Abstract
Eosinophils have broad and extensive immunomodulatory capacity; recent studies have focused on the roles of distinct eosinophil subsets in specific tissue microenvironments. Ly6G is a GPI-linked leukocyte surface antigen understood primarily as a marker of mouse neutrophils, although its full function is not known. Here, we show that Ly6G/Gr1, detected by monoclonal antibodies 1A8 (anti-Ly6G) and RB6–8C5 (anti-Gr1) is detected prominently on a significant fraction of eosinophils from mouse bone marrow and bone marrow-derived culture, with fractions expressing this antigen increasing in IL-5-enriched microenvironments. Among our findings, we identified SiglecF+Gr1+ eosinophils in bone marrow from naïve, allergen-challenged and IL-5 transgenic mice; SiglecF+Gr1+ eosinophils were also prominent ex vivo, in bone marrow-derived eosinophils (bmEos) in IL-5-enriched culture. Reducing the IL-5 concentration 20-fold had no impact on the rate of generation of SiglecF+ bmEos but did result in a marked increase in the Gr1− fraction (from 17.4 ± 2 to 30 ± 2.3%, ***p < 0.005). Reducing the IL-5 concentration also enhanced chemotaxis; SiglecF+Gr1− bmEos were considerably more responsive to eotaxin-1 than were their SiglecF+Gr1+ counterparts. These results suggest that (1) IL-5 regulates the expression of Ly6G/Gr1, either directly or indirectly, in cells of the eosinophil lineage, (2) eosinophils generated in response to high concentrations of IL-5 can be distinguished from those generated under homeostatic conditions by expression of the Ly6G/Gr1 cell surface antigen, and (3) expression of Ly6G/Gr1 may have an impact on function, directly or indirectly, including their potential to undergo chemotaxis in response to eotaxin-1.
Keywords: Cytokine, Interleukin-5, Eotaxin-1, Hematopoiesis, Chemotaxis
Graphical Abstract

INTRODUCTION
Eosinophils are granulocytic leukocytes that develop in the bone marrow from pluripotential progenitor cells [1, 2]. While much of the older literature on eosinophils focuses on their role as cytotoxic effector cells, this limited view has been largely supplanted by findings that have revealed their broad and extensive immunomodulatory potential [3 - 5]. Recent studies that explore the role of eosinophils at homeostasis in the gastrointestinal tract [6, 7], that examine their distinct eosinophil-mediated antimicrobial activities [7, 8] and that define the properties of eosinophil-specific subtypes [5, 9, 10], all suggest that there are new and profound complexities remaining to be resolved.
Studies of eosinophils in and from mouse models of Th2-mediated allergy have ultimately led to our current understanding of phenotype-specific asthma and provide targets for novel therapies [11, 12]. However, while human eosinophils can be easily isolated by negative-selection technologies [13], there are no readily-available tools that facilitate isolation of live, untouched eosinophils from wild-type mice. To date, the major source of mouse eosinophils has been peripheral blood and tissues from interleukin-5 transgenic (IL5tg) strains [14, 15] in which profound eosinophilia results from cytokine overproduction in vivo. Another method used to generate eosinophils is based on ex vivo culture [16] in which unselected cells from mouse bone marrow are manipulated via cytokine stimulation to generate pure cultures of phenotypically mature eosinophils, known as bmEos. While this latter method has been used extensively to generate eosinophils from various wild-type and gene-deleted mouse strains, this culture system as currently designed [16] and related methods [17] are also dependent on high concentrations of IL-5.
Here we examine expression of the GPI-linked cell surface antigen, Ly6G, as a feature of mouse eosinophilic leukocytes and their development in vivo and ex vivo. While Ly6G has been used primarily to identify neutrophils in flow cytometric profiles and to execute depletion strategies [18, 19], we found that this antigen is also readily detected on mouse eosinophils, with cell surface expression increasing in association with elevated concentrations of IL5 in mouse serum or in tissue culture medium. Furthermore, while primarily understood as an eosinophil chemoattractant, eotaxin-1 has a clear but under-appreciated role in promoting eosinophil hematopoiesis [20, 21]. As such, in addition to modulation of IL-5 concentrations, we also explored the role of eotaxin-1 (CCL11) in promoting eosinophil maturation and expression of Ly6G in ex vivo culture.
MATERIALS AND METHODS
Mice.
Wild-type BALB/c and C57BL/6 mice (6 – 8 weeks old) were purchased from Charles River Laboratories, Frederick, MD. Interleukin (IL) 5 overexpressing-transgenic mice [14], IL5 gene-deleted mice [22], and eotaxin-1 gene-deleted mice [20] were maintained on-site in the 14BS vivarium and the NIAID Taconic Consortium. All mice were bred and maintained under pathogen-free conditions at an American Association for the Accreditation of Laboratory Animal Care accredited animal facility at the NIAID and housed in accordance with the procedures outlined in the Guide for the Care and Use of Laboratory Animals under an animal study proposal approved by the NIAID Animal Care and Use Committee, approved all the experimental procedures as per protocols LAD 7 and LAD 8E.
Harvesting and culture of mouse bone marrow-derived eosinophils (bmEos).
BmEos were generated from unselected bone marrow cells from BALB/c and C57BL/6 mice as previously described [16]. Briefly, bone marrow cells were harvested under sterile conditions and cultured in base medium consisting of RPMI 1640 medium with containing 20% FBS, 10 μg/mL streptomycin, 100 IU/mL penicillin, 2 mM glutamate, 1X non-essential amino acids, 1 mM sodium pyruvate, and 50 μM β-mercaptoethanol with 100 μg/mL stem cell factor (SCF) and 100 μg/mL FLT3 ligand (FLT-3L) for four days. On day 4, the growth medium was removed and replaced with base media. The standard method includes addition of recombinant murine (rm)IL-5 at 10 ng/mL (Peprotech; [16]); in some experiments, we varied the concentration of rmIL-5 from 0.1 to 10 ng/mL. Some experiments included recombinant murine eotaxin-1 at 0.5 ng/mL (Peprotech). Every second day, half of the media was removed and replaced, maintaining the cell concentration at 106/mL. All evaluations were performed on eosinophils at day 12 of culture unless otherwise indicated.
Analysis of surface receptors via flow cytometry.
BmEos at days of culture as indicated were washed in phosphate buffered saline (PBS) and stained with Live-Dead Aqua for 30 minutes at 4°C followed by addition of specific or isotype control antibody in PBS with 0.1% bovine serum albumin (BSA) for 30 min also at 4°C. Samples were evaluated on the BD LSRII flow cytometer and analyzed using FlowJo software. Abs used in the study included anti-CD16/CD32, PE-anti-mouse SiglecF, APC-Gr1 (anti-Ly6G/Ly6C), AF700-anti-mouse Ly6G (clone 1A8), FITC-anti-mouse Ly6C (clones AL-21, HK1.4), AF700-anti-mouse CD11c, PE-Cy7-anti-mouse MHCII, PerCP-Cy5.5-anti-mouse CCR3, BV605-anti-mouse CD125, and FITC-anti-mouse CD34.
Sensitization and challenge with A. fumigatus extract.
BALB/c mice were sensitized via intraperitoneal injection with an extract of the fungal allergen Aspergillus fumigatus (HollisterStier) as previously described [23] with some minor changes. Briefly, 20 μg was administered as a 1:1 admixture in ImjectAlum (Pierce) on days 0 and 7 followed by intranasal administration of 25 μg A. fumigatus in PBS with 0.1% BSA on days 15, 16 and 17 as previously described. Bone marrow, spleen and lung tissues and bronchoalveolar lavage fluid (BAL) were isolated for analysis on day 19.
Evaluation of peripheral blood eosinophils.
Retro-orbital bleeding into EDTA-coated microvette tubes (Sarstedt) was performed on anesthetized mice. Leukocyte counts and differentials were determined using the HEMAVET® HV 950Fs Analyzer via the NCI Pathology and Hematology Laboratory, Frederick, MD.
