Abstract
Reproductive isolation between different host populations is often based on intraspecific sex pheromone differences. The mechanisms underlying these differences have not been thoroughly elucidated to date. Previous studies suggested that Chilo suppressalis has differentiated into rice and water-oat host populations, and these two populations manifest clear differences in sex pheromone titer and mating rhythm. Hence, this moth is an ideal model to investigate the endogenous mechanisms of intraspecific reproductive isolation. Here, we identified a series of putative genes associated with sex pheromone biosynthesis based on the C. suppressalis pheromone gland transcriptome data. Transcripts of most genes were at higher level in the rice population. Then we obtained 11 pivotal differentially expressed genes (DEGs). The expression levels of these DEGs exhibited a distinct increase in the rice population. Moreover, we also observed the expression rhythm of these DEGs is discrepant between two host populations. Our study offers a new understanding to elucidate the mechanisms of intraspecific reproductive isolation.
Subject terms: Evolution, Molecular biology
Introduction
Reproductive isolation is generally regarded as an important step to promote sympatric speciation1–3. In many moths, reproductive isolation is common to maintain reproductive isolation between host populations through intraspecific differences in sex pheromone communication4–8. Previous studies revealed that in addition to the differences in the ratios of sex pheromone compounds, variations in sex pheromone titer can also lead to strong reproductive isolation among different populations9–11. In insects, the production of sex pheromone often exhibits a diel rhythm, which is closely related to the mating activity rhythm12,13. When moths differ in their sex pheromone production rhythms, they are selected to be sexually active at different times, which can cause extensive communication interference, eventually resulting in intraspecific reproductive isolation5,14–16. Hence, understanding the endogenous mechanisms of the variations in sex pheromone would help to elucidate the occurrence of intraspecific reproductive isolation.
In several moths, the regulation of sex pheromone biosynthesis has been shown to involve a series of biochemical enzymes17–19. Hence, the divergence in these enzymes may lead to variations of pheromone production. With the development of molecular biology techniques, a large number of putative genes involved in sex pheromone biosynthesis have been identified, including acetyl-CoA carboxylase, fatty acid synthase, fatty acid transport proteins, acyl-CoA oxidases, desaturases, and aldehyde reductase20–24. Several studies showed that these genes generally exhibited diel expression rhythms in moths25,26. Thus, the difference in expression levels and rhythm of genes related to sex pheromone biosynthesis probably cause the variations in sex pheromone production among different populations.
Chilo suppressalis Walker (Lepidoptera: Crambidae) is an economically important pest in Asia, which appeared in rice and water-oat populations27–30. As early as the 1970s-1980s, Japanese scholars identified three active components in the sex pheromone gland extract of females, including Z9-16Ald, Z11-16Ald, and Z13-18Ald10. They also demonstrated the sex pheromone titer was of a higher level in the rice population than in the water-oat population10. Meanwhile, the two host populations showed significant differences in diel mating rhythm16, which may be dominated by variation in sex pheromones titer rhythms22,31. Thus, C. suppressalis is an ideal model to investigate intraspecific differences in sex pheromone communication. We proposed that the differential expression of putative sex pheromone biosynthetic genes might lead to divergence in sex pheromone production.
In this study, we obtained the putative genes related to sex pheromone biosynthesis and analyzed expression patterns of the key differentially expressed genes (DEGs) in the two host populations, basing on the pheromone gland transcriptome data. There were divergences in expression patterns between the two populations of C. suppressalis, which is consistent with sex pheromone titer and mating rhythm between these two populations in the previous researches. Our study provides a new insight into the endogenous mechanism of intraspecific reproductive isolation.
Results
Sex pheromone gland transcriptome assembly, functional annotation and DEG analysis in C. suppressalis
To elucidate the molecular basis of the variation in sex pheromone, we performed RNA-Seq to quantify gene expression between the rice and water-oat populations. After removing low-quality reads, 23 to 33 million clean reads from each sample were obtained for further analysis (Table 1). The final non-redundant dataset contained 169477 unigenes with an N50 of 1830 bp and a mean length of 1115 bp (Table S1). The sequences of all the reads have been deposited in the NCBI SRA database with the accession number, SRP162187. Unigenes in the transcriptome were annotated according to seven databases. A total of 61,422 (36.24%), 28,805 (16.99%), 21,036 (12.41%), 42,162 (24.87%), 47,893 (28.25%), 48,322 (28.51%), and 30,528 (18.01%) unigenes could be mapped to the NR, NT, KO, Swiss-Prot, PFAM, KOG and GO databases, respectively (Table S2). The percent of annotated genes assigned to the biological processes, cellular components and molecular function categories was 48.5%, 30.4% and 21.0%, respectively, based on the GO database32 (Fig. S1).
Table 1.
Summary of C. suppressalis pheromone gland sequencing reads and quality metrics.
