Abstract
The triggering receptor expressed on myeloid cells (TREM) protein family forms a class of type I transmembrane proteins expressed in immune cells that play important roles in innate and adaptive immune responses. The TREM family member TREM-like transcript 1 (TLT-1, also TREML1) is expressed in megakaryocytes and packaged into platelet granules. TLT-1 binds fibrinogen and plays a role in bleeding initiated by inflammatory insults. Here, we describe a proteomics screen that maps the TLT-1 interactome in resting and activated human platelets. Several identified TLT-1 interactors are involved in cell adhesion and migration, as well as platelet activation. Select interactors, including β3-integrin, RACK1, GRB2, and Rabs 5A, 7, and 11A, were additionally characterized in co-immunoprecipitation/immunoblotting experiments. Finally, several phosphorylation sites were found on immunoprecipitated TLT-1, including Thr280, a novel, regulated site on a conserved residue near the TLT-1 ITIM regulatory sequence.
Keywords: TLT1, TREML1, platelet activation, platelet aggregation, LC-MS/MS, fibrinogen
1. Introduction
Platelets are anucleate blood cells derived from megakaryocytes that are essential for regulating hemostasis and immune responses. Platelet function relies on the secretion of active molecules from intracellular vesicles, including dense (δ) granules, alpha (α) granules, and lysosomes. These granules contain soluble and membrane-bound proteins that are essential for platelet aggregation, coagulation reactions, and pathogen defense mechanisms. Platelet activation is induced by secreted factors, including thrombin or adenosine diphosphate (ADP), as well as exposure to extracellular matrix components in the vascular wall (i.e. collagen or podoplanin). Following platelet activation, platelet granules are rapidly mobilized to the plasma membrane, to which they fuse, exposing their membrane-tethered factors and releasing their soluble cargo into the extracellular environment [1–3].
The triggering receptor expressed on myeloid cells (TREM) protein family forms a class of single-pass transmembrane proteins expressed in a variety of immune cells that function to modulate innate and adaptive immune responses [4]. The TREM family member TREM-like transcript 1 (TLT-1, also TREML1) is expressed in megakaryocytes and platelets, and is known to play essential roles in platelet aggregation and hemostasis [5–7]. TLT-1 resides in platelet α-granules, prepackaged from megakaryocytes, and is rapidly transported to the membrane following platelet activation [5, 8]. However, a portion of TLT-1 reserves are sequestered in the platelet center, called the granulomere, distinct from α-granules, in a region hypothesized to be the contracted microtubule bundle, or the marginal band, which forms during platelet activation [5].
The majority of TREM family members interact with DNAX-activating protein 12 (DAP-12) following various external stimuli [4]. DAP-12 contains an immunoreceptor tyrosine-based activation motif (ITAM; YXXI/LX6–12YXXI/L) [9]. Phosphorylation of the DAP-12 ITAM tyrosine activates signaling cascades that result in cell adhesion, survival, and calcium mobilization [4]. TLT-1 possesses an extracellular Ig superfamily V-set domain and, unlike other TREM family members, houses two intracellular immunoreceptor tyrosine-based inhibitory motifs (ITIMs; S/I/V/LXYXXI/V/L) within its C-terminal tail [5, 8]. These tyrosine-based motifs surround the only two tyrosine residues within the TLT-1 C-terminal tail, Tyr265 and Tyr281. The SH2 domain-containing protein phosphatases SHP-1 and SHP-2 have been shown to co-immunoprecipitate with TLT-1 following induction of tyrosine phosphorylation via pervanadate treatment, likely through an SH2–phosphotyrosine interaction [8, 10]. Barrow, et. al. demonstrated that phosphorylation of both Tyr265 and Tyr281 was regulated by tyrosine phosphatases, however, only phosphorylation of Tyr281 induced the TLT-1/SHP-2 interaction [8]. The classical ITIM motif is thought to bind SHP-2 and reverse the effect of src family kinases, given its role in recruiting tyrosine phosphatases to signaling hubs following phosphorylation of the ITIM tyrosine [11]. However, phosphorylation of Tyr281 within the TLT-1 ITIM is required for FcεRI-mediated calcium mobilization through SHP-2 [8]. Additionally, a soluble form of TLT-1 that is released upon platelet activation is on its own capable of inducing platelet aggregation in vitro [6, 7, 12]. Intriguingly, TLT-1 signaling following platelet activation appears to be distinct from that of other TREM family members [4, 5]. However, our knowledge of TLT-1 mechanistic signaling is limited.
In order to expand our understanding of TLT-1 signaling, we conducted a proteomics analysis to define TLT-1 protein-protein interactions in both resting and activated platelets. TLT-1 was immunoprecipitated from biological replicates of resting human platelets and platelets activated with collagen. Proteins in immune complex with TLT-1 were identified via liquid chromatography tandem mass spectrometry (LC-MS/MS) (Figure 1), revealing a TLT-1 interactome that is functionally enriched in platelet degranulation and cell motility. This work presents several novel TLT-1 interacting partners that were validated biochemically in 293 cells and/or platelets, including GRB2, RACK1, β3-integrin, and Rabs 5a, 7, and 11a, which may coordinate with TLT-1 prior to or during platelet activation.
Figure 1.
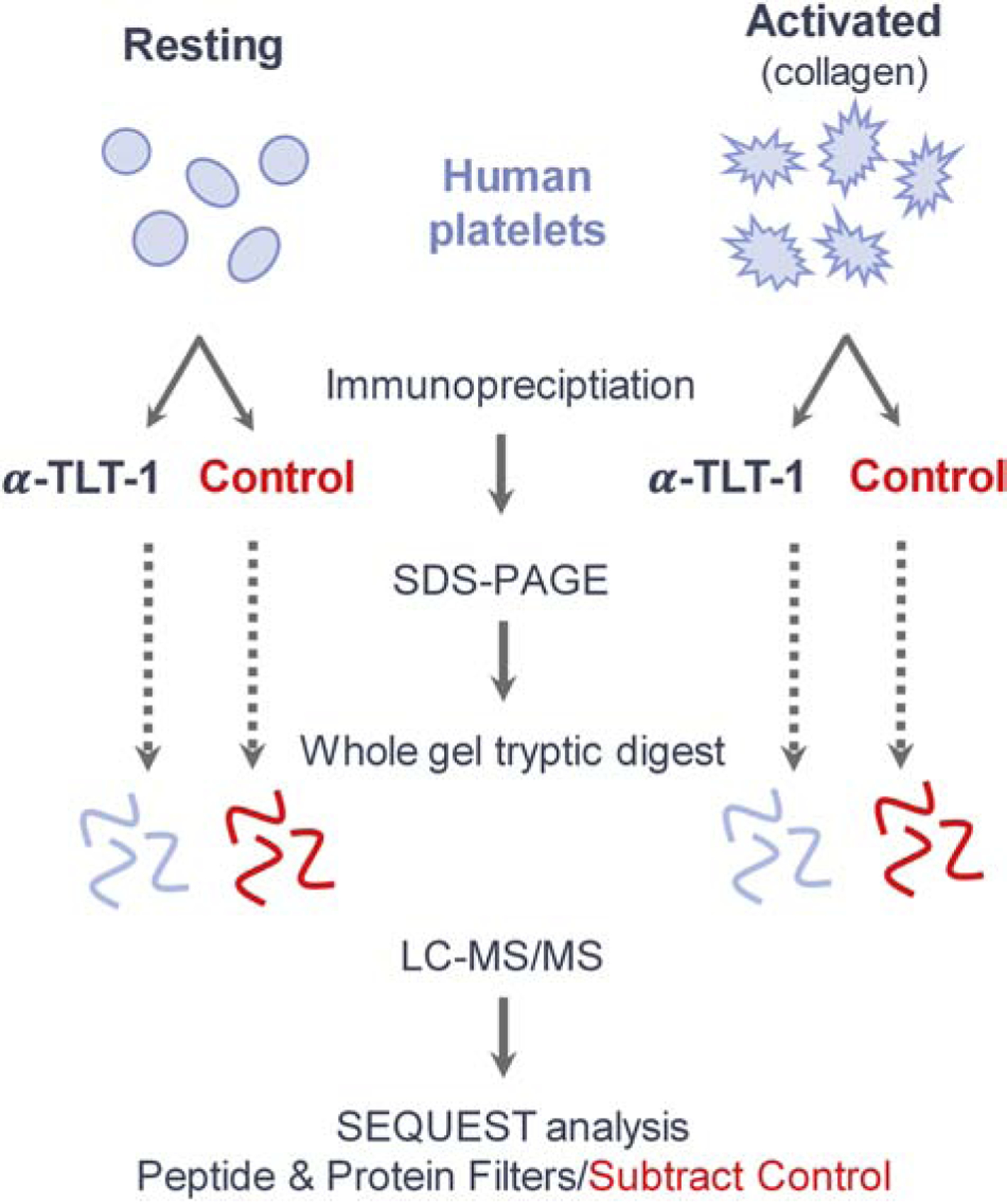
Work flow for the identification of TLT-1 interactors in resting and collagen-activated human platelets. Platelets obtained from two healthy donors were divided into two treatment groups, each. Platelets in the “Resting” group were left untreated, and those in the “Activated” group were treated with collagen. Each treatment group was split for immunoprecipitation with α-TLT-1 or beads alone as a control. Immunoprecipitations were subjected to SDS-PAGE and silver staining. Proteins in each treatment group were divided into 12 regions by molecular weight prior to tryptic digest and analysis via LC-MS/MS. Acquired spectra were searched against a human proteome using the SEQUEST algorithm. Proteins and peptide filters are described in the methods. Proteins identified in the control immunoprecipitation (IgG) were subtracted from the experimental dataset unless 5X overrepresented in the experimental set.
2. Materials and methods
2.1. Ethics statement
The University of Puerto Rico Institutional Review Board (Federalwide Assurance no. FWA00000944) approved the study, and donors provided written informed consent before enrollment (protocol no. 1213–126; primary investigator, A. Washington). All donors, genders unnoted, were between 21 and 60 years of age and had not taken aspirin, non-steroidal anti-inflammatory agents, or anti-platelet drugs within one month of giving blood.
2.2. Materials
Penicillin/Streptomycin 100X solution and Dulbecco’s Modified Eagle Medium (DMEM) were obtained from Mediatech (Manassas, VA, USA). Fetal bovine serum (FBS) and cosmic calf serum (CCS) were purchased from Hyclone (Logan, Utah, USA). The ProFection® Mammalian Transfection System kit for calcium phosphate transfections and the trypsin used in enzymatic digests prior to LC-MS/MS analysis were from Promega (Madison, WI, USA). Protein G resin was obtained from G-Biosciences (St. Louis, MO, USA). Protein A resin and enhanced chemiluminescence (ECL) reagents were purchased from Pierce (Rockford, IL, USA), and x-ray film was from Denville Scientific (Metuchen, NJ, USA). Packing material used for HPLC was 5 μm C18-coated silica beads, 200 Å pore size, purchased from Michrom Bioresources Inc. (Auburn, CA, USA). Nitrocellulose membranes were from GVS Life Sciences (Sanford, ME, USA). All additional reagents were purchased from Sigma (St. Louis, MO, USA) unless otherwise noted.
2.3. Plasmids and antibodies
Mammalian expression constructs for full-length mouse TLT-1 (MR224649), RAB5A (MR202355), RAB7 (MR202190), and RAB11A (MR202384) in pCMV6-Entry, tagged with MYC and FLAG sequences at C-termini, were obtained from Origene (Rockville, MD, USA). pLX304 V5-tagged human TLT-1 (HsCD00436793) and RACK-1 (HsCD00441268) constructs were obtained from the DNASU Plasmid Repository (Tempe, AZ, USA). ITGB3-MYC-His (pcDNA3.1-beta-3, #27289, originally from the lab of Timothy Springer) and SHP-2-MYC (pIRES2-EGFP-SHP2WT, #12283, originally from the lab of Anton Bennett) were obtained from Addgene (Cambridge, MA, USA).