Evaluation of eosinophils in bone marrow.
Bone marrow was harvested from the femurs and tibiae of mice as previously described [24] and washed in PBS with 0.1% BSA. Red blood cells were lysed using sterile distilled water followed by addition of 10x concentrated PBS. The cell pellet was stained with Live-Dead Aqua for 30 mins at 4°C, followed by the addition of specific antibodies or isotype controls in PBS with 0.1% BSA for 30 mins also at 4°C. Samples were evaluated on the BD LSRII flow cytometer and analyzed using FlowJo software. Abs used in the study include anti-CD16/32, anti-mouse SiglecF-PE, anti-mouse Gr1-APC, PerCP-Cy5.5-anti-mouse CCR3, and BV605-anti-mouse CD125.
FACS isolation of Gr1− and Gr1+ eosinophils and re-introduction into bmEos culture.
On day 11 standard culture as described above (with rmIL-5 at 10 ng/mL), cells were washed once with PBS with 0.1% BSA and incubated for 30 min at 4°C with anti-CD16/32 (Mouse BD Fc Block) and APC-anti-Gr1 (RB6–8C5; BD Biosciences, Franklin Lakes, NJ, USA). After incubation, cells were washed with PBS with 0.1% BSA and resuspended at 20 × 106 cells / mL in PBS with 0.1% BSA. The bmEos were sorted into Gr1− and Gr1+ populations for analysis via FACSAria (BD Biosciences) into PBS with 0.1% BSA. Aliquots were returned as separate populations to bmEos culture at a final concentration of 106 cells / mL.
RNA isolation, cDNA synthesis and qPCR analysis.
Mouse bmEos from day 11 cultures separated by FACS into Gr1− and Gr1+ populations as described above were sorted directly into TRIzol LS Reagent (Life Technologies) and stored at −20°C until use. Chloroform (0.2x volume) was added to the samples in TRIzol LS Reagent and the aqueous phase containing RNA was isolated following centrifugation. Isolated RNA was purified with the RNeasy MinElute Cleanup Kit (Qiagen) and resuspended in diethyl-pyrocarbonate (DEPC) treated water. RNA concentrations were measured via NanoDrop One (Thermo Scientific). Approximately 1μg of RNA was treated with DNase I (Invitrogen) and reverse transcribed to synthesize cDNA via First Strand cDNA Synthesis Kit for RT-PCR (AMV) (Roche Diagnostics). Quantitative PCR (qPCR) was carried out using standard methods on a GeneAmp® PCR System 9700 Thermocycler (Applied Biosystems) using ABI TaqMan 2X Reagent (ThermoFisher) and FAM-labeled primer/probes for GATA-1 (ABI: Mm00484678_m1), EPX (ABI: Mm00514768_m1), MBP (ABI: Mm00435905_m1), CCR3 (ABI: Mm00515543_s1) and IL-5Ra (ABI: Mm00434284_m1). Mouse GAPDH (VIC-labeled, ABI #43058313) was used as the housekeeping gene for all experiments. No template and no reverse transcriptase controls were included in all experiments.
Chemotaxis Assay.
Chemotaxis assay was performed using an AP48 Neuro Probe Multiwell Chamber with a 5.0-μm filter pore size with total, Gr1+ or Gr1− bmEos. Chemotaxis media was prepared using RPMI 1640, 1.0% FBS, and 10 mM HEPES. Recombinant murine eotaxin-1 was added to the chemotaxis media at final concentrations of 0.1 ng/mL, 1.0 ng/mL, 10 ng/mL, 100 ng/mL, and 1000 ng/mL in the lower chamber and 5.2 × 104 bmEos from cultures as indicated were added to the upper chamber. Chemokinesis and vehicle controls were included. The chamber was incubated at 37°C for 2 hours in 5% CO2 to allow for transmigration between the wells. After 2 hours, the cells were removed from the lower chamber and migrated eosinophils were identified by their forward and side scatter properties on a BD LSRII flow cytometer and analyzed on FlowJo software. Chemotactic fraction was calculated as number of eosinophils migrated in response to chemoattractant divided by number of eosinophils migrated in response to diluent.
Determination of serum IL-5 and eotaxin-1 concentrations.
Whole blood samples were collected from mice by retro-orbital bleeding. After 30 minutes of clotting time, serum was collected after centrifugation at 3000 rpm for 10 min at 4°C. IL5 and eotaxin-1 concentrations were determined using ELISA DuoSets as described above.
Cytokine-free survival of bmEos.
BmEos cultured as described above were washed twice with PBS with 0.1% BSA. Cells were replaced in culture in base medium with no cytokines at 37°C and 5% CO2. Survival was monitored on an ongoing basis by trypan blue exclusion and visual inspection.
Statistical evaluation.
Statistical evaluations were performed using Mann-Whitney u-test, 1- or 2-way ANOVA as appropriate using algorithms within GraphPad PRISM 7.0.
RESULTS AND DISCUSSION
Expression of the cell surface antigen, Ly6G on mouse eosinophils.
Ly6G is a 25 kDa GPI-linked cell surface antigen detected by the monoclonal antibody anti-Gr1 (clone RB6–8C5); it is a member of a group of leukocyte antigens that share conserved LU (three-fold repeat Ly6 antigen / urokinase-type plasminogen activator receptor) domains [25] and are encoded by genes that reside on mouse chromosome 15 [18, 26]. Ly6G is a universal marker for mouse neutrophils, although its function remains uncertain [18]. While mouse eosinophils (and basophils) are typically described as Gr1− or Gr1lo, the Ly6G/Gr1 antigen has been detected at high levels (i.e., Gr1+/hi) on wild-type mouse eosinophils in specific microenvironments. Notably, Jordan, McKee and colleagues [27, 28] identified a population of IL-4+Gr1+ cells that were essential for generation of antigen-specific B cells in the spleens of alum-primed wild-type mice; Wang and Weller [29] identified these critical effector cells as Gr1+IL4+ eosinophils. Likewise, our group identified a small but distinct population of eosinophils in the lungs of allergen-challenged mice that were SiglecF+Gr1+/hi/Ly6G+ [23]; these eosinophils contained a unique subset of proinflammatory mediators that distinguished them from the majority SiglecF+Gr1lo population. Just recently, Mack and colleagues [30] presented findings on mice conditionally-deficient in the Tribbles family pseudo-kinase, Trib1 (Trib1ΔHSC mice) in which eosinophils are constitutively Ly6G+.
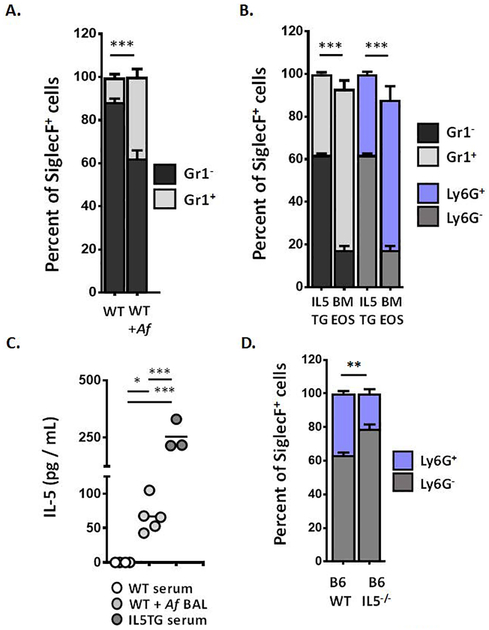
As shown in Fig. 1A, most of the eosinophils (SiglecF+ cells [31]) isolated directly from bone marrow of wild-type (WT) BALB/c mice at homeostasis were Gr1−; a relatively small fraction of SiglecF+ eosinophils from blood and bone marrow were Gr1+ (11 ± 1.6 %). By contrast, the fraction of SiglecF+Gr1+ eosinophils in the bone marrow of BALB/c mice sensitized and challenged with an extract of Aspergillus fumigatus was substantially larger (at 38 ± 3.7%, ***p < 0.005 vs. WT, Fig. 1A); no Gr1+ eosinophils were detected in spleen of Af sensitized and challenged mice, and, as reported previously [23], only ~1% of the eosinophils in the lung were Gr1+ (data not shown). The fraction of Gr1+ eosinophils (SiglecF+ cells) in bone marrow from IL5 transgenic mice is likewise elevated [Fig. 1B] when compared wild-type counterparts as shown in Fig. 1A; bone marrow-derived eosinophils (bmEos) generated under standard conditions (with 10 ng/mL IL-5) and evaluated at day 12 of culture displayed an even larger fraction of SiglecF+Gr1+ eosinophils, at 76 ± 4.0% (***p < 0.005 vs. IL5TG).
