Abstract
Objective(s):
Plasmid-mediated quinolone resistance (PMQR) determinants and integrons have a considerable contribution to bacterial drug resistance in Gram-negative pathogens. We studied the prevalence of PMQR genes and integron carriage in multidrug-resistant community isolates of Klebsiella spp.
Materials and Methods:
Two hundred and fifty Klebsiella spp. isolates were collected from outpatient specimens between August 2015 and October 2017 in Yazd central Laboratory, Iran. Antibiotic susceptibility was determined against 17 antibiotics and minimum inhibitory concentration (MIC) of ciprofloxacin was measured by E-test. Polymerase chain reaction (PCR) was employed for detection of qnrA, qnrB, qnrS, aac(6’)-Ib-cr, oqxAB and qepA genes.
Results:
Disc diffusion results showed that 17 isolates (6.8%) were multidrug resistant (MDR), two of which were Klebsiella oxytoca and 15 were Klebsiella pneumoniae. MIC measurements revealed 11 ciprofloxacin-resistant isolates (including the two K. oxytoca), three intermediately-resistant and three ciprofloxacin-susceptible isolates. All ciprofloxacin-resistant and intermediately-resistant isolates carried at least one and up to four PMQR genes. The most prevalent PMQR gene was oqxAB (93.75%) followed by aac(6’)-ib-cr (50.0%), qnrB (25.0%) and qnrS (18.75%) but qnrA and qepA were not detected. Class 1 integron was observed in 14 (82.3%) isolates including nine ciprofloxacin-resistant, two intermediately-resistant, and three susceptible isolates. Class 2 and 3 integrons were not observed.
Conclusion:
Presence of MDR, multiple PMQR determinants as well as class 1 integron in community isolates of Klebsiella spp. can be an important source of transmission of these opportunistic pathogens.
Key Words: Community isolates, Integron, Klebsiella spp., MDR, Plasmid-mediated quinolone- resistance, PMQR
Introduction
Klebsiella pneumoniae is an opportunistic pathogen and the cause of a significant number of nosocomial infections including pneumonia, wound, blood, intra-abdominal and urinary tract infections (1). Over the past few years, the most common treatment for these infections has been the use of extended-spectrum β-lactam antibiotics and fluoroquinolones (2). However, multidrug resistant (MDR) isolates are rapidly spreading not only in nosocomial infections but also more recently in the community isolates (2, 3). Previously, fluoroquinolone resistance was thought to be solely due to the mutations occurring within its DNA gyrase target gene (4). However, the discovery of plasmid-mediated quinolone resistance (PMQR) in clinical isolates of Enterobacteriaceae has shown to play an important role in quinolone resistance (5). PMQR genes, frequently found in clinical isolates of Enterobacteriaceae, include: qnrA, qnrB, qnrS, qnrC and qnrD which their pentapeptide protein products protect quinolones from inhibition by DNA gyrase and topoisomerase IV, as well as the aminoglycoside acetyltransferase (Aac(6’)-Ib-cr) which is responsible for enzymatic modification of fluoroquinolones (5). Furthermore, fluoroquinolone-resistance is often observed in extended spectrum β-lactamase (ESBL) producing K. pneumoniae (6, 7). Over-expression of efflux pumps such as QepA, AcrAB and OqxAB also contribute to quinolone and fluoroquinolone resistance (8-10). Among the efflux pumps discovered in Gram-negative bacteria, the oqxAB was initially detected on a conjugative plasmid in Escherichia coli isolated from swine manure (11). Dissemination of oqxAB, a PMQR determinant, was later shown in clinical isolates of K. pneumoniae from Spain, South Korea and China (10, 12, 13). Furthermore, presence of PMQR genes on mobile genetic elements such as antibiotic resistant integrons, often carried by conjugative plasmids, has been shown to contribute to the dissemination of multidrug resistance in pathogens as well as environmental bacteria (14, 15). Among these mobile genetic elements, class 1 integron is the most commonly detected among Gram-negative MDR clinical isolates followed by class 2 integron (16, 17). However, there are few studies on integron carriage by community acquired isolates of Klebsiella spp. The aim of this study was to investigate the presence of multiple antibiotic resistance determinants in Klebsiella spp. isolates from outpatient specimens and the possible correlation between quinolone-resistance, PMQR and integron carriage in these organisms.
