Abstract
Depleting regulatory T cells (Treg cells) to counteract immunosuppressive features of the tumor microenvironment (TME) is an attractive strategy for cancer treatment; however, autoimmunity due to systemic impairment of their suppressive function limits its therapeutic potential. Elucidating approaches that specifically disrupt intratumoral Treg cells is direly needed for cancer immunotherapy. We found CD36 was selectively up-regulated in intrautumoral Treg cells as a central metabolic modulator. CD36 fine-tuned mitochondrial fitness via PPAR-β signaling, programming Treg cells to adapt to a lactic acid-enriched TME. Genetic ablation of Cd36 in Treg cells suppressed tumor growth accompanied by a decrease in intratumoral Treg cells and enhancement of anti-tumor activity in tumor-infiltrating lymphocytes without disrupting immune homeostasis. Furthermore, CD36 targeting elicited additive anti-tumor responses with anti-PD-1 therapy. Our findings uncover the unexplored metabolic adaptation that orchestrate survival and functions of intratumoral Treg cells, and the therapeutic potential of targeting this pathway for reprogramming the TME.
Introduction
Regulatory T (Treg) cells are found at high frequencies in both mouse and human cancers1–3, where they represent a major barrier to anti-tumor immunity and cancer immunotherapy4,5. While strategies depleting Treg cells increase anti-tumor responses6–8, the severe autoimmunity caused by systemic loss of Treg cells and the unwanted depletion of effector T cells limit the therapeutic potential of Treg-targeting approaches. In addition, systemic impairment of suppressive functions in Treg cells upon treatments targeting immune checkpoints, such as OX40, GITR and CTLA-4, expressing in Treg cells also hampers the application of Treg cell-targeting approaches in cancer treatment9–11. To date, the search for effective targeting approaches that selectively demolish intratumoral Treg cells remains a challenge for cancer immunotherapy12.
Progressive adaptation in transcriptome in Treg cells migrating to barrier tissues has been revealed13 and tissue context-dependent signals have been proposed to drive tissue-specific adaptation in Treg cells without detailed understanding14–20. It also remains elusive whether the tissue context-dependent adaptation is required for proper functioning of peripheral Treg cells. Emerging evidence reveals that metabolic machinery and nutrient-sensing mechanisms play critical roles to fine-tune proliferation, survival, suppressive function and lineage stability in Treg cells21–27. Since the tumor microenvironment (TME) can impose a variety of types of metabolic stress on infiltrating immune cells28, including acidosis, hypoxia, and nutrient deprivation, it is likely that intratumoral Treg cells must adjust their metabolic preferences in response to these conditions as a consequence of adaptation to the TME. We therefore speculate that the metabolic adaptation engaged by intratumoral Treg cells orchestrate signal pathways to support survival and suppressive activity.
We report here that intratumoral Treg cells up-regulate CD36 expression to support mitochondrial fitness and biogenesis via a PPAR-β-dependent mechanism. Genetic ablation of Cd36 in Treg cells selectively abrogated the abundance and suppressive activity of intratumoral Treg cells. Importantly, mice with genetic ablation of Cd36 in Treg cells did not elicit autoimmunity and CD36-deficient splenic Treg cells remained effective on restricting T cell transfer-induced colitis. Our results revealed that CD36-PPAR-β signal sustains survival and functional fitness in intratumoral Treg cells by modulating mitochondrial fitness and NAD levels, which are critical for metabolizing lactate in the TME. We further provide proof-of-concept evidence that targeting CD36 with a monoclonal antibody induces superior anti-tumor immunity and elicits additive anti-tumor responses with anti-PD-1 treatment. These results highlight the unexplored CD36-PPAR-β-modulated metabolic adaptation, which allows intratumoral Treg cells to utilize lactate in tumors, and suggest that targeting metabolic adaptation in intratumoral Treg cells would be a promising strategy for reprogramming the TME without perturbing systemic immune and tissue homeostasis.
Results
Intratumoral Treg cells increase lipid metabolism and CD36 expression
To elucidate whether intratumoral Treg cells preferentially engage specific metabolic pathways, we first analyzed RNA-sequencing results from intratumoral and circulating Treg cells obtained from breast cancer patients in a previously published study29. Gene pathway analysis, with a particular focus on metabolic pathways, revealed that intratumoral Treg cells highly expressed metabolic genes responsible for lipid metabolism when compared to circulating Treg cells (Fig. 1a,b), suggesting that intratumoral Treg cells may increase their lipid metabolism. Indeed, when we compared peripheral blood mononuclear cells (PBMCs) and intratumoral Treg cells from non-small-cell lung carcinoma (NSCLC) patients, intratumoral Treg cells internalized a higher amount of a green fluorescent fatty acid, Bodipy FL C12, and contained a higher neutral lipid content based on the staining of Bodipy (Extended Data Fig. 1a,b). By examining Treg cells in PBMCs and tumor-infiltrated lymph nodes (TILN) from melanoma patients, we also confirmed that intratumoral Treg cells from the majority of patients internalized a higher amount of Bodipy FL C12 (Extended Data Fig. 1c); however, Treg cells in both PBMCs and tumor-infiltrated lymph nodes (TILNs) contained similar lipid content (Extended Data Fig. 1d). To further explore these phenotypes, we used melanoma cell engraftment models to assess lipid metabolism in Treg cells residing in tumors and other peripheral tissues. Intratumoral Treg cells displayed an elevated ability to take up fatty acids (Fig. 1c) and had a higher lipid content (Fig. 1d) compared to Treg cells from other tissues of YUMM1.7 melanoma-bearing mice. Similarly, intratumoral Treg cells of B16 melanoma-bearing mice and MC38 colon carcinoma-bearing mice also exhibited enhanced fatty acid uptake (Extended Data Fig. 1e,f). These observations suggest that the increase in lipid metabolism, especially lipid uptake, by intratumoral Treg cells is a conserved phenotype in both human and murine models. Of note, among genes controlling lipid uptake, Cd36, encoding a scavenger receptor responsible for long chain fatty acid and oxidized low-density lipoprotein uptake, was significantly up-regulated in intratumoral Treg cells compared to circulating Treg cells from breast cancer patients29. By examining Treg cells in PBMCs and TILNs from melanoma patients, we confirmed that intratumoral Treg cells from the majority of patients expressed higher amounts of CD36 (Fig. 1e). In addition, we found that intratumoral Treg cells, but not Treg cells residing in other peripheral tissues or secondary lymphoid organs from Yumm1.7 melanoma-bearing mice, expressed abundant CD36 (Fig. 1f). The increased expression of CD36 found in intratumoral Treg cells was also observed in NSCLC patients, as well as in the mouse B16 melanoma model, a genetically engineered Braf/PTEN melanoma mouse model, a K-rasLSL-G12D/+/p53fl/fl conditional mouse model of NSCLC, and a MC38 colon carcinoma model (Extended Data Fig. 1g-k). Furthermore, we found that culturing inducible Treg cells (iTreg cells) in conditioned medium obtained from cancer cell cultures substantially increased CD36 expression, while hypoxia and lactic acid failed to induce CD36 expression in Treg cells (Extended Data Fig. 1l). Lipid removal abolished the effects of cancer cell-conditioned media on stimulating CD36 expression in Treg cells (Extended Data Fig. 1m). Together, our results suggest that the TME can stimulate CD36 expression in Treg cells, which might support the demands for metabolic adaptation in intratumoral Treg cells.
Fig. 1. Intratumoral Treg cells elevates expression of CD36 and genes involved in lipid metabolism.
a, Pathway enrichment analysis focusing on metabolic machineries for RNA expression in Treg cells from breast cancers (n=4) and PBMC (n=6) of breast cancer patients. Pathways with significant differential expression between intratumoral and PBMC Treg cells (Nominal p value<0.05) were presented based on gene hit size. b, Enrichment plots of fatty acid metabolic process (top) and lipid binding (bottom) pathways in intratumoral Treg cell compared to PBMC Treg cells, identified by gene set enrichment analysis (GSEA) computational method. Columns indicate individual samples, and rows are each gene. Red represents a high expression level, and blue indicates a low expression level. c, d, Representative histogram (left) and quantitative results of geometric mean (GeoMean) fluorescent intensity (right) of Bodipy FL C12 (n=6, one outlier was removed from the thymus group) (c) and Bodipy 493/503 (Bodipy) (n=5) (d) in Treg cells from indicated tissues of Yumm1.7 melanoma-bearing B6 mice. DLN: draining lymph node; spleen; thymus; tumor. e, f, Representative histogram (left) and quantitative results of geometric mean (GeoMean) fluorescent intensity of CD36 surface staining in Treg cells from PBMC and tumor infiltrated lymph nodes (TILNs) of melanoma patients (n=12) (e) and from indicated tissues of Yumm1.7 melanoma-bearing B6 mice (DLN, n=15; spleen, n=15; thymus, n=9; tumor, n=15) (f). TILs: tumor-infiltrating lymphocytes. Data are representative results of three independent experiments with similar results (c, d) or cumulative results from three independent experiments (e, f). Each symbol represents one individual. ES: enrichment score; NES: normalized enrichment score; FDR: false discovery rate; NOM p-val: Nominal p value. Data are mean ± S.D. and were analyzed by two-tailed, unpaired Student’s t-test (c, d, f) or by two-tailed, paired Student’s t-test (e).
