FtsZ (17, 42). Therefore, it is likely that excess FtsA is toxic because it displaces ZipA from FtsZ, because it overwhelms the ability of ZipA to disassemble FtsA minirings, or some combination of both.
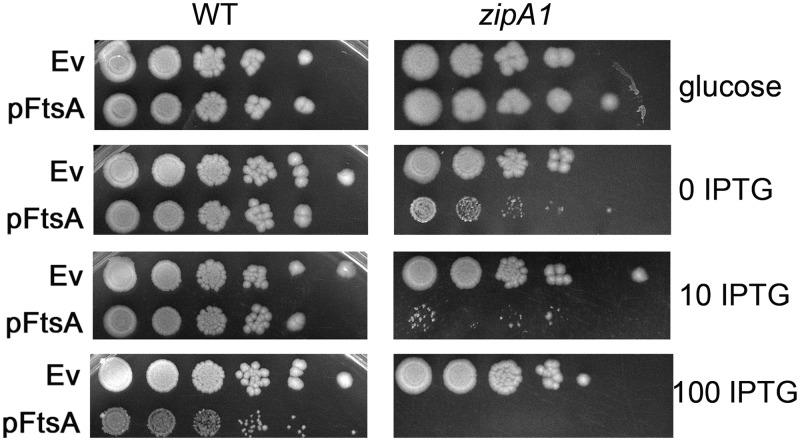
Consistent with these ideas, we found that cells carrying the zipA1 allele are significantly more sensitive to higher FtsA protein levels than cells with WT zipA. Whereas 100 μM IPTG was needed to significantly inhibit growth of WT cells expressing FtsA from pDSW210F, uninduced levels of FtsA expression from the same plasmid severely reduced viability in zipA1 cells, and 10 μM IPTG abolished growth (Fig. 6). Full viability of zipA1 cells carrying pDSW210F-FtsA was achieved only when glucose was added to the growth medium to repress promoter leakage (Fig. 6).
FIG 6.
FtsA is more toxic in zipA1 cells than in WT cells. WM1074 or WM5337 (MG1655 zipA1) cells carrying either pDSW210F empty vector (Ev) or pDSW210F-FtsA under IPTG control were serially diluted 10-fold, spotted on plates containing either glucose (to suppress gene expression) or the indicated amounts of IPTG (in micromolar), and incubated overnight at 30°C.
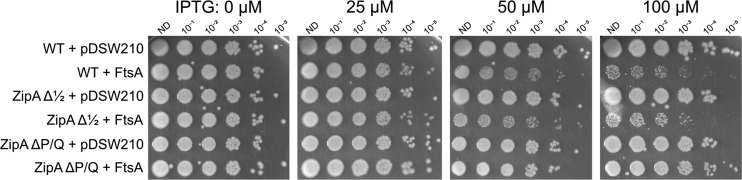
To test whether deletion of ZipA’s linker might alter its ability to resist the toxic effects of excess FtsA, we introduced pDSW210F-FtsA in strains expressing native chromosomal copies of WT zipA, zipAΔ1/2, or zipAΔP/Q. As expected, FtsA was toxic to WT cells at IPTG levels above 50 μM, and the same was observed for cells with ZipAΔ1/2 (Fig. 7). In contrast, ZipAΔP/Q conferred strong resistance to excess FtsA at 50 or 100 μM IPTG (Fig. 7). We observed similar FtsA resistance when both ZipAΔP/Q and FtsA were expressed from compatible plasmids in a zipA1 strain at the 42°C nonpermissive temperature (see Fig. S6 in the supplemental material), indicating that this effect is not a result of a strain-specific suppressor. These results suggest that deletion of the entire P/Q linker alters the ZipA protein’s structure in a way that enhances ZipA binding to FtsZ and more effectively compensates for excess FtsA, potentially by bringing FtsZ protofilaments closer to the membrane. It is notable that the ΔP/Q deletion, but not the Δ1/2 deletion, both confers FtsA resistance and rescues normal protein migration in SDS-PAGE, suggesting that the same structural features enable both effects.
FIG 7.
Overproduced FtsA is less toxic in zipAΔP/Q cells than in WT cells or in cells with zipAΔ1/2. WM1074 (WT) or WM1074 derivatives with chromosomal zipA replaced with either zipAΔP/Q or zipAΔ1/2 carrying either pDSW210F empty vector or pDSW210F-FtsA under IPTG control were serially diluted 10-fold, spotted on plates containing the indicated amounts of IPTG (in micromolar), and incubated overnight at 30°C.
Deletions of the ZipA linker do not significantly suppress or exacerbate other divisome defects.