| Sample | Raw Reads | Clean Reads | Clean Bases | Q20 | GC Content |
|---|---|---|---|---|---|
| R0a_1 | 33,299,218 | 32,148,426 | 4.82 G | 97.16% | 43.04% |
| R0_2 | 29,545,158 | 28,379,906 | 4.26 G | 97.60% | 42.88% |
| R0_3 | 30,256,192 | 29,104,104 | 4.37 G | 97.58% | 42.71% |
| R3_1 | 33,268,874 | 31,460,310 | 4.72 G | 96.88% | 42.98% |
| R3_2 | 26,126,644 | 25,183,518 | 3.78 G | 96.85% | 43.15% |
| R3_3 | 27,058,962 | 26,096,298 | 3.91 G | 97.14% | 42.74% |
| R6_1 | 29,501,788 | 28,422,308 | 4.26 G | 97.08% | 42.50% |
| R6_2 | 32,295,978 | 31,014,090 | 4.65 G | 97.03% | 43.83% |
| R6_3 | 24,932,768 | 23,954,566 | 3.59 G | 97.02% | 42.32% |
| W0_1 | 29,814,248 | 28,002,098 | 4.20 G | 96.57% | 44.59% |
| W0_2 | 27,343,088 | 25,831,456 | 3.87 G | 96.66% | 42.56% |
| W0_3 | 27,862,866 | 26,854,950 | 4.03 G | 97.04% | 42.88% |
| W3_1 | 29,736,018 | 28,114,434 | 4.22 G | 96.79% | 43.22% |
| W3_2 | 27,946,514 | 26,971,444 | 4.05 G | 97.19% | 43.07% |
| W3_3 | 28,831,070 | 27,203,918 | 4.08 G | 96.82% | 42.60% |
| W6_1 | 30,226,446 | 28,957,384 | 4.34 G | 97.19% | 44.26% |
| W6_2 | 27,521,754 | 26,507,980 | 3.98 G | 97.14% | 42.85% |
| W6_3 | 28,076,838 | 27,099,430 | 4.06 G | 97.28% | 42.97% |
aR and W stands for rice population and water-oat population; 0, 3 and 6 stand for 0 h, 3 h and 6 h after the onset of scotophase.
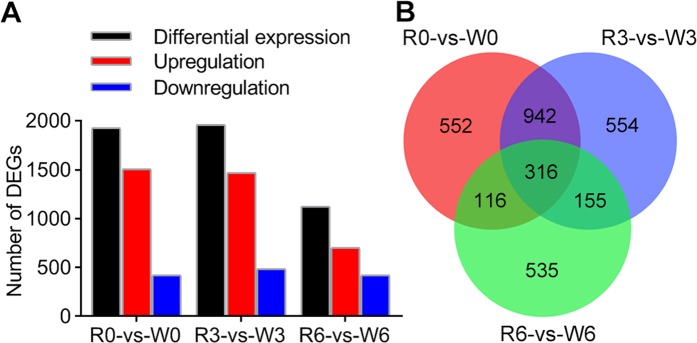
Among the DEGs between the two host populations of C. suppressalis, 1926 genes were differentially regulated between R0 and W0, including the upregulation of 1505 genes and the downregulation of 421 genes. Similarly, a total of 1957 DEGs, including 1471 upregulated and 486 downregulated, were detected when comparing R3 and W3 (Fig. 1A, Table S3). A significant decrease in the DEGs between R6 and W6 was observed, with 702 genes upregulated and 420 genes downregulated (Fig. 1A, Table S3). These results showed that most DEGs are upregulated in the rice population. Among these DEGs, 316 unigenes were commonly differentially expressed in response to the three comparison groups (Fig. 1B). Meanwhile, through KEGG pathway enrichment analysis, we found that the amino sugar and nucleotide sugar metabolism pathways and steroid hormone biosynthesis were significantly enriched (Table 2). This suggests that energy regulation may be an important factor in the process of sex pheromone production.
Figure 1.
Analysis of DEGs in the pheromone gland transcriptome between rice (R) and water-oat populations (W) of C. suppressalis. (A) Distribution of upregulated and downregulated DEGs in each comparison; (B) Venn diagram of DEGs in different comparisons among the groups. Upregulations indicate that the genes have a higher expression in the rice population compared to the water-oat population.
Table 2.
KEGG pathway enrichment of DEGs in the pheromone gland transcriptome between the rice and water-oat populations of C. suppressalis.
| Enriched KEGG pathway | Gene number | P-valueb |
|---|---|---|
| R0-vs-W0a | ||
| Ribosome | 40 | 5.85E-03 |
| Purine metabolism | 33 | 9.83E-05 |
| Pyrimidine metabolism | 28 | 6.39E-07 |
| Cell cycle | 23 | 4.53E-05 |
| Glutathione metabolism | 10 | 4.81E-02 |
| Protein export | 9 | 9.85E-04 |
| Arginine and proline metabolism | 9 | 4.10E-02 |
| R3-vs-W3 | ||
| Cell cycle | 6 | 1.61E-03 |
| Purine metabolism | 6 | 2.51E-02 |
| Pyrimidine metabolism | 5 | 1.05E-02 |
| Cysteine and methionine metabolism | 4 | 5.03E-03 |
| Nicotinate and nicotinamide metabolism | 3 | 7.14E-03 |
| TGF-beta signaling pathway | 3 | 1.96E-02 |
| Protein export | 2 | 4.55E-02 |
| R6-vs-W6 | ||
| Retinol metabolism | 7 | 3.88E-04 |
| Cell cycle | 6 | 4.67E-02 |
| Steroid hormone biosynthesis | 5 | 2.29E-03 |
| Amino sugar and nucleotide sugar metabolism | 5 | 2.76E-02 |
| Arginine and proline metabolism | 4 | 4.68E-02 |
| Linoleic acid metabolism | 3 | 3.66E-03 |
| Protein export | 3 | 3.54E-02 |
| Nicotinate and nicotinamide metabolism | 3 | 4.86E-02 |
| D-Glutamine and D-glutamate metabolism | 2 | 6.64E-03 |
| Mucin type O-Glycan biosynthesis | 2 | 3.26E-02 |
aR and W stands for rice population and water-oat population; 0, 3 and 6 stand for 0 h, 3 h and 6 h after the onset of scotophase. bP-value <0.05 is considered significant.