Materials and Methods
Bacterial isolates and identification
Two hundred and fifty non-duplicate community acquired Klebsiella isolates were collected from outpatient specimens in Yazd Central Laboratory between August 2015 and October 2017 (mean age between 35 and 81 years old). Conventional biochemical tests were used for identification of the isolates which were then maintained in Tryptic Soy Broth (TSB; Merck, Germany) containing 4% glycerol at −70 °C.
Antimicrobial susceptibility testing
Susceptibility testing against various antibiotics was performed by disc diffusion according to the CLSI guidelines (2017) using commercially available discs (Mast, UK) including: amoxicillin (AMX, 10 μg), cefalotin (CF, 30 μg), ceftazidime (CAZ, 30 μg), cefotaxime (CTX, 30 μg), gentamicin (GM, 10 μg), nalidixic acid (NA, 30 μg), kanamycin (KM, 30 μg), amikacin (AN, 30 μg), trimethoprim/sulfamethoxazole (SXT, 23.75/1.25 μg), ciprofloxacin (CIP, 30 μg), ofloxacin (OFX, 5 μg), norfloxacin (NOR, 10 μg), nitrofurantoin (NF, 300 μg), meropenem (MEM, 10 μg), ertapenem (ETP, 10 μg), tetracycline (TE, 30 μg) chloramphenicol (CM, 30 μg) (18). K. pneumoniae ATCC 10031 was used as the susceptible control.
Determination of minimum inhibitory concentrations
Minimum inhibitory concentrations (MICs) were measured for ciprofloxacin-resistant and intermediately-resistant isolates by E-test (Liofilchem, Italy) according to the CLSI 2017 guidelines (18).
DNA Extraction and PCR amplification
DNA extraction was carried out by boiling. Briefly, a loopful of bacteria grown overnight on MacConkey agar (Merck, Germany) were resuspended in 500 ml sterile double distilled water, boiled at 100 ºC for 10 min, and were centrifuged at 10,000 g for 10 min. The supernatant used as DNA template for detection of qnr genes (qnrA, qnrB and qnrS) as well as the presence of integrase genes (Int1, Int2 and Int3) by PCR using specific protocols and primers shown in Table 1 (3, 10, 19, 20). Each PCR reaction mixture (25 μl) contained 1.5 μl DNA template, 1.5 mM MgCl2, 0.25 mM of dNTP mix (Cinnagen, Iran), 1 unit of DFS-Taq DNA polymerase (Bioron, Germany), and 20 pmol of each primer (Faza Biothec, Iran). PCR amplifications were carried out in a thermo cycler (Applied Biosystems, USA). PCR products were separated on 1.5% agarose gels and visualized using gel a documentation system (ATP, Iran). PCR products were sequenced (Pishgam company, Tehran, Iran) to determine the subclasses of qnr genes as well as confirmation of integrase amplification products.
Table 1.