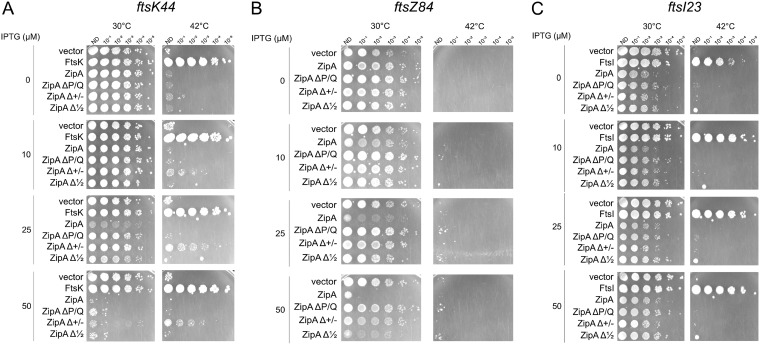
As ZipAΔP/Q could resist excess FtsA, which is a feature of gain-of-function alleles such as the FtsA* and FtsZ* alleles, we asked whether ZipAΔP/Q or ZipAΔ1/2 might be able to suppress the thermosensitivity of fts cell division mutants. We tested ftsK44 and ftsI23 thermosensitive alleles, both of which can be suppressed by FtsA* or FtsZ* (41, 43). However, neither WT ZipA, ZipAΔP/Q, nor ZipAΔ1/2 could rescue viability at the nonpermissive temperature of 42°C (Fig. 8A and C). This suggests that although ZipAΔP/Q can resist FtsA overproduction, it is not able to overcome multiple divisome defects in the way that FtsA* or FtsZ* can. Curiously, production of ZipAΔ+/−, which is largely nonfunctional under normal conditions, modestly suppressed thermosensitivity of ftsK44 at 42°C. How this suppression occurs might be interesting but is presently unclear.
FIG 8.
Neither ZipA nor the linker deletions of ZipA confer significant suppression of several key fts mutants. MG1655 derivatives containing various pDSW210 plasmid derivatives of ZipA or ZipA linker mutants in strains with the ftsK44 (A), ftsZ84 (B), or ftsI23 (C) thermosensitive allele were grown at 30°C, serially diluted 10-fold, and spotted onto plates at the permissive (30°C) or nonpermissive (42°C) temperature with various levels of IPTG to induce the ZipA constructs.
We also compared the abilities of ZipA derivatives to affect viability of cells carrying the thermosensitive ftsZ84 allele, which decreases FtsZ’s GTPase activity and inhibits normal Z-ring dynamics (44–47). Although it was reported that excess ZipA could suppress ftsZ84 (48), we observed no suppression of ftsZ84 by ZipA or any of the three linker deletion mutants at 42°C in no-salt LB medium, which is the most stringent test of ftsZ84 function (Fig. 8B). As in other strain backgrounds, we found that at the permissive temperature, WT ZipA was more toxic to ftsZ84 cells when modestly overproduced at low IPTG induction levels than ZipAΔP/Q or ZipAΔ1/2 at equivalent induction levels.
Loss of the ZipA P/Q domain exacerbates effects of weakening FtsA activity.
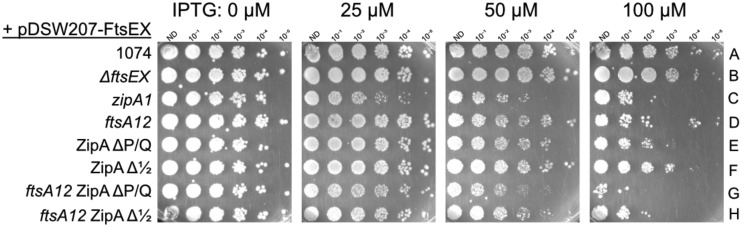
FtsX, helped by its FtsZ-binding partner FtsE, interacts with FtsA to provide a checkpoint for septum formation in response to periplasmic cues (49). When the FtsEX complex is overproduced, the specific interaction between FtsX and FtsA preferentially displaces FtsA from the Z ring, forcing cells to rely on ZipA as the main membrane anchor for FtsZ (42). Although we have shown here that removal of the ZipA linker domains has no detectable deleterious effect on ZipA activity, we reasoned that displacing FtsA from the Z ring might expose defects of the ZipA linker deletions. To test this idea, we expressed FtsEX from pDSW207, which has a stronger pTrc promoter than pDSW210 and fuses GFP to the N terminus of FtsE. These plasmids were introduced into strains carrying either WT zipA or the zipA1, zipAΔP/Q, or zipAΔ1/2 allele. In addition, we also introduced these plasmids into strains carrying ftsA12 zipAΔP/Q or ftsA12 zipAΔ1/2, which might amplify any negative effects.
In spot viability tests at the permissive temperature of 30°C, overproduction of GFP-FtsEX with up to 100 μM IPTG had no detectable toxicity in WT cells (WM1074) or in control cells harboring a deletion of ftsEX (Fig. 9, rows A and B). In contrast, cells with the ftsA12 allele became sensitive to GFP-FtsEX induction at 100 μM IPTG, and cells with the zipA1 allele were still more sensitive, with cells showing a viability decrease even at 50 μM IPTG (Fig. 9, rows C and D). This increased sensitivity to FtsEX levels caused by ftsA12, and further by zipA1, was also observed previously (42).
