Abstract
The United States–Affiliated Pacific Islands (USAPI) are part of the US National Tuberculosis (TB) Surveillance System and use laboratory services contracted through a cooperative agreement with the Centers for Disease Control and Prevention (CDC). In 2004, the CDC established the National Tuberculosis Genotyping Service, a system to genotype 1 isolate from each culture-confirmed case of TB. To describe the molecular epidemiology of TB in the region, we examined all Mycobacterium tuberculosis isolates submitted for genotyping from January 1, 2004, to December 31, 2008. Over this time period, the USAPI jurisdictions reported 1339 verified TB cases to the National Tuberculosis Surveillance System. Among 419 (31%) reported culture-confirmed TB cases, 352 (84%) had complete genotype results. Routine TB genotyping allowed, for the first time, an exploration of the molecular epidemiology of TB in the USAPI.
Keywords: genotype, molecular epidemiology, Pacific Islands (trust territory), tuberculosis
Introduction
The United States–Affiliated Pacific Islands (USAPI) include 3 US territories (American Samoa, the Commonwealth of the Northern Marianas Islands, and Guam) and 3 independent nations (Federated States of Micronesia [FSM], Republic of Marshall Islands [RMI], and Palau) associated with the United States through the 1986 US Compact of Free Association.1 The compact mandates certain economic provisions, including economic support and technical assistance by the US Centers for Disease Control and Prevention (CDC) for the tuberculosis (TB) control programs in the 6 USAPI jurisdictions.
Since 2002, the USAPI have reported each confirmed TB case to the CDC via the National Tuberculosis Surveillance System (NTSS).2 TB case rates in the USAPI have been 2- to 50-fold higher than in the US states with the highest case rates (Hawaii, with a case rate of 9.2/100 000 population in 2009).2 USAPI jurisdictions face unique challenges with limited health care infrastructure, highly mobile populations, varied economic and social conditions, and population centers separated by vast expanses of ocean. TB programs in the USAPI have struggled to diagnose and manage TB patients according to the World Health Organization DOTS Plus strategy.3 For example, radiography services were often inaccessible, and diagnosis was based primarily on acid-fast bacilli sputum smear results, with the ability to obtain culture not consistently available until 2007. Public health programs also lacked human resources to provide treatment of TB disease via directly observed therapy.
In 2004, the CDC established the National TB Genotyping Service (NTGS) to genotype at least one Mycobacterium tuberculosis isolate from every culture-confirmed TB case reported in the United States, including the USAPI.4 USAPI’s initial participation in the NTGS was limited due to logistical challenges and the high cost of processing and shipping isolates from the USAPI to the United States. In 2007, the CDC established a contractual agreement with a regional laboratory in Hawaii to provide shipping, culture in liquid media, and drug susceptibility testing for all isolates from the USAPI. While the application of TB genotyping has utility in low-burden, high-resource settings,5,6 whether TB genotyping could be implemented or could have program-matic relevance in a high-burden, low-resource setting, such as the USAPI, was unclear.
The molecular epidemiology of TB in the USAPI had not previously been described. In this descriptive analysis, we described the implementation and results of TB genotyping in the USAPI, including one of its initial applications in the region, when genotyping assisted in the investigation of an outbreak.
Methods
The USAPI jurisdictions performed Ziehl-Nielson smear microscopy to make a preliminary TB diagnosis and then shipped sputum specimens to the regional laboratory in Hawaii, for additional testing by Truant auramine-rhodamine staining followed by fluorescent microscopy and liquid media culture.7 For genotyping, primary cultures positive for M tuberculosis were subcultured in a liquid medium (10% glycerol in Dubos Davis broth with Tween and albumin). Subcultures were incubated at 37°C, with visual monitoring for growth.8 On culture growth, isolates were batched and shipped to the NTGS laboratory in California for genotyping.