The mouse α-FLAG (M2) antibody was used for Western blotting at a concentration of 0.5 μg/mL. Cell Signaling Technologies, Inc. (Danvers, MA, USA) was the source of the α-FLAG (M2, rabbit mAb), which was used at a dilution of 1:1000 for Western blotting. The rabbit α-pT280TLT-1 antibody (used at 1:500) was raised against the sequence CSKPVpTYATVIFPGGNK (human TLT-1 peptide) by Bon Opus Biosciences (Millburn, NJ, USA). Our in-house antibody, AB69 (mouse IgG2b mAb with reactivity to the TLT-1 extracellular domain) [13], was used for immunoprecipitations and Western blotting of human TLT-1 from platelets. The phosphotyrosine antibody, α-pY (4G10; 1:1000), was from EMD Millipore (Billerica, MA, USA). All primary antibodies for use in Western blotting of 293 cell extracts were diluted in 1.5% BSA in Tris-Buffered Saline (0.9% NaCl, 0.4% Tris-HCl, and 0.1% Tris-base) with 0.05% Tween 20 (TBS-T) and containing 0.005% sodium azide. For Western blotting of platelet extracts, antibodies were diluted in 5% BSA in Tris-Buffered Saline. Horseradish peroxidase (HRP)-conjugated secondary antibodies were obtained from EMD Millipore and used at the following concentrations: goat α-mouse IgG-HRP (1:5,000) and goat α-rabbit IgG-HRP (1:15,000). All secondary antibodies were diluted in TBS-T.
2.4. Platelet preparation, activation and lysis
Platelet preparation and isolation protocols were obtained from [14]. Blood was obtained by venipuncture and whole blood was collected in acid citrate dextrose (1:7) from healthy donors and centrifuged at 800 × g for 20 min to remove red blood cells. Platelets were isolated by centrifugation at 2,100 × g for 10 min and resuspended in Tyrode’s solution (2 mM MgCl2, 137 mM NaCl, 2.68 mM KCl, 3 mM NaH2PO4, 0.1% glucose, 5 mM HEPES, pH 7.35) at a final concentration of 3.2×108 cells/mL. Platelets were incubated at room temperature for 30 min prior to use. Prostaglandin E1 (20 nM) and apyrase (0.02 U/mL) were added at the stage of PRP and to the final prep. Platelets were activated with Ca2+ (2 mM) and thrombin (0.2 U/mL). Collagen activation was the same as thrombin except it was added at 5 μg/mL.
2.5. HEK 293 cell culture, transfection, and lysis
HEK 293 cells were cultured in DMEM supplemented with 5% each of FBS and CCS, 50 U/mL penicillin, and 50 μg/mL streptomycin at 37 °C in 5% atmospheric CO2. For transfection, cells were grown to 60% of confluence and transfected with 5 μg DNA per plasmid per 10-cm dish via calcium phosphate precipitation. Six hours post-transfection, cells were washed with phosphate-buffered saline (PBS) and returned to full medium overnight before lysis. Hydrogen peroxide (H2O2) treatments were conducted at 8.8 mM H2O2 for 15 min at standard culture conditions prior to lysis. Calyculin A treatments were conducted at 100 nM calyculin A in dimethyl sulfoxide (DMSO) for 30 min at standard culture conditions. Following treatment, cells were lysed on ice with Brain Complex Lysis Buffer (BCLB: 25 mM Tris pH 7.2, 137 mM NaCl, 10% glycerol, 1% igepal, 25 mM NaF, 10 mM Na2H2P2O7, 1 mM Na3VO4, 1 mM phenylmethylsulfonyl fluoride (PMSF), 10 μg/mL each leupeptin and pepstatin-A). Lysates were centrifuged and the supernatant was reserved for analysis.
2.6. Immunoprecipitation, SDS-PAGE and Western blotting
For immunoprecipitations from 293 cell lysates, protein levels were normalized, and lysates were incubated with α-FLAG Affinity Gel, or the α-V5 or α-MYC antibodies with a 50/50 mixture of protein A and G resin, rocking overnight at 4 °C. The beads were washed and bound proteins were eluted and denatured in 25 μL sample buffer (150 mM Tris pH 6.8, 2% SDS, 5% β-mercaptoethanol, 7.8% glycerol, 0.25 ng/mL bromophenol blue) at 95 °C for 5 min. For Western blotting of whole cell extracts (WCEs), samples were denatured in sample buffer and 15 μg total protein was loaded per lane. Immunoprecipitations and WCEs were separated on 10% acrylamide gels (30% w/v and 37.5:1 acrylamide:bis-acrylamide) following stacking over 4.2% acrylamide gels, unless otherwise indicated. Proteins were transferred to a nitrocellulose membrane in a submersible transfer unit at 4 °C. Membranes were blocked with 5% non-fat dry milk in TBS-T prior to incubation with primary antibody solutions overnight at 4 °C and HRP-conjugated secondary antibody solutions for three hours at 25 °C. Membranes were incubated with ECL reagents prior to exposure to x-ray film.
Platelets were lysed in a buffer containing 1% triton (1% triton-X, 150 mM NaCl, 50 mM Tris pH 7.4, 1 mM EDTA, 1 mM PMSF, 20 mM NaF, 10 mM NaVO4 and 1x protease inhibitor). Immunoprecipitations from platelet lysates (3 × 108 platelets/mL) were conducted with 5 μg α-TLT-1 (clone AB69; 50 % slurry of Protein A and G resin) in 500 μL platelet lysate. Immunoprecipitations and WCEs were separated on 12% acrylamide gels (30% w/v and 37.5:1 acrylamide:bis-acrylamide) following stacking over 4.2% acrylamide gels. Proteins were then transferred to a PVDF membrane, and membranes were blocked and incubated with primary and secondary antibodies as described above.
2.7. Sample preparation for liquid chromatography/tandem mass spectrometry (LC-MS/MS)
Following SDS-PAGE, acrylamide gels containing 293 and platelet immunoprecipitates were stained with Coomassie Brilliant Blue (293 immunoprecipitates) or silver staining (platelet immunoprecipitates) and divided into eight and ten sections, respectively, per lane. For Coomassie staining, gels were rocked in 40% methanol/20% acetic acid with 0.1% Coomassie Brilliant Blue for 1 hour and room temperature, and destained overnight in 30% methanol/10% acetic acid. For silver staining, gels were stained and destained with a FOCUS FASTsilver kit (G-Biosciences; St. Louis, MO, USA). Excised sections were diced to 1-mm cubes and transferred to separate microcentrifuge tubes. Gel pieces were de-stained according to the respective staining protocols and subjected to tryptic digest (4–6 ng/μLtrypsin in 50 mM NH4HCO3) and incubated overnight at 37 °C. Peptides were extracted from gel pieces dried prior to LC-MS/MS analysis.
2.8. LC-MS/MS spectral acquisition and analysis
Dried peptides were re-suspended in 2.5% MeCN/0.15% FA (Solvent A) and separated on a reverse-phase high performance liquid chromatography (HPLC) column packed in house with 5-μm C18 beads (column length was 12 cm × 100 μm (i.d.), bead pore size was 200 Å).
Platelet immunoprecipitations (two biological replicates) were analyzed on the LTQ-Orbitrap Discovery mass spectrometer fitted with a Finnigan Surveyor Pump Plus and Micro AS autosampler (Thermo Electron; San Jose, CA, USA) and controlled with Xcalibur™ 2.1 Software (Thermo Fisher Scientific, Inc.; Waltham, MA, USA). Following a 15-min loading phase onto the C18 column in Solvent A, peptides were eluted with a 0–50% gradient of Solvent B (99.85% MeCN, 0.15% FA) over 38 min and electrosprayed (2.1 kV) into the mass spectrometer. This gradient was followed by 7 min at 100% Solvent B prior to a 10-min equilibration in 100% Solvent A. The flow rate was maintained at 500 nL/min throughout the analysis. The precursor scan (360–1700 m/z) was followed by ten low energy collision-induced dissociation (CID) tandem mass spectra (normalized collision energy (NCE) was 35%). CID spectra were acquired for the top ten most abundant ions in the precursor scan (isolation width was ± 2 m/z). All mass spectra were obtained in centroid with precursor ion spectra acquired in the orbitrap (resolution=3×104, scan speed was 1 Hz) and fragment ion spectra in the linear ion trap (positive ion mode, dynamic exclusion disabled).
The single 293 immunoprecipitation was analyzed on the LTQ-XL mass spectrometer. The 15-min loading phase (Solvent A) was followed by a 0–50% gradient of Solvent B over 43 min, 7 min at 100% Solvent B, and 10 min in 100% Solvent A. The flow rate was maintained at 500 nL/min over the column throughout the analysis. The precursor scan (400–1600 m/z) was followed by ten low energy CID MS2 spectra (NCE was 35%) for the top ten most abundant ions in the MS scan (positive ion mode, dynamic exclusion settings: repeat count=2, repeat duration=30 sec, exclusion list size=180, exclusion duration=60 sec, isolation width=±1.5 m/z).
For targeted analyses of TLT-1 ITIM-containing species in human platelets, cysteine residues were reduced and alkylated with dithiothreitol and iodoacetamide prior to tryptic digest, given the presence of cysteine in the ITIM-containing peptide (VLVCSKPVTYATVIFPGGNK). Peptides were separated via HPLC using the Easy n-LC 1200 prior to MS/MS analysis on the Q Exactive Plus mass spectrometer fitted with a Nanospray Flex ion source (Thermo Fisher Scientific, Inc.; Waltham, MA). Chromatography columns (30 cm long, 100 μm inner diameter) were packed in-house with 1.8 μm C18 packing material. Peptides were eluted using a 0–50% gradient of Solvent B (80% acetonitrile, 0.15% formic acid) over 175 min, followed by 10 min at 100% Solvent B. Each precursor scan (scan range = 400–1500 m/z, resolution = 7.0×104, AGC = 1.0×106, maximum ion time = 100 ms) was followed by targeted MS2 of the human TLT-1 ITIM-containing peptide VLVCSKPVTYATVIFPGGNK in phosphorylated (m/z=744.0496, z=3+) and unphosphorylated (m/z=717.3941, z=3+) states (resolution = 3.5×104, AGC = 5.0×104, maximum ion time = 50 ms, isolation window = ±0.4 m/z, collision energy = 26%). Note that the masses given include the added mass of cysteine carbamidomethylation.
SEQUEST (version 28) searches for TLT-1 interactors were performed using a forward and reverse 2011 Uniprot Human Protein database [15] requiring tryptic peptides and permitting the following modifications: phosphorylation of serine, threonine and tyrosine (+79.966 Da), oxidation of methionine (+15.995 Da), and acrylamidation of cysteine (+71.037 Da) with parent ion mass tolerances of ± 4 ppm for LTQ-Orbitrap and Q Exactive Plus data and ± 2 Da for LTQ-XL data, fragment ion tolerances of ± 1 Da and ± 0.01 Da for MS2 data acquired in linear ion trap (LTQ-XL and LTQ-Orbitrap) and orbitrap (Q Exactive Plus) analyzers, respectively, and a unique ΔCorr of ≥ 0.2, resulting in a false positive rate <1%. Peptides identified in the control lanes were removed from the corresponding experimental dataset unless they were present in the experimental set at an abundance five times greater than in the control set. A complete list of identified proteins from each experimental condition and replicate can be found in Supplementary Tables 1 & 2.