Figure 1. Mouse eosinophils express the Ly6G/Gr1 antigen.
A. Percent eosinophils (SiglecF+ cells) from bone marrow of wild-type (WT) BALB/c mice at baseline or sensitized and challenged with Aspergillus fumigatus (+Af) that are Gr1+ (light bar) vs. Gr1− (dark bar) as determined by flow cytometry with the mAb RB6–8C5; n = 3 – 5 per group, ***p < 0.005, Mann-Whitney u-test. B. Percent eosinophils (SiglecF+ cells) from bone marrow of BALB/c interleukin-5 transgenic (IL5TG) mice and from ex vivo cultures of from bone marrow-derived progenitors from BALB/c mice (BMEOS) that are Gr1+ (light bar) vs. Gr1− (dark bar). The cell surface antigen detected by RB6–8C5 was confirmed as Ly6G using mAbs 1A8 (anti-Ly6G, blue bar); n = 3 per group, ***p < 0.005, 1-way ANOVA; see also Suppl. Fig. 1. C. IL-5 (pg/mL) detected in serum and BAL of mice indicated in A. and B; *p < 0.05, ***p < 0.005, 1-way ANOVA. D. Percent eosinophils (SiglecF+ cells) from bone marrow of wild-type (WT) C57BL/6 (B6) and C57BL/6 IL-5 gene-deleted mice (B6 IL5−/−) that are Ly6G+ (blue bar) vs. Ly6G− (grey bar); n = 3 – 6 per group, **p < 0.01, Mann-Whitney u-test.
While the anti-Gr1 (RB6–8C5) monoclonal antibody detects primarily the Ly6G antigen, there is at least one report noting that this antibody may cross-react with the related antigen, Ly6C [32], a cell surface antigen that is expressed widely, but best known for its role in differentiating between proinflammatory and surveillance monocytes [33]; as such, we confirmed that the SiglecF+Gr1+ eosinophils were virtually all Ly6G+ (either Ly6G+ alone or double positive Ly6G+Ly6C+) by independent flow cytometric evaluation with monoclonal antibodies that were specific for anti-Ly6G (clone 1A8) and anti-Ly6C (using both clones HK1.4 and AL-21, in separate analyses; see [Suppl. Fig. 1A]). As an aside, our results for BALB/c bmEos obtained here are different from those reported previously for bmEos generated from C57BL/6 progenitors [23], which are predominantly Ly6C+ [Suppl. Fig. 1B]. Whether the C57BL/6 bmEos ultimately transition to Ly6G+ over time in the presence of IL5 is not known; of note, there is a substantial SiglecF+Ly6G+ fraction in bone marrow of wild-type C57BL/6 mice, as considered below. The remaining work in this manuscript features bmEos from the BALB/c mouse strain. Going forward, and for clarity and accuracy, we will refer to this cell surface antigen by both names (Ly6G/Gr1) in the text. We will refer to specific findings generated using the RB6–8C5 monoclonal antibody as Gr1+ vs. Gr1−, and findings generated using the 1A8 monoclonal antibody as Ly6G+ vs. Ly6G−; the data in Fig. 1B demonstrate clearly that the results generated with these of two antibodies are functionally equivalent in this case.
Our findings thus far suggest that Ly6G/Gr1 expression on eosinophils from bone marrow or from bone marrow culture may relate, directly or indirectly, to the ambient levels of IL-5. While no IL-5 was detected in the serum of wild-type mice, IL-5 was prominent in BAL from allergen challenged mice (67 ± 24 pg / mL), and in sera from the BALBc-IL5tg mice (254 ± 66 pg / mL) [Fig. 1C]. Of note, serum IL-5 levels in the BALB/c-IL5tg strain are elevated, they are within physiologic (or more specifically, pathophysiologic) parameters, as they are within range of that reported in BALB/c mouse S. mansoni infection [34] and likewise in response to infection and/or challenge with parasitic pathogens in both mouse models and human subjects; see [35 – 38] for additional examples.
As previously reported, C57BL/6 IL5−/− mice maintain normal numbers of eosinophils in the bone marrow and peripheral blood [Suppl. Fig. 2], although they are incapable of mounting a response to Th2 stimuli [22]. We found that, compared to the fraction of SiglecF+Ly6G+ eosinophils in bone marrow of C57BL/6 wild-type mice (37 ± 1.4%), the fraction of these eosinophils in the C57BL/6 IL5−/− mice was significantly reduced (21 ± 2.6%, **p < 0.01 [Fig. 1D].
Taken together, these results suggest that (1) SiglecF+ eosinophils in bone marrow generated in the presence of elevated levels of IL5 can be distinguished from homeostatic eosinophils that develop in the absence of this cytokine by expression of Ly6G/Gr1 and (2) IL5 may regulate the expression of Ly6G/Gr1, directly or indirectly, in cells of the eosinophil lineage.
Expression of Ly6G/Gr1 on bone marrow-derived eosinophils.
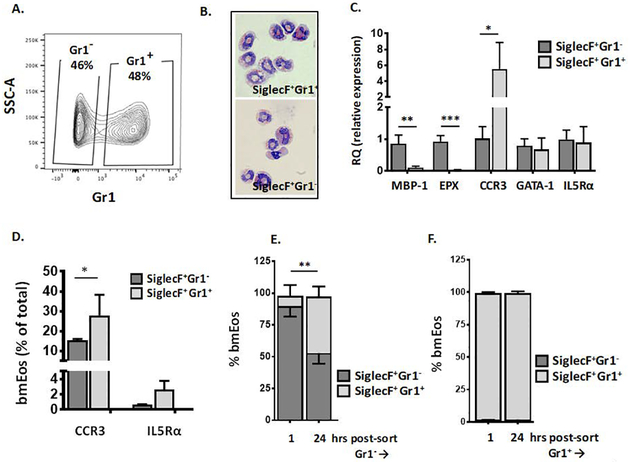
Given the findings in Fig. 1, we used the bmEos culture method to explore expression of Ly6G/Gr1 in response to IL-5. As described in the Methods, unselected bone marrow cells are initially cultured in factors that support developing progenitors (rmSCF and rmFLt3L); on day 4, the cells are washed and placed into growth medium with containing rmIL-5. As shown in Fig. 2A, at day 11 of culture, all viable cells were SiglecF+ and were equally divided between cells that were Gr1− and Gr1+. Both SiglecF+Gr1− and SiglecF+Gr1+ cells were clearly eosinophils by morphology; both displayed heteromorphic nuclei and red-staining cytoplasm with modified Giemsa stain [Fig. 2B]. Nonetheless, the two subsets displayed distinct patterns of gene expression; SiglecF+Gr1− bmEos express comparatively higher levels of transcripts encoding MBP-1 and EPX, granule proteins which are typically detected at the eosinophil promyelocyte stage [39], and relatively lower levels of transcript encoding the chemokine receptor, CCR3; transcripts encoding GATA-1 and IL5Rα were evenly distributed [Fig. 2C]. Differential expression of CCR3 (CD193) was confirmed by flow cytometry; likewise, IL5Rα (CD125) was detected on both populations at indistinguishable levels [Fig 2D]. Most intriguing, when SiglecF+Gr1− and SiglecF+Gr1+ bmEos were returned to culture independently of one another, each in the presence of 10 ng/mL IL-5, the SiglecF+Gr1− bmEos rapidly converted to the SiglecF+Gr1+ state [Fig. 2E]. By contrast, SiglecF+Gr1+ cells returned to culture maintain the SiglecF+Gr1+ profile along with full viability for the ensuing 24 hrs [Fig. 2F]; see Suppl. Fig. 3A and 3B for cell number and viability for both cell populations.