Primers and thermocycler conditions used for amplification of plasmid-mediated quinolone resistance (PMQR) and integron genes in ciprofloxacin-resistant community isolates of Klebsiella spp
| Gene | Primer Sequence |
PCR Product Size
(bp) |
Thermocycler conditions
|
Ref | |||
|---|---|---|---|---|---|---|---|
| Denaturation | Annealing | Extension | Number of cycles | ||||
|
qnrA-F qnrA-R |
5’-TTCTCACGCCAGGATTTGAG-3’ 3’-TGCCAGGCACAGATCTTGAC-5’ |
571 | 94º 1 min |
57º 1 min |
72º 1 min |
30 | |
|
qnrB-F qnrB-R |
5’-TGGCGAAAAAATTGAACAGAA-3’ 3’-GAGCAACGATCGCCTGGTAG-5’ |
594 | 94º 1 min |
57º 1 min |
72º 1 min |
30 | |
|
qnrS-F qnrS-R |
5’-GACGTGCTAACTTGCGTGAT-3’ 3’-AACACCTCGACTTAAGTCTGA-5’ |
388 | 94º 1 min |
57º 1 min |
72º 1 min |
30 | |
|
aac(6’)-Ib
-
cr-F aac(6’)-Ib-cr-R |
5’-TTGCGATGCTCTATGAGTGGCTA-3’ 3’-CTCGAATGCCTGGCGTGTTT-5’ |
482 | 94º 1 min |
54º 1 min |
72º 1 min |
30 | |
|
oqxA-F oqxA-R |
5’-CTCGGCGCGCGATGATGCT-3’ 3’-CCACTCTTCACGGGAGACGA-5’ |
392 | 94º 45 sec |
57º 45 sec |
68º 1 min |
34 | |
|
oqxB-F oqxB-R |
5’-TTCTCCCCCGGCGGGAAGTAC-3’ 3’-CTCGGCCATTTTGGCGCGTA-3’ |
512 | 94º 45 sec |
64º 45 sec |
72º 1 min |
32 | |
|
qepA-F qepA-R |
5’-CTGCAGGTACTGCGTCATG-3’ 3’-CGTGTTGCTGGAGTTCTTC-5’ |
709 | 94º 45 sec |
56º 45 sec |
72º 45 sec |
30 | |
| Int1-F Int1-R |
5’-CCTCCCGCACGATGATC-3’ 3’-TCCACGCATCGTCAGGC-5’ |
280 | 94º 1 min |
60º 1 min |
72º 1 min |
35 | |
| Int2-F Int2-R |
5’-TTATTGCTGGGATTAGGC-3’ 3’-ACGGCTACCCTCTGTTATC-5’ |
233 | 94º 1 min |
60º 1 min |
72º 1 min |
35 | |
| Int3-F Int3-R |
5’-AGTGGGTGGCGAATGAGTG-3’ 3’-TGTTCTTGTATCGGCAGGTG-5’ |
600 | 94º 1 min |
60º 1 min |
72º 1 min |
35 | |
Results
Of the 250 Klebsiella outpatient isolates, nine were K. oxytoca (3.6%) and the rest were K. pneumoniae. The majority of the isolates were urinary (98%) including all K. oxytoca and 96.4% of K. pneumoniae isolates. Three isolates (1.2%) were from wounds and two (0.8%) were from sputum specimens. Disc diffusion results showed that 17 isolates (6.8%) were resistant to at least three antibiotic classes and were considered multidrug resistant (MDR) (Table 2). Fifteen MDR isolates were obtained from urine of female subjects over 60 years of age. One isolate was recovered from the sputum of a female subject and one from the urine of a male subject, both also over 60 years old. Among the MDR isolates, two were identified as K. oxytoca and 15 were K. pneumoniae. MIC measurements revealed 11 ciprofloxacin-resistant (including the two K. oxytoca isolates), with MICs, 6 to >32 μg/ml, three intermediately-resistant (MIC, 1.5 to 3.0 μg/ml) and three ciprofloxacin-susceptible (MIC, 0.25 to 1.0 μg/ml) isolates.
Table 2.