The expression patterns of putative genes associated with sex pheromone biosynthesis between the rice and water-oat populations in C. suppressalis
The enzymatic reactions that occur during sex pheromone biosynthesis include various processes21,33. Based on functional annotation and homology analysis, we identified a series of putative genes associated with sex pheromone biosynthesis and investigated the overall expression kinetics of these genes. These genes were involved four biochemical processes, including fatty acid synthesis, desaturation, β-oxidation and reduction reactions. Transcripts of these genes were have higher levels in the rice populations (Fig. 2), which is in agreement with the sex pheromone titer data10. Next, we obtained 11 key putative DEGs associated with sex pheromone biosynthesis, including 2 ELO, 1 ACC, 1 FATP, 1 ALF, 2 ALR, 1 ACD, 1 3-KCT and 2 DES genes (Table 3). The results showed that the expression levels of 11 genes exhibited a significant elevation in the rice population (Fig. 3). Moreover, ELO7, ACC, FATP, ALR2, 3-KCT, DES1 and DES7, had higher expression at 0 h or 3 h after the onset of scotophase in the rice population (Fig. 3A–D). Additionally, ELO7, FATP, ACD and DES1 had higher expression at 3 h or 6 h after the onset of scotophase in the water-oat population (Fig. 3A,B,D). This suggests the different expression peaks of these genes may lead to the differentiation in mating rhythm between the two populations in C. suppressalis.
Figure 2.
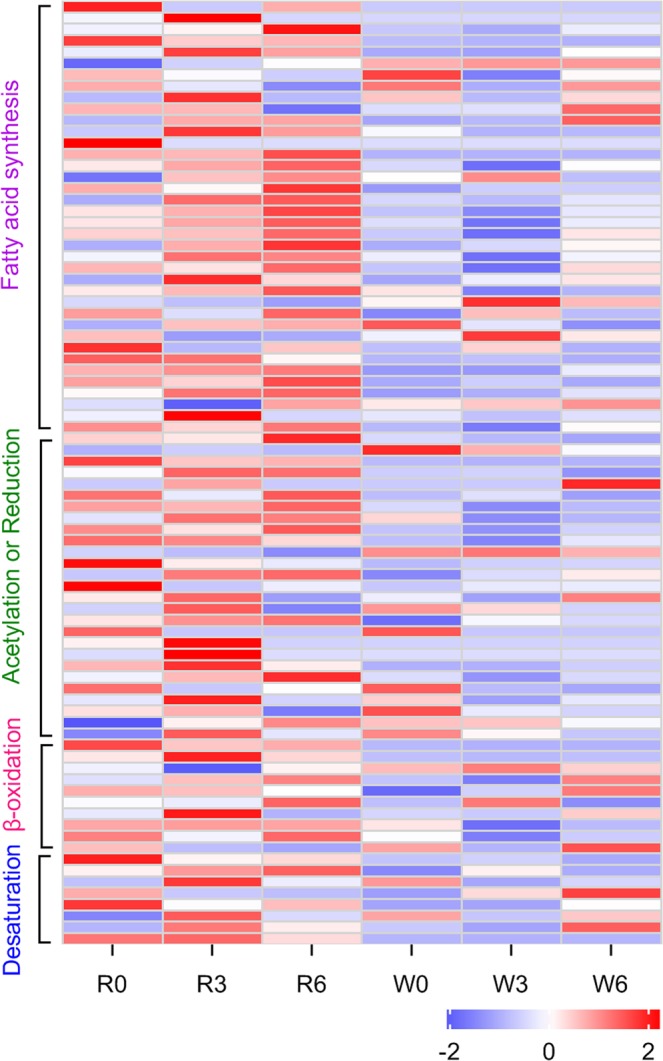
Heatmap analysis of expression patterns of putative genes associated with sex pheromone biosynthesis at 0 h, 3 h and 6 h after the onset of scotophase between rice (R) and water-oat populations (W) in C. suppressalis. Prior to analysis, FPKM values of the genes were Z-score standardized. The heatmap was plotted using GraphPad Prism 8 (GraphPad Software Inc., San Diego, CA, https://www.graphpad.com). Blue and red colors in the heatmap correspond to low and high relative gene expression level, respectively.
Table 3.
The putative DEGs associated with sex pheromone biosynthesis in C. suppressalis.
| Gene (Abbreviation) | Query ID | R0-vs-W0a | R3-vs-W3 | R6-vs-W6 | |||
|---|---|---|---|---|---|---|---|
| | log2 ratio | | P-valueb | | log2 ratio | | P-value | | log2 ratio | | P-value | ||
| Elongation of very long chain fatty acids 1 (ELO1) | Cluster-28055.77797 | Infinity | 1.00 | Infinity | 2.70 E-02 | Infinity | 0.37 |
| Elongation of very long chain fatty acids 7 (ELO7) | Cluster-28055.94024 | Infinity | 1.52E-03 | 0.96 | 1.00 | 2.25 | 1.00 |
| Acetyl-CoA Carboxylase (ACC) | Cluster-28055.115852 | 3.64 | 1.00E-08 | 3.37 | 1.36E-04 | 4.02 | 0.13 |
| Fatty acid transport protein (FATP) | Cluster-28055.89390 | 2.50 | 2.12E-04 | 2.06 | 0.99 | 1.81 | 1.00 |
| Acetyltransferase1 (ALF) | Cluster-28055.50331 | 1.01 | 1.00 | 1.59 | 1.00 | 3.26 | 2.11E-02 |
| Aldo-Ketose Reductase2 (ALR2) | Cluster-28055.89156 | 3.68 | 1.16E-12 | 3.61 | 7.87E-07 | 3.78 | 0.21 |
| Aldo-Ketose Reductase3 (ALR3) | Cluster-28055.76800 | 0.77 | 1.00 | 1.52 | 0.7741 | 3.43 | 2.09E-03 |
| Acyl-CoA dehydrogenase (ACD) | Cluster-28055.81963 | 3.79 | 1.00 | Infinity | 1.76E-08 | 3.99 | 0.66 |
| 3-ketoacyl-CoA thiolase (3-KCT) | Cluster-28055.79228 | 3.85 | 2.12E-14 | 3.73 | 1.91E-07 | 3.87 | 0.15 |
| Desaturase 1 (DES1) | Cluster-28055.80420 | 1.74 | 2.35E-02 | 0.60 | 1.00 | 1.35 | 0.86 |
| Desaturase 7 (DES7) | Cluster-28055.25897 | 4.29 | 2.86E-03 | 4.94 | 1.42 E-06 | 3.32 | 1.00 |
aR and W stands for rice population and water-oat population; 0, 3 and 6 stand for 0 h, 3 h and 6 h after the onset of scotophase. bP-value <0.05 is considered significant.
