Abstract
RHS elements are components of conserved toxin-delivery systems, wide-spread within the bacterial kingdom and some of the most positively selected genes known. However, very little is known about how Rhs toxins affect bacterial biology. Salmonella Typhimurium contains a full-length rhs gene and an adjacent orphan rhs gene, which lacks the conserved delivery part of the Rhs protein. Here we show that, in addition to the conventional delivery, Rhs toxin-antitoxin pairs encode for functional type-II toxin-antitoxin (TA) loci that regulate S. Typhimurium proliferation within macrophages. Mutant S. Typhimurium cells lacking both Rhs toxins proliferate 2-times better within macrophages, mainly because of an increased growth rate. Thus, in addition to providing strong positive selection for the rhs loci under conditions when there is little or no toxin delivery, internal expression of the toxin-antitoxin system regulates growth in the stressful environment found inside macrophages.
Author summary
Bacteria that reside and multiply inside of phagocytic cells are hard to treat with common antibiotics, partly because subpopulations of bacteria are non-growing. Very little is known about how bacteria regulate their growth in the phagocytic vesicle. We show that RHS elements, previously known to function as mobilizable toxins that inhibit growth of neighboring bacteria, also function as internally expressed toxin-antitoxin systems that regulate Salmonella Typhimurium growth in macrophages. RHS elements were discovered more than 30 years ago, but their role in biology has long remained unclear even though they are some of the most positively selected genes known. Our results suggest an explanation to why rhs genes are under such strong positive selection in addition to suggesting a novel function for these toxins in regulating bacterial growth.
Introduction
How bacteria regulate their growth during infection is of fundamental interest for bacterial physiology and for development of new improved treatment regimens for bacterial infections. Previous studies show that toxin-antitoxin (TA) modules are important for regulating the growth of bacteria within phagocytic vacuoles in immune cells [1, 2]. TA-systems are divided into six classes (I-VI) depending on the nature of the toxin and antitoxin (protein or RNA) and the way the antitoxin mediates protection (binding, degradation or regulation of expression) (reviewed in [3]). The toxin and antitoxin of type II TA-systems are both proteins and the antitoxin protect against toxicity of the cognate toxin by binding and blocking its activity. The antitoxins of type II TA-systems are actively targeted by cellular proteases and therefore less stable, i.e. have a shorter protein half-live than their cognate toxins [4, 5]. Degradation of antitoxins results in free unbound toxins that are able to corrupt essential cellular processes, which ultimately results in growth arrest [6, 7]. TA-modules were initially considered to function as “addiction modules” or “selfish-genetic elements” found on low-copy-number plasmids, where they provide plasmid stability through post-segregational distortion [8–10]. Daughter cells that lose the plasmid stop proliferating because the unstable antitoxin needs to be continuously produced from the plasmid to prevent self-intoxication by the more stable toxin [8–10]. Type II TA-systems were later also found to be abundant in bacterial genomes and have since been linked to bacterial virulence in many bacteria, including Escherichia coli [11], Mycobacterium tuberculosis [12], Staphylococcus aureus [13] and Salmonella enterica [14].
Salmonella enterica serovar Typhimurium (hereafter S. Typhimurium) is a facultative intracellular pathogen that causes mild self-limiting gastro-intestinal disease in humans [15] but also typhoid-like disease in many hosts including humans [15, 16]. S. Typhimurium has a number of virulence factors allowing it to survive and replicate within phagocytic cells and to cause systemic disease [17]. Previous findings suggest that S. Typhimurium cells enter a non-growing state upon entry to host macrophages, through a mechanism dependent on type-II TA-modules found in the S. Typhimurium chromosome [1].
Bacterial contact-dependent growth inhibition (CDI) systems comprise functional toxin-antitoxin modules, but where typical TA-modules are expressed inside the bacterial cell, the toxins of CDI systems are delivered between cells leaving the antitoxin inside the CDI producing cell. Rhs proteins are widely distributed CDI toxins, common throughout β-, γ- and δ-proteobacteria and with distantly related YD-peptide repeat proteins found also in Gram-positive bacteria and in higher vertebrates [18, 19]. In Gram-negative bacteria, Rhs toxins are secreted and mobilized by the type VI secretion system (T6SS) [20–22], which forms a needle-like cell-puncturing device that penetrates the membrane of the target cell and can deliver multiple effectors to both prokaryotic and eukaryotic cells [23, 24]. Effectors can be delivered through the needle-tube or by attachment to the tip of the puncturing device. The tip of the T6S devise is formed by e.g. the PAAR-domain containing Rhs toxins [25, 26]. Rhs proteins contain a delivery part, which folds into a cone-like structure forming a shell that encapsulates the highly variable C-terminal toxin domains demarcated by the PxxxxDPxGL motif [27, 28]. These C-terminal toxins are delivered to target cells upon cell-cell contact [20]. To protect themselves from auto-inhibition, bacteria with Rhs toxins also encode small, highly specific antitoxins or immunity proteins, that bind and block the toxic activity of their cognate toxins [20]. The activities of the various C-terminal domains range from nucleases to ADP-ribosylating toxins and many of the toxic activities remain unknown [20, 29–31].
S. Typhimurium contains a single rhs locus with a full-length rhs gene and one adjacent orphan toxin, which lacks the Rhs shell, hereafter referred to as the delivery part of the Rhs protein (Fig 1A). The rhs locus is found adjacent to the genes encoding a T6SS on Salmonella Pathogenicity Island 6 (SPI-6) [32, 33]. In addition to their role in antagonistic interactions [20, 34], Rhs proteins have been shown to modulate host inflammatory responses [35] and S. Typhimurium SL1344 mutants lacking rhs genes are completely attenuated in pig and cattle infection models [36], indicating a strong role for these systems in bacterial pathogenesis. In addition, removal of only the orphan toxin alone reduced S. Typhimurium proliferation in mice [33].
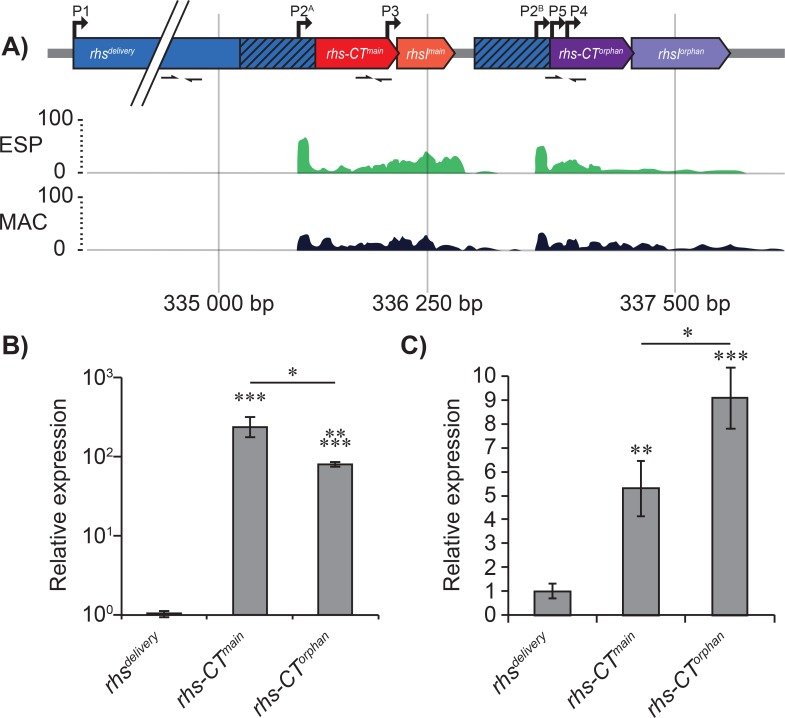
Fig 1. Transcriptional activity of the rhs locus.
A) Illustration showing transcriptional activity from the SalCom database. Predicted transcription start-sites are shown as arrows above the schematic gene. B-C) Relative expression of rhsdelivery, rhs-CTmain and rhs-CTorphan determined by qPCR in B) LB broth or C) in RAW 264.7 macrophages. Error-bars are SEM. Statistical significance was determined using Mann-Whittney test where * = P<0.05, ** = P<0.01 and *** = P<0.001, **** = P<0.0001, ***** = P<0.00001.
We were interested in finding out more about the role of Rhs toxins in S. Typhimurium biology. According to a genome-wide survey of the transcriptional response to infection-relevant conditions, multiple promoters were identified in the rhs locus (Fig 1A)[37], suggesting that while the full-length rhs gene was expressed rarely or not at all, strong expression of the main and orphan C-terminal toxins occurred at most conditions. We therefore set out to investigate if this expression resulted in functional toxin(s) being produced and if these toxins had a biological function in the cell. Here we show that main and orphan rhs genes encode functional type-II TA-modules that regulate S. Typhimurium proliferation inside macrophages.