Antibiotic resistance profiles, plasmid-mediated quinolone resistance (PMQR) and class 1 integron carriage in 17 community Klebsiella isolates
| Isolate No. | CIP MIC (μg/ml) |
PMQR
genes |
Class 1 Integron |
Antibiotic resistance profile
(disc diffusion) |
|---|---|---|---|---|
| Kp 31 | 0.25 | qnrB, oqxAB | + | AMX, CF, TE, CAZ, SXT, CIPI |
| Kp 143 | 1.0 | oqxAB | + | AMX, NA, TE, SXT, CIPI |
| Kp 172 | 1.0 | qnrS, oqxB | + | AMX, CF, CIP, OFL, NA, TE, SXT |
| Kp 11 | 1.5 | qnrB, aac(6’)-ib-cr | + | AMX, CF, CTX, CAZ, MEP, GM, AK, NA, TE, SXT, CIPI, OFLI |
| Kp 9 | 3.0 | oqxAB, aac(6’)-ib-cr | - | AMX, CF, CTX, CM, NA, TE, SXT CIPI, OFLI |
| Kp 63 | 3.0 | qnrB, qnrS , aac(6’)-ib-cr, oqxA | + | AMX, CF, NF, TE, SXT, CIPI, OFLI |
| Kp 102 | 6.0 | oqxAB | + | AMX, CF, NF, TE, SXT, CIPI, OFLI |
| Ko 141 | 12 | oqxAB | + | AMX, CF, NA, CIP, OFL, NF, TE |
| Kp 182 | 12 | - | - | AMX, CIP, OFL, NOR, NA, NF, GM, SXT |
| Kp 42 | >32 | qnrB, oqxAB, aac(6’)-ib-cr | + | AMX, CF, CAZ, CTX, CIP, OFL, NA, GM, AK, KM, NF, TE, SXT |
| Ko 55 | >32 | oqxAB, aac(6’)-ib-cr | + | AMX, CF, CAZ, CTX, CIP, OFL, NA, KM, NF, TE, SXT |
| Kp 142 | >32 | qnrS , aac(6’)-ib-cr, oqxA | + | AMX, CF, CAZ, CIP, OFL, NA, KM, NF, CM, TE, SXT |
| Kp 192 | >32 | oqxA | + | AMX, CF, CTX, CIP, OFL, NA, NF, SXT |
| Kp 229 | >32 | aac(6’)-ib-cr | + | AMX, CF, CIP, OFL, NA, GM, NF, SXT |
| Kp 231 | >32 | oqxAB | - | AMX, CIP, OFL, NA, NF, TE, SXT |
| Kp 234 | >32 | oqxAB, aac(6’)-ib-cr | + | AMX, CF, CAZ, CTX, CIP, OFL, NA, GM, KM, NF, TE, SXT |
| Kp 236 | >32 | oqxAB | + | AMX, CIP, OFL, NOR, NA, GM, NF, TE, SXT |
AMX: Amoxicillin; CF: Cefalotin; CAZ: Ceftazidime; CTX: Cefotaxon; GM: Gentamicin; AN: Amikacin; KM: Kanamycin; NA: Nalidixic acid; CP: Ciprofloxacin; OFX: Ofloxacin; NOR: Norfloxacin; NF: Nitrofurantoin; MEM: Meropenem; CM: Chloramphenicol; Te: Tetracycline; SXT: Trimethoprim-Sulfamethaxazole; Ko: Klebsiella oxytoca; Kp: Klebsiella pneumoniae. All isolates were obtained from urine specimens of female subjects except for Kp 9 (sputum of a female subject and Kp 172 (urine of a male subject)
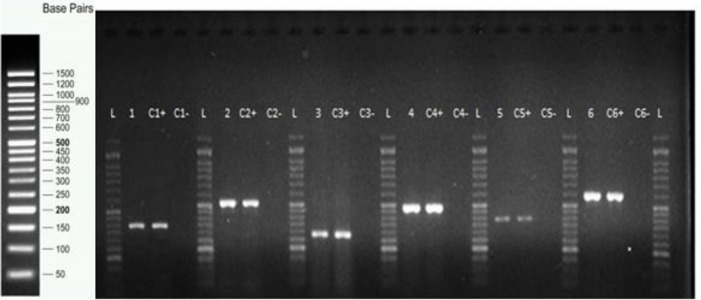
Figure 1 shows representatives of the PMQR and int1 gene amplification products. Of the 11 ciprofloxacin-resistant isolates, four carried the oqxAB genes and one had oqxA alone, two had oqxAB/aac(6’)-ib-cr genes, one carried aac(6’)-ib-cr, one had qnrB/oqxAB/aac(6’)-ib-cr, one harbored qnrS/oqxA/aac(6’)-ib-cr and finally one did not carry any of the tested genes (Table 2). Among the three intermediately ciprofloxacin-resistant isolates, one carried qnrB/aac(6’)-ib-cr, one had oqxAB/aac(6’)-ib-cr and one carried qnrB/qnrS/oqxA/aac(6’)-ib-cr genes. Of the three ciprofloxacin-susceptible isolates which were resistant to β-lactam antibiotics, one carried the oqxAB, and two harbored qnr genes (qnrS/oqxB and qnrB/oqxAB). Interestingly, the aac(6’)-ib-cr gene was not observed in the ciprofloxacin-susceptible isolates. None of the isolates carried qnrA or qepA. Class 1 integron was present in nine of the 11 ciprofloxacin-resistant isolates, one did not have any of the PMQR genes and one carried only the oqxAB
Figure 1.