Figure 3.
Expression levels of putative DEGs associated with sex pheromone biosynthesis between the rice populations (R) and water-oat populations (W) in C. suppressalis, as determined by FPKM values. FPKM values are represented as the means ± standard error of mean (SEM) based on three biological replicates.
To verify the reliability of this transcriptome database, we randomly select 5 DEGs, ELO1, ALF, ACD, DES1 and DES7, to be validated through RT-qPCR (Fig. 4). This validation result is coincided with the RNA sequencing regarding the gene expression levels at the 3 stages.
Figure 4.
Relative expression of putative sex pheromone biosynthetic genes between the rice populations (R) and water-oat populations (W) in C. suppressalis, as determined by RT-qPCR. Relative expression values are represented as the means ± standard error of mean (SEM) based on three biological replicates. Significance analysis of the relative gene expression between the two populations was carried out by independent sample t-tests, *P <0.05, **P <0.01.
Discussion
In moths, variations in sex pheromone are an important component in the evolution of the sex pheromone communication system10, which is an initial step towards pre-mating reproductive isolation in host populations5. In this study, we revealed the differences in the expression patterns of putative sex pheromone biosynthetic genes in rice and water-oat populations of C. suppressalis, which is of great significance for our understanding of the endogenous mechanisms of intraspecific reproductive isolation
Here we obtained 11 key putative DEGs involved in sex pheromone biosynthesis with higher expression in the rice population (Table 3). Among them, ELO, ACC and FATP are all highly important enzymes in the fatty acid synthesis, which are essential precursors in the process of sex pheromones production34–37. For example, ACC as an important regulator mediated de novo biosynthesis of fatty acid from acetyl-CoA38, and it has been proved to be a key component involved in sex pheromone biosynthesis in Helicoverpa armigera39. The extracellular fatty acids can also be derived, transported, absorbed and activated by FATP40. Additionally, acetyl-CoA can be used as substrate and fatty acids can continue to extend the carbon chain under the action of ELO36,41. The increased expression of these genes in the rice population suggested that the long-chain fatty acids were derived not only from intracellular sources but were also supplied energy from extracellular substrates at the same time (Fig. 3A), so that there was more increased substrate content for sex pheromone biosynthesis in the rice population. Desaturases are key enzymes that introduce double bonds into pheromone molecules42,43. It has been confirmed in studies of B. mori44, Plutella xylostella45, Ostrinia moths46,47 and Thaumetopoea pityocampa48 that desaturases play an important role in sex pheromone biosynthesis. In the sibling species of leafroller moths, Ctenopseustis and Planotortrix, differential expression of the desaturase genes resulted in interspecific sex pheromone differences49. In our research, we found that the expressions of DES1 and DES7 in rice population were higher than that in water-oat population (Fig. 3D). This result showed that desaturation would be more effective in increase of sex pheromone biosynthesis in the rice population. ACD catalyses the β-oxidation (shortening of carbon chains) of long-chain fatty acids and ALR can catalyse fatty acids into fatty aldehydes33,50. In our study, we speculate that the high expression of ALR2, ALR3 and ACD could expedite the β-oxidation or reduction process and elevate the sex pheromone biosynthesis in the rice population. The sex pheromone titer measurement would be required to further validate.
On the other hand, we found that there were differences in the expression rhythms over time of the putative genes related to sex pheromone synthesis between the two host populations (Fig. 3). Among them, ACC, FATP, ELO7 and DES1 were synchronously highly expressed in the early stages of scotophase in rice population and in the late stages of scotophase in water-oat population (Fig. 3A,D). This difference potentially gives rise to variations in sex pheromone production, which adapts to the courtship and mating rhythm of different populations50,51.
Conclusions
Taken together, these results revealed that there are divergences in expression levels and rhythms of putative genes associated with sex pheromone biosynthesis in the two host populations. Further explorations would be needed to assess the specific functions of these genes in sex pheromone production. Our study provides a genetic basis for understanding mechanism of intraspecific reproductive isolation.
Materials and Methods
Insects
Approximately 1000 and 700 overwintering larvae of C. suppressalis were collected from rice (113° 57′E, 30° 29′N) and water-oat fields (114° 16′E, 30° 28′N), respectively, in Wuhan City, China in December 2015. The collected overwintering larvae of C. suppressalis were reared at 28 ± 1 °C, with LD 15: 9 h (light: dark photoperiod) and RH = 80 ± 5% in an insectarium52. The mating rhythm of emerged moths in the overwintering generation was observed to identify the rice and water-oat populations16,53,54. The offspring of the two host populations were fed on an artificial diet55, and the 2-day-old virgin female moths were used in the subsequent experiments.
