Abstract
WW domain binding protein 5 (WBP5), also known as Transcriptional Elongation Factor A like 9 (TCEAL9) has been proposed as a candidate oncogene for human colorectal cancers with microsatellite instability and as a predictive indicator of small cell lung cancers. Furthermore, several independent studies have proposed WBP5, and its association with Wilms Tumor-1 (WT1) expression, as part of a gene expression-based risk score for predicting survival and clinical outcome in patients with Acute Myeloid Leukaemia (AML). To date, the prognostic significance of the sole WBP5 expression and its impact on the survival outcome in AML patients remains largely understudied. In the present study, we have made use of publicly available patient expression arrays and have developed an unbiased approach to classify AML patients into low versus high WBP5 expressers and to balance them for known mutations and cytogenetic findings. Interestingly, we found that patients characterized by high WBP5 expression displayed inferior overall and event-free survival rates. Notably, gene expression profiling showed that patients with high WBP5 had elevated expression of several HOX cluster genes, such as HOXA5, HOXA7, HOXA9 and HOXA10, and several of their partner proteins, such as MEIS1 and FOXC1, which have been demonstrated to be causative for AML. Taken together, our data suggest that WBP5 expression level could serve as an indicator for prognosis and survival outcome in patients with AML.
Subject terms: Acute myeloid leukaemia, Data mining
Introduction
Acute myeloid leukaemia (AML) is a hierarchically-organized myeloproliferative disorder that is caused by stepwise acquisition of different mutations that prime malignant transformation and affect normal maturation of myeloid precursor cells1,2. Despite concerted efforts in the development of new treatments, many patients are refractory to current therapeutic approaches or have a high relapse rate, with the overall long-term survival of patients being below 40% and more than 60% of the patients over 65 years of age succumbing to the disease within one year of diagnosis3. In current medical practice, the diagnosis, prognosis, and therapeutic choices are dictated by detection of genetic mutations and the measurement of specific biomarkers that are used to classify patients into risk categories. However, due to the heterogeneous nature of the disease, prognosis within these categories is highly variable.
Aside from chromosome lesions, such as those involving MLL (i.e. MLL-AF9, MLL-ENL)4,5 or RUNX1 (i.e. RUNX1-ETO, RUNX1-EVI1)6,7 translocations, common prognostic and categorization factors involve mutations in the genes encoding signalling proteins (FLT3, RAS and KIT)8,9, transcription factors (CEBPA, PU.1 and GATA2)10–12 and DNA methylation related genes (TET2, DNMT3A, IDH1 and IDH2)13–16. Patient stratification has been further refined by the advent of next-generation whole-genome and transcriptome sequencing technologies; however, the identification of new reliable biomarkers is still required in clinical practice for use as prognostic factors and as new actionable therapeutic targets.
WW domain binding protein 5 (WBP5), a novel upstream regulator of the Hippo pathway17, has been recently associated with a variety of cancers, such as advanced gastric cancer with aggressive lymph node metastasis18, colorectal cancers with microsatellite instability19 and in small cell lung cancers20 where it has been reported to influence tumour growth by promoting cell proliferation and inhibiting apoptosis. A link of WBP5 with leukaemia has become evident in recent years through the generation of gene expression prognostic signatures for predicting clinical outcomes in patients with AML. In fact, by making use of cDNA microarrays, Metzeler and co-workers presented WBP5 as part of a gene expression-based signature that comprises an 86-probe set (66 genes), which was used for predicting survival outcome in patients with cytogenetically normal AML (CN-AML)21. Subsequently, Bou Samra et al. developed an independent and further refined gene expression-based risk score in which WBP5 was part of a 22-gene signature that displayed a strong prognostic value in 2 independent cohorts of CN-AML patients22. Recently, Niavarani et al. have reported WBP5 as part of a 17-probe set signature to predict unfavourable outcome in association with high levels of WT1 in AML patients23. To date, whether the sole expression of WBP5 could serve as a prognostic indicator and whether its expression has any impact on the establishment and maintenance of myeloid diseases has not been assessed.
In the present study, we have taken an unbiased bioinformatic approach to identify new molecular biomarkers by making use of publicly available patient gene expression arrays in which the whole AML patient cohort was ranked according to the expression of every gene probe into high and low expressers and used to determine the impact of this classification on the overall and event-free survival outcome and on the gene expression profiles. Our approach identified WBP5 as one of the most significant genes and showed that high expression of WBP5 is associated with a markedly inferior outcome and with an elevation of leukaemia associated HOX gene clusters expression. The prognostic value of WBP5 was validated in five independent AML gene expression datasets, thus suggesting WBP5 to be a new reliable molecular biomarker and a new potential therapeutic candidate for AML patients.
Materials and Methods
Patient profiling arrays information
The overall survival (OS) and event-free survival (EFS) scores were determined using non-parametric Kaplan-Meyer estimates; comparison of survival between the low and high WBP5 subgroups was based on two-sided log rank test.
Data processing
GSE689124, GSE1543425, GSE1320426, GSE115927, GSE2284528 microarray raw data were downloaded from NCBI Gene Expression Omnibus (GEO). For each probe in each dataset, expression was scaled from 0 to 1. Then high and low expressing patient groups were established using 0.7–1 or 0–0.3 expression, respectively. Once subgroups were determined, raw CEL data was downloaded for each patient and expression values were calculated, background corrected, log2 transformed and quantile normalized in R (version 3.6.0) using affy package (version 1.62.0) and the rma function. Differential gene expression was carried out using the limma package (version 3.40.2) by fitting a linear model of high vs low patients for WBP5 expression.
Subgroup balancing
After selecting high and low expressing patients as separate subgroups, we checked if there was a significant proportion of each subgroup that contained patients with specific cytogenetic abnormality, disease marker expression, age or gender using Fisher exact test. If we found a significant imbalance, we adopted a randomization strategy to balance the groups. Patients were shuffled in and out of the high and low expressing subgroups until there was no significant proportion of a specific cytogenetic abnormality, disease marker expression, age or gender.