For phosphorylation analysis of TLT-1, raw data for each experiment were separately searched via SEQUEST (version 28) against a forward and reverse mouse or human TLT-1 sequence including common contaminants with no enzyme specified, permitting the same modifications and mass tolerances specificed above. No enzyme was specified in order to expand the single-sequence database and reduce the false discovery rate. Resulting peptide tables were filtered to include only tryptic peptides (Supplementary Tables 3 and 4). Phosphopeptides resulting from this search were manually validated to confirm positive database hits (Supplementary Figure 2).
Protein functions were assigned based on annotations in PhosphoSitePlus [16] and UniProt [17]. Known TLT-1 interactors were identified by The Biogrid curations [18] and primary literature searches. Gene ontology (GO) enrichment analyses were conducted with the Metascape Enrichment tool using default settings with the following databases selected: GO Biological Process, GO Molecular Function, GO Cellular Component [19]. Protein-level identifications of TLT-1 interactors in platelets were obtained from The PlateletWeb [20].
3. Results
3.1. Identification of novel TLT-1 binding partners in resting and activated platelets
As a first step in elucidating the mechanism of TLT-1 in platelet aggregation, we conducted a proteomics screen that aimed to identify novel TLT-1 interactors in resting and activated platelets. Briefly, TLT-1 was immunoprecipitated from resting and collagen-activated human platelets and immune complexes were analyzed via SDS-PAGE (Figure 1). Control immunoprecipitations using only beads were conducted for each condition from the same platelet lysates (Figure 1). Experimental and control lanes were each divided into ten sections, subjected to in-gel tryptic digestion, and extracted peptides were subjected to LC-MS/MS analysis (Figure 1). Two biological replicates were performed for each condition.
Proteins identified in immune complex with TLT-1 in resting and activated platelets, common to both biological replicates and therefore representing strong candidate interactors with importance in TLT-1 signaling, are summarized in Tables 1 & 2, respectively. Full tables containing TLT-1 peptides and those of its interactors identified in each biological replical are included in Supplementary Tables 1 & 3. A total of eight proteins were unique to activated platelets and 31 were found only in resting platelets. Only two TLT-1 interactors were common to both platelet environments: Histone H1E and RAB7A. In addition to RAB7A, several additional Rab family members were found to interact with TLT-1 uniquely in resting or activated platelets. TLT-1 is known to be rapidly mobilized to the membrane from α-granules following platelet activation, which likely involves a change in the Rab protein population on the granule surface.
Table 1.
TLT-1 binding partners identified in resting platelets. TLT-1 was immunoprecipitated from untreated human platelets from two healthy donors, and subjected to SDS-PAGE. Gel regions were subjected to in-gel tryptic digestion prior to LC-MS/MS analysis of extracted peptides (Figure 1). Spectral counts of proteins identified in both biological replicates using the SEQUEST algorithm are indicated.
| Gene Symbol | Uniprot Ace | Protein Name | Total # peptides | |
|---|---|---|---|---|
| Donor 1 | Donor 2 | |||
| ACTN4 | actinin alpha 4 | 22 | 35 | |
| AHCYL2 | adenosylhomocysteinase like 2 | 4 | 3 | |
| ARHGAP6 | Rho GTPase activating protein 6 | 4 | 9 | |
| ATP5A1 | ATP synthase F1 subunit alpha | 4 | 42 | |
| AZGP1 | alpha-2-glycoprotein 1, zinc-binding | 5 | 5 | |
| CYB5R3 | cytochrome b5 reductase 3 | 4 | 4 | |
| DBN1 | drebrin 1 | 11 | 7 | |
| EIF3D | eukaryotic translation initiation factor 3 subunit D | 3 | 5 | |
| EPB49 | dematin actin binding protein | 6 | 8 | |
| FHL1 | four and a half LIM domains 1 | 3 | 10 | |
| GIT1 | GIT ArfGAP 1 | 6 | 4 | |
| HIST1H1E | histone cluster 1 H1 family member e | 8 | 8 | |
| ITGB3 | integrin subunit beta 3 | 20 | 25 | |
| MMRN1 | multimerin 1 | 3 | 15 | |
| MYO18A | myosin XVIIIA | 17 | 5 | |
| NAP1L4 | nucleosome assembly protein 1 like 4 | 3 | 6 | |
| OSBPL8 | oxysterol binding protein like 8 | 6 | 5 | |
| PPP1R9B | protein phosphatase 1 regulatory subunit 9B | 5 | 20 | |
| RAB11A | RAB11A, member RAS oncogene family | 9 | 12 | |
| RAB13 | RAB13, member RAS oncogene family | 3 | 4 | |
| RAB1A | RAB1A, member RAS oncogene family | 6 | 3 | |
| RAB27B | RAB27B, member RAS oncogene family | 12 | 5 | |
| RAB6A | RAB6A, member RAS oncogene family | 9 | 3 | |
| RAB7A | RAB7A, member RAS oncogene family | 7 | 3 | |
| RDH11 | retinol dehydrogenase 11 | 8 | 14 | |
| SLC2A3 | solute carrier family 2 member 3 | 5 | 4 | |
| SYNPO2 | synaptopodin 2 | 3 | 4 | |
| TGFB1I1 | transforming growth factor beta 1 induced transcript 1 | 4 | 15 | |
| TMED8 | transmembrane p24 trafficking protein family member 8 | 6 | 73 | |
| TUBB | tubulin beta class I | 45 | 63 | |
| USO1 | USO1 vesicle transport factor | 3 | 3 | |
| USP15 | ubiquitin specific peptidase 15 | 7 | 3 | |
| VPS41 | VPS41, HOPS complex subunit | 5 | 3 | |
| TLT1 | TREM-like transcript 1 | 126 | 144 | |
Table 2.
TLT-1 binding partners identified in active platelets. TLT-1 was immunoprecipitated from human platelets from two healthy donors, activated in vitro with collagen, and subjected to SDS-PAGE. Gel regions were subjected to in-gel tryptic digestion prior to LC-MS/MS analysis of extracted peptides (Figure 1). Spectral counts of proteins identified in both biological replicates using the SEQUEST algorithm are indicated.
| Gene Symbol | Uniprot Ace | Description | Total # peptides | |
|---|---|---|---|---|
| Donor 1 | Donor 2 | |||
| GSTP1 | glutathione S-transferase pi 1 | 12 | 3 | |
| HIST1H1E | histone cluster 1 H1 family member e | 3 | 3 | |
| MOBKL1A | MOB kinase activator 1B | 6 | 4 | |
| MYO5A | myosin VA | 3 | 4 | |
| RAB37 | RAB37, member RAS oncogene family | 10 | 7 | |
| RAB5A | RAB5A, member RAS oncogene family | 8 | 4 | |
| RAB5C | RAB5C, member RAS oncogene family | 11 | 3 | |
| RAB7A | RAB7A, member RAS oncogene family | 17 | 10 | |
| RAN | RAN, member RAS oncogene family | 9 | 6 | |
| USP9X | ubiquitin specific peptidase 9 X-linked | 4 | 3 | |
| TLT1 | TREM-like transcript 1 | 113 | 71 | |
Functional classification of TLT-1 interactors in resting and active platelets are summarized in Figure 2A. Proteins involved in trafficking, cell adhesion, and cytoskeletal dynamics were highly represented in immune complex with TLT-1 (Figure 2A). A gene ontology (GO) term enrichment analysis (GO cellular component, GO molecular function, GO biological process) conducted via the Metascape platform (www.metascape.org) [19] revealed an enrichment (FDR > 0.01) of terms related to Rab proteins and vesicular trafficking, as well as cell spreading, adhesion, and migration (Figure 2B, Supplemental Table 5). Importantly, “Platelet Degranulation” (FDR = 2.8 × 10−6) was considered enriched in the data set. All proteins annotated with this term originated from the TLT-1 immune complex in active platelets, with the exception of RAB5A and USP9X (Supplemental Table 5).
Figure 2.
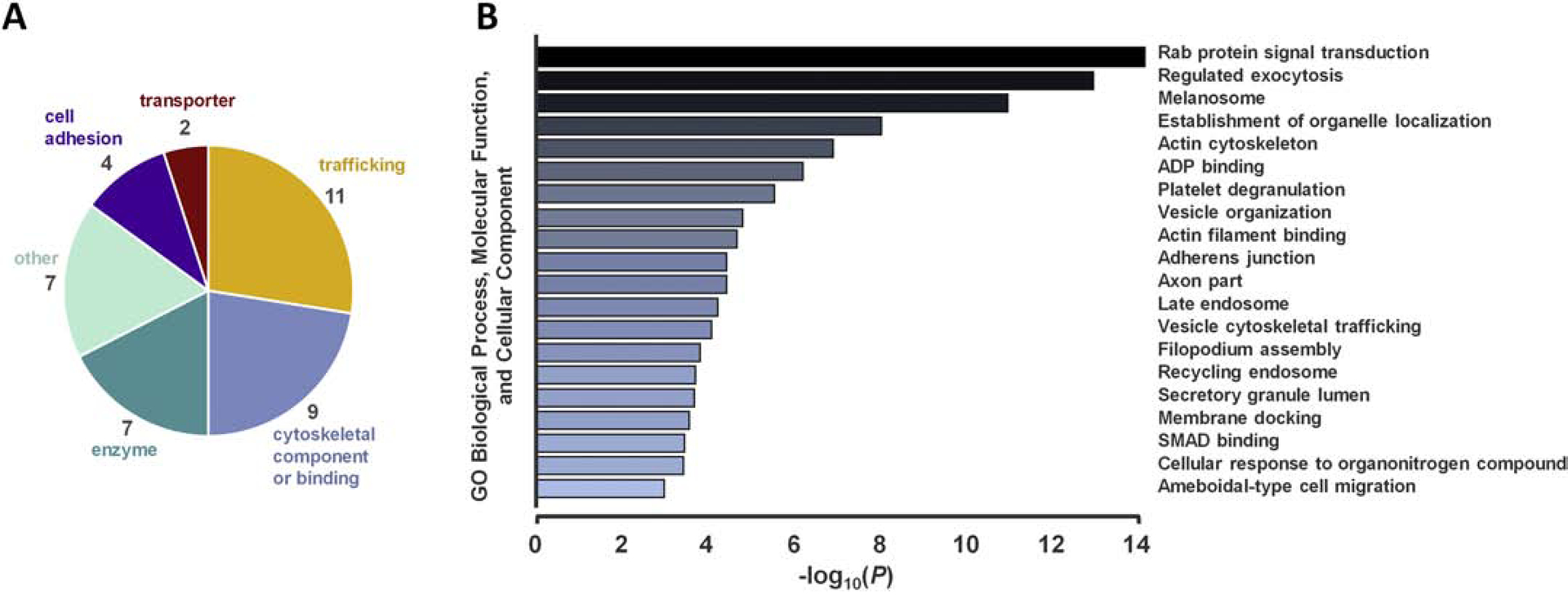
Functional characterization and gene ontology (GO) enrichment analysis of TLT-1 interactors in human platelets. A) TLT-1 interactors are categorized by general function. B) A GO enrichment analysis was conducted using the Metascape platform [19]. GO terms with P < 10−3 are shown. Genes within each enriched group are tabulated in Supplementary Table 5.