FIG 9.
Increased sensitivity of the ZipA linker mutants to the effects of excess FtsEX. WT (WM1074) or various mutant derivatives of WM1074 carrying the IPTG-inducible pDSW207-FtsEX plasmid were grown to mid-exponential phase, diluted 10-fold, spotted onto LB plates supplemented with various concentrations of IPTG, and incubated overnight at 30°C.
Most significantly, cells carrying zipAΔP/Q were more sensitive to FtsEX overproduction than WT cells, with a drop in viability nearly as severe as for cells with zipA1 at 50 or 100 μM IPTG (Fig. 9, compare row A with row E). In addition, cells with both ftsA12 and zipAΔP/Q alleles were more sensitive to GFP-FtsEX than cells with the single alleles (compare row D or E with row G), with nearly no viability at 100 μM IPTG. Interestingly, cells with the zipAΔ1/2 allele were also more sensitive to GFP-FtsEX than WT cells, but the effects were much milder than with zipAΔP/Q. This is consistent with the larger ΔP/Q deletion having a more significant effect than the smaller Δ1/2 deletion (compare rows F and H with rows A, D, E, and G).
Conclusions and implications.
The linker domains of FtsA and ZipA from various bacterial species are quite variable in terms of amino acid sequence and length. Nonetheless, we were surprised to find that deletions of large proportions of the linker regions in both FtsA and ZipA of E. coli had only subtle effects on cell division and viability, at least under the conditions used in this study. The unstructured linker in E. coli FtsA is relatively short compared to those in FtsZ, ZipA, or some other bacterial FtsAs, such as that from S. pneumoniae. It seems logical that such a short peptide would not have an extensive functional role beyond structural support for the membrane-bound oligomer, and our data support this hypothesis.
In contrast, we anticipated that deletions in the much longer linker in ZipA would have some aberrant effect on cell division, especially considering that distinctive linker sequence motifs are generally conserved across other species. Although deletions of the E. coli ZipA linker regions have little to no effect in otherwise WT E. coli cells even under several stress conditions tested, we found that loss of the P/Q region in particular renders cells more resistant to excess levels of FtsA, and either loss of P/Q or loss of half of the linker results in more sensitivity to excess levels of FtsEX. In addition, overproduction of ZipAΔP/Q or ZipAΔ1/2 is generally less toxic to cells than equivalent levels of WT ZipA (Fig. 4 and 8A and B).
What clues could these phenotypes provide for the function of the ZipA linker region? Although the mechanism underlying toxicity of excess FtsA is not completely understood, it is likely that excess FtsA displaces ZipA binding to FtsZ protofilaments, and excess FtsZ or more FtsZ bundling provides more (or more usable) substrate for ZipA and FtsA binding, thus alleviating the competition. It should be noted here that ZipA likely binds to FtsZ with higher avidity than FtsA (42, 50). Based on this model, one hypothesis is that ZipAΔP/Q binds more efficiently to FtsZ than does WT ZipA and hence competes even more effectively with FtsA. This would also explain the enhanced sensitivity of ZipAΔP/Q to excess FtsEX compared with WT ZipA, as cells with ZipAΔP/Q should be more sensitive to FtsEX-mediated displacement of FtsA from the Z ring than cells with WT ZipA (42). In other words, if ZipAΔP/Q binds FtsZ better than WT ZipA, then it should compete with FtsA more effectively, and thus any further weakness in FtsA-FtsZ interactions should have a synthetic toxic effect on cell division. However, the generally decreased toxicities of ZipAΔP/Q or ZipAΔ1/2 relative to ZipA do not support the idea that the linker mutants bind FtsZ more efficiently and displace FtsA more than WT ZipA, unless ZipA toxicity also involves mechanisms other than displacing FtsA. Given that ZipA interacts with FtsZ, FtsA, PBPs, and probably other proteins, such additional mechanisms are likely. Future experiments with mutants that uncouple the multiple functions of ZipA will be needed to dissect these effects and gain more insights into its roles in cell division. Although it is clear from the present study that a long flexible linker is not required for ZipA to function in cell division, it will be interesting to determine how spatial separation of FtsZ filaments from the membrane affects their interactions with their membrane tethers and overall activity in cell division.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The strains and plasmids used in this study are listed in Tables 1 and 2. All strains were grown in lysogeny broth (LB) agar or liquid medium (with 0.5% NaCl) at 30°C except where indicated otherwise. LB medium was supplemented with the following as needed: ampicillin (50 μg ml−1), tetracycline (10 μg ml−1), stabilized 3% hydrogen peroxide, glucose (0.2%), or IPTG.
TABLE 1.
