Abstract
Plant-inhabiting microorganisms interact directly with each other affecting disease progression. However, the role of host plant and plant habitat in shaping pathobiome composition and their implication for host susceptibility/resistance to a particular disease are currently unknown. For the elucidation of these questions, both epiphytic and endophytic bacterial communities, present in asymptomatic and symptomatic twigs from olive cultivars displaying different susceptibilities to olive knot (OK) disease, were investigated using culturing methods. OK disease was the main driver of the bacterial community, causing changes on their diversity, abundance and composition. OK disease effect was most notorious on OK-susceptible cultivar and when considering the endophytic communities. Plant habitat (epiphytes vs. endophytes) also contributed to the bacterial community assembling, in particular on symptomatic twigs (knots) of OK-susceptible cultivar. In contrast, host cultivar had little effect on the bacterial community composition, but OK-symptomatic twigs (knots) revealed to be more affected by this driver. Overall, the pathobiome seems to result from an intricate interaction between the pathogen, the resident bacteria, and the plant host. Specific bacterial genera were associated to the presence or absence of OK disease in each cultivar. Their ability to trigger and/or suppress disease should be studied in the future.
Subject terms: Bacteria, Microbial communities
Introduction
It is now well established that plants harbor a complex microbial community (microbiota) that provides numerous health benefits1. From the various mechanisms employed by microbes to improve host plant health, microbe-microbe interactions seem to play fundamental roles2. Indeed, there are some studies indicating that within plant microbiota, pathogens can establish multiple interactions, either positive or negative, with other microorganisms that may trigger or influence the disease process3,4. Such microbial consortium, which play a direct role on the progression of disease, has been recently termed as pathobiome3. Although in this concept the pathogenic agent has been regarded as integrated within this biotic environment3, the host plant role in shaping the pathobiome and its implication for host susceptibility/resistance to a particular disease have not yet been studied. As the structure of plant-associated microbiota is plant genotype dependent5,6, we hypothesized that distinct microbial compositions among plant genotypes with diverse microbial interactions may lead to different pathobiomes. Another critical question is whether the pathobiome composition depends on plant habitat. Plant-associated microorganisms have the ability to colonize the surface (epiphytic) or the internal (endophytic) plant tissues1. However, whether the microbial interactions in the plant tissue surface may lead to different pathobiomes than those from the interior of the same plant tissues is still largely unknown. Microbiota comparisons (either epiphytic or endophytic) between healthy and diseased plant tissues, present in cultivars with contrasting susceptibility to diseases, could be helpful to elucidate these questions7. Such approach, besides providing new insights on the potential role of microbiota in plant resistance, could additionally contribute for the identification of microbial strains that could be used in the future as “probiotic”. The application of such probiotic microorganisms could drive the plant microbiota towards a pathogen-resistant microbial composition. In humans for instance, the faecal microbial transplantation has been largely recognized as a promising therapy to treat gastrointestinal diseases8.
Pseudomonas savastanoi pv. savastanoi (Pss) is the causal agent of the olive knot (OK) disease, which is one of the major threats to olive tree (Olea europaea L.) production in most olive growing regions of the world, in particular Mediterranean region9,10. Pss lives epiphytically on the surface of olive organs9, and under favorable weather conditions, Pss population increase and penetrate into olive tissues, leading to the formation of tumorous overgrowths10. These knots are deeply colonized by Pss microcolonies and comprise the main symptoms of OK disease, occurring mostly on olive tree twigs, branches and trunks11. Several non-pathogenic bacterial species from these knots have been reported to cooperate with the Pss for increasing disease severity12–14. Therefore, these knots can provide an excellent model system for studying the impact of plant host and plant habitat on the pathobiome structure. So far, no olive tree genotype has been found to be completely resistant to OK, but different olive cultivars exhibit different susceptibilities to OK disease15. For instances, among the most important Portuguese commercial olive cultivars, cv. Cobrançosa is less susceptible to OK than cv. Verdeal Transmontana16. Both cultivars can simultaneously display asymptomatic twigs and symptomatic twigs (with knots) in the same olive tree, also making this system suitable for studying the impact of host genotype on the “health” vs. “disease” (i.e., pathobiome) microbiota structure.
Here, we investigated the epiphytic and endophytic bacterial community of asymptomatic and symptomatic (knots) twigs, present in olive cultivars of different susceptibilities to OK (cvs. Cobrançosa and Verdeal Transmontana), using culturing methods followed by sequencing of the PCR amplicons of bacterial isolates. Such approach would allow to capture the complex pathogen-microbe-plant interactions and predict links between the cultured microbial community and disease/healthy states. The possible contribution of such links for the different susceptibilities of cultivars to OK disease could also be elucidated. Therefore, with this work we aim to answer the following questions: (i) May host cultivar shape the associated pathobiome community? (ii) Is the pathobiome composition dependent on the plant habitat (epiphytic vs. endophytic)? (iii) Is there any bacterial consortium specifically associated to asymptomatic (“healthy-promoting microbiota”) twigs or to knots (“disease-promoting microbiota”)? (iv) Are “healthy- or disease-promoting microbiota” linked to cultivar susceptibility to OK disease? The isolation of bacteria with a potential role on plant resistance to OK disease could be a first step for envisaging a biocontrol strategy for the inhibition of olive knot disease.
Results
The isolation of bacterial epiphytes and endophytes from asymptomatic and OK-symptomatic (knots) twigs from the 28 olive trees of both cultivars (Cobrançosa and Verdeal Transmontana) resulted in a total of 312 isolates. All isolates, corresponding to 66 operational taxonomic units (OTUs), belong to 31 genera and 17 families, mostly from the Proteobacteria and Actinobacteria phyla (76.3% and 18.2% of the total bacterial isolates, respectively) (Fig. S1). Considering all OTUs, 68.2% were found on the surface and 56.1% in the interior of plant tissues. The epiphytic bacterial communities were predominantly dominated by members belonging to genera Pseudomonas and Curtobacterium accounting together for 83.2% of total epiphytes, whereas Pseudomonas and Pantoea were dominant in the endophytic community, accounting together for 71.3% of the total endophytic isolates. Although all surveyed environments were colonized by Pss, the pathogen abundance was significantly higher in knots (OK-symptomatic twigs) than in asymptomatic twigs, either for epiphytic (2.0-fold, p < 0.001) or endophytic (2.1-fold, p < 0.001) communities (Table S1). Curiously, the more resistant cv. Cobrançosa presented higher abundance of Pss than the OK-susceptible cv. Verdeal Transmontana (1.9-fold, p < 0.001) within epiphytic community, but within the endophytic community was observed the opposite (a reduction of 1.4-fold, p < 0.001).