CD36 controls the accumulation and suppressive function of intratumoral Treg cells
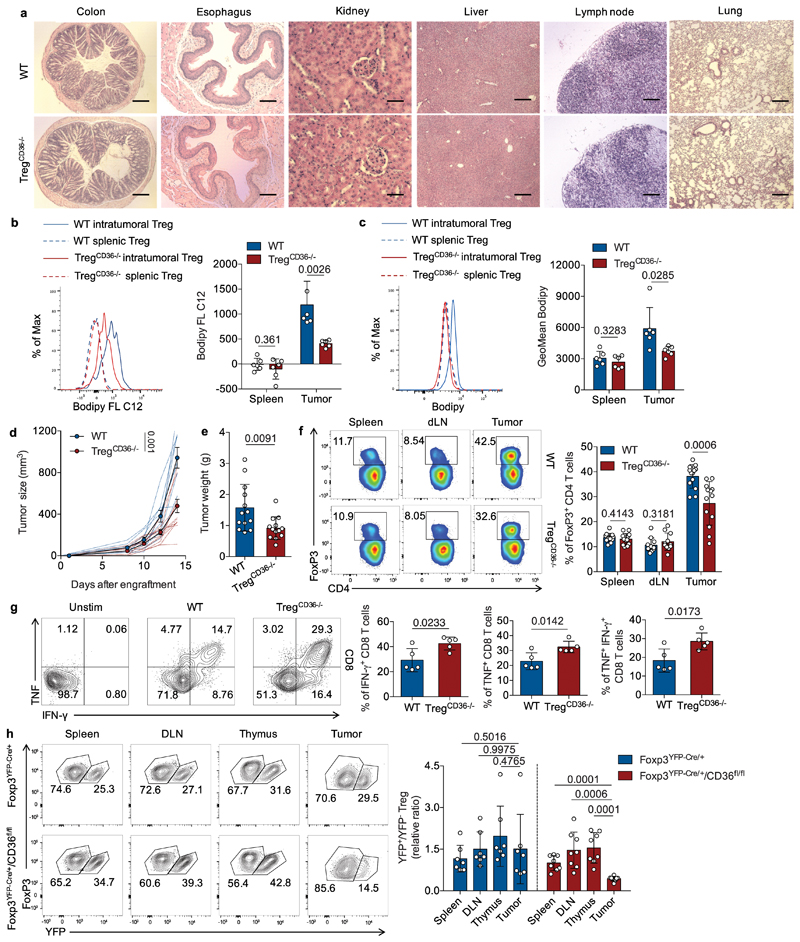
To investigate whether the expression of CD36 modulates Treg behavior in tumors, we generated Treg cell-specific CD36-deficient mice (designated TregCd36-/-) by crossing Cd36fl/fl mice with Foxp3YFP-Cre mice. Given that genetic ablation of critical regulators in Treg cells could lead to systemic activation of T lymphocytes and autoimmunity due to impairment of Treg cell suppressive functions, we first examined whether deficiency of CD36 in Treg cell impacts immune homeostasis. We found that aged TregCd36-/- mice (21-23 weeks) displayed comparable body weights to Foxp3YFP-Cre mice (referred to wild-type mice throughout this study) in both genders (Extended Data Fig. 2a). TregCd36-/- mice also contained a similar proportion of effector or memory population (CD44hiCD62Llo) in both CD4+ and CD8+ T cell compartments compared to wild-type mice (Extended Data Fig. 2b). Moreover, TregCd36-/- mice showed neither abnormal infiltration of lymphocytes and myeloid cells in various organs nor severe systemic inflammatory disorders (Fig. 2a and Extended Data Fig. 2c), suggesting that CD36 is not required for Treg cells to maintain immune homeostasis. We then engrafted YUMM1.7 melanoma cells into wild-type and TregCd36-/- mice. We found that genetic ablation of Cd36 in Treg cells severely decreased lipid uptake and content in intratumoral Treg cells, but not splenic Treg cells (Fig. 2b,c), indicating that intratumoral Treg cells rely on CD36 expression to support enhanced lipid uptake. We also observed growth deceleration of engrafted YUMM1.7 melanoma (Fig. 2d,e), B16 melanoma (Extended Data Fig. 2d), and MC38 colon carcinoma (Extended Data Fig. 2e) in TregCd36-/- mice. Moreover, TregCd36-/- mice had a profound loss of intratumoral Treg cells, but not Treg cells in spleen and draining lymph nodes at the end point of analyses (Fig. 2f and Extended Data Fig. 2f). This was accompanied by a significant increase in the frequency of CD8+ tumor-infiltrating lymphocytes (TILs) as well as the ratio of intratumoral CD8+ to Treg cells (Extended Data Fig. 2g,h), a favorable parameter associated with strong anti-tumor responses. In addition, a higher frequency of CD8+ TILs and CD4+FoxP3– TILs in TregCd36-/- mice produced anti-tumor effector cytokines, including interferon-γ (IFN-γ) and tumor necrosis factor (TNF) (Fig. 2g and Extended Data Fig. 2i), suggesting that the TME of TregCd36-/- mice is less immunosuppressive. To further validate the dependency of CD36 on supporting accumulation of intratumoral Treg cells, we generated heterozygous Foxp3YFP-Cre/+Cd36fl/fl female mice, which simultaneously harbor a wild-type Treg population and a CD36-deficient Treg population driven by FoxP3 expression mediated by X chromosome inactivation. The wild-type and CD36-deficient Treg cells can be detected on the basis of the expression of yellow fluorescent protein (YFP). To exclude the potential toxicity induced by Cre recombinase, we also generated heterozygous Foxp3YFP-Cre/+ female mice as control mice. By comparing YFP+ populations among FoxP3+ Treg cells in both tumor-bearing Foxp3YFP-Cre/+Cd36fl/fl and Foxp3YFP-Cre/+ female mice, we only detected a reduction in frequencies of CD36-deficient Treg cells (Cre+ population in Foxp3YFP-Cre/+Cd36fl/fl mice) in tumors, whereas the frequency ratios between Cre+ and Cre- Treg cells in both spleens and draining lymph nodes were comparable between Foxp3YFP-Cre/+Cd36fl/fl and Foxp3YFP-Cre/+ mice (Fig. 2h). This result suggested that loss of CD36 expression selectively perturb accumulation of intratumoral Treg cells via intrinsic regulation. Altogether, these results reveal a crucial role for CD36 in selectively endowing Treg cells with the ability to accumulate in the TME.
Fig. 2. Disruption of CD36 selectively impairs the accumulation and suppressive function of intratumoral Treg cells.
a, Representative images of haematoxylin and eosin (H&E) staining for indicated tissues from wild-type (WT) or TregCd36-/- mice at the age of 21-23 weeks. Scale bar, 200 µm. b, c, Representative histogram (left) and quantitative results of geometric mean (GeoMean) fluorescent intensity (right) of Bodipy FL C12 (b) and Bodipy 493/503 (c) in splenic and intratumoral Treg cells from Yumm1.7 melanoma-bearing WT or TregCD36-/- mice (n=6 per group). d, e, Tumor growth (WT, n=9; TregCd36-/-, n=10) (d) and tumor weight (n=13) (e) of YUMM1.7 melanoma from WT or TregCd36-/- mice. Foxp3YFP-Cre mice were used as WT mice. f, Representative plots (left) and percentage of FoxP3+ Treg cells among CD4+ T cells in indicated tissues of tumor-bearing WT and TregCd36-/- mice (spleen and tumor n=13; dLN, n=11). g, Representative plots (left) and percentage of indicated cytokine-producing CD8+ T cells among total tumor-infiltrating CD8+ T cells from indicated mice (right) (n=5 per group). h, Representative plots (left) and quantitative ratios (right) of CD36-deficient/WT Treg cells in tumors and indicated tissues from either Foxp3YFP-Cre/+/Cd36fl/fl female mice (n=8) or Foxp3YFP-Cre/+ female mice (n=7). Data are representative result of two independent experiments with similar results (b, c, g) or cumulative results from at least two independent experiments (d, e, f, h). Each symbol represents one individual. Data are mean ± S.D. (b, c, e, f, g, h) or ± S.E.M. (d) and were analyzed by two-tailed, unpaired Student’s t-test (b, c, d, e, f, g, h).
CD36 is dispensable in Treg cells for maintaining periphery homeostasis
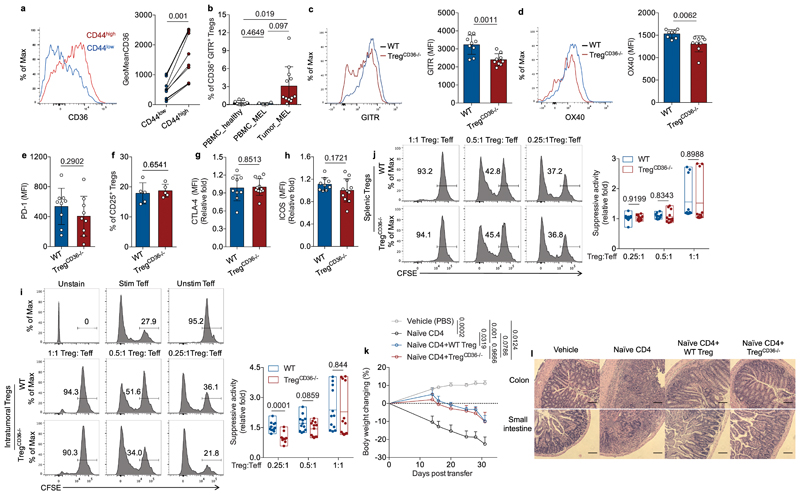
Intriguingly, we also found that intratumoral effector Treg cells (CD44hiCD62Llo) exhibited higher expression of CD36 compared to intratumoral CD44lo Treg cells (resting Treg cells) in murine melanoma model (Fig. 3a). Similarly, a higher percentage of tumor-infiltrating GITR+CD25+ effector Treg cells, the most suppressive subset of effector Treg cells30,31, from TILs of melanoma patients expressed CD36 compared to GITR+CD25+ effector Treg cells in PBMCs from melanoma patients and healthy donors (Fig. 3b). We next examined the expression of immunomodulatory receptors in intratumoral Treg cells and found that CD36-deficient Treg cells reduced the expression of glucocorticoid-induced TNFR-related protein (GITR) and OX40 (Fig. 3c,d), but not programmed cell death protein 1 (PD-1), CD25, cytotoxic T-lymphocyte-associated protein 4 (CTLA-4), or inducible T-cell costimulator (ICOS), compared to wild-type Treg cells (Fig. 3e-h). These results suggest that CD36 expression contributes to the suppressive functions of effector Treg cells. In support of this notion, we found that intratumoral Treg cells from TregCd36-/- mice displayed reduced suppressive capacity compared to wild-type intratumoral Treg cells in the ex vivo suppressive assay (Fig. 3i). However, wild-type and CD36-deficient splenic Treg cells displayed comparable suppressive capacity (Fig. 3j), suggesting that CD36 is only required for supporting suppressive activity of intratumoral Treg cells, but not splenic Treg cells. To further examine if CD36 is dispensable for Treg cells to restraint peripheral inflammation, we analyzed the ability of CD36-deficient Treg cells on suppressing T cell transfer-induced colitis. Disease onset such as weight loss was detected at two weeks post naïve CD4+ T cell transfer. However, co-transferring CD36-deficient Treg cells was able to ameliorate weight loss in the recipient mice comparably to wild-type Treg cells (Fig. 3k). Moreover, several organs were removed at the study endpoint and processed for further evaluation by histology. Compared with naïve CD4+ T cell transfer, which triggered massive infiltration of lymphocytes and myeloid cells, co-transfer of either wild-type or CD36-deficient Treg cells impeded infiltration of lymphocytes and myeloid cells as well as morphology changes in colon and small intestine, colon shortening, and enlargement of spleen in the recipient mice (Fig. 3l and Extended Data Fig. 3a,b). Furthermore, we found that genetic ablation of Cd36 neither affects the expression of activation markers, including CD44, CD103 and KLRG1 (Extended Data Fig. 3c-e), nor FoxP3 expression (indicated by the fluorescence intensity of YFP) (Extended Data Fig. 3f) in intratumoral Treg cells. However, CD36-deficient intratumoral Treg cells slightly enhanced the production of pro-inflammatory cytokines IFN-γ and TNF (Extended Data Fig. 3g-i), suggesting that CD36 restraints the ability to produce pro-inflammatory cytokine in intratumoral Treg cells. Together, our results suggest that CD36 expression specifically supports intratumoral Treg cells suppressive functions.