3.2. TLT-1 interacting partners and phosphorylation near the ITIM are dynamically regulated
Platelet activation is known to stimulate a variety of signaling cascades. The observed changes in TLT-1 binding partners across resting and active platelets could result from changes in subcellular localization of either TLT-1 or its interactors, or through regulated post-translational modification. We considered that TLT-1 phosphorylation, in particular, would be a strong candidate modifier of TLT-1 interacting partners. We chose 293 cells as a simple model in which to express TLT-1 at high levels to assist in the identification of phosphorylation sites with potential relevance to platelet biology. Regulated TLT-1 tyrosine phosphorylation has been demonstrated in the presence/absence of pervanadate, a tyrosine phosphatase inhibitor [8, 10]. H2O2 on its own is also a strong tyrosine phosphatase inhibitor. To determine whether H2O2 would alter TLT-1 phosphorylation, 293 cells transiently expressing the mouse allele of TLT-1 were stimulated with H2O2. The mouse allele was chosen given its availability with C-terminal MYC and FLAG epitope tags and its relevance to studies involving the TLT-1−/− mouse [6]. Immunoprecipitated TLT-1 (α-FLAG) was subjected to SDS-PAGE and Western blotting (α-FLAG and α-pY; Figure 3A). Strong phosphotyrosine signals were observed in the immunoprecipitation from H2O2-treated cells in the highest molecular weight protein product of TLT-1 (45–50 kDa), as well as a species of ~30 kDa (Figure 3A). Interestingly, these two bands of lower molecular weight are apparent only following H2O2 stimulation (Figure 3A), suggesting that the inhibition of tyrosine phosphatases or the cellular response to oxidative stress leads to TLT-1 truncation, as opposed to its normal alternative splicing [6, 7, 12]. TLT-1 shedding is known to be regulated at least in part by ADAM17 [21]. Given the FLAG epitope tag is located on the TLT-1 C-terminus, the immunoprecipitated truncation could represent the C-terminal cleavage product of a phosphotyrosine-dependent proteolytic event.
Figure 3.
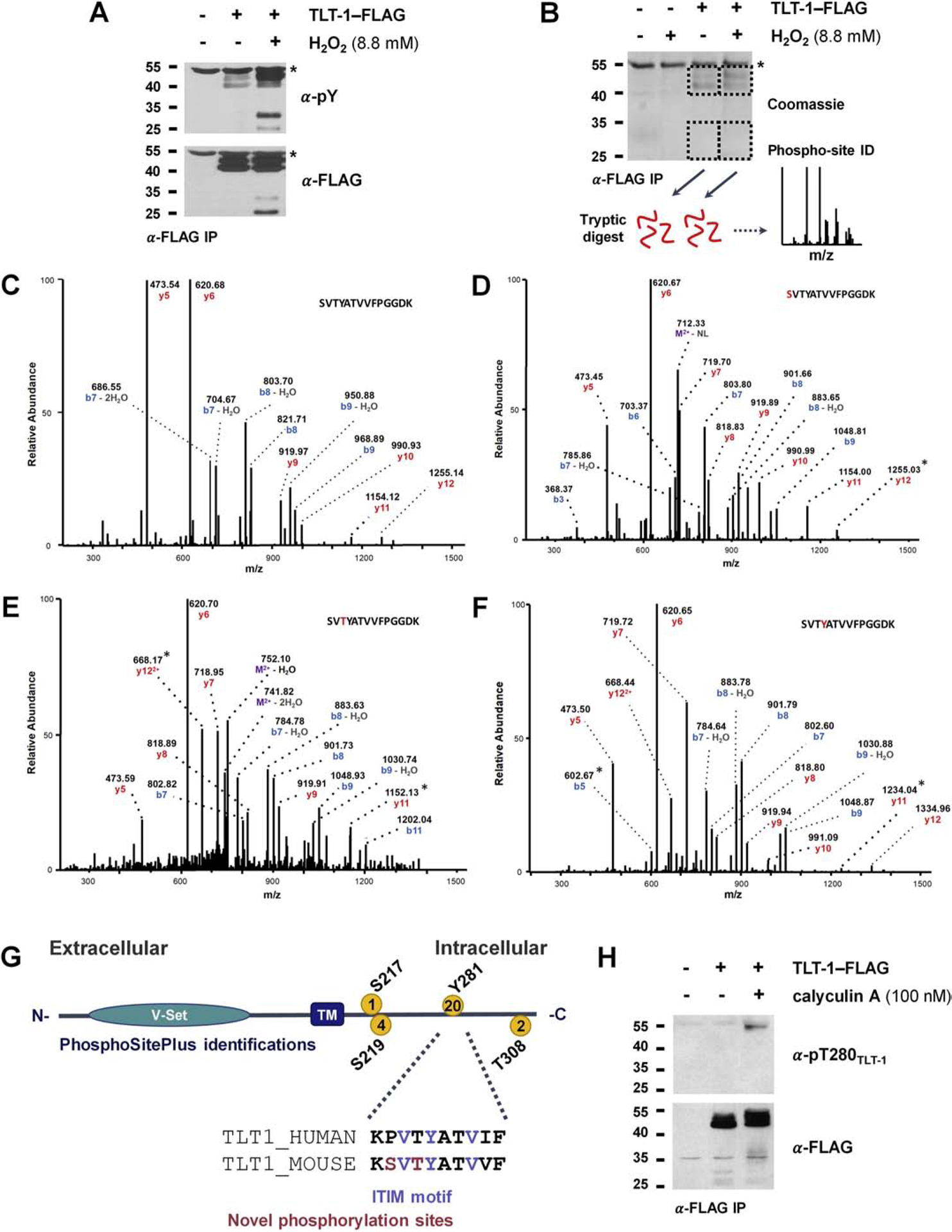
Identification of a regulated phosphorylation site on conserved TLT-1 Thr280 near ITIM. A) TLT-1 tyrosine phosphorylation is inducible by H2O2. TLT-1 was immunoprecipitated from 293 cells expressing mouse TLT-1 with a FLAG C-terminal tag. Cells were either left untreated or were treated with a tyrosine phosphatase inhibitor (H2O2) for 15 min prior to lysis. Immunoprecipitations were subjected to SDS-PAGE and immunoblotting for TLT-1 (α-FLAG) and phosphotyrosine (α-pY). B) Coomassie-stained immunoprecipitation and schematic for the identification of TLT-1 phosphorylation sites. 293 cells transiently expressing TLT-1-FLAG (mouse) were treated with H2O2 for 15 min prior to lysis. α-FLAG immunoprecipitations were subjected to SDS-PAGE and Coomassie staining. Bands containing α-FLAG signals in A were excised and digested with trypsin prior to LC-MS/MS analysis. C–E) Fragmentation spectra of tryptic peptide containing the ITIM. b and y ions are annotated. Asterisks denoted fragments that define phosphorylated residue. G) TLT-1 domain structure, phosphorylation site identifications curated on phosphosite.org, conservation of ITIM and surrounding residues. H) Regulated phosphorylation of TLT-1 Thr280. 293 cells transiently expressing TLT-1-FLAG (mouse) were treated with a serine/threonine phosphatase inhibitor (calyculin A) prior to lysis. Immunoprecipitations (α-FLAG) were subjected to SDS-PAGE and immunoblotting with an antibody raised against a TLT-1 peptide (human sequence) containing phosphorylated T280.
The single known tyrosine phosphorylation site (PhosphoSite.org [16]) falls within the ITIM at Tyr281 of the human sequence (Figure 3G), which is hypothesized to be important to the role of TLT-1 in platelet biology. This is one of only two intracellular tyrosines (Supplementary Figure 1), which suggests that the strong induction of tyrosine phosphorylation originated from one or both of these sites (Figure 3A). We employed mass spectrometry to identify TLT-1 phosphorylation sites in the stimulated and unstimulated states observed in Figure 3A. TLT-1 was immunoprecipitated from unstimulated cells and H2O2-treated cells as described above and analyzed by SDS-PAGE and Coomassie staining (Figure 3B). Bands containing TLT-1 were excised and subjected to tryptic digest prior to LC-MS/MS analysis of extracted peptides (Figure 3B).
TLT-1 phosphorylation sites identified across conditions in 293 cells are summarized in Supplementary Table 6 with fragmentation spectra included Supplementary Figure 2. For comparison, we have included the phosphorylation sites identified on human TLT-1 in resting and active platelets (Supplementary Table 6, Supplementary Figure 2), from the proteomics screen described above. All but one site we identified on the mouse sequence in 293 cells were conserved in the human sequence (Figure 3G, Supplementary Figure 1). We identified one novel phosphorylation site on a conserved residue, Thr280, near the ITIM sequence (Figure 3G). Fragmentation spectra of various phosphorylation states of the peptide containing the ITIM sequence are shown in Figures 3C–F. Asterisks denote fragments that demonstrate the accurate identification of each phosphorylation site. Noteably, all TLT-1 peptides were identified in the band containing full-length TLT-1; no peptides were found in the band containing the assumed cleaved C-terminal peptided observed in Figure 3B. Phosphorylation of Tyr281 was identified only in the stimulated cells, while peptides housing phosphorylated Ser278 and Thr280 were present in both stimulated and unstimulated conditions (Supplementary Table 6). Given the conservation and novelty of Thr280 (Figure 3G) and its proximity to the ITIM tyrosine, we generated a phospho-specific antibody to this site. To determine whether Thr280 phosphorylation was regulated, we treated cells transiently-expressing TLT-1 with calyculin A, a serine/threonine phosphatase inhibitor. Immunoprecipitates (α-FLAG) were subjected to SDS-PAGE and Western blotting for phosphorylation of TLT-1 Thr280 (α-pThr280TLT-1, Figure 3H). The induction of phosphorylation at Thr280 following calyculin A treatment suggests that this is a regulated phosphorylation site. The close proximity of Thr280 to the ITIM tyrosine (Tyr281) indicates that this may be an important regulatory region within the TLT-1 intracellular domain. Interestingly, the peptide containing the ITIM was not identified with multiple sites phosphorylated, suggesting that phosphorylation of these sites could be counterregulatory to one another. However, it remains possible that multiply-phosphorylated peptides may be present at low abundance, but remain undetected by LC-MS/MS.
To determine whether the ITIM-proximal phosphorylation sites found in 293 cells could potentially be relevant to platelet biology, we employed a targeted approach to identify these phosphorylation sites in platelets. TLT-1 was immunoprecipitated (α-TLT1) from activated human platelets or platelets treated with H2O2, and immunoprecipitates were analyzed by SDS-PAGE and Coomassie staining. TLT-1 bands were excised, subjected to reduction/alkylation of cysteine residues and digested with trypsin. Peptides were analyzed via LC-MS/MS, targeting the mass of the human peptides containing the ITIM tyrosine (Y281) and proximal threonine (T280) that were found phosphorylated in 293 cells (Figure 3C–F). Peptides containing the phosphorylated Y281 were identified in both H2O2 treatments and activated platelets (Supplementary Figure 3). The annotated spectrum of the unphosphorylated peptide (Supplementary Figure 3A) is shown alongside spectra of the phosphopeptides identified in H2O2 treated and activated platelets (Supplementary Figure 3B–C). Due to a co-eluting ion within the isolation window used for fragmentation, considerable background was observed in the spectrum of the phosphopeptide obtained from activated platelets (Supplementary Figure 3C). However, the presence of characteristic fragments observed in clean spectra of the unphosphorylated and phosphorylated ITIM-containing peptides (Supplementary Figure 3A–B) confirms that the phosphopeptide was correctly identified. Diagnostic fragments demonstrating phosphorylation site localization on the ITIM tyrosine (Y281) in Supplementary Figure 3B are denoted by boxed fragment ion annotations. Fragments distinguishing between phosphorylation of Y281, T280, and S276 were not apparent from the phosphopeptide ion in activated platelets (Supplementary Figure 3C), however, the elution time of the phosphopeptide identified in activated platelets was close to the confirmed pY281-containing species in H2O2-treated platelets (Supplementary Figure 3D–E). Given that the various ITIM-containing phosphopeptides from mouse (Figure 3D–F) exhibited markedly different elution times (the peptide containing phosphotyrosine eluted at 35% MeCN, phosphoserine at 40% MeCN, and phosphothreonine at 45% MeCN), the phosphorylated species identified in activated platelets likely contains pY281.