Comparison of bacterial communities
The abundance and diversity of bacteria differed between asymptomatic and OK-symptomatic (knots) twigs, depending also on the host cultivar and plant habitat (Figs. 1 and S2). Although a significant reduction (p < 0.001) in epiphytes abundance was detected from asymptomatic to OK-symptomatic twigs (up to 6.9-fold), an opposite result was observed for endophytes (an increase of 3.2-fold). This increase on endophytic abundance was significantly (p < 0.05) greater on cv. Cobrançosa (83.3%) when compared to cv. Verdeal Transmontana (52.9%). The bacterial diversity (determined by the species richness and Shannon-Wiener diversity index) was only significantly different between asymptomatic and OK-symptomatic twigs for the endophytic community. In cv. Cobrançosa, the richness of endophytes increased significantly (up to 2.2-fold, p < 0.001) from asymptomatic to OK-symptomatic twigs, whereas an opposite result was observed in cv. Verdeal Transmontana when considering Shannon-Wiener diversity index (a decrease up to 1.4-fold, p < 0.001).
Figure 1.
Comparison of bacterial diversity between asymptomatic and OK-symptomatic (knots) twigs, either within endophytic or epiphytic communities from each olive tree cultivar (Cobrançosa and Verdeal Transmontana). Diversity at community level was evaluated by determining bacterial abundance, richness and by using Shannon–Wiener index. Box plots depict medians (central horizontal lines), the inter-quartile ranges (boxes), 95% confidence intervals (whiskers), and outliers (black dots). Significant differences between pairs of values are represented over horizontal lines.
The whole bacterial community composition significantly differs between asymptomatic and OK-symptomatic (knots) twigs, as revealed by the non-metric multidimensional scaling (NMDS) plots and analysis of similarities (ANOSIM; R = 0.255, p < 0.001) based on Bray-Curtis index (Fig. 2). These differences were higher on the OK-susceptible cv. Verdeal Transmontana, either within epiphytic (R = 0.671, p < 0.001) or endophytic (R = 0.865, p < 0.001) communities, than in the more resistant cv. Cobrançosa (R = 0.497 and R = 0.416 with p < 0.001, respectively). In addition, the dissimilarity found on bacterial composition between epiphytic and endophytic communities was always greater in OK-symptomatic twigs (R = 1.000 and R = 0.999 with p < 0.001, for cv. Cobrançosa and Verdeal Transmontana, respectively) than in asymptomatic twigs (R = 0.253 and R = 0.523 with p < 0.001, respectively).
Figure 2.
Nonmetric multidimensional scaling (NMDS) plots and ANOSIM tests for the bacterial assemblages in twigs of olive trees from cvs. Cobrançosa and Verdeal Transmontana, considering the presence of OK disease symptoms (asymptomatic vs. OK-symptomatic) and plant habitat (epiphytic vs. endophytic). Bray-Curtis coefficient was used as a measure of similarity between populations and Kruskal’s stress values are presented (values less than 0.2 represent good ordination plots). ANOSIM test showed the R-statistics (R) and the statistical significance, which is denoted by asterisks (*p < 0.05; **p < 0.01; ***p < 0.001).
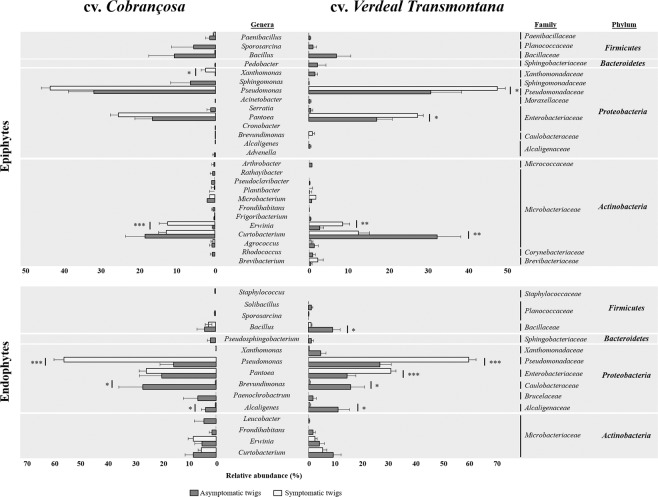
The taxonomic differences of epiphytes and endophytes in asymptomatic and OK-symptomatic twigs were evaluated by comparing differences on the relative abundances at genus, family and phylum levels (Fig. 3). Overall, both asymptomatic and OK-symptomatic twigs (from both olive cultivars) were dominated by bacterial isolates belonging to Pseudomonadaceae family (Proteobacteria phylum), accounting for 26.8% and 51.7% of the total number of isolates obtained in each twig type, respectively. Microbacteriaceae and Enterobacteriaceae were the second most representative families of asymptomatic and OK-symptomatic twigs, representing together 24.4% and 27.3% of the total isolates in each sample type, respectively. As previously revealed, the bacterial communities of each cultivar were differently affected by OK disease. In cv. Cobrançosa, a significant increase in Xanthomonas (up to 106.4-fold), Erwinia (25.5-fold) and Pseudomonas (3.6-fold), as well as a significant decrease on Brevundimonas (140.0-fold) and Alcaligenes (16.4-fold) were observed in OK-symptomatic twigs (knots) in relation to asymptomatic twigs. In contrast, slighter changes occurred in cv. Verdeal Transmontana, where minor (but significant) increases were detected in the abundance of Erwinia (up to 3.2-fold), Pseudomonas (up to 1.5-fold) and Pantoea (up to 1.6-fold), as well as a significant decrease on Curtobacterium (up to 2.6-fold). However, the number of bacterial genera that disappeared with OK disease was greater in cv. Verdeal Transmontana (in total 19) when compared to cv. Cobrançosa (in total 15). Furthermore, in the OK-susceptible cultivar (cv. Verdeal Transmontana), Bacillus and Alcaligenes genera were only present on asymptomatic twigs, while Brevundimonas was only isolated from OK-symptomatic tissues.
Figure 3.
Comparison of the relative abundance of bacterial epiphytes and endophytes, between asymptomatic and OK-symptomatic twigs of olive trees from cvs. Cobrançosa and Verdeal Transmontana. Comparisons are made considering different taxonomic levels (genus, family and phylum). Each value is expressed as mean ± standard error (n = 14, corresponding to 2 olive orchards × 7 trees). Statistically differences between pairs of values are showed by asterisks (*p < 0.05; **p < 0.01; ***p < 0.001).
Contribution of different drivers for bacterial community shaping
For determining the relative contribution of host cultivar, occurrence of OK-symptoms and plant habitat (epi- or endophytic) in shaping the bacterial community, a variation partitioning analysis was performed (Table S2). Results revealed that bacterial composition in twigs was mainly explained by the absence/presence of OK-symptoms (responsible for 7.3% of the total variation) and plant habitat (epi- or endophytic, 7.1% of the total variation), contrasting with host cultivar that only explained 3.6% of the total community variation. The amount of variance explained by the occurrence of OK-symptoms was greater in cv. Verdeal Transmontana (20.5%) and endophytic (11.7%) communities. Plant habitat mainly affected the bacterial composition in cv. Verdeal Transmontana and symptomatic twigs, explaining 13.8% and 26.8% of species composition variance, respectively. Host cultivar had a higher influence on OK-symptomatic (24.4%) twigs and epiphytic (14.7%) communities.