TB genotypes were determined by using a standardized protocol for spacer oligonucleotide typing (spoligotyping) and a panel of 12-locus mycobacterial interspersed repetitive unit-variable number of tandem repeats (MIRU-VNTR).9–11 To facilitate communication of genotype data, the national “PCR Type” naming convention was used, where “PCR” is followed by 5 digits, which are assigned sequentially to each unique spoligotype and 12-locus MIRU-VNTR combination identified in the United States and its affiliated territories.12
The study population included all verified TB cases reported from January 2004 to December 2008. We determined the total number of TB cases reported to the NTSS (ie, with surveillance records), the number and proportion of those that were culture-confirmed, and the number and proportion of total reported culture-confirmed TB cases with a valid genotype. NTGS results were linked to individual NTSS case records using a standardized case identification number and a unique laboratory accession number, thus forming discrete genotype/surveillance patient-level records in a linked data set.2 These lists are not name based or maintained in one location, making the process of linking records challenging. We examined linking inconsistencies to determine the proportion of eligible culture-positive TB cases with valid genotype results.
When multiple isolates were genotyped for the same individual in the same surveillance year, the first genotype result was included in analysis. Two patients with discordant genotyping results were excluded from analysis.
A TB genotype cluster was defined as at least 2 cases with matching PCR Types (ie, indistinguishable spoligotype and 12-locus MIRU-VNTR results) reported from the same USAPI jurisdiction during the study period.
Univariate analyses were used to determine demographic and epidemiologic factors associated with genotype clustering. We estimated the relative genetic diversity of isolates collected in the USAPI. Methods for assigning phylogenetic lineages and subfamily were primarily based on spoligotype and in some instances, 12-locus MIRU-VNTR results, as described by Kato-Maeda et al.13
Results