Representatives of polymerase chain reaction (PCR) amplification products of plasmid-mediated quinolone resistance (PMQR) and int1 genes in outpatient isolates of Klebsiella spp. Lane 1, oqxA (392 bp); lane 2, oqxB (512 bp); lane 3, int1 (280 bp); lane 4, aac(6’)-Ib-cr (482 bp); lane 5, qnrS (388 bp); lane 6, qnrB (594 bp). L; 50 bp DNA ladder; C-, negative control; C+, positive control
gene complex (Table 2). Finally, the three ciprofloxacin-susceptible and two of the intermediately-resistant isolates also carried class 1 integron (Table 2). Class 2 and 3 integrons were not observed. Sequencing results showed that among the four qnrB positive isolates, three were qnrB1 and one was qnrB4. The five qnrS positive strains all were identified as qnrS1 (Table 3).
Table 3.
Identification of qnr genes detected in outpatent,s isolates of Klebsiella spp
| GenBank accession number | GenBank No. | Qnr gene type | Isolate No. |
|---|---|---|---|
|
MH369800 MH369801 MH369802 MH369803 MH369804 MH333285 MH369805 MH369806 MH369807 |
Y63S Y118S Y142S Y172S Y230S Y11B Y31B Y42B Y63B |
qnrS1 qnrS1 qnrS1 qnrS1 qnrS1 qnrB1 qnrB4 qnrB1 qnrB1 |
KP 63 KP 118* KP 142 KP 172 KP 230* KP 11 KP 31 KP 42 KP 63 |
* Ciprofloxacin susceptible isolates. The nucleotide sequences of qnrB and qnrS genes were analyzed by Chromas Pro version 1.7.5 Technelysium
Kp: Klebsiella pneumoniae
Discussion
In this study, 17/250 (6.8%) of the outpatient’s isolates were not only resistant to ciprofloxacin but were also MDR. The most prevalent PMQR gene among these isolates was oqxAB (93.75%) followed by aac(6’)-ib-cr (50.0%), qnrB (25.0%) and qnrS (18.75%). None of the isolates carried qnrA or qepA. We also found that 82.3% of the ciprofloxacin-resistant isolates harbored class 1 integron but class 2 and 3 integrons were not observed. Among the few Iranian studies, Hashemi et al. reported that 4.1% of community isolates of Enterobacteriaceae in Hamadan, Iran, were resistant to ciprofloxacin (21). Seyedpour et al. reported that 13.5% of the community isolates collected from outpatients, harbored PMQR genes among which, aac(6’)-Ib-cr, qnrB and qnrS were predominant, respectively (3).