Although pT280 was not identified in the TLT-1 immunoprecipitate from human platelets via LC-MS/MS, we concluded that it may be present at levels below our limit of detection, and turned to Western blotting with the α-p280TLT1 probe in an attempt to visualize T280 phosphorylation platelets. TLT-1 was immunprecipitated from human platelets after various activation times and subjected to Western blotting for α-TLT-1 and α-p280TLT1. Phosphorylation of T280 was visible following 10 minutes of platelet activation (Supplementary Figure 4). Given the identification of the two ITIM-proximal phosphorylation sites observed in both platelets and 293 cells, we concluded that 293 cells were an appropriate model in which to study TLT-1 signaling.
The most robust induction of phosphorylation observed from the mass spectrometric studies and Western blotting was observed following H2O2 treatment. We asked whether any of the proteins identified as TLT-1 interactors in resting and active platelets might be induced to associate or dissociate upon TLT-1 tyrosine phosphorylation. We compared proteins that were bound to TLT-1 in the immunoprecipitation shown in Figure 3B to those identified in platelets. Each gel lane was divided into eight sections and excised bands were digested with trypsin prior to analysis of extracted peptides by LC-MS/MS. As controls, 293 cells that were not expressing TLT-1 were treated with or without H2O2 were subjected to the same procedures for immunoprecipitation and LC-MS/MS.
Proteins identified in immune complex with TLT-1 in 293 cells are summarized in Supplementary Table 7. A total of 19 proteins were unique to cells treated with H2O2 and 31 were identified only in untreated cells, while 13 proteins identified in complex with TLT-1 in both conditions. Surprisingly, the TLT-1 interactors identified in 293 cells did not appreciably overlap with the two biological replicates obtained from platelets, with the exception of ATP51A. Notably, a known tyrosine phosphorylation-dependent TLT-1 interactor in platelets, PTPN11 (SHP2), was identified as a H2O2-induced binding partner (Supplementary Table 7). Of the proteins identified as TLT-1 binding partners in 293 cells, 81% have been detected at the protein level in platelets (Supplementary Figure 5A) [20]. General functions and enriched GO terms of the TLT-1 interactors identified in 293 cells are summarized in Supplementary Figures 5B & 5C (Supplementary Table 8). Given the lack of overlap in TLT-1 interactors, this experiment was not replicated. However, proteins identified as TLT-1 interactors in 293 cells with known roles in platelet biology were considered for biochemical validation.
3.3. Biochemical validation of known and novel TLT-1 interactors
Several of the novel TLT-1 interactors identified in the two biological replicates in platelets and complimentary study in 293 cells were of biological interest for further biochemical validation. The 293 cells were chosen as a simple system in which to validate the interactions identified via LC-MS/MS. Using transiently expressed proteins in this manner, we were easily able to test several potential interactions and their ability to be regulated in a phosphorylation-dependent manner. We were particularly interested in the enrichment of functional terms related to cell motility and platelet aggregation (Figure 2B). Notably, we identified several Rab proteins that differentially bound to TLT-1 in resting and active platelets, suggesting a potential role of the TLT-1/Rab interaction in vesicle trafficking prior to or following platelet activation. Three family members identified in immune complex with TLT-1 in platelets (Rabs 5A, 7, and 11A) were chosen to confirm biochemically in 293 cells. In addition, proteins that are known to play roles in platelet activation and possess phosphotyrosine-binding domains were confirmed: the Src-homology 2 (SH2) domain-containing proteins SHP2 (PTPN11, a known TLT-1 interactor), growth factor receptor bound protein 2 (GRB2), and receptor of activated protein C kinase 1 (RACK1), which were all identified in the TLT-1 immunoprecipitation from 293 lysates. The TLT-1/GRB2 interaction was additionally tested in resting and active platelets. Also essential to platelet activation, the β3 integrin subunit (ITGB3), which bound TLT-1 in platelets, was validated biochemically in both platelets and 293 cells.
For co-immunoprecipitation experiments in 293 cells, mammalian expression constructs of SHP-2, RACK1, ITGB3, and RAB5A, RAB7, and RAB11A were each co-expressed individually with TLT-1-FLAG-MYC (mouse) or TLT-1-V5 (human). Either TLT-1 or the co-expressed interactor was immunoprecipitated, and immunoprecipitates were subjected to SDS-PAGE and immunoblotting for the other protein in the presence/absence of H2O2 (Figure 4A–D). To confirm the interaction between TLT-1 and growth factor receptor-bound protein 2 (GRB2), TLT-1 was expressed alone in 293 cells, and immunoprecipitates (α-FLAG) were blotted for endogenous GRB2 (α-GRB2) in the presence and absence of H2O2 (Figure 4E). The phosphorylation-dependent TLT-1/SHP-2 interaction, previously demonstrated [8], was recapitulated here with H2O2 treatment (Figure 4A). Other interactors that demonstrated H2O2-induced interactions were the Rab family members (Figure 4B) and the SH2-domain containing adaptor GRB2 (Figure 4E). RACK-1 and β3-integrin exhibited phosphorylation-independent interactions with TLT-1 (Figure 4C,D). The TLT-1/β3-integrin and TLT-1/GRB2 interactions was also validated by co-immunoprecipitation in human platelets. Immunoprecipitations from active and resting platelets revealed β3-integrin in immune complex with TLT-1 (α-TLT-1) in resting, but not active platelets (Figure 4F), in agreement with the LC-MS/MS data (Tables 1 & 2). GRB2, however, was induced to bind to TLT-1 upon platelet activation (Figure 4G).
Figure 4.
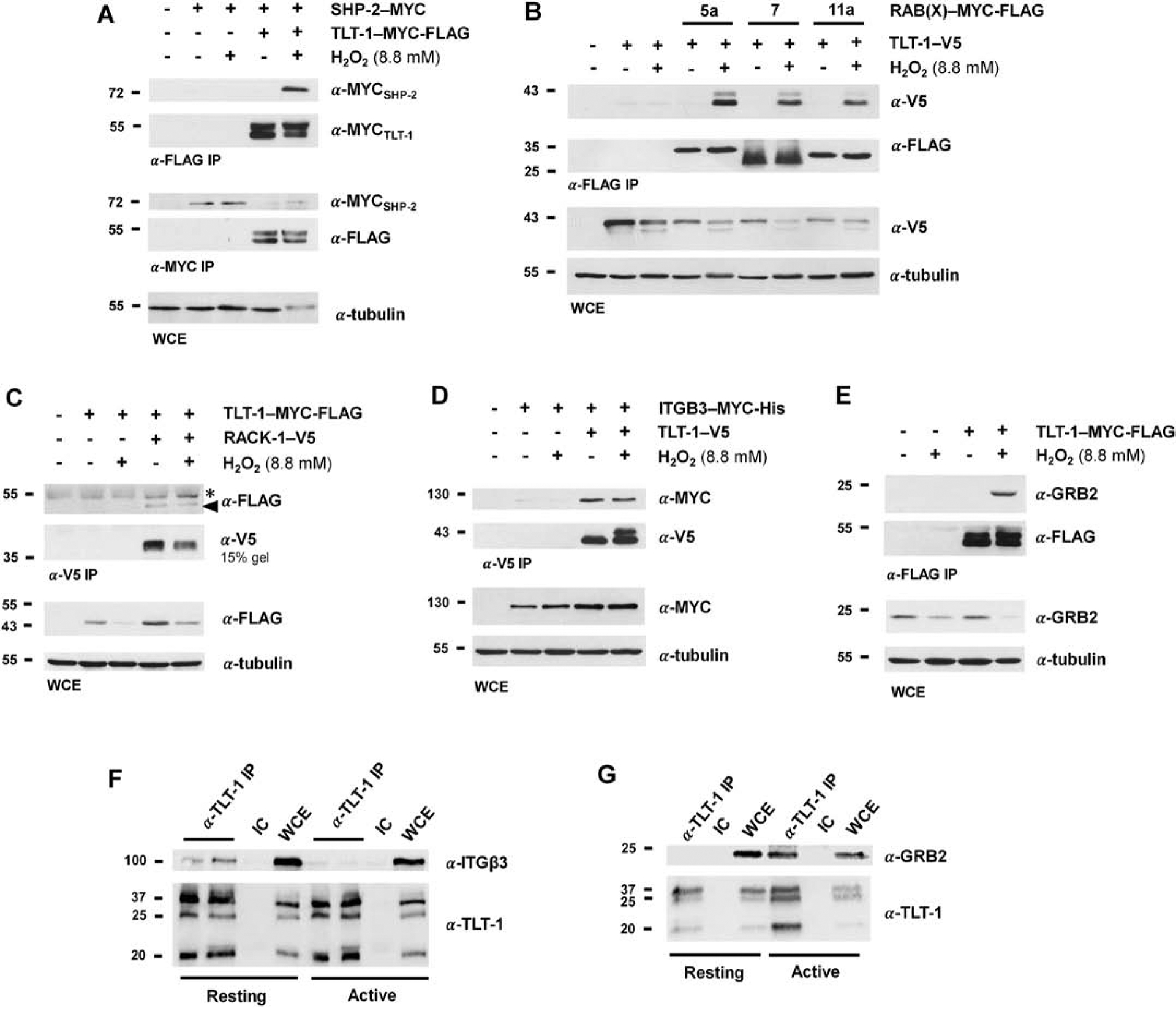
Biochemical validation of TLT-1 interactors. MYC-FLAG- (mouse) or V5-tagged (human) TLT-1 was transiently expressed alone or with SHP-2-MYC (A), RAB(X)-MYC-FLAG (5A, 7, or 11A) (B), RACK-1-V5 (C), or ITGB3-MYC-His (D) in 293 cells. Cells were either left untreated or were treated with H2O2 (8.8 mM) prior to lysis. Immunoprecipitates (IPs) of the indicated epitope tag were subjected to SDS-PAGE analysis and Western blotting for the immunoprecipitated and co-expressed proteins. Whole cell extracts (WCEs) were immunoblotted for co-expressed proteins, unless otherwise indicated, and tubulin as a loading control. A) SHP-2, a known TLT-1 interactor, exhibits H2O2-induced binding to TLT-1. SHP-2 expression levels are shown in the α-MYC IP, as the signal was not observed in the WCE. B) TLT-1 exhibits H2O2-induced binding to RABs 5A, 7, and 11A. C) The TLT-1/RACK-1 interaction is independent of H2O2 stimulation. TLT-1 signal in the α-FLAG blot of the α-V5 IP is indicated by a triangle. The asterisk denotes a background signal from the heavy chain of the antibody. The α-V5 blot of the α-V5 IP was obtained on a 15% gel to separate the signal from the light chain of the antibody. D) The TLT-1/ITGB3 (β3-integrin) interaction is independent of H2O2 stimulation. E) GRB2 binds to TLT-1 in a H2O2-dependent manner. TLT-1-MYC-FLAG was expressed alone in 293 cells, and immunoprecipitates (α-FLAG) were blotted for endogenous GRB2 (α-GRB2). F) TLT-1 and ITGB3 interact in resting platelets. TLT-1 was immunoprecipitated (α-TLT-1) from resting and active (+ collagen) platelets. An isotype control (IC) was employed to ensure ITGB3 was binding in a specific manner. G) TLT-1 and GRB2 interact in active platelets. TLT-1 was immunoprecipitated (α-TLT-1) from resting and active (+ collagen) platelets.
4. Discussion
Herein, we report the identification of several novel TLT-1 interactors in resting and active platelets as well as 293 cells, revealing several proteins that are known to play important roles in platelet aggregation (Figure 2, Tables 1 & 2). Of these, we confirmed seven TLT-1 interactors, all known to be essential to platelet function during activation, through co-immunoprecipitation and immunoblotting (Figure 4). These included three Rab proteins, the adaptors GRB2 and RACK1, and β3-integrin (Figure 4B–E), in addition to the known TLT-1 interactor SHP-2 (Figure 4A). Interestingly, several of these novel interactors exhibited regulated binding to TLT-1 in the presence/absence of H2O2, suggesting that TLT-1 post-translational modification (i.e. phosphorylation) may be required to induce their interaction.