Fig. 2. Characterization of SiglecF+Gr1− and SiglecF+Gr1+ bone marrow-derived eosinophils (bmEos).
Unselected bone marrow cells were grown under standard conditions for four days with 100 μg/mL SCF and 100 μg/mL FLT3L. At day 4, these factors are replaced with IL-5 (10 ng/mL) as described in the Methods. A. Flow plot demonstrating the distribution of bmEos at day 11 of culture into SiglecF+Gr1− and SiglecF+Gr1+ fractions. B. Modified Giemsa-stained SiglecF+Gr1− and SiglecF+Gr1+ bmEos, original magnification, 64x. C. Relative expression (RQ) of transcripts encoding eosinophil major basic protein-1 (MBP-1), eosinophil peroxidase (EPX), CCR3, GATA-1, and interleukin-5 receptor alpha chain (IL5Rα) in SiglecF+Gr1− vs. SiglecF+Gr1+ eosinophils as in A.; n = 3 per group, *p < 0.05, **p < 0.01, ***p < 0.005; 1-way ANOVA. D. Percent of SiglecF+Gr1− vs. SiglecF+Gr1+ bmEos that express cell surface receptors for CCR3 and IL5Rα; n = 3 per group, *p < 0.05, Mann-Whitney u-test. E. SiglecF+Gr1− bmEos separated as shown in A., returned to culture and evaluated 1 hr and 24 hrs later; SiglecF+Gr1+ (light bar; above), SiglecF+Gr1− (dark bar; below; see also Suppl. Fig. 3 for total cell counts and viability), n = 3 per group, **p < 0.01 Mann-Whitney u-test. F. SiglecF+Gr1+ bmEos separated as in A., returned to culture and evaluated 1 hr and 24 hrs later; SiglecF+Gr1+ (light bar; see also Suppl. Fig. 3 for total cell counts and viability), n = 3 per group.
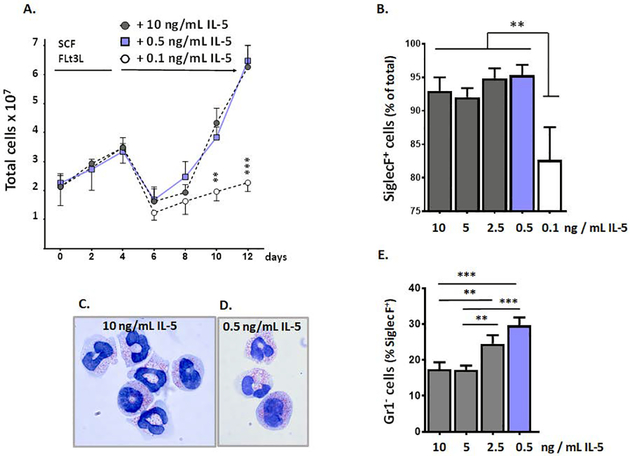
Reducing IL-5 concentration had no impact on generation of eosinophils but limited the fraction of SiglecF+Gr1+ cells in a dose-dependent fashion.
In contrast to results obtained with naïve wild-type and IL5tg mice, which maintain homeostatic (wild-type) or elevated but still physiologically relevant concentrations of IL5 (IL5tg), respectively, the standard protocol for generating bone marrow-derived eosinophils ex vivo [16] employs supraphysiologic concentrations of IL-5, at 10 ng/mL. As such, we determined the impact of reducing the concentration of IL-5 on eosinophil development and expression of the Ly6G/Gr1 antigen ex vivo. As shown in Fig. 3A, we found that the concentration of IL-5 used in this protocol could be reduced 20-fold, from 10 ng/mL to the more physiologic 0.5 ng/mL (500 pg/mL) with no impact on generation of eosinophils. This represents a critical protocol improvement, as 0.5 ng/mL (or 500 pg/mL) is within the same order of magnitude as concentrations detected in serum of IL5tg mice (at 254 ± 66 pg/mL) and, importantly, is within range of those identified in serum of mice infected with the helminthic parasite, Schistosoma mansoni (at 100 – 500 pg/mL; [34]) and also in other examples of parasitic infection and challenge [35 – 38]. Reducing the concentration of IL-5 in the bmEos culture to 0.4, 0.3, 0.2, or 0.1 ng/mL did not fully eliminate the effectiveness of this protocol but did reduce the final cell counts in a dose-dependent fashion as shown in Suppl. Fig. 4. As shown in Fig. 3B, cells in culture were nearly completely SiglecF+ at day 12 in response to IL-5 concentrations above 0.5 ng/mL; differentiation was not complete (~80 – 85% SiglecF+) at this time point in response to 0.1 ng/mL IL-5.
Figure 3. IL-5 concentration and Ly6G/Gr1 expression on bone marrow-derived eosinophils (bmEos).
A. Bone marrow-derived eosinophil cultures grown from days 0 to day 4 with SCF and FLT3L as indicated; at day 4, these factors are replaced with IL5, at either the standard concentration (10 ng/mL, filled circles) or one of two lower concentrations (0.5 or 0.1 ng/mL, filled squares or open circles, respectively). Total cells on each day are as shown; n = 3 – 5 per point, **p < 0.01, ***p < 0.001; see also Suppl. Fig. 4 for counts on day 12 with intermediate concentrations (0.2, 0.3, 0.4 ng/mL). B. SiglecF+ cells (% of total cells) at day 12 of bmEos cultures grown in concentrations of IL-5 as indicated; n = 3 per point, **p < 0.01, 1-way ANOVA. C. Modified Giemsa-stained bmEos from cultures grown in 10 ng/mL IL-5, original magnification 100x; D. as in C., bmEos from cultures grown in 0.5 ng/mL IL5, original magnification 100x; E. Gr1− cells (% of SiglecF+) in bmEos cultures grown in concentrations of IL-5 as indicated; n = 3 per point **p < 0.01; ***p < 0.005, 1-way ANOVA.
Eosinophils generated in cultures with 0.5 ng/mL IL-5 were indistinguishable at the light microscopic level from those produced in response to the standard 10 ng/mL; all cells had heteromorphic nuclei and cytoplasmic granules that stained red with eosin-dye [Fig. 3C and 3D]. As anticipated, the fraction of SiglecF+Gr1− bmEos increased in inverse proportion to the concentration IL-5 in the culture medium, decreasing from 10 to 0.5 ng/mL. As shown in Fig. 3E, the SiglecF+Ly6G− fraction reached 30 ± 2.3% in 0.5 ng/mL IL-5 (***p < 0.005 vs. 10 ng/mL IL5).
Taken together, these findings are consistent with our earlier hypothesis, that Ly6G/Gr1 expression on mouse eosinophilic leukocytes is regulated, directly or indirectly, by the actions of the cytokine, IL-5. This is particularly intriguing not only because Ly6G has been characterized as a specific neutrophil marker, but because its function, even on mouse neutrophils, remains for the most part uncertain. On the one hand, Wang and colleagues [40] showed that direct administration of the 1A8 anti-Ly6G antibody prevented neutrophil migration, notably toward the chemoattractant LTB4 in studies performed ex vivo. However, in experiments performed with the recently-described Catchup mouse, in which the Ly6G locus was disrupted with the gene encoding fluorescent Td-tomato, Hasenberg and colleagues [41] reported that the resulting Ly6G-deficient neutrophils functioned normally in vivo in response to both sterile and infectious inflammatory stimuli. Interestingly, these authors identified a small fraction of SiglecF+ eosinophils that were Td-tomato-positive in mouse blood under homeostatic conditions. Introduction of the IL-5 transgene into this mouse model might confirm and extend our findings on cytokine modulation of Ly6G expressed by mouse eosinophils in vivo, notably those in bone marrow. Likewise, it would be intriguing to determine whether IL-5 modulates expression of Ly6G on mouse neutrophils, given reports of functional IL-5 receptors on these cells [42, 43]. Toward this end, we identified a substantial fraction (~25%) of neutrophils (SiglecF−Ly6G+ cells) in the bone marrow of both naïve wild-type and BALB/c-IL5tg mice that expressed CD125 (IL5Rα). In contrast to what we observed in (SiglecF+) eosinophils from bone marrow, we detected no difference in percent Ly6G+ positive cells [Suppl. Fig. 5A], although the MFI declines somewhat when comparing neutrophils from the wild-type bone marrow to those from IL5tg mice [Suppl. Fig. 5B]. This observation might be the subject of a more complete study using appropriate cytokines and/or bacterial or viral pathogens promote neutrophil hematopoiesis and to recruit neutrophils to systemic circulation and/or the airways, respectively.