Sex pheromone gland sample preparation for RNA extraction
Our previous study have shown that the mating activity reached different peaks at 3 h and 6 h after the onset of scotophase in the rice and water-oat populations, respectively16. We thus prepared pheromone glands samples for RNA-Seq at 0 h (R0, W0), 3 h (R3, W3) and 6 h (R6, W6) after the onset of scotophase in two host populations. The pheromone glands were extruded by gently exerting force on the abdomen of 2-day-old virgin female moths randomly for samples. Pheromone glands were extracted and immediately frozen in liquid nitrogen and stored at −80 °C for RNA extraction. Samples of C. suppressalis pheromone glands were collected from groups of 30 females, and three biological replicates were used for each analysis. Total RNA was extracted using the Trizol reagent (Invitrogen, Life Technologies, USA) following the manufacturer’s instructions.
cDNA library construction and RNA sequencing
A total of 1.5 µg of RNA per sample was used as the input material for the RNA sample preparations. Sequencing libraries were generated using NEBNext Ultra RNA Library Prep Kit for Illumina (NEB, USA) following the manufacturer’s recommendations, and index codes were added to attribute the sequences to each sample. The concentration and size of the cDNA library was determined with a Qubit 2.0 Fluorometer (Invitrogen, Carlsbad, CA, US), and library quality was assessed on the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). After cDNA sample quality inspection, the library preparations were sequenced on an Illumina HiSeq. 2500 system (Illumina, San Diego, CA, USA) with paired-end reads. The base periods and mass fractions resulting from the high–throughput sequencing were recorded in the FASTQ format.
De novo transcriptome assembly and functional annotation
The raw reads were filtered to remove adaptors, low-quality reads (less than 50% ratio of greater than 20 bases), and reads showing an N ratio (unknown sequences) greater than 5% using the FastX programme (version 0.0.13). After filtering, we obtained and evaluated the clean reads. A pool of reads was formed by merging eighteen samples of sequencing data. We performed de novo transcriptome assembly of the clean reads to obtain the final unigenes using Trinity56 with min_kmer_cov set to 2 by default and all other parameters set to default. Gene function was annotated based on the following databases: NR (NCBI non-redundant protein sequences), Nt (NCBI non-redundant nucleotide sequences), Swiss-Prot (A manually annotated and reviewed protein sequence database), PFAM (Protein family), KOG/COG (Clusters of Orthologous Groups of proteins), KEGG (Kyoto Encyclopedia of Genes and Genomes) and GO (Gene Ontology).
Differentially expressed gene (DEG) profiling of rice and water-oat populations of C. suppressalis
By combining and splicing the mRNA sequences of the 18 samples from the two populations of C. suppressalis, we obtained transcriptome databases and used them as a reference sequence. We matched the clean reads of each sample in the reference sequence using RSEM software57. Default values were used for the Bowtie 2 parameters used in the RSEM software.
For the differential gene expression analysis, we obtained the key genes with differentially expressed counts from the different samples and performed GO functional analysis and KEGG pathway analysis. The P-value was calculated as described by Robinson and Rivals58,59. The FPKM (Fragments Per Kilobase of transcript sequence per Millions base pairs sequenced) method was used to calculate the unigene expression counts and differential expression levels between different samples for an individual gene60. This method eliminates the influence of differences in lengths and number of sequences on the expression level. In addition, we used DESeq to screen for differentially expressed genes among the two populations. Genes with a |log2 ratio| ≥ 1 and an adjusted P-value <0.05 were used as the parameters for DEGs.
Identification of putative genes associated with sex pheromone biosynthesis and DEGs analysis
We focused on the putative target genes involved in moth sex pheromone production. Candidate unigenes were selected according to their NR annotation. To compare the expression levels of sex pheromone biosynthetic genes between rice and water-oat populations, a profiling analysis of the DEGs was conducted. The FPKM value of each gene was used for comparing expression levels.
Quantitative real-time PCR (RT-qPCR) validation
To further validate the differences in gene expression levels between the rice and water-oat populations, we randomly selected 5 genes and quantified their expression by ABI QuantStudio 6 Flex (Thermo Fisher Scientific, Massachusetts, USA), with three biological and technical replicates. Briefly, total RNA was extracted using the RNAiso Plus reagent (TAKARA, 9109, Dalian, China). The PrimeScript RT Reagent Kit with gDNA Eraser (TAKARA, RR047, Dalian, China) were used to reverse transcribe and synthesize cDNA. Premier 5.0 (Premier Biosoft International, Palo Alto, CA) was used to design the primers (Table S4). The β-actin and GAPDH genes of C. suppressalis were used as internal reference genes24. The melting curves and standard curves of β-actin, GAPDH, ELO1, ALF and ACD were analyzed by qTOWER 2.0 Real-Time PCR (Analytik Jena, Jena, Germany), and DES1 and DES7 were determined by ABI QuantStudio 6 Flex (Fig. S2). RT-qPCR efficiency of primers were determined by a slope analysis with a linear regression model. Calculating the corresponding RT-qPCR efficiency (E) according to the following formula: E = (10[−1/slope]−1) × 100%61. Relative expression was analyzed via the 2−ΔΔCT method62 and normalized the geometric mean of two reference genes expression63,64.
Data analysis
Heatmap analysis is based on normalised FPKM via Z-score method65. The relative gene expression values were analysed by SPSS 11.5 software (SPSS Inc., Chicago, IL, USA, https://www.ibm.com/analytics/spss-statistics-software) with independent sample t-tests. All the results were visualised using GraphPad Prism 8 (GraphPad Software Inc., San Diego, CA, https://www.graphpad.com) software packages. The FPKM values and relative expression values were represented as the means ± standard error of mean (SEM) based on three biological replicates.
Supplementary information
Acknowledgements
We thank Rong Chen and Yuan Zhou for their generous help in collecting and rearing insects. We also thank Novogene (Novogene, Beijing, China) for sequencing consultation and support. This study was funded by grants from the Hubei Provincial Natural Science Foundation of China (Grant number 2016CFA068).
Author contributions
S.G. and Z.T. contributed equally to this work. S.G., Z.T., W.L.Q., W.L. and X.P.W. conceived and designed this study. S.G., Z.T., W.L.Q. and D.S. performed the experiment and data analyses. S.G., Z.T. and W.L.Q. contributed materials/ analysis tools. S.G. and Z.T. wrote the manuscript with help from all the authors. All authors reviewed manuscript.