Rab proteins are small G proteins that orchestrate vesicular trafficking, a mechanism that is central to the release of active molecules from intracellular granules to drive platelet aggregation, coagulation, and defense against pathogens. Several Rab proteins exhibited differential binding across resting and active platelets (Tables 1 & 2), likely the result of a Rab shift from the population coating intracellular granules to Rabs involved in granule exocytosis following platelet activation. Rabs 5, 7, and 11 are all pre-packaged into platelets from megakaryocytes, although RAB11 and RAB7 are thought to be represented at higher levels than RAB5 [22]. RAB11 is involved in the recycling of vesicles to the plasma membrane, while RAB5 family members (A–C) regulate endocytosis and trafficking through early endosomes and RAB7A traffics late endosomes and multivesicular bodies [22–25].
Interestingly, RAB 11 and 7 were found in complex with TLT-1 in resting platelets, while 7 and 5 were bound to TLT-1 in activated platelets. This may reflect the new paradigm of granule release which suggests that few granules actually fuse with the plasma membrane. Granules have been demonstrated to fuse to other granules upon activation. Cargo from the fused granules is released though a small neck or tube-like structure of the few granules that fuse to the plasma membrane. Thus, trafficking in platelets may have slightly different dynamic than seen in with the constitutive secretion pathways of cell.
The RAB/TLT-1 interactions tested in 293 cells revealed H2O2-regulated binding to TLT-1. Given that Rab proteins do not possess phosphorylation-dependent binding domains, these are not likely direct interactions between TLT-1 phosphorylated residues and Rab family members. Whether these interactions are phosphorylation-dependent or the result of the activation of multiple signaling pathways following H2O2 treatment will require further investigation in future studies.
The β3 subunit of the β3-integrin family was identified as a TLT-1 binding partner in resting platelets. Although both β3 heterodimers, αvβ3 and αIIbβ3, are packaged into platelets, the αIIbβ3 is represented at higher levels and serves as a major mediator of platelet aggregation following activation [26, 27]. In resting platelets, granules serve as a reservoir of a subset of the αIIbβ3 heterodimer population, although the majority of the αIIbβ3 pool is localized to the plasma membrane. Upon platelet activation, granular αIIbβ3 integrins are rapidly transported to the membrane to increase surface localization of αIIbβ3, amplifying platelet aggregation [28, 29]. Similarly, TLT-1 is known to play a role in platelet aggregation [5–7]. The identification of the TLT-1/β3 interaction in resting platelets may indicate a concerted role of TLT-1 and β3-containing integrin heterodimers in facilitating platelet aggregation following activation. This interaction was also validated in 293 cells, but was found to be phosphorylation-independent (Figure 4D).
RACK1 (also GNB2L1), which was also identified in immune complex with TLT-1 in 293 cells (Supplementary Table 7), is known to associate with β3-integrins and facilitate signal transduction through β3-integrins during platelet aggregation [30]. RACK1 possesses seven WD repeats, which complex phosphoserine/threonine residues on target proteins. This allows RACK1 to act as an adaptor protein, facilitating protein complex assembly at signaling hubs. Canonically, RACK1 binds phosphorylated residues of various active protein kinase C (PKC) isoforms, bringing PKC in proximity to its substrates. RACK1 has been shown to constitutively bind β3-integrins, linking integrin complexes to the β isoform of activated PKC (PKCβ) [30]. The β3/RACK1/PKCβ complex is required for platelet spreading during platelet aggregation [30]. Like β3-integrin, RACK1 did not exhibit differential binding to TLT-1 in the presence and absence of H2O2 (Figure 4C). RACK1 could act as an adaptor between TLT-1 and other interactors, for example β3-integrin, given the identification of intracellular TLT-1 phosphoserine and phosphothreonine residues, which remained phosphorylated in the presence and absence of H2O2 (Supplementary Table 6).
GRB2, another protein identified in immune complex with TLT-1 in activated platelets (Figure 4G) and H2O2-stimulated 293 cells (Figure 4E, Supplementary Table 7), is also known to associate with β3-integrin, as well as the TLT-1 interactor SHP1 [31]. GRB2 is an adaptor protein containing one SH2 domain flanked by dual N- and C-terminal SH3 domains that is involved in a variety of cellular processes related to Ras signaling [32]. The GRB2 SH3 domains bind signaling molecules such and the Ras-GEF Son of Sevenless (SOS), transporting these GEFs to phosphotyrosine residues through the GRB2 SH2-binding motif, which induces proximal Ras-GTPase activity and downstream signaling [33–36]. GRB2 is involved in thrombin-induced platelet aggregation, where it forms complexes with β3-integrin via the GRB2 SH2 domain, bringing SH3-bound signaling proteins to the integrin complex [31]. The phospho-dependent recruitment of GRB2 to the TLT-1 complex further supports the participation of TLT-1 in the β3-integrin complex, along with RACK1 and SHP2, forming a functional complex in platelet activation.
Previously, the TLT-1 interactors moesin, ezrin, and radixin were identified bound to TLT-1 in platelets following two minutes of activation [6]. Interestingly. these proteins were not found in immune complex with TLT-1 in this study. However, the activation times prior to platelet lysis were markedly different between the two studies; platelet activation in the previous study was only two minutes [6], in comparison to the ten-minute activation employed here. Moesin, ezrin, and radixin are likely important TLT-1 interactors at early time points following platelet activation, and dissociate between two and ten minutes of platelet activation.
Given the lack of TLT-1 enzymatic activity, post-translational modification of TLT-1 is likely central to its function in platelet aggregation. Here, we report multiple phosphorylation sites on TLT-1 in 293 cells and platelets, as well as a novel threonine phosphorylation site near the C-terminal ITIM. Phosphorylation of Thr280 was found to be regulated by serine/threonine phosphatases (Figure 3H), which is conserved in mouse and human sequences (Supplementary Figure 1). The proximity of this conserved residue to the ITIM tyrosine suggests that the TLT-1 C-terminal tail could be an important regulatory region of the protein. Phosphorylation of Thr280 could act to promote or inhibit interactions of ITIM-binding proteins, given its location immediately N-terminal to the ITIM tyrosine. In addition to phosphorylation, we observed a truncated form of TLT-1 in H2O2-treated cells via Western blotting (Figure 3A), signifying a potential cleavage product. A secreted form of TLT-1 with functional implications in platelet activation has been described [6, 7, 12], and the observations reported here could demonstrate a phosphorylation-regulated release mechanism of the TLT-1 ectodomain. Peptides from the truncated form were not identified with mass spectrometry, although this likely reflects the differences in sensitivity between Western blotting and LC-MS/MS. Shedding of the TLT-1 ectodomain has is at least partially regulated by the metalloprotease ADAM17 [21], which may indicate that TLT-1 tyrosine phosphorylation is an important step in the release of soluble TLT-1 from the membrane-bound precursor.
Conclusions
Using LC-MS/MS methodology, we have mapped the TLT-1 interactome in resting and active platelets. The set of proteins found to interact with TLT-1 was enriched in proteins involved in cell adhesion and migration, as well as platelet activation (Figure 2). Select interactors were validated via immunoblotting in co-immunoprecipitation experiments (Figure 4). Specifically, we have demonstrated H2O2-regulated binding of novel TLT-1 interactors GRB2, RAB5A, RAB7, and RAB11A (Figure 4B), as well as the known TLT-1 phosphotyrosine-dependent interactor SHP2 (Figure 4A), and H2O2-indifferent binding of RACK1 and β3-integrin (Figure 4C & D) in 293 cells. We have also validated the TLT-1/β3-integrin and TLT-1/GRB2 interactions in resting and active platelets using Western blotting (Figure 4E). Additionally, we have identified Thr280 as a novel, regulated TLT-1 phosphorylation on a conserved residue near the TLT-1 ITIM sequence (Figure 3), suggesting it may be an important regulator of TLT-1 function, likely through affecting the binding of TLT-1 interactors. This study defines several novel TLT-1 interactors with likely signaling implications in platelet aggregation dynamics.
Supplementary Material
Supplementary Figure 1. Alignment of human and mouse TLT-1 sequences. Mouse and human TLT-1 sequences are 64.64% conserved. The extracellular V-set domain and the transmembrane domain regions are underlined, and the ITIM motif is highlighted in the purple box. Numbers under residues in bold denote PhosphoSitePlus (phosphosite.org) curations of experiments in which the given residue was found phosphorylated in large- and small-scale proteomics experiments. Alignments were generated with the Uniprot alignment tool.
Supplementary Figure 2. Fragmentation spectra of TLT-1 phosphorylation sites on human and mouse sequences, as well as the corresponding unphosphorylated peptides. (A-I) Post-translationally modified sites are indicated as red residues in the peptides sequences. Highlighted methionines are oxidized (+15.9949 Da), cysteines have acrylamide adducts (+71.0371 Da), and serine, threonine, or tyrosine residues are phosphorylated (+ 79.9663 Da).
Supplementary Figure 3. TLT-1 Y281 is phosphorylated in activated platelets and platelets treated with H2O2. TLT-1 was immunoprecipitated from activated human platelets or platelets treated with 8 mM H2O2. Immunoprecipitates were analyzed by SDS-PAGE and Coomassie staining. TLT-1 bands were excised and proteins were subjected to reduction and alkylation prior to tryptic digest. Peptides were analyzed on a Q Exactive Plus mass spectrometer, targeting the mass of the human peptides containing the ITIM tyrosine (Y281) and proximal threonine (T280) with the sequences VLVC^SKPVTYATVIFPGGNK (m/z=717.3941, z=3+) and VLVC^SKPVTY@ATVIFPGGNK ((m/z=744.0496, z=3+) where ^ and @ denote carbamidomethylation and phosphorylation, respectively. An annotated spectrum of the unphosphorylated peptide (A) is shown alongside spectra of the phosphopeptides identified in H2O2 treated platelets (B) as well as activated platelets (C). Notably, there was considerable background in the spectrum of the phosphopeptide obtained from activated platelets (C) due to a co-eluting ion within the isolation window used for fragmentation. Nonetheless, the presence of characteristic fragments confirm that the phosphopeptide is correctly identified. Fragments confirming phosphorylation site localization on the ITIM tyrosine (Y281) in (B) are denoted by boxed fragment annotations. Although fragments distinguishing between phosphorylation of Y281, T280, and S276 were not identified in the spectrum of the phosphopeptide in activated platelets (C), the elution time of the phosphopeptide in activated platelets was the same as the confirmed pY281-containing species in H2O2-treated platelets (D & E). Given our previous identifications of the various ITIM-containing phosphopeptides from mouse, which exhibited markedly different elution times (pY at 35% MeCN, pS at 40% MeCN, and pT at 45% MeCN), we conclude that the phosphorylated species identified in activated platelets contains pY281.
Supplementary Figure 4. TLT-1 T280 is phosphorylated in platelets following platelet activation. TLT-1 was immunoprecipitated from platelets at 0, 1, 3, 5, and 10 minutes following activation. Immunoprecipitates were analyzed by SDS-PAGE and Western blotting for TLT-1 (α-TLT1) and TLT-1 pT280 (α-T280TLT1). Phosphorylation of TLT-1 T280 is detected by Western blotting after five minutes of platelet activation and becomes more robust after 10 minutes of activation.
Supplementary Figure 5. Functional characterization and gene ontology (GO) enrichment analysis of TLT-1 interactors in 293 cells. A) 81% of proteins identified in complex with TLT-1 in 293 cells are present in platelets. TLT-1 interactors were searched in the Platelet Web database [20] for expression detected at the protein levels in platelets. B) TLT-1 interactors are categorized by general function. C) A GO enrichment analysis was conducted using the Metascape platform [19]. GO terms with P < 10−3 are shown. Genes within each enriched group are tabulated in Supplementary Table 8.