Survival analysis
For the GSE6891, GSE1241721 and GSE3764229 datasets, the high expressing subgroup and the low expressing subgroup were compared for each probe using overall survival data. Event-free survival was analysed for GSE6891 dataset only. A p-value was calculated to determine significant differences using Wilcoxon rank-sum test.
Kaplan-Meier plots were generated using Python (version 3.5.5) lifelines package (version 0.14.6). P-values represent Wilcoxon rank-sum test results comparing high and low expressing patients.
Unsupervised hierarchical clustering
The hierarchical clustering of patients shown in this study was unsupervised and was performed on normalized data using Pearson correlation Euclidian distance metric with complete linkage agglomeration method. Hierarchical clustering of genes was done using the z-score values of the genes based on Euclidian distance metric with complete linkage agglomeration method.
Statistical analysis
Statistical analysis throughout this study was determined by performing t-test for pairwise comparison and the p-values are indicated where appropriate.
Results
Generation of a workflow for identification of new molecular biomarkers
To search for new potential molecular biomarkers for AML, we retrieved 5 independent publicly available microarray datasets GSE689124 (461 samples), GSE1543425 (251 samples), GSE1320426 (542 samples), GSE115927 (285 samples) and GSE2284528 (154 samples) from the Gene Expression Omnibus (GEO) public repository. Firstly, we applied our pipeline described in Fig. 1 to the GSE6891 dataset as it encompasses patients with different types of AML and every patient within the cohort is annotated with a comprehensive mutational and survival analysis. We developed a computer algorithm to systematically screen every gene probe within the dataset by ranking all patients from the lowest to the highest and classifying them into low (0–30% of the expression range) and high expressers (70–100%) for each given probe. For each gene, a comparison between the low and high subgroups was then made by screening survival data to identify those that have a statistically significant impact on both overall and event-free survival, i.e. those demonstrating a p-value lower than 0.05 as calculated by Wilcoxon rank-sum test. Upon identification of a positive hit, low and high expresser subgroups were adjusted for age, sex, cytogenetic findings, and mutational status and the survival analysis was repeated. After balancing, the list of genes that demonstrated a significant survival impact was then trimmed based on the involvement or deregulation of those genes in cancer; the genes that were positive by all those criteria were then surveyed for their influence on global gene expression and further validated in the other independent datasets. WBP5, a gene that has recently been associated with a variety of cancers and haematological malignancies, passed all criteria and became the main subject of this study.
Figure 1.
Flow chart showing the sequential steps for the identification of new molecular biomarkers from AML patient expression arrays.
High WBP5 expression is associated with inferior survival in AML patients
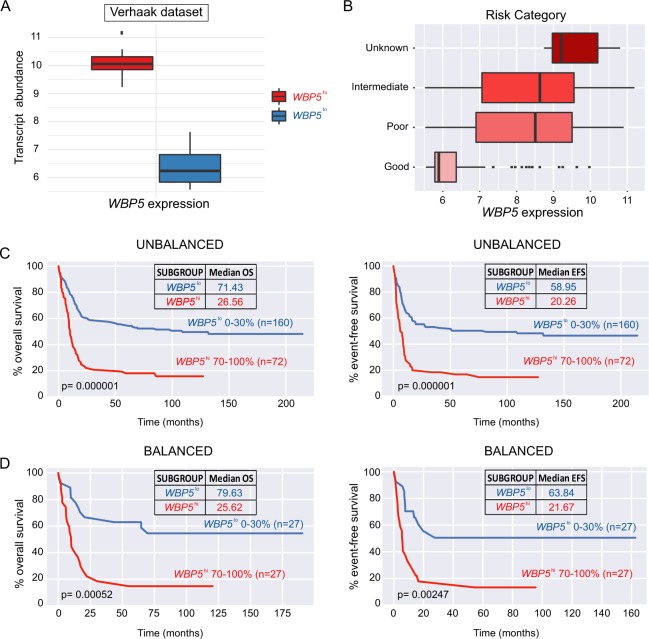
In the patient cohort described by Verhaak et al.24 we ranked patients according to their WBP5 expression levels and selected the bottom 30% of the whole expression range as low expressers (WBP5low, n = 160) and the top 30% as high expressers (WBP5high, n = 72) as indicated in Fig. 2A. We found a large association of high WBP5 levels with poor cytogenetic risk, while a large proportion of low expressers belonged to the good risk subgroup (Fig. 2B). Next, we compared the survival outcomes for the whole cohort and observed WBP5high patients to be associated with a significantly unfavourable overall survival (median OS = 26.56 vs 71.43, p = 0.000001) and event-free survival outcome (median EFS = 20.26 vs 58.95, p = 0.00001) (Fig. 2C).
Figure 2.
High WBP5 expression is associated with unfavourable outcome in patients with AML. (A) Boxplot representation of WBP5 expression boundaries for patient samples from the Verhaak et al. dataset (0–30% low expression, 70–100% high expression). (B) Boxplot depicting the association of WBP5 expression with risk group categorization (poor, intermediate, good and unknown). (C) Kaplan-Meier representation of overall (OS) and event-free (EFS) survival for WBP5low and WBP5high AML patients prior to or (D) after balancing for age, sex, FAB and cytogenetic findings. The number of patients and the median OS and EFS are indicated in the plot. P-value was calculated using Wilcoxon rank-sum test.
Furthermore, we tested how WBP5 expression could be modulated in different AML subtypes by analysing the mutational status of the two subgroups (WBP5high vs WBP5low); this approach demonstrated a correlation of high WBP5 expression with adverse cytogenetic findings such as NPM1c and FLT3-ITD, these being found in 43% vs 15% (p = 0.0000089) and in 40.2% vs 13.7% (p = 0.000013), respectively. Moreover, we found EVI1 to be more frequent in the WBP5high group when compared to the WBP5low group (15.27% vs 1.25%, p = 0.000063); conversely, CEBPA mutations, that are normally associated with favourable prognosis, were exclusively found in WBP5low patients (11.25% vs 0%, p = 0.0011). Furthermore, we observed the low expressers to be largely associated with idt(16) (14.37% vs 1.38%, p = 0.0018) and t(8;21) translocation (22% vs 0%, p = 0.0000046), a genetic lesion normally associated with good overall survival. No relationship between WBP5 expression and age or sex was observed (Table 1).