Fig. 3. CD36 expression selectively supports suppressive activity of intratumoral Treg cells.
a, Representative histogram (left) and quantitative results of geometric mean (GeoMean) fluorescent intensity (right) of CD36 in CD44lo and CD44hi intratumoral Treg cells from Yumm1.7 melanoma-bearing WT mice (n=10). b, Percentage of CD36+ Treg cells among GITR+/FoxP3+ Treg cells of PBMCs from healthy donors (n=9) or melanoma patients (n=4) or TILs from melanoma patients (n=12). c, d, Representative histograms and quantitative results of geometric mean fluorescent intensity (MFI) of GITR (c) and OX40 (d) in intratumoral Treg cells from indicated mice (n=9 per group, one outlier was removed from OX40_WT group). e, f, g, h, Expression of PD-1 in intratumoral Treg cells (n=9 per group) (e), percentage of CD25+/FoxP3+ Treg cells among total intratumoral Treg cells population (n=5 per group) (f), and expression of CTLA-4 (g) and ICOS (h) (Foxp3YFP-Cre, n=9; TregCd36-/-, n=11) in intratumoral Treg cells of YUMM1.7 melanoma-bearing WT and TregCD36-/- mice. i, j, Ex vivo suppression of CFSE-labeled WT naïve CD8+ T cell proliferation by WT and TregCd36-/- Treg cells sorted from tumors (i) or spleens (j) with annotated ratios (n=12). k, Body weight measurement in Rag1-/- mice receiving naïve CD4+ T cell alone or in combination with either WT or TregCD36-/- Treg cells (vehicle, n=4; naïve CD4, n=7; WT Treg, n=6; TregCd36-/-, n=6). l, Representative images of haematoxylin and eosin (H&E) staining of colon and small intestine from indicated group of Rag1-/- mice. Scale bar, 200 µm. Data are representative result of three independent experiments with similar results (l, f) or cumulative results from at least two independent experiments (a, b, c, d, e, g, h, i, j, k). Each symbol represents one individual. Data are mean ± S.D. (a, b, c, d, e, f, g, h, i, j) or ± S.E.M. (k) and were analyzed by two-tailed, unpaired Student’s t-test (b, c, d, e, f, g, h, i, j, k) or two-tailed, paired Student’s t-test (a).
CD36 deficiency stimulates apoptosis in intratumoral Treg cells
To investigate the underlying basis for the reduced cellularity of CD36-deficient Treg cells in the TME, we first examined proliferative ability by staining Ki67 (Extended Data Fig. 3j) and found that CD36 deficiency did not alter proliferation in intratumoral Treg cells. However, we observed higher amounts of cleaved caspase-3 (Fig. 4a) and Annexin V staining (Extended Data Fig. 3k) in CD36-deficient intratumoral Treg cells. CD36 deficiency did not enhance cleaved caspase-3 abundance in Treg cells from thymus and other secondary lymphoid organs, except a slight increase in draining lymph nodes (Extended Data Fig. 3l), indicating that intratumoral Treg cells require CD36-mediated regulation to prevent apoptosis. Since mitochondrial metabolism and fitness have been suggested to modulate Treg cell suppressive function and survival22,25,32,33, we then examined whether CD36-deficient intratumoral Treg cells fail to sustain mitochondrial fitness. Strikingly, compared to wild-type Treg cells, CD36-deficient intratumoral Treg cells, but not Treg cells in other tissues, displayed reduced mitochondrial membrane potential as measured by MitoTracker DeepRed staining (Fig. 4b). This observation was further supported by electron microscopy analysis in which we found that intratumoral Treg cells from TregCd36-/- mice had fewer mitochondria (Fig. 4c) as well as fewer cristae in each mitochondrion (Fig. 4d). Nevertheless, wild-type and CD36-deficient splenic Treg cells displayed comparable numbers of mitochondria and crista density. By examining the transcriptome of YFP+ intratumoral Treg cells in both tumor-bearing Foxp3YFP-Cre/+Cd36fl/fl and Foxp3YFP-Cre/+ female mice, we found that CD36-deficient Treg cells (Cre+ population in Foxp3YFP-Cre/+Cd36fl/fl mice) in tumors had less expression of mitochondrial genes encoding mitochondrial tRNAs, which control translation of proteins for electron transport chain, compared to wild-type intratumoral Treg cells (Fig. 4e). This result further supports that CD36-deficient intratumoral Treg cells fail to maintain mitochondrial fitness. To further elucidate the impact of CD36 on Treg mitochondrial metabolism, we treated iTreg cells generated from either wild-type or CD36-deficient CD4+ T cells with cancer cell-conditioned medium to induce CD36 expression as shown before. We then performed the Seahorse extracellular flux assays and found that CD36-deficient Treg cells demonstrated reduced oxygen consumption rates (OCR) (Fig. 4f and Extended Data Fig. 4a) but increased glycolytic rates (Extended Data Fig. 4b-c). These results suggest that CD36 ablation can impair oxidative phosphorylation (OXPHOS) and skew the metabolic preference of Treg cells toward aerobic glycolysis. In light of these observations, we speculated that enhanced CD36 expression in intratumoral Treg cells may support Treg metabolic flexibility via modulation of mitochondrial fitness in response to the TME-imposed metabolic stress28,34,35. We therefore examined whether CD36-deficient Treg cells displayed survival defects in response to metabolic challenges. In contrast to normal culture conditions (RMPI plus 10% FBS; designated RMPI), CD36-deficient Treg cells exposed to cancer cell-conditioned medium exhibited reduced viability (Extended Data Fig. 4d). Since lactic acid concentrations can be higher in cancer cell-conditioned medium and the accumulation of lactic acid is a common feature of the TME, we thus postulated that CD36-deficient Treg cells might fail to sustain survival in this condition due to concentrations of lactic acid. In support of this postulate, we found that CD36-deficient Treg cells displayed a profound survival defect in response to increasing concentrations of lactic acid (Fig. 4g). Interestingly, a recent study has suggested that elevated electron transport chain activity results in an increased NAD/NADH ratio to support lactic acid conversion into pyruvate in Treg cells26, which can support survival of Treg cells in a lactic acid-enriched condition. Thus, we speculated that CD36-deficient Treg cells may have a lower NAD/NADH ratio compared to wild-type Treg cells due to reduced mitochondrial fitness and OXPHOS. Indeed, CD36-deficient Treg cells had a lower NAD/NADH ratio compared to wild-type Treg cells (Fig. 4h), and supplementation with nicotinamide riboside (NR) to replenish NAD36 partially rescued the viability of CD36-deficient Treg cells exposed to cancer cell-conditioned medium (Fig. 4i). Hence, the failure of CD36-deficient intratumoral Treg cells to persist in vivo might result from reduced mitochondrial fitness and OXPHOS, which allow Treg cells to survive in lactic acid-enriched conditions via an NAD-regulated metabolic process.
Fig. 4. CD36 deficiency stimulates apoptosis in intratumoral Treg cells.
a, Representative histograms (left) and quantitative analysis (right) of cleaved caspase-3 levels in intratumoral Treg cells from WT (n=13) and TregCd36-/- tumor-bearing mice (n=14). b, Representative histograms (left) and quantitative analysis (right) of MitoTracker Deep Red (MDR) staining in Treg cells of spleen, non-draining lymph node (LN), draining lymph node (DLN), blood, thymus, and tumor from tumor-bearing WT and TregCd36-/- mice (n=11 per group). c, d, Representative electron microscope images (left) and quantitative plots (right) of mitochondrion number (c) and crista density (d) in splenic and intratumoral Treg cells from tumor-bearing WT and TregCd36-/- mice. Photo of individual cells was taken with EM for the mitochondrial calculation (WT spleen, n=15; WT tumor, n=16, TregCd36-/- spleen, n=14; TregCd36-/- tumor, n=17). Scale bars: 500 nm in (c) and 200 nm in (d). e, Expression of genes encoding mitochondrial tRNAs in WT and TregCd36-/- intratumoral Treg cells, assessed by RNA-seq (n=3 per group). f, OCR of indicated iTreg cell cultured in cancer cell-conditioned medium for 48 hrs (n=3 per group). g, The viability of either WT or TregCd36-/- iTreg cell cultured in cancer cell-conditioned medium as above and then treated with indicated concentration of lactic acids for another 72 hrs (n=4 in RPMI group; n=5 in other groups). h, NAD/NADH ratio of indicated iTreg cell cultured in cancer cell-conditioned medium for 48 hrs (n=16). i, Relative viability of either WT or TregCd36-/- iTreg cells treated with cancer cell-conditioned medium with or without NR (400 μM) for 72 hrs (n=11 per group). Results were normalized to the survival cells of control treatment in indicated group. NR: nicotinamide riboside. Data are representative result of three independent experiments with similar results (f, g) or cumulative results of three independent experiments (a, b, e, h, i). Data are mean ± S.D. and were analyzed by two-tailed, unpaired Student’s t-test.