Potential bacteria consortia for olive tree susceptibility/resistance
One goal of this study was the identification of a set of bacterial genera associated to asymptomatic twigs or knots (OK-symptomatic twigs) and elucidate if these bacterial consortia could explain differences in OK disease susceptibility of olive tree cultivars. To more accurately predict such relationships, a random forest analysis was employed to rank the importance of bacterial genera in distinguishing either asymptomatic from OK-symptomatic twigs (Fig. S3) and cv. Cobrançosa from cv. Verdeal Transmontana (Fig. S4). According to their Gini coefficient value (higher the value, greater its importance)17, ten and nine different bacterial genera were selected as the most important in discriminating twigs with/without OK disease symptoms and host cultivar, respectively (Figs. S3 and S4). These bacterial genera were used to perform a multiple factor analysis (MFA), in order to find relationships between bacterial genera and presence/absence of OK symptoms and/or susceptibility/resistance of cultivar to OK disease (Fig. 4). In this analysis, the first dimension revealed a clear opposition between bacteria present in asymptomatic and in knots (OK-symptomatic twigs), either within epiphytic (Fig. 4a) or endophytic (Fig. 4b) bacterial communities. Pseudomonas, Erwinia and Pantoea were positively correlated with the presence of OK disease in the epiphytic community, as well as in the endophytic community (except Erwinia). This result is corroborated by the significantly positive correlation of these genera with Pss abundance (Table S3). On the other hand, Alcaligenes and Bacillus were positively correlated with asymptomatic twigs, either in the epiphytic and endophytic communities. In addition, Arthrobacter and Curtobacterium (in epiphytic community) and Brevundimonas, Frondihabitans, and Xanthomonas (in endophytic community) were positively correlated with asymptomatic twigs. Some of these bacterial genera were also found to be negatively correlated with Pss abundance (Bacillus, Curtobacterium, and Brevundimonas; Table S3). The second dimension of the MFA ordination of both epiphytic and endophytic bacterial communities clearly separated both olive cultivars (Fig. 4). Brevibacterium and Alcaligenes in epiphytic community, as well as Xanthomonas, Alcaligenes and Pseudomonas in endophytic community, were positively correlated with cv. Verdeal Transmontana. Erwinia was the only genus found to be specifically associated to cv. Cobrançosa.
Figure 4.
Circle plot of multiple factor analysis (MFA) correlations of bacterial abundance among olive cultivars (Cobrançosa and Verdeal Transmontana) and the presence of OK disease symptoms (asymptomatic and OK-symptomatic), when applied to epiphytic (a) and endophytic (b) bacterial communities. On the right, the correlation table between each dimension and bacteria genera is shown.
Discussion
With this work, we attempted to disclose the role that host plant-microbe-pathogen interactions may have in the development of OK disease and reveal the underlying pathobiome. For this, epiphytic and endophytic bacterial communities from asymptomatic and OK-symptomatic olive twigs, taken from olive tree cultivars with contrasting susceptibilities to OK disease, were compared. Greater differences on bacterial abundance, diversity and composition were detected for the OK-susceptible cv. Verdeal Transmontana compared to OK-resistant cultivar. Thus, the olive cultivar seems to influence the establishment of pathobiome communities in olive knots. Accordingly, previous studies have suggested that differences on microbial abundance and diversity between asymptomatic and symptomatic tissues were possibly related to the susceptibility of the plant host to a certain disease16,18–21. We hypothesized that the detected greater differences in the bacterial composition of OK-susceptible cv. Verdeal Transmontana, when compared to cv. Cobrançosa, may be due to differential metabolite alterations occurring on both cultivars upon Pss infection. Indeed, plants can defend themselves against pathogens by a variety of mechanisms that enable the detection of pathogen invasion and activation of a defense response22. This defensive response is highly complex and involves a cellular reprogramming that is characterized by an altered plant metabolism with the biosynthesis of defensive compounds23. Although there are still gaps in understanding the dynamism and complexity of such metabolic alterations23, recent studies have indicated that this response is cultivar dependent24. Our hypothesis is further reinforced by the Varpart analysis, which showed a greater contribution of host cultivar to the bacterial assemblage in OK-symptomatic twigs (24.4%) when compared to asymptomatic twigs (2.5%), as similarly observed for the fungal community in the same olive tree cultivars16. The role of host plant in structuring both rhizosphere and root endosphere bacterial communities in response to a pathogen attack was already reported25,26. In these studies, plants subjected to pathogen attack were proposed to recruit protective bacteria for suppressing pathogens in the rhizosphere25. We hypothesize that the same could be occurring in the olive tree phyllosphere upon Pss infection. However, further studies should be conducted to confirm such effect in our pathosystem, as differences on bacterial composition between olive tree cultivars may also be due to Pss itself. For example, mammalian bacterial pathogens have been described to change their environment/habitat in their favor, producing a specific niche or creating a barrier to competing microbes27. In the rhizosphere, bacteria have also been described to alter the soil environment in such a way that certain microbial species are in advantage over others28. In our study, differential changes observed among cultivars upon Pss infection, may also reflect changes made by the pathogen Pss to the microhabitat. Probably, Pss could interact differently with the resident bacteria (pathogen-bacteria interaction) of each olive cultivar. Indeed, both cultivars had a distinct initial bacterial community, which after Pss interaction may probably lead to higher community fluctuation in cv. Verdeal Transmontana than in cv. Cobrançosa. Although the results presented here are in accordance with the accepted idea that host microbiome is a key for plant capability to overcome a pathogen attack29,30, this assumption still needs to be confirmed with further work.
Changes in bacterial diversity and composition between asymptomatic and OK-symptomatic twigs were greater for endophytes than for epiphytes, thus suggesting a greater sensitivity of endophytes to Pss infection. While no comparative studies are available considering the effect of a plant disease in host bacterial epiphytic and endophytic community composition, Gomes et al.16 obtained an opposite result with fungal communities, being epiphytes more affected by bacterial disease than endophytes. Furthermore, we have detected distinct changes on endophytic communities from both cultivars. While OK symptoms (knots) increased the diversity of endophytic bacterial in cv. Cobrançosa, the opposite was detected in cv. Verdeal Transmontana. Altogether, the results suggest that the interaction established between pathogen-host plant or pathogen-native bacterial community could benefit or inhibit specific bacterial endophytes. In fact, the capacity of Pseudomonas to affect the growth and density of other interacting bacteria has been described in different organisms, including plants31–33. This effect has been reported to be a consequence of the cooperation and competition of Pseudomonas species with other microorganisms31,34. Endophytically, Pss cells are organized in clusters, forming also biofilm layers35. The formation of this biofilm provides several advantages to certain bacteria, such as social cooperation, resource capture and protection from antimicrobials36, which may explain the increase of bacterial abundance in OK symptomatic twigs. Based on our results, such pathogen effect seems to have greater impact on the endophytic bacterial community when compared to epiphytic community.