There are a limited number of studies on community isolates of K. pneumoniae worldwide. In a study performed in 2011 in Morocco, 2/36 (5.6%) of ESBL producing K. pneumoniae isolates from outpatients carried the aac (6’)-Ib-cr gene and qnrS1 (22). In a larger scale study two years later, the same group showed that among 34 ESBL producing K. pneumoniae community isolates, aac (6’)-Ib-cr, was the most prevalent PMQR gene followed by qnrB1, qnrS1, qnrB2 and qnrA6 (23). Except for oqxAB and qnrA, our results were similar to the latter report. A study from Vietnam, showed that among 45 K. pneumoniae community isolates from healthy individuals, the prevalence of qnrS was 33.3% followed by aac (6’)-Ib-cr (2.2%) (24). In the present study, the majority of the quinolone resistant outpatient isolates carried an oqxAB gene. Rodriguez-Martinez et al. showed that a high level expression of this efflux pump decreased quinolone susceptibility in ESBL producing K. pneumoniae (10). The majority of studies report the presence of oqxA in clinical isolates of K. pneumoniae. However, human and veterinary strains can easily spread in communities. Hence, as a PMQR determinant, oqxA can have a major contribution in dissemination of fluoroquinolone resistance along with other antibiotic-resistance genes among the community isolates especially if they are located on integron/plasmid. In fact, Dakic et al. showed that 17.8% of community-acquired urinary tract isolates of Enterobacteriaceae in Greece carried integrons (mostly class 1) and observed a significant association between integron carriage and reduced susceptibility to a range of antibiotics including fluoroquinolones (25). In a recent study from Brazil, PMQR genes (55.5%) as well as frequent presence of the class 1 integron were detected in the community isolates of K. pneumoniae and E. coli (26). Despite the belief that integron harboring MDR clinical isolates are the source of community acquired infections, Leverstein-van Hall et al. showed the spread of antibiotic resistance genes from integron positive community strains of Enterobacteriaceae into the hospital strains (27). Interestingly, Kaplan et al. showed high percentages of ciprofloxacin-resistant Enterobacteriaceae in raw sewage and activated sludge from a waste water plant, the majority of which harbored at least one PMQR determinant (59.6% and 75%, respectively). They also found that PMQR gene presence did not correlate with class 1 integron carriage (28). In this research, class 1 integron was observed in the majority of the MDR isolates regardless of ciprofloxacin susceptibility or PMQR gene carriage. The predominance of class 1 integron has been shown in Gram-positive and Gram-negative clinical isolates as well as a number of different environmental bacteria (15).
Conclusion
Presence of multidrug resistance, multiple PMQR determinants as well as class 1 integron in community isolates of Klebsiella spp. is alarming and presents an important source for cross transmission of these opportunistic pathogens among community members and hospitalized patients.
Acknowledgment
The authors are grateful to Shahid Beheshti University in Tehran and Shahid Sadoughi University of Medical Sciences in Yazd, Iran, for the financial support of this study. The results described in this paper were part of a PhD thesis of MR Malek Jamshidi.
Conflicts of Interest
All contributing authors declare no conflicts of interest.