Supplementary Table 1. Peptides identified in each experimental replicate from protein complexes in anti-TLT-1 immunoprecipitations from human platelets. RAW spectra were searched against a forward and reverse human protein database via SEQUEST requiring tryptic cleavages and peptides were filtered by mass accuracy (± 4 ppm) and unique deltaCorr (≥ 0.15). All peptides from proteins present in the control immunoprecipitations were removed, unless spectral counts were enriched in experimental conditions over the control by a factor of five or more. Colors indicate experimental condition (see key to the right). Post-translational modifications are given by the following characters: * = oxidation of methionine, ^ = acrylamidation of cysteine, and @ = phosphorylation of serine, threonine, or tyrosine.
Supplementary Table 2. Peptides identified in each experimental replicate from protein complexes in anti-FLAG immunoprecipitations from 293 cells expressing mouse TLT-1-FLAG. RAW spectra were searched against a forward and reverse human protein database via SEQUEST requiring tryptic cleavages and peptides were filtered by mass accuracy (± 4 ppm) and unique deltaCorr (≥ 0.15). All peptides from proteins present in the control immunoprecipitations were removed, unless spectral counts were enriched in experimental conditions over the control by a factor of five or more. Colors indicate experimental condition (see key to the right). Post-translational modifications are given by the following characters: * = oxidation of methionine, ^ = acrylamidation of cysteine, and @ = phosphorylation of serine, threonine, or tyrosine.
Supplementary Table 3. Human TLT-1 peptides identified in each experimental replicate from anti-TLT-1 immunoprecipitations from human platelets. RAW spectra were searched against a forward and reverse human TLT-1 protein sequence via SEQUEST with no enzyme specified. Peptides were filtered by mass accuracy (± 4 ppm) and to include only tryptic cleavages. Colors indicate experimental condition (see key to the right). Post-translational modifications are given by the following characters: * = oxidation of methionine, ^ = acrylamidation of cysteine, and @ = phosphorylation of serine, threonine, or tyrosine.
Supplementary Table 4. Mouse TLT-1 peptides identified in each experimental replicate from anti-FLAG immunoprecipitations from 293 cells expressing mouse TLT-1-FLAG. RAW spectra were searched against a forward and reverse mouse TLT-1 protein sequence via SEQUEST with no enzyme specified. Peptides were filtered by mass accuracy (± 4 ppm) and to include only tryptic cleavages. Colors indicate experimental condition (see key to the right). Post-translational modifications are given by the following characters: * = oxidation of methionine, ^ = acrylamidation of cysteine, and @ = phosphorylation of serine, threonine, or tyrosine.
Supplementary Table 5. Enriched GO terms in TLT-1 protein complexes in platelets. GO enrichment analyses were conducted via the Metascape (metascape.org) platform.
Supplementary Table 6. Phosphorylation sites identified on mouse and human TLT-1. Spectra obtained from LC-MS/MS runs of TLT-1 immunoprecipitations from platelets and 293 cells were subjected to a second pass through the SEQUEST algorithm, in which the human (platelets) or mouse (ectopic expression in 293 cells) TLT-1 sequence with no enzyme specified. Annotated spectra are included in Figure 3 and Supplementary Figure 2. NT = no treatment, @ = phosphorylation (+ 79.966 Da), * = oxidation (+ 15.995 Da), ^ = acrylamidation.
Supplementary Table 7. TLT-1 binding partners identified in stimulated (H2O2) and unstimulated 293 cells. Spectral counts of TLT-1 interactors identified in treated and untreated conditions were obtained from peptide tables in Supplementary Table 2. TLT-1 peptides can be found in Supplementary Table 4.
Supplementary Table 8. Enriched GO terms in TLT-1 protein complexes in 293 cells. GO enrichment analyses were conducted via the Metascape (metascape.org) platform.
Significance.
Platelet function relies on the secretion of active molecules from intracellular vesicles, or granules, which contain soluble and membrane-bound proteins that are essential for platelet aggregation, coagulation reactions, and pathogen defense mechanisms. TLT-1 is sequestered in α-granules and transported to the plasma membrane, where it plays a unique role in hemostasis after inflammatory insults. Despite the known importance of TLT-1 in platelet biology, our knowledge of TLT-1 mechanistic signaling is limited. This study defines the TLT-1 interactome in resting and active human platelets, identifying several novel TLT-1 interactors, as well as TLT-1 phosphorylation sites, all with likely signaling implications in platelet aggregation dynamics.
Highlights.
Novel TLT-1 binding partners were identified in resting and activated human platelets.
The interaction of GRB2, RACK1, β3-integrin, and Rabs 5, 7, and 11 with TLT-1 were confirmed by co-immunoprecipitation and immunoblotting.
Regulated phosphorylation, including the novel Thr280 site near the ITIM were identified.
Funding
This work was supported by the U.S. National Science Foundation (NSF) grants IOS 1656510 and DBI REU 1262786; the Vermont Genetics Network through U. S. National Institutes of Health (NIH) Grant 8P20GM103449 from the INBRE program of the NIGMS; and U.S. NIH Grant 5P20RR016435 from the COBRE program of the NIGMS, and Grant HL090933. Additional support was provided by the University of Puerto Rico Clinical Cancer Center and U.S. NIH Grants R21 HL140268, P20RR016470 and P20GM103475 from the INBRE program of NIGMS, U54 MD007600 and U54NS043011 from the Research Centers for Minority Institutions (RCMI) program of the NIMHD and the Research Initiative for Scientific Enhancement (RISE) 2R25GM061838 and 8G12MD007600 of NIGMS. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Abbreviations
- ADP
adenosine diphosphate
- FcεRI
high affinity immunoglobulin epsilon receptor
- GRB2
growth factor receptor-bound protein 2
- HEK
human embryonic kidney
- GEF
guanine exchange factor
- i.d.
inner diameter
- Ig
immunoglobulin
- ITAM
immunoreceptor tyrosine-based activating motif
- ITGB3
β3-integrin
- ITIM
immunoreceptor tyrosine-based inhibitory motif
- LC-MS/MS
liquid chromatography/tandem mass spectrometry
- PKC
protein kinase C
- RAB
Ras-related protein
- RACK1
receptor of activated protein C kinase 1
- SDS-PAGE
sodium dodecyl sulfate–polyacrylamide gel electrophoresis
- SH2
Src homology 2
- SHP1
SH2 domain-containing tyrosine phosphatase 1
- SHP2
SH2 domain-containing tyrosine phosphatase 2
- TLT-1
TREM-like transcript 1
- TREM
triggering receptor expressed of myeloid cells
- TREML1
TREM-like 1
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest
None declared.
REFERENECES
- [1].Golebiewska EM, Poole AW, Secrets of platelet exocytosis–what do we really know about platelet secretion mechanisms?, British journal of haematology 165(2) (2014) 204–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Blair P, Flaumenhaft R, Platelet α-granules: basic biology and clinical correlates, Blood reviews 23(4) (2009) 177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Estevez B, Du X, New concepts and mechanisms of platelet activation signaling, Physiology 32(2) (2017) 162–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Ford JW, McVicar DW, TREM and TREM-like receptors in inflammation and disease, Current opinion in immunology 21(1) (2009) 38–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Washington AV, Schubert RL, Quigley L, Disipio T, Feltz R, Cho EH, McVicar DW, A TREM family member, TLT-1, is found exclusively in the α-granules of megakaryocytes and platelets, Blood 104(4) (2004) 1042–1047. [DOI] [PubMed] [Google Scholar]
- [6].Washington AV, Gibot S, Acevedo I, Gattis J, Quigley L, Feltz R, De La Mota A, Schubert RL, Gomez-Rodriguez J, Cheng J, TREM-like transcript-1 protects against inflammation-associated hemorrhage by facilitating platelet aggregation in mice and humans, The Journal of clinical investigation 119(6) (2009) 1489–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Giomarelli B, Washington VA, Chisholm MM, Quigley L, McMahon JB, Mori T, McVicar DW, Inhibition of thrombin-induced platelet aggregation using human single-chain Fv antibodies specific for TREM-like transcript-1, Thrombosis and haemostasis 97(06) (2007) 955–963. [PubMed] [Google Scholar]
- [8].Barrow AD, Astoul E, Floto A, Brooke G, Relou IA, Jennings NS, Smith KG, Ouwehand W, Farndale RW, Alexander DR, Cutting edge: TREM-like transcript-1, a platelet immunoreceptor tyrosine-based inhibition motif encoding costimulatory immunoreceptor that enhances, rather than inhibits, calcium signaling via SHP-2, The Journal of Immunology 172(10) (2004) 5838–5842. [DOI] [PubMed] [Google Scholar]
- [9].Campbell KS, Colonna M, DAP12: a key accessory protein for relaying signals by natural killer cell receptors, The international journal of biochemistry & cell biology 31(6) (1999) 631–636. [DOI] [PubMed] [Google Scholar]
- [10].Washington AV, Quigley L, McVicar DW, Initial characterization of TREM-like transcript (TLT)–1: a putative inhibitory receptor within the TREM cluster, Blood 100(10) (2002) 3822–3824. [DOI] [PubMed] [Google Scholar]
- [11].Billadeau DD, Leibson PJ, ITAMs versus ITIMs: striking a balance during cell regulation, The Journal of clinical investigation 109(2) (2002) 161–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Morales J, Villa K, Gattis J, Castro W, Colon K, Lubkowski J, Sanabria P, Hunter R, Washington AV, Soluble TLT-1 modulates platelet-endothelial cell interactions and actin polymerization, Blood coagulation & fibrinolysis: an international journal in haemostasis and thrombosis 21(3) (2010) 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Glembotsky AC, Sliwa D, Bluteau D, Balayn N, Oyarzún CPM, Raimbault A, Bordas M, Droin N, Pirozhkova I, Washington V, Downregulation of TREM-like transcript-1 and collagen receptor α2 subunit, two novel RUNX1-targets, contributes to platelet dysfunction in familial platelet disorder with predisposition to acute myelogenous leukemia, haematologica 104(6) (2019) 1244–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Cazenave J-P, Ohlmann P, Cassel D, Eckly A, Hechler B, Gachet C, Preparation of washed platelet suspensions from human and rodent blood, Platelets and megakaryocytes, Springer2004, pp. 13–28. [DOI] [PubMed] [Google Scholar]
- [15].Consortium U, UniProt: a worldwide hub of protein knowledge, Nucleic acids research 47(D1) (2018) D506–D515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Hornbeck PV, Zhang B, Murray B, Kornhauser JM, Latham V, Skrzypek E, PhosphoSitePlus, 2014: mutations, PTMs and recalibrations, Nucleic acids research 43(D1) (2014) D512–D520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Apweiler R, Bairoch A, Wu CH, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, UniProt: the universal protein knowledgebase, Nucleic acids research 32(suppl_1) (2004) D115–D119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Chatr-Aryamontri A, Oughtred R, Boucher L, Rust J, Chang C, Kolas NK, O’Donnell L, Oster S, Theesfeld C, Sellam A, The BioGRID interaction database: 2017 update, Nucleic acids research 45(D1) (2017) D369–D379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Tripathi S, Pohl MO, Zhou Y, Rodriguez-Frandsen A, Wang G, Stein DA, Moulton HM, DeJesus P, Che J, Mulder LC, Meta-and orthogonal integration of influenza “OMICs” data defines a role for UBR4 in virus budding, Cell host & microbe 18(6) (2015) 723–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Boyanova D, Nilla S, Birschmann I, Dandekar T, Dittrich M, PlateletWeb: a systems biologic analysis of signaling networks in human platelets, Blood 119(3) (2012) e22–e34. [DOI] [PubMed] [Google Scholar]