CD36-PPAR-β signaling orchestrates metabolic adaptation in intratumoral Treg cells
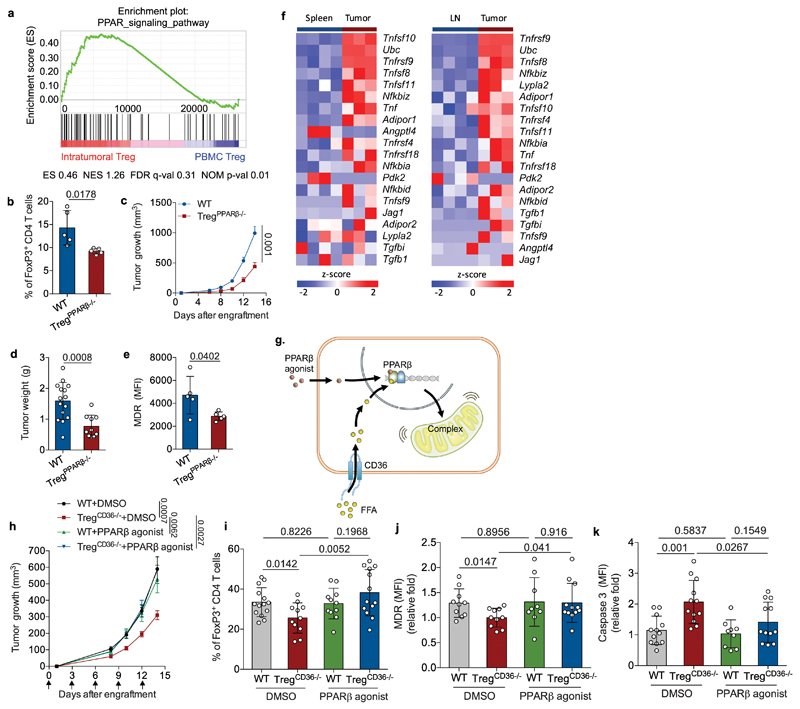
To understand how CD36 stimulates mitochondrial fitness in intratumoral Treg cells, we assessed changes in the transcriptomes of intratumoral and circulating Treg cells from breast cancer patients29. As expected, intratumoral Treg cells up-regulated genes controlling mitochondrial functions and biogenesis (Extended Data Fig. 5a). Moreover, we found that intratumoral Treg cells displayed increased expression of genes involved in the PPAR signaling pathway (Fig. 5a). Since CD36 has been suggested to support metabolic flexibility in metabolic tissues by boosting PPAR-β- and PPAR-γ-dependent regulation of mitochondrial activity and biogenesis37–39, we postulated that CD36-induced metabolic reprogramming might promote mitochondrial fitness in intratumoral Treg cells by providing lipid signals to adjust PPAR transcriptional regulation. To test this, we crossed both Pparbfl/fl and Ppargfl/fl mice with Foxp3YFP-Cre mice to obtain Treg-specific PPAR-β-deficient mice (designated TregPPARβ-/-) and PPAR-γ-deficient mice (designated TregPPARγ-/-), respectively. We observed that genetic ablation of PPAR-γ in Treg cells impaired neither accumulation of intratumoral Treg cells nor YUMM1.7 melanoma growth (Extended Data Fig. 5b-d). In contrast, TregPPARβ-/- mice recapitulated the characteristic features of TregCd36-/- mice, including reduced intratumoral Treg accumulation (Fig. 5b), growth deceleration of engrafted YUMM1.7 melanoma (Fig. 5c,d), and an increase in CD8+ TILs (Extended Data Fig. 5e). Similar to CD36-deficient intratumoral Treg cells, PPAR-β-deficient intratumoral Treg cells displayed reduced mitochondrial membrane potential compared to wild-type intratumoral Treg cells (Fig. 5e). PPAR-β-deficient intratumoral Treg cells expressed less CD36 compared to wild-type intratumoral Treg cells (Extended Data Fig. 5f). We further found that intratumoral Treg cells up-regulated expression of multiple target genes of PPAR-β compared to Treg cells in spleens and draining lymph nodes of tumor-bearing mice (Fig. 5f), suggesting that intratumoral Treg cells might increase PPAR-β activity. To elucidate the relationship between CD36 and PPAR-β activation and their roles on supporting accumulation of intratumoral Treg cells, we speculated that treating PPAR-β selective agonist (GW501516) could bypass the requirement of CD36 to sustain mitochondrial fitness and survival in CD36-deficient intratumoral Treg cells (Fig. 5g). We then treated YUMM1.7 melanoma-engrafted wild-type mice and TregCd36-/- mice with either GW501516 or control vehicle for two weeks and found that treatment with GW501516 restored tumor growth as well as intratumoral Treg cell abundance in TregCd36-/- mice (Fig. 5h,i). In addition, intratumoral Treg cells from GW501516-treated TregCd36-/- mice had increased mitochondrial membrane potentials and less cleaved caspase-3 compared to DMSO- treated TregCd36-/- mice (Fig. 5j,k). In parallel, GW501516 treatment in CD36-deficient Treg cells increased NAD/NADH ratios (Extended Data Fig. 5g), indicating that CD36-controlled lipid uptake activates PPAR-β pathways to support mitochondrial fitness and enhancement of NAD/NADH ratios in intratumoral Treg cells. Moreover, the activation of PPAR-β pathways may further amplify CD36-mediated metabolic adaptation in intratumoral Treg cells by enhancing CD36 expression. Altogether, these results reveal that CD36-PPAR-β signaling orchestrate metabolic programs to support Treg persistence in the TME.
Fig. 5. PPAR-β signaling is required for metabolic adaptation in intratumoral Treg cells.
a, Enrichment plots of PPAR signaling pathways in intratumoral Treg cells (n=4) compared to PBMC Treg cells (n=6), identified by GSEA computational method. ES: enrichment score; NES: normalized enrichment score; FDR: false discovery rate; NOM p-val: Nominal p value. b, Percentage of FoxP3+ Treg cells among CD4+ tumor-infiltrating T lymphocytes from tumor-bearing WT and TregPPARβ-/- mice (n=5). c, d, Tumor growth (c) and tumor weight (d) of YUMM1.7 melanoma from WT or TregPPARβ-/- mice (WT, n=15; TregCd36-/-, n=10). Foxp3YFP-Cre mice were used as WT mice. e, Quantitative result of geometric MFI of MDR staining in intratumoral Treg cells from WT and TregPPARβ-/- mice (n=5 per group). f, Expression of target genes of PPAR-β in Treg cells from spleens, draining lymph nodes (LN) and tumor, assessed by RNA-seq. g, Illustration of postulated signaling cascade and the rationale for applying PPAR-β agonist in TregCd36-/- mice. h, i, WT and TregCd36-/- mice were engrafted with YUMM1.7 melanoma cells and then treated with either DMSO or PPARβ agonist as described in methods. Tumor growth (WT+DMSO, n=6; TregCd36-/-+DMSO, n=11; WT+PPAR-β agonist, n=5; TregCd36-/-+PPAR-β agonist, n=9) (h) and percentage of FoxP3+ Treg cells among CD4+ tumor-infiltrating T lymphocytes (WT+DMSO, n=14; TregCd36-/-+DMSO, n=11; WT+PPAR-β agonist, n=11; TregCd36-/-+PPAR-β agonist, n=13) (i) were analyzed. j, k, Quantitative analysis of MitoTracker Deep Red (MDR) staining (DMSO, n=10; PPAR-β agonist, n=12) (j) and expression of cleaved caspase-3 (k) in intratumoral Treg cells of WT and TregCd36-/- mice treated with indicated treatments (n=9 in WT+PPAR-β agonist group; n=12 in other groups). Data are representative result of three independent experiments with similar results (b, e) or cumulative results from at least three independent experiments (c, d, f, h, i, j, k). Data are mean ± S.D. (b, d, e, i, j, k) or ± S.E.M. (c, h) and were analyzed by two-tailed, unpaired Student’s t-test.
CD36-targeting reinforces antitumor immunity by impairing intratumoral Treg cells
Next, we hypothesized that blocking CD36-mediated metabolic adaptation could specifically disturb intratumoral Treg cells without systemic loss of Treg cells and global impairment of Treg suppressive functions. To test this, we treated Yumm1.7 melanoma-engrafted mice with an anti-CD36 monoclonal antibody (mAb), which interferes with CD36-mediated fatty acid and oxidized low-density lipoprotein uptake40. We found that anti-CD36 mAb treatment reduced tumor growth (Fig. 6a) and was accompanied by reduced accumulation of intratumoral Treg cells, while the Treg cell population was sustained in spleen and draining lymph nodes (Fig. 6b and Extended Data Fig. 6a). Similar to genetic ablation of Cd36 in Treg cells, anti-CD36 mAb treatment promoted apoptosis in intratumoral Treg cells (Fig. 6c), and led to a significant increase in tumor infiltration of CD8+ T cells (Extended Data Fig. 6b). In addition, treating mice with an anti-CD36 mAb improved the production of anti-tumor effector cytokines in CD8+ and CD4+ TILs (Extended Data Fig. 6c-d). Since CD36 expression can support metabolic flexibility and metastasis in cancer cells as well as other immune cells40,41, the anti-tumor responses induced by anti-CD36 mAb may be Treg cell-independent. To this end, we performed the same treatment using TregCd36-/- mice as recipients. Our results showed that anti-CD36 mAb treatment was incapable of suppressing tumor progression in TregCd36-/- mice (Fig. 6d), indicating that the anti-tumor responses induced by anti-CD36 mAb treatment might mainly result from targeting CD36 expressed in Treg cells instead of other CD36-expressing cells.
Fig. 6. CD36-targeting impairs intratumoral Treg cells and primes tumors to PD-1 blockade.
a, b, c, Tumor growth (a) (Ctrl, n=4; α-CD36 Ab, n=6), percentage of FoxP3+ Treg cells among CD4+ T cells of indicated tissues (n=4) (b), and expression of cleaved caspase-3 in Treg cells isolated from indicated tissues (n=4) (c) of YUMM1.7 melanoma-bearing B6 mice treated with the indicated treatments. d, Tumor growth of YUMM1.7 melanoma-bearing WT and TregCd36-/- mice treated with either control vehicle (Ctrl) or α-CD36 mAb (WT+Ctrl, n=12; WT+α-CD36 Ab, n=10; TregCd36-/-+Ctrl, n=11; TregCd36-/-+α-CD36 Ab, n=10). e, f, Tumor growth (e) and survival curves (f) of YUMM1.7 melanoma-bearing WT and TregCd36-/- mice treated with the indicated treatments (WT, n=5; TregCd36-/-, n=4; WT+α-PD1, n=4; TregCd36-/-+α-PD1, n=4). Comparison between TregCd36-/- and TregCd36-/-+α-PD1 was done based on the readouts on day 16 and other comparisons were done based on the readouts on day 14. g, Tumor growth of inducible Braf/Pten melanoma-bearing mice treated with indicated treatments (Ctrl, n = 10; α-PD1, n = 11; α-CD36, n = 11; α-CD36 + αPD-1, n = 11). Arrows indicate the date of treatment. Data are representative result of at least two independent experiments with similar results (a, b, c, e, f) or cumulative results from at least two independent experiments (d, g). Each symbol represents one individual. Data are mean ± S.D. (b, c) or ± S.E.M. (a, d, e) and were analyzed by two-tailed, unpaired Student’s t-test. Difference between survival curves was analyzed by Log-rank (Mantel-Cox) test (f).
As T cell exhaustion may limit the therapeutic outcomes of Treg cell-targeting interventions31, we postulated that reinvigorating exhausted T cells with PD-1 blockade may potentiate the anti-tumor effects of CD36 blockade to restrain tumor progression. Indeed, we found that anti-PD-1 mAb more effectively limited tumor progression and prolonged survival in tumor-bearing TregCd36-/- mice compared to wild-type mice (Fig. 6e,f). In addition to genetic ablation of CD36 in Treg cells, we observed that anti-PD-1 mAb also potentiated the anti-tumor responses of anti-CD36 mAb in both the genetically engineered Braf/PTEN melanoma mouse model (Fig. 6g) and the YUMM1.7 engraftment model (Extended Data Fig. 6e,f). These results demonstrate that targeting CD36 in Treg cells might reprogram the TME towards more immunostimulatory conditions, which may therapeutically complement the effect of PD-1 blockade to counteract T-cell exhaustion. This points to CD36 blockade as a new potential immunotherapeutic intervention with reduced side effects caused by systemic impairment of Treg cells.
Discussion
Exploiting the regulatory circuits by which metabolic processes orchestrate immune responses in immune cells is an attractive strategy for fine-tuning host immunity in diseases. Here we uncover that CD36-PPAR-β signaling sustains survival and functional fitness in intratumoral Treg cells by modulating mitochondrial fitness and NAD levels. Owing to the uniqueness of the metabolic stress occurring in the TME and the selectivity of CD36-PPAR-β signaling for intratumoral Treg cells, targeting CD36 may provide broad therapeutic potential with limited negative impact on immune and peripheral tissue homeostasis in cancer patients. Moreover, the additive anti-tumor effects elicited by combined treatment with PD-1 blockade and CD36 targeting further warrant the development of CD36 inhibition approaches as potential cancer treatments.