Data availability
All the data presented here will be available under request to the scientific community.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Shuang Guo and Zhong Tian.
Supplementary information
is available for this paper at 10.1038/s41598-020-60529-x.
References
- 1.Diehl S, Bush G. An evolutionary and applied perspective of insect biotypes. Annu. Rev. Entomol. 1984;29:471–504. doi: 10.1146/annurev.en.29.010184.002351. [DOI] [Google Scholar]
- 2.Salt RGW. The evolution of reproductive isolation as a correlated character under sympatric conditions: experimental dvidence. Evolution. 1990;44:1140–1152. doi: 10.1111/j.1558-5646.1990.tb05221.x. [DOI] [PubMed] [Google Scholar]
- 3.Bolnick DI, Fitzpatrick BM. Sympatric speciation: models and empirical evidence. Annu. Rev. Ecol. Evol. S. 2007;38:459–487. doi: 10.1146/annurev.ecolsys.38.091206.095804. [DOI] [Google Scholar]
- 4.Anglade P, Stockel J, Cooperators IWGO. Intraspecific sex-pheromone variability in the European corn borer, Ostrinia nubilalis Hbn. (Lepidoptera, Pyralidae) Agronomie. 1984;4:183–187. doi: 10.1051/agro:19840209. [DOI] [Google Scholar]
- 5.Groot AT, et al. Host strain specific sex pheromone variation in Spodoptera frugiperda. Front. Zool. 2008;5:20. doi: 10.1186/1742-9994-5-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smadja C, Butlin RK. On the scent of speciation: the chemosensory system and its role in premating isolation. Heredity. 2009;102:77. doi: 10.1038/hdy.2008.55. [DOI] [PubMed] [Google Scholar]
- 7.Astridt G, Melanie M, Davidg H, Gerhard S. The roles and interactions of reproductive isolation mechanisms in fall armyworm (Lepidoptera: Noctuidae) host strains. Ecol. Entomol. 2010;35:105–118. doi: 10.1111/j.1365-2311.2009.01138.x. [DOI] [Google Scholar]
- 8.Duménil C, et al. Intraspecific Variation in Female Sex Pheromone of the Codling Moth Cydia pomonella. Insects. 2014;5:705–721. doi: 10.3390/insects5040705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ming QL, Yan YH, Wang CZ. Mechanisms of premating isolation between Helicoverpa armigera (Hübner) and Helicoverpa assulta (Guenée) (Lepidoptera: Noctuidae) J. Insect Physiol. 2007;53:170–178. doi: 10.1016/j.jinsphys.2006.11.007. [DOI] [PubMed] [Google Scholar]
- 10.Kiritani, K. & Tatsuki, S. The Rice stem borer, Chilo suppressalis: A history of applied entomology in Japan. Tokyo: University of Tokyo Press (2009).
- 11.Lassance JM, et al. Allelic variation in a fatty-acyl reductase gene causes divergence in moth sex pheromones. Nat. 2010;466:486–489. doi: 10.1038/nature09058. [DOI] [PubMed] [Google Scholar]
- 12.Xiang YY, Yang MF, Li ZZ. Calling behavior and rhythms of sex pheromone production in the black cutworm moth in China. J. Insect Behav. 2010;23:35–44. doi: 10.1007/s10905-009-9193-0. [DOI] [Google Scholar]
- 13.Liu HX, Zhao WM, Yang MH, Liu JL, Zhang JT. Diel rhythms of sexual behavior and pheromone titers in Isoceras sibirica Alpherakey (Lepidoptera, cossidae) Arch. Insect Biochem. Physiol. 2013;84:15–26. doi: 10.1002/arch.21109. [DOI] [PubMed] [Google Scholar]
- 14.Kamimura M, Tatsuki S. Diel rhythms of calling behavior and pheromone production of oriental tobacco budworm moth, Helicoverpa assulta (Lepidoptera: Noctuidae) J. Chem. Ecol. 1993;19:2953–2963. doi: 10.1007/BF00980595. [DOI] [PubMed] [Google Scholar]
- 15.Mazor M, Dunkelblum E. Circadian rhythms of sexual behavior and pheromone titers of two closely related moth species autographa gamma and Cornutiplusia circumflexa. J. Chem. Ecol. 2005;31:2153–2168. doi: 10.1007/s10886-005-6082-7. [DOI] [PubMed] [Google Scholar]
- 16.Quan WL, et al. Do differences in life-history traits and the timing of peak mating activity between host-associated populations of Chilo suppressalis have a genetic basis? Ecol. Evol. 2016;6:4478–4487. doi: 10.1002/ece3.2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roelofs W, et al. Sex pheromone production and perception in European corn borer moths is determined by both autosomal and sex-linked genes. Proc. Natl Acad. Sci. USA. 1987;84:7585–7589. doi: 10.1073/pnas.84.21.7585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dopman EB, Bogdanowicz SM, Harrison RG. Genetic mapping of sexual isolation between E and Z pheromone strains of the Eeuropean corn Borer (Ostrinia nubilalis) Genet. 2004;167:301–309. doi: 10.1534/genetics.167.1.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Groot AT, Dekker T, Heckel DG. The genetic basis of pheromone evolution in moths. Annu. Rev. Entomol. 2016;61:99–117. doi: 10.1146/annurev-ento-010715-023638. [DOI] [PubMed] [Google Scholar]
- 20.Strandh M, Johansson T, Ahren D, Lofstedt C. Transcriptional analysis of the pheromone gland of the turnip moth, Agrotis segetum (Noctuidae), reveals candidate genes involved in pheromone production. Insect Mol. Biol. 2008;17:73–85. doi: 10.1111/j.1365-2583.2008.00782.x. [DOI] [PubMed] [Google Scholar]