During the 2004 to 2008 study period, 533 M tuberculosis isolates from the 6 USAPI jurisdictions were genotyped. Figure 1 shows the number of genotyped isolates within each jurisdiction by year. Regionally the number of patients with isolates sent for genotyping has increased over the last 5 years from <50 in 2004 to 191 in 2008.
Figure 1.
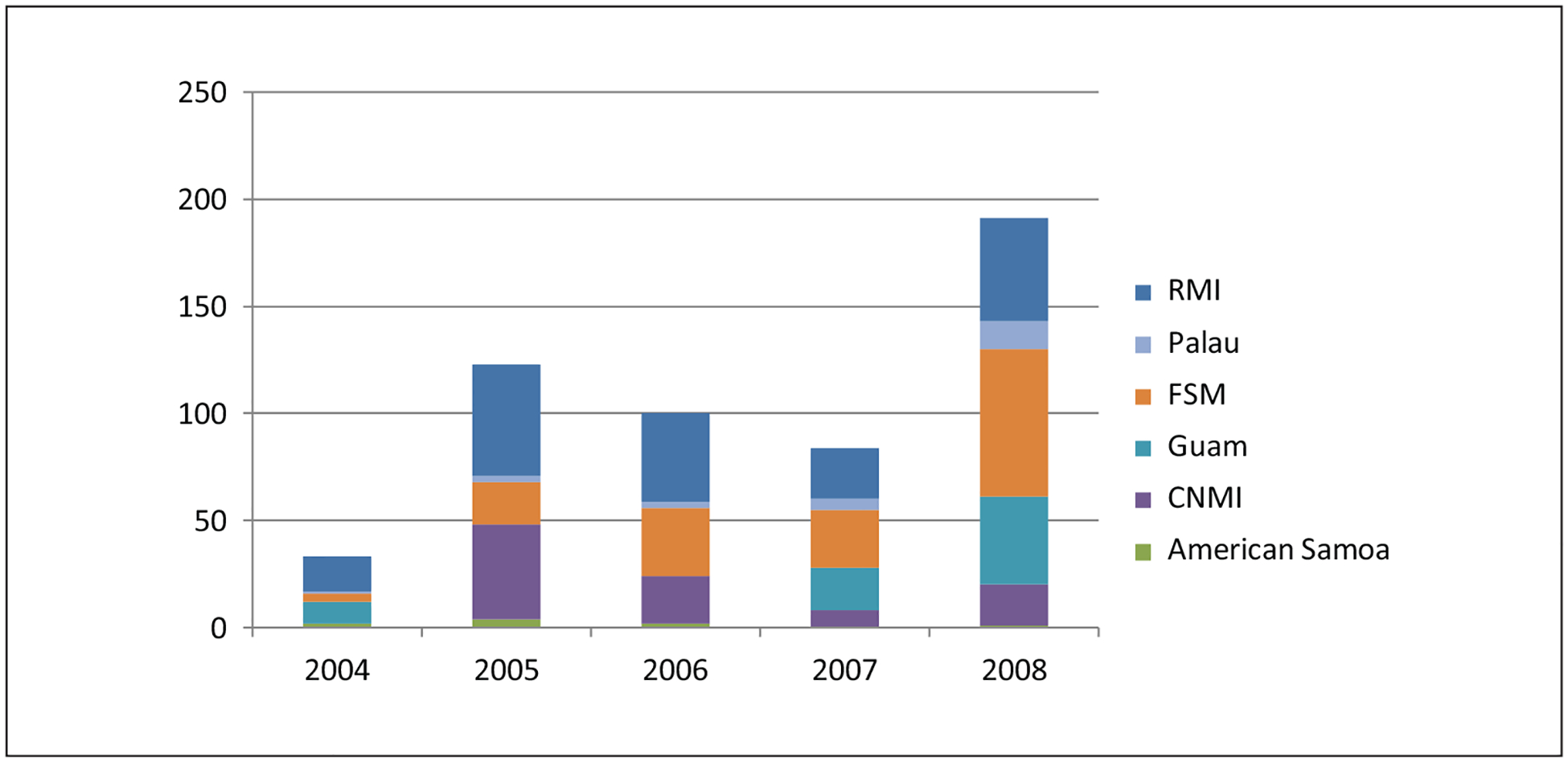
Number of genotyped isolates in each jurisdiction by year—USAPI, 2004–2008.
During the same time period, the USAPI jurisdictions reported 1339 verified TB cases to the NTSS. Among those the number of reported cases that linked to a genotyped isolate was 352 (84%; Table 1). There were 2 patients with discordant genotype results and thus were excluded from final analysis.
Table 1.
Number of TB Cases Reported, With Culture Confirmation, and With Genotype Results by Jurisdiction—USAPI, 2004–2008.
| Jurisdiction | No. of TB Cases Reported to Surveillance System, N | No. of Culture-Confirmed Cases in Surveillance Records, n (%)a | No. of Isolates Submitted to Genotyping Laboratory, n | No. of Genotyped Cases Linked to Surveillance Records, n |
|---|---|---|---|---|
| American Samoa | 16 | 6 (38) | 9 | 8 |
| CNMI | 201 | 64 (32) | 93 | 77 |
| FSM | 468 | 66 (14) | 152 | 98 |
| Guam | 350 | 163 (47) | 71 | 55 |
| Palau | 54 | 11 (20) | 25 | 21 |
| RMI | 250 | 109 (44) | 182 | 93 |
| Total | 1339 | 419 (31) | 532 | 352 |
Abbreviations: TB, tuberculosis; USAPI, United States–Affiliated Pacific Islands; CNMI, Commonwealth of the Northern Marianas Island; FSM, Federated States of Micronesia; RMI, Republic of the Marshall Islands.
percentages are using the number of surveillance records as the denominator.
A number of isolates with complete TB genotype results did not correspond to a patient that had been reported to NTSS. However, the discrepancies were not unidirectional. As shown in Figure 2, the NTGS genotyped 532 nonduplicate isolates and the NTSS reported 419 culture-confirmed cases. Of the 419, 253 (60%) had genotyping results. There were 180 isolates with genotyping results that were not successfully linked to a reported TB case. There were 14 genotyped isolates that linked to cases that were not reported as culture-confirmed and 85 where culture data were missing.
Figure 2.