In this study, a number of bacterial genera were found to be specifically associated with asymptomatic or symptomatic twigs of each cultivar. Among the genera most associated to OK-symptomatic twigs, both Pantoea and Erwinia have already been reported to occur in olive knots and suggested to be crucial for the development of OK disease13. Indeed, Pantoea agglomerans and Erwinia toletana have been frequently associated with olive knots12–14,37. Furthermore, when inoculated together with Pss they both promoted tumors size increases in olive trees37,38. Although it is not known how exactly P. agglomerans and E. toletana modulates the OK-disease severity, a number of studies provided evidences of a crosstalk between Pss, P. agglomerans and E. toletana that could have a role on Pss virulence12,39. Interestingly, apart from Pss, Pantoea and Erwinia, other Pseudomonas species were found to be associated with knots, both as an epiphyte and endophyte. A Pseudomonas spp. complex has been previously reported to be associated to plant diseases in different crops, such as Solanum lycopersicum40, Prunus41, citrus42 and mango43. In our study, the frequent occurrence of a Pseudomonas sp. with Pss in olive knots suggests that this consortium is stable and both organisms probably benefit from the presence of each other. Further work needs to be done to establish whether Pseudomonas microflora contributes to the OK disease caused by Pss.
Although a broader and complete view of bacterial communities could have been achieved by using culture-independent methods44, such as metabarcoding approaches using 16S rDNA barcode, we have decided to identify bacterial communities using culture-dependent approach for obtaining bacterial isolates that could be used in the future. Although more limited in depicting whole bacterial communities, results have clearly revealed that host genotype and habitat strongly influence plant pathobiome. Therefore, by understanding OK pathobiome and its relation with disease susceptibility, an additional purpose of this work was to obtain bacterial isolates that could be used for envisaging new biological control methods against olive diseases. The occurrence of bacteria specifically associated with asymptomatic twigs, particularly in the resistant cv. Cobrançosa, may give hints about their role in disease control. Among the genera associated to asymptomatic twigs (either as an epiphyte and endophyte), Bacillus have been identified as the most promising in improving plant growth and controlling plant diseases45. In fact, there are many studies indicating the ability of Bacillus spp. to inhibit microbial pathogen growth either in soil or in plant tissues45–47. In olive, a few number of species belonging to this genus were described to have a high antagonistic potential, not only against Verticillium dahliae48,49, but also against Pss50,51. Bacillus spp. isolated from olive leaves were tested against Pss, revealing promising results in both in vitro and in planta assays50,51. In the present study, other genera (Alcaligenes, Brevundimonas, Curtobacterium and Arthrobacter) were also strongly correlated with asymptomatic tissues. Although Alcaligenes include clinically relevant strains52,53, some studies have been reporting the bacteriostatic and fungistatic activity (biocontrol activity) of some members against an array of plant pathogens54–56. Species belonging to Brevundimonas were previously described to confer fitness advantages to host plants, being indicated as potential soil bioremediators57 and plant growth promotors58. However, members of this genus are frequently known as causing severe infections in humans59,60, compromising their use in the control of plant diseases. Members of Curtobacterium have been mainly described as plant pathogens61 and have been found as endophytes on some woody plant species, such as coffee62, orange and tangerine63. However, there are also reports showing the biocontrol potential of some members of this genus. For example, C. flaccumfaciens revealed to inhibit the phytopathogens Xyllela fastidiosa18,64 and V. dahliae65. Arthrobacter includes a large number of widespread species, in particular in soil, and with great importance in environmental and industrial applications66–68. Apart from nitrogen fixation69, members of Arthrobacter genus also revealed antagonism towards several plant pathogens and capacity to inhibit plant diseases70,71. The role of these bacterial genera associated to asymptomatic twigs, on the defense of olive trees against OK disease remains a topic for further study.
In summary, with this work we revealed that olive bacterial communities change with OK disease. This effect was most notorious within endophytes than within epiphytes and was dependent on host cultivar. Indeed, we observed a greater effect of OK disease on bacterial community assemblage associated with cv. Verdeal Transmontana (more susceptible) than with cv. Cobrançosa (more resistant). Overall, the composition of bacterial community in olive knots seems to result from complex interactions between host plant-Pss-native bacteria. Our work also identified key bacterial genera (especially Bacillus and Brevundimonas) that could play an important role in the susceptibility/resistance of cultivars to OK disease. Understanding the mechanisms of interaction (cooperation vs. competition) and communication of these bacteria with Pss will shed light on the role of these bacteria on the process of OK disease development.
Material and Methods
Asymptomatic and diseased twigs sampling
Sample collection was performed during spring 2015, in two olive orchards located in Mirandela (northeast of Portugal), at coordinates N41°32.593′; W07°07.445′ (orchard 1) and N41°32.756′; W07°07.590′ (orchard 2). These orchards contain two olive cultivars of varying susceptibilities to OK disease (i.e., cv. Verdeal Transmontana is more susceptible than cv. Cobrançosa16), growing together within 7 m of each other, under identical environmental conditions and management practices (integrated production guidelines). In each orchard, seven olive trees of each cultivar were randomly selected. Both asymptomatic and OK-symptomatic twigs (with knots) were collected from the same branch, at mid-canopy height, using sterilized shears and gloves. The collected samples were individually placed into sterile roll bags, brought to the lab on ice, and then stored at 4 °C until bacterial isolation, which was performed within one week.
Epiphytic and endophytic bacterial isolation
After removing the leaves from twigs, the epiphytic bacteria were isolated from pieces of five asymptomatic twigs or knots (with ca. 1-gram weight) cut from symptomatic twigs. These plant segments were individually immersed in 9 mL peptone water (10 g/L peptone, 5 g/L sodium chloride) and shaken for one hour, at 100 rpm at room temperature. Aliquots of 1 ml of the bacterial suspension were then incorporated in triplicate onto 10 mL of Luria Bertani (LB) agar medium (10 g/L peptone, 5 g/L yeast extract, 5 g/L sodium chloride, 10 g/L agar) and incubated at 25 °C, in the dark until bacterial growth. Daily observations were performed in order to isolate and count bacterial colonies (CFU, Colony Forming Units). For isolation, single colonies were picked up, cultured in sterile LB plates and stored at 4 °C when full growth was observed. The abundance of epiphytes was expressed as log CFU/cm2, representing the number of colonies per cm2 of twig/knot surface. Surface of asymptomatic twigs and knots were measured based on cylinder (A = 2πrh + 2πr2) and sphere (A = 4πr2) area equations, respectively, where A is the area, r is the radius and h is the height of plant segments. The average twig and knot segments area were 11.0 ± 3.6 and 2.9 ± 1.3 cm2, respectively, for cv. Cobrançosa, and 11.0 ± 2.3 and 2.9 ± 1.2 cm2, respectively, for cv. Verdeal Transmontana. A total of 280 plant segments (2 olive orchards × 2 olive cultivars × 7 olive trees × 5 twigs or 5 knots) were used for isolating epiphytes.