Results
rhs toxins are expressed from internal promoters in the rhs ORF
According to a genome-wide survey of the transcriptional response to an array of 22 infection-relevant conditions encountered by S. Typhimurium 4/74, the main rhs gene (STM0291) encoding the Rhs shell and the associated T6SS genes are not transcribed during any condition (Fig 1A) [37]. However, the C-terminal toxin domains of both the main and orphan rhs genes, rhs-CT, as well as their cognate immunity genes, rhsI, were transcribed in several of the conditions assayed, including Early Stationary Phase (ESP), a condition which stimulates expression of the Salmonella pathogenicity island -1 (SPI-1) virulence genes and during intra-macrophage growth (MAC) (Fig 1A). To validate this expression, we performed quantitative real-time PCR using oligos that bind within the rhsdelivery, the main rhs-CT (rhs-CTmain) or the orphan rhs-CT (rhs-CTorphan) (Fig 1A, Table A in S1 Text). Contrary to the transcriptomics data, the qPCR results suggest a small but significant expression of rhsdelivery. The rhs-CTmain and rhs-CTorphan are, however, expressed 100-fold higher than rhsdelivery in ESP and 5 to 9-fold higher respectively in MAC conditions (after 8h of growth) (Fig 1B and 1C).
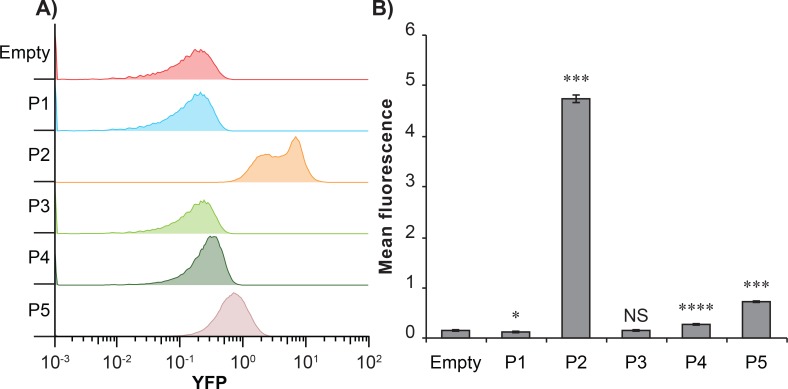
Transcriptional profiling revealed four transcriptional start sites (TSS’s) in the rhs locus region (here called P2-P5) (Fig 1A). Two of these TSS’s are located in a conserved region found in both the main and orphan rhs genes (P2A and P2B). These TSS’s are located 89 nt upstream of the sequence encoding the conserved PxxxxDPXGL motif, which suggests that they could hypothetically generate an mRNA that would allow for the translation of a functional toxin and antitoxin. The other three TSS’s identified were unique to either the main or orphan rhs genes. To test whether these TSS’s (P2- P5) and an additional hypothetical TSS, upstream from the start codon of the rhsdelivery gene (P1) allow for transcription, we constructed transcriptional fusions with sYFP2 to these promoters on a pSC101 vector backbone (S1 Fig). We defined the promoter region as -100 nt from the hypothetical TSS, except for P1 where we decided to include 200 nt in the transcriptional constructs. These constructs were transformed into S. Typhimurium 14028S and fluorescent activity was assayed in over-night cultures (grown for 16–18 h) by flow cytometry. In line with the published transcriptomic dataset we found the highest fluorescent activity for the P2 constructs in LB (Fig 2A and 2B). Also promoters P4 and P5 were active in LB, whereas we could detect no activity for P1 and P3 (Fig 2A and 2B). We chose to focus the rest of this study on the P2A/P2B and their associated main and orphan rhs transcripts. The identified P2A/P2B TSS was confirmed by 5’RACE and bioinformatic analyses identified putative -10 and -35 regions upstream of the TSS (S2 Fig). In addition, putative ribosome binding sites (RBS's) preceding several putative translation start sites (CTG (ORF1) or ATG (ORF2)) were identified in both the main and orphan transcripts (S2A and S2B Fig). These data suggest that the Rhs toxins could be expressed internally, similarly to type II TA-modules.
Fig 2. Transcriptional activity of P1-P5 promoters using transcriptional fusions.
Transcriptional activity of P1-P5 promoters measured using fluorescence reporters with P1-P5 promoters to sYFP2. A) Representative flow graphs or B) mean YFP fluorescence of strains carrying pSK945-949 (P1-P5) or pEH167 (empty). Error-bars are SEM. Statistical significance was determined using two-tailed students T-test where * = P<0.05, ** = P<0.01, *** = P<0.001 and **** = P<0.0001.
Internal transcription results in toxic proteins
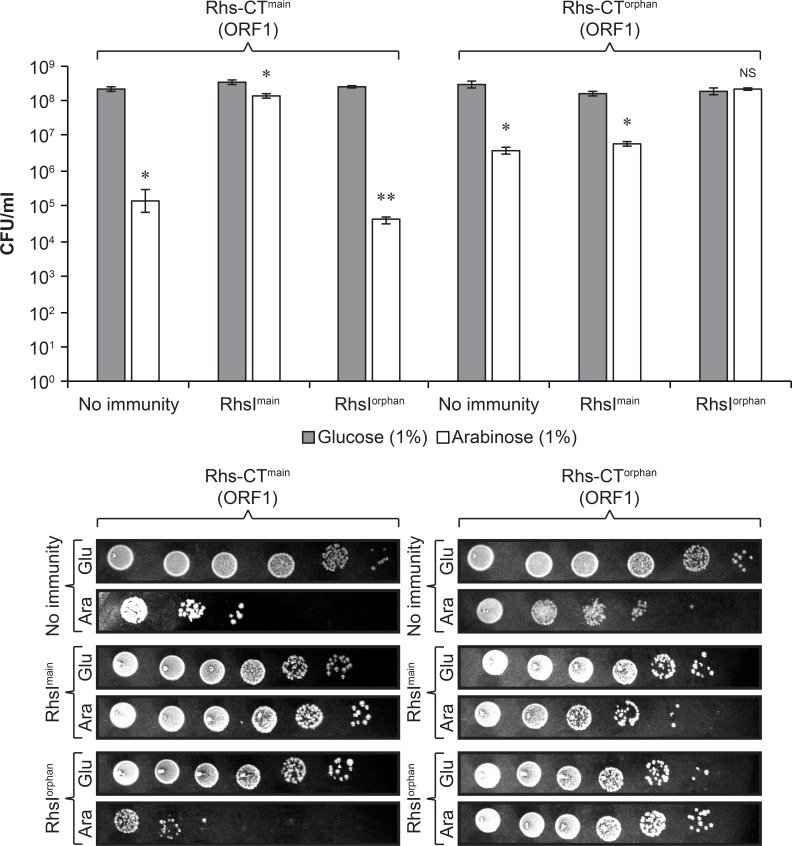
Transcriptional activity from the rhs-CTmain and rhs-CTorphan from P2A and P2B does not necessarily infer that a functional toxic protein is generated from these transcripts. For a functional toxin to be expressed, a translation start site is required. The downstream region of the confirmed TSS´s contained two potential translation start sites (S2A and S2B Fig). To investigate if either of these translation start sites produced a toxic protein, we cloned the putative ORFs (from the predicted start codon to the stop codon) under an arabinose inducible promoter on a p15A vector backbone. Toxicity of the internal transcripts was determined by transformation into NEB high-efficiency chemically competent cells in the presence or absence of the inducer (L-arabinose) and the ability of the cognate immunity protein to rescue this toxic effect. NEB cells transformed with plasmids expressing the longer ORF1 of Rhs-CTmain or Rhs-CTorphan (from CTG) grew unaffectedly in glucose, with or without expression of their cognate immunity proteins, whereas 3- and 1.5- orders of magnitude less growth was observed in L-arabinose in the absence of cognate immunity proteins for the Rhs-CTmain and Rhs-CTorphan toxin, respectively (Fig 3). In contrast, the growth of NEB cells transformed with plasmids expressing the shorter ORF2 of Rhs-CTmain or Rhs-CTorphan was unaffected in the absence of cognate immunity when grown in either arabinose or glucose (S3 Fig). We concluded that translation of a functional toxin, originating from the CTG start codon (ORF1) was possible from the P2A transcript and that this toxin could be inactivated by the presence of its cognate immunity protein in the cell.
Fig 3. Toxicity of Rhs-CTmain and Rhs-CTorphan.
Colony forming unit (CFU) values of NEB 5-α cells with or without immunity plasmid (No immunity/rhsImain pDAL923 /rhsIorphan pDAL938) 21h after transformation with plasmids encoding arabinose inducible rhsCTmain pSK1913 /rhsCTorphan pSK1914 (ORF1) grown on either 1% glucose or 1% arabinose. n = 3, Error-bars are SEM. Statistical significance was determined using two-tailed students t-test where * = P<0.05 and ** = P<0.01.