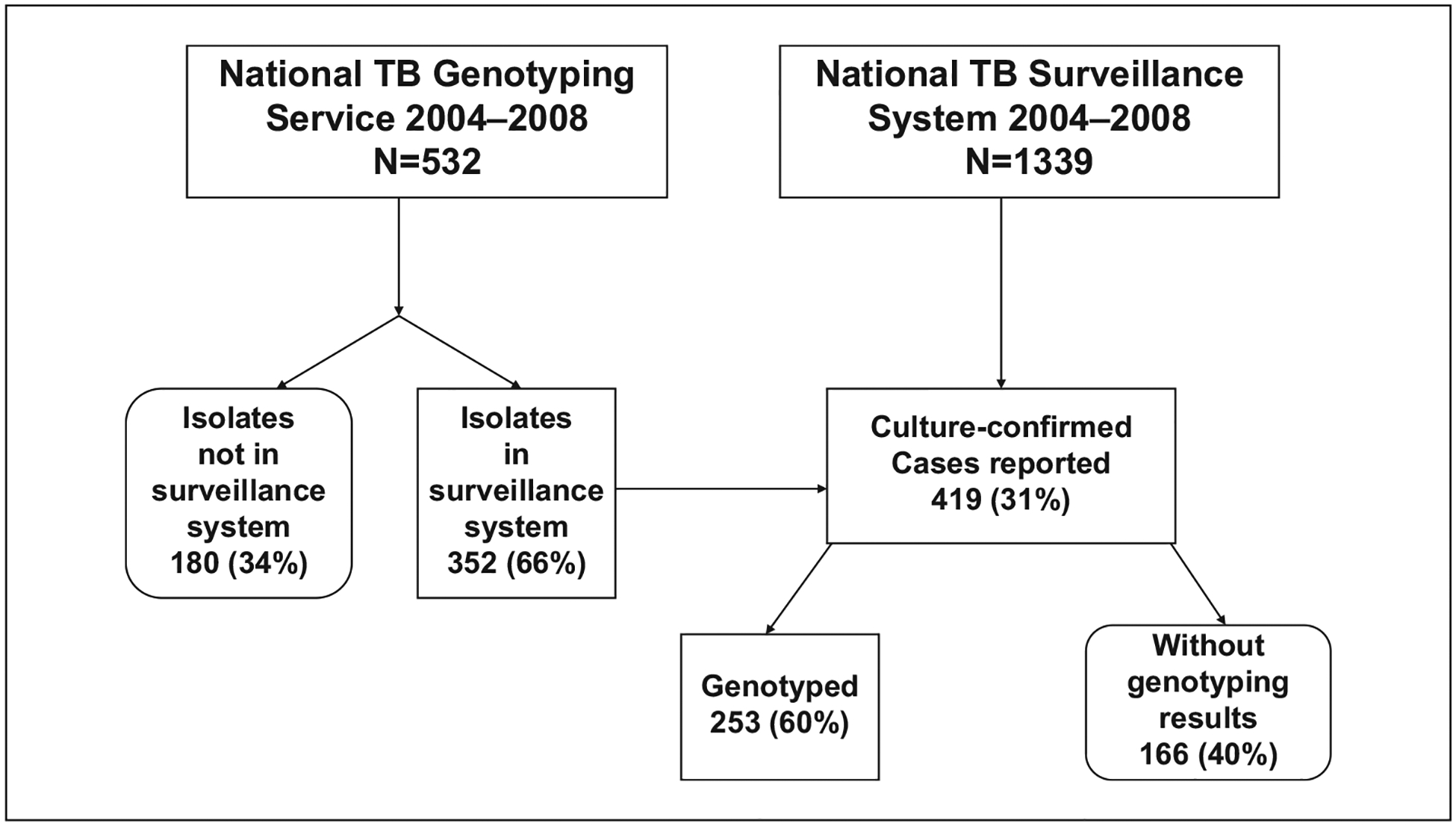
Number of surveillance records, culture-confirmed cases, and genotyped isolates, 2004–2008.
Among the 6 USAPI jurisdictions, FSM and RMI reported the largest number of TB cases. These 2 jurisdictions also had a higher proportion of cases that clustered within each jurisdiction. In FSM and RMI, 85% of cases were in genotype clusters. Table 2 shows the percentage of cases within the genotype clusters by jurisdiction.
Table 2.
Genotype Clustering by Jurisdiction—USAPI, 2004–2008.
| Jurisdiction | Number of Genotype Clustersa | Range of Number of Cases Within Each Cluster | Percentage of Cases Within a Genotype Cluster |
|---|---|---|---|
| American Samoa | 1 | 2 | 25 |
| CNMI | 4 | 2–19 | 46 |
| Guam | 11 | 2–15 | 65 |
| FSM | 15 | 2–19 | 85 |
| Palau | 3 | 3–4 | 45 |
| RMI | 9 | 2–56 | 85 |
Abbreviations: USAPI, United States–Affiliated Pacific Islands; CNMI, Commonwealth of the Northern Marianas Island; FSM, Federated States of Micronesia; RMI, Republic of the Marshall Islands.
Genotype cluster is at least 2 cases in the same jurisdiction with isolates that have matching spoligotype and 12-locus MIRU-VNTR (ie, PCR Type).