References
- 1.Paczosa MK, Mecsasb J. Klebsiella pneumoniae: Going on the offense with a strong defense. Microbiol Mol Biol Rev. 2016;80:629–661. doi: 10.1128/MMBR.00078-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jadhav S, Misra R, Gandham N, Ujagare M, a Ghosh P, Angadiand K, et al. Increasing incidence of multidrug resistant Klebsiella pneumoniae infections in hospital and community settings. Inter J Microbiol Res. 2012;4:253–257. [Google Scholar]
- 3.Seyedpour SM, Eftekhar F. Quinolone susceptibility and detection of qnr and aac(6’)-Ib-cr genes in community isolates of Klebsiella pneumoniae. Jundishapur J Microbiol. 2014;7:1–4. doi: 10.5812/jjm.11136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hooper DC. Mechanisms of quinolone resistance. Drug Resist updates. 2003;2:38–55. doi: 10.1054/drup.1998.0068. [DOI] [PubMed] [Google Scholar]
- 5.Jacoby GA, Strahilevitz J, Hooper DC. Plasmid-mediated quinolone resistance. Microbiol Spectrum . 2014;2:1–24. doi: 10.1128/microbiolspec.PLAS-0006-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raei F, Eftekhar F, Feizabadi MM. Prevalence of quinolone resistance among extended-spectrum β-lactamase producing uropathogenic Klebsiella pneumoniae. Jundishapur J Microbiol. 2014;7:e10887. doi: 10.5812/jjm.10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Briales A, Rodríguez-Martínez JM, Velasco C, de Alba PD, Rodríguez-Baño J, Martínez-Martínez L, et al. Prevalence of plasmid-mediated quinolone resistance determinants qnr and aac(6’)-Ib-cr in Escherichia coli and Klebsiella pneumoniae producing extended-spectrum. Int J Antimicrob Agents . 2012;39:431–434. doi: 10.1016/j.ijantimicag.2011.12.009. [DOI] [PubMed] [Google Scholar]
- 8.Rahman Z, Islam A, Rashid MU, Johura FT, Monira S, Watanabe H, et al. Existence of a novel qepA variant in quinolone resistant Escherichia coli from aquatic habitats of Bangladesh. Gut Pathol. 2017;9:58–61. doi: 10.1186/s13099-017-0207-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pakzad I, Karin MZ, Taherikalani M, Boustanshenas M, Lari AR. Contribution of AcrAB efflux pump to ciprofloxacin resistance in Klebsiellapneumoniae isolated from burn patients. GMS Hyg Infec Cont. 2013;8:1–6. doi: 10.3205/dgkh000215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rodríguez-Martínez JM, Díaz de Alba P, Briales A, Machuca J, Lossa M, Fernández-Cuenca F, et al. Contribution of OqxAB efflux pumps to quinolone resistance in extended-spectrum-β-lactamase-producing Klebsiella pneumoniae. J Antimicrob Chemother. 2013;68:68–73. doi: 10.1093/jac/dks377. [DOI] [PubMed] [Google Scholar]
- 11.Sorensen AH, Hansen LH, Johannesen E, Sorensen SJ. Conjugative plasmid conferring resistance to olaquindox. Antimicrob Agents Chemother. 2003;47:798–799. doi: 10.1128/AAC.47.2.798-799.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park KS, Kim MH, Park TS, Nam YS, Lee HJ, Suh JT. Prevalence of the plasmid-mediated quinolone resistance genes, aac(6’)-Ib-cr, qepA, and oqxAB in clinical isolates of extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli and Klebsiella pneumoniae in Korea. Ann Clin Lab Sci. 2012;42:191–197. [PubMed] [Google Scholar]
- 13.Yuan J, Xu X, Guo Q, Zhao X, Ye X, Guo Y, et al. Prevalence of the oqxAB gene complex in Klebsiella pneumoniae and Escherichia coli clinical isolates. J Antimicrob Chemother. 2012;67:1655–1659. doi: 10.1093/jac/dks086. [DOI] [PubMed] [Google Scholar]
- 14.Navon-Venezia S, Kondratyeva K, Carattoli A. Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol Rev. 2017;41:252–275. doi: 10.1093/femsre/fux013. [DOI] [PubMed] [Google Scholar]