Although our results reveal that CD36-mediated metabolic adaptation, including mitochondrial fitness and NAD production, support intratumoral Treg survival and suppressive functions, it remains unknown whether the suppressive functions of intratumoral Treg cells are modulated by lipid-derived signals or the enhanced usage of lactic acids. While OXPHOS is thought to promote Treg immunosuppression via undefined mechanisms42, it remains unexplored whether distinct fuels driving OXPHOS may differentially impact suppressive activity of Treg cells. Given that Treg cells could convert lactic acid into pyruvate that might be further utilized for production of acetyl-CoA, α-ketoglutartae, succinate and fumurate, which can be used for modulating transcriptome and function in immune cells via epigenetic regulations43,44, it is possible that CD36-mediated metabolic adaptation could instruct intratumoral Treg-specific transcriptome that support unique suppressive activities. On the other hand, aerobic glycolysis has also been suggested to support expansion of intratumoral Treg cells45. Since accumulation of NADH can impede glycolytic activity, it is also possible that the declined mitochondrial fitness in CD36-deficient intratumoral Treg cells could hamper expansion and survival by restricting glycolysis. Of note, in this study, the authors report that intratumoral Treg cells increased lipid content as a result of elevated glycolysis that supports fatty acid synthesis and cell expansion45. In contrast to our conclusion that intratumoral Treg cells increase lipid uptake, the author does not observe increased lipid uptake in intratumoral Treg cells by using systemic injection of Bodipy-conjugated lipids into tumor-bearing mice. We reason that the injection of limited amounts of Bodipy-conjugated lipids may not be sensitive enough to detect enhanced lipid uptake ability in intratumoral Treg cells. However, in agreement with our observation, both studies suggest that intratumoral Treg cells engage distinct metabolic processes that could be used to fuel survival and suppressive activity. Thus, elucidation of the mechanistic link between metabolic adaptation and specialized transcriptome dictating suppressive activity and tumor-specific behavior in intratumoral Treg cells is therefore an important avenue for future research. A better understanding of these questions may allow us to incorporate interventions targeting lactic acid production in cancer cells and lactic acid uptake in Treg cells for cancer treatment.
Although Treg-specific ablation of CD36 or PPAR-β does not compromise formation of Treg cells in thymus and secondary lymphoid organs, it remains likely that the induction of CD36-PPAR-β signal in particular circumstance, such as tumor, might synergize with FoxP3-mediated metabolic program to boost mitochondrial activity and oxidative phosphorylation in Treg cells. The robust increase of OXPHOS in this circumstance may further support DNA methylation-mediated chromatin remodeling that is critical for the expression of FoxP3 target genes via generation of metabolites controlling DNA demethylase activity. Furthermore, as exposure of cancer cell-conditioned medium stimulates expression of CD36 in Treg cells, while PPAR-β-deficient intratumoral Treg cells express lower levels of CD36, it is likely that lipids produced by tumor cells serve as immunosuppressive regulators to support the engagement of CD36-dependent metabolic adaptation in intratumoral Treg cells. Future studies elucidating the identity of these lipids can shed light on the impact of tumor metabolism on the TME.
Methods
Mice
C57BL/6/J, Foxp3YFP-Cre, Rag1–/– (B6.129S7-Rag1tm1Mom/J) mice were purchased from Jackson Laboratories. Cd36fl/fl mice were generated as previously described46 and provided by I. J. Goldberg at New York University. Ppargfl/fl and Pparbfl/fl mice were generated and provided by B. Desvergne at University of Lausanne47. BRafCA; Tyr::CreER; Ptenlox4-5 (Braf/Pten) was obtained from M. Bonsenburg at Yale University. K-rasLSL-G12D/+/p53fl/fl conditional mouse model of NSCLC was obtained from E. Meylan at Ecole Polytechnique Fédérale de Lausanne. Animals were housed in specific-pathogen-free facilities at the University of Lausanne and all experimental studies were approved and performed in accordance with guidelines and regulations implemented by the Swiss Animal Welfare Ordinance.
Cell lines and in vitro cultures
YUMM1.7 melanoma cell line was kindly provided by M.Bosenberg48. YUMM1.7 and B16-ova melanoma cell lines were cultured in DMEM with 10% fetal bovine serum and 1% penicillin-streptomycin and used for experiments when in exponential growth phase. MC38 colon adenocarcinoma cell line was provided by M. De Palma at Ecole Polytechnique Fédérale de Lausanne and was maintained in IMDM with 10% fetal bovine serum and 1% penicillin-streptomycin.
Cancer cell-conditioned medium and iTreg cell culture
iTreg cells were generated by activating naïve CD4+ T cells with Dynabeads conjugated with anti-CD3 and anti-CD28 mAbs (ThermoFisher) in RMPI media supplemented with 10% FBS, 10 ng/ml TGF-β and 50U/ml IL-2 for three days. Then activated CD4+ T cells were maintained in RPMI media plus 10% FBS and 50 U/ml IL-2 for another 2 days. Differentiated iTreg cells were firstly sorted by using FACS cell sorter and then incubated in indicated culture condition for 48 h. The survival and NAD/NADH levels of iTreg cells were then determined by live/dead staining and ELISA kits, respectively. For NAD/NADH measurement in CD36-deficient iTreg cells treated with PPAR-β agonist, sorted iTreg cells were cultured in cancer cell-conditioned medium in the presence of DMSO or GW50156 for 48 h. Control RPMI for iTreg cell in vitro treatment was prepared with RPMI 1640 medium (Biological Industries) supplementing with 2 mM Glucose, 10 mM Glutamine, 10% dialyzed FBS, 0.1% β-mercaptoethanol, and indicated concentrations of lactic acids. YUMM1.7 cancer cell-conditioned medium was collected by incubating YUMM1.7 cells (70-80% density) with control RPMI described above for 18 h. Then, culture medium was collected and centrifuged at 2000 rpm for 15 min to remove debris and cancer cells as cancer cell-conditioned medium. YUMM1.7 cancer cell-conditioned medium collected as described above was treated with Cleanascite™ reagent (Biotech Support Group) prior to Treg cell culture at a volume ratio of 1:5 according to the manufacturer’s instruction.
Ex vivo suppression assay
CD8+ T cells from spleen of Ly5.1 mice were enriched using negative selection kit (MojoSort Mouse CD8 T cell isolation kit, BioLegend), and stained with CellTrace™ CFSE cell proliferation kit (ThermoFisher) for 15 min at 37 °C. 1 × 104 CD8+ cells were seeded into 96-well round plate in RPMI medium consisting of 50 U/ml IL-2. CD44+YFP+ Treg cells (CD45.2+) isolated from splenocytes or TILs of FoxP3YFP-Cre or TregCD36-/- mice were added according to indicated ratios for Treg:Teff. Then, anti-CD3–CD28-conjugated Dynabeads (ThermoFisher) were supplemented into cultures except for negative control groups. Cells were incubated at 37 °C, 5% CO2 for 72 h and then the proliferation of CD8+ T cells were determined by CFSE dilution with flow cytometry analysis.
Tumor engraftment and treatment of tumor-bearing mice
For tumor induction, 3-week-old Braf/Pten mice were treated with 4-hydroxytamoxifen on the skin surface as described before to induce tumor formation49. For tumor engraftment, 5 × 104 cells YUMM1.7, B16-OVA, or one million1 × 106 MC38 tumor cells were injected subcutaneously in 50 μl PBS. Tumors were measured every 2-3 days post tumor engraftment or indicated treatments and calculated. Tumor volume was calculated by volume = (length × width2)/2 for engrafted tumor or volume = (length × width × height) for inducible tumor. For in vivo treatment, Yumm1.7-bearing mice were administrated every 3 days with either DMSO or PPAR-β agonist (GW 501516) (1 mg per kg of body weight, Cayman Chemical) by intraperitoneal injection. For antibody-based treatment, tumor-bearing mice were treated with anti-PD-1 antibody (200 μg per injection, BioXcell, clone 29F.1A12) and anti-CD36 antibody (200 μg per injection, clone CRF D-2712 50, provided by R. Silverstein at Medical College of Wisconsin) according to indicated combination by intraperitoneal injection. For antibody treatment in the Braf/Pten mouse model, four weeks after tumor induction, tumor-bearing Braf/Pten mice were treated with anti-CD36 antibody and/or anti-PD-1 antibody as indicated above for a period of 10 days. All experiments were conducted according to Swiss federal regulations.
Tumor digestion and cell isolation
Tumors were minced into small pieces in RPMI containing 2% FBS, 1% penicillin-streptomycin (p/s), DNase I (1 µg/ml, Sigma-Aldrich), and collagenase (0.5 mg/ml, Sigma-Aldrich) and kept for digestion for 40 min at 37 °C, followed by the filtration with a 70 μm cell strainer. Filtered cells were incubated with ACK lysis buffer (Invitrogen) to lyse red blood cells and then washed with fluorescent activated cell sorter (FACS) buffer (phosphate-buffered saline with 2% fetal bovine serum and 2 mM EDTA). Tumor-infiltrating leukocytes were further enriched by percoll density gradient centrifugation (800xg, 30 min) at room temperature as described before51.
Flow cytometry, cell sorting and antibodies
Single cell suspensions were incubated with Fc receptor-blocking anti-CD16/32 (93) and anti-CD351 (TX61) antibodies (BioLegend) on ice for 10 min before staining. Cell suspensions were first stained with LIVE/DEAD® Fixable Violet Dead Cell Stain Kit (ThermoFisher) at 37 °C for 10 min. Aftrer washing, surface proteins were stained for 30 min at 4 °C. To detect cytokine production upon ex-vivo re-stimulation, cell suspensions were re-suspended in RPMI 1640 with 10% FBS and then added to plates coated with 1 μg/ml anti-CD3 antibody (clone 145-2C11, BioLegend) and anti-CD28 antibody (clone 37.51, BioLegend) for another 5 h at 37 °C in the presence 2.5 μg/ml Brefeldin A Solution (BFA) (BioLegend). Cells were processed for surface marker staining as described above and then intracellular cytokine staining. Samples were analyzed on LSRII flow cytometers (BD Biosciences) and data were analyzed with FlowJo. Cells were sorted either on FACSAria™ III sorter (BD Biosciences) or SH800S Cell Sorter (Sony). Human Treg cells were defined by the following staining: Live/Dead-neg/CD45RA-neg/CD3-pos/CD4-pos/CD8-neg/CD25-pos/FoxP3-pos; Mouse non Treg CD4 T cells were defined by the following staining: CD45-pos/CD3-pos/CD4-pos/CD8-neg/FoxP3-neg; CD8 T cells were defined by the following staining: CD45-pos/CD3-pos/CD8-pos/CD4-neg; Treg cells were defined by the following staining: CD45-pos/CD3-pos/CD4-pos/CD8-neg/FoxP3-pos. The following antibodies against mouse proteins were used: anti-CD45 (30-F11), anti-CD3ε (17A2), anti-CD4 (RM4-5), anti-CD8α (53.6.7), anti-CD44 (IM7), anti-62L (Mel-14), anti-PD1 (RMP1-30), anti-CD134 (OX40) (OX-86), anti-CD357 (GITR) (DTA-1), anti-CD36 (CRF D-2712), anti-IgA (mA-6E1), anti-FoxP3 (MF-14), anti-GFP/YFP (FM264G), anti-IFN-γ (XMG1.2), anti-TNF (MP6-XT22), anti-IL-17A (TC11-18H10.1), anti-Ki67 (16A8), anti-CD278 (ICOS) (15F9), anti-CD152 (CTLA4) (UC10-4B9), cleaved Caspase-3 (Asp175). The following antibodies against human proteins were used: Anti-CD45 (2D1), anti-CD3 (SK7), anti-CD4 (SK3), anti-CD25 (BC96), anti-CD8 (RPA-TP), anti-CD36 (TR9), anti-PD1 (E12.1), anti-GITR (eBIOAITR), and anti-FoxP3 (150D). These antibodies were purchased from BioLegend, eBiosciences and Cell Signaling.