Eotaxin-1 is detected in serum at homeostasis in normal mice and has an impact on eosinophil development.
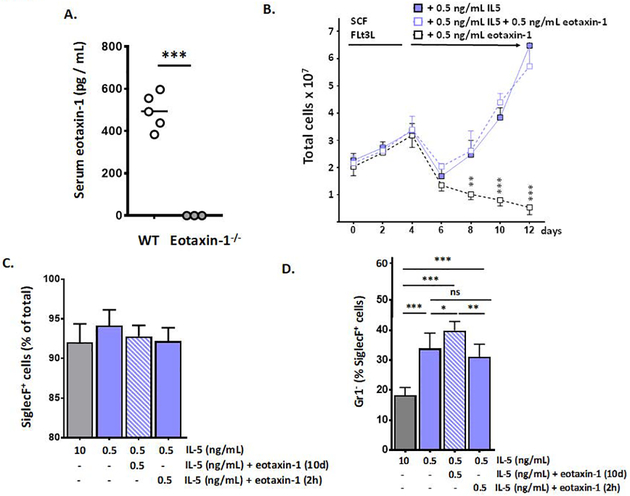
Eotaxin-1 (CCL11) was initially identified as an eosinophil-specific chemoattractant in a guinea pig model of airway inflammation [44]; it is produced constitutively by numerous tissues and signals via the chemoattractant receptor, CCR3 [45]. Eotaxin-1 is best known for its actions in promoting eosinophil recruitment in response to allergic inflammatory stimuli and maintaining homeostatic levels of eosinophils in the gastrointestinal tract [6, 45]. Although not fully appreciated as a hematopoietic factor, Matthews and colleagues [20] reported diminished levels of peripheral blood eosinophils in eotaxin-1 gene-deleted mice in the absence of allergen provocation. Likewise, Lamkhioued and colleagues [21] reported that a small fraction of human cord blood CD34+ progenitors had receptors for eotaxin-1 (CCR3) and generated eosinophil-peroxidase (EPX)-positive eosinophils in culture in response to eotaxin-1 alone, in the absence of IL-5. Given these findings, we asked whether eotaxin-1 might have an impact on eosinophil development and on expression of Ly6G/Gr1 in the presence or absence of IL-5.
As shown in Fig. 4A, wild-type BALB/c mice at baseline display serum eotaxin-1 levels of 494 ± 86 pg/mL (or ~ 0.5 ng/mL). As such, we selected this concentration for use in our revised bmEos culture protocol. We found that addition of recombinant murine eotaxin-1 at 0.5 ng/mL to the bmEos culture together with reduced IL-5 (at 0.5 ng/mL) had no impact on the generation of SiglecF+ eosinophils; interestingly, and in contrast to the studies noted above performed by Lamkhioued and colleagues with human CD34+ progenitors [21], eotaxin-1 alone did not support eosinophil expansion [Fig. 4B and 4C]. However, adding eotaxin-1 together with IL-5 at this lower concentration had a small but significant impact on the distribution of SiglecF+Gr1− vs. SiglecF+Gr1+ cells. Addition of eotaxin-1 to the bmEos culture resulted in an increase in the fraction of SiglecF+Gr1− cells, to 40 ± 3.0% (*p < 0.05 vs. 0.5 ng/mL IL-5 alone). By contrast, adding eotaxin-1 for 2 hrs only just prior to harvest had no impact over culture in 0.5 ng/mL IL-5 alone, a result suggesting that eotaxin-1 was acting on these cells over time and not simply promoting brief cell activation [Fig. 4D]. Addition of eotaxin-1 to cultures grown in IL-5 also resulted in a significant increase in the fraction of eosinophils that expressed IL5Rα (CD125; 49 ± 9.7% vs. 30 ± 4.7% ***p < 0.005; Fig. 4E) and CD34 (49 ± 5.2% vs. 28 ± 3.8% ***p < 0.005; Fig. 4F). By contrast, addition of eotaxin-1 has no impact on expression of CCR3+ [Fig. 4G]. Interestingly, only 15 – 30% of the SiglecF+ cells were also CCR3+ at the day 12 time point; these results are distinct from those reported by Schollaert and colleagues [17] who initiated these cultures with low density bone marrow progenitors and found that virtually all SiglecF+ cells generated in response to 10 ng/mL IL5 were also CCR3+.
Figure 4. Eotaxin-1 limits expression of Ly6G/Gr1 on bone marrow-derived eosinophils (bmEos).
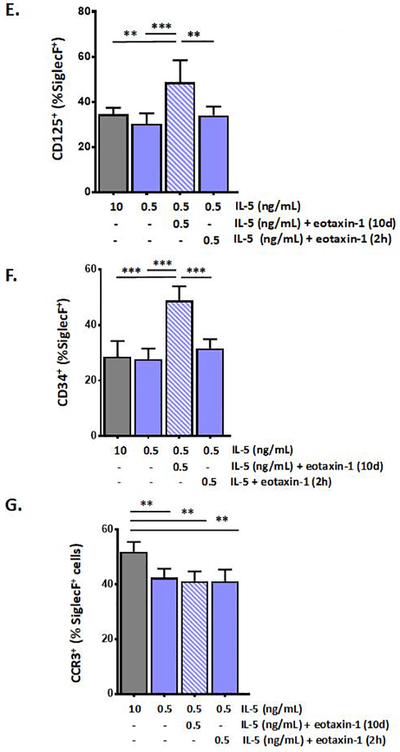
A. Serum eotaxin-1 in WT (wild-type) and eotaxin-1 gene-deleted mice; n = 3 – 5 mice, ***p < 0.005, Mann-Whitney u-test. B. Bone marrow-derived eosinophils cultures initiated under standard conditions as indicated; at day 4, these factors are replaced with 0.5 ng/mL IL-5 alone, 0.5 ng/mL IL-5 with 0.5 ng/mL eotaxin-1, or 0.5 ng/mL eotaxin-1 alone; total cells in culture on each day are as shown; n = 3 – 5 per point, ***p < 0.005. C. SiglecF+ cells (% of total) at day 12 of culture under growth conditions indicated; n = 5 – 8 per point; no significant differences detected between groups. D. Gr1− cells (% of SiglecF+) in bmEos cultures under growth conditions indicated; E. CD125+ (IL5-Rα+) cells (% of SiglecF+) in bmEos cultures as above. F. CD34+ cells (% of SiglecF+) in bmEos cultures grown as above. G. CCR3+ cells (% of SiglecF+) in bmEos cultures as above; for D. – G., n = 5 – 8 per point, *p < 0.05, **p < 0.01; ***p < 0.005, ns, not significant, 1-way ANOVA.