Strains used in this study
| Strain | Description | Source or reference |
|---|---|---|
| WM1072 | MG1655 ΔrecA | G. M. Weinstock |
| WM1074 | MG1655 ΔlacU169 | Lab collection |
| WM1115 | MG1655 ftsA12 | 16 |
| WM1125 | MG1655 ftsZ84 | 53 |
| WM2101 | MG1655 ftsK44 ycaD::Tn10 | 41 |
| WM1281 | PB103 ftsA null recA56-srlD::Tn10(pSC101ts, ftsA) | 54 |
| WM2530 | DY330 {W3110 ΔlacU169 gal490 pglD8 [λcI857 Δ(cro-bioA)]} ftsA* leuO::Tn10 | 55 |
| WM4649 | MG1655 ftsI23 | 34 |
| WM5337 | MG1655 zipA1 ΔnupC::Tn10 | 34 |
| WM5521 | EC1215 ΔftsEX | 56 |
| WM6483 | WM1074 zipAΔP/Q ΔnupC::Tn10 | This study |
| WM6392 | WM1074 zipAΔ1/2 ΔnupC::Tn10 | This study |
| WM6433 | CR201 cya::kan pBAD-ccdB | 57 |
| WM6491 | WM1074 zipAΔ1/2 ΔnupC::Tn10 ftsA12 | This study |
| WM6492 | WM1074 zipAΔP/Q ΔnupC::Tn10 ftsA12 | This study |
TABLE 2.
Plasmids used in this study
| Plasmid | Description | Source or reference |
|---|---|---|
| pDSW210 | Ptrc-gfp pBR322 derivative | 58 |
| pKD46 | Expresses λRed functions | 51 |
| pWM2784 | pDSW210F (with N-terminal FLAG tag) | 59 |
| pWM2785 | pDSW210F-ftsA | 59 |
| pWM5741 | pDSW210F-ftsA recloned WT linker | This study |
| pWM5742 | pDSW210F-ftsAΔ10 | This study |
| pWM5743 | pDSW210F-ftsAΔ11 | This study |
| pWM5744 | pDSW210F-ftsAΔ9–10 | This study |
| pWM5745 | pDSW210F-ftsAΔ11–12 | This study |
| pWM5746 | pDSW210F-ftsAΔ8–10 | This study |
| pWM5747 | pDSW210F-ftsAΔ11–13 | This study |
| pWM5749 | pDSW210F-ftsAΔ11–14 | This study |
| pWM5870 | pDSW210F-ftsAΔ6–10 | This study |
| pWM5871 | pDSW210F-ftsAΔ8–13 | This study |
| pWM5872 | pDSW210F-ftsAΔ8–15 | This study |
| pWM6031 | pDSW210F-ftsAΔ1–20 | This study |
| pWM6032 | pDSW210F-ftsA scrambled linker sequence | This study |
| pWM6059 | pDSW210-zipAΔP/Q | This study |
| pWM6060 | pDSW210-zipAΔ+/− | This study |
| pWM6061 | pDSW210-zipAΔ1/2 | This study |
| pWM6062 | pDSW210F-ftsAΔ5–15 | This study |
| pWM6063 | pDSW210F-ftsAΔ5–19 | This study |
| pDSW207 | pDSW210 derivative with stronger trc promoter | 58 |
| pWM2799 | pDSW207-ftsEX | Lab collection |
| pKG116 | pACYC184 derivative with nahG promoter | 60 |
| pWM5779 | pKG116-ftsA | Lab collection |
Construction of strains and plasmids.
All oligonucleotide primers are listed in Table S1 in the supplemental material. The cloned WT linker and smaller FtsA linker deletions (Δ10 through Δ8–15) were assembled in the pDSW210F plasmid as follows. Mutant oligonucleotides were synthesized by Genewiz as AscI-PstI flanked fragments. Following AscI-PstI digestion, the fragments were ligated into AscI-PstI-digested pDSW210F-FtsA using the NEB Quick Ligation kit. FtsA inserts were verified by sequencing before being used in experiments, using pDSW210 vector primers 1157 and 1225.
The larger FtsA linker deletions (Δ5–15, Δ5-19, Δ1-20, and the scrambled linker sequence) were constructed in pDSW210F (pWM2784) with primer pairs 2188/2189, 2196/2197, 2159/2160, and 2163/2164, using the NEB Q5 site-directed mutagenesis kit. All mutants were confirmed by sequencing with pDSW210 vector primers 1157 and 1225 before being used in experiments.
The ZipA linker deletions were created using the NEB Q5 site-directed mutagenesis kit similarly to the FtsA linker deletions. Primers 2190/2191, 2192/2193, and 2194/2195 were used to remove the P/Q, charged, and 1/2 linker domains from ZipA-GFP in pDSW210. Deletions were confirmed using pDSW210 vector primers 1157 and 1158.