Endophytes were isolated from the same twig/knot segments used to isolate epiphytes. For this, plant segments were surface sterilized by immersion in ethanol 70% (v/v) for 1 min, followed by sodium hypochlorite 3% (v/v) for 1 min, and then rinsed three times in sterile distilled water (1 min, each). After drying, each twig/knot was cut into segments (ca. 4–5 mm). Five sterilized segments per twig/knot were aseptically transferred onto LB medium, in quintuplicate, and incubated at 25 °C in the dark, until bacterial growth. Cultures were daily monitored and single colonies emerging from tissues segments were counted and subcultured into LB medium in order to obtain pure cultures. Endophytes were isolated from the same 280 plant segments used for epiphytes isolation, corresponding to a total of 7,000 inoculated plant pieces (280 plant segments × 25 pieces per segment).
DNA isolation and 16S rDNA sequencing
The taxonomic identification of bacterial isolates was performed by using both morphological and molecular approaches. Bacterial isolates were firstly grouped based on the cultural features of their colonies, such as colony color, size, shape, opacity, elevation, and margin surface. From each morphotype, two isolates were selected for molecular identification, raising a total of 294 bacteria isolates from twigs and tumors of both Cobrançosa (78 and 66 isolates, respectively) and Verdeal Transmontana (92 and 58 isolates, respectively) cultivars. Bacterial DNA was extracted using REDExtract-N-Amp™ Plant PCR kit (Sigma, Poole, UK), following manufacturer instructions, and used for PCR amplification of V1–V4 regions from 16S rRNA. For PCR reaction, 3 µL of extracted DNA was used in a 50 µL reaction mixture, containing 0.25 µL of each dNTP at 10 mM, 7 µL of 10x buffer, 2.5 µL of 25 mM MgCl2, 0.25 µL of DFS-Taq DNA Polymerase (5 units/µL) and 1 µl of each primer at 10 µM (V1F: 5′- AGAGTTTGATCCTGGCTCAG-3′; V4R: 5′-TACNVGGGTATCTAATCC-3′)72. Amplifications occurred in a MyCycler™ Thermocycler (Bio-Rad), using the following PCR program: 94 °C for 5 min, followed by 35 cycles of 94 °C for 50 sec, 45 °C for 30 sec, 72 °C for 90 sec, with a final extension of 72 °C for 5 min. PCR product was sequenced by Macrogen Inc. (Madrid) and taxonomic identification was performed by using the NCBI database (http://www.ncbi.nlm.nih.gov) and BLAST algorithm. Operational taxonomic units (OTUs) displaying the lowest E-value and the highest identity score were identified to bacterial species (when identity presented a value >98%) or genus (when presenting 95% to 97% identity). For sequence identities <95%, OTUs were labelled as ‘unknown’. Identified bacterial isolates were preserved in the culture collection of the Mountain Research Centre (CIMO), Instituto Politécnico de Bragança.
Data analysis
The cultivable epiphytic or endophytic bacterial community of asymptomatic and symptomatic (i.e., knots) twigs, present in both cvs. Cobrançosa and Verdeal Transmontana, were compared for determining if their variation is affected by plant host and plant habitat. Pss abundance was always excluded from analyses, for determining the true bacterial community changes and not be affected by the overabundance of the pathogen Pss in the symptomatic twigs (knots).
Bacterial diversity analyses
Diversity of bacterial communities was assessed by evaluating the abundance (relative number of isolates per tree), richness (number of OTUs per tree) and Shannon-Wiener Index (H’), determined by the R software73 and using an OTUs abundance matrix. The percentual changes of these diversity parameters occurring on OK-symptomatic twigs in relation to asymptomatic twigs was calculated by using the following formula: percentual changes (%) = [(symptomatic twigs − asymptomatic twigs)/symptomatic twigs] × 100. Estimated changes are presented as the mean of replicates (i.e., tree = 14, corresponding to 2 olive orchards × 7 trees) and respective SE values). Differences among means were determined by an analysis of variance (ANOVA) with R software, where the means were compared using Tukey’s test (p < 0.05).
Comparison of bacterial communities composition
Multivariate statistical analyses were performed to describe differences on the bacterial communities’ composition, among twig status (asymptomatic vs. OK-symptomatic) and plant habitat (epiphytic vs. endophytic). All statistical analyses were performed using the R software73. Non-metric multidimensional scaling (NMDS) was performed using Bray-Curtis index obtained from a normalized abundance OTU matrix, in order to calculate the dissimilarity in the composition of bacterial communities. Kruskal’s stress was used to estimate the model’s goodness of fit (commonly acceptable when lower than 0.2)74. A one-way analysis of similarity (ANOSIM) was also performed using Bray–Curtis distance matrices, for finding significant differences between the bacterial community groups observed in NMDS ordination. This analysis generates a R-value, which range from 0 (completely similar) to 1 (completely different), associated to a p-value (significant when lower than 0.05)75. Both NMDS and ANOSIM analyses were performed using the “vegan”76 package (metaMDS and anosim functions, respectively). For determining the main taxonomic changes that occur in the bacterial communities among samples, the mean percentage of bacterial abundance at different taxonomic levels (phylum, family and genus) was calculated across asymptomatic and OK-symptomatic (knots) twigs. Differences among means were determined by ANOVA with R software73, and means were compared using the Tukey’s test (p < 0.05). A log10 transformation of bacterial abundance was used for meeting ANOVA assumptions.
Factors driving the shaping of bacterial communities
In order to assess the contribution of distinct factors [host cultivar (cv. Cobrançosa vs. cv. Verdeal Transmontana), disease (asymptomatic vs. OK-symptomatic) and plant habitat (epiphytic vs. endophytic)] to differences detected on bacterial communities, a variation partitioning analysis was performed. For this, the varpart function included in the “vegan” package of R software73 and a normalized abundance OTU matrix were used. The significance of each fraction was tested using the anova.cca function.
Identification of bacterial consortium associated to each host cultivar and twig status
A multiple factor analysis (MFA) was used to identify bacterial genera associated to a specific host cultivar and disease symptoms. For this analysis, only the epiphytic and endophytic bacterial genera with the greatest power to separate asymptomatic from OK-symptomatic and cv. Cobrançosa from cv. Verdeal Transmontana twigs were used. These bacterial genera were identified by using a random forest analysis, which was computed with the “RandomForest” package17 from R. The importance of bacterial genera for distinguishing communities was measured by considering the decrease in mean Gini coefficient, where a higher decrease will imply a higher importance77. MFA was computed by using the “FactoMineR”78 package from R. Bacterial genera and variables were graphically represented by the first two dimensions. Then, Spearman correlations were performed through the “corrplot”79 package from R, to check the correlation of pre-selected epiphytic and endophytic bacterial genera with the relative abundance of Pss.