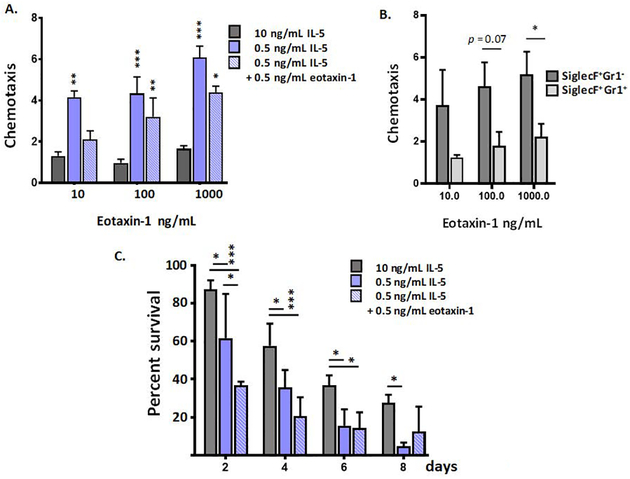
Reducing IL-5 with or without eotaxin-1 augments chemotactic responses and limits cytokine-free survival of bone-marrow derived eosinophils.
The ability to undergo chemotaxis to biochemical ligands is a critical feature of eosinophilic leukocytes. We and others have shown that bmEos respond to chemotactic ligands, both ex vivo and when introduced in vivo [16, 17, 46, 47]. We demonstrate here that a 20-fold reduction in IL-5 generates eosinophils with significantly greater capacity to undergo chemotaxis to eotaxin-1 across a substantial range of concentrations (**p < 0.01, **p < 0.001; Fig. 5A). This observation is largely due to the responses of the Gr1− eosinophils, which are significantly more chemotactic to eotaxin-1 than are their Gr1+ counterparts [Fig. 5B]. Interestingly, this observation is not directly related to the number of CCR3+ cells; as shown in Fig. 2D, there are more CCR3+ cells among the SiglecF+Gr1+ group. We also examined survival of bmEos after removal of exogenous cytokine support. This property of eosinophils is particularly intriguing, as we and others have found that eosinophils from allergen-challenged humans [48, 49] and wild-type mice [50] can survive for prolonged periods of time ex vivo in the absence of pro-survival cytokines. We have not previously determined whether bone marrow-derived eosinophils were capable of sustained survival ex vivo in the absence of cytokine support. As shown, in Fig. 5C, we found that eosinophils grown in the higher concentrations of IL-5 (10 ng/mL) were comparatively sustained ex vivo after removal from growth medium; these cells remained 28 ± 4% viable at 8 days after their removal from IL-5-enriched growth medium. By contrast, bmEos grown in the lower IL5 concentration (0.5 ng/mL) underwent cell death at a more rapid rate, comparable to that of isolated human eosinophils [51]. This finding may be related to a technical aspect of the experimental design, i.e., higher residual concentrations of IL5 may remain associated with the cells in the former cultures even after extensive washing. Growth in eotaxin-1 has no impact on the rate of subsequent cytokine-free cell death.
Figure 5. Interleukin-5 and eotaxin-1 contribute to the functional phenotype of bone marrow derived eosinophils.
A. Chemotaxis in response to increasing concentrations of eotaxin-1 (10 to 1000 ng/mL). Values shown are ratios of cells migrated in response to eotaxin-1 to cells migrated in response to medium alone. Growth conditions for bmEos cultures (10 ng/mL IL-5, 0.5 ng/mL IL-5, 0.5 ng/mL IL5 with 0.5 ng/mL eotaxin-1 (10 days) are as indicated; **p < 0.01, ***p < 0.005 vs. 10 ng/mL IL5 cultures, 2-way ANOVA. B. BmEos grown for 10 days in 0.5 ng/mL IL-5 with 0.5 ng/mL eotaxin-1 were separated into SiglecF+Gr1− and SiglecF+Gr1+ fractions and chemotaxis in response to increasing concentrations of eotaxin-1 (10 – 1000 ng/mL) was evaluated. Ratios as in A. are shown; n = 5 per group, *p < 0.05, 1-way ANOVA. C. Percent survival of bmEos grown in culture as above, n = 3 per point, *p < 0.05, **p < 0.01, ***p < 0.005, 2-way ANOVA.
Conclusions
To summarize, we examined the impact of physiologic concentrations of IL-5 and eotaxin-1 used to promote eosinophil growth and development ex vivo. As a direct result of this study, we found that substantial fractions of SiglecF+ eosinophils isolated directly from mice as well as eosinophils generated ex vivo were Ly6G+/Gr1+ and that the fraction eosinophils expressing this antigen increases in association with increasing concentrations of IL-5. From a purely technical standpoint, these results may need to be considered when using anti-Ly6G strategies to detect or deplete neutrophils, particularly in the setting of a profound Th2 response. However, from a larger perspective, our results also suggest that a broader focus on eosinophils and classic IL5-mediated responses may provide some clues towards identifying the otherwise elusive function of this cell surface antigen. Toward this end, we found that reducing the concentration of IL5 in bone marrow-derived eosinophil cultures into the physiologic range (500 pg/mL) had no impact on the growth and expansion from progenitors, but, interestingly, the bmEos generated were more responsive to eotaxin-1-mediated chemotaxis overall, and, notably, the Gr1− bmEos were the cells that maintained this functionality. Likewise, given observations regarding eotaxin-1 and its potential role in hematopoiesis, we examined the impact of this mediator when introduced at physiologic levels to the bone marrow derived eosinophil culture. We found that while eotaxin-1 alone at 500 pg/mL did not support the generation of eosinophils from progenitors, but addition of eotaxin-1 together with IL-5 further limited expression of the Ly6G/Gr1 surface antigen.
Supplementary Material
Suppl. Fig. 1. Characterization of bone marrow-derived eosinophils from BALB/c and C57BL/6 progenitors. By day 11 of culture under standard conditions virtually all cells are SiglecF+. A. BmEos from BALB/c progenitors are predominantly Ly6G+ (over 80%; see red box), distributed between single Ly6G+ and double Ly6G+Ly6C+ subsets. There is no single Ly6C+ population. By contrast, and confirming our earlier result [23], B. bmEos from C57BL/6 progenitors grown under identical conditions are virtually all Ly6C+ (> 90%), with a larger Ly6C+ single and smaller double Ly6G+Ly6C+ population (upper panel in red box); there is no single Ly6G+ single population.
Suppl. Fig.2. Peripheral blood eosinophils from wild-type and IL-5−/− mice. Eosinophil counts from the peripheral blood of wild-type C57BL/6 mice (WT; white bar) and C57BL/6 IL-5−/− mice (grey bar). No significant differences (ns) were detected, n = 8 per group.
Suppl. Fig. 3. Total cell number and percent viability of SiglecF+Gr1− and SiglecF+Gr1+ bone marrow-derived eosinophils (bmEos). In Fig. 2E and 2F, we demonstrate that isolated SiglecF+Gr1− cells rapidly convert to SiglecF+Gr1+ cells upon return to culture; by contrast, SiglecF+Gr1+ cells retain their cell surface phenotype throughout. As shown here, A. the total cell number remains constant, as does B. percent viability; day 0, day of isolation; day 1, first day after returning to culture. The viability of those first isolated as SiglecF+Gr1+ seems to trend downward, but this does not reach statistical significance; n = 3 per group.
Suppl. Fig. 4. IL-5 dependence of bmEos protocol. Unselected progenitors are cultured for four days in rmSCF (100 μg/mL) and rmFLT3L (100 μg/mL); at day 4, the cells are transferred to medium with IL-5 alone (0.5 to 10 ng/mL). At IL-5 concentrations below 0.5 ng/mL, the yield (total eosinophils) at day 12 drops in a dose-dependent fashion; no eosinophils are obtained from this protocol in the absence of IL-5 (see Figs. 4B).
Suppl Fig. 5. IL-5Rα (CD125) positive neutrophils in mouse bone marrow. A. Detection of IL-5Rα neutrophils (SiglecF−Ly6G+ CD125+) in bone marrow of wild-type (WT) and IL5tg mice. B. MFI of IL-5Rα on these cells; n = 6 per group, **p < 0.01.
ACKNOWLEDGEMENTS
This work was supported by funds from NIAID Division of Intramural Research (Z01-AI000941 to HFR).