In total, 38 TB genotype clusters were identified with 263 (75%) TB cases belonging to a cluster. The most prevalent genotype patterns were East-Asian, Beijing (PCR00803 [22%] and PCR00002 [8%]), IndoOceanic, Manila (PCR00041 [12%] and PCR00017 [5%]),13,14 and Euro-American, Latin American Mediterranean (PCR03135 [4%]; Table 3).
Table 3.
Most Common Genotype Patterns—USAPI, 2004–2008.
| PCR Type (Lineage) | Spoligotype/12-Locus MIRU-VNTR | Number of Cases | Median Age in Years (Range) |
|---|---|---|---|
| PCR00803 (East-Asian, Beijing) | 000000000003771/222325173533 | 78 | 40 (8–73) |
| PCR00041 (IndoOceanic, Manila) | 677777477413771/254326223432 | 43 | 50 (25–82) |
| PCR00002 (East-Asian, Beijing) | 000000000003771/223325173533 | 29 | 32 (1–73) |
| PCR00017 (IndoOceanic, Manila) | 677777477413771/254326223422 | 11 | 25 (0–54) |
| PCR03135 (Euro-American, Latin American-Mediterranean) | 577777607760771/124326163326 | 9 | 21 (10–45) |
Abbreviation: USAPI, United States–Affiliated Pacific Islands.
Three genotype patterns were predominantly found within 1 USAPI jurisdiction. All PCR03135 cases and nearly all PCR00017 (82%) cases were identified in FSM. PCR00803 and other East-Asian, Beijing genotypes were common among the RMI cases, but PCR00803 was rarely seen in the other 5 USAPI jurisdictions.
Discussion
Reliable laboratory diagnosis is one of the cornerstones of TB control. New laboratory resources in USAPI, established in 2007, increased confirmed TB diagnoses with culture results and TB genotyping. Routine TB genotyping allowed, for the first time, an exploration of the molecular epidemiology of TB in the USAPI.
The 6 USAPI jurisdictions increased the number of M tuberculosis isolates submitted for genotyping between 2004 and 2008. Certain genotype patterns (eg, PCR00803 in RMI) appear to be more prevalent within certain jurisdictions. Genotype clustering has been used to estimate disease attributable to recent transmission in other populations, but it is unclear whether that can be applied to the USAPI given the predominance of a few highly prevalent strains and the low proportion of culture-positive cases among reported cases. These factors make it difficult to discern endemic circulating strains from novel strains, which may be a result of recent TB transmission events. As the proportion of culture-positive cases with subsequent genotyping results increases, the expected prevalence of each genotype will be better understood and TB controllers may more readily recognize unusual clustering that might be attributable to recent transmission, or potential outbreaks.
This was the first analysis of the molecular epidemiology in the USAPI; however, this study does have limitations. First, the implementation of genotyping in the USAPI was dependent on consistent support for shipping of specimens to a regional laboratory for culture and subsequent testing; therefore, there were limited data from the first 3 years of the study period. Second, the surveillance system remained paper-based through 2008, making the linking of data between the NTSS and the NTGS difficult and likely leading to missed linkages between the 2 data sets. Finally, because genotyping results were not available for many TB cases, the degree of clustering and inferences about the proportion of cases attributed to recent transmission may be distorted.