The zipAΔP/Q and zipAΔ1/2 alleles were recombineered into the native zipA locus of the E. coli WM1074 chromosome using standard λRed recombineering (35). The chromosomal zipAΔP/Q allele was engineered in two steps. First, PCR was used to amplify a cya::kan pBAD-ccdB cassette flanked by the zipA sequence immediately upstream and downstream of the PQ domain from a genomic DNA preparation from strain CR201 using primers 2280/2281. The resulting PCR fragment was electroporated into DY330 ftsA* cells induced for λRed recombinase functions. Cells were selected on LB plus kanamycin (25 μg ml−1) plus 0.2% glucose. Insertion of the cassette at the native zipA locus was verified by colony PCR with primers 2364 and 2365 and sequencing. In the second step, the entire zipAΔP/Q sequence was amplified from pDSW210F-ZipAΔP/Q using primers 265 and 2366 and electroporated into the zipA cya::kan pBAD-ccdB cells induced for λRed recombinase functions (51). Cells in which the cya::kan pBAD-ccdB cassette in zipA was replaced with ΔP/Q were selected on LB agar plates with 0.2% arabinose. The loss of the cya::kan pBAD-ccdB cassette was verified by colony PCR at the native zipA locus and DNA sequencing. The zipAΔ1/2 allele was recombineered into the chromosome in a single step by electroporating the entire zipAΔ1/2 sequence amplified from pDSW210F-ZipAΔ1/2 into a zipA1 strain carrying plasmid pKD46 induced for λRed recombinase functions. Recombinants were selected by recovering the cells in LB and overnight plating at 42°C. Cells were screened by colony PCR at the native zipA locus using primers 2364/2365 and sequencing. A phage P1 lysate was created from the zipAΔP/Q and zipAΔ1/2 mutants to move them out of the recombineering strain into a clean WM1074 background, and deletions were confirmed by colony PCR at the native zipA locus.
Plasmid pDSW207-ftsEX was constructed by PCR amplifying the chromosomal ftsE-ftsX genes from WM1074 using the forward primer GFP-ftsEX-F-EcoRI (CCG GAA TTC ATG ATT CGC TTT GAA CAT) and reverse primer GFP-ftsEX-R-HindIII (CGG AAG CTT TTA TTC AGG CGT AAA GTG). The PCR product was digested with EcoRI and HindIII and cloned into the EcoRI-HindIII site of pDSW207, resulting in pDSW207-ftsEX.
Serial dilution plating assays.
Overnight cultures were grown in LB and 0.2% glucose at 30°C, back-diluted 1:200 into fresh medium, and grown to an optical density (OD) of 0.3 to 0.4. The cell density measurement was used to normalize the concentration of cells for each strain in each serial dilution plate. Tenfold dilutions were created, and cells were spotted onto prewarmed LB agar supplemented with antibiotic and IPTG as necessary using a 48-pin cell replicator. Plates were incubated for 16 to 20 h at the indicated temperature before imaging.
Western blot analysis.
Cell lysates were collected and proteins separated using 12% SDS-polyacrylamide gel electrophoresis. Following transfer to a nitrocellulose membrane and blocking with 5% bovine serum albumin (BSA), proteins were detected using the lab collection of affinity-purified rabbit anti-ZipA, polyclonal rabbit anti-GFP (Invitrogen), or monoclonal M2 mouse anti-FLAG (Sigma-Aldrich) antibodies at a 1:1,000 dilution in phosphate-buffered saline (PBS) plus 2% BSA. Immunstar goat anti-rabbit IgG conjugated to horseradish peroxidase secondary antibody (Bio-Rad) or goat anti-mouse IgG conjugated to horseradish peroxidase (Sigma-Aldrich) was used at a dilution of 1:10,000 in PBS plus 2% BSA. The Pierce ECL Western blotting substrate detection kit (Thermo Scientific) was used, and blots were developed with an ImageQuant LAS 4000 mini-image analyzer.
Cell imaging.
Overnight cell cultures were back-diluted 1:200 in fresh LB medium and grown to mid-exponential phase. Cells were imaged with either phase-contrast or differential interference contrast (DIC) optics on an Olympus BX63 microscope with a Hamamatsu C11440 ORCA-Spark digital CMOS camera. Image data were acquired and analyzed with cellSens software (Olympus) or ImageJ (52).
Supplementary Material
ACKNOWLEDGMENTS
We thank Yipeng Wang, Marcin Krupka, Steven Distelhorst, Todd Cameron, and Peter Christie for contributing plasmids and valuable ideas, David Weiss for the gift of the EC1512 strain, Nick De Lay for the gift of the CR201 strain, and Yan Wang for help with strain construction.