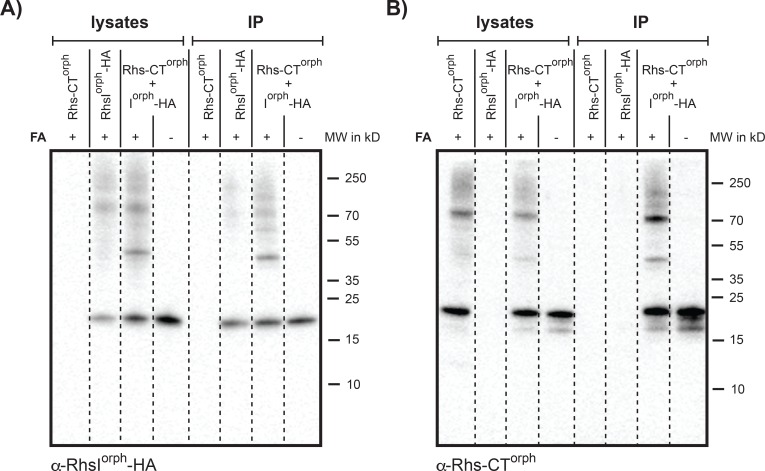
Cells expressing adequate levels of cognate immunity protein are protected from the toxicity of C-terminally encoded toxins of CdiA and Rhs molecules [20, 29, 34]. For CdiA toxins, the immunity proteins neutralize the toxins by binding the toxin directly, similarly to known type-II-TA-systems [38]. To investigate if also RhsI mediates protection through direct interaction with the toxin we used co-immunoprecipitation. HA-tagged RhsIorphan was detected in cell lysates of cells expressing rhsI orphan-HA as well as in immunoprecipitated samples using anti-HA magnetic beads (Fig 4A). Similarly, Rhs-CTorphan was present in all whole cell lysates of strains expressing rhs-CTorphan, but could only be detected in the immunoprecipitated fractions of cells expressing rhsI orphan-HA (Fig 4B). Co-immunoprecipitation of Rhs-CTorphan and RhsIorphan-HA was observed both in cell lysates treated with formaldehyde prior to lysis and in untreated cell lysates. This suggests that the Rhs orphan immunity neutralizes Rhs orphan toxin through direct interaction as observed for other CDI and type II-TA-systems.
Fig 4. RhsCTorph and RhsIorph-HA interact upon co-expression.
Immunoblot analysis of cleared cell lysates and immunoprecipitated samples (IP) using anti-HA magnetic beads with (+) or without (-) formaldehyde treatment (FA). Cells contain either RhsCTorphan expressed from an arabinose inducible plasmid (pSK502) or RhsCTorphan and RhsIOrphan-HA expressed from an arabinose inducible plasmid (pSK4251). A) RhsIOrphan-HA was detected with anti-HA antibodies. B) RhsCTorphan was detected using purified anti-RhsCTOrphan antiserum.
The immunoprecipitations were done with a plasmid where the entire rhsCT-Iorphan locus was cloned with its native translation starts. To be able to detect RhsCTorphan by western blot, we had to over-express an inactive mutant of this toxin from a previously described vector [34]. This is not surprising as native levels of expression (from the CTG start codon) are expected to be much lower than from the artificial ATG start included in the over-expression construct. To see if the internal transcription also was able to produce protein from the native P2A and P2B transcripts and the CTG translation start site, we constructed translational reporter fusions where sYFP2 was inserted 3 aa downstream of the CTG start codon in ORF1, shown to give a toxic product (Fig 3). Protein levels were investigated by single molecule fluorescence microscopy. Using this construct, a small, yet significant level of YFP was detected from Rhs-CTorphan, but not Rhs-CTmain. (S4A Fig), suggesting that both internal transcripts could result in protein although at levels below the detection limit for the native Rhs-CTmain. To further investigate if we could observe a signal from Rhs-CTmain we repeated the experiment at a higher laser voltage and longer exposure time. At these settings, a significant level of expression could be observed from the native Rhs-CTmain (S4B Fig).
Rhs toxins repress S. Typhimurium proliferation within host macrophages
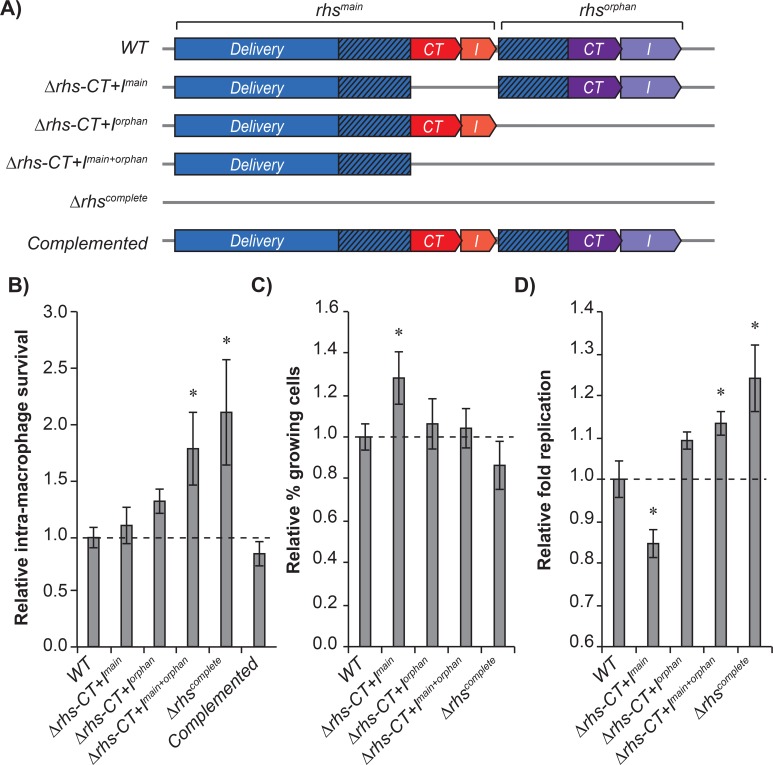
Previous studies have shown that TA-systems are important for S. Typhimurium growth regulation within macrophages [1]. Our results suggest that the Rhs-CTmain and Rhs-CTorphan toxins are expressed intracellularly similar to type-II TA-systems, but whether this has an effect on bacterial physiology is unclear. To test if Rhs toxins were important for S. Typhimurium proliferation within macrophages, we infected RAW264.7 macrophage cells with wild type or mutant S. Typhimurium bacteria lacking rhs-CTmain (Δrhs-CTmain), rhs-CTorphan (Δrhs-CTorphan), rhs-CTmain and rhs-CTorphan (Δrhs-CTmain+orphan) or the entire rhs-locus (Δrhscomplete) (Fig 5A). To reduce fluctuation between biological replicates due to variation in the number of RAW 264.7 cells in individual wells, we labelled wild type S. Typhimurium with one antibiotic resistance marker (CAM) and wild type/mutant with another antibiotic resistance marker (KAN) and infected each well with a mixture of wild type/wild type or wild type/mutant S. Typhimurium. The survival of wild type and wild type/mutant bacteria was monitored by plating cell lysates on LB agar plates containing either KAN or CAM antibiotics. This allowed us to calculate an accurate intramacrophage survival of our mutants relative to wild type for each biological replicate. Relative intramacrophage survival was calculated as the ratio of colony forming units (CFU) on KAN/CAM at 15h divided by the same ratio at 0h. To avoid competition and potential Rhs toxin delivery between wild type and mutants during the infection experiment, we used a low multiplicity of infection (MOI = 10) to ensure that only one infecting bacterium would be found per macrophage. Interestingly, we find that the Δrhs-CTmain+orphan and Δrhscomplete mutants proliferate approximately 2-times better within the macrophages than wild type S. Typhimurium (Fig 5B), suggesting that the Rhs toxins slow down or arrest growth of S. Typhimurium within macrophages. To control that no other mutations (that could have accumulated during strain construction) affected the growth of S. Typhimurium in our assay, we whole genome sequenced the Δrhscomplete mutant. (The Δrhscomplete mutant was generated by P22 transduction into the Δrhs-CTmain+orphan mutant, which in turn was generated by P22 transduction into the rhs-CTorphan mutant. Thus, the rhs-CTorphan and Δrhs-CTmain+orphan mutant should not contain additional mutations). Whole genome sequencing revealed 3 additional SNPs in wild type S. Typhimurium LT2 (listed in Table B in S1 Text). These mutations reflect the fact that our genetic engineering was performed in S. typhiumurium LT2 after which the constructs were moved to 14028S by P22 transduction. To ensure that these SNPs were not contributing to the observed phenotype of our mutants, we constructed a complemented strain where the entire Rhs-locus was reconstituted in its native genetic location in the Δrhscomplete mutant (carrying all three SNPs). No increase in survival could be observed in this mutant (Fig 5B).
Fig 5. Internal expression of rhs-CTmain and rhs-CTorphan reduce S. Typhimurium growth in RAW 264.7 macrophages.
A) Illustration showing the genetic mutants used in B-D. B) Relative intra-macrophage survival of wild type, Δrhs-CTmain, Δrhs-CTorphan, Δrhs-CTmain+orphan, Δrhs complete mutants after 16 h. Relative survival was calculated as ratio of mutant/wild type at 16h as compared to the same ratio at 0 h. C) Average wild type, Δrhs-CTmain, Δrhs-CTorphan, Δrhs-CTmain+orphan, Δrhscomplete mutants that starts to replicate after 16 h in RAW 264.7 macrophages relative to wild type. D) Relative growth rate of wild type and rhs mutants during 16 h infection. Error-bars are SEM. Statistical significance was determined using Mann-Whittney test where * = P<0.05.