Mitochondrion, fatty acid uptake and lipid content assay
For measuring mitochondrial membrane potential, cells were washed and incubated with pre-warmed (37 °C) staining solution (RPMI with 2% FBS) containing MitoTracker® Deep Red FM (ThermoFisher) and MitoTracker® Green FM (ThermoFisher) at the working concentrations of 10nm and 100nM for 15min, respectively. After staining, the cells were washed and resuspended in fresh FACS buffer for surface marker staining as described above. For measuring fatty acid uptake, cells were incubated in RPMI medium (or human T cell culture medium) containing C1-BODIPY® 500/510 C12 (Life Technologies) at final concentration of 0.5 μM for 15 min at 37 °C. After incubation, cells were washed with FACS buffer for surface staining. For lipid content detection, after permeablization and fixation, cells were stained using BODIPY® 493/503 (Life Technologies) at a final concentration of 500 ng/ml together with other intracellular proteins.
RNA sequencing and bioinformatics analysis
500-600 viable CD4+CD44+YFP+ intratumoral Treg cells from heterozygous Foxp3YFP-Cre/+ female mice or heterozygous Foxp3YFP-Cre/+Cd36fl/fl female mice were isolated by FACS cell sorters (with at least 99% purity) directly into 4 μl lysis buffer consisting of 0.2 % (vol/vol) Triton X-100 solution (MgBCH-Axon Lab) and RNase inhibitor (Clontech). Plate containing samples were sealed, flash-frozen and kept at −80 °C before further processing following a version of the Smart-Seq2 protocol described before52. The RNA-sequencing raw data were processed through the standard RNA-seq analysis pipeline. Briefly, read alignment was examined using tophat2 v2.1.0 and then compared to the Mus musculus GRCm38.p4 genome version. Following the alignment, reads mapped to each gene were annotated using HTseq count. The differential expression analyses were conducted based on the DESeq2 R library. The differential expression test and visualization were then examined by the START Web-based RNA-seq analysis resources53. Gene Set Enrichment Analysis (GSEA) was performed using GSEA software.
Electron microscopy analysis and histology analysis
For electron microscopy analysis, sorted cells were fixed in glutaraldehyde 2.5% (EMS) and osmium tetroxide 1% (EMS) overnight at 4 °C, followed by several washes with water and acetone (Sigma), and embedded in Epon (Sigma) resin the following day. Before imaging, 50 nm slides were prepared by using a Leica Ultracut microtome, and were contrasted using uranyl acetate (Sigma) and Reynolds lead citrate (Sigma). Electron microscope images were taken with a transmission electron microscope Philips CM100 at an acceleration of 80kV with a TVIPS TemCam-F416 digital camera with a magnification of 4800x and 11’000x. Image analysis and quantification were carried out using EMMENU, 3dmod (University of Colorado) and Fifi (ImageJ) software. To quantify mitochondria per sorted cell, a grid was applied and each intersection was defined as being part of the nucleus, cytoplasm or mitochondria and the length of each crista was measured divided by the mitochondrial area for determination of the crista density. For histology analysis, organs were trimmed and placed in the labeled cassettes and fixed in formalin for 24h for further embedding in molten paraffin wax. Paraffin sections at a thickness of 3-5 μm was stained with hematoxylin and eosin according to standard procedures. Images were taken and exported on a Nikon Eclipse Ti-S inverted microscope.
Human patient assessment
This study was performed in accordance with the guidelines of ethic regulation for human samples under approved protocol at Lausanne University Hospital (protocol 87/06), University Hospital Basel (EK128/13) and Memorial Sloan Kettering Cancer Center (00-144 and 19-101). Human samples were analyzed following safety regulation and stained with antibodies for FACS analysis as described above.
T cell transfer model of colitis
Wild-type and CD36-deficient Treg cells were sorted from the spleens of either Foxp3YFP-Cre mice or TregCD36-/- mice and naïve CD4+ T cells were harvested using a combination of negative magnetic selection (MojoSort Mouse CD4 T cell isolation kit, BioLegend) and FACS sorting (>98% purity). To induce colitis, naïve CD4+ cells (5 × 105 cells) were transferred intravenously into Rag1−/− recipients. In some recipients, 4 × 105 CD44+YFP+ Treg cells isolated from splenocytes of Foxp3YFP-Cre mice or TregCD36-/- mice were co-transferred with naïve CD4+ T cells. Recipient mice were monitored and weighted every two or three days after transferring for signs of diseases such as weight loss. Diseases onset usually occurs at 4-5 weeks post-transfer. The endpoints of this study included the determination of body weight loss, colitis length, and diarrhea. In addition, colons and small intestines were collected and processed for further evaluation by Hemotoxylin and Eosin staining.
NAD and NADH measurement
The ratio of nicotinamide adenine dinucleotide (NAD) and nicotinamide adenine dinucleotide hydrate (NADH) were measured by using the commercial NAD/NADH Quantification Kit (Sigma-Aldrich MAK037). The cells were first de-proteinized to prevent the NAD and NADH consumption by enzymes. After washing with cold PBS, cell pellets suspended in NADH/NAD extraction buffer (200 μl) were treated with two repetitive freeze-thaw cycles and then spun at 13,000×g for 5 min at 4 °C. The supernatant was then divided into two aliquots, one was for NADtotal detection, and the other one was heated at 60° C for 30 min for NAD decomposition. The samples were then transferred to a 96-well plate for measuring the absorbance of 450 nm. The amount of oxidized NAD (NAD+) was presented subtracting NADH from NADtotal. The ratio of NAD/NADH in a sample could be determined by the following equation: ratio = (NADtotal – NADH)/NADH.
Seahorse extracellular flux analyses
Extracellular flux analysis was performed with an XF96 Seahorse Extracellular Flux Analyzer as described before54 with minor modifications. Cells were treated with oligomycin (0.5 μM, Sigma-Aldrich), FCCP (2 μM, Sigma-Aldrich), rotenone (0.5 μM, Sigma-Aldrich), antimycin A (0.5 μM, Sigma-Aldrich), glucose (10 mM, Sigma-Aldrich) and 2-DG (50 mM, Sigma-Aldrich). Each condition was performed with 3–6 replicates in each single experiment.
Statistical analyses
Statistical analyses were performed using the two-tailed, unpaired, Student’s t-test. Log-rank (Mantel–Cox) test was used for survival curve analysis. Each point represented a biological replicate and all data were presented as the mean ± SD, or mean ± SEM as indicated. The P values were represented as follows: ***P < 0.001, **P < 0.01 and *P < 0.05. P <0.05 was considered statistically significant.
Extended Data
Extended Data Fig. 1. Lipid accumulation and increased CD36 expression in intratumoral Treg cells.
a, b, Quantitative results of geometric mean (GeoMean) fluorescent intensity of Bodipy FL C12 (a) and Bodipy 493/503 (Bodipy) (b) in Treg cells from paired TILs and PBMCs of non-small cell lung cancer (NSCLC) patients (n=6 per group). c, d, Quantitative results of GeoMean fluorescent intensity of Bodipy FL C12 (c) and Bodipy 493/503 (d) in Treg cells from paired PBMC and tumor infiltrated lymph nodes (TILNs) of melanoma patients (n=19). e, f, Quantitative results of fluorescent intensity of Bodipy FL C12 in Treg cells from indicated tissues of B16 melanoma-bearing mice (dLN, n=7; Spleen, n=6; Thymus, n=7; Tumor, n=6) (e), and MC38 colon carcinoma-bearing mice (n=8) (f). g, Quantitative result of GeoMean fluorescent intensity of CD36 surface staining in Treg cells from paired TILs and PBMCs of NSCLC patients (n=6) h, i, j, k, Quantitative results of surface expression of CD36 in Treg cells of indicated tissues from B16-OVA melanoma-bearing B6 mice (n=6, one outlier was removed from dLN) (h), inducible Braf/Pten melanoma-bearing mice (n=9) (i), K-rasLSL-G12D/+/p53fl/fl mouse model of NSCLC (Blood, n=8; Tumor, n=13) (j), and MC38 colon cancer (n=8) (k). l, CD36 expression in iTreg cells cultured in different indicated conditions for 48h. (RPMI: normal cell culture RPMI 1640 medium indicated in methods; CM, cancer cell conditioned medium, n=4 per group). m, CD36 expression in iTreg cells cultured in cancer cell-conditioned medium treated with control procedure or lipid removal procedure for 48h. (n=6 per group). Data are representative result of at least two independent experiments with similar results (l, m) or cumulative results from at least two independent experiments (a, b, c, d, e, f, g, h, i, j, k). Each symbol represents one individual. Data are mean ± S.D. and were analyzed by two-tailed, unpaired Student’s t-test (e, f, h, i, j, k, l, m) or two-tailed, paired Student’s t-test (a, b) or one-tailed, paired Student’s t-test (c, d, g).
Extended Data Fig. 2. CD36 expression supports the accumulation and suppressive function of intratumoral Treg cells.
a, Body weight of WT and TregCd36-/- mice at the age of 21-23 weeks (WT male, n=4; TregCd36-/- male, n=6; WT female, n=6; TregCd36-/- female, n=5;). b, Representative plots (left) and quantitative frequency of CD44hi/CD62Llow CD4+ or CD8+ T cells (right) in aged WT and TregCd36-/- mice (n=7 per group). c, Representative images of hematoxylin and eosin (H&E) staining for indicated tissues from WT or TregCd36-/- mice at the age of 21-23 weeks. Scale bar, 200 µm. d, e, Tumor growth of B16-OVA melanoma (n=7 per group) (d) or MC38 colon carcinoma (n=6 per group) (e) from WT or TregCd36-/- mice. f, g, h, Absolute number of FoxP3+ Treg cells per gram tumor (f), percentage of CD8+ T cells out of CD3+ T cells among tumor-infiltrating T cells (g), and the ratio of CD8+ to Treg cell TIL density (h) of YUMM1.7 melanoma-bearing WT and TregCd36-/- mice (n=11 per group). i, Representative plots (left) and percentage of indicated cytokine-producing CD4+ T cells among total tumor-infiltrating CD4+ T cells from indicated mice (right) (n=5 per group). Data are representative result of at least two independent experiments with similar results (c, d, e, i) or cumulative results from at least two independent experiments (a, b, f, g, h) Each symbol represents one individual. Data are mean ± S.D. (a, b, f, g, h, i) or ± S.E.M. (d, e) and were analyzed by two-tailed, unpaired Student’s t-test.