Genotyping is an important adjunct to epidemiologic investigations. Contact investigations around contagious TB patients are important to identify and treat others who might have active TB disease or recent TB infection. However, when contact investigations are incomplete, it can be difficult to ascertain transmission venues and identify persons at highest risk of latent TB infection, or progression to active disease. In these situations, particularly, genotyping can help a TB program identify additional patients who may be in the same chain of transmission.15
The overall utility of genotyping to the region was limited not only by the proportion of culture-confirmed cases that have isolates submitted for genotyping but also by the proportion of total cases that are culture-confirmed, which was only 31% of reported TB cases during the study period. In addition, although the relevance of phylogenetic lineage is not fully understood, recent evidence suggests that lineages are associated with clinical manifestations and might have implications on diagnosis and treatment.14
The completeness and accuracy of surveillance systems directly affect the utility of genotyping to understand the molecular epidemiology of TB in a geographic region. Because TB genotyping can only be performed on culture-confirmed cases (ie, with a M tuberculosis isolate), our finding in 5 USAPI jurisdictions that the number of genotyped isolates exceeded the number of culture-confirmed cases is counterintuitive. This discrepancy highlights an important gap in the reporting of TB cases to the NTSS. Recent implementation of an electronic reporting system has aided jurisdictions in improving all data management activities in USAPI, specifically surveillance efforts in reporting all culture-confirmed cases. Finally, CDC’s newly introduced Tuberculosis Genotyping Information Management System12 will improve the USAPI’s access to genotyping results, more easily linking them to surveillance records, thus shortening the time interval before genotyping findings can be applied to routine TB control activities.16
Conclusion
Descriptions of the implementation and utilization of TB genotyping in low-resource settings are few in number. Molecular methods when applied universally to all culture-confirmed isolates can help provide information about chains of transmission, discover epidemiologic links that would otherwise be missed, and identify outbreaks. As TB surveillance systems in the USAPI improve and more isolates are sent for culture and subsequent genotyping, molecular methods will add to the ability of TB control programs to understand transmission within and between jurisdictions.
A Case Study: Application of Genotyping in an Outbreak Investigation in the Federated States of Micronesia (FSM).
One of the most common uses of molecular epidemiology is to detect and confirm outbreaks.16,17 In July 2008, Chuuk State, FSM, experienced the emergence of multidrug-resistant (MDR) TB. FSM requested onsite assistance from CDC to investigate these cases.18,19 This outbreak investigation gave context to using TB genotyping data in USAPI and prompted the initial examination of the molecular epidemiology of M tuberculosis in these jurisdictions.
Molecular epidemiology helped demonstrate 2 distinct outbreaks where there had been thought to be only one. Three patients had 5-drug resistant MDR TB, with an East-Asian, Beijing genotype (PCR00002: spoligotype: 000000000003771; 12-locus MIRU-VNTR: 223325173533). The investigation determined that the index patient was likely infected with a PCR00002 isolate while working abroad alongside other foreign nationals in garment factories. Two other patients with 3-drug resistant MDR TB and a distinct genotype (PCR00286; spoligotype: 777777777760771; 12-locus MIRU-VNTR: 223325143323) both lived in the same village where 22 other cases of TB had been diagnosed during the preceding 3 years. Prior to the MDR TB cases, all the culture-confirmed cases associated with that village had isolates with the same PCR00286 genotype pattern, a potential 2-drug resistance profile that was a precursor of the MDR TB outbreak genotype. The index patient in this second outbreak had a history of TB and acquired MDR TB over the course of treatment, likely due to lack of adherence and poor directly observed therapy practices.
The genotype in the first outbreak, PCR00002, accounts for approximately 15% of TB cases in FSM. The combination of this genotype and drug susceptibility pattern has only been seen in patients who acquired TB disease through the chain of transmission initiated by the index patient.
The genotype in the second outbreak, PCR00286, had only been seen in patients epidemiologically linked to others living in the same village. Transmission of this genotype had been documented over the preceding 3 years in this village with both a 2- and 3-drug-resistant profile.
The genotyping data proved to be critical in identifying not only 1 outbreak, but 2 outbreaks that were occurring simultaneously, on the central island of Weno, Chuuk State. The subsequent diagnosis and active case-finding activities for both of these outbreaks targeted 2 different villages, with slightly different strategies to TB screening and treatment of presumed MDR latent TB infection.
Acknowledgments
We are grateful to the laboratory personnel at Hawaii’s Diagnostic Laboratory Services, the National TB Genotyping Service scientists at the California Department of Health Services, and to the local and state TB programs who participate in surveillance and genotyping activities. The authors would like to particularly acknowledge the TB program directors and staff from each of the USAPI jurisdictions. Grace Lin, Steven Yu, Claire Ying, Matt Bankowski, Heidi Soeters, Lori Armstrong, Risa Bukbuk, Mayleen Ekiek, Dorina Fred, Krista Powell, and Richard Brostrom have been particularly helpful in reviewing the genotyping data and providing vital information on how surveillance systems function in the USAPI.
Funding
The author(s) received no financial support for the research, authorship, and/or publication of this article.
Footnotes
Declaration of Conflicting Interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article. Also, the findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
References
- 1.Department of Interior, US Government. Compact of free association. http://www.fsmlaw.org/compact/index.htm. Accessed November 15, 2012.