Previous studies indicate that only a subpopulation of S. Typhimurium cells will start to replicate during infection and that a large fraction remains non-growing [39]. Thus, the observed outgrowth could be the result of two different scenarios where either; i) a larger fraction of mutant bacteria resume growth after entry into the macrophages, ii) the bacteria that resume growth proliferate faster within the macrophage or a combination of the above. To test which of these scenarios was the case we used a previously described fluorescent dilution assay, where the bacteria were transformed with a plasmid constitutively expressing GFP and where RFP (dsRed) expression is under the control of an arabinose inducible promoter. By expressing RFP prior to infection (by growing the bacteria in media supplemented with arabinose), the fraction of growing S. Typhimurium cells within the macrophages can be detected as the RFP signal will be diluted by 50% with each cell division [39]. The fraction of growing cells was calculated as 1-fraction non-growing cells (S5 Fig) and adjusted to account for the number of generations the growing bacteria had undergone during the experiment (for calculations see methods section). Surprisingly, no significant differences could be observed in the population of bacteria that resumed growth after entry into the macrophages in wild type or mutant S. Typhimurium (Fig 5C). In contrast, 27% more bacteria started to replicate in macrophages in S. Typhimurium lacking rhs-CTmain (Δrhs-CTmain), suggesting that this mutation affected the proportion of cells resuming growth. The increased proliferation observed for the other mutants could, however, not be accounted for by an increased fraction of cells resuming growth. To investigate if instead the mutant bacteria grew faster within the macrophage, we calculated the fold replication and number of generations the bacteria underwent during the time-frame of the experiment. The fold replication bacteria undergo during 16h of growth within macrophages can be calculated as the change in median RFP intensity between the initial population (0h) and the growing population after 16h (S5 Fig). The relative fold replication was then calculated as the fold replication of the mutant over wild type. S. Typhimurium cells lacking the entire rhs operon grew ~30% faster than wild type during the experiment, allowing the mutant bacteria to divide one extra time during the experiment (~7 generations compared to ~6 generation) (Fig 5D). One extra division could account for the 2-fold increase in CFU observed in Fig 5B. Similarly, for the single rhs-CTorphan and the double CT deletion mutants grew 20% faster than wild type (Fig 5D). For the rhs-CTmain deletion mutant no increase in growth rate could be observed, suggesting that the increase in fraction of cells resuming growth must account for the increase in CFU. Taken together, our results suggest that the internal expression of the Rhs toxins contribute mainly to the growth rate of S. Typhimurium after entry into macrophages and not the fraction of cells resuming growth.
RhsI antitoxins are degraded by proteases
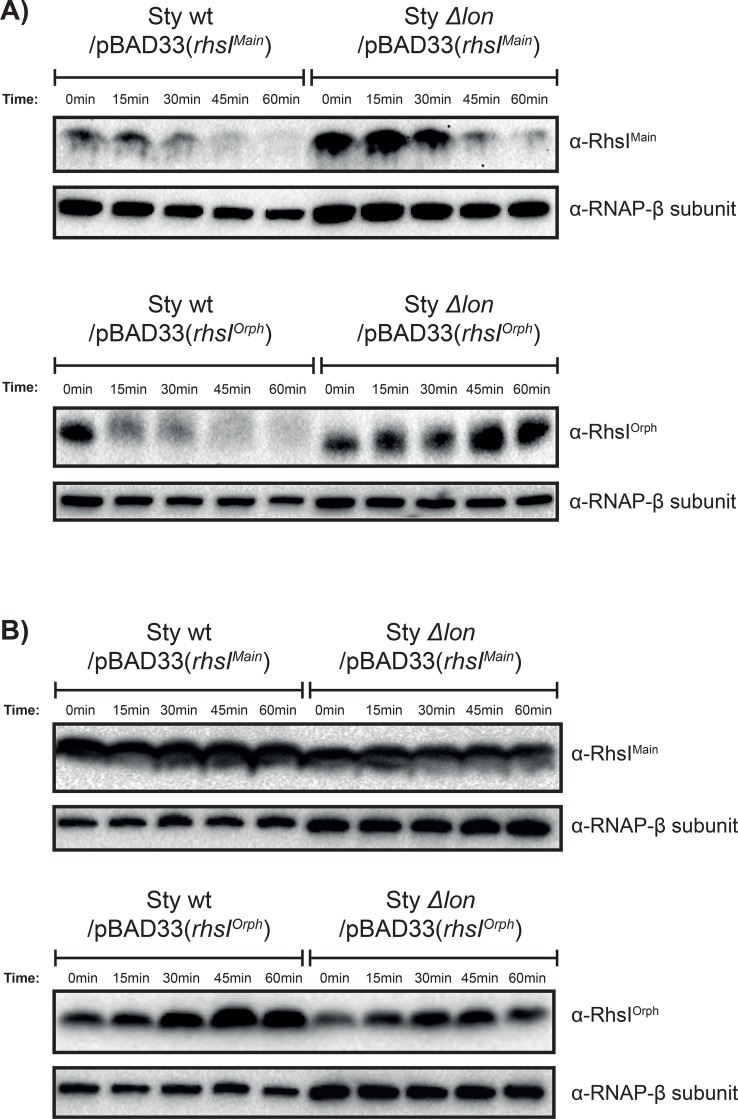
A remaining question is how Rhs toxins arrest growth. A plausible hypothesis is that they function similarly to toxins belonging to type II TA-systems and CdiA toxins, where the antitoxin is degraded by a stress-induced protease; Lon [40, 41]. Lon is up-regulated in S. Typhimurium after entry into macrophages [42], suggesting a faster turnover of antitoxin degradation in this condition. This could result in that cells with the Rhs TA-system arrest growth because they lack sufficient levels of antitoxin, whereas mutant cells lacking the TA-system can grow less hindered. To test if RhsI was susceptible to degradation by the Lon protease, we cloned C-terminally HA-tagged versions of the RhsI proteins under an arabinose inducible promoter expressed from the pBAD33 vector. Protein stability was measured in wild type and Δlon bacteria using western blot with anti-HA antibodies after the addition of either DL-Serine hydroxamate (SHX), a seryl-tRNA synthetase inhibitor, or the macrolide antibiotic Erythromycin, an inhibitor of protein synthesis, to cultures expressing RhsImain or RhsIorphan. The RhsIorphan protein was stable over the course of 1h in Δlon bacteria compared to wild type where degradation occurred already after 15 min after SHX addition (Fig 6A, lower panel). In contrast, the RhsImain protein was degraded 30 min after SHX addition in both wild type and Δlon bacteria, suggesting that another protease is responsible for the degradation of this protein (Fig 6A, upper panel). Both proteins were completely stable in both wild type and Δlon background upon Erythromycin treatment (Fig 6B). These results suggest that Rhs immunities do not have a fast turn-over under favorable conditions but are unstable and subject to proteolytic degradation when the cell is stressed. From these results it appears that Rhs toxins could arrest growth upon entry to macrophages through stress-mediated activation of cellular proteases, resulting in RhsI antitoxin degradation.
Fig 6. RhsImain and RhsIorphan are degraded by proteases (including Lon) upon stringent-response activation.
Immunoblot analysis of RhsI-HAmain or RhsI-HAorphan protein (11.6 and 18.5 kDa respectively) stability in the presence (Sty wt) and absence (Sty Δlon) of Lon protease before and after A) SHX or B) Erythromycin treatment for 1 h. Levels of the RNA polymerase β-subunit are shown as control.
Discussion
RHS elements have previously been described as weapons used in antagonistic bacterial interactions. Here we show that Rhs toxins are not only being delivered between cells but are also expressed and functional within a bacterial cell. We also show that Rhs toxins affect proliferation of Salmonella cells during macrophage infection by changing the growth rate of S. typhimurium within macrophages. Thus, Rhs toxins comprise functional type-II TA loci that repress S. Typhimurium growth in vivo. Previous analysis of more than 150 Salmonella genomes showed that all Salmonella serovars contain at least one Rhs toxin, with some strains having up to 11 different orphan toxins [34]. Bioinfomatic analyses of all genomes on NCBI showed that the P2 promoter is found upstream of at least 50 different Rhs toxins in Salmonella spp. but also in E. coli, Shigella, Pseudomonas and Erwinia spp, suggesting that also these toxins could be expressed internally as seen in S. Typhimurium (S6 Fig). This raises an interesting question; does the presence of other Rhs toxins affect Salmonella growth in vivo and, if so, what are the consequences of having up to 12 toxins for Salmonella’s lifestyle and ability to replicate in macrophages? On the other hand, it is not obvious why S. Typhimurium has two Rhs toxins, when one alone (the orphan) has a strong effect on intramacrophage replication. A possible explanation could be that the two toxins are used for different things. For example, the main Rhs toxin can be delivered between bacteria, whereas mobilization of the orphan toxin requires a pre-existing duplication of the locus followed by a recombination event [34]. Our results here suggest that the orphan toxin mainly functions in controlling intramacrophage replication, whereas very little contribution can be seen from the main Rhs toxin. It is also possible that the contribution of rhsCT main cannot be fully evaluated in the single mutant as removal of rhsCT main allows for a recombination event that will fuse rhsCT orphan to the rhs-delivery mechanism [34]. Such recombination events between hundreds of base pairs of exact homology are very frequent [43], but are prevented in wild type Salmonella by the internal expression of RhsCT/I main (which upon loss would cause a growth defect to that cell). Changing the chromosomal organization of the locus could result in a number of changes including altered expression levels of the RhsCTorphan as well as potential delivery of the toxin between cells, which could affect proliferation in our infection assay.