Extended Data Fig. 3. Effects of CD36 in expression of activation markers and stability of intratumoral Treg cells.
a, Representative images of guts (a) and spleens (b) from indicated group of Rag1-/- mice. c, d, e, Expression of CD44 (n=17) (c), CD103 (n=5) (d), and KLRG1 (n=5) (e) in intratumoral Treg cells of YUMM1.7 melanoma-bearing WT and TregCd36-/- mice. f, The expression of YFP in intratumoral Treg cells of YUMM1.7 melanoma-bearing WT and TregCd36-/- mice (n=15). g, h, i, Representative plots of IFNγ and TNF production among total intratumoral Treg cells from indicated mice (g), and quantitative result of percentage of IFNγ-producing (n=19 per group) (h) and TNF-producing (n=18, one outlier was removed from TregCd36-/-) (i) Treg cells among total intratumoral Treg cells of indicated mice. j, Expression of Ki67 in intratumoral Treg cells of YUMM1.7 melanoma-bearing WT and TregCd36-/- mice (n=10 per group). k, Representative histograms (left) and quantitative analysis (right) of Annexin V staining in intratumoral Treg cells from WT and TregCd36-/- tumor-bearing mice (n=14 per group). l, Quantitative analysis of cleaved caspase-3 levels in Treg cells of indicated tissues from WT (n=13 per group except for thymus, n=9) and TregCd36-/- (n=14 per group except for thymus, n=9) tumor-bearing mice. LN: non-draining lymph node; DLN: draining lymph node. Data are representative results of two independent experiments with similar results (a, b, d, e) or cumulative results from at least three independent experiments (c, f, g, h, i, j, k, l). Each symbol represents one individual. Data are mean ± S.D. (c, d, e, f, h, i, j, k, l) and were analyzed by two-tailed, unpaired Student’s t-test.
Extended Data Fig. 4. CD36-deficiency results in a metabolic shift and elevated apoptosis in Treg cells.
a, Indicated iTreg cells cultured in cancer cell-conditioned medium for 48 hrs (n=3 per group). Oxygen consumption rate (OCR) of indicated iTreg cells was measured and then followed by treatment with oligomycin, FCCP, and antimycin A plus Rotenone (n≥4 per group). b, Indicated iTreg cells cultured in cancer cell-conditioned medium for 48 hrs (n≥4 per group) and then media were refreshed with Seahorse Flux assay media without glucose. Basel extracellular acidification rate (ECAR) of indicated iTreg cells was measured and then followed by treatment with glucose, oligomycin, FCCP and 2-DG (n=4 per group). c, Quantitative result of glycolysis and glycolytic capacity based on the measurement of b. d, The viability of either WT or TregCd36-/- iTreg cells cultured under indicated conditions for 72 hrs (n=6 per group). Data are representative results of three independent experiments with similar results (a, b, c, d). Data are mean ± S.D. and were analyzed by two-tailed, unpaired Student’s t-test (c, d).
Extended Data Fig. 5. Intratumoral Treg cells require PPAR-β, not PPAR-γ, signaling for metabolic adaptation.
a, Enrichment plots of signals controlling mitochondrial matrix (left) and mitochondrial envelope in intratumoral Treg (n=4) compared to PBMC Treg cells (n=6), identified by GSEA computational method. ES: enrichment score; NES: normalized enrichment score; FDR: false discovery rate; NOM p-val: Nominal p value. b, c, d, Percentage of FoxP3+ Treg cells among CD4+ tumor-infiltrating T lymphocytes (n=5) (b), tumor growth (n=5) (c) and tumor weight (n=7) (d) from tumor-bearing WT and TregPPARγ-/- mice. e, Percentage of CD8+ T cells among tumor-infiltrating T cells from tumor-bearing WT and TregPPARβ-/- mice (n=10). f, Quantitative result of CD36+ intratumoral Treg cells from YUMM1.7 melanoma-bearing WT and TregPPARβ-/- mice (WT, n=14; TregPPARβ-/-, n=11). g, NAD/NADH ratio of indicated iTreg cells cultured in cancer cell-conditioned medium with DMSO or PPAR-β agonist for 48h (DMSO, n=8; PPAR-β agonist, n=10). Data are representative results of at least two independent experiments with similar results (b, c, d) or cumulative results from at least two independent experiments (e, f, g). Each symbol represents one individual. Data are mean ± S.D. (b, d, e, f, g) or ± S.E.M. (c) and were analyzed by two-tailed, unpaired Student’s t-test.
Extended Data Fig. 6. CD36-targeting unleashes host antitumor immunity.
a, b, c, d, Absolute number of FoxP3+ Treg cells per gram tumor (n=10 per group) (a), percentage of CD8+ T cells among tumor-infiltrating T cells (n=10 per group) (b) and representative plots and percentage of indicated cytokine-producing CD8+ T cells among total tumor-infiltrating CD8+ T cells (c) and CD4+ T cells among total tumor-infiltrating CD4+ T cells (d) from YUMM1.7 melanoma-bearing mice treated with indicated treatments (n=10 per group). e, f, Tumor growth (e) and survival curves (f) of YUMM1.7 melanoma-bearing B6 mice treated with indicated treatments (Ctrl, n = 10; α-PD1, n = 10; α-CD36, n = 11; α-CD36 + αPD-1, n = 11). Arrows indicate the date of treatment. Dotted lines indicate the tumor volume of 800 mm3. Data are cumulative results from at least two independent experiments. Each symbol represents one individual. Data are mean ± S.D. and were analyzed by two-tailed, unpaired Student’s t-test (a, b, c, d). Difference between survival curves was analyzed by Log-rank (Mantel-Cox) test (f).
Acknowledgments
We thank L.-F. Lu and W.-L. Lo for critical reading and comments for this study. We also thank Y. Maeda and H. Nishikawa for helpful discussion. P.-C.H. was supported in part by the SNSF project grants (31003A_163204 and 31003A_182470), the Swiss Cancer Foundation (KFS-3949-08-2016), the Swiss Institute for Experimental Cancer Research (ISREC 26075483), European Research Council Staring Grant (802773-MitoGuide), the Cancer Research Institute-Clinic and Laboratory Integration Program award, and the SITC-MRA Young Investigator Award. C.J. is supported by SNSF project grants (PMPDP3_129022 and PZ00P3_161459). A.Z. is supported by SNSF project grant (320030_162575) and Cancer League Switzerland (KFS-3394-02-2014). R.S. and I.G. are supported by NIH funding (P01 HL46403, P01 HL087018 and R01 HL142152 to R.S. and HL45095 and HL073029 to I.G.). E.M. acknowledges funding from the Swiss Cancer Research Foundation (KFS-3681-08-2015-R). S.M.F. acknowledges funding from FWO grant and projects as well as KU Leuven Methusalem co-funding. J.F.G. is supported by a FWO postdoctoral fellowship. J.D.W and T.M. are supported by NIH funding (P30 CA008748 and R01 CA 056821), the Swim Across America, Ludwig Institute for Cancer Research, Parker Institute for Cancer Immunotherapy and Breast Cancer Research Foundation. R.Z. is supported by the Parker Institute for Cancer Immunotherapy Bridge Scholar Award. We also appreciate the support provided by Electron Microscopy Facility at University of Lausanne and Biomedical Sequencing Facility at Research Center for Molecular Medicine of the Austrian Academy of Sciences.
Footnotes
Reporting Summary. Detailed information on experimental design is available in the Life Sciences Reporting Summary linked to this article.
Data availability
The RNA-seq data for intratumoral Treg cells are available in the Gene Expression Omnibus database under accession code (GSE139325). All relevant data are available from the corresponding author upon request.
Author Contributions
H.W. and P.-C.H. contributed to overall project design and wrote manuscript. H.W., F.F., Y.-C.T., C.-H.T., and F.P. performed in vitro and in vivo animal works and data analysis. X.X. and S.R.M. performed analysis of RNA sequencing results. H.W., M.P.T., R.Z., J.D.W., T.M., C.J., I.S., and A.Z. conducted collection and flow cytometry analysis of human samples. J.F.G. and S.-M.F. supported metabolomite analysis. R.S. and I.G. provided the hybridoma clone for anti-CD36 antibody production and CD36flox mice, respectively. E.M. provided samples of NSCLC murine models.
Competing Interests Statement
H.W. and P.-C.H. are inventors of patent application related to targeting CD36 in cancer immunotherapy. P.-C.H. is serving as a member of scientific advisory board for Elixiron Immunotherapeutics and receiving research grants from Roche and Idorsia. J.D.W. is serving as a consultant for Adaptive Biotech; Advaxis; Amgen; Apricity; Array BioPharma; Ascentage Pharma; Astellas; Bayer; Beigene; Bristol Myers Squibb; Celgene; Chugai; Elucida; Eli Lilly; F Star; Genentech; Imvaq; Janssen; Kyowa Hakko Kirin; Kleo Pharma; Linneaus;MedImmune; Merck; Neon Therapuetics; Northern Biologics; Ono; Polaris Pharma; Polynoma; Psioxus; Puretech; Recepta; Takara Bio; Trieza; Sellas Life Sciences; Serametrix; Surface Oncology; Syndax; Synthologic. J.D.W. received research support from Bristol Myers Squibb; Medimmune; Merck Pharmaceuticals; Genentech and has equity in Potenza Therapeutics; Tizona Pharmaceuticals; Adaptive Biotechnologies; Elucida; Imvaq; Beigene; Trieza; Linneaus. P.C.H. received honorarium from Pfizer and Chugai and J.D.W received honorarium from Esanex.