- 21.Vogel H, Heidel AJ, Heckel DG, Groot AT. Transcriptome analysis of the sex pheromone gland of the noctuid moth Heliothis virescens. BMC Genomics. 2010;11:29. doi: 10.1186/1471-2164-11-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gu SH, et al. Identification of genes expressed in the sex pheromone gland of the black cutworm Agrotis ipsilon with putative roles in sex pheromone biosynthesis and transport. BMC Genomics. 2013;14:636. doi: 10.1186/1471-2164-14-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Antony B, et al. Genes involved in sex pheromone biosynthesis of Ephestia cautella, an important food storage pest, are determined by transcriptome sequencing. BMC Genomics. 2015;16:532. doi: 10.1186/s12864-015-1710-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xia YH, Zhang YN, Hou XQ, Li F, Dong SL. Large number of putative chemoreception and pheromone biosynthesis genes revealed by analyzing transcriptome from ovipositor-pheromone glands of Chilo suppressalis. Sci. Rep. 2015;5:7888. doi: 10.1038/srep07888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Du M, et al. Identification of lipases Involved in PBAN stimulated pheromone production in Bombyx mori using the DGE and RNAi approaches. Plos One. 2012;7:e31045. doi: 10.1371/journal.pone.0031045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang S, et al. Identification of differentially expressed genes in the pheromone glands of mated and virgin Bombyx mori by digital gene expression profiling. Plos One. 2014;9:e111003. doi: 10.1371/journal.pone.0111003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maki Y, Yamashita M. Ecological difference of rice stem borer, Chilo suppressalis Walker in the various host plants. Bull. Hyogo Pref. Agric. Exp. Stn. 1956;3:47–50. [Google Scholar]
- 28.Samudra I. Temporal differences in mating behavior between rice- and water-oat-populations of the striped stem borer, Chilo suppressalis (Walker) (Lepidoptera: Crambidae) Appl. Entomol. Zool. 2002;37:257–262. doi: 10.1303/aez.2002.257. [DOI] [Google Scholar]
- 29.Hou ML, Lin W, Han YQ. Seasonal changes in supercooling points and glycerol content in overwintering larvae of the Asiatic rice borer from rice and water-oat plants. Env. Entomol. 2009;38:1182–1188. doi: 10.1603/022.038.0427. [DOI] [PubMed] [Google Scholar]
- 30.Zhou Y, et al. Divergence in larval diapause induction between the rice and water-oat populations of the striped stem borer, Chilo suppressalis (Walker) (Lepidoptera: Crambidae) Env. Sci. Pollut. Res. 2018;25:29715–29724. doi: 10.1007/s11356-018-2930-1. [DOI] [PubMed] [Google Scholar]
- 31.Quan WL, et al. Difference in diel mating time contributes to assortative mating between host plant-associated populations of Chilo suppressalis. Sci. Rep. 2017;7:45265. doi: 10.1038/srep45265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Götz S, et al. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008;36:3420–3435. doi: 10.1093/nar/gkn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang YN, Xia YH, Zhu JY, Li SY, Dong SL. Putative pathway of sex pheromone biosynthesis and degradation by expression patterns of genes identified from female pheromone gland and adult antenna of Sesamia inferens (Walker) J. Chem. Ecol. 2014;40:439–451. doi: 10.1007/s10886-014-0433-1. [DOI] [PubMed] [Google Scholar]
- 34.Raina AK. Neuroendocrine control of sex pheromone biosynthesis in Lepidoptera. Annu. Rev. Entomol. 1993;38:329–349. doi: 10.1146/annurev.en.38.010193.001553. [DOI] [PubMed] [Google Scholar]
- 35.Tillman JA, Seybold SJ, Jurenka RA, Blomquist GJ. Insect pheromones-an overview of biosynthesis and endocrine regulation. Insect Biochem. Mol. Biol. 1999;29:481–514. doi: 10.1016/S0965-1748(99)00016-8. [DOI] [PubMed] [Google Scholar]
- 36.Chertemps T, et al. A female-biased expressed elongase involved in long-chain hydrocarbon biosynthesis and courtship behavior in Drosophila melanogaster. Proc. Natl Acad. Sci. USA. 2007;104:4273–4278. doi: 10.1073/pnas.0608142104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Groot AT. Circadian rhythms of sexual activities in moths: a review. Front. Ecol. Evol. 2014;2:1–6. doi: 10.3389/fevo.2014.00043. [DOI] [Google Scholar]
- 38.Choi JW, Da Silva NA. Improving polyketide and fatty acid synthesis by engineering of the yeast acetyl-CoA carboxylase. J. Biotechnol. 2014;187:56–59. doi: 10.1016/j.jbiotec.2014.07.430. [DOI] [PubMed] [Google Scholar]
- 39.Du M, et al. Calcineurin-mediated dephosphorylation of Acetyl-coA carboxylase is required for pheromone biosynthesis activating neuropeptide (PBAN)-induced sex pheromone biosynthesis in Helicoverpa armigera. Mol. Cell Proteom. 2007;16:2138–2152. doi: 10.1074/mcp.RA117.000065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anderson CM, Stahl A. SLC27 fatty acid transport proteins. Asp. Med. 2013;34:516–528. doi: 10.1016/j.mam.2012.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oh CS, Toke DA, Mandala S, Martin CE. ELO2 and ELO3, homologues of the Saccharomyces cerevisiae ELO1 gene, function in fatty acid elongation and are required for sphingolipid formation. J. Biol. Chem. 1997;272:17376–17384. doi: 10.1074/jbc.272.28.17376. [DOI] [PubMed] [Google Scholar]
- 42.Zhao C, Löfstedt C, Wang X. Sex pheromone biosynthesis in the Asian corn borer Ostrinia furnacalis (II): Biosynthesis of (E)- and (Z)-12-tetradecenyl acetate involves Δ14 desaturation. Arch. Insect Biochem. Physiol. 1990;15:57–65. doi: 10.1002/arch.940150106. [DOI] [Google Scholar]