So, what could be the benefit of maintaining these Rhs toxins in Salmonella genomes? One possible explanation is that there is no benefit, but that Rhs toxins represent selfish elements and are expressed internally simply to maintain selection of themselves. This fits nicely with the observation that these internal promoters are expressed under all conditions tested and could also explain why Rhs proteins are some of the most positively selected genes known [44]. Selfish genetic elements can either consist of sequences that frequently transfer between strains but can also consist of elements that provide a benefit to the cell, or a combination of both [45]. These selfish genetic elements may have been domesticated or co-opted by the bacteria to provide positive effects for the cell, like defense against phage, or ability to compete with other bacteria. The Rhs toxins in S. Typhimurium can be mobilized and used to outcompete bacteria lacking these systems [34]. But, the toxins are found on the H-NS silenced SPI-6 [46], previously shown to be important for colonization of mice [33]. Thus, it is possible that the internal expression of Rhs toxins provides a selection to maintain the full-length Rhs toxin and/or SPI-6 in conditions when they are not expressed. Another possibility is that growth arrest after entry into macrophages could be important for S. Typhimurium in vivo. Replication in macrophages occurs mainly during systemic spread of the disease within the lymph nodes, liver and spleen. Interestingly, a previous study showed that Rhs-CTorphan, but not RhsImain is important for systemic dissemination in mice [33], suggesting that internal expression could be important for virulence. Salmonella gastroenteritis infections are often self-limiting and do not proceed beyond the lamnia propria. However, during typhoid fever, the Salmonella infected phagocytes gain access to the lymphnodes, liver and spleen [47]. It is possible that regulation of growth upon entry to phagocytes is an important factor in deciding whether the infection stays local or becomes systemic.
The full-length Rhs toxins or the adjacent T6SS genes are not expressed during any of the conditions assayed in Kröger et al. [37]. Previous findings suggest that SPI-6 is strongly silenced by H-NS and that only in conditions where H-NS expression is decreased or absent can T6SS activity be observed [32, 46]. Other contact-dependent competition systems have been suggested to contribute to kin selection by exclusion of others (by inhibiting the growth of non-self bacteria) [48]. Rhs toxins have also been shown to be important during intestinal colonization [32, 49]. As the intra-macrophage population most likely is clonal, there is probably no competition with non-kin cells in this niche. Thus, expression of a system that is only used as a competition system, in these conditions, could be considered a waste of resources. We cannot, however, exclude the possibility that the ability to deliver toxins between isogenic strains adds to the growth regulation observed in our experiments. We have previously shown that self-delivery of CdiA toxins increases the fraction of non-growing cells in the population [41], so it is possible that Rhs toxins expressed at a low level (undetectable by RNA seq) are delivered between isogenic cells within the phagocytic vesicle. This could potentially explain the increased effect observed in the complete mutant as compared to the one lacking both toxins.
A remaining question is how Rhs toxins arrest growth. Our results suggest that both RhsI proteins are susceptible protease mediated degradation under conditions when translation is arrested by induction of the stringent response, but not when translation is arrested by antibiotics. For one of the immunity proteins, RhsIorphan, this degradation was completely abolished in cells lacking Lon protease. Lon protease becomes activated by poly-phosphate under conditions when stringent response is active [50, 51], suggesting that activated Lon is required for degradation of RhsIorphan. As Lon is up-regulated in S. Typhimurium after entry into macrophages [42], it is likely that antitoxin turnover is increased in this condition. This would result in that cells with the Rhs TA-system arrest growth because they lack sufficient levels of antitoxin, whereas mutant cells lacking the Rhs TA-system can grow unhindered. Previous findings show similar results where lack of certain toxin-antitoxin pairs decrease the non-growing Salmonella population within macrophages [1]. It is likely that each individual toxin adds a little to the effect and it would be interesting to see how a combined mutant lacking both type II-TA-systems and Rhs toxins would behave during macrophage infection. Interestingly, we observe a stronger phenotype in vivo for the Rhsorphan toxin even though it is less toxic than Rhsmain when expressed from an arabinose inducible promoter (as seen in Fig 3). A possible explanation is that the two immunity proteins are not degraded to the same extent or at the same time in the macrophages. RhsIorphan is degraded by the Lon protease, whereas another protease, possibly not activated to the same extent or at the same time within the macrophage, degrades RhsImain (as seen in Fig 6). Protease mutants are non-viable in macrophages [52], making it difficult to test how protease mediated degradation of the antitoxin affects Salmonella growth within macrophages.
An interesting and logical follow up question is of course how the growth arrested cells are able to resume growth? As with many type II toxins, CDI toxins are rarely lethal, depending on their toxic activity. It is easy to envision that cells undergone toxicity that results in degraded RNA molecules or dissipation of the proton motive force, easily can resume growth once the toxicity has been removed through degradation or neutralization of the toxin by proteases or newly synthesized antitoxin respectively. The toxic activity of the Rhs toxins in S. Typhimurium is not known. Structural or functional predictions suggest no membrane association for either the toxins or immunities, implying that the Rhs toxins do not act on the membrane. PFAM database search reveals a known toxin domain (NTox) in the Rhsmain toxin, suggesting that this protein is an RNAse [53]. For Rhsorphan toxin, no clues to its activity could be revealed from its sequence.
Orphan toxins that lack the secretion component of the toxin are not only found for Rhs toxins, but are also often present in cdiBAI loci [54]. A fundamental question is how these orphan modules are maintained in bacterial populations. One possible selection pressure could be that these toxins provide a silent arsenal of novel toxins that can be mobilized by bacteria and used in future competitions against neighboring bacteria. For example, this would facilitate a clonal selection in a population of genetically identical bacteria [34]. Our findings that orphan Rhs toxins represent functional TA-modules expressed inside the bacteria provides an additional explanation to how these orphan toxins can be maintained when not used for competition. The remaining question is whether also CdiA toxins, colicins, microcins and other T6SS toxic effector proteins also contain internal transcription and translation start sites allowing them to be expressed internally, and if that expression would have a function on said bacteria’s physiology.
Methods
Bacterial strains and growth conditions
Bacterial strains used in the study are derivatives of either Salmonella enterica subsp. Enterica serovar Typhimurium strain LT2 or strain 14028S (ATCC 14028) and are listed in Table C in S1 Text. Liquid media was Luria-Broth (LB) (Miller, 1972). Over-night cultures were grown for 16–18 hr at 37°C and 200 rpm. For solid media LB was supplemented with 10 g/L agar. Antibiotics were used at the following concentrations; 50 mg/l Kanamycin (KAN), 15 mg/L Tetracycline (TET), 12.5 mg/L Chloramphenicol (CAM), mg/L Carbenicillin (CARB) and 100 mg/L Ampicillin (AMP). Genetic mutations in the chromosome were made using λ red recombineering [55]. Markers were moved between Salmonella strains using P22 transductions. Transductants were purified on EBU plates [56] and clones were confirmed by PCR and sequencing. Antibiotic resistance markers were excised using Flp recombinase expressed from the pcp20 vector as described previously [55]. More detailed information about strain constructions can be found in the S1 Text.
Expression of transcriptional fluorescent protein reporters measured through flow cytometry
Bacteria from over-night cultures were washed 2x with 1 ml sterile-filtered PBS and diluted to 1x106 CFU/ml in PBS. Flow cytometry was performed using a MACSQuant VYB (Miltenyi Biotec), measuring 100,000 events. YFP was excited with a blue laser (488 nm; bandpass filter 525/50 channel B1), dsRed was excited with a yellow laser (561 nm; bandpass filter 615/20 nm channel Y2). The MACSQuantifyTM Software (Miltenyi Biotec) was used to aquire the data and the FlowJo Software (FlowJo, LLC) for analysis.
5’RACE
5’ RACE (rapid amplification of cDNA ends) was carried out with or without treatment by 5´→3´ exoribonuclease XRN-1 (NEB), using DNase I-digested total RNA isolated from early stationary phase (ESP). RNA was ligated with an RNA adaptor (5’ CGACUGGAGCACGAGGACACUGACAUGGACUGAAGGAGUAGAAA) and the product was reverse transcribed to cDNA using gene-specific oligo SK332 and Maxima first strand cDNA Synthesis Kit for RT-qPCR (K1641, Thermo Scientific). The DNA fragment was amplified in two consecutive rounds of PCR with DreamTaq DNA polymerase (ThermoScientific), first step with adapter-specific oligo FS17 and in the second, nested step with PCR oligo FS354, both in combination with gene specific reverse oligo SK332. The XRN-1-enriched DNA fragments were gel purified and cloned into TOPO vector pCR-BLUNT-II (Thermofisher), and 10 clones were sequenced to validate the transcriptional start site.