References
- 1.Roychoudhuri R, Eil RL, Restifo NP. The interplay of effector and regulatory T cells in cancer. Current opinion in immunology. 2015;33:101–111. doi: 10.1016/j.coi.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 2.Delgoffe GM, et al. Stability and function of regulatory T cells is maintained by a neuropilin-1-semaphorin-4a axis. Nature. 2013;501:252–256. doi: 10.1038/nature12428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saito T, et al. Two FOXP3(+)CD4(+) T cell subpopulations distinctly control the prognosis of colorectal cancers. Nat Med. 2016;22:679–684. doi: 10.1038/nm.4086. [DOI] [PubMed] [Google Scholar]
- 4.Rech AJ, et al. CD25 blockade depletes and selectively reprograms regulatory T cells in concert with immunotherapy in cancer patients. Sci Transl Med. 2012;4:134ra162. doi: 10.1126/scitranslmed.3003330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sutmuller RP, et al. Synergism of cytotoxic T lymphocyte-associated antigen 4 blockade and depletion of CD25(+) regulatory T cells in antitumor therapy reveals alternative pathways for suppression of autoreactive cytotoxic T lymphocyte responses. J Exp Med. 2001;194:823–832. doi: 10.1084/jem.194.6.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marabelle A, et al. Depleting tumor-specific Treg cells at a single site eradicates disseminated tumors. The Journal of clinical investigation. 2013;123:2447–2463. doi: 10.1172/JCI64859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Teng MW, et al. Conditional regulatory T-cell depletion releases adaptive immunity preventing carcinogenesis and suppressing established tumor growth. Cancer Res. 2010;70:7800–7809. doi: 10.1158/0008-5472.CAN-10-1681. [DOI] [PubMed] [Google Scholar]
- 8.Walter S, et al. Multipeptide immune response to cancer vaccine IMA901 after single-dose cyclophosphamide associates with longer patient survival. Nature medicine. 2012;18:1254–1261. doi: 10.1038/nm.2883. [DOI] [PubMed] [Google Scholar]
- 9.Nishikawa H, Sakaguchi S. Regulatory T cells in tumor immunity. International journal of cancer. 2010;127:759–767. doi: 10.1002/ijc.25429. [DOI] [PubMed] [Google Scholar]
- 10.Simpson TR, et al. Fc-dependent depletion of tumor-infiltrating regulatory T cells co-defines the efficacy of anti-CTLA-4 therapy against melanoma. J Exp Med. 2013;210:1695–1710. doi: 10.1084/jem.20130579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Curtin JF, et al. Treg depletion inhibits efficacy of cancer immunotherapy: implications for clinical trials. PLoS One. 2008;3:e1983. doi: 10.1371/journal.pone.0001983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arce Vargas F, et al. Fc Effector Function Contributes to the Activity of Human Anti-CTLA-4 Antibodies. Cancer Cell. 2018;33:649–663 e644. doi: 10.1016/j.ccell.2018.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miragaia RJ, et al. Single-Cell Transcriptomics of Regulatory T Cells Reveals Trajectories of Tissue Adaptation. Immunity. 2019;50:493–504 e497. doi: 10.1016/j.immuni.2019.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arpaia N, et al. A Distinct Function of Regulatory T Cells in Tissue Protection. Cell. 2015;162:1078–1089. doi: 10.1016/j.cell.2015.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burzyn D, et al. A special population of regulatory T cells potentiates muscle repair. Cell. 2013;155:1282–1295. doi: 10.1016/j.cell.2013.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cipolletta D, et al. PPAR-gamma is a major driver of the accumulation and phenotype of adipose tissue Treg cells. Nature. 2012;486:549–553. doi: 10.1038/nature11132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Panduro M, Benoist C, Mathis D. Tissue Treg cells. Annu Rev Immunol. 2016;34:609–633. doi: 10.1146/annurev-immunol-032712-095948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vignali DA, Collison LW, Workman CJ. How regulatory T cells work. Nat Rev Immunol. 2008;8:523–532. doi: 10.1038/nri2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chaudhry A, Rudensky AY. Control of inflammation by integration of environmental cues by regulatory T cells. J Clin Invest. 2013;123:939–944. doi: 10.1172/JCI57175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Campbell DJ, Koch MA. Phenotypical and functional specialization of FOXP3+ regulatory T cells. Nat Rev Immunol. 2011;11:119–130. doi: 10.1038/nri2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeng H, et al. mTORC1 couples immune signals and metabolic programming to establish T(reg)-cell function. Nature. 2013;499:485–490. doi: 10.1038/nature12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang K, et al. Homeostatic control of metabolic and functional fitness of Treg cells by LKB1 signalling. Nature. 2017;548:602–606. doi: 10.1038/nature23665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huynh A, et al. Control of PI(3) kinase in Treg cells maintains homeostasis and lineage stability. Nat Immunol. 2015;16:188–196. doi: 10.1038/ni.3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gerriets VA, et al. Foxp3 and Toll-like receptor signaling balance Treg cell anabolic metabolism for suppression. Nat Immunol. 2016;17:1459–1466. doi: 10.1038/ni.3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weinberg SE, et al. Mitochondrial complex III is essential for suppressive function of regulatory T cells. Nature. 2019;565:495–499. doi: 10.1038/s41586-018-0846-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Angelin A, et al. Foxp3 Reprograms T Cell Metabolism to Function in Low-Glucose, High-Lactate Environments. Cell Metab. 2017;25:1282–1293 e1287. doi: 10.1016/j.cmet.2016.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang H, Franco F, Ho PC. Metabolic Regulation of Treg cells in Cancer: Opportunities for Immunotherapy. Trends Cancer. 2017;3:583–592. doi: 10.1016/j.trecan.2017.06.005. [DOI] [PubMed] [Google Scholar]
- 28.Li X, et al. Navigating metabolic pathways to enhance antitumour immunity and immunotherapy. Nat Rev Clin Oncol. 2019 doi: 10.1038/s41571-019-0203-7. [DOI] [PubMed] [Google Scholar]
- 29.Plitas G, et al. Regulatory T Cells Exhibit Distinct Features in Human Breast Cancer. Immunity. 2016;45:1122–1134. doi: 10.1016/j.immuni.2016.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Piconese S, et al. Human OX40 tunes the function of regulatory T cells in tumor and nontumor areas of hepatitis C virus-infected liver tissue. Hepatology. 2014;60:1494–1507. doi: 10.1002/hep.27188. [DOI] [PubMed] [Google Scholar]
- 31.Zappasodi R, et al. Rational design of anti-GITR-based combination immunotherapy. Nat Med. 2019;25:759–766. doi: 10.1038/s41591-019-0420-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He N, et al. Metabolic control of regulatory T cell (Treg) survival and function by Lkb1. Proc Natl Acad Sci U S A. 2017;114:12542–12547. doi: 10.1073/pnas.1715363114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beier UH, et al. Essential role of mitochondrial energy metabolism in Foxp3(+) T-regulatory cell function and allograft survival. FASEB J. 2015;29:2315–2326. doi: 10.1096/fj.14-268409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ho PC, et al. Phosphoenolpyruvate Is a Metabolic Checkpoint of Anti-tumor T Cell Responses. Cell. 2015;162:1217–1228. doi: 10.1016/j.cell.2015.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Siska PJ, Rathmell JC. T cell metabolic fitness in antitumor immunity. Trends Immunol. 2015;36:257–264. doi: 10.1016/j.it.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vannini N, et al. The NAD-Booster Nicotinamide Riboside Potently Stimulates Hematopoiesis through Increased Mitochondrial Clearance. Cell Stem Cell. 2019;24:405–418 e407. doi: 10.1016/j.stem.2019.02.012. [DOI] [PubMed] [Google Scholar]
- 37.Ravnskjaer K, et al. PPARdelta is a fatty acid sensor that enhances mitochondrial oxidation in insulin-secreting cells and protects against fatty acid-induced dysfunction. J Lipid Res. 2010;51:1370–1379. doi: 10.1194/jlr.M001123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morino K, et al. Regulation of mitochondrial biogenesis by lipoprotein lipase in muscle of insulin-resistant offspring of parents with type 2 diabetes. Diabetes. 2012;61:877–887. doi: 10.2337/db11-1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fan W, et al. PPARdelta Promotes Running Endurance by Preserving Glucose. Cell Metab. 2017;25:1186–1193 e1184. doi: 10.1016/j.cmet.2017.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pascual G, et al. Targeting metastasis-initiating cells through the fatty acid receptor CD36. Nature. 2017;541:41–45. doi: 10.1038/nature20791. [DOI] [PubMed] [Google Scholar]
- 41.Ladanyi A, et al. Adipocyte-induced CD36 expression drives ovarian cancer progression and metastasis. Oncogene. 2018;37:2285–2301. doi: 10.1038/s41388-017-0093-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miska J, et al. HIF-1alpha Is a Metabolic Switch between Glycolytic-Driven Migration and Oxidative Phosphorylation-Driven Immunosuppression of Treg cells in Glioblastoma. Cell Rep. 2019;27:226–237 e224. doi: 10.1016/j.celrep.2019.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Phan AT, Goldrath AW, Glass CK. Metabolic and Epigenetic Coordination of T Cell and Macrophage Immunity. Immunity. 2017;46:714–729. doi: 10.1016/j.immuni.2017.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu PS, Ho PC. Mitochondria: A master regulator in macrophage and T cell immunity. Mitochondrion. 2018;41:45–50. doi: 10.1016/j.mito.2017.11.002. [DOI] [PubMed] [Google Scholar]
- 45.Pacella I, et al. Fatty acid metabolism complements glycolysis in the selective regulatory T cell expansion during tumor growth. Proc Natl Acad Sci U S A. 2018;115:E6546–E6555. doi: 10.1073/pnas.1720113115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Son NH, et al. Endothelial cell CD36 optimizes tissue fatty acid uptake. J Clin Invest. 2018;128:4329–4342. doi: 10.1172/JCI99315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dammone G, et al. PPARgamma Controls Ectopic Adipogenesis and Cross-Talks with Myogenesis During Skeletal Muscle Regeneration. International journal of molecular sciences. 2018;19 doi: 10.3390/ijms19072044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meeth K, Wang JX, Micevic G, Damsky W, Bosenberg MW, et al. The YUMM lines: a series of congenic mouse melanoma cell lines with defined genetic alterations. Pigment cell & melanoma research. 2016;29:590–597. doi: 10.1111/pcmr.12498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ho PC, et al. Immune-based antitumor effects of BRAF inhibitors rely on signaling by CD40L and IFNgamma. Cancer Res. 2014;74:3205–3217. doi: 10.1158/0008-5472.CAN-13-3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Driscoll WS, Vaisar T, Tang J, Wilson CL, Raines EW. Macrophage ADAM17 deficiency augments CD36-dependent apoptotic cell uptake and the linked anti-inflammatory phenotype. Circulation research. 2013;113:52–61. doi: 10.1161/CIRCRESAHA.112.300683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cheng WC, et al. Uncoupling protein 2 reprograms the tumor microenvironment to support the anti-tumor immune cycle. Nat Immunol. 2019;20:206–217. doi: 10.1038/s41590-018-0290-0. [DOI] [PubMed] [Google Scholar]
- 52.Picelli S, et al. Full-length RNA-seq from single cells using Smart-seq2. Nature protocols. 2014;9:171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- 53.Nelson JW, Sklenar J, Barnes AP, Minnier J. The START App: a web-based RNAseq analysis and visualization resource. Bioinformatics. 2017;33:447–449. doi: 10.1093/bioinformatics/btw624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu PS, et al. alpha-ketoglutarate orchestrates macrophage activation through metabolic and epigenetic reprogramming. Nat Immunol. 2017;18:985–994. doi: 10.1038/ni.3796. [DOI] [PubMed] [Google Scholar]