- [21].Fong KP, Barry C, Tran AN, Traxler EA, Wannemacher KM, Tang H-Y, Speicher KD, Blair IA, Speicher DW, Grosser T, Deciphering the human platelet sheddome, Blood 117(1) (2011) e15–e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Walsh TG, Li Y, Wersäll A, Poole AW, Small GTPases in platelet membrane trafficking, Platelets (2018) 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Takahashi S, Kubo K, Waguri S, Yabashi A, Shin H-W, Katoh Y, Nakayama K, Rab11 regulates exocytosis of recycling vesicles at the plasma membrane, J Cell Sci 125(17) (2012) 4049–4057. [DOI] [PubMed] [Google Scholar]
- [24].Rastogi R, Verma JK, Kapoor A, Langsley G, Mukhopadhyay A, Rab5 isoforms specifically regulate different modes of endocytosis in Leishmania, Journal of Biological Chemistry 291(28) (2016) 14732–14746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Bastin G, Heximer SP, Rab family proteins regulate the endosomal trafficking and function of RGS4, Journal of Biological Chemistry 288(30) (2013) 21836–21849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Bennett J, Berger B, Billings P, The structure and function of platelet integrins, Journal of Thrombosis and Haemostasis 7 (2009) 200–205. [DOI] [PubMed] [Google Scholar]
- [27].Bennett JS, Structure and function of the platelet integrin α IIb β 3, The Journal of clinical investigation 115(12) (2005) 3363–3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Niiya K, Hodson E, Bader R, Byers-Ward V, Koziol JA, Plow EF, Ruggeri ZM, Increased surface expression of the membrane glycoprotein IIb/IIIa complex induced by platelet activation. Relationship to the binding of fibrinogen and platelet aggregation, Blood 70(2) (1987) 475–483. [PubMed] [Google Scholar]
- [29].Durrant TN, van den Bosch MT, Hers I, Integrin αIIbβ3 outside-in signaling, Blood 130(14) (2017) 1607–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Buensuceso CS, Obergfell A, Soriani A, Eto K, Kiosses WB, Arias-Salgado EG, Kawakami T, Shattil SJ, Regulation of outside-in signaling in platelets by integrin-associated protein kinase Cβ, Journal of Biological Chemistry 280(1) (2005) 644–653. [DOI] [PubMed] [Google Scholar]
- [31].Abdelhafid S, Wang-Qing L, Vidal M, Garbay C, Rendu F, Bachelot-Loza C, Differential effect of the inhibition of Grb2–SH3 interactions in platelet activation induced by thrombin and by Fc receptor engagement, Biochemical Journal 363(3) (2002) 717–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Lowenstein E, Daly R, Batzer A, Li W, Margolis B, Lammers R, Ullrich A, Skolnik E, Bar-Sagi D, Schlessinger J, The SH2 and SH3 domain-containing protein GRB2 links receptor tyrosine kinases to ras signaling, Cell 70(3) (1992) 431–442. [DOI] [PubMed] [Google Scholar]
- [33].Buday L, Downward J, Epidermal growth factor regulates p21ras through the formation of a complex of receptor, Grb2 adapter protein, and Sos nucleotide exchange factor, Cell 73(3) (1993) 611–620. [DOI] [PubMed] [Google Scholar]
- [34].Baltensperger K, Kozma LM, Cherniack AD, Klarlund JK, Chawla A, Banerjee U, Czech MP, Binding of the Ras activator son of sevenless to insulin receptor substrate-1 signaling complexes, Science 260(5116) (1993) 1950–1952. [DOI] [PubMed] [Google Scholar]
- [35].Gale NW, Kaplan S, Lowenstein EJ, Schlessinger J, Bar-Sagi D, Grb2 mediates the EGF-dependent activation of guanine nucleotide exchange on Ras, Nature 363(6424) (1993) 88. [DOI] [PubMed] [Google Scholar]
- [36].Li N.a., Batzer A, Daly R, Yajnik V, Skolnik E, Chardin P, Bar-Sagi D, Margolis B, Schlessinger J, Guanine-nucleotide-releasing factor hSos1 binds to Grb2 and links receptor tyrosine kinases to Ras signalling, Nature 363(6424) (1993) 85. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Alignment of human and mouse TLT-1 sequences. Mouse and human TLT-1 sequences are 64.64% conserved. The extracellular V-set domain and the transmembrane domain regions are underlined, and the ITIM motif is highlighted in the purple box. Numbers under residues in bold denote PhosphoSitePlus (phosphosite.org) curations of experiments in which the given residue was found phosphorylated in large- and small-scale proteomics experiments. Alignments were generated with the Uniprot alignment tool.
Supplementary Figure 2. Fragmentation spectra of TLT-1 phosphorylation sites on human and mouse sequences, as well as the corresponding unphosphorylated peptides. (A-I) Post-translationally modified sites are indicated as red residues in the peptides sequences. Highlighted methionines are oxidized (+15.9949 Da), cysteines have acrylamide adducts (+71.0371 Da), and serine, threonine, or tyrosine residues are phosphorylated (+ 79.9663 Da).
Supplementary Figure 3. TLT-1 Y281 is phosphorylated in activated platelets and platelets treated with H2O2. TLT-1 was immunoprecipitated from activated human platelets or platelets treated with 8 mM H2O2. Immunoprecipitates were analyzed by SDS-PAGE and Coomassie staining. TLT-1 bands were excised and proteins were subjected to reduction and alkylation prior to tryptic digest. Peptides were analyzed on a Q Exactive Plus mass spectrometer, targeting the mass of the human peptides containing the ITIM tyrosine (Y281) and proximal threonine (T280) with the sequences VLVC^SKPVTYATVIFPGGNK (m/z=717.3941, z=3+) and VLVC^SKPVTY@ATVIFPGGNK ((m/z=744.0496, z=3+) where ^ and @ denote carbamidomethylation and phosphorylation, respectively. An annotated spectrum of the unphosphorylated peptide (A) is shown alongside spectra of the phosphopeptides identified in H2O2 treated platelets (B) as well as activated platelets (C). Notably, there was considerable background in the spectrum of the phosphopeptide obtained from activated platelets (C) due to a co-eluting ion within the isolation window used for fragmentation. Nonetheless, the presence of characteristic fragments confirm that the phosphopeptide is correctly identified. Fragments confirming phosphorylation site localization on the ITIM tyrosine (Y281) in (B) are denoted by boxed fragment annotations. Although fragments distinguishing between phosphorylation of Y281, T280, and S276 were not identified in the spectrum of the phosphopeptide in activated platelets (C), the elution time of the phosphopeptide in activated platelets was the same as the confirmed pY281-containing species in H2O2-treated platelets (D & E). Given our previous identifications of the various ITIM-containing phosphopeptides from mouse, which exhibited markedly different elution times (pY at 35% MeCN, pS at 40% MeCN, and pT at 45% MeCN), we conclude that the phosphorylated species identified in activated platelets contains pY281.
Supplementary Figure 4. TLT-1 T280 is phosphorylated in platelets following platelet activation. TLT-1 was immunoprecipitated from platelets at 0, 1, 3, 5, and 10 minutes following activation. Immunoprecipitates were analyzed by SDS-PAGE and Western blotting for TLT-1 (α-TLT1) and TLT-1 pT280 (α-T280TLT1). Phosphorylation of TLT-1 T280 is detected by Western blotting after five minutes of platelet activation and becomes more robust after 10 minutes of activation.
Supplementary Figure 5. Functional characterization and gene ontology (GO) enrichment analysis of TLT-1 interactors in 293 cells. A) 81% of proteins identified in complex with TLT-1 in 293 cells are present in platelets. TLT-1 interactors were searched in the Platelet Web database [20] for expression detected at the protein levels in platelets. B) TLT-1 interactors are categorized by general function. C) A GO enrichment analysis was conducted using the Metascape platform [19]. GO terms with P < 10−3 are shown. Genes within each enriched group are tabulated in Supplementary Table 8.
Supplementary Table 1. Peptides identified in each experimental replicate from protein complexes in anti-TLT-1 immunoprecipitations from human platelets. RAW spectra were searched against a forward and reverse human protein database via SEQUEST requiring tryptic cleavages and peptides were filtered by mass accuracy (± 4 ppm) and unique deltaCorr (≥ 0.15). All peptides from proteins present in the control immunoprecipitations were removed, unless spectral counts were enriched in experimental conditions over the control by a factor of five or more. Colors indicate experimental condition (see key to the right). Post-translational modifications are given by the following characters: * = oxidation of methionine, ^ = acrylamidation of cysteine, and @ = phosphorylation of serine, threonine, or tyrosine.
Supplementary Table 2. Peptides identified in each experimental replicate from protein complexes in anti-FLAG immunoprecipitations from 293 cells expressing mouse TLT-1-FLAG. RAW spectra were searched against a forward and reverse human protein database via SEQUEST requiring tryptic cleavages and peptides were filtered by mass accuracy (± 4 ppm) and unique deltaCorr (≥ 0.15). All peptides from proteins present in the control immunoprecipitations were removed, unless spectral counts were enriched in experimental conditions over the control by a factor of five or more. Colors indicate experimental condition (see key to the right). Post-translational modifications are given by the following characters: * = oxidation of methionine, ^ = acrylamidation of cysteine, and @ = phosphorylation of serine, threonine, or tyrosine.
Supplementary Table 3. Human TLT-1 peptides identified in each experimental replicate from anti-TLT-1 immunoprecipitations from human platelets. RAW spectra were searched against a forward and reverse human TLT-1 protein sequence via SEQUEST with no enzyme specified. Peptides were filtered by mass accuracy (± 4 ppm) and to include only tryptic cleavages. Colors indicate experimental condition (see key to the right). Post-translational modifications are given by the following characters: * = oxidation of methionine, ^ = acrylamidation of cysteine, and @ = phosphorylation of serine, threonine, or tyrosine.
Supplementary Table 4. Mouse TLT-1 peptides identified in each experimental replicate from anti-FLAG immunoprecipitations from 293 cells expressing mouse TLT-1-FLAG. RAW spectra were searched against a forward and reverse mouse TLT-1 protein sequence via SEQUEST with no enzyme specified. Peptides were filtered by mass accuracy (± 4 ppm) and to include only tryptic cleavages. Colors indicate experimental condition (see key to the right). Post-translational modifications are given by the following characters: * = oxidation of methionine, ^ = acrylamidation of cysteine, and @ = phosphorylation of serine, threonine, or tyrosine.
Supplementary Table 5. Enriched GO terms in TLT-1 protein complexes in platelets. GO enrichment analyses were conducted via the Metascape (metascape.org) platform.
Supplementary Table 6. Phosphorylation sites identified on mouse and human TLT-1. Spectra obtained from LC-MS/MS runs of TLT-1 immunoprecipitations from platelets and 293 cells were subjected to a second pass through the SEQUEST algorithm, in which the human (platelets) or mouse (ectopic expression in 293 cells) TLT-1 sequence with no enzyme specified. Annotated spectra are included in Figure 3 and Supplementary Figure 2. NT = no treatment, @ = phosphorylation (+ 79.966 Da), * = oxidation (+ 15.995 Da), ^ = acrylamidation.
Supplementary Table 7. TLT-1 binding partners identified in stimulated (H2O2) and unstimulated 293 cells. Spectral counts of TLT-1 interactors identified in treated and untreated conditions were obtained from peptide tables in Supplementary Table 2. TLT-1 peptides can be found in Supplementary Table 4.
Supplementary Table 8. Enriched GO terms in TLT-1 protein complexes in 293 cells. GO enrichment analyses were conducted via the Metascape (metascape.org) platform.