Western blot
500 μl of an OD600 = 0.7 culture of exponentially growing S. Typhimurium cells, induced with 0.2% arabinose at OD600 = 0.3, were harvested by centrifugation at 4°C for 5 min at 21000 x g. Cells were re-suspended in 100 μl of sample buffer (50mM Tris pH 6.8, 1% SDS (w/v), 1% Triton X-100, 10% Glycerol, 0.2% bromophenol blue) and boiled for 5min at 95°C. 150mM DTT was then added to each sample followed by the pelleting of membrane debris and undigested genomic DNA at 21000 x g for 5 min before the supernatant were separated on a Mini-PROTEAN TGX gel (Biorad, USA). PageRuler Prestained Protein Ladder (Thermo Scientific, USA) was used as size marker. Proteins were then transferred to a Trans-Blot Turbo Mini 0.2 μm PVDF membrane (BioRad, USA) using the Trans-Blot Turbo system (Biorad, USA). RhsImain-HA and RhsIoprhan-HA proteins were detected using anti-HA antibody (11867423001, Sigma-Aldrich, Germany). Equal loading was confirmed by probing for RNA polymerase β-subunit using an anti-RNAP β-subunit antibody (ab191598, Abcam, United Kingdom). Secondary antibody towards anti-HA antibodies was anti-rat IgG coupled to HRP (GENA935, Sigma-Aldrich, Germany) and secondary antibody towards anti-RNAP antibodies was anti-rabbit IgG coupled to HRP (A1949, Sigma-Aldrich, Germany). Bands were visualized with Clarity Western ECL Substrate (Biorad, USA) and a ChemiDoc MP system (Biorad, USA).
Co-Immunopurification (Co-IP)
Exponentially growing cultures of SK502 (pCH450::rhsCTorphan (H208A)), SK4251(pBAD24::rhsCT-Iorphan-HA), SK4292 (pCH450::rhsCTorphan (H208A) and pBAD24::rhsCT-Iorphan-HA) (OD600 ~0.9–1.2) were induced with 0.5% L-arabinose (f.c.) at OD600 = 0.3 for ~1h. 90 ml of cells were harvested by centrifugation and washed once with 1 x PBS. Cells were resuspended in 5 ml 1 x PBS supplemented with 2% formaldehyde (f.c.) and incubated for 10 min at RT. Residual formaldehyde was quenched by addition of 5 ml 1M Tris-Cl pH 8.0 and two subsequent washes with 0.5 M Tris-Cl pH 8.0. Cells were resuspended in lysis buffer (20 mM Tris-Cl pH 7.5, 0.1% Tween-20, 0.25% Triton-X-100, 1x Halt-TM EDTA-free protease inhibitor mix) and broken by 6 cycles of sonication for 10 sec at 50% amplitude on ice. Debris and remaining cells were removed by centrifugation at 21.000 x g for 15 min at 4°C. Cleared cell lysates were incubated with PierceTM anti-HA magnetic beads (Thermo Scientific) according to the manufacturer’s instructions. Bound proteins were eluted with 50 μl 0.2 M glycine pH 2. Cleared cell lysates and eluates from the magnetic beads were dissolved in NuPAGE LDS sample buffer (Thermo Fisher Scientific) supplemented with 0.1 M dithiotreitol (f.c.) and incubated at 60°C for 10 min prior to loading. Of each cleared cell lysate, a volume corresponding to 1ml of bacteria with OD600 = 0.14 (for detection of RhsIorph-HA) or OD600 = 0.014 (for detection of RhsCTorph) was separated on a 10% NuPAGE Bis-Tris gels (Novagen) using MES running buffer supplemented with antioxidant. Proteins were transferred to a PVDF-membrane using pre-assembled iBlotTM transfer stacks (Thermo Fisher Scientific). RhsCTorph was detected using purified anti-RhsCTorph antiserum [34] and anti-rabbit IgG coupled to HRP (Sigma). RhsIorph-HA was detected using anti-HA-antibody as described above.
RAW264.7 culturing conditions
The murine macrophage-like cell line RAW264.7 (ATCC) was cultured in 1x Dulbecco’s Modified Eagle Media (DMEM, GIBCO) supplemented with 1% GlutaMAX (GIBCO), 1% non-essential amino acids (Sigma), 10% heat inactivated Fetal Bovine Serum (HI-FBS) (GIBCO) at 37°C with 10% CO2 in T-25 bottles (Sarstedt). Cells were passaged when confluent or every 48 h.
RAW264.7 infection assay
1x105/ml RAW264.7 cells were seeded in 24-well plates (Sarstedt) in pH buffered DMEM (high glucose, HEPES, no phenol red, GIBCO) supplemented with, 1% non-essential amino acids (Sigma) and 10% HI-FBS (GIBCO) and grown at 37°C with 10% CO2 for 48 h before the infection. Over-night cultures of S. Typhimurium 14028S were centrifuged at 6000 RPM and washed with sterile 1x PBS before being diluted to OD600 = 0.4. Bacteria were then opsonized in Balb/cAnCrl mouse serum for 30 min. The old growth medium was removed and fresh pH buffered DMEM without FBS was added before infection at MOI 10. The infection was synchronized by centrifuging the cells for 5 min at 1200 rpm followed by 45 min incubation at 37°C with 10% CO2. The wells were washed with 1x PBS before adding fresh pH buffered DMEM + gentamicin (100 mg/l). Cells were incubated 45 min at 37°C to kill off extracellular bacteria followed by washing with 1x PBS before being lysed with 0.1% Triton-X solution (Sigma). Lysates were immediately plated for CFU counts of the 0 h time point. Fresh pH buffered DMEM + gentamicin (20 mg/l) was added to the remaining wells for later time points and incubated at 37°C with 10% CO2. Relative intra-macrophage survival was calculated as the ratio of WT and mutant CFU/ml at 16 h divided by the ratio of WT and mutant CFU/ml at 0 h.
Fluorescence dilution assay
S. Typhimurium 14028S (ATCC) transformed with pDiGc (Helaine et al., 2010) was used for infection according to infection protocol above. After lysing the RAW264.7 cells the lysates were washed once with sterile filtered PBS followed by analysis by flow cytometry (MACS, Miltenyi). Samples were gated for bacteria, GFP and dsRed (see “Fluorescent reporter assays” for settings). The % of bacteria that started to replicate was calculated by dividing the fraction of cells which lost RFP signal with at least 50% (S5 Fig) by 2n where n is the number of generations the bacteria underwent during the experiment. The number of generations was determined by taking the log2 of the relative change in mean RFP value between the initial non-divided population and the dividing population (S5 Fig).
Toxicity assay
Electro-competent SK4287, SK4288 and SK4289 were transformed with pSK1913, pSK1914, pSK1915 or pSK1918 by electroporation at 2.5 kV, 200 Ω, 25 μF and resuspended in 1 ml LB directly after. The transformed cells were incubated at 37°C for 1 h to recover. After recovery the transformed samples were split in two to which glucose or arabinose was added to a final concentration of 1% alongside with appropriate antibiotics for plasmid selection. Cultures were grown for 20 h at 30°C and plated for CFU counts on selective LA plates, supplemented with 1% glucose, by spotting.
Quantitative RT-PCR
Bacteria for RNA extraction were grown over-night in LB or for 8h in RAW264.7 macrophages. LB cultures were diluted 1:1000 in LB broth and grown until OD600 = 2.0 (ESP). RNA degradation and transcription were stopped by the addition of 2/5 v/v 95% ethanol and 5% phenol (pH 4.3) on ice for 30 min—2 h. Cells pellets were stored at -80°C until RNA extraction was performed using TRIzol (Invitrogen). DNA was removed using Turbo DNase I (Thermofisher) according to the manufacturer’s instructions. Successful DNA removal was confirmed by PCR using oligos SK411/SK412 (40 cycles), and RNA integrity vas validated on 2% agarose in 1xTEB, run at 80V. Total RNA was diluted to 5 ng/μl and RT-qPCR was performed using Brilliant Ultra-Fast SYBR Green QRT-PCR Master Mix (Agilent) with oligos SK362/SK363 (delivery) SK3/SK4 (rhs-CTmain) and SK5/SK6 (rhs-CTorphan). The reactions were run on a StepOnePlus Real-Time PCR System (Applied biosystems). Relative gene expression was calculated using the 2-ΔΔCT method, normalizing the amount of target mRNA to expression of house-keeping gene recA amplified with oligos SK7/SK8 and wild type [57].
Bioinformatic analyses
P2 promoter sequence from S. Typhimurium (CGGCTGCCGGGGCAGTATTTTGACGACGAGACAGGGCTGCATTACAATCTGTTCAGATATTATGCACCGGAGTGTGGACGGTTTGTCAGTCAGGATCCGATCGGGCTG, 108bp) was used as the query for BLASTN search against the nt database (2019-04-29 release). 1147 hits with percent identity > = 80%, alignment length > = 95 bp and e-value < = 1e-10 were kept, corresponding to 800 different genomes. Full-length genomes were then retrieved from NCBI, and sub-sequences matching the given promoter were obtained for further analysis, as well as an additional 138 bp downstream sequence directly after the promoter region. Promoter and the downstream sequences were aligned separately using MAFFT [58] v7.407 with the L-INS-i option, and the alignments were then merged and ordered after the similarity of the aligned promoter sequences.
Supporting information
(PDF)
Overview of the A) P2A or B) P2B promoter sequences. -35 (yellow) and -10 (blue) regions were predicted using the softberry BPROM software. Transcription start sites (TSS´s) (bold and underlined) were determined using C) 5’RACE. Potential start codons are in green, RBS’s in orange and in-frame stop codons in pink.
(PDF)
CFU counts from transformations of plasmids encoding arabinose inducible rhs-CTmain/rhs-CTorphan (ORF2) into NEB 5-α harboring no immunity or rhsImain/rhsIorphan on a plasmids grown in either 1% glucose or 1% arabinose. n = 3, Error-bars are SEM. Statistical significance was determined using two-tailed students t-test where *** = P<0.005.