ABBREVIATIONS
- Af
Aspergillus fumigatus
- BMEOS
Bone marrow-derived cultured eosinophils
- CCR3
CC chemokine receptor 3
- EPX
Eosinophil peroxidase
- IL-5
Interleukin-5
- MBP-1
Major basic protein-1
- MFI
Mean fluorescence index
- TG
Transgenic
Footnotes
CONFLICT OF INTEREST DISCLOSURE
The authors of this manuscript have no conflicts of interest to disclose with respect to the research results reported herein.
REFERENCES
- 1.Rosenberg HF, Dyer KD, Foster PS 2013. Eosinophils: changing perspectives in health and disease. Nat Rev Immunol 13: 9–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Furuta GT, Atkins FD, Lee NA, Lee JJ 2014. Changing roles of eosinophils in health and disease. Ann Allergy Asthma Immunol. 113: 3–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee JJ, Jacobsen EA, McGarry MP, Schleimer RP, Lee NA 2010. Eosinophils in health and disease: the LIAR hypothesis. Clin Exp Allergy 40: 563–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davoine F, Lacy P 2014. Eosinophil cytokines, chemokines, and growth factors: emerging roles in immunity. Front Immunol. 5: 570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weller PF, Spencer LA 2017. Functions of tissue-resident eosinophils. Nat Rev Immunol 17: 746–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schroeder S, Masterson JC, Fillon S, Furuta GT 2013. Eosinophils as regulators of gastrointestinal physiological homeostasis, 1st ed. In Eosinophils in Health and Disease. (Lee JJ, Rosenberg HF, eds.) Academic Press, Waltham, MA, 341–345. [Google Scholar]
- 7.Rosenberg HF, Masterson JC, Furuta GT Eosinophils, probiotics and the microbiome. J Leukoc Biol. 100: 881–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yousefi S, Stojkov D, Germic N, Simon D, Wang X, Benarafa C, Simon HU 2019. Untangling “NETosis” from NETs. Eur J Immunol 49: 221–227. [DOI] [PubMed] [Google Scholar]
- 9.Abdala-Valencia H, Coden ME, Chiarella SE, Jacobsen EA, Bochner BS, Lee JJ, Berdnikovs S 2018. Shaping eosinophil identity in the tissue contexts of development, homeostasis and disease. J Leukoc Biol. 104: 95–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mesnil C, Raulier S, Paulissen G, Xiao X, Birrell MA, Pirottin D, Janss T, Starkl P, Ramery E, Henket M, Schleich FN, Radermecker M, Thielemans K, Gillet L, Thiry M, Belvisi MG, Louis R, Desmet C, Marichal T, Bureau F 2016. Lung-resident eosinophils represent a distinct regulatory eosinophil subset. J Clin Invest 126: 3279–3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foster PS, Maltby S, Rosenberg HF, Tay HL, Hogan SP, Collison AM, Yang M, Kaiko GE, Hansbro PM, Kumar RK, Mattes J 2017. Modeling TH2 responses and airway inflammation to understand fundamental mechanisms regulating the pathogenesis of asthma. Immunol Rev. 278: 20–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spencer LA, Weller PF 2010. Eosinophils and Th2 immunity: contemporary insights. Immunol Cell Biol. 88: 250–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Son K, Mukherjee M, McIntyre BAS, Eguez JC, Radford K, LaVigne N, Ethier C, Davoine F, Janssen L, Lacy P, Nair P 2017. Improved recovery of functionally active eosinophils and neutrophils using novel immunomagnetic technology. J Immunol Methods 449: 44–55. [DOI] [PubMed] [Google Scholar]
- 14.Dent LA, Strath M, Mellor AL, Sanderson CJ 1990. Eosinophilia in transgenic mice expressing interleukin 5. J Exp Med 172: 1425–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee NA, McGarry MP, Larson KA, Horton MA, Kristensen AB, Lee JJ 1997. Expression of IL-5 in thymocytes/T cells leads to the development of a massive eosinophilia, extramedullary eosinophilopoiesis, and unique histopathologies. J Immunol 158: 1332–1344. [PubMed] [Google Scholar]
- 16.Dyer KD, Moser JM, Czapiga M, Siegel SJ, Percopo CM, Rosenberg HF 2008. Functionally competent eosinophils differentiated ex vivo in high purity from normal mouse bone marrow. J Immunol 181: 4004–4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schollaert KD, Stephens MR, Gray JK, Fulkerson PC 2014. Generation of eosinophils from cryopreserved murine bone marrow cells. PLoS One. 9:e116141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee PY, Wang J-X, Parisini E, Dascher CC, Nigrovic PA 2013. Ly6 family proteins in neutrophil biology. J Leukoc Biol 94: 585–594. [DOI] [PubMed] [Google Scholar]
- 19.Rose S, Misharin A, Perlman H 2012. A novel/Ly6G-based strategy to analyze the mouse splenic myeloid compartment. Cytometry A. 81: 343–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matthews AN, Friend DS, Zimmermann N, Sarafi MN, Luster AD, Pearlman E, Wert SE, Rothenberg ME 1998. Eotaxin is required for the baseline level of tissue eosinophils. Proc Natl Acad Sci USA 95: 6273–6278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lamkhioued B, Abdelilah SG, Hamid Q, Mansour N, Delespesse G, Renzi PM 2003. The CCR3 receptor is involved in eosinophil differentiation and is up-regulated by Th2 cytokines in CD34+ progenitor cells. J Immunol 170: 537–547. [DOI] [PubMed] [Google Scholar]
- 22.Kopf M, Brombacher F, Hodgkin PD, Ramsay AJ, Milbourne EA, Dai WJ, Ovington KS, Behm CA, Kohler G, Young IG, Matthaei KI 1996. IL-5-deficient mice have a developmental defect in CD5+ B-1 cells and lack eosinophilia but have normal antibody and cytotoxic T cell responses. Immunity 4: 15–24. [DOI] [PubMed] [Google Scholar]
- 23.Percopo CM, Brenner TA, Ma M, Kraemer LS, Hakeem RM, Lee JJ, Rosenberg HF 2017. SiglecF+Gr1hi eosinophils are a distinct subpopulation within the lungs of allergen-challenged mice. J Leukoc Biol. 101:321–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bystrom J, Wynn TA, Domachowske JB, Rosenberg HF 2004. Gene microarray analysis reveals interleukin-5-dependent transcriptional targets in mouse bone marrow. Blood 103: 868–877. [DOI] [PubMed] [Google Scholar]
- 25.Loughner CL, Bruford EA, McAndrews MS, Delp EE, Swamynathan S, Swamynathan SK 2016. Organization, evolution and functions of the human and mouse Ly6/uPAR family genes. Human Genomics 10: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.LeClair KP, Rabin M, Nesbitt MN, Pravtcheva D, Ruddle FH, Palfree RG, Bothwell A 1987. Murine Ly6 mutigene family is located on chromosome 15. Proc Natl Acad Sci USA 84: 1638–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jordan MB, Mills DM, Kappler J, Marrack P, Cambier JC 2004. Promotion of B cell immune responses via an alum-induced myeloid cell population. Science 304: 1808–1810. [DOI] [PubMed] [Google Scholar]
- 28.McKee A MacLeod M, White J, Crawford F, Kappler J, Marrack P 2008. Gr1+IL4-producing innate cells are induced in response to Th2 stimuli and suppress Th1-dependent antibody responses. Int Immunol 20: 659–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang H-B, Weller PF 2008. Pivotal advance: Eosinophils mediate early alum adjuvant-elicited B cell priming and IgM production. J Leukoc Biol 83: 817–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mack EA, Stein SJ, Rome KS, Xu L, Wertheim GB, Meio RC, Pear WS 2019. Trib1 regulates eosinophil lineage commitment and identity by restraining the neutrophil program. Blood 133: 2413–2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dyer KD, Garcia-Crespo KE, Killoran KE, Rosenberg HF 2011. Antigen profiles for the quantitative assessment of eosinophils in mouse tissues by flow cytometry. J Immunol Methods. 369: 91–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wojtasiak M, Pickett DL, Tate MD, Londrigan SL, Bedoui S, Brooks AG, Reading PC 2010. Depletion of Gr1+ but not Ly6G+ immune cells exacerbates virus replication and disease in an intranasal model of herpes simplex virus type 1 infection. J Gen Virol 91: 2158–2166. [DOI] [PubMed] [Google Scholar]