This work was supported by grant GM131705 from the National Institutes of Health, the John and Rebekah Harper Fellowship, and the Graduate School of Biomedical Sciences.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Haeusser DP, Margolin W. 2016. Splitsville: structural and functional insights into the dynamic bacterial Z ring. Nat Rev Microbiol 14:305–319. doi: 10.1038/nrmicro.2016.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Du S, Lutkenhaus J. 2017. Assembly and activation of the Escherichia coli divisome. Mol Microbiol 105:177–187. doi: 10.1111/mmi.13696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang X, Lyu Z, Miguel A, McQuillen R, Huang KC, Xiao J. 2016. GTPase activity-coupled treadmilling of the bacterial tubulin FtsZ organizes septal cell-wall synthesis. Science 77610:744–747. doi: 10.1126/science.aak9995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bisson-Filho AW, Hsu Y-P, Squyres GR, Kuru E, Wu F, Jukes C, Sun Y, Dekker C, Holden S, VanNieuwenhze MS, Brun YV, Garner EC. 2017. Treadmilling by FtsZ filaments drives peptidoglycan synthesis and bacterial cell division. Science 355:739–743. doi: 10.1126/science.aak9973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pichoff S, Lutkenhaus J. 2002. Unique and overlapping roles for ZipA and FtsA in septal ring assembly in Escherichia coli. EMBO J 21:685–693. doi: 10.1093/emboj/21.4.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pichoff S, Lutkenhaus J. 2005. Tethering the Z ring to the membrane through a conserved membrane targeting sequence in FtsA. Mol Microbiol 55:1722–1734. doi: 10.1111/j.1365-2958.2005.04522.x. [DOI] [PubMed] [Google Scholar]
- 7.Rico AI, Krupka M, Vicente M. 2013. In the beginning, Escherichia coli assembled the proto-ring: an initial phase of division. J Biol Chem 288:20830–20836. doi: 10.1074/jbc.R113.479519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krupka M, Margolin W. 2018. Unite to divide: oligomerization of tubulin and actin homologs regulates initiation of bacterial cell division. F1000Res 7:235. doi: 10.12688/f1000research.13504.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schoenemann KM, Krupka M, Rowlett VW, Distelhorst SL, Hu B, Margolin W. 2018. Gain-of-function variants of FtsA form diverse oligomeric structures on lipids and enhance FtsZ protofilament bundling. Mol Microbiol 109:676–693. doi: 10.1111/mmi.14069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pichoff S, Shen B, Sullivan B, Lutkenhaus J. 2012. FtsA mutants impaired for self-interaction bypass ZipA suggesting a model in which FtsA’s self-interaction competes with its ability to recruit downstream division proteins. Mol Microbiol 83:151–167. doi: 10.1111/j.1365-2958.2011.07923.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pichoff S, Du S, Lutkenhaus J. 2018. Disruption of divisome assembly rescued by FtsN-FtsA interaction in Escherichia coli. Proc Natl Acad Sci U S A 115:E6855–E6862. doi: 10.1073/pnas.1806450115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krupka M, Rowlett VW, Morado DR, Vitrac H, Schoenemann KM, Liu J, Margolin W. 2017. Escherichia coli FtsA forms lipid-bound minirings that antagonize lateral interactions between FtsZ protofilaments. Nat Commun 8:15957. doi: 10.1038/ncomms15957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Busiek KK, Eraso JM, Wang Y, Margolin W. 2012. The early divisome protein FtsA interacts directly through its 1c subdomain with the cytoplasmic domain of the late divisome protein FtsN. J Bacteriol 194:1989–2000. doi: 10.1128/JB.06683-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pichoff S, Du S, Lutkenhaus J. 2015. The bypass of ZipA by overexpression of FtsN requires a previously unknown conserved FtsN motif essential for FtsA-FtsN interaction supporting a model in which FtsA monomers recruit late cell division proteins to the Z ring. Mol Microbiol 95:971–987. doi: 10.1111/mmi.12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vega DE, Margolin W. 2018. Direct interaction between the two Z ring membrane anchors FtsA and ZipA. J Bacteriol 201:e00579-18. doi: 10.1128/JB.00579-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geissler B, Elraheb D, Margolin W. 2003. A gain of function mutation in ftsA bypasses the requirement for the essential cell division gene zipA in Escherichia coli. Proc Natl Acad Sci U S A 100:4197–4202. doi: 10.1073/pnas.0635003100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herricks JR, Nguyen D, Margolin W. 2014. A thermosensitive defect in the ATP binding pocket of FtsA can be suppressed by allosteric changes in the dimer interface. Mol Microbiol 94:713–727. doi: 10.1111/mmi.12790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Potluri L-P, Kannan S, Young KD. 2012. ZipA is required for FtsZ-dependent preseptal peptidoglycan synthesis prior to invagination during cell division. J Bacteriol 194:5334–5342. doi: 