(PDF)
Single molecule fluorescence of strains with translational fusions of Rhs-CTmain and Rhs-CTorphan ORF1’s (CTG) to sYFP2. Strains were grown in M9-glucose to reduce background fluorescence. A) YFP fluorescence (au). B) Repeated experiment at higher laser voltage. Error-bars are SEM. Statistical significance was determined using two-tailed students t-test where *** = P<0.001 and ***** = P<0.00001.
(PDF)
Representative graph of flowcytometric data used to identify the dividing population of bacteria after 16h of growth in RAW264.7 macrophages. The growing population is determined as the cells where the dsRed fluorescent signal is decreased to levels below half of the mean fluorescent signal of the non-growing population.
(PDF)
Alignment showing P2 promoter-like sequences in bacterial genomes from NCBI. Downstream sequences illustrates different types of toxins the P2 promoters are found adjacent to. Homologous residues are shown in blue. Promoter sequences (-10, -35, TSS) and the conserved PxxxDPxGL motif are annotated above the sequences and demarcated with a black box.
(PDF)
Supplementary methods and Tables A-C.
(PDF)
(XLSX)
Acknowledgments
We thank Prune Leroy for the help with fluorescent microscopy and data analysis, Erik Holmqvist for the generous gift of the pEH167 plasmid and Fredrik Söderbom for the FS17 and FS354 oligos used in the study.
Data Availability
The data underlying the results presented in the study are available in S1 Data.
Funding Statement
This study was supported by grants from the Swedish Foundation of Strategic Research (ICA12-0025) (https://strategiska.se/), the Swedish research council (E0239301)(https://www.vr.se/), the European research council (ERC-2018-STG-804068)(https://erc.europa.eu/) and the Wenner-Gren foundations (https://www.swgc.org/) to S.K. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Helaine S, Cheverton AM, Watson KG, Faure LM, Matthews SA, Holden DW. Internalization of Salmonella by macrophages induces formation of nonreplicating persisters. Science. 2014;343(6167):204–8. Epub 2014/01/11. 10.1126/science.1244705 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Korch SB, Contreras H, Clark-Curtiss JE. Three Mycobacterium tuberculosis Rel toxin-antitoxin modules inhibit mycobacterial growth and are expressed in infected human macrophages. J Bacteriol. 2009;191(5):1618–30. Epub 2008/12/31. 10.1128/JB.01318-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Page R, Peti W. Toxin-antitoxin systems in bacterial growth arrest and persistence. Nat Chem Biol. 2016;12(4):208–14. Epub 2016/03/19. 10.1038/nchembio.2044 . [DOI] [PubMed] [Google Scholar]
- 4.Muthuramalingam M, White JC, Bourne CR. Toxin-Antitoxin Modules Are Pliable Switches Activated by Multiple Protease Pathways. Toxins (Basel). 2016;8(7). Epub 2016/07/15. 10.3390/toxins8070214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilbaux M, Mine N, Guerout AM, Mazel D, Van Melderen L. Functional interactions between coexisting toxin-antitoxin systems of the ccd family in Escherichia coli O157:H7. J Bacteriol. 2007;189(7):2712–9. Epub 2007/01/30. 10.1128/JB.01679-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keren I, Shah D, Spoering A, Kaldalu N, Lewis K. Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J Bacteriol. 2004;186(24):8172–80. Epub 2004/12/04. 10.1128/JB.186.24.8172-8180.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vazquez-Laslop N, Lee H, Neyfakh AA. Increased persistence in Escherichia coli caused by controlled expression of toxins or other unrelated proteins. J Bacteriol. 2006;188(10):3494–7. Epub 2006/05/05. 10.1128/JB.188.10.3494-3497.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ogura T, Hiraga S. Mini-F plasmid genes that couple host cell division to plasmid proliferation. Proc Natl Acad Sci U S A. 1983;80(15):4784–8. Epub 1983/08/01. 10.1073/pnas.80.15.4784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerdes K, Bech FW, Jorgensen ST, Lobner-Olesen A, Rasmussen PB, Atlung T, et al. Mechanism of postsegregational killing by the hok gene product of the parB system of plasmid R1 and its homology with the relF gene product of the E. coli relB operon. EMBO J. 1986;5(8):2023–9. Epub 1986/08/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerdes K, Rasmussen PB, Molin S. Unique type of plasmid maintenance function: postsegregational killing of plasmid-free cells. Proc Natl Acad Sci U S A. 1986;83(10):3116–20. Epub 1986/05/01. 10.1073/pnas.83.10.3116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Norton JP, Mulvey MA. Toxin-antitoxin systems are important for niche-specific colonization and stress resistance of uropathogenic Escherichia coli. PLoS Pathog. 2012;8(10):e1002954 Epub 2012/10/12. 10.1371/journal.ppat.1002954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stewart GR, Patel J, Robertson BD, Rae A, Young DB. Mycobacterial mutants with defective control of phagosomal acidification. PLoS Pathog. 2005;1(3):269–78. Epub 2005/12/03. 10.1371/journal.ppat.0010033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bukowski M, Lyzen R, Helbin WM, Bonar E, Szalewska-Palasz A, Wegrzyn G, et al. A regulatory role for Staphylococcus aureus toxin-antitoxin system PemIKSa. Nat Commun. 2013;4:2012 Epub 2013/06/19. 10.1038/ncomms3012 . [DOI] [PubMed] [Google Scholar]
- 14.De la Cruz MA, Zhao W, Farenc C, Gimenez G, Raoult D, Cambillau C, et al. A toxin-antitoxin module of Salmonella promotes virulence in mice. PLoS Pathog. 2013;9(12):e1003827 Epub 2014/01/05. 10.1371/journal.ppat.1003827 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Haraga A, Ohlson MB, Miller SI. Salmonellae interplay with host cells. Nat Rev Microbiol. 2008;6(1):53–66. Epub 2007/11/21. 10.1038/nrmicro1788 . [DOI] [PubMed] [Google Scholar]
- 16.Haselbeck AH, Panzner U, Im J, Baker S, Meyer CG, Marks F. Current perspectives on invasive nontyphoidal Salmonella disease. Curr Opin Infect Dis. 2017;30(5):498–503. Epub 2017/07/22. 10.1097/QCO.0000000000000398 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Linehan SA, Holden DW. The interplay between Salmonella typhimurium and its macrophage host—what can it teach us about innate immunity? Immunol Lett. 2003;85(2):183–92. Epub 2003/01/16. 10.1016/s0165-2478(02)00227-4 . [DOI] [PubMed] [Google Scholar]
- 18.Tucker RP, Beckmann J, Leachman NT, Scholer J, Chiquet-Ehrismann R. Phylogenetic analysis of the teneurins: conserved features and premetazoan ancestry. Molecular biology and evolution. 2012;29(3):1019–29. Epub 2011/11/03. 10.1093/molbev/msr271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Foster SJ. Molecular analysis of three major wall-associated proteins of Bacillus subtilis 168: evidence for processing of the product of a gene encoding a 258 kDa precursor two-domain ligand-binding protein. Molecular microbiology. 1993;8(2):299–310. Epub 1993/04/01. 10.1111/j.1365-2958.1993.tb01574.x . [DOI] [PubMed] [Google Scholar]
- 20.Koskiniemi S, Lamoureux JG, Nikolakakis KC, T'Kint de Roodenbeke C, Kaplan MD, Low DA, et al. Rhs proteins from diverse bacteria mediate intercellular competition. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(17):7032–7. Epub 2013/04/11. 10.1073/pnas.1300627110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whitney JC, Beck CM, Goo YA, Russell AB, Harding BN, De Leon JA, et al. Genetically distinct pathways guide effector export through the type VI secretion system. Mol Microbiol. 2014;92(3):529–42. 10.1111/mmi.12571 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alcoforado Diniz J, Coulthurst SJ. Intraspecies Competition in Serratia marcescens Is Mediated by Type VI-Secreted Rhs Effectors and a Conserved Effector-Associated Accessory Protein. Journal of bacteriology. 2015;197(14):2350–60. Epub 2015/05/06. 10.1128/JB.00199-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hood RD, Singh P, Hsu F, Guvener T, Carl MA, Trinidad RR, et al. A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe. 2010;7(1):25–37. Epub 2010/02/02. 10.1016/j.chom.2009.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma AT, Mekalanos JJ. In vivo actin cross-linking induced by Vibrio cholerae type VI secretion system is associated with intestinal inflammation. Proc Natl Acad Sci U S A. 2010;107(9):4365–70. Epub 2010/02/13. 10.1073/pnas.0915156107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Russell AB, Peterson SB, Mougous JD. Type VI secretion system effectors: poisons with a purpose. Nat Rev Microbiol. 2014;12(2):137–48. Epub 2014/01/05. 10.1038/nrmicro3185 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shneider MM, Buth SA, Ho BT, Basler M, Mekalanos JJ, Leiman PG. PAAR-repeat proteins sharpen and diversify the type VI secretion system spike. Nature. 2013;500(7462):350–3. Epub 2013/08/09. 10.1038/nature12453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jackson AP, Thomas GH, Parkhill J, Thomson NR. Evolutionary diversification of an ancient gene family (rhs) through C-terminal displacement. BMC genomics. 2009;10:584 Epub 2009/12/09. 10.1186/1471-2164-10-584 PubMed Central PMCID: PMC2935791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Busby JN, Panjikar S, Landsberg MJ, Hurst MR, Lott JS. The BC component of ABC toxins is an RHS-repeat-containing protein encapsulation device. Nature. 2013;501(7468):547–50. Epub 2013/08/06. 10.1038/nature12465 . [DOI] [PubMed] [Google Scholar]