Supplementary information
Acknowledgements
The authors are grateful to the FEDER and Foundation for Science and Technology (FCT, Portugal) under Programme PT2020 for financial support to CIMO (UIDB/00690/2020) and BioISI (UID/MULTI/04046/2013), as well as the Horizon 2020, the European Union’s Framework Programme for Research and Innovation, for financial support the project PRIMA/0002/2018 INTOMED - Innovative tools to combat crop pests in the Mediterranean. D. Mina thanks FCT, POPH-QREN and FSE for SFRH-BD-105341/2014 grant.
Author contributions
P.B. conceived the experiments; sample collection was carried out by J.A.P., D.M. and P.B.; laboratory work was carried out by D.M., T.L.-N., and P.B.; data analyses were conducted by D.M. and P.B.; All authors reviewed and contributed to manuscript writing.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-60596-0.
References
- 1.Compant S, Samad A, Faist H, Sessitsch A. A review on the plant microbiome: Ecology, functions, and emerging trends in microbial application. J. Adv. Res. 2019;19:29–37. doi: 10.1016/j.jare.2019.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim YC, et al. The multifactorial basis for plant health promotion by plant-associated bacteria. Appl. Environ. Microbiol. 2011;77:1548–1555. doi: 10.1128/AEM.01867-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vayssier-Taussat M, et al. Shifting the paradigm from pathogens to pathobiome: new concepts in the light of meta-omics. Front Cell Infect. Microbiol. 2014;4:1–7. doi: 10.3389/fcimb.2014.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jakuschkin B, et al. Deciphering the pathobiome: intra- and interkingdom interactions involving the pathogen Erysiphe alphitoides. Microb. Ecol. 2016;72:870–880. doi: 10.1007/s00248-016-0777-x. [DOI] [PubMed] [Google Scholar]
- 5.Bodenhausen N, Bortfeld-Miller M, Ackermann M, Vorholt JA. A synthetic community approach reveals plant genotypes affecting the phyllosphere microbiota. PLoS Genet. 2014;10:e1004283. doi: 10.1371/journal.pgen.1004283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wagner MR, et al. Host genotype and age shape the leaf and root microbiomes of a wild perennial plant. Nat. Commun. 2016;7:12151. doi: 10.1038/ncomms12151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wille L, Messmer MM, Studer B, Hohmann P. Insights to plant–microbe interactions provide opportunities to improve resistance breeding against root diseases in grain legumes. Plant Cell. Environ. 2019;42:20–40. doi: 10.1111/pce.13214. [DOI] [PubMed] [Google Scholar]
- 8.Weingarden A, et al. Dynamic changes in short- and long-term bacterial composition following fecal microbiota transplantation for recurrent Clostridium difficile infection. Microbiome. 2015;3:10. doi: 10.1186/s40168-015-0070-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Quesada JM, et al. Dissemination of Pseudomonas savastanoi pv. savastanoi populations and subsequent appearance of olive knot disease. Plant Pathol. 2010;59:262–269. doi: 10.1111/j.1365-3059.2009.02200.x. [DOI] [Google Scholar]
- 10.Quesada, J. M., Penyalver, R. & López, M. M. Epidemiology and control of plant diseases caused by phytopathogenic bacteria: the case of olive knot disease caused by Pseudomonas savastanoi pv. savastanoi in Plant Pathology (ed. Cumagun, C. J. R.) 299–326 (IntechOpen, 2012).
- 11.Ramos C, Matas M, Bardaji L, Aragón IM, Murillo J. Pseudomonas savastanoi pv. savastanoi: some like it knot. Mol. Plant Pathol. 2012;13:998–1009. doi: 10.1111/j.1364-3703.2012.00816.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hosni T, et al. Sharing of quorum-sensing signals and role of interspecies communities in a bacterial plant disease. ISME J. 2011;5:1857–1870. doi: 10.1038/ismej.2011.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Passos da Silva D, et al. Bacterial multispecies studies and microbiome analysis of a plant disease. Microbiology. 2014;160:556–566. doi: 10.1099/mic.0.074468-0. [DOI] [PubMed] [Google Scholar]
- 14.Buonaurio R, et al. The olive knot disease as a model to study the role of interspecies bacterial communities in plant disease. Front. Plant Sci. 2015;6:434. doi: 10.3389/fpls.2015.00434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Penyalver R, et al. Factors affecting Pseudomonas savastanoi pv. savastanoi plant inoculations and their use for evaluation of olive cultivar susceptibility. Phytopathol. 2006;96:313–319. doi: 10.1094/PHYTO-96-0313. [DOI] [PubMed] [Google Scholar]
- 16.Gomes T, Pereira JA, Lino-Neto T, Bennet AE, Baptista P. Bacterial disease induced changes in fungal communities of olive tree twigs depend on host genotype. Sci. Rep. 2019;9:5882. doi: 10.1038/s41598-019-42391-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cutler DR, et al. Random Forests for classification in ecology. Ecol. Appl. 2007;88:2783–2792. doi: 10.1890/07-0539.1. [DOI] [PubMed] [Google Scholar]
- 18.Lacava PT, Araujo WL, Marcon J, Maccheroni W, Azevedo JL. Interaction between endophytic bacteria from citrus plants and the phytopathogenic bacteria Xylella fastidiosa, causal agent of citrus-variegated chlorosis. Lett. Appl. Microbiol. 2004;39:55–59. doi: 10.1111/j.1472-765X.2004.01543.x. [DOI] [PubMed] [Google Scholar]
- 19.Suhaimi NSM, et al. Diversity of microbiota associated with symptomatic and nonsymptomatic bacterial wilt-diseased banana plants determined using 16S rRNA metagenome sequencing. World J. Microbiol. Biotechnol. 2017;33:168. doi: 10.1007/s11274-017-2336-0. [DOI] [PubMed] [Google Scholar]
- 20.Zhao Y, et al. Endosphere microbiome comparison between symptomatic and asymptomatic roots of Brassica napus infected with Plasmodiophora brassicae. PLOS ONE. 2017;12:e0185907. doi: 10.1371/journal.pone.0185907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hamonts K, et al. Field study reveals core plant microbiota and relative importance of their drivers. Environ. Microbiol. 2018;20:124–140. doi: 10.1111/1462-2920.14031. [DOI] [PubMed] [Google Scholar]