- 43.Liénard MA, et al. Elucidation of the sex-pheromone biosynthesis producing 5, 7-dodecadienes in Dendrolimus punctatus (Lepidoptera: Lasiocampidae) reveals Δ11- and Δ9-desaturases with unusual catalytic properties. Insect Biochem. Mol. Bio. 2010;40:440–452. doi: 10.1016/j.ibmb.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 44.Moto K, et al. Involvement of a Bifunctional Fatty-Acyl Desaturase in the Biosynthesis of the Silkmoth, Bombyx mori, Sex Pheromone. Proc. Natl Acad. Sci. USA. 2004;101:8631. doi: 10.1073/pnas.0402056101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.He P, et al. A reference gene set for sex pheromone biosynthesis and degradation genes from the diamondback moth, Plutella xylostella, based on genome and transcriptome digital gene expression analyses. BMC Genomics. 2017;18:219. doi: 10.1186/s12864-017-3592-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sakai R, Fukuzawa M, Nakano R, Tatsuki S, Ishikawa Y. Alternative suppression of transcription from two desaturase genes is the key for species-specific sex pheromone biosynthesis in two Ostrinia moths. Insect Biochem. Mol. Bio. 2009;39:62–67. doi: 10.1016/j.ibmb.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 47.Fujii T, et al. Sex pheromone desaturase functioning in a primitive Ostrinia moth is cryptically conserved in congeners’ genomes. Proc. Natl Acad. Sci. USA. 2011;108:7102–7106. doi: 10.1073/pnas.1019519108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arsequell G, Fabriàs G, Camps F. Sex pheromone biosynthesis in the processionary moth Thaumetopoea pityocampa, by delta-13 desaturation. Arch. Insect Biochem. Physiol. 1990;14:47. doi: 10.1002/arch.940140106. [DOI] [PubMed] [Google Scholar]
- 49.Albre J, et al. Sex pheromone evolution is associated with differential regulation of the same desaturase gene in two genera of leafroller moths. Plos Genet. 2012;8:e1002489. doi: 10.1371/journal.pgen.1002489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Strandh M, Johansson T, Löfstedt C. Global transcriptional analysis of pheromone biosynthesis-related genes in the female turnip moth, Agrotis segetum (Noctuidae) using a custom-made cDNA microarray. Insect Biochem. Mol. Biol. 2009;39:484–489. doi: 10.1016/j.ibmb.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 51.Lin X, Wang B, Du Y. Key genes of the sex pheromone biosynthesis pathway in female moths are required for pheromone quality and possibly mediate olfactory plasticity in conspecific male moths in Spodoptera litura. Insect Mol. Bio. 2017;27:8–21. doi: 10.1111/imb.12335. [DOI] [PubMed] [Google Scholar]
- 52.Xu S, et al. Relationships between body weight of overwintering larvae and supercooling capacity; diapause intensity and post-diapause reproductive potential in Chilo suppressalis Walker. J. Insect Physiol. 2011;57:653–659. doi: 10.1016/j.jinsphys.2010.12.010. [DOI] [PubMed] [Google Scholar]
- 53.Konno Y. Mating-choice and host preference tests in the rice-feeding and water-oat-feeding types of the rice stem borer, Chilo suppressalis Walker (Lepidoptera: Pyralidae) Ann. Rept Plant. Prot. North. Jpn. 1998;49:102–104. [Google Scholar]
- 54.Ishiguro N, Yoshida K, Tsuchida K. Genetic differences between rice and water-oat feeders in the rice stem borer, Chilo suppressalis (Walker) (Lepidoptera: Crambidae) Appl. Entomol. Zool. 2006;41:585–593. doi: 10.1303/aez.2006.585. [DOI] [Google Scholar]
- 55.Han LZ, Li SB, Liu PL, Peng YF, Hou ML. New artificial diet for continuous rearing of Chilo suppressalis (Lepidoptera: Crambidae) Ann. Entomol. Soc. Am. 2012;105:253–258. doi: 10.1603/AN10170. [DOI] [Google Scholar]
- 56.Grabherr MG, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinforma. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Robinson MD, Grigull J, Mohammad N, Hughes TR. FunSpec: a web-based cluster interpreter for yeast. BMC Bioinforma. 2002;3:35. doi: 10.1186/1471-2105-3-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rivals I, Personnaz L, Taing L, Potier MC. Enrichment or depletion of a GO category within a class of genes: which test? Bioinforma. 2007;23:401–407. doi: 10.1093/bioinformatics/btl633. [DOI] [PubMed] [Google Scholar]
- 60.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C (T) method. Nat. Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 63.Nailis H, et al. Development and evaluation of different normalization strategies for gene expression studies in Candida albicans biofilms by real-time PCR. BMC Mol. Biol. 2006;7:25. doi: 10.1186/1471-2199-7-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cao KX, et al. Cold exposure induces the acquisition of brown adipocyte gene expression profiles in cattle inguinal fat normalized with a new set of reference genes for qRT-PCR. Res. Vet. Sci. 2017;114:1–5. doi: 10.1016/j.rvsc.2017.02.021. [DOI] [PubMed] [Google Scholar]
- 65.Kang WH, et al. Transcriptome profiling of abiotic responses to heat, cold, salt, and osmotic stress of Capsicum annuum L. Sci. Data. 2020;7:17. doi: 10.1038/s41597-020-0352-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data presented here will be available under request to the scientific community.