- 15.Wen Y, Pu X, Zheng W, Hu G High prevalence of plasmid-mediated quinolone resistance and IncQ plasmids carrying qnrS2 gene in bacteria from rivers near hospitals and aquaculture in China. PLoS ONE. 2016;11:e0159418. doi: 10.1371/journal.pone.0159418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Akya A, Elahi A, Chegenelorestani R, Rezaee M. Dissemination of multidrug-resistant, class I and II integrons and molecular typing of CTX-M-producing Klebsiella pneumoniae. Int’l J Appl Basic Med Res. 2018;8:100–105. doi: 10.4103/ijabmr.IJABMR_333_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deng Y, Bao X, Ji L, Chen L, Liu J, Miao J, et al. Resistance integrons: class 1, 2 and 3 integrons. Ann Clin Microbiol Antimicrob. 2015;14:45–55. doi: 10.1186/s12941-015-0100-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing. 27th Informational Supplement. M100-S17, Clinical and Laboratory Standards Institute, Wayne, PA. 2017. [Google Scholar]
- 19.Chen X, Zhang W, Pan W, Yin J, Pan Z, Gao S, et al. Prevalence of qnr, aac(6’)-Ib-cr, qepA, and oqxAB in Escherichia coli isolates from humans, animals, and the environment. Antimicrob Agents Chemother. 2012;56:3423–3427. doi: 10.1128/AAC.06191-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khoramrooz SS, Sharifi A, Yazdanpanah M, Malek Hosseini SA, Emaneini M, Gharibpour F, et al. High frequency of class 1 integrons in Escherichia coli isolated from patients with urinary tract infections in Yasuj, Iran. Iran Red Crescent Med J. 2016;18:e26399. doi: 10.5812/ircmj.26399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hashemi SH, Esna-Ashari F, Tavakoli S, Mamani M. The prevalence of antibiotic resistance of Enterobacteriaceae strains isolated in community and hospital-acquired infections in teaching hospitals of Hamadan West of Iran. J Res Health Sci. 2013;13:75–80. [PubMed] [Google Scholar]
- 22.Barguigua A, EI Otmani F, Talmi M, Bourjilat F, Haouzane F, Zerouali K, et al. Characterization of extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae isolates from the community in Morocco. J Med Microbiol. 2011;60:1344–1352. doi: 10.1099/jmm.0.032482-0. [DOI] [PubMed] [Google Scholar]
- 23.Barguigua A, EI Otmani F, Talmi M, Reguig A, Jamali L, Zerouali K, et al. Prevalence and genotypic analysis of plasmid-mediated beta-lactamases among urinary Klebsiella pneumoniae isolates in Moroccan community. J Antibiotics. 2013;66:11–16. doi: 10.1038/ja.2012.91. [DOI] [PubMed] [Google Scholar]
- 24.Le TM, Baker S, Le TP, Le TP, Cao TT, Tran TT, et al. High prevalence of plasmid-mediated quinolone resistance determinants in commensal members of the Enterobacteriaceae in Ho Chi Minh City, Vietnam. J Med Microbiol. 2009;58:1585–1592. doi: 10.1099/jmm.0.010033-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dakic I, Lilakos D, Svabic-Vlahovic M, Christofilopoulou S, Poggas N, Charvalos E. Integron-associated antimicrobial resistance in isolates of members of Enterobacteriaceae causing community-acquired urinary infections. Microb Ecol Health Dis. 2007;19:201–206. [Google Scholar]
- 26.Araújo BF, de Campos PA, Royer S, Ferreira ML, Gonçalves IR, da Fonseca Batistão DW. High frequency of the combined presence of QRDR mutations and PMQR determinants in multidrug-resistant Klebsiella pneumoniae and Escherichia coli isolates from nosocomial and community-acquired infections. J Med Microbiol. 2017;66:1144–1150. doi: 10.1099/jmm.0.000551. [DOI] [PubMed] [Google Scholar]
- 27.Leverstein-van Hall MA, Paauw A, Box AT, Blok HE, Verhoef J Fluit AC. Presence of integron associated resistance in the community is widespread and contributes to multidrug resistance in the hospital. J Clin Microbiol. 2002;40:3038–3040. doi: 10.1128/JCM.40.8.3038-3040.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaplan E, Ofek M, Jurkevitch E, Cytryn E. Characterization of fluoroquinolone resistance and qnr diversity in Enterobacteriaceae from municipal biosolids. Front Microbiol. 2013;4:144–150. doi: 10.3389/fmicb.2013.00144. [DOI] [PMC free article] [PubMed] [Google Scholar]