Table 1.
Genetic and karyotypic characteristics of the WBP5high and WBP5low patient subgroups.
| WBP5low (n = 160) | WBP5hi (n = 72) | p-value | |
|---|---|---|---|
| Molecular abnormalities | |||
| IDH1 | 7 | 4 | 0.74231192 |
| IDH2 | 5 | 6 | 0.10035424 |
| NPM1c | 24 | 31 | 8.95E-06 |
| FLT3-ITD | 22 | 29 | 1.38E-05 |
| FLT3-TKD | 17 | 7 | 1 |
| NRAS | 22 | 4 | 0.07474453 |
| KRAS | 4 | 0 | 0.31345029 |
| EVI1 | 2 | 11 | 6.37E-05 |
| CEBPA | 18 | 0 | 0.00112984 |
| Karyotypic abnormalities | |||
| +8 | 10 | 4 | 1 |
| −5/7q | 6 | 10 | 0.00925701 |
| −9q | 5 | 0 | 0.32772068 |
| 11q23 | 5 | 3 | 0.70598866 |
| complex | 8 | 0 | 0.06052875 |
| NN | 50 | 31 | 0.10135058 |
| Other | 14 | 9 | 0.47627351 |
| abn(3q) | 0 | 1 | 1 |
| failure | 1 | 4 | 0.03330118 |
| idt(16) | 23 | 1 | 0.00180154 |
| t(15;17) | 1 | 0 | 0.1433 |
| t(6;9) | 0 | 1 | 0.31034483 |
| t(8;21) | 32 | 0 | 4.68E-06 |
| t(9;22) | 1 | 0 | 1 |
| FAB classification | |||
| M0 | 8 | 3 | 1 |
| M1 | 25 | 27 | 0.00054777 |
| M2 | 39 | 20 | 0.62606826 |
| M4 | 36 | 8 | 0.04664368 |
| M5 | 39 | 8 | 0.0216892 |
| M6 | 0 | 1 | 0.31034483 |
| unknown | 3 | 0 | 0.55419693 |
| Sex | |||
| Male | 80 | 31 | 0.39421991 |
| Female | 80 | 41 | 0.39421991 |
The table shows data for the 232 patients from the Verhaak dataset (GSE6891), including the occurrence of common karyotypic lesions and molecular aberrations, FAB classification and sex of patients.
Considering that NPM1c and FLT3-ITD, often found together, are indicators of bad prognosis while CEBPA and t(8;21) predict a more favourable outcome, we postulated that this observation could account for the inferior overall and event-free survival observed in the cohort of WBP5high patients. To rule out this possibility, the subgroups of low and high expressers were adjusted for age, cytogenetics findings, FAB classification and mutational status to avoid a potential bias to a specific driver mutation or subtype.
Surprisingly, survival analysis after balancing revealed that WBP5high patients (n = 27) still displayed inferior overall (median OS = 25.62 vs 79.63, p = 0.00052) and event-free survival (median EFS = 21.67 vs 63.84, p = 0.00247) when compared to WBP5low cohort (n = 27), thus suggesting WBP5 expression to be a strong disease indicator independently of its association with specific oncogenic mutations. WBP5 was also reported in three independent studies to be part of gene expression risk scores that predict adverse outcomes in AML patients;21–23 to further determine the validity of WBP5 as a potential biomarker and to assess whether WBP5 expression alone could predict the inferior outcome determined in those cohorts by the use of those risk panels, we retrieved those datasets and performed survival analysis comparing WBP5 low and high expressers. In agreement with the results from our pipeline, high expressers demonstrated lower survival rates in both cohorts after balancing (median OS = 347.25 vs 691.14, p = 0.01621 and 496.33 vs 1180.73, p = 0.01478) (Fig. S1), thus suggesting that the sole expression of WBP5 could serve as a reliable inferior predictor for AML.
WBP5high AML patients display a specific gene expression profile
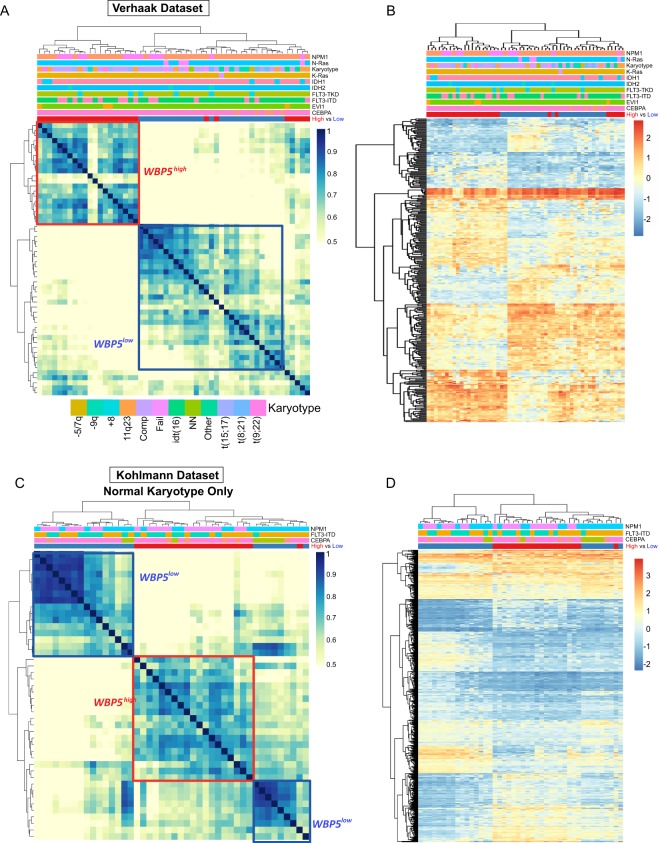
In order to investigate the molecular profiles associated with different levels of WBP5, we sought to identify similarities and differences between the low and high WBP5 expressers by performing differential gene expression analysis between the two groups. Differentially expressed genes were considered as those displaying a log fold change (FC) >1 and an adjusted p-value < 0.05. These genes were used to hierarchically cluster the patients using Pearson correlation coefficient. This analysis showed that most of the high expressers (20 out of 27 patients) formed a discrete cluster with high correlation coefficient, with the exception of a subgroup of 5 WBP5high patients that clustered separately and more closely with the low WBP5 cohort (Fig. 3A). This could be due to those patients being characterized by a specific mutation that was not screened for this analysis.