- 29.Aoki SK, Pamma R, Hernday AD, Bickham JE, Braaten BA, Low DA. Contact-dependent inhibition of growth in Escherichia coli. Science. 2005;309(5738):1245–8. Epub 2005/08/20. 10.1126/science.1115109 . [DOI] [PubMed] [Google Scholar]
- 30.Aoki SK, Diner EJ, de Roodenbeke CT, Burgess BR, Poole SJ, Braaten BA, et al. A widespread family of polymorphic contact-dependent toxin delivery systems in bacteria. Nature. 2010;468(7322):439–42. Epub 2010/11/19. 10.1038/nature09490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ting SY, Bosch DE, Mangiameli SM, Radey MC, Huang S, Park YJ, et al. Bifunctional Immunity Proteins Protect Bacteria against FtsZ-Targeting ADP-Ribosylating Toxins. Cell. 2018. Epub 2018/10/23. 10.1016/j.cell.2018.09.037 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sana TG, Flaugnatti N, Lugo KA, Lam LH, Jacobson A, Baylot V, et al. Salmonella Typhimurium utilizes a T6SS-mediated antibacterial weapon to establish in the host gut. Proc Natl Acad Sci U S A. 2016;113(34):E5044–51. Epub 2016/08/10. 10.1073/pnas.1608858113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mulder DT, Cooper CA, Coombes BK. Type VI secretion system-associated gene clusters contribute to pathogenesis of Salmonella enterica serovar Typhimurium. Infection and immunity. 2012;80(6):1996–2007. Epub 2012/04/12. 10.1128/IAI.06205-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koskiniemi S, Garza-Sanchez F, Sandegren L, Webb JS, Braaten BA, Poole SJ, et al. Selection of Orphan Rhs Toxin Expression in Evolved Salmonella enterica Serovar Typhimurium. PLoS Genet. 2014;10(3):e1004255 Epub 2014/03/29. 10.1371/journal.pgen.1004255 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kung VL, Khare S, Stehlik C, Bacon EM, Hughes AJ, Hauser AR. An rhs gene of Pseudomonas aeruginosa encodes a virulence protein that activates the inflammasome. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(4):1275–80. Epub 2012/01/11. 10.1073/pnas.1109285109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chaudhuri RR, Morgan E, Peters SE, Pleasance SJ, Hudson DL, Davies HM, et al. Comprehensive assignment of roles for Salmonella typhimurium genes in intestinal colonization of food-producing animals. PLoS genetics. 2013;9(4):e1003456 Epub 2013/05/03. 10.1371/journal.pgen.1003456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kroger C, Colgan A, Srikumar S, Handler K, Sivasankaran SK, Hammarlof DL, et al. An infection-relevant transcriptomic compendium for Salmonella enterica Serovar Typhimurium. Cell Host Microbe. 2013;14(6):683–95. Epub 2013/12/18. 10.1016/j.chom.2013.11.010 . [DOI] [PubMed] [Google Scholar]
- 38.Morse RP, Nikolakakis KC, Willett JL, Gerrick E, Low DA, Hayes CS, et al. Structural basis of toxicity and immunity in contact-dependent growth inhibition (CDI) systems. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(52):21480–5. Epub 2012/12/14. 10.1073/pnas.1216238110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Helaine S, Thompson JA, Watson KG, Liu M, Boyle C, Holden DW. Dynamics of intracellular bacterial replication at the single cell level. Proc Natl Acad Sci U S A. 2010;107(8):3746–51. Epub 2010/02/06. 10.1073/pnas.1000041107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harms A, Brodersen DE, Mitarai N, Gerdes K. Toxins, Targets, and Triggers: An Overview of Toxin-Antitoxin Biology. Mol Cell. 2018;70(5):768–84. Epub 2018/02/06. 10.1016/j.molcel.2018.01.003 . [DOI] [PubMed] [Google Scholar]
- 41.Ghosh A, Baltekin O, Waneskog M, Elkhalifa D, Hammarlof DL, Elf J, et al. Contact-dependent growth inhibition induces high levels of antibiotic-tolerant persister cells in clonal bacterial populations. EMBO J. 2018. Epub 2018/03/25. 10.15252/embj.201798026 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takaya A, Tomoyasu T, Tokumitsu A, Morioka M, Yamamoto T. The ATP-dependent lon protease of Salmonella enterica serovar Typhimurium regulates invasion and expression of genes carried on Salmonella pathogenicity island 1. J Bacteriol. 2002;184(1):224–32. Epub 2001/12/14. 10.1128/JB.184.1.224-232.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lovett ST, Hurley RL, Sutera VA Jr., Aubuchon RH, Lebedeva MA. Crossing over between regions of limited homology in Escherichia coli. RecA-dependent and RecA-independent pathways. Genetics. 2002;160(3):851–9. Epub 2002/03/20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Petersen L, Bollback JP, Dimmic M, Hubisz M, Nielsen R. Genes under positive selection in Escherichia coli. Genome research. 2007;17(9):1336–43. Epub 2007/08/07. 10.1101/gr.6254707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Werren JH. Selfish genetic elements, genetic conflict, and evolutionary innovation. Proc Natl Acad Sci U S A. 2011;108 Suppl 2:10863–70. Epub 2011/06/22. 10.1073/pnas.1102343108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brunet YR, Khodr A, Logger L, Aussel L, Mignot T, Rimsky S, et al. H-NS Silencing of the Salmonella Pathogenicity Island 6-Encoded Type VI Secretion System Limits Salmonella enterica Serovar Typhimurium Interbacterial Killing. Infect Immun. 2015;83(7):2738–50. Epub 2015/04/29. 10.1128/IAI.00198-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vazquez-Torres A, Jones-Carson J, Baumler AJ, Falkow S, Valdivia R, Brown W, et al. Extraintestinal dissemination of Salmonella by CD18-expressing phagocytes. Nature. 1999;401(6755):804–8. Epub 1999/11/05. 10.1038/44593 . [DOI] [PubMed] [Google Scholar]
- 48.Virtanen P, Waneskog M, Koskiniemi S. Class II contact-dependent growth inhibition (CDI) systems allow for broad-range cross-species toxin delivery within the Enterobacteriaceae family. Mol Microbiol. 2019. Epub 2019/02/03. 10.1111/mmi.14214 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ma J, Sun M, Dong W, Pan Z, Lu C, Yao H. PAAR-Rhs proteins harbor various C-terminal toxins to diversify the antibacterial pathways of type VI secretion systems. Environ Microbiol. 2017;19(1):345–60. Epub 2016/11/22. 10.1111/1462-2920.13621 . [DOI] [PubMed] [Google Scholar]
- 50.Kuroda A. A polyphosphate-lon protease complex in the adaptation of Escherichia coli to amino acid starvation. Biosci Biotechnol Biochem. 2006;70(2):325–31. Epub 2006/02/24. 10.1271/bbb.70.325 . [DOI] [PubMed] [Google Scholar]
- 51.Nomura K, Kato J, Takiguchi N, Ohtake H, Kuroda A. Effects of inorganic polyphosphate on the proteolytic and DNA-binding activities of Lon in Escherichia coli. J Biol Chem. 2004;279(33):34406–10. Epub 2004/06/10. 10.1074/jbc.M404725200 . [DOI] [PubMed] [Google Scholar]
- 52.Takaya A, Suzuki M, Matsui H, Tomoyasu T, Sashinami H, Nakane A, et al. Lon, a stress-induced ATP-dependent protease, is critically important for systemic Salmonella enterica serovar typhimurium infection of mice. Infect Immun. 2003;71(2):690–6. Epub 2003/01/24. 10.1128/IAI.71.2.690-696.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang D, de Souza RF, Anantharaman V, Iyer LM, Aravind L. Polymorphic toxin systems: Comprehensive characterization of trafficking modes, processing, mechanisms of action, immunity and ecology using comparative genomics. Biology direct. 2012;7:18 Epub 2012/06/27. 10.1186/1745-6150-7-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Poole SJ, Diner EJ, Aoki SK, Braaten BA, t'Kint de Roodenbeke C, Low DA, et al. Identification of functional toxin/immunity genes linked to contact-dependent growth inhibition (CDI) and rearrangement hotspot (Rhs) systems. PLoS genetics. 2011;7(8):e1002217 Epub 2011/08/11. 10.1371/journal.pgen.1002217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(12):6640–5. Epub 2000/06/01. 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.B B. Curing bacterial cells of lysogenic viruses by using UCB indicator plates. BioTechniques. 1984;2:234–40. [Google Scholar]
- 57.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–8. Epub 2002/02/16. 10.1006/meth.2001.1262 . [DOI] [PubMed] [Google Scholar]
- 58.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–80. Epub 2013/01/19. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]