- 2.Centers for Disease Control and Prevention. Reported TB in the United States, 2009. Atlanta, GA: US Department of Health and Human Services; 2010. [Google Scholar]
- 3.World Health Organization. 2010. Global Plan to Stop TB 2006–2015. http://whqlibdoc.who.int/publications/2006/924159487X_eng.pdf. Accessed November 15, 2012.
- 4.Centers for Disease Control and Prevention. New CDC program for rapid genotyping of Mycobacterium tuberculosis isolates. MMWR Morb Mortal Wkly Rep. 2005;54(2):47. [Google Scholar]
- 5.Clark CM, Driver CR, Munsiff SS, et al. Universal genotyping in tuberculosis control program, New York City, 2001–2003. Emerg Infect Dis. 2006;12:719–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haddad MB, Diem LA, Cowan LS, et al. Tuberculosis genotyping in six low-incidence states, 2000–2003. Am J Prev Med. 2007;32:239–243. [DOI] [PubMed] [Google Scholar]
- 7.Kommareddi S, Abramowsky C, Swinehart G, Hrabak L. Nontuberculous mycobacterial infections: comparison of the fluorescent auramine-O and Ziehl-Neelsen techniques in tissue diagnosis. Hum Pathol. 1984:15:1085–1089. [DOI] [PubMed] [Google Scholar]
- 8.Dubos RJ, Davis BD, et al. The effect of water soluble lipids on the growth and biological properties of tubercle bacilli. Am Rev Tuberc. 1946;54:204–212. [DOI] [PubMed] [Google Scholar]
- 9.Kamerbeek J, Schouls L, Kolk A, et al. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol. 1997;35:907–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Supply P, Mazars E, Lesjean S, Vincent V, Gicquel B, Locht C. Variable human minisatellite-like regions in the Mycobacterium tuberculosis genome. Mol Microbiol. 2000;36:762–771. [DOI] [PubMed] [Google Scholar]
- 11.Cowan LS, Diem L, Monson T, et al. Evaluation of a two-step approach for large-scale, prospective genotyping of Mycobacterium tuberculosis isolates in the United States. J Clin Microbiol. 2005;43:688–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghosh S, Moonan PK, Cowan L, Grant J, Kammerer S, Navin TR. Tuberculosis genotyping information management system: enhancing tuberculosis surveillance in the United States. Infect Genet Evol. 2012;12:782–788. [DOI] [PubMed] [Google Scholar]
- 13.Kato-Maeda M, Gagneux S, Flores LL, et al. Strain classification of Mycobacterium tuberculosis: congruence between large sequence polymorphisms and spoligotypes. Int J Tuberc Lung Dis. 2011;15:131–133. [PMC free article] [PubMed] [Google Scholar]
- 14.Click ES, Moonan PK, Winston CA, Cowan LS, Oeltmann JE. Relationship between Mycobacterium tuberculosis phylogenetic lineage and clinical site of disease. Clin Infect Dis. 2012;54:211–219. [DOI] [PubMed] [Google Scholar]
- 15.McElroy PD, Sterling TR, Driver CR, et al. Use of DNA fingerprinting to investigate a multiyear, multistate tuberculosis outbreak. Emerg Infect Dis. 2002;8:1252–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McNabb SJ, Kammerer JS, Hickey AC, et al. Added epidemiologic value to tuberculosis prevention and control of the investigation of clustered genotypes of Mycobacterium tuberculosis isolates. Am J Epidemiol. 2004;160:589–597. [DOI] [PubMed] [Google Scholar]
- 17.Barnes PF, Cave MD. Molecular epidemiology of tuberculosis. N Engl J Med. 2003;349:1149–1156. [DOI] [PubMed] [Google Scholar]
- 18.Centers for Disease Control and Prevention. Two simultaneous outbreaks of multidrug-resistant tuberculosis—Federated States of Micronesia, 2007–2009. MMWR Morb Mortal Wkly Rep. 2009;58(10): 253–256. [PubMed] [Google Scholar]
- 19.Brostrom R, Fred D, Heetderks A, et al. Islands of hope: building local capacity to manage an outbreak of multidrug-resistant TB in the Pacific. Am J Public Health. 2011;101:14–18. [DOI] [PMC free article] [PubMed] [Google Scholar]