- 33.Mildner A, Marinkovic G, Jung S 2016. Murine monocytes: origins, subsets, fates and function. Microbiol Spectr 4: 5. [DOI] [PubMed] [Google Scholar]
- 34.Swartz JM, Dyer KD, Cheever AW, Ramalingam T, Pesnicak L, Domachowske JB, Lee JJ, Lee NA, Foster PS, Wynn TA, Rosenberg HF 2006. Schistosoma mansoni infection in eosinophil lineage-ablated mice. Blood 108: 2420–2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kan H, Ogata T, Taniyama A, Migita M, Matsuda M, Matsuda I, Nawa Y 1995. Extraordinarily high eosinophilia and elevated serum interleukin-5 level observed in a patient infected with Paragonimus westermani. Pediatrics 96: 351–354. [PubMed] [Google Scholar]
- 36.Evans CAW, Garcia HH, Hartnell A, Gilman RH, Jose PJ, Martinez M, Remick DG, Williams TJ, Friedland JS 1998. Elevated concentrations of eotaxin and interleukin-5 in human neurocysticercosis. Inf Immun 66: 4522–4525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Finlay CM, Stefanska AM, Walsh KP, Kelly PJ, Boon L, Lavelle EC, Walsh PT, Mills KHG 2016. Helminth products protect against autoimmunity via innate type w cytokines IL-5 and IL-33 which promote eosinophilia. J Immunol 196:703–714. [DOI] [PubMed] [Google Scholar]
- 38.Hepworth MR, Danilowica-Luebert E, Rausch S, Metz M, Klotz C, Maurer M, Hartmann S 2012. Mast cell orchestration of Th2 immunity. Proc Natl Acad Sci USA 109: 6644–6649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Acharya KR, Ackerman SJ 2014. Eosinophil granule proteins: form and function. J Biol Chem 289: 17406–17415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang JX, Bair AM, King SL, Shnayder R, Huang YF, Shieh CC, Soberman RJ, Fuhlbrigge RG, Nigrovic PA 2012. Ly6G ligation blocks recruitment of neutrophils via a beta2-integrin-dependent mechanism. Blood 120: 1489–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hasenberg A, Hasenberg M, Mann L, Neumann F, Borkenstein L, Stecher M, Kraus A, Engel DR, Klingberg A, Seddigh P, Abdullah Z, Klebow S, Engelmann S, Reinhold A, Brandau S, Seeling M, Waisman A, Schraven B, Gothert JR, Nimmerjahn F, Gunzer M 2015. Catchup: a mouse model for imaging-based tracking and modulation of neutrophil granulocytes. Nature Methods 12: 445–452. [DOI] [PubMed] [Google Scholar]
- 42.Linch SN, Danielson ET, Kelly AM, Tamakawa RA, Lee JJ, Gold JA 2012. Interleukin 5 is protective during sepsis in an eosinophil-independent manner. Am J Respir Crit Care Med 186: 246–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gorski S, Braciale T 2013. IL-5 produced by natural helper cells suppresses neutrophil function during influenza infection (P4239). J Immunol 190 (1 Suppl.) 130.19. [Google Scholar]
- 44.Jose PJ, Griffiths-Johnson DA, Collins PD, Walsh DT, Moqbel R, Totty NF, Truong O, Hsuan JJ, Williams TJ 1994. Eotaxin: a potent eosinophil chemoattractant cytokine detected in a guinea pig model of allergic airways inflammation. J Exp Med 179: 881–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Williams TJ 2015. Eotaxin-1 (CCL11). Front Immunol 6: 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sturm EM, Dyer KD, Percopo CM, Heinemann A, Rosenberg HF 2013. Chemotaxis of bone marrow derived eosinophils in vivo: a novel method to explore receptor-dependent trafficking in the mouse. Eur J Immunol 43: 2217–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wen T, Besse JA, Mingler MK, Fulkerson PC, Rothenberg ME 2013. Eosinophil adoptive transfer system to directly evaluate pulmonary eosinophil trafficking in vivo. Proc Natl Acad Sci USA 110: 6067–6072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sedgwick JB, Calhoun WJ, Vrtis RF, Bates ME, McAllister PK, Busse WW 1992. Comparison of airway and blood eosinophil function after in vivo antigen challenge. J Immunol. 149: 3710–3718. [PubMed] [Google Scholar]
- 49.Yamamoto H, Sedgwick JB, Vrtis RF, Busse WW 2000. The effect of transendothelial migration on eosinophil function. Am J Respir Cell Mol Biol. 23: 379–388. [DOI] [PubMed] [Google Scholar]
- 50.Geslewitz WE, Percopo CM, Rosenberg HF 2018. Eosinophil persistence in vivo and sustained viability ex vivo in response to respiratory challenge with fungal allergen. Clin Exp Allergy 48: 29–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Percopo CM, Dyer KD, Killoran KE, Rosenberg HF. 2010. Isolation of human eosinophils: microbead method has no impact on IL-5 sustained viability. Exp Dermatol. 19: 467–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Suppl. Fig. 1. Characterization of bone marrow-derived eosinophils from BALB/c and C57BL/6 progenitors. By day 11 of culture under standard conditions virtually all cells are SiglecF+. A. BmEos from BALB/c progenitors are predominantly Ly6G+ (over 80%; see red box), distributed between single Ly6G+ and double Ly6G+Ly6C+ subsets. There is no single Ly6C+ population. By contrast, and confirming our earlier result [23], B. bmEos from C57BL/6 progenitors grown under identical conditions are virtually all Ly6C+ (> 90%), with a larger Ly6C+ single and smaller double Ly6G+Ly6C+ population (upper panel in red box); there is no single Ly6G+ single population.
Suppl. Fig.2. Peripheral blood eosinophils from wild-type and IL-5−/− mice. Eosinophil counts from the peripheral blood of wild-type C57BL/6 mice (WT; white bar) and C57BL/6 IL-5−/− mice (grey bar). No significant differences (ns) were detected, n = 8 per group.
Suppl. Fig. 3. Total cell number and percent viability of SiglecF+Gr1− and SiglecF+Gr1+ bone marrow-derived eosinophils (bmEos). In Fig. 2E and 2F, we demonstrate that isolated SiglecF+Gr1− cells rapidly convert to SiglecF+Gr1+ cells upon return to culture; by contrast, SiglecF+Gr1+ cells retain their cell surface phenotype throughout. As shown here, A. the total cell number remains constant, as does B. percent viability; day 0, day of isolation; day 1, first day after returning to culture. The viability of those first isolated as SiglecF+Gr1+ seems to trend downward, but this does not reach statistical significance; n = 3 per group.
Suppl. Fig. 4. IL-5 dependence of bmEos protocol. Unselected progenitors are cultured for four days in rmSCF (100 μg/mL) and rmFLT3L (100 μg/mL); at day 4, the cells are transferred to medium with IL-5 alone (0.5 to 10 ng/mL). At IL-5 concentrations below 0.5 ng/mL, the yield (total eosinophils) at day 12 drops in a dose-dependent fashion; no eosinophils are obtained from this protocol in the absence of IL-5 (see Figs. 4B).
Suppl Fig. 5. IL-5Rα (CD125) positive neutrophils in mouse bone marrow. A. Detection of IL-5Rα neutrophils (SiglecF−Ly6G+ CD125+) in bone marrow of wild-type (WT) and IL5tg mice. B. MFI of IL-5Rα on these cells; n = 6 per group, **p < 0.01.