10.1128/JB.00859-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pazos M, Peters K, Casanova M, Palacios P, VanNieuwenhze M, Breukink E, Vicente M, Vollmer W. 2018. Z-ring membrane anchors associate with cell wall synthases to initiate bacterial cell division. Nat Commun 9:5090. doi: 10.1038/s41467-018-07559-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gardner K, Moore DA, Erickson HP. 2013. The C-terminal linker of Escherichia coli FtsZ functions as an intrinsically disordered peptide. Mol Microbiol 89:264–275. doi: 10.1111/mmi.12279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ortiz C, Natale P, Cueto L, Vicente M. 2016. The keepers of the ring: regulators of FtsZ assembly. FEMS Microbiol Rev 40:57–67. doi: 10.1093/femsre/fuv040. [DOI] [PubMed] [Google Scholar]
- 22.Buske PJ, Mittal A, Pappu RV, Levin PA. 2015. An intrinsically disordered linker plays a critical role in bacterial cell division. Semin Cell Dev Biol 37:3–10. doi: 10.1016/j.semcdb.2014.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sundararajan K, Miguel A, Desmarais SM, Meier EL, Casey Huang K, Goley ED ED. 2015. The bacterial tubulin FtsZ requires its intrinsically disordered linker to direct robust cell wall construction. Nat Commun 6:7281. doi: 10.1038/ncomms8281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buske PJ, Levin PA. 2013. A flexible C-terminal linker is required for proper FtsZ assembly in vitro and cytokinetic ring formation in vivo. Mol Microbiol 89:249–263. doi: 10.1111/mmi.12272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pichoff S, Lutkenhaus J. 2007. Identification of a region of FtsA required for interaction with FtsZ. Mol Microbiol 64:1129–1138. doi: 10.1111/j.1365-2958.2007.05735.x. [DOI] [PubMed] [Google Scholar]
- 26.Krupka M, Cabré EJ, Jiménez M, Rivas G, Rico AI, Vicente M. 2014. Role of the FtsA C Terminus as a switch for polymerization and membrane association. mBio 5:e02221-14. doi: 10.1128/mBio.02221-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Szwedziak P, Wang Q, Freund SV, Löwe J. 2012. FtsA forms actin-like protofilaments. EMBO J 31:2249–2260. doi: 10.1038/emboj.2012.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hale CA, de Boer PA. 1997. Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell 88:175–185. doi: 10.1016/s0092-8674(00)81838-3. [DOI] [PubMed] [Google Scholar]
- 29.Ohashi T, Hale CA, de Boer PAJ, Erickson HP. 2002. Structural evidence that the P/Q domain of ZipA is an unstructured, flexible tether between the membrane and the C-terminal FtsZ-binding domain. J Bacteriol 184:4313–4315. doi: 10.1128/jb.184.15.4313-4315.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohashi T, Galiacy SD, Briscoe G, Erickson HP. 2007. An experimental study of GFP-based FRET, with application to intrinsically unstructured proteins. Protein Sci 16:1429–1438. doi: 10.1110/ps.072845607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.López-Montero I, López-Navajas P, Mingorance J, Rivas G, Vélez M, Vicente M, Monroy F. 2013. Intrinsic disorder of the bacterial cell division protein ZipA: coil-to-brush conformational transition. FASEB J 27:3363–3375. doi: 10.1096/fj.12-224337. [DOI] [PubMed] [Google Scholar]
- 32.Shiomi D, Margolin W. 2008. Compensation for the loss of the conserved membrane targeting sequence of FtsA provides new insights into its function. Mol Microbiol 67:558–569. doi: 10.1111/j.1365-2958.2007.06085.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yim L, Vandenbussche G, Mingorance J, Rueda S, Casanova M, Ruysschaert JM, Vicente M. 2000. Role of the carboxy terminus of Escherichia coli FtsA in self-interaction and cell division. J Bacteriol 182:6366–6373. doi: 10.1128/jb.182.22.6366-6373.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vega DE, Margolin W, Vega DE, Margolin W. 2018. Suppression of a thermosensitive zipA cell division mutant by altering amino acid metabolism. J Bacteriol 200:e00535-17. doi: 10.1128/JB.00535-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thomason LC, Sawitzke JA, Li X, Costantino N, Court DL. 2014. Recombineering: genetic engineering in bacteria using homologous recombination. Curr Protoc Mol Biol 106:1.16.1–1.16.39. [DOI] [PubMed] [Google Scholar]