Figure 3.
WBP5high and WBP5low subgroups have distinct global gene expression profiles. (A) Hierarchical clustering of the Pearson correlation coefficient of mRNA expression showing distinct clusters for high and low WBP5 expressers from the Verhaak cohort. The mutational status of every patient is indicated by a colour coded graph on the top of the clustering map. A legend indicating each genetic subgroup is placed below the clustering map. (B) Heatmap indicating the gene expression profiles of WBP5low and WBP5high subgroups. (C) Hierarchical clustering of the Pearson correlation coefficient and (D) heatmap of mRNA values from the WBP5low and WBP5high subgroups of the patient cohort reported by Kohlmann et al. The cluster of low and high WBP5 expressers are indicated with blue and red boxes, respectively.
Next, we studied the identified differential genes that are specifically deregulated in the WBP5high subgroup and observed that the high expresser subgroup displayed a distinct gene expression profile, as indicated in the heatmap in Fig. 3B.
To validate our observations, we made use of another independent gene expression profiling dataset reported by Kohlmann et al.25. For this dataset, which comprises 251 AML patients with normal karyotype, we applied the same classification criterion for ranking them into high (n = 23) and low WBP5 expressers (n = 21) and balancing for sex, age, FAB and cytogenetic findings. Although this cohort does not encompass patients carrying karyotypic lesions, our analysis showed a similar trend, with most of the high expressers forming a defined cluster (Fig. 3C). Moreover, those patients again displayed a distinct gene expression profile compared to the low expressers as previously seen for the Verhaak dataset, thus strengthening the validity of our observations (Fig. 3D). We further confirmed this trend in three other independent cohorts, such as those reported by Valk et al.27, Haferlach et al.26 and Taskesen et al.28. Similarly, in the Valk and Taskesen datasets we observed that those patient subgroups formed moderate clusters with good correlation coefficients, with the exception of the Haferlach dataset in which, despite clustering together, both high and low expressers seemed to form rather smaller sub-clusters (Fig. 2), possibly due to the fact that those patients could not be balanced for the lack of information on their cytogenetic and mutational statuses.
High WBP5 expression is associated with an elevated HOX cluster gene expression
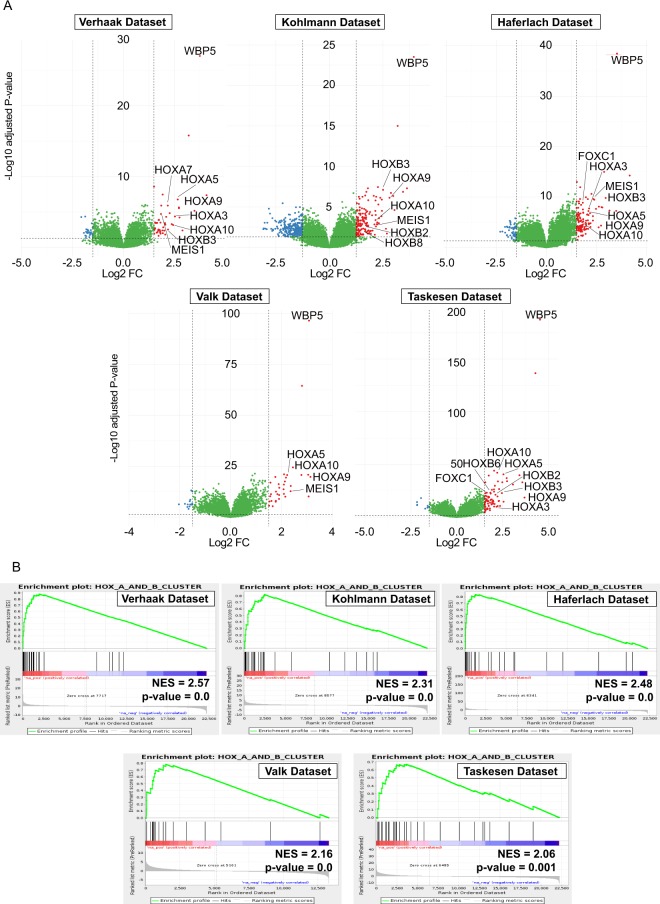
By performing differential gene expression analysis on all the five independent datasets, within the core of the high WBP5 expressers we considered as more highly expressed genes with an average log2 fold change value of at least 1.5 above and downregulated genes with values at least 1.5 below that of low expressers. Note that in the 15 genes showing highest differential expression in most of the cohorts (above 2 log2 FC) we observed several members of the HOXA and HOXB gene clusters, among which HOXA9, HOXA5 (with the exception of Kohlmann dataset), HOXA10, HOXB2 and HOXB3 (Fig. 4A,B). The full expression data for all the HOXA and HOXB cluster genes are provided in Supplementary Fig. 3. Intriguingly, several genes that are known partners of HOXA9, such as MEIS1, were also found to be differentially expressed. This has profound implications as those genes play key roles in the progression and maintenance of different types of myeloid diseases, and the combination of HOXA9 and MEIS1 has been shown to be causal for the development of AML30–32.
Figure 4.
Correlation of high WBP5 expression with HOXA and HOXB clusters gene signatures. (A) Volcano plots indicating the most differentially more highly (red dots, above 1.5 log2 FC) and lowly expressed genes (blue dots, below −1.5 log2 FC) in the GSE6891 (Verhaak), GSE15434 (Kohlmann), GSE13204 (Haferlach), GSE1159 (Valk) and GSE22845 (Taskesen) datasets. The most highly differentially expressed HOXA, HOXB and HOX-TALE genes are indicated in each plot. (B) Gene Set Enrichment Analysis (GSEA) plots show enrichment of HOXA and HOXB clusters genes ranked using a signal-to-noise metric according to expression in WBP5 low versus high expressers.
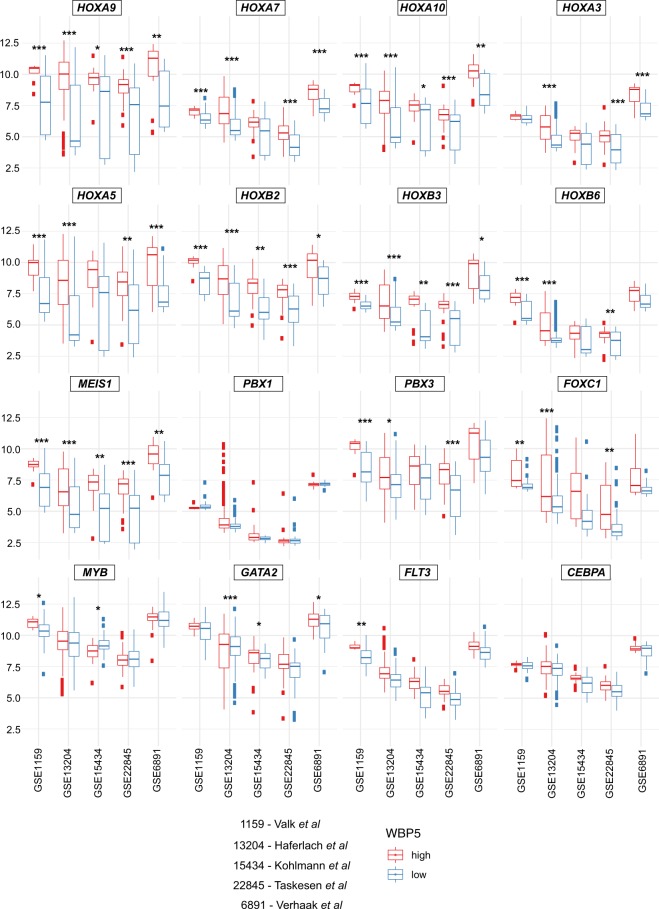
We also observed a moderate correlation of WBP5 expression with the levels of HOXA7, another gene that was reported to influence leukaemia latency and phenotype and is required for efficient immortalization of myeloid cells by MLL-ENL fusions33. Analysis of the HOX-Tale partner proteins also revealed higher levels of PBX3 transcript in all cohorts, but not that of PBX1 or PBX2.
This analysis also pointed to the higher expression of several other genes normally deregulated in a wide spectrum of leukaemias, including genes that act as transcriptional regulators (PRDM16 and NFIX) and a gene that influences proliferative advantage and survival (IGFBP2). Moreover, we observed higher expression of a number of genes that are part of the FLT3-ITD molecular signature, such as HOXB2, HOXB3, HOXB4, HOXB5, HOXB6, CRNDE, CLU, CTSG, COL4A5 and KRT18, among others (Fig. S4)34,35.
We also looked at the expression of genes that have been shown to be directly involved in HOXA-driven leukemogenesis and observed a significant increase in the expression of GATA236, while no difference was observed for FLT39,37,38, MYB39,40 or CEBPA41. Intriguingly, we also noted increased expression of FOXC142, although in this cohort it failed to reach statistical significance (Fig. 5).
Figure 5.
Elevated expression of HOXA, HOXB and HOX partner genes across five different independent gene expression datasets. Expression of representative HOXA (HOXA9, HOXA7, HOAX10 and HOXA5) and HOXB (HOXB2, HOXB3, HOXB6) cluster genes and HOX partners (MEIS1, PBX1, PBX3 and FOXC1) or downstream effectors (MYB, GATA2, FLT3 and CEBPA). Data are represented as boxplots in which high expressers are indicated in red and low expressers are indicated in blue. Every plot presents a color-coded boxplot showing median and interquartile ranges. The statistical analysis reported indicates p-value adjusted for false discovery rates. ***<0.001, **<0.01, *<0.05.
To confirm these observations, we calculated the average mRNA levels of WBP5high and WBP5low patients from all the other independent patient gene expression datasets; notably, we observed a gene expression pattern similar to the one obtained from the Verhaak cohort, that is, a large elevation of several HOXA and HOXB cluster members, such HOXA9, HOXA10 and HOX-TALE partner MEIS1 (Figs. 4A,B, 5 and S3). Several genes which influence gene expression, cell growth or that are part of FLT3-ITD signature were confirmed in virtually all cohorts as more highly expressed genes, including CPA3, PRDM16, IGFBP2 and CTSG (Fig. S4).
In a recent study by Naivarani et al., WBP5 was included in a 17-probe set signature that is used as predictor of long-term prognosis in AML patients characterized by high WT1 levels23. To test this, we probed for this relationship in our pipeline and observed a strong association between WBP5 and WT1 transcripts in all the dataset tested. (Fig. S3).
In summary, these analyses highlight WBP5 as a reliable prognostic and stratification biomarker and the association between high WBP5 expression and the elevation of HOX cluster genes levels.
Discussion
AML is among the most malignant cancers of the blood and due to its heterogeneous nature and complex biological behaviours there are currently limited therapeutic approaches.
The ability to diagnose and prevent myeloid leukaemias is hampered by the lack of quantifiable, reliable, and easily measurable biomarkers that correlate with disease progression. Steady improvements in survival rate and disease control have been made in the past decades, but despite the efforts aimed at developing new personalized and sensitive therapeutic approaches for patient therapy, AML is still associated with high morbidity and mortality. Recent advances in transcriptomic and epigenomic analysis have been fundamental in providing a comprehensive insight into the mechanism of leukaemia progression and in identifying new molecular biomarkers for prognosis, disease control, and therapeutic stratification.
In this study, we have developed an unbiased bioinformatic pipeline to screen five independent patient gene expression arrays through which we show that higher expression of WBP5 can serve to predict inferior outcome in patients with AML, thus providing a new potential stratification for therapy choices.
WBP5 has been previously proposed as a candidate oncogene in colorectal cancers with microsatellite instability and its expression is associated with advanced gastric cancers with lymph node metastasis18,19. Moreover, Guo and co-workers were the first to report WBP5 expression to be significantly elevated in drug resistant small cell lung cancer patients and to be correlated with shorter survival time and advanced clinical stage. In this latter study, the authors showed that WBP5 modulates multidrug resistance both in vitro and in vivo, and that it does so by regulating the Hippo pathway20,43. Those studies reported that high expression of WBP5 might be a predictor of unfavourable disease progression in many different cancers. Importantly, three independent studies provided a strong link between WBP5 expression and adverse clinical outcome in leukaemia21,22. In fact, Metzeler and co-workers were the first to report WBP5 as a risk factor by developing an 86-probe set gene expression signature to predict inferior outcome in patients with CN-AML. In this study, the authors showed patients that are characterized by an elevation of those genes displayed a strong association with FLT3-ITD mutation21. This work was followed by another study in which Bou Samra et al. proposed WBP5 as part of a further refined prediction risk score that consisted of a panel of 22 genes, those being associated with poor prognosis in CN-AML patients. Importantly, the authors showed WBP5 to rank first within this prediction panel according to the hazard ratio22. In line with those findings, our study demonstrates that high WBP5 expressers have significantly lower overall and event-free survival and are associated with an unfavourable prognosis. We found that high WBP5 expressers have a higher frequency of FLT3-ITD and NPM1c, which are often found together and are indicators of inferior outcome44,45. Conversely, we observed the WBP5low sub-group to correlate with CEBPA and RUNX1-ETO lesions, which are generally associated with favourable outcome46. This is consistent with the reports that WBP5 is associated with inferior survival in AML patients by displaying the highest hazard ratio and that its high expression correlates with advanced clinical stage and poor survival in lung cancers.
In order to gain an insight into the biological influence of aberrant WBP5 expression in AML, we performed gene expression analysis in five independent patient expression arrays. Notably, we found that high WBP5 levels correlated with elevated levels of several genes belonging to the HOXA and HOXB clusters; specifically, we observed a strong association with HOXA9 and HOXA10 and a moderate correlation with HOXA7. Many studies have demonstrated the importance of those genes in regulating the proliferation of haematopoietic cells and how their deregulation is paramount in driving the onset of myeloid leukaemias in vivo47–50. Importantly, we also observed a strong association of high WBP5 levels with HOXA9 oncogenic partners, among which MEIS1, PBX3 and FOXC142,51,52. We also observed elevated expression of a number of cancer-associated genes that have been reported to influence the proliferative advantage and the survival of leukaemic cells and genes that were reported to be part of the FLT3-ITD molecular signature, such as CRNDE, CLU, CTSG, IGFBP2, CPA3 and PRDM1634,35. Notably, we also observed a strong association of WBP5 with WT1, in agreement with previous reports. Our work suggests that a major mechanism of WBP5 influencing leukaemia behaviour might directly or indirectly act through regulating these genes. To date, WBP5 has been considered within risk prediction signatures to infer on clinical outcome but the importance of the sole WBP5 expression as a reliable biomarker has not been investigated. We did so by performing survival analysis comparing low and high WBP5 expressers in the cohort of patients from which those gene expression prognostic signatures have been determined and observed that WBP5 alone was able to predict the same inferior clinical outcome in those patients. As such, our data suggest that it would be fruitful to perform a more elaborate study to further validate the importance of those findings in vitro and in vivo.
Taken together, our bioinformatic approach shows that WBP5 expression is a reliable and powerful indicator of inferior outcome for AML and that it may be a candidate target for developing new therapeutic approaches.
Supplementary information
Acknowledgements
C.W. is supported by a Zhujiang Overseas Young Talents Postdoctoral Fellowship, a Chinese Academy of Sciences President’s International Fellowship Initiative for Postdoctoral Researchers (2019PB0177) and a National Natural Science Foundation of China Research Fund for International Young Scientists grant (31950410553). This work was supported by the National Natural Science Foundation of China (31671537, awarded to MAE) and through funding provided by the College of Medical and Dental Sciences of the University of Birmingham.
Author contributions
C.W. and G.V. conceived and designed the research; C.W. and P.C. performed the bioinformatic analysis; MAE, P.G. and J.F. provided relevant advice and financial support and edited the manuscript; G.V. supervised the research; C.W. and G.V. wrote and edited the manuscript.
Code availability
The full code to reproduce the data presented in this manuscript is available at https://github.com/doncarlos999/WBP5_analysis.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-60480-x.
References
- 1.Kelly LM, Gilliland DG. Genetics of myeloid leukemias. Annu. Rev. Genomics Hum. Genet. 2002;3:179–198. doi: 10.1146/annurev.genom.3.032802.115046. [DOI] [PubMed] [Google Scholar]
- 2.Moore MA. Converging pathways in leukemogenesis and stem cell self-renewal. Exp. Hematol. 2005;33:719–737. doi: 10.1016/j.exphem.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 3.De Kouchkovsky I, Abdul-Hay M. ‘Acute myeloid leukemia: a comprehensive review and 2016 update’. Blood Cancer J. 2016;6:e441. doi: 10.1038/bcj.2016.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat. Rev. Cancer. 2007;7:823–833. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- 5.Aplan PD. Chromosomal translocations involving the MLL gene: molecular mechanisms. DNA Repair. 2006;5:1265–1272. doi: 10.1016/j.dnarep.2006.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Metzeler KH, Bloomfield CD. Clinical Relevance of RUNX1 and CBFB Alterations in Acute Myeloid Leukemia and Other Hematological Disorders. Adv. Exp. Med. Biol. 2017;962:175–199. doi: 10.1007/978-981-10-3233-2_12. [DOI] [PubMed] [Google Scholar]
- 7.Loke J, et al. RUNX1-ETO and RUNX1-EVI1 Differentially Reprogram the Chromatin Landscape in t(8;21) and t(3;21) AML. Cell Rep. 2017;19:1654–1668. doi: 10.1016/j.celrep.2017.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tenen DG. Disruption of differentiation in human cancer: AML shows the way. Nat. Rev. Cancer. 2003;3:89–101. doi: 10.1038/nrc989. [DOI] [PubMed] [Google Scholar]
- 9.Volpe G, et al. C/EBPalpha and MYB regulate FLT3 expression in AML. Leukemia. 2013;27:1487–1496. doi: 10.1038/leu.2013.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rosenbauer F, Tenen DG. Transcription factors in myeloid development: balancing differentiation with transformation. Nat. Rev. Immunol. 2007;7:105–117. doi: 10.1038/nri2024. [DOI] [PubMed] [Google Scholar]
- 11.Nerlov C. C/EBPalpha mutations in acute myeloid leukaemias. Nat. Rev. Cancer. 2004;4:394–400. doi: 10.1038/nrc1363. [DOI] [PubMed] [Google Scholar]
- 12.Volpe Giacomo, Cauchy Pierre, Walton David S, Ward Carl, Blakemore Daniel, Bayley Rachael, Clarke Mary L, Schmidt Luisa, Nerlov Claus, Garcia Paloma, Dumon Stéphanie, Grebien Florian, Frampton Jon. Dependence on Myb expression is attenuated in myeloid leukaemia with N-terminal CEBPA mutations. Life Science Alliance. 2019;2(2):e201800207. doi: 10.26508/lsa.201800207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weissmann S, et al. Landscape of TET2 mutations in acute myeloid leukemia. Leukemia. 2012;26:934–942. doi: 10.1038/leu.2011.326. [DOI] [PubMed] [Google Scholar]
- 14.Bacher U, et al. TET2 deletions are a recurrent but rare phenomenon in myeloid malignancies and are frequently accompanied by TET2 mutations on the remaining allele. Br. J. Haematol. 2012;156:67–75. doi: 10.1111/j.1365-2141.2011.08911.x. [DOI] [PubMed] [Google Scholar]
- 15.Patel JP, et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N. Engl. J. Med. 2012;366:1079–1089. doi: 10.1056/NEJMoa1112304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cancer Genome Atlas Research, N. et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu W, et al. Regulation of neuronal cell death by c-Abl-Hippo/MST2 signaling pathway. PLoS One. 2012;7:e36562. doi: 10.1371/journal.pone.0036562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suh YS, et al. Overexpression of Plasminogen Activator Inhibitor-1 in Advanced Gastric Cancer with Aggressive Lymph Node Metastasis. Cancer Res. Treat. 2015;47:718–726. doi: 10.4143/crt.2014.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gylfe AE, et al. Identification of candidate oncogenes in human colorectal cancers with microsatellite instability. Gastroenterology. 2013;145:540–543 e522. doi: 10.1053/j.gastro.2013.05.015. [DOI] [PubMed] [Google Scholar]
- 20.Tang R, et al. WW domain binding protein 5 induces multidrug resistance of small cell lung cancer under the regulation of miR-335 through the Hippo pathway. Br. J. Cancer. 2016;115:243–251. doi: 10.1038/bjc.2016.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Metzeler KH, et al. An 86-probe-set gene-expression signature predicts survival in cytogenetically normal acute myeloid leukemia. Blood. 2008;112:4193–4201. doi: 10.1182/blood-2008-02-134411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bou Samra E, Klein B, Commes T, Moreaux J. Development of gene expression-based risk score in cytogenetically normal acute myeloid leukemia patients. Oncotarget. 2012;3:824–832. doi: 10.18632/oncotarget.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Niavarani A, et al. A 4-gene expression score associated with high levels of Wilms Tumor-1 (WT1) expression is an adverse prognostic factor in acute myeloid leukaemia. Br. J. Haematol. 2016;172:401–411. doi: 10.1111/bjh.13836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Verhaak RG, et al. Prediction of molecular subtypes in acute myeloid leukemia based on gene expression profiling. Haematologica. 2009;94:131–134. doi: 10.3324/haematol.13299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kohlmann A, et al. Gene expression profiling in AML with normal karyotype can predict mutations for molecular markers and allows novel insights into perturbed biological pathways. Leukemia. 2010;24:1216–1220. doi: 10.1038/leu.2010.73. [DOI] [PubMed] [Google Scholar]
- 26.Haferlach T, et al. Clinical utility of microarray-based gene expression profiling in the diagnosis and subclassification of leukemia: report from the International Microarray Innovations in Leukemia Study Group. J. Clin. Oncol. 2010;28:2529–2537. doi: 10.1200/JCO.2009.23.4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Valk PJ, et al. Prognostically useful gene-expression profiles in acute myeloid leukemia. N. Engl. J. Med. 2004;350:1617–1628. doi: 10.1056/NEJMoa040465. [DOI] [PubMed] [Google Scholar]
- 28.Taskesen E, et al. Prognostic impact, concurrent genetic mutations, and gene expression features of AML with CEBPA mutations in a cohort of 1182 cytogenetically normal AML patients: further evidence for CEBPA double mutant AML as a distinctive disease entity. Blood. 2011;117:2469–2475. doi: 10.1182/blood-2010-09-307280. [DOI] [PubMed] [Google Scholar]
- 29.Li Z, et al. Identification of a 24-gene prognostic signature that improves the European LeukemiaNet risk classification of acute myeloid leukemia: an international collaborative study. J. Clin. Oncol. 2013;31:1172–1181. doi: 10.1200/JCO.2012.44.3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zeisig BB, et al. Hoxa9 and Meis1 are key targets for MLL-ENL-mediated cellular immortalization. Mol. Cell Biol. 2004;24:617–628. doi: 10.1128/mcb.24.2.617-628.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thorsteinsdottir U, et al. Overexpression of the myeloid leukemia-associated Hoxa9 gene in bone marrow cells induces stem cell expansion. Blood. 2002;99:121–129. doi: 10.1182/blood.v99.1.121. [DOI] [PubMed] [Google Scholar]
- 32.Thorsteinsdottir U, Kroon E, Jerome L, Blasi F, Sauvageau G. Defining roles for HOX and MEIS1 genes in induction of acute myeloid leukemia. Mol. Cell Biol. 2001;21:224–234. doi: 10.1128/MCB.21.1.224-234.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ayton PM, Cleary ML. Transformation of myeloid progenitors by MLL oncoproteins is dependent on Hoxa7 and Hoxa9. Genes. Dev. 2003;17:2298–2307. doi: 10.1101/gad.1111603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cauchy P, et al. Chronic FLT3-ITD Signaling in Acute Myeloid Leukemia Is Connected to a Specific Chromatin Signature. Cell Rep. 2015;12:821–836. doi: 10.1016/j.celrep.2015.06.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Volpe G, et al. Prognostic significance of high GFI1 expression in AML of normal karyotype and its association with a FLT3-ITD signature. Sci. Rep. 2017;7:11148. doi: 10.1038/s41598-017-11718-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dang J, et al. AMKL chimeric transcription factors are potent inducers of leukemia. Leukemia. 2017;31:2228–2234. doi: 10.1038/leu.2017.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Staffas A, et al. Upregulation of Flt3 is a passive event in Hoxa9/Meis1-induced acute myeloid leukemia in mice. Oncogene. 2017;36:1516–1524. doi: 10.1038/onc.2016.318. [DOI] [PubMed] [Google Scholar]
- 38.Wang GG, Pasillas MP, Kamps MP. Meis1 programs transcription of FLT3 and cancer stem cell character, using a mechanism that requires interaction with Pbx and a novel function of the Meis1 C-terminus. Blood. 2005;106:254–264. doi: 10.1182/blood-2004-12-4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hess JL, et al. c-Myb is an essential downstream target for homeobox-mediated transformation of hematopoietic cells. Blood. 2006;108:297–304. doi: 10.1182/blood-2005-12-5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dasse E, et al. Distinct regulation of c-myb gene expression by HoxA9, Meis1 and Pbx proteins in normal hematopoietic progenitors and transformed myeloid cells. Blood Cancer J. 2012;2:e76. doi: 10.1038/bcj.2012.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Collins C, et al. C/EBPalpha is an essential collaborator in Hoxa9/Meis1-mediated leukemogenesis. Proc. Natl Acad. Sci. USA. 2014;111:9899–9904. doi: 10.1073/pnas.1402238111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Somerville TD, et al. Frequent Derepression of the Mesenchymal Transcription Factor Gene FOXC1 in Acute Myeloid Leukemia. Cancer Cell. 2015;28:329–342. doi: 10.1016/j.ccell.2015.07.017. [DOI] [PubMed] [Google Scholar]
- 43.Guo L, et al. Gene expression profiling of drug-resistant small cell lung cancer cells by combining microRNA and cDNA expression analysis. Eur. J. Cancer. 2010;46:1692–1702. doi: 10.1016/j.ejca.2010.02.043. [DOI] [PubMed] [Google Scholar]
- 44.Verhaak RG, et al. Mutations in nucleophosmin (NPM1) in acute myeloid leukemia (AML): association with other gene abnormalities and previously established gene expression signatures and their favorable prognostic significance. Blood. 2005;106:3747–3754. doi: 10.1182/blood-2005-05-2168. [DOI] [PubMed] [Google Scholar]
- 45.How J, et al. The prognostic impact of FLT3-ITD and NPM1 mutations in patients with relapsed acute myeloid leukemia and intermediate-risk cytogenetics. Blood Cancer J. 2013;3:e116. doi: 10.1038/bcj.2013.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Renneville A, et al. The favorable impact of CEBPA mutations in patients with acute myeloid leukemia is only observed in the absence of associated cytogenetic abnormalities and FLT3 internal duplication. Blood. 2009;113:5090–5093. doi: 10.1182/blood-2008-12-194704. [DOI] [PubMed] [Google Scholar]
- 47.Bach C, et al. Leukemogenic transformation by HOXA cluster genes. Blood. 2010;115:2910–2918. doi: 10.1182/blood-2009-04-216606. [DOI] [PubMed] [Google Scholar]
- 48.Bijl J, et al. Evidence for Hox and E2A-PBX1 collaboration in mouse T-cell leukemia. Oncogene. 2008;27:6356–6364. doi: 10.1038/onc.2008.233. [DOI] [PubMed] [Google Scholar]
- 49.Grier DG, et al. The pathophysiology of HOX genes and their role in cancer. J. Pathol. 2005;205:154–171. doi: 10.1002/path.1710. [DOI] [PubMed] [Google Scholar]
- 50.Bijl J, et al. Analysis of HSC activity and compensatory Hox gene expression profile in Hoxb cluster mutant fetal liver cells. Blood. 2006;108:116–122. doi: 10.1182/blood-2005-06-2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Garcia-Cuellar MP, Steger J, Fuller E, Hetzner K, Slany RK. Pbx3 and Meis1 cooperate through multiple mechanisms to support Hox-induced murine leukemia. Haematologica. 2015;100:905–913. doi: 10.3324/haematol.2015.124032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kroon E, et al. Hoxa9 transforms primary bone marrow cells through specific collaboration with Meis1a but not Pbx1b. EMBO J. 1998;17:3714–3725. doi: 10.1093/emboj/17.13.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The full code to reproduce the data presented in this manuscript is available at https://github.com/doncarlos999/WBP5_analysis.