- 22.Andersen E, Ali S, Byamukama E, Yen Y, Nepal M. Disease resistance mechanisms in plants. Genes. 2018;9:339. doi: 10.3390/genes9070339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tugizimana F, Mhlongo M, Piater L, Dubery I. Metabolomics in plant priming research: the way forward? Int. J. Mol. Sci. 2018;19:1759. doi: 10.3390/ijms19061759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tugizimana F, Djami-Tchatchou AT, Steenkamp PA, Piater LA, Dubery IA. Metabolomic analysis of defense-related reprogramming in sorghum bicolor in response to Colletotrichum sublineolum infection reveals a functional metabolic web of phenylpropanoid and flavonoid pathways. Front. Plant Sci. 2019;9:1840. doi: 10.3389/fpls.2018.01840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berendsen RL, Pieterse CMJ, Bakker PAHM. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012;17:478–486. doi: 10.1016/j.tplants.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 26.Zamioudis C, Pieterse CMJ. Modulation of host immunity by beneficial microbes. Mol. Plant Microbe In. 2012;25:139–150. doi: 10.1094/MPMI-06-11-0179. [DOI] [PubMed] [Google Scholar]
- 27.Wilson NG, Hernandez-Leyva A, Kau AL. The ABCs of wheeze: Asthma and bacterial communities. PLoS Pathog. 2019;15:e1007645. doi: 10.1371/journal.ppat.1007645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scharf BE, Hynes MF, Alexandre GM. Chemotaxis signaling systems in model beneficial plant–bacteria associations. Plant Mol. Biol. 2016;90:549–559. doi: 10.1007/s11103-016-0432-4. [DOI] [PubMed] [Google Scholar]
- 29.Rodriguez PA, et al. Systems biology of plant-microbiome interactions. Mol. Plant. 2019;12:804–821. doi: 10.1016/j.molp.2019.05.006. [DOI] [PubMed] [Google Scholar]
- 30.Vannier N, Agler M, Hacquard S. Microbiota-mediated disease resistance in plants. PLoS Pathog. 2019;15:e1007740. doi: 10.1371/journal.ppat.1007740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tashiro Y, Yawata Y, Toyofuku M, Uchiyama H, Nomura N. Interspecies interaction between Pseudomonas aeruginosa and other microorganisms. Microbes Environ. 2013;28:13–24. doi: 10.1264/jsme2.ME12167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pandey SK, Chandel SCR. Efficacy of Pseudomonas as biocontrol agent against plant pathogenic fungi. Int. J. Curr. Microbiol. App. Sci. 2014;3:493–500. [Google Scholar]
- 33.Zohara F, Akanda MAM, Paul NC, Rahman M, Islam MT. Inhibitory effects of Pseudomonas spp. on plant pathogen Phytophthora capsici in vitro and in planta. Biocatal. Agric. Biotechnol. 2016;5:69–77. doi: 10.1016/j.bcab.2015.12.009. [DOI] [Google Scholar]
- 34.Dandekar AA, Chugani S, Greenberg EP. Bacterial quorum sensing and metabolic incentives to cooperate. Science. 2012;338:264–266. doi: 10.1126/science.1227289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Temsah M, Hanna L, Saad AT. Anatomical pathogenesis of Pseudomonas savastanoi on olive and genesis of knots. J. Plant Pathol. 2008;90:225–232. [Google Scholar]
- 36.Flemming HC, et al. Biofilms: an emergent form of bacterial life. Nat. Rev. Microbiol. 2016;14:563–575. doi: 10.1038/nrmicro.2016.94. [DOI] [PubMed] [Google Scholar]
- 37.Marchi G, et al. Interaction between Pseudomonas savastanoi pv. savastanoi and Pantoea agglomerans in olive knots. Plant Pathol. 2006;55:614–624. doi: 10.1111/j.1365-3059.2006.01449.x. [DOI] [Google Scholar]
- 38.Hosni, T. Interaction between Pseudomonas savastanoi pv. savastanoi, the causal agent of olive knot, and the endophytic bacterial species associated with the knot. Ph.D. thesis. University of Perugia, Perugia, Italy (2010).
- 39.Caballo-Ponce E, et al. Quorum sensing in Pseudomonas savastanoi pv. savastanoi and Erwinia toletana: role in virulence and interspecies interactions in the olive knot. Appl. Environ. Microbiol. 2018;84:e00950–18. doi: 10.1128/AEM.00950-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kůdela V, Krejzar V, Pánková I. Pseudomonas corrugata and Pseudomonas marginalis associated with the collapse of tomato plants in rockwool slab hydroponic culture. Plant Prot. Sci. 2010;46:1–11. doi: 10.17221/44/2009-PPS. [DOI] [Google Scholar]
- 41.Ruinelli M, Blom J, Smits THM, Pothier JF. Comparative genomics and pathogenicity potential of members of the Pseudomonas syringae species complex on Prunus spp. BMC Genomics. 2019;20:1471–2164. doi: 10.1186/s12864-019-5555-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Beiki F, et al. New Pseudomonas spp. are pathogenic to citrus. PLOS ONE. 2016;11:e0148796. doi: 10.1371/journal.pone.0148796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gutiérrez-Barranquero JA, Cazorla FM, de Vicente A. Pseudomonas syringae pv. syringae associated with mango trees, a particular pathogen within the “Hodgepodge” of the Pseudomonas syringae complex. Front. Plant Sci. 2019;10:570. doi: 10.3389/fpls.2019.00570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hug LA. Sizing up the uncultured microbial majority. mSystems. 2018;3:e00185–18. doi: 10.1128/mSystems.00185-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Radhakrishnan R, Hashem A, Abd Allah EF. Bacillus: a biological tool for crop improvement through bio-molecular changes in adverse environments. Front. Physiol. 2017;8:667. doi: 10.3389/fphys.2017.00667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Caulier S, et al. Versatile antagonistic activities of soil-borne Bacillus spp. and Pseudomonas spp. against Phytophthora infestans and other potato pathogens. Front. Microbiol. 2018;9:143. doi: 10.3389/fmicb.2018.00143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shafi J, Tian H, Ji M. Bacillus species as versatile weapons for plant pathogens: a review. Biotechnol. Biotec. Eq. 2017;31:446–459. doi: 10.1080/13102818.2017.1286950. [DOI] [Google Scholar]
- 48.Muller H, et al. Plant genotype-specific archaeal and bacterial endophytes but similar Bacillus antagonists colonize Mediterranean olive trees. Front. Microbiol. 2015;6:138. doi: 10.3389/fmicb.2015.00138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cabanás CG, et al. Bacillales members from the olive rhizosphere are effective biological control agents against the defoliating pathotype of Verticillium dahliae. Agriculture. 2018;8:1–23. [Google Scholar]
- 50.Krid S, et al. Pseudomonas savastanoi endophytic bacteria in olive tree knots and antagonistic potential of strains of Pseudomonas fluorescens and Bacillus subtilis. J. Plant Pathol. 2010;92:335–341. [Google Scholar]
- 51.Krid S, Triki MA, Gargouri A, Rhouma A. Biocontrol of olive knot disease by Bacillus subtilis isolated from olive leaves. Ann. Microbiol. 2012;62:149–154. doi: 10.1007/s13213-011-0239-0. [DOI] [Google Scholar]
- 52.Sonnenberg GF, et al. Innate lymphoid cells promote anatomical containment of lymphoid-resident commensal bacteria. Science. 2012;336:1321–1325. doi: 10.1126/science.1222551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tena D, Fernández C, Lago MR. Alcaligenes faecalis: an unusual cause of skin and soft tissue infection. Japanese J. Infect. Dis. 2015;68:128–130. doi: 10.7883/yoken.JJID.2014.164. [DOI] [PubMed] [Google Scholar]
- 54.Sayyed RZ, Chincholkar SB. Siderophore-producing Alcaligenes faecalis exhibited more biocontrol potential vis-à-vis chemical fungicide. Curr. Microbiol. 2008;58:47–51. doi: 10.1007/s00284-008-9264-z. [DOI] [PubMed] [Google Scholar]
- 55.Kavroulakis N, et al. Antagonistic bacteria of composted agro-industrial residues exhibit antibiosis against soil-borne fungal plant pathogens and protection of tomato plants from Fusarium oxysporum f.sp. radicis-lycopersici. Plant Soil. 2010;333:233–247. doi: 10.1007/s11104-010-0338-x. [DOI] [Google Scholar]
- 56.Yokoyama S, Adachi Y, Asakura S, Kohyama E. Characterization of Alcaligenes faecalis strain AD15 indicating biocontrol activity against plant pathogens. J. Gen. Appl. Microbiol. 2013;59:89–95. doi: 10.2323/jgam.59.089. [DOI] [PubMed] [Google Scholar]
- 57.Singh N, et al. Brevundimonas diminuta mediated alleviation of arsenic toxicity and plant growth promotion in Oryza sativa L. Ecotoxicol. Environ. Saf. 2016;125:25–34. doi: 10.1016/j.ecoenv.2015.11.020. [DOI] [PubMed] [Google Scholar]
- 58.Kumar V, Gera R. Isolation of a multi-trait plant growth promoting Brevundimonas sp. and its effect on the growth of Bt-cotton. Biotech. 2014;4:97–101. doi: 10.1007/s13205-013-0126-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ryana MP, Pembroke JT. Brevundimonas spp: Emerging global opportunistic pathogens. Virulence. 2018;9:480–493. doi: 10.1080/21505594.2017.1419116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Swain B, Rout S. Brevundimonas diminuta: an unusual cause for bacteraemia at a teaching hospital. The Antiseptic. 2017;114:27–28. [Google Scholar]
- 61.Silva Júnior TAF, Negrão DR, Itako AT, Maringoni AC. Pathogenicity of Curtobacterium flaccumfaciens pv. flaccumfaciens to several plant species. J. Plant Pathol. 2012;94:427–430. [Google Scholar]
- 62.Veja FE, Pava-Ripoll M, Posada F, Buyer JS. Endophytic bacteria in Coffea arabica L. J. Basic Microbiol. 2005;45:371–380. doi: 10.1002/jobm.200410551. [DOI] [PubMed] [Google Scholar]
- 63.Araújo WL, et al. Diversity of endophytic bacterial populations and their interaction with Xylella fastidiosa in citrus plants. Appl. Environ. Microbiol. 2002;68:4906–4914. doi: 10.1128/AEM.68.10.4906-4914.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Azevedo JL, Araújo WL, Lacava PT. The diversity of citrus endophytic bacteria and their interactions with Xylella fastidiosa and host plants. Genet. Mol. Biol. 2016;39:476–491. doi: 10.1590/1678-4685-gmb-2016-0056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Berg G, et al. Endophytic and ectophytic potato-associated bacterial communities differ in structure and antagonistic function against plant pathogenic fungi. FEMS Microb. Ecol. 2005;51:215–229. doi: 10.1016/j.femsec.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 66.Zhang C, Li M, Xu X, Liu N. Effects of carbon nanotubes on atrazine biodegradation by Arthrobacter sp. J. Hazardous Mater. 2015;287:1–6. doi: 10.1016/j.jhazmat.2015.01.039. [DOI] [PubMed] [Google Scholar]
- 67.He T, Xie D, Li Z, Ni J, Sun Q. Ammonium stimulates nitrate reduction during simultaneous nitrification and denitrification process by Arthrobacter arilaitensis Y-10. Bioresour. Technol. 2017;239:66–73. doi: 10.1016/j.biortech.2017.04.125. [DOI] [PubMed] [Google Scholar]
- 68.Prum C, Dolphen R, Thiravetyan P. Enhancing arsenic removal from arsenic-contaminated water by Echinodorus cordifolius −endophytic Arthrobacter creatinolyticus interactions. J. Environ. Manage. 2018;213:11–19. doi: 10.1016/j.jenvman.2018.02.060. [DOI] [PubMed] [Google Scholar]
- 69.Fernández-González AJ, et al. The rhizosphere microbiome of burned holm-oak: potential role of the genus Arthrobacter in the recovery of burned soils. Sci. Rep. 2017;7:6008. doi: 10.1038/s41598-017-06112-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Velázquez-Becerra C, et al. The rhizobacterium Arthrobacter agilis produces dimethylhexadecylamine, a compound that inhibits growth of phytopathogenic fungi. in vitro. Protoplasma. 2013;250:1251–1262. doi: 10.1007/s00709-013-0506-y. [DOI] [PubMed] [Google Scholar]
- 71.Zhang J, et al. Characterization of diazotrophic growth-promoting rhizobacteria isolated from ginger root soil as antagonists against Ralstonia solanacearum. Biotechnol. Biotec. Eq. 2018;32:1447–1454. doi: 10.1080/13102818.2018.1533431. [DOI] [Google Scholar]
- 72.Cai L, Ye L, Tong AHY, Lok S, Zhang T. Biased diversity metrics revealed by bacterial 16S pyrotags derived from different primer sets. PLOS ONE. 2013;8:e53649. doi: 10.1371/journal.pone.0053649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, URL http://www.R-project.org/ (2018).
- 74.McCune, B. P. & Grace, J. B. Analysis of ecological communities. MjM Software Design. Gleneden Beach, Oregon (2002).
- 75.Clarke, K. R. & Gorley, R. N. PRIMER v7: User Manual/Tutorial. PRIMER-E. Plymouth, UK (2015).
- 76.Oksanen, J. et al. Vegan: Community ecology package. R package version 2.5-2, https://cran.rproject.org/web/packages/vegan/index.html (2018).
- 77.Breiman L. Random forests. Mach. Learn. 2001;45:5–32. doi: 10.1023/A:1010933404324. [DOI] [Google Scholar]
- 78.Le S, Josse J, Husson F. FactoMineR: An R package for multivariate analysis. J. Stat. Softw. 2008;25:1–18. doi: 10.18637/jss.v025.i01. [DOI] [Google Scholar]
- 79.Wei, T. & Simko, V. R package corrplot: Visualization of a correlation matrix. R package version 0.84, https://github.com/taiyun/corrplot (2017).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.