- 36.Addinall SG, Bi E, Lutkenhaus J. 1996. FtsZ ring formation in fts mutants. J Bacteriol 178:3877–3884. doi: 10.1128/jb.178.13.3877-3884.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Samaluru H, SaiSree L, Reddy M. 2007. Role of SufI (FtsP) in cell division of Escherichia coli: evidence for its involvement in stabilizing the assembly of the divisome. J Bacteriol 189:8044–8052. doi: 10.1128/JB.00773-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carlsson J, Carpenter VS. 1980. The recA+ gene product is more important than catalase and superoxide dismutase in protecting Escherichia coli against hydrogen peroxide toxicity. J Bacteriol 142:319–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dai K, Lutkenhaus J. 1992. The proper ratio of FtsZ to FtsA is required for cell division to occur in Escherichia coli. J Bacteriol 174:6145–6151. doi: 10.1128/jb.174.19.6145-6151.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dewar SJ, Begg KJ, Donachie WD. 1992. Inhibition of cell division initiation by an imbalance in the ratio of FtsA to FtsZ. J Bacteriol 174:6314–6316. doi: 10.1128/jb.174.19.6314-6316.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haeusser DP, Rowlett VW, Margolin W. 2015. A mutation in Escherichia coli ftsZ bypasses the requirement for the essential division gene zipA and confers resistance to FtsZ assembly inhibitors by stabilizing protofilament bundling. Mol Microbiol 97:988–1005. doi: 10.1111/mmi.13081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Du S, Henke W, Pichoff S, Lutkenhaus J. 2019. How FtsEX localizes to the Z ring and interacts with FtsA to regulate cell division. Mol Microbiol 112:881–895. doi: 10.1111/mmi.14324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Geissler B, Margolin W. 2005. Evidence for functional overlap among multiple bacterial cell division proteins: compensating for the loss of FtsK. Mol Microbiol 58:596–612. doi: 10.1111/j.1365-2958.2005.04858.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.de Boer P, Crossley R, Rothfield L. 1992. The essential bacterial cell-division protein FtsZ is a GTPase. Nature 359:254–256. doi: 10.1038/359254a0. [DOI] [PubMed] [Google Scholar]
- 45.Raychaudhuri D, Park JT. 1992. Escherichia coli cell-division gene ftsZ encodes a novel GTP-binding protein. Nature 359:251–254. doi: 10.1038/359251a0. [DOI] [PubMed] [Google Scholar]
- 46.Stricker J, Maddox P, Salmon ED, Erickson HP. 2002. Rapid assembly dynamics of the Escherichia coli FtsZ-ring demonstrated by fluorescence recovery after photobleaching. Proc Natl Acad Sci U S A 99:3171–3175. doi: 10.1073/pnas.052595099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arjes HA, Lai B, Emelue E, Steinbach A, Levin PA. 2015. Mutations in the bacterial cell division protein FtsZ highlight the role of GTP binding and longitudinal subunit interactions in assembly and function. BMC Microbiol 15:209. doi: 10.1186/s12866-015-0544-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Raychaudhuri D. 1999. ZipA is a MAP-Tau homolog and is essential for structural integrity of the cytokinetic FtsZ ring during bacterial cell division. EMBO J 18:2372–2383. doi: 10.1093/emboj/18.9.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pichoff S, Du S, Lutkenhaus J. 2019. Roles of FtsEX in cell division. Res Microbiol 170:374–380. doi: 10.1016/j.resmic.2019.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Du S, Park K-T, Lutkenhaus J. 2015. Oligomerization of FtsZ converts the FtsZ tail motif (CCTP) into a multivalent ligand with high avidity for partners ZipA and SlmA. Mol Microbiol 95:173–188. doi: 10.1111/mmi.12854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu XC, Margolin W. 2000. Deletion of the min operon results in increased thermosensitivity of an ftsZ84 mutant and abnormal FtsZ ring assembly, placement, and disassembly. J Bacteriol 182:6203–6213. doi: 10.1128/jb.182.21.6203-6213.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hale CA, de Boer PA. 1999. Recruitment of ZipA to the septal ring of Escherichia coli is dependent on FtsZ and independent of FtsA. J Bacteriol 181:167–176. doi: 10.1128/JB.181.1.167-176.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu D, Ellis HM, Lee EC, Jenkins NA, Copeland NG, Court DL. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc Natl Acad Sci U S A 97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Arends SJR, Kustusch RJ, Weiss DS. 2009. ATP-binding site lesions in FtsE impair cell division. J Bacteriol 191:3772–3784. doi: 10.1128/JB.00179-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fan Y, Evans CR, Barber KW, Banerjee K, Weiss KJ, Margolin W, Igoshin OA, Rinehart J, Ling J. 2017. Heterogeneity of stop codon readthrough in single bacterial cells and implications for population fitness. Mol Cell 67:826–836. doi: 10.1016/j.molcel.2017.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weiss DS, Chen JC, Ghigo JM, Boyd D, Beckwith J. 1999. Localization of FtsI (PBP3) to the septal ring requires its membrane anchor, the Z ring, FtsA, FtsQ, and FtsL. J Bacteriol 181:508–520. doi: 10.1128/JB.181.2.508-520.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shiomi D, Margolin W. 2007. Dimerization or oligomerization of the actin-like FtsA protein enhances the integrity of the cytokinetic Z ring. Mol Microbiol 66:1396–1415. doi: 10.1111/j.1365-2958.2007.05998.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Han X-S, Parkinson JS. 2014. An unorthodox sensory adaptation site in the Escherichia coli serine chemoreceptor. J Bacteriol 196:641–649. doi: 10.1128/